Abstract
Ferritin, a ubiquitous iron storage protein, has a crucial role in innate immunity in arthropods, which have no adaptive immune system. Arthropods are thought to have two types of ferritin molecules: the secreted type and the cytosolic type. Here, we present the first crystal structure of ferritin from crustacean, kuruma prawn (Marsupenaeus japonicus), at 1.16 Å resolution. This shrimp ferritin (MjFer) is the cytosolic type, and its structure shows well‐conserved ferritin fold composed of a 4‐helix bundle that assembles into a cage‐like 24‐mer. The structure of MjFer was more similar to those of human and vertebrate ferritins than to that of the secreted‐type arthropod ferritin from an insect. MjFer possesses both a ferroxidase site and a nucleation site, which are the main characteristics of vertebrate H and L chain ferritins, respectively. The first crystal structure of crustacean ferritin, MjFer, has exceptionally high quality that provides the detailed structural information of metal moving pathway in ferritin.
Keywords: ferritin, crustacean, iron, ferroxidase site, nucleation site
Short abstract
PDB Code(s): http://firstglance.jmol.org/fg.htm?mol=6A4U
Introduction
Iron is an essential element in virtually all living species. However, excess iron in the living cell is harmful due to its highly reactive nature. Ferritin is a ubiquitous iron storage protein that sequesters iron in a nontoxic and biologically available form in its spherical cage‐like oligomer.1, 2 Needless to say, pathogenic organisms, such as bacteria and fungi, also require iron to proliferate in hosts. Accordingly, an iron‐withholding strategy is one of the major strategies in immune systems, especially in innate immunity.3, 4, 5 To defend themselves from pathogenic organisms, arthropods depend solely on innate immunity because they lack adaptive immunity. Therefore, ferritin has been identified as a major component of innate immunity system and a stress‐inducible protein in arthropods because it can accumulate iron effectively and, as a result, it takes away usable iron from pathogens. Since 1996, when ferritin protein and its cDNA were first identified in crayfish,6 cDNA sequences from crustaceans with great market value have been identified and their expression patterns were investigated. A 2005 three‐dimensional structural study determined the crystal structure of a secreted‐type ferritin from an insect (Trichoplusia ni).7 In contrast to insects, in which most of the ferritins identified are the secreted type,8 most crustacean ferritins identified do not contain signal peptides, suggesting that they may be cytosolic proteins,6, 9, 10, 11, 12, 13, 14, 15 although a few secreted‐type ferritins have also been reported in crustaceans.16 The expressions of these ferritin genes are generally upregulated by the presence of a pathogenic organism or its component and by an overload of iron or another heavy metal. Furthermore, exogenous injection of purified crustacean ferritin protects the hosts from microbial infection.9
In general, vertebrate ferritins can be classified into several groups, such as those of H, L, and M chains or as mitochondrial ferritins. The ferritin H chain (FtH) possesses the ferroxidase site that catalyzes the rapid oxidation of ferrous iron, whereas the L chain (FtL) does not have this catalytic site but does have a nucleation site, which facilitates the iron core formation in the inner cavity of ferritin. In mammals, these two ferritin chains are mixed to form heteropolymers in various ratios depending on the organs, and these two chains have cooperative roles in iron sequestration.17 In contrast, most crustacean ferritins have both sites in their primary sequences. Here, we present the first crystal structure of ferritin from a crustacean (kuruma prawn, Marsupenaeus japonicus), which is cytosolic ferritin similar to its vertebrate counterpart. Its crystal structure has among the highest resolution ever reported for ferritin structures in various species. It provides the detailed structural features of “hybrid‐type ferritins” of FtH and FtL.
