Abstract
The present study has taken advantage of publicly available cell type specific mRNA expression databases in order to identify potential genes participating in the development of retinal AII amacrine cells. We profile two such genes, Delta/Notch-like EGF repeat containing (Dner) and nuclear factor I/A (Nfia), that are each heavily expressed in AII amacrine cells in the mature mouse retina, and which conjointly identify this retinal cell population in its entirety when using antibodies to DNER and NFIA. DNER is present on the plasma membrane, while NFIA is confined to the nucleus, consistent with known functions of each of these two proteins. DNER also identifies some other subsets of retinal ganglion and amacrine cell types, along with horizontal cells, while NFIA identifies a subset of bipolar cells as well as Muller glia and astrocytes. During early postnatal development, NFIA labels astrocytes on the day of birth, AII amacrine cells at postnatal (P) day 5, and Muller glia by P10, when horizontal cells also transiently exhibit NFIA immunofluorescence. DNER, by contrast, is present in ganglion and amacrine cells on P1, also labeling the horizontal cells by P10. Developing AII amacrine cells exhibit accumulating DNER labeling at the dendritic stalk, labeling which becomes progressively conspicuous by P10, as it is in maturity. This developmental time course is consistent with a prospective role for each gene in the differentiation of AII amacrine cells.
Keywords: Delta/Notch-like EGF repeat containing, Nuclear factor I/A, Prox1, RRID:IMSR_JAX:000664, RRID:SCR_007303, RRID:AB_355202, RRID:AB_1854422, RRID:AB_10064230, RRID:AB_90755, RRID:AB_213554, RRID:AB_2187701, RRID:AB_568862, RRID:AB_10013783, RRID:AB_397879, RRID:AB_476889, RRID:AB_2079751, RRID:AB_2556746, RRID:AB_2340854, RRID:AB_2340819, RRID:AB_2336933, RRID:AB_142628, RRID:AB_2340437, RRID:AB_2313584, RRID:AB_2534016, RRID:AB_2492288, RRID:AB_141362
Introduction
The mammalian retina is a thin neural tissue that lines the back of the eye and is responsible for the transmission of incoming light signals to higher visual regions of the brain. Within the retina, this information is already beginning to be processed as separate parallel visual pathways. This is accomplished by many different cell types: current estimates suggest that there are approximately 85 unique neuronal populations in the retina (Masland, 2012; Macosko et al., 2015; Shekhar et al., 2016; Baden et al., 2016). One such cell type is the AII amacrine cell, a narrow-field amacrine cell initially reported to have a unique radial orientation and apparent separation of its dendrites into pre- and post-synaptic compartments (Famiglietti and Kolb, 1974; Strettoi et al., 1992). Subsequent anatomical and physiological studies have not only confirmed, but expounded on this compartmentalization, revealing a stunning complexity in how this cell is integrated into the retinal circuitry: the AII amacrine cell forms synapses with no less than 28 retinal cell types (Marc et al., 2014), participating in multiple retinal circuits, with a particularly critical role in mediating scotopic vision (Demb and Singer, 2012). Little is known, however, about the developmental processes and genetic underpinnings that establish this neuronal population, create their complex dendritic arbors, and integrate these cells into the retinal circuitry.
Understanding the mechanisms underlying AII amacrine cell development requires a knowledge of the molecular players expressed by these cells at different ages. Many proteins have been identified to be expressed by mature AII amacrine cells, including Calretinin, Parvalbumin, Disabled1, and Prox1 (Wässle et al., 1993; Massey and Mills, 1999; Rice and Curran, 2000; Dyer et al., 2003). Only a few proteins, however, have been shown to be important for developing AII amacrine cells, such as Dscaml1 and NeuroD2, both implicated in the growth of AII neurites (Fuerst et al., 2009; Cherry et al., 2011). The present study has sought to identify other novel proteins that are expressed by developing and mature AII amacrine cells, by taking advantage of recent advances in molecular profiling, particularly in single cell RNA-sequencing, that have begun to differentiate the subtleties that exist between different cell types through their transcriptome-wide molecular signatures (Kay et al., 2012; Siegert et al., 2012; Macosko et al., 2015; Shekhar et al., 2016). These gene expression profiles offer a comprehensive look into the proteins that must be critical for the development and function of specific cell types in the retina, including the AII amacrine cell.
In this study, we have used recently published expression profiles of retinal cell types to identify genes that may have roles critical for the development of AII amacrine cells. We found two genes, Delta/Notch-like EGF repeat containing (Dner) and nuclear factor I/A (Nfia), that are highly expressed in these cells, both in development and adulthood. We subsequently validated the expression of their protein products using immunofluorescent techniques, and determined that probing for both proteins conjointly is a reliable method for identifying the population of AII amacrine cells. As both genes are known to play a role in neural development outside the retina, they are prime candidates for understanding the mechanisms that underlie the development of the AII amacrine cell.
Materials and Methods
Candidate Gene Search
Three publicly-available gene expression datasets were analyzed to identify novel proteins expressed by AII amacrine cells (NCBI GEO series GSE63473, GSE81904, and GSE35077; www.ncbi.nlm.nih.gov/geo/; RRID:SCR_007303). Two different single cell RNA-seq experiments (one using dissociated cells from whole retinas and another analyzing single cells after enriching for bipolar cells) were combined to assess the levels of gene expression within different cell types of the retina, including the AII amacrine cells (Macosko et al., 2015; Shekhar et al., 2016). Cells were organized into cell clusters (as defined by the original authors) and, for every gene, the total number of transcripts sequenced across all cells within a cluster was tabulated; from these data, the average number of transcripts expressed per cell was calculated. Genes that were highly expressed in AII amacrine cells were then compared against all other cell populations to determine the specificity of expression. The same approach was used to assess the transcriptome of AII amacrine cells at postnatal day 7 (day of birth = P1), using microarray data derived from purified populations of retinal cell types (Kay et al., 2012). For gene transcripts identified by multiple microarray probes, the maximum expression value across the probes was used. When analyzing the transcriptome within AII amacrine cells, gene expression was normalized to the highest expressed gene; when comparing across cell types, gene expression was normalized to that within the AII amacrine cell population.
Tissue Preparation
Retinas were collected from adult (>P30) or developing (P1, P5, P10) C57Bl/6J (B6/J; RRID:IMSR_JAX:000664) mice that were originally obtained from The Jackson Laboratories (Bar Harbor, ME) and maintained at UCSB. Adult mice were given a lethal injection of sodium pentobarbital (>120mg/kg, i.p.; Euthasol, #710101; Vibrac, Fort Worth, TX), and, once deeply anesthetized (as verified by a lack of response to a tail pinch), intracardially perfused with approximately 2 ml of 0.9% saline via syringe, followed by approximately 50 ml of 4% paraformaldehyde in 0.1 M sodium phosphate buffer (PB, pH = 7.3) administered over 15 mins via gravity, all as recently described in greater detail (Keeley et al., 2017). Young postnatal mice were given a similar injection of sodium pentobarbital, but once anesthetized, eyes were removed from the orbit and directly immersed in 4% paraformaldehyde for a total of 30 mins. Whole retinas were then dissected free from the eyes, and four relieving cuts were made to allow the retinas to lie flat. Some of these retinal wholemounts were embedded in 5% agarose in 0.1M PB and then sectioned radially into 150 μm sections on a Vibratome (The Vibratome Company, St Louis, MO).
