Abstract
Chlamydial major outer membrane protein (MOMP) is the major protein constituent of the bacterial pathogen Chlamydia trachomatis. Chlamydia trachomatis Serovars D–K are the leading cause of genital tract infections which can lead to infertility or ectopic pregnancies. A vaccine against Chlamydia is highly desirable but currently not available. MOMP accounts for ~ 60% of the chlamydial protein mass and is considered to be one of the lead vaccine candidates against C. trachomatis. We report on the spectroscopic analysis of C. trachomatis native MOMP Serovars D, E, F, and J as well as C. muridarum MOMP by size exclusion chromatography multi angle light scattering (SEC MALS), circular dichroism (CD) and attenuated total reflectance Fourier transform infrared spectroscopy (ATR‐FTIR). MOMP was purified from the native bacterium grown in either adherent HeLa cells or in different suspension cell lines. Our results confirm that MOMP forms homo‐trimers in detergent micelles. The secondary structure composition of C. trachomatis MOMP was conserved across serovars, but different from composition of C. muridarum MOMP with a 13% (CD) to 18% (ATR‐FTIR) reduction in β‐sheet conformation for C. trachomatis MOMP. When Serovar E MOMP was isolated from suspension cell lines the α‐helix content increased by 7% (CD) to 13% (ATIR‐FTIR). Maintenance of a native‐like tertiary and quaternary structure in subunit vaccines is important for the generation of protective antibodies. This biophysical characterization of MOMP presented here serves, in the absence of functional assays, as a method for monitoring the structural integrity of MOMP.
Keywords: major outer membrane protein; Chlamydia trachomatis; Chlamydia muridarum; Serovar D, E, J, and F; β‐barrel protein; CD spectroscopy; ATR FTIR spectroscopy; transmission electron microscopy; multi‐angle light scattering
Introduction
Chlamydia trachomatis is an obligate intracellular, Gram‐negative bacterial pathogen that belongs to the genus Chlamydophila. Other members of the genus Chlamydophila are C. psittaci and C. pneumoniae, all of which are responsible for human infections.1, 2 C. trachomatis is categorized into biovars based on tissue tropism and serovars based on the immune response to major outer membrane protein (MOMP). The trachoma biovar (Serovars A–C) is the leading cause of non‐congenital blindness in developing nations.3 The genital tract biovar (Serovars D–K) as well as the lymphogranuloma venereum biovar (Serovars L1–L3) are sexually transmitted. Genital tract biovars cause inflammatory diseases such as pelvic inflammatory disease and tubal factor infertility, whereas lymphogranuloma venereum biovars cause infection of lymphatics and lymph nodes leading to invasive urogenital or anorectal infection.4 With almost 150 million new cases occurring per year worldwide, C. trachomatis infection is the most common bacterial sexually transmitted disease in the world, and the number of diagnosed cases of chlamydial infection has been increasing over the past 10 years.5 Most C. trachomatis genital tract infections are asymptomatic, and as a result, an estimated 70–80% of all infections are not treated. The symptoms of chlamydial genital tract infection are chronic abdominal pain, ectopic pregnancy and infertility.6 It has been reported that 50% of all cases of infertility in the Western world are caused by Chlamydia infection.7 Infection with C. trachomatis has been reported to be associated with cervical cancer8 and also facilitates the transmission of HIV. Treatment of chlamydial infection can reduce the transmission of HIV and viral shedding.9 Chlamydia infection can be treated with antibiotics, but a vaccine against Chlamydia remains elusive.