Results and Discussion
Overall structure of MjFer and its channel structures
X‐ray diffraction data were collected up to 1.16 Å resolution at SPring‐8 BL26B1. The I4 crystal of MjFer contained six monomers in its asymmetric unit (PDB ID: http://firstglance.jmol.org/fg.htm?mol=6A4U). The crystallographic and refinement statistics are shown in Table 1. Similar to other ferritin family proteins, MjFer forms a spherical oligomer composed of 24 subunits [Fig. 1(A,B)]. This structure of MjFer would be a functional form of crustacean ferritin, because Huang et al. demonstrated that the purified crayfish ferritin, which has quite similar primary sequence to MjFer (~70%), forms an oligomer that has a comparable molecular weight with vertebrate ferritin.6 It shows the general characteristics of ferritin structures, such as four‐, three‐, and twofold symmetry axes and channels along with the three‐ and fourfold axes. The rms deviation of the main chain between MjFer and both FtH (PDB ID: http://firstglance.jmol.org/fg.htm?mol=3ajo)18 and FtL (PDB ID: http://firstglance.jmol.org/fg.htm?mol=2ffx)19 are 0.69 Å for 171 amino acid residues, and that between MjFer and bullfrog M ferritin (PDB ID: http://firstglance.jmol.org/fg.htm?mol=3ka3) is 0.62 Å for 171 residues, while that between MjFer and insect secreted‐type ferritin (PDB ID: http://firstglance.jmol.org/fg.htm?mol=1z6o)7 is 1.37 Å for 171 amino acid residues, suggesting that MjFer is more closely related to the vertebrate ferritin than to the insect secreted‐type ferritin, despite the evolutional distance. However, among the three‐dimensional structures of ferritin family proteins available in PDB, ferritin from the marine worm, Chaetopterus sp (ChF)20 (PDB ID: http://firstglance.jmol.org/fg.htm?mol=5wpn), has the closest structure with MjFer. ChF has been identified as secreted type of ferritin from marine worm. The amino acid sequence identity between MjFer and ChF is 66.5% and the main chain rmsd is 0.478 Å. The detailed structure around the threefold channel is shown in Figure 1(C,E). Generally, the threefold channel forms a funnel‐like pore that is lined with negatively charged amino acid side chains of glutamate and aspartate, rendering it hydrophilic and suitable for metal entry. For example, the threefold channel of human H ferritin homo polymer has two acidic side chains, Asp131 and Glu134,18, 21, 22 both of which are highly conserved among ferritins from vertebrates, plants,23 and algae,24 while they are substituted in the bacterial type of ferritin.25, 26 The threefold symmetry channel of MjFer is lined with Glu133, corresponding to the conserved Glu134, and with Asp134. The highly conserved Asp131 of FtH is substituted to lysine in MjFer, although this channel still has a negatively charged surface because of the two acidic residues (Glu133 and Asp134) [Fig. 1(C)]. In contrast to the threefold channel, the channels along the fourfold symmetry axes form a hydrophobic and tightly packed channel lined with the side chains of Leu162 and Met166, similar to the case of vertebrate ferritins [Fig. 1(D,F)].
Table 1.
Crystallographic Statistics
| Data collection | |
|---|---|
| X‐ray source | SPring‐8 BL26B1 |
| Detector | R‐AXIS V |
| Wave length (Å) | 0.8 |
| Resolution (Å) | 50.00–1.16 (1.18–1.16) |
| Space group | I4 |
| Unit cell parameters | |
| a, b (Å) | 124.872 |
| c (Å) | 175.683 |
| Total reflections | 1,405,342 |
| Unique reflections | 456,412 (22,818) |
| Completeness (%) | 98.8 (99.0) |
| Redundancy | 3.1 (2.9) |
| Mean I/σ (I) | 26.9 (3.08) |
| R merrge | 0.056 (0.464) |
| R pim | 0.036 (0.327) |
| CC1/2 (highest resolution shell) | 0.729 |
| Refinement | |
| R work/R free | 0.1377/0.1566 |
| No. of atoms/Bfactors (Å2) | |
| Protein | 9351/11.63 |
| Ligands | |
| Ethylene glycol | 200/31.20 |
| Magnesium | 9/37.07 |
| sulfate | 65/67.49 |
| Water | 1007/26.40 |
| RMSD from ideal geometry | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.838 |
| Coordinate error (Å) | 0.09 |
| Ramachandran plot | |
| Favored regions (%) | 98.5 |
| Allowed regions (%) | 1.50 |
| Outliners (%) | 0 |
| PDB code | http://firstglance.jmol.org/fg.htm?mol=6A4U |
Figure 1.