Antibodies
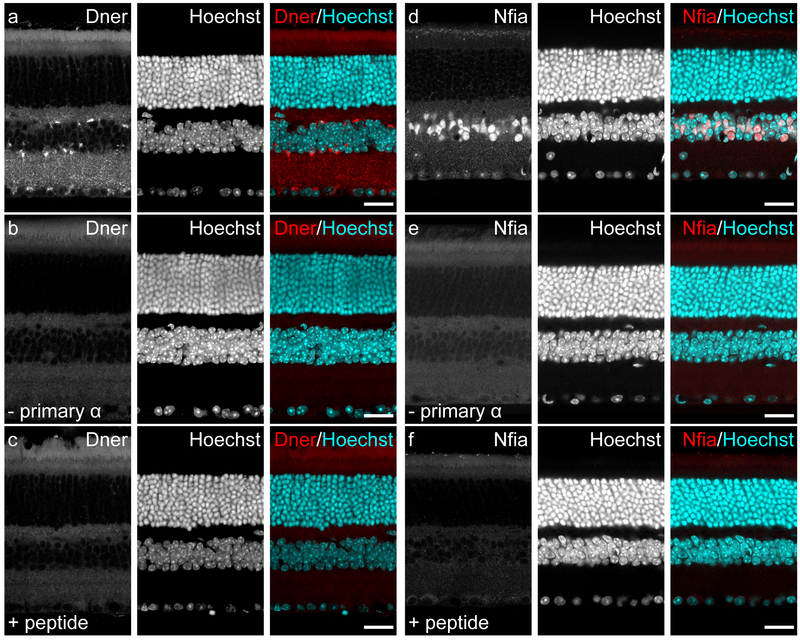
We used two antibodies in this study, the staining patterns of which had never been characterized in the mouse retina. The antibody used to detect DNER (1:500; #AF2254; R&D Systems, Minneapolis, MN; RRID:AB_355202) was a goat polyclonal IgG raised against recombinant mouse DNER (aa26–637) derived from the mouse myeloma cell line NS0, which was confirmed by the manufacturer to detect mouse DNER in ELISA and Western blot assays, as well as label Purkinje cells of the mouse cerebellum, as expected. This antibody also labeled cells in the chick cochlea in a manner that was consistent with RNA expression data (Kowalik and Hudspeth, 2011). The antibody used to detect NFIA (1:500; #HPA006111; Sigma, St. Louis, MO; RRID:AB_1854422) was a rabbit polyclonal IgG raised against recombinant human NFIA (aa278–378). It was developed in conjunction with the Human Protein Atlas project (www.proteinatlas.org), which validated that the staining pattern of this antibody in human tissues matched known expression data and subcellular localization. While this antibody was raised against the human NFIA protein, the immunogen used is 98% homologous with the mouse NFIA protein, suggesting a high likelihood that this antibody would be useful in identifying NFIA in mouse tissue. Indeed, immunostaining of the mouse cortex revealed nuclear staining consistent with known expression patterns (Nagao et al., 2016). Serial dilutions of each antibody were initially run with adult retinal sections to determine optimal concentrations for immunofluorescence. Additionally, two controls were performed to determine their specificity: first, each antibody was incubated with an eight-fold excess of its immunogenic peptide (Recombinant Mouse DNER Fc Chimera, #2254-DN-050, R&D Systems; PrEST Antigen NFIA, #APrEST70957, Sigma) at room temperature for two hours prior to use; second, PBS-TX was substituted for the primary antibody itself. Both controls yielded no trace of labeling on these sections (figure 1).
Figure 1: Validation of the DNER and NFIA antibodies.
(a) When used to label retinal sections from the adult mouse, the primary antibody to DNER revealed a staining pattern that appeared membranous in the nuclear layers and punctate in the plexiform layers. (b) This labeling was nonexistent in sections incubated with only the secondary antibody solution. (c) This labeling was also not observed when the primary antibody was preincubated with the immunizing peptide, instead revealing a staining pattern resembling that seen with the secondary antibody alone. (d) The primary antibody to NFIA labeled nuclei in the INL and GCL of the adult mouse retina. (e) This nuclear staining was never observed in sections that were incubated with only the secondary antibody. (f) Preincubation of the NFIA primary antibody with the immunizing peptide also failed to label nuclei in these sections, with all fluorescent signal appearing to mimic the labeling seen in the secondary antibody only condition. Scale bars = 25 μm.
All other primary antibodies used in this study detect proteins which have been well characterized in the mouse retina, and immunofluorescence labeling using these antibodies was consistent with the staining patterns initially reported for these antigens. The details for each of these antibodies are listed in Table 1. Evidence for the specificity of these antibodies to their specific targets in the mouse retina is as follows. 1) The antibody to Prox1 has been used previously to label the nuclei of AII amacrine cells (Keeley et al., 2014; Pérez de Sevilla Müller et al., 2017), consistent with the known localization of the protein in these cells (Dyer et al., 2003). Prox1 is also found in the nuclei of bipolar cells and horizontal cells (Dyer et al., 2003), which are also labeled using this antibody. 2) The antibody to TH detects a single band on immunoblots between 50–65 kDa when using lysates from mouse brain (Millipore, #AB1542 datasheet) and PC12 cells (Haycock and Waymire, 1982). This antibody labels the somata and processes of the dopaminergic amacrine cells using immunofluorescence, as expected (Versaux-Botteri et al., 1984). 3) The antibody to Calb recognizes the Calbindin D-28K protein on immunoblots when using lysates from rat hippocampus (Millipore, #PC253L datasheet), and we have used this antibody previously to label the somata and processes of horizontal cells (Raven et al., 2005); additionally, it labels amacrine cells and some ganglion cells. This is consistent with the originally described staining pattern (Pochet et al., 1991). 4) The antibody to VGluT3 labels the plasma membrane of a single population of amacrine cells in the inner nuclear layer (INL), as well as processes that stratify in the middle of the inner plexiform layer (IPL), as expected (Haverkamp and Wässle, 2004). Additionally, this antibody labels VGluT3+ amacrine cells that have been genetically encoded to express a fluorescent reporter protein (data not shown). 5) The antibody to PKC labels the membranes of rod bipolar cells and some amacrine cells, a staining pattern that is consistent with that originally observed (Pinto et al., 1994). 6) The antibody to Syt2 (alternatively named znp-1 and developed as part of a library of monoclonal antibodies to detect proteins in zebrafish embryos; Trevarrow et al., 1990) recognizes a 60 kDa protein on immunoblots when using lysates from mouse cerebellum, which has been confirmed to be synaptotagmin 2 via mass spectrometry (Fox and Sanes, 2007). This antibody labels synaptic puncta in the outer plexiform layer (OPL), as well as the membranes of type 2 and type 6 bipolar cells, the former being much more strongly-labeled than the latter (Fox and Sanes, 2007). 7) The antibody to GS detects a single 45 kDa protein on immunoblots using rat cerebrum lysate, and labels astrocytes in the rat cerebrum (BD Biosciences, #610517 datasheet). This antibody labels the processes of Müller glia in the retina, as expected (Linser et al., 1984). 8) The antibody to GFAP reacts specifically with GFAP protein in immunoblotting assays and labels astrocytes, Bergmann glia, and chondrocytes of elastic cartilage in immunohistochemical staining (Sigma, #C9205 datasheet). Consistent with the reported expression of GFAP in mature mouse retina (Bromberg and Schachner, 1978), this antibody labels astrocytic processes in the nerve fiber layer. 9) The antibody to ChAT detects a single band ~68–70 kDA in size when using mouse brain lysate (Millipore, #AB144P datasheet), and immunofluorescence reveals two populations of amacrine cells, one residing in the INL and one residing in the ganglion cell layer (GCL) (Kang et al., 2004), as expected (Tatton et al., 1990).
Table 1:
Primary antibodies used in this study.
| Antigen | Abbreviation | Type | Immunogen | Supplier / Cat# / RRID | Dilution | Structures Labeled in Mouse Retina |
|---|---|---|---|---|---|---|
| Delta/Notch-like EGF-related Receptor | Dner | Goat Polyclonal | Mouse Dner, peptide mapping to amino acids 26–637. | R&D
Systems AF2254 RRID:AB_355202 |
1:250 | Determined in this study |
| Nuclear Factor 1A-type | Nfia | Rabbit Polyclonal | Human NFIA, peptide mapping to amino acids 301–401. | Atlas
Antibodies HPA006111 RRID:AB_1854422 |
1:500 | Determined in this study |
| Prospero Homeobox 1 | Prox1 | Rabbit Polyclonal | Mouse Prox1, peptide mapping to amino acids 723–737. | Covance PRB-238C RRID:AB_10064230 |
1:1,000 | AII amacrine cells Bipolar cells Horizontal cells |
| Tyrosine Hydroxylase | TH | Sheep Polyclonal | Rat TH, purified from a pheochromocytoma. | Millipore AB1542 RRID:AB_90755 |
1:1,000 | Dopaminergic amacrine cells |
| Calbindin 1 | Calb | Rabbit Polyclonal | Bovine Calb D-28K, purified from cerebellum. | Millipore PC253L RRID:AB_213554 |
1:10,000 | Horizontal cells Amacrine cells Ganglion cells |
| Vesicular Glutamate Transporter 3 | VGluT3 | Goat Polyclonal | Human VGLUT3, peptide mapping near the N-terminus. | Santa Cruz
Biotechnology SC26031 RRID:AB_2187701 |
1:500 | VGluT3+ amacrine cells |
| Protein Kinase C | PKC | Mouse Monoclonal | Human PKCγ, peptide mapping to amino acids 499–697. | Millipore 05–983 RRID:AB_568862 |
1:500 | Rod bipolar cells Amacrine cells |
| Synaptotagmin 2 | Syt2 | Mouse Monoclonal | Homogenized whole zebrafish. | ZIRC ZDB-ATB-081002–25 RRID:AB_10013783 |
1:100 | Type 2 bipolar cells Type 6 bipolar cells |
| Glutamine Synthetase | GS | Mouse Monoclonal | Human GS, peptide mapping to amino acids 1–373. | BD
Biosciences 610517 RRID:AB_397879 |
1:500 | Müller glia |
| Glial Fibrillary Acidic Protein | GFAP | Mouse Monoclonal pre-conjugated to Cy3 | Pig GFAP, purified from spinal cord. | Sigma C9205 RRID:AB_476889 |
1:500 | Retinal astrocytes |
| Choline Acetyltransferase | ChAT | Goat Polyclonal | Human ChAT, purified from placenta. | Millipore AB144P RRID:AB_2079751 |
1:250 | Cholinergic amacrine cells |
Immunofluorescence
The following protocol was used to label retinal wholemounts or sections with antibodies: tissue was first incubated in 5% Normal Donkey Serum (#D9663; Sigma, St Louis, MO) in phosphate buffered saline with 1% TritonX-100 (PBS-TX) for a total of three hours. The samples were then rinsed with PBS three times for 10 mins each, then transferred to a solution of primary antibodies diluted in PBS-TX and incubated for three nights. After another set of three rinses in PBS, the tissue was incubated in a solution of secondary antibodies conjugated to fluorescent dyes (Table 2) in PBS-TX overnight. For some sections, Hoechst 33342 (1:1000; #H3570; Thermo Fisher Scientific, Waltham, MA) was added with secondary antibodies to label cell nuclei. After a final three rinses in PBS, wholemount retinas or sections were mounted on glass slides with Fluoro-Gel (#17985–10; EMS, Hatfield, PA). All incubation steps were performed with slight agitation, using an orbital shaker, and at 4°C. 3D image stacks were captured using an Olympus Fluoview1000 laser scanning confocal microscope, and all micrographs presented here are maximum projection images (between 1 μm and 3 μm thick) derived from these stacks, with a few exceptions that are noted in the figure legends.
Table 2:
Secondary antibodies used in this study.
| Antigen | Type | Fluorophore | Supplier / Cat# / RRID | Dilution |
|---|---|---|---|---|
| Mouse IgG | Donkey Polyclonal | DyLight488 | Thermo Fisher
Scientific SA5–10166RRID:AB_2556746 |
1:200 |
| Mouse IgG | Donkey Polyclonal | AlexaFluor594 | Jackson
ImmunoResearch 715–585-150 RRID:AB_2340854 |
1:200 |
| Mouse IgG | Donkey Polyclonal | Cy5 | Jackson
ImmunoResearch 715–175-150 RRID:AB_2340819 |
1:200 |
| Goat IgG | Donkey Polyclonal | AlexaFluor488 | Jackson
ImmunoResearch 705–545-147 RRID:AB_2336933 |
1:200 |
| Goat IgG | Donkey Polyclonal | AlexaFluor546 | Thermo Fisher
Scientific A-11056 RRID:AB_142628 |
1:200 |
| Goat IgG | Donkey Polyclonal | AlexaFluor647 | Jackson
ImmunoResearch 705–605-147 RRID:AB_2340437 |
1:200 |
| Rabbit IgG | Donkey Polyclonal | AlexaFluor488 | Jackson
ImmunoResearch 711–545-152 RRID:AB_2313584 |
1:200 |
| Rabbit IgG | Donkey Polyclonal | AlexaFluor546 | Thermo Fisher
Scientific A-10040 RRID:AB_2534016 |
1:200 |
| Rabbit IgG | Donkey Polyclonal | AlexaFluor647 | Jackson
ImmunoResearch 711–605-152 RRID:AB_2492288 |
1:200 |
| Sheep IgG | Donkey Polyclonal | AlexaFluor488 | Thermo Fisher
Scientific A-11015 RRID:AB_141362 |
1:200 |
Cell Number Quantification
Six retinas were used to estimate the total number of DNER+/NFIA+ AII amacrine cells in B6/J retinas, to compare with previously reported estimates quantifying Prox1+ AII amacrine cells (Keeley et al., 2014). Four mid-eccentric locations were chosen, one per retinal quadrant, where all double-labeled amacrine cells were counted; each sampled field was 25,192μm2 in area. Average densities were calculated, then multiplied by retinal area to estimate the total number of AII amacrine cells per retina.
Results
Dner and Nfia are expressed in the postnatal and adult retina, and are particularly highly expressed in AII amacrine cells.