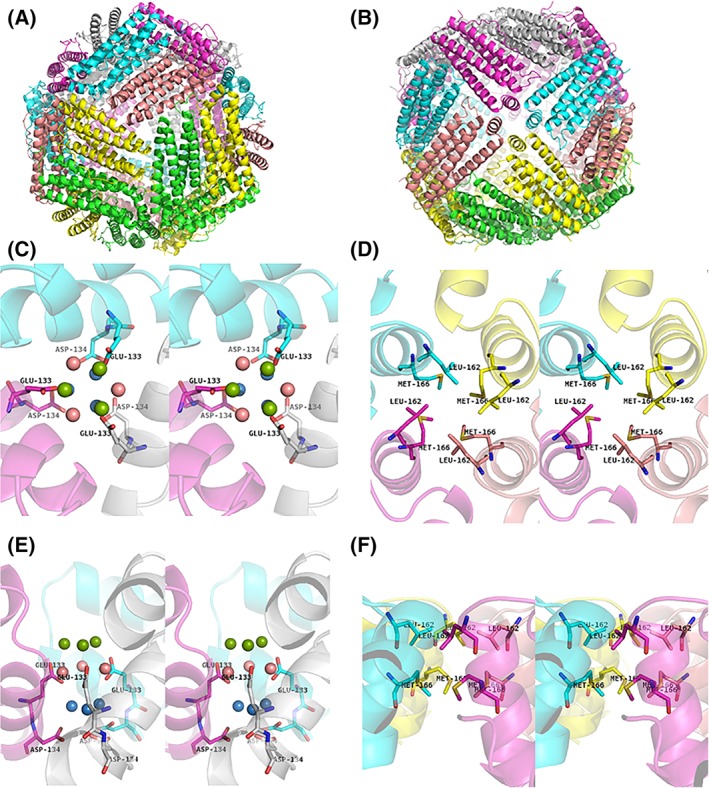
Overall structure of MjFer viewed from the (A) threefold symmetry axis and (B) fourfold symmetry axis. (C) An enlarged image of the threefold metal entry channel of MjFer viewed from the threefold symmetry axis (stereo image). The acidic side chains (Glu133 and Asp134) around this channel are shown as sticks. The water molecules seen around the channel are shown as balls. The water molecules located in the outer, middle, and inner parts of the channel are shown in green, pink, and blue, respectively. (D) An enlarged image of the channel around the fourfold symmetry axis (stereo image). The amino acid residues (Leu162 and Met166) that form the narrow channel are shown as sticks. (E,F) The cross‐sectional view of the (C) threefold and (D) fourfold channels, respectively.
Structure of the ferroxidase site and nucleation site
Ferroxidation is the crucial process of iron incorporation into the ferritin shell. The ferroxidase site has been identified and characterized in FtH, bullfrog M ferritin, and other molecules. The ferroxidase site in FtH is composed of Glu27, Tyr34, Glu62, His65, Glu107, and Gln141,21, 27 while the corresponding residues are Glu24, Tyr31, Glu60, His63, Glu106, and Gln140 in MjFer [Fig. 2(B)]. Among the amino acid side chains in the ferroxidase site, the side chains of His63 in MjFer have double conformation [Fig. 2(B)]. However, no clear electron density of metal ligands is visible around the site. Instead, site A of the ferroxidase center is occupied by a water molecule [water No. 1, Fig. 2(B)]. This site is generally occupied by metal ligands, for example, magnesium in H chain ferritin (3ajo) and zinc in ChF.20 In MjFer structure, the distance from the water 1 to the adjacent carbonyl oxygen of glutamate side chain is ~2 Å, which is closer when compared with the distance of the ordinary hydrogen bond. It is possible that this site is occupied by magnesium with very low occupancy. We assigned water here, because putting magnesium here produced prominent negative F o − F c electron density map. The corresponding site of water 2 in MjFer [Fig. 2(B)] is occupied by a metal ligand in frog M ferritin.28 Magnesium ions are positioned mainly on outer surface of MjFer.