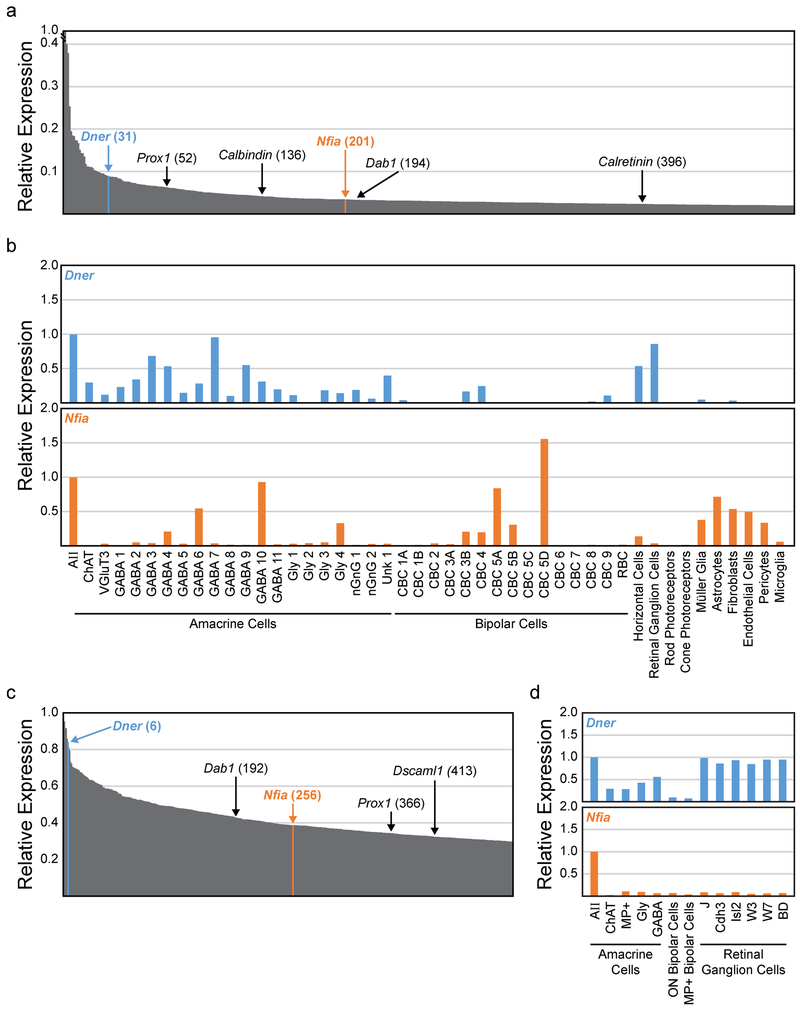
We consulted three available databases reporting retinal cell type specific transcriptomes, including that for the AII cells (Kay et al., 2012; Macosko et al., 2015; Shekhar et al., 2016). Single cell RNA-seq of adult retinal cells, with the individual profiles subsequently clustered into prospective cell types based on expression patterns, revealed two genes that were strongly expressed in AII amacrine cells: Dner and Nfia, being the 31st and 201st highest expressed genes in these cells, respectively (figure 2a). Additionally, both genes were expressed at higher levels within the AII amacrine cells than in any other cell cluster (with the one exception of Nfia expression in a type of cone bipolar cell), and no other cell cluster showed expression of both genes at levels appreciable to that observed in the AII cells (figure 2b). This same pattern of expression was found using a microarray analysis of purified populations of cells from early postnatal mouse retina (P6): AII amacrine cells displayed high levels of expression of both genes (Dner and Nfia were the 6th and 256th highest expressed genes at this time point, respectively), and were the only cell type to express both genes at such high levels (figure 2c and 2d). Dner encodes the Delta/Notch-like EGF-related receptor, a single pass transmembrane receptor that has been shown to be important for development of neuron branching and synaptogenesis, presumably through its interaction with the Notch signaling pathway (Saito and Takeshima, 2006; Fukazawa et al., 2008). Nfia encodes the nuclear factor 1 A-type, one of three NFI transcription factors shown to be critical for brain development (das Neves et al., 1999). This transcriptome analysis led us to believe that 1) both Dner and Nfia are expressed by AII amacrine cells, 2) double-labeling with antibodies to DNER and NFIA should reliably identify the AII amacrine cell population, and 3) the developmental time course of their expression may portend a role in the differentiation of AII amacrine cells.
Figure 2: Transcriptome analysis of AII amacrine cells.
(a) Relative expression of the top 500 genes in adult AII amacrine cells, with known AII marker genes and new candidate marker genes highlighted with their relative position in parentheses (derived from Macosko et al., 2015 and Shekhar et al., 2016). (b) Expression of two candidate marker genes, Dner (blue top histogram) and Nfia (orange bottom histogram), across several cell populations, normalized to their expression in AII amacrine cells. (c) Relative expression of the top 400 genes in postnatal (P7) AII amacrine cells, with the position of several AII genes highlighted (derived from Kay et al., 2012). (d) Expression of Dner and Nfia in AII amacrine cells compared to other cell types/classes at P7.
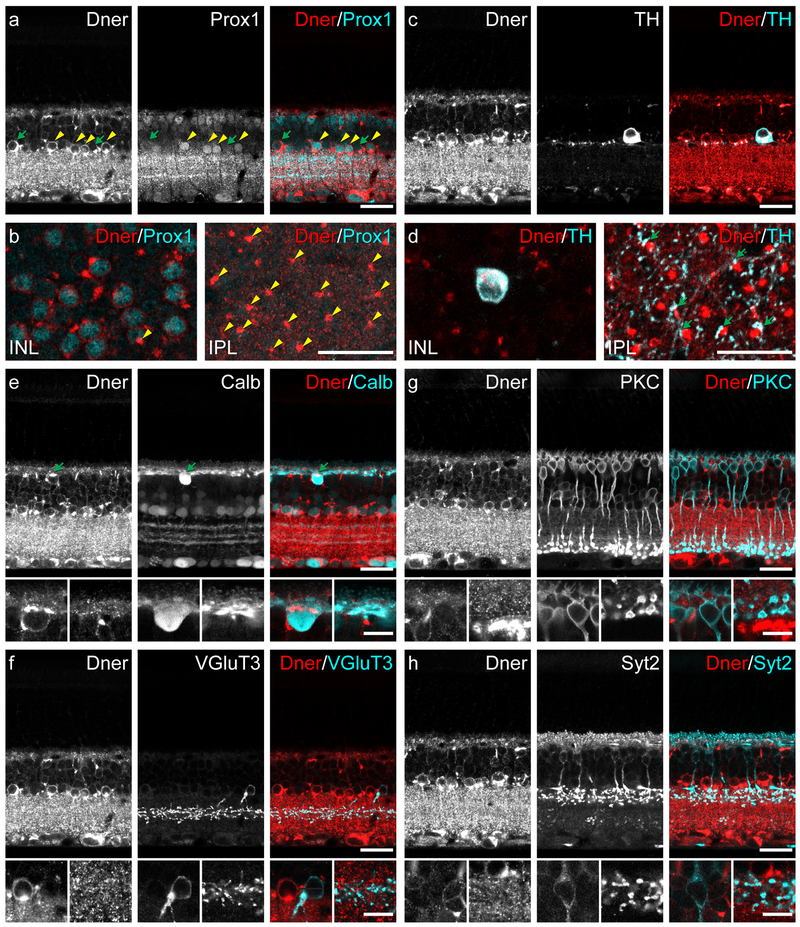
DNER is localized to the somatic membrane of cells in the inner nuclear layer and ganglion cell layer, and is found throughout the plexiform layers.
As shown in figure 3a, antibodies to DNER revealed strong punctate labeling throughout both the IPL and OPL, and many, but not all, cell bodies were labeled in the GCL and INL. We did not detect any labeling in the outer nuclear layer (ONL). This pattern of labeling appears to be consistent with the expression profile analysis, with broad expression in amacrine, ganglion, and horizontal cells, and a lack of expression in bipolar and photoreceptor cells (figure 2b). Additionally, the labeling appeared to be membranous, as expected of a transmembrane protein; indeed, previous studies overexpressing DNER in cell culture found the protein in the plasma membrane of the soma and dendritic compartments, as well as in the membrane of cytoplasmic endosomes (Eiraku et al., 2002).
Figure 3: Localization of DNER protein in adult retina.
(a) Co-labeling of DNER and Prox1, the latter being a known marker of AII amacrine cells, revealed a population of double-labeled neurons along the inner margin of the INL (yellow arrowheads), although not all DNER+ cells in this location were Prox1+ (green arrows). (b) Each DNER+/Prox+ amacrine cell gave rise to an intensely DNER-immunoreactive dendritic stalk (yellow arrowheads) projecting into the IPL. (c) TH+ dopaminergic amacrine cells in the INL also have strong DNER expression. (d) Dopaminergic amacrine cells stratify their processes in S1 of the IPL, at the same depth as the DNER+ dendritic stalks of the presumptive AII amacrine cells. These two cell types are known to make synapses at this stratum of the IPL; consistent with this, many TH+ puncta could be seen in close apposition with DNER+ stalks (green arrows). (e) Calbindin+ cells were often DNER+ as well, including the horizontal cells in the outer edge of the INL (green arrow) and several types of amacrine cells. (f) VGlut3+ amacrine cells were not DNER+. (g) PKC+ rod bipolar cells were not DNER+. (h) Syt2+ type 2 bipolar cells were not DNER+. Higher magnification panels in (e), (f), (g) and (h) are one micron thick optical sections and illustrate the somata and processes of horizontal cells, VGluT3+ amacrine cells, rod bipolar cells and type 2 bipolar cells, respectively. Scale bars = 10 μm for high magnification panels, 25 μm for all other panels.
Cells in the INL varied in their intensity of DNER labeling. In particular, there was an intensely immunopositive population of cells in the INL, residing adjacent to the IPL. The somal membrane is labeled, particularly along the basal side of the cell, giving rise to a thick DNER+ dendritic stalk extending into the IPL, where it quickly disappears in a dense thicket of labeling that fills the plexiform layer (figure 3a, 3b). Note that many of these labeled cells are also Prox1+, with the DNER labeling encircling the Prox1+ nucleus (figure 3a, 3b), suggesting that a subset of these cells are indeed the AII amacrine cells. While every Prox1+ amacrine cell exhibited DNER labeling and had a heavily labeled dendritic stalk (figure 3b, yellow arrowheads), other cells at this depth of the INL are also DNER+, some of which include the dopaminergic amacrine cells, readily distinguished by their larger size, shape, and tyrosine hydroxylase (TH) immunoreactivity (figure 3c, 3d). These cells are known to make synaptic contacts with the AII amacrine cells at the very outer limit of the IPL (Marc et al., 2014), and consistent with this, we found many instances of TH+ puncta directly juxtaposed with the DNER+ dendritic stalks (figure 3d, green arrows). Many of the remaining DNER+ cells in the INL were also calbindin+, particularly the horizontal cells, which displayed a conspicuous concentration of DNER adjacent to the OPL where the dendrites emerge from the soma (figure 3e, green arrow). Additionally, some other calbindin+ amacrine cells were also DNER+, likely to include the cholinergic amacrine cells (figure 3e). DNER did not appear to be present in the VGluT3+ amacrine cells (figure 3f), nor the rod bipolar cells (figure 3g) or type 2 bipolar cells (figure 3h), nor the Muller glia (not shown).
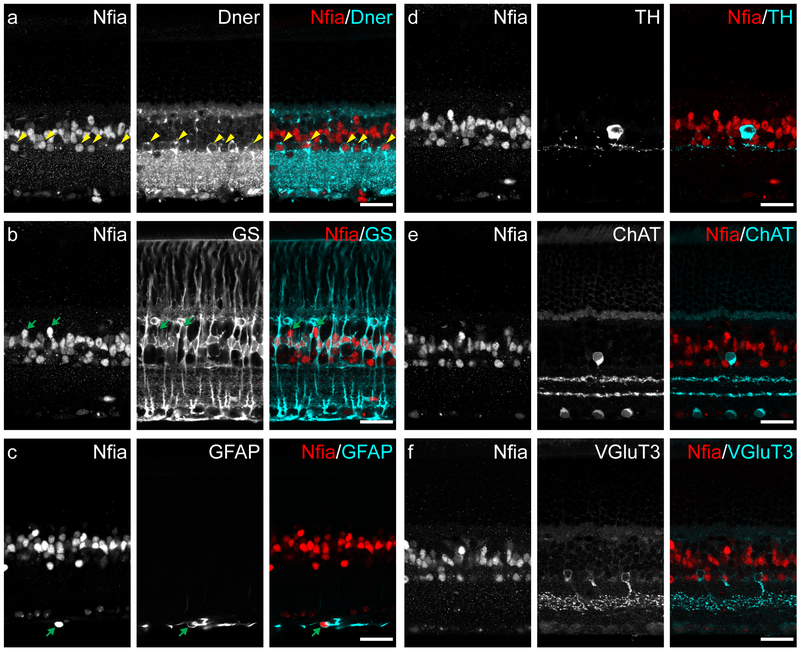
NFIA is localized to the nuclei of cells in the inner nuclear layer and nerve fiber layer.
Antibodies to NFIA labeled the nuclei of several populations of cells in the retina, residing in the amacrine cell layer and middle of the INL, as well as sparse nuclei in the nerve fiber layer (figure 4a). When double-labeling with the antibody to DNER, many of the NFIA+ cells in the amacrine cell layer, although not all, colocalized with the intensely labeled DNER+ profiles indicative of the AII amacrine cells (figure 4a, yellow arrowheads). Most of the NFIA+ cells in the middle of the INL were immunopositive for glutamine synthetase (GS), a marker of Müller glia (figure 4b), while those cells that were not GS+ had a location and shape consistent with being cone bipolar cells (figure 4b, green arrows), perhaps being the Type 5A and/or 5D cone bipolar cell as suggested by the expression profile data (figure 2b). The NFIA+ nuclei in the nerve fiber layer were enveloped by the intermediate filament GFAP (green arrow, figure 4c), suggesting that the population of retinal astrocytes also express this transcription factor, being consistent with the expression data (figure 2b). The NFIA+ cells in the amacrine cell layer that were not the AII amacrine cells were also neither the dopaminergic amacrine cells (figure 4d), the cholinergic amacrine cells (figure 4e), nor the VGluT3+ amacrine cells (figure 4f), and therefore likely to be one or two of the currently unidentified GABAergic amacrine cell types (figure 2b).
Figure 4: Localization of NFIA protein in adult retina.
(a) NFIA+ cell nuclei were found in the INL, both in the middle and along the inner edge. Many NFIA+ nuclei closest to the IPL (yellow arrowheads) were also DNER+, potentially indicative of the AII amacrine cell population. (b) Most NFIA+ nuclei in the middle of the INL were also GS+, and thus the population of Muller glia; the remaining NFIA+ profiles in this layer appeared to be a sparse type or types of bipolar cell (green arrows). (c) NFIA+ nuclei in the inner most edge of the retina, the nerve fiber layer, were surrounded by GFAP labeling (green arrow), indicative of being retinal astrocytes. (d) TH+ amacrine cells were not NFIA+. (e) ChAT+ amacrine cells were not NFIA+. (f) VGluT3+ amacrine cells were not NFIA+. Scale bars = 25 μm.