Figure 2.
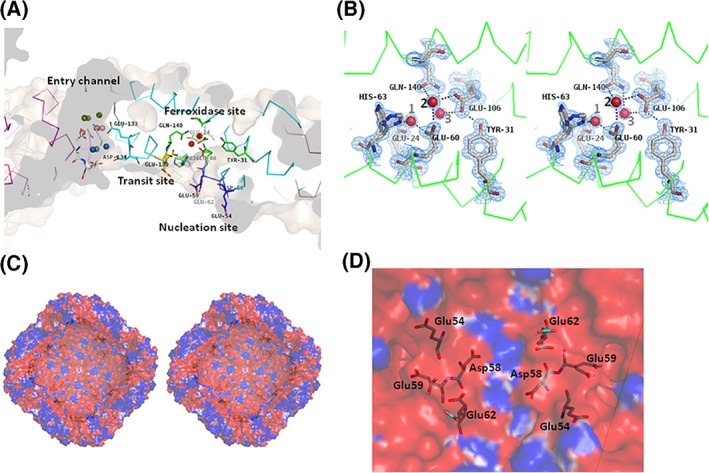
(A) The locations of the threefold metal entry channel (cyan), transit site (yellow), ferroxidase site (green), and nucleation site (blue) in the MjFer protein shell. The color codes of water molecules around the threefold entry channel are the same as in Figure 1(C). (B) The detailed structure of the ferroxidase site of MjFer (stereo view). The electron density of the 2|F o| − |F c| map for this site is contoured at 1.5σ and is shown in blue mesh. Water molecules that are localized in the vicinity of the previously reported iron binding site are shown as red balls. The broken lines indicate possible hydrogen bonds. (C) The cross‐section image of the inner cavity of the MjFer 24‐mer viewed from the fourfold symmetry axis. The amino acid residues of the putative nucleation sites (Glu54, Asp58, Glu59, and Glu62) are shown as sticks. The surface electrostatic potential of MjFer 24‐mer was rendered from the −1 (red; electronegative) to +1 kT (blue, electropositive) range, where k is the Boltzmann constant and T is the absolute temperature (K). (D) An enlarged image of the inner cavity of MjFer [Fig. 2(C)] around the nucleation site viewed from a twofold symmetry axis. The amino acid residues that form the nucleation site are shown as sticks.
It has been suggested that iron atoms enter from the threefold symmetry channel and subsequently are translocated to the ferroxidase site via the conserved glutamate side chain: Glu173 in soybean ferritin,23 Glu140 in FtH,18 and Glu136 in bullfrog M ferritin.28 Previously, we designated this glutamate residue a “transit site.” The corresponding residue, Glu139, whose side chain shows double conformation, is also present in the structure of MjFer [Fig. 2(A)]. This side‐chain flexibility would enable iron to translocate from the entry channel to the ferroxidase site. The side chain of His63, which is an indispensable member of the ferroxidase site, also has double conformation, which would facilitate iron translocation from Glu139 to the ferroxidase site [Fig. 2(A,B)].
On the other hand, four acidic amino acid residues of the putative nucleation site (Glu54, Asp58, Glu59, and Glu62), which correspond, respectively, to Glu57, Glu60, Glu61, and Glu64 of FtL,29 are present facing the inner cavity of MjFer 24mer [Fig. 2(A,D)]. Although Glu60 of FtL is substituted by Asp58 in MjFer, the positions of these residues are well conserved and the negatively charged inner surface is also formed in MjFer [Fig. 2(C,D)]. However, it is possible that this substitution influences the rate of nucleation, because Glu60 in FtL, corresponding to Asp58 in MjFer, is one of the amino acid residues that binds the metal directly in iron core formation.29
Thus, we have determined the first crystal structure of a crustacean ferritin, MjFer, at 1.16 Å resolution. MjFer would be a sophisticated model for studying the iron storage mechanism of ferritin, because it provides all the components required for iron incorporation and storage and it produces excellent crystals for structural studies. Further comprehensive studies are required to elucidate the detailed mechanisms underlying the iron storage by ferritin.