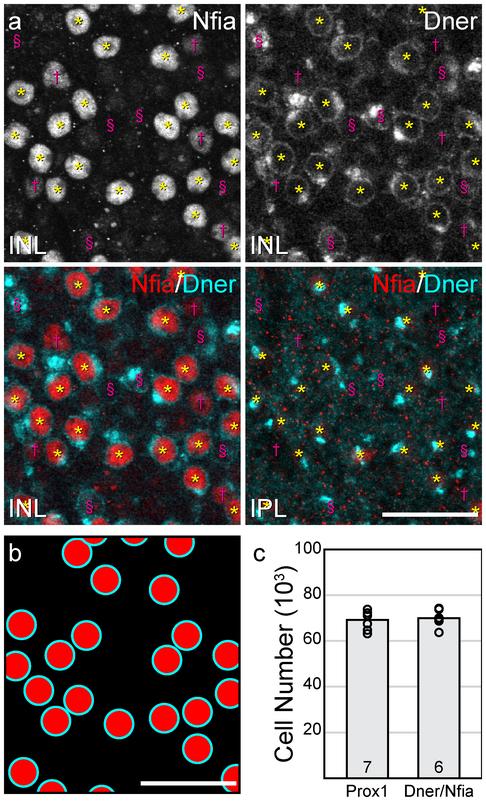
Double-labeling for NFIA and DNER allows for the accurate identification of AII amacrine cells.
When double-labeling the retina in wholemount preparations for both NFIA and DNER, a set of intensely labeled NFIA+ nuclei colocalizes with the conspicuous DNER+ dendritic stalk (figure 5a, yellow asterisks), while other cells show fainter NFIA-labeling though lack DNER (figure 5a, magenta daggers), while still others are clearly DNER+, but show no trace of NFIA-labeling (figure 5a, magenta section symbols). The coincidence of these two labels identifying a population of double-labeled cells (e.g. figure 5b) is present at all locations across the retina, in densities that appear similar to the population of Prox1+ AII amacrine cells (Keeley et al., 2014). Their local distribution (e.g. figure 5b) also mimics the random somal patterning shown to be present in the Prox1+ amacrine cell population in the mouse retina (Keeley and Reese, in press). Because our Prox1 antibody was raised in the same species to that used for generating the NFIA antibody, we could not directly compare the double-labeled population to the Prox1+ population. Instead, we have estimated the total number of DNER+/ NFIA+ double-labeled cells at the inner margin of the INL, and compared this directly with estimates of the number of Prox1+ cells at this depth. The total population of AII amacrine cells, labeled with Prox1 antibodies, in the B6/J strain of mouse has been previously estimated to be 69,223 ± 1,566 (mean ± SEM) cells per retina (figure 5c; Keeley et al., 2014). Our estimated total number of DNER+/ NFIA+ double-labeled cells, conducted in this very strain, was 69,998 ± 1,572 cells per retina (figure 5c). We note that the variance is both low, as well as comparable, between the labeling methods, the coefficient of variation being 0.06 in both cases. We therefore conclude that this double-labeling strategy is effective in unambiguously identifying AII amacrine cells.
Figure 5: Quantification of double-labeled AII amacrine cells.
(a) Wholemount retinas stained for NFIA and DNER allow for the identification of the AII amacrine cell population, as these cells have nuclei that are NFIA+ residing at the inner margin of the INL and have a DNER+ stalk extending into the IPL (yellow asterisks). NFIA+ but DNER- populations are present (magenta daggers), as are DNER+ but NFIA- populations (magenta section symbols). (b) The distribution of DNER+/NFIA+ AII amacrine cells, identified in (a), illustrates the method of counting cells to estimate total cell number. (c) Estimates of the total number of AII amacrine cells, derived from B6/J retinas in which Prox1+ amacrine cells were counted (Keeley et al., 2014), were strikingly similar to estimates from B6/J retinas in which DNER+/NFIA+ amacrine cells were counted. n = number of retinas. Scale bars = 25 μm.
DNER and NFIA are expressed in the early postnatal retina, suggesting a developmental role for both proteins.
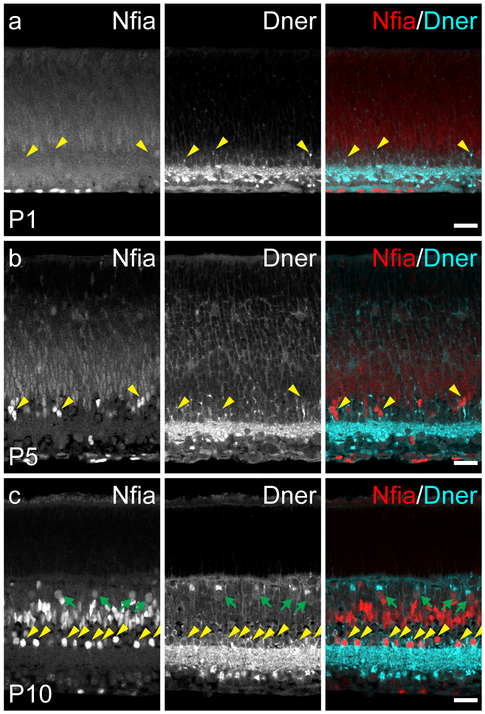
As predicted by the transcriptome analysis (e.g. figure 2c, 2d), both genes were expressed during early postnatal development. On the day of birth, NFIA-labeling is only present in nuclei residing on the inner surface of the retina. The positioning and elongated morphology of these cells are both consistent with their being the population of retinal astrocytes that invade the retina from the optic nerve during the perinatal period; there is no NFIA present in cells endogenous to the retina at this age (figure 6a). DNER, by contrast, is expressed throughout the GCL and IPL in a similar pattern as seen in adulthood, with punctate labeling in the plexiform layer and strong localization of protein in somata in the GCL (figure 6a). DNER+ puncta, likely presaging the intense labeling of AII amacrine cell dendritic stalks, can also be seen forming in the nascent INL (figure 6a, yellow arrowheads), although the intensity of labeling is much weaker. At postnatal day 5, intensely-labeled NFIA+ nuclei appear in the INL, many of which are adjacent to DNER+ stalks extending to the IPL, indicative of differentiating AII amacrine cells (figure 6b, yellow arrowheads). By postnatal day 10, the AII amacrine cell population is clearly revealed by double-labeling with these two markers, as a tightly stratified population of NFIA+ and DNER+ cells is present at the inner edge of the INL (figure 6c, yellow arrowheads). Also present are the populations of NFIA+ nuclei in the middle of the INL (figure 6c), with a comparable density and distribution that is seen in adulthood, with one notable exception: faintly labeled NFIA+ nuclei residing in the outer half of the INL from which bright DNER+ profiles extend in the OPL (figure 6c, green arrows). These cells are likely the population of horizontal cells, given their infrequency and positioning, despite their lack of NFIA expression in adulthood. Taken together, these results indicate that expression of NFIA is gradually elevated during postnatal development of the retina, first in immigrating astrocytes, and then in AII amacrine cells, followed by Müller glia, bipolar cells, and horizontal cells, with the latter expressing the protein only transiently. DNER, by contrast, is already present in the retina by the day of birth, likely originating from the populations of retinal ganglion cells and AII amacrine cells, with expression expanding throughout the INL and OPL over time. Neither protein was found in the neuroblastic layer at any postnatal time point, suggesting that these genes most likely exert their influence on postmitotic, differentiating cells of the retina.
Figure 6: DNER and NFIA expression in postnatal retinal development.
(a) On the day of birth (P1), NFIA was only expressed in nuclei along the innermost margin of the retina, indicative of retinal astrocytes. DNER, by contrast, was expressed in the GCL and IPL, with some DNER+ stalks observed in the developing INL (yellow arrowheads). (b) At P5, NFIA was found in nuclei of cells in the INL, some of which also appeared to be juxtaposed to strong DNER labeling (yellow arrowheads). (c) At P10, NFIA was expressed in a population of amacrine cells along the inner margin of the INL, many cells in the middle of the INL, and a population of cells near the outer margin of the INL. DNER expression was maintained in the GCL and IPL, and now also expressed by cells in the amacrine cell division of the INL, identifying the population of AII amacrine cells by virtue of their coincident expression of NFIA (yellow arrowheads), and to the population of horizontal cells, which were also transiently NFIA+ at this age (green arrows). Scale bars = 25 μm.
Discussion
In this study, we used publicly available expression datasets of individual retinal cell types (Kay et al., 2012; Macosko et al., 2015; Shekhar et al., 2016) to identify two genes, Dner and Nfia, that are highly expressed in AII amacrine cells. We verified that the proteins are detectable in these cells, and while each is not exclusive to the AII amacrine cells, labelling for both proteins allows for accurate identification of this entire population. In addition, these genes are expressed in early postnatal development, being upregulated in AII amacrine cells during this time, suggesting a potential role of these two genes in their differentiation. Each of these genes has been previously studied with respect to the development of structures elsewhere in nervous system, which may prove relevant for understanding the development of the AII amacrine cell.
DNER is expressed in various brain regions, including the developing and mature cortex, cerebellum, and hippocampus. Initial reports indicated that the protein is localized to the plasma membrane of the dendritic compartment, but is excluded from the axonal compartment (Eiraku et al., 2002), while cell culture experiments confirmed that DNER expression on the plasma membrane plays a promotive role in neuritogenesis (Fukazawa et al., 2008). Knockout mice showed impairments in motor function considered a consequence of abnormal cerebellar development, where a delay in the morphogenesis of this structure is thought to disrupt the refinement of Purkinje cell connectivity in adulthood (Tohgo et al., 2006; Saito and Takeshima, 2006). The localization of DNER to the dendritic compartment of neurons, and its role in neuronal development in vitro and in vivo, has implications for the development of the AII amacrine cell. The arbor of this cell exhibits an exquisite compartmentalization into four regions, here listed emerging from the soma: the neck, where it receives inputs from the dopaminergic amacrine cells; the lobular appendages, the site of conventional glycinergic output synapses with OFF cone bipolar cells and retinal ganglion cells; the waist, mainly a transitional zone; and the arboreal dendrites, where the cell receives input from rod bipolar cells, and is electrically coupled to all ON cone bipolar cells as well as other AII amacrine cells (Marc et al., 2014). We have shown that DNER is accumulated at the neck of the AII amacrine cells, and that this pattern is present during early postnatal development, suggesting a potential role in the differentiation of AII amacrine cell morphology and connectivity.
The function of Dner in the zebrafish has been characterized as well, shown to induce the differentiation of neurons and glia at the expense of proliferation when overexpressed in neural progenitor cells, and these effects were determined to work through both Notch-dependent and Notch-independent mechanisms (Hsieh et al., 2013). Notch signaling is involved in cell fate decisions in the retina (Livesey and Cepko, 2001), particularly in the creation of Müller glia (Vetter and Moore, 2001). As we found no evidence of DNER expression in the neuroblastic layer of the postnatal retina, however, this seems an unlikely function for it in the developing mouse retina. It should be noted as well that a recent report has indicated that this protein is not a direct ligand of Notch, as previously assumed (Greene et al., 2016), complicating the interpretation of how DNER might play a role in these processes.
NFIA is also widely expressed throughout the developing nervous system. It is one of four NFI transcription factors, all of which display unique, but overlapping, patterns of expression during development and in the adult mouse (Gronostajski et al., 2000). All of these transcription factors are found in the brain, and play a critical role in neural development, as mice deficient in these proteins exhibit many behavioral dysfunctions (Mason et al., 2009). One well studied role for NFIA, in particular, is in gliogenesis, as knockout mice show an absence of important glial structures at the midline leading to a failure of the corpus callosum to form (das Neves et al., 1999; Shu et al., 2003). As a key player in the differentiation of glial populations, NFIA interacts with many other known transcription factors that participate in this process, such as Hes1, Lhx2, Sox9, and Sox10, as well as influencing the Notch pathway in progenitor cells (Piper et al., 2010; Subramanian et al., 2011; Kang et al., 2012; Glasgow et al., 2014). It is interesting to note that many of these genes are also active in the developing retina (de Melo et al., 2016), and that we have found NFIA to be expressed in both Müller glia and astrocytes in the developing and mature retina, suggesting a similar role for NFIA in gliogensis in the retina. Indeed, NFIA overexpression in early postnatal retinal progenitor cells promotes the formation of cells with Müller glia-like radial morphology (de Melo et al., 2016), although overexpression itself is not sufficient to create mature glial cells.
NFIA does not just affect glial populations, however, as it appears to play a role in neuronal development as well (Planchez et al., 2009). NFIA is expressed by cerebellar granule cells and is involved in their migration, dendritic arborization, and axon guidance (Wang et al., 2007). Abnormalities in these processes were observed in knockout mice, as was a decrease in the expression of several cell adhesion molecules, two of which were confirmed to have NFI binding sites upstream of their promoters, suggesting direct regulation of these genes by NFIA (Wang et al., 2007). It is possible that similar mechanisms may be at play in the AII amacrine cell population, as these cells express several cell adhesion molecules in development, such as Cadherin1, EphrinA5, and EphA7 (Kay et al., 2012). The Dner gene, itself coding for an adhesion molecule, contains a NFI binding motif in the first intron, hinting at a possible regulation by NFIA. Future studies that target these genes in the retina, including knockout and misexpression experiments, should help elucidate the role of these two molecules in the development of the retina, and in the population of AII amacrine cells themselves.
Acknowledgements
This work was supported by grants from the National Institutes of Health (EY019968; S10 OD010610). We thank Caroline Ackley for conducting the counts of double-labeled AII amacrine cells.