Materials and Methods
Molecular cloning and expression of MjFer
Oligonucleotide primers were designed according to the deposited sequence of ferritin from M. japonicus (GenBank accession no. KC556901).10 The sequences of the primer set were Fw (5′‐GACTCCATGGCCAGCCAAGTCC‐3′) and Rv (5′‐GATCGGATCCTAGTTGAGCTCTTTGTC‐3′), which respectively contained the NcoI and BamHI restriction sites. Total RNA was extracted from the hepatopancreas of live kuruma prawn using Sepasol reagent (Nacalai Tesque, Kyoto, Japan), followed by cDNA synthesis by Primescript RTase (Takara, Ohtsu, Japan). MjFer cDNA was amplified by PCR using the primer set. The PCR fragment was digested by NcoI and BamHI (Takara) and inserted into the NcoI/BamHI restriction site of pET 21d (Novagen, San Diego, CA). The expression plasmids were transformed into the Escherichia coli strain BL21(DE3) (Novagen). Protein expressions were performed at 20°C after isopropyl‐β‐d‐thiogalactopyranoside induction. The cells were harvested and disrupted by sonication, followed by protein extraction with phosphate‐buffered saline containing a protease inhibitor cocktail (Nacalai). MjFer was expressed as a soluble protein and was further purified by ammonium sulfate precipitation, anion exchange chromatography by Q‐sepharose (GE Healthcare, Piscataway, NJ) and finally, size exclusion chromatography using a Superdex 200 pg 16/60 column (GE Healthcare) which had been equilibrated by 10 mM Tris–HCl pH 7.5 containing 0.15 M NaCl. Concentrations of purified proteins were adjusted by absorbance at 280 nm. The molar extinction coefficient was calculated to be 18,910 M−1 cm−1 by ProtParam program (https://web.expasy.org/protparam).
Crystallization and data collection
Initial crystals were obtained in a week using crystallization screening kits. The crystallization condition was optimized using the hanging‐drop vapor‐diffusion method. The optimized crystallization drops were prepared by mixing equal volumes of mother liquid (35% MPD, 100 mM imidazole‐HCl pH 8.0 and 200 mM MgCl2) and protein solution containing 15 mg ml−1 of recombinant MjFer at 20°C. Diffraction data on MjFer crystals were collected to 1.16 Å resolution at 100 K at SPring‐8 BL26B1 after flush cooling with liquid nitrogen. Ethylene glycol (10% v/v) was used as a cryoprotectant. Diffraction data were processed, merged, and scaled with HKL‐2000 (HKL Research, Charlottesville, CA). Data processing statistics are shown in Table 1.
Structure determination and refinement
The structure of MjFer was determined by molecular replacement using the Molrep30 program in CCP4i suite31 1.4.4 using the structure of a subunit of FtH (PDB ID: http://firstglance.jmol.org/fg.htm?mol=3ajo) as a search model. Refinement was performed using the REFMAC532 and PHENIX33 programs including anisotropic B‐factor refinement. The structure was visualized and rebuilt using COOT 0.7.2.1 and further modified on σ‐weighted (2|F o| − |F c|) and (|F o| − |F c|) electron density maps. The final refinement was carried out using PHENIX that also calculated the maximum‐likelihood based coordinate error. The coordinate error, R work, and R free of the final model were calculated as 0.09, 0.1377, and 0.1566, respectively. Figures were produced by PyMOL (DeLano Scientific, San Carlos, CA). The secondary structure was assigned using the DSSP program.34 Images of the electrostatic potential of the protein surface were generated using the APBS application [Fig. 2(C,D)].35
Acknowledgments
This work was supported by JSPS KAKENHI to T. M. (Grant number JP 15K07574) and the National Natural Science Foundation of China to G. Z. (No. 31730069). The crystal data collection was carried out at the beamlines of SPring‐8 BL26B1 under the proposal numbers of 2015A1052, 2015A1063, and 2015A6539.