References
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, & Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature. 529(7586):345–50. doi: 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JS, & Schachner M (1978). Localization of nervous system antigens in retina by immunohistology. Invest Ophthalmol Vis Sci. 17(9):920–4. [PubMed] [Google Scholar]
- Cherry TJ, Wang S, Bormuth I, Schwab M, Olson J, & Cepko CL (2011). NeuroD factors regulate cell fate and neurite stratification in the developing retina. J Neurosci. 31(20):7365–79. doi: 10.1523/JNEUROSCI.2555-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves L, Duchala CS, Tolentino-Silva F, Haxhiu MA, Colmenares C, Macklin WB, Campbell CE, Butz KG, & Gronostajski RM (1999). Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc Natl Acad Sci USA. 96(21):11946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Clark BS, & Blackshaw S (2016). Multiple intrinsic factors act in concert with Lhx2 to direct retinal gliogenesis. Sci Rep. 6:32757. doi: 10.1038/srep32757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb JB, & Singer JH (2012). Intrinsic properties and functional circuitry of the AII amacrine cell. Vis Neurosci. 29(1):51–60. doi: 10.1017/S0952523811000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, & Oliver G (2003). Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 34(1):53–8. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Hirata Y, Takeshima H, Hirano T, & Kengaku M (2002). Delta/notch-like epidermal growth factor (EGF)-related receptor, a novel EGF-like repeat-containing protein targeted to dendrites of developing and adult central nervous system neurons. J Biol Chem. 277(28):25400–7. [DOI] [PubMed] [Google Scholar]
- Famiglietti EV, & Kolb H (1975). A bistratified amacrine cell and synaptic circuitry in the inner plexiform layer of the retina. Brain Res. 84(2):293–300. [DOI] [PubMed] [Google Scholar]
- Fox MA, & Sanes JR (2007). Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol. 503(2):280–96. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, & Burgess RW (2009). DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 64(4):484–97. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa N, Yokoyama S, Eiraku M, Kengaku M, & Maeda N (2008). Receptor type protein tyrosine phosphatase zeta-pleiotrophin signaling controls endocytic trafficking of DNER that regulates neuritogenesis. Mol Cell Biol. 28(14):4494–506. doi: 10.1128/MCB.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Zhu W, Stolt CC, Huang TW, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, & Deneen B (2014). Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 17(10):1322–9. doi: 10.1038/nn.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene M, Lai Y, Pajcini K, Bailis W, Pear WS, & Lancaster E (2016). Delta/Notch-Like EGF-Related Receptor (DNER) Is Not a Notch Ligand. PLoS One. 11(9):e0161157. doi: 10.1371/journal.pone.0161157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM (2000). Roles of the NFI/CTF gene family in transcription and development. Gene. 249(1–2):31–45. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, & Wässle H (2004). Characterization of an amacrine cell type of the mammalian retina immunoreactive for vesicular glutamate transporter 3. J Comp Neurol. 468(2):251–63. doi: 10.1002/cne.10962. [DOI] [PubMed] [Google Scholar]
- Haycock JW, & Waymire JC (1982). Activating antibodies to tyrosine hydroxylase. J Biol Chem. 257(16):9416–23. [PubMed] [Google Scholar]
- Hsieh FY, Ma TL, Shih HY, Lin SJ, Huang CW, Wang HY, & Cheng YC (2013). Dner inhibits neural progenitor proliferation and induces neuronal and glial differentiation in zebrafish. Dev Biol. 375(1):1–12. doi: 10.1016/j.ydbio.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Kang TH, Ryu YH, Kim IB, Oh GT, & Chun MH (2004). Comparative study of cholinergic cells in retinas of various mouse strains. Cell Tissue Res. 317(2):109–15. doi: 10.1007/s00441-004-0907-5. [DOI] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, & Deneen B (2012). Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 74(1):79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Chu MW, & Sanes JR (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 483(7390):465–9. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, & Reese BE (2017). The somal patterning of the AII amacrine cell mosaic in the mouse retina is indistinguishable from random simulations matched for density and constrained by soma size Vis Neurosci. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Whitney IE, Madsen NR, St John AJ, Borhanian S, Leong SA, Williams RW, & Reese BE (2014). Independent genomic control of neuronal number across retinal cell types. Dev Cell. 30(1):103–9. doi: 10.1016/j.devcel.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Whitney IE, & Reese BE (2017). Genomic control of retinal cell number: challenges, protocol, and results. Methods Mol Biol. 1488:365–390. doi: 10.1007/978-1-4939-6427-7_17. [DOI] [PubMed] [Google Scholar]
- Kowalik L & Hudspeth AJ (2011). A search for factors specifying tonotopy implicates DNER in hair-cell development in the chick’s cochlea. Dev Biol. 354(2):221–31. doi: 10.1016/j.ydbio.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linser PJ, Sorrentino M, & Moscona AA (1984). Cellular compartmentalization of carbonic anhydrase-C and glutamine synthetase in developing and mature mouse neural retina. Dev Brain Res. 13(1):65–71. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, & Cepko CL (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2(2):109–18. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, & McCarroll SA (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161(5):1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc RE, Anderson JR, Jones BW, Sigulinsky CL, & Lauritzen JS (2014). The AII amacrine cell connectome: a dense network hub. Front Neural Circuits. 8:104. doi: 10.3389/fncir.2014.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH (2102). The neuronal organization of the retina. Neuron. 76(2):266–80. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S, Piper M, Gronostajski RM, & Richards LJ (2009). Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 39(1):10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- Massey SC, & Mills SL (1999). Antibody to calretinin stains AII amacrine cells in the rabbit retina: double-label and confocal analyses. J Comp Neurol. 411(1):3–18. [PubMed] [Google Scholar]
- Nagao M, Ogata T, Sawada Y, & Gotoh Y (2016). Zbtb20 promotes astrocytogenesis during neocortical development. Nat Commun. 7:11102. doi: 10.1038/ncomms11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Sevilla Müller L, Azar SS, de Los Santos J, & Brecha NC (2017). Prox1 is a marker for AII amacrine cells in the mouse retina. Front Neuroanat. 11:39. doi: 10.3389/fnana.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Grünert U, Studholme K, Yazulla S, Kirsch J, & Becker CM (1994). Glycine receptors in the retinas of normal and spastic mutant mice. Invest Ophthalmol Vis Sci. 35(10):3633–9. [PubMed] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, Tole S, Gronostajski RM, Bailey TL, & Richards LJ (2010). NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 30(27):9127–39. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, Osinski JM, Gronostajski RM, & Richards LJ (2008). Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 508(3):385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- Pochet R, Pasteels B, Seto-Ohshima A, Bastianelli E, Kitajima S, & Van Eldik LJ (1991). Calmodulin and calbindin localization in retina from six vertebrate species. J Comp Neurol. 314(4):750–62. doi: 10.1002/cne.903140408. [DOI] [PubMed] [Google Scholar]
- Raven MA, Stagg SB, Nassar H, & Reese BE (2005). Developmental improvement in the regularity and packing of mouse horizontal cells: Implications for mechanisms underlying mosaic pattern formation. Vis Neurosci. 22(5):569–73. doi: 10.1017/S095252380522504X [DOI] [PubMed] [Google Scholar]
- Rice DS, & Curran T (2000). Disabled-1 is expressed in type AII amacrine cells in the mouse retina. J Comp Neurol. 424(2):327–38. [DOI] [PubMed] [Google Scholar]
- Saito SY, & Takeshima H (2006). DNER as key molecule for cerebellar maturation. Cerebellum. 5(3):227–31. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, & Sanes JR Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 166(5):1308–1323. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, & Richards LJ (2003). Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 23(1):203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, & Roska B (2012). Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 15(3):487–95. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- Strettoi E, Raviola E, & Dacheux RF (1992). Synaptic connections of the narrow-field, bistratified rod amacrine cell (AII) in the rabbit retina. J Comp Neurol. 325(2):152–68. [DOI] [PubMed] [Google Scholar]
- Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, & Tole S (2011). Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc Natl Acad Sci USA. 108(27):E265–74. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton WG, Kwan MM, Verrier MC, Seniuk NA, & Theriault E (1990). MPTP produces reversible disappearance of tyrosine hydroxylase-containing retinal amacrine cells. Brain Res. 527(1):21–31. [DOI] [PubMed] [Google Scholar]
- Tohgo A, Eiraku M, Miyazaki T, Miura E, Kawaguchi SY, Nishi M, Watanabe M, Hirano T, Kengaku M, & Takeshima H (2006). Impaired cerebellar functions in mutant mice lacking DNER. Mol Cell Neurosci. 31(2):326–33. [DOI] [PubMed] [Google Scholar]
- Trevarrow B, Marks DL, & Kimmel CB (1990). Organization of hindbrain segments in the zebrafish embryo. Neruon. 4(5):669–79. [DOI] [PubMed] [Google Scholar]
- Versaux-Botteri C, Nguyen-Legros J, Vigny A, & Raoux N (1984). Morphology, density and distribution of tyrosine hydroxylase-like immunoreactive cells in the retina of mice. Brain Res. 301(1):192–7. [DOI] [PubMed] [Google Scholar]
- Vetter ML, & Moore KB (2001). Becoming glial in the neural retina. Dev Dyn. 221(2):146–53. [DOI] [PubMed] [Google Scholar]
- Wässle H, Grünert U, & Röhrenbeck J (1993). Immunocytochemical staining of AII-amacrine cells in the rat retina with antibodies against parvalbumin. J Comp Neurol. 332(4):407–20. [DOI] [PubMed] [Google Scholar]
- Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, & Kilpatrick DL (2007). Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 27(23):6115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]