References
- 1. Theil EC, Matzapetakis M, Liu X (2006) Ferritins: iron/oxygen biominerals in protein nanocages. J Biol Inorg Chem 11:803–810. [DOI] [PubMed] [Google Scholar]
- 2. Arosio P, Ingrassia R, Cavadini P (2009) Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta 1790:589–599. [DOI] [PubMed] [Google Scholar]
- 3. Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A (2004) Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432:917–921. [DOI] [PubMed] [Google Scholar]
- 4. Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK (2002) The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore‐mediated iron acquisition. Mol Cell 10:1033–1043. [DOI] [PubMed] [Google Scholar]
- 5. Ong ST, Ho JZ, Ho B, Ding JL (2006) Iron‐withholding strategy in innate immunity. Immunobiology 211:295–314. [DOI] [PubMed] [Google Scholar]
- 6. Huang TS, Law JH, Soderhall K (1996) Purification and cDNA cloning of ferritin from the hepatopancreas of the freshwater crayfish Pacifastacus leniusculus . Eur J Biochem 236:450–456. [DOI] [PubMed] [Google Scholar]
- 7. Hamburger AE, West AP Jr, Hamburger ZA, Hamburger P, Bjorkman PJ (2005) Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains. J Mol Biol 349:558–569. [DOI] [PubMed] [Google Scholar]
- 8. Missirlis F, Kosmidis S, Brody T, Mavrakis M, Holmberg S, Odenwald WF, Skoulakis EM, Rouault TA (2007) Homeostatic mechanisms for iron storage revealed by genetic manipulations and live imaging of Drosophila ferritin. Genetics 177:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen XX, Li YY, Chang XJ, Xie XL, Liang YT, Wang KJ, Zheng WY, Liu HP (2018) A CqFerritin protein inhibits white spot syndrome virus infection via regulating iron ions in red claw crayfish Cherax quadricarinatus . Dev Comp Immunol 82:104–112. [DOI] [PubMed] [Google Scholar]
- 10. Feng WR, Zhang M, Su YQ, Wang J, Wang YT, Mao Y (2014) Identification and analysis of a Marsupenaeus japonicus ferritin that is regulated at the transcriptional level by WSSV infection. Gene 544:184–190. [DOI] [PubMed] [Google Scholar]
- 11. Hsieh SL, Chiu YC, Kuo CM (2006) Molecular cloning and tissue distribution of ferritin in Pacific white shrimp (Litopenaeus vannamei). Fish Shellfish Immunol 21:279–283. [DOI] [PubMed] [Google Scholar]
- 12. Maiti B, Khushiramani R, Tyagi A, Karunasagar I, Karunasagar I (2010) Recombinant ferritin protein protects Penaeus monodon infected by pathogenic Vibrio harveyi . Dis Aquat Org 88:99–105. [DOI] [PubMed] [Google Scholar]
- 13. Ye T, Wu X, Wu W, Dai C, Yuan J (2015) Ferritin protect shrimp Litopenaeus vannamei from WSSV infection by inhibiting virus replication. Fish Shellfish Immunol 42:138–143. [DOI] [PubMed] [Google Scholar]
- 14. Zhang J, Gui T, Wang J, Xiang J (2015) The ferritin gene in ridgetail white prawn Exopalaemon carinicauda: cloning, expression and function. Int J Biol Macromol 72:320–325. [DOI] [PubMed] [Google Scholar]
- 15. Zhang J, Li F, Wang Z, Zhang X, Zhou Q, Xiang J (2006) Cloning, expression and identification of ferritin from Chinese shrimp, Fenneropenaeus chinensis . J Biotechnol 125:173–184. [DOI] [PubMed] [Google Scholar]
- 16. Kong P, Wang L, Zhang H, Zhou Z, Qiu L, Gai Y, Song L (2010) Two novel secreted ferritins involved in immune defense of Chinese mitten crab Eriocheir sinensis . Fish Shellfish Immunol 28:604–612. [DOI] [PubMed] [Google Scholar]
- 17. Harrison PM, Arosio P (1996) Ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275:161–203. [DOI] [PubMed] [Google Scholar]
- 18. Masuda T, Goto F, Yoshihara T, Mikami B (2010) The universal mechanism for iron translocation to the ferroxidase site in ferritin, which is mediated by the well conserved transit site. Biochem Biophys Res Commun 400:94–99. [DOI] [PubMed] [Google Scholar]
- 19. Wang Z, Li C, Ellenburg M, Soistman E, Ruble J, Wright B, Ho JX, Carter DC (2006) Structure of human ferritin L chain. Acta Cryst D62:800–806. [DOI] [PubMed] [Google Scholar]
- 20. De Meulenaere E, Bailey JB, Tezcan FA, Deheyn DD (2017) First biochemical and crystallographic characterization of a fast‐performing ferritin from a marine invertebrate. Biochem J 474:4193–4206. [DOI] [PubMed] [Google Scholar]
- 21. Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P, Cesareni G, Thomas CD, Shaw WV, Harrison PM (1991) Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349:541–544. [DOI] [PubMed] [Google Scholar]
- 22. Pozzi C, Di Pisa F, Bernacchioni C, Ciambellotti S, Turano P, Mangani S (2015) Iron binding to human heavy‐chain ferritin. Acta Cryst D71:1909–1920. [DOI] [PubMed] [Google Scholar]
- 23. Masuda T, Goto F, Yoshihara T, Mikami B (2010) Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin. J Biol Chem 285:4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masuda T, Morimoto SI, Mikami B, Toyohara H (2012) The extension peptide of plant ferritin from sea lettuce contributes to shell stability and surface hydrophobicity. Protein Sci 21:786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Brun NE, Crow A, Murphy ME, Mauk AG, Moore GR (2010) Iron core mineralisation in prokaryotic ferritins. Biochim Biophys Acta 1800:732–744. [DOI] [PubMed] [Google Scholar]
- 26. Marchetti A, Parker MS, Moccia LP, Lin EO, Arrieta AL, Ribalet F, Murphy MEP, Maldonado MT, Armbrust EV (2009) Ferritin is used for iron storage in bloom‐forming marine pennate diatoms. Nature 457:467–470. [DOI] [PubMed] [Google Scholar]
- 27. Lawson DM, Treffry A, Artymiuk PJ, Harrison PM, Yewdall SJ, Luzzago A, Cesareni G, Levi S, Arosio P (1989) Identification of the ferroxidase centre in ferritin. FEBS Lett 254:207–210. [DOI] [PubMed] [Google Scholar]
- 28. Tosha T, Ng HL, Bhattasali O, Alber T, Theil EC (2010) Moving metal ions through ferritin‐protein nanocages from three‐fold pores to catalytic sites. J Am Chem Soc 132:14562–14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pozzi C, Ciambellotti S (2017) Chemistry at the protein‐mineral interface in L‐ferritin assists the assembly of a functional (mu(3)‐oxo)Tris[(mu(2)‐peroxo)] triiron(III) cluster. Proc Natl Acad Sci U S A 114:2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025. [Google Scholar]
- 31. Potterton E, Briggs P, Turkenburg M, Dodson E (2003) A graphical user interface to the CCP4 program suite. Acta Cryst D59:1131–1137. [DOI] [PubMed] [Google Scholar]
- 32. Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Cryst D53:240–255. [DOI] [PubMed] [Google Scholar]
- 33. Adams PD, Grosse‐Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Cryst D58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 34. Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen‐bonded and geometrical features. Biopolymers 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 35. Holst M, Saied F (1993) Multi grid solution of the poisson‐boltzmann equation. J Comput Chem 14:105–113. [Google Scholar]