MOMP is a 40 kDa cysteine‐rich surface protein that is encoded by the ompA gene and contributes almost 60% to the chlamydial total protein mass. MOMP is embedded in the bacterial outer membrane and its primary sequence consists of five constant domains that alternate with four immunodominant variable domains (VS1–4). Epitopes within VS1–4 provide serovar specificity.10, 11 MOMP has been shown in multiple studies to be an immunodominant antigen of natural C. trachomatis infection12, 13 and has been proposed by many investigators as an important vaccine component.14, 15, 16, 17, 18 Sequence diversity within the variable domains of MOMP enables immune evasion, and recombination events within MOMP VS4 are thought to play a role in tissue tropism.19, 20 Moreover, neutralizing monoclonal antibodies against VS1, VS2, and VS4 have been shown to prevent chlamydial infection by inhibiting host cell attachment.21
The MOMP sequence appears specific to the genus of Chlamydia and has not been detected in the genome of other bacterial genuses. The primary structure of MOMP contains 8–10 Cysteine residues, which is unusual for membrane proteins. It has been shown that cysteine residues in MOMP form inter‐ and intra‐molecular disulfide bonds with other cysteine rich proteins. The disulfide bonds form an extensive intermolecular network that increases the structural rigidity of the outer membrane.22, 23, 24 The intermolecular network at the outer membrane is called chlamydial outer membrane complex (COMC) and is thought to substitute for the very limited amount of chlamydial cell wall peptidoglycan, a component that provides rigidity and structural strength against osmotic pressure to the wall of Gram‐negative bacteria.22 The major constituents of COMC are MOMP and two cysteine rich proteins OmcA and OmcB.25, 26 MOMP is part of the outer shell of two distinct chlamydial life‐cycle manifestations: (1) elementary bodies (EB), which are non‐replicating but infectious and (2) reticulate bodies (RB) which are the non‐infectious, metabolically active, replicating form of the bacteria. RBs replicate inside the host cell in organelle‐like inclusions that are formed by a distinct membrane. Multiple RBs are present within a single inclusion. Disulfide cross‐links in the COMC are reduced in RBs which leads to an increase in diameter and allows membrane fluidity to facilitate replication.27
To date, there is no X‐ray or NMR structure of MOMP available. Structural elucidation of MOMP has been hampered by the requirement for detergents to keep the membrane protein in solution during purification, as well as the lack of a recombinant expression system that preserves MOMP in its native conformation. Despite increasing efforts to express recombinant MOMP,28 purification and reconstitution remain problematic, and extracting MOMP in its native form from Chlamydia preparations appears to be the optimal solution for the purpose of structure determination.
MOMP has been shown to function as an ion channel and is considered to be a porin with a beta barrel structure.29 Beta barrel proteins are composed of antiparallel β‐strands spanning the outer membrane, tight β‐turns extending into the periplasmic space, and flexible loops reaching beyond the extracellular surface.30 Several groups have proposed models of MOMP based on the hypothesis that MOMP exists in a 16 stranded anti‐parallel β‐barrel topology. MOMP topology models have been developed for Serovars C,31 D,10 F,32 and the mouse‐specific species C. muridarum.33 The differences in MOMP primary sequences of C. trachomatis Serovars C, D, F, and C. muridarum are almost exclusively located in the variable domains. Although the sequence homology within the different C. trachomatis serovars and C. muridarum is between 87.5% and 96.4% (Supplement ST1), the predicted locations of the 16 β‐strands are different in all four models. As pointed out by Feher et al., current prediction algorithms for the location and number of strands in MOMP allow for equal probability of 10, 12, 14, or 16 membrane spanning strands of the β barrel.31 This uncertainty is reflected in a 14 stranded beta barrel 3D model of C. pneumoniae MOMP, which has 76.4% sequence identity with C. trachomatis Serovar D MOMP.34
Here we report on the purification and spectroscopic structural characterization of C. trachomatis Serovars D, E, F, and J native MOMP (nMOMP). Chlamydia muridarum nMOMP has been extensively investigated and described in the literature24, 35, 36, 37, 38 and was, therefore, included in the analysis as a comparison. Size exclusion multi angle light scattering (SEC MALS) was applied to characterize protein‐detergent micelles of nMOMP solubilized in zwitterionic detergent. Circular dichroism (CD) and attenuated total reflection Fourier transform infrared spectroscopy (ATR‐FTIR) measurements were performed to assess the secondary structure composition of C. trachomatis Serovars D, E, F, and J nMOMP obtained from chlamydial bodies grown in adherent cervical cancer cells (HeLa 229). Adherent polarized HeLa cells are frequently used in Chlamydia infection models because of their good infection rates. However, bacterial growth in adherent cell lines becomes limiting for larger scale production of chlamydial bodies. Infection of non‐adherent cell lines with Chlamydia provides a potential means to overcome this limitation. Therefore, we purified C. trachomatis serotype E nMOMP obtained from chlamydial infection of adherent and non‐adherent HeLa cells, as well as three more suspension cell lines. The resulting purified protein preparations were then used to determine the structural characteristics of nMOMP and to identify variations arising from the cell substrate used in the infection model.
Results
nMOMP from C. trachomatis Serovars D, E, J, and F as well as C. muridarum, was purified from chlamydial bodies and prepared as protein‐detergent micelles in Zwittergent 3–14. Chlamydial bodies of the different serovars were extracted from infected adherent HeLa 229 cells. In order to gain insight into the impact of cell substrate on the isolation, purification, and structure of nMOMP, serovar E chlamydial bodies were obtained from four different suspension mammalian cell lines: CAP, CLDK, CHO, and HeLa. Structural characterizations of all nMOMP preparations were performed using SEC MALS, CD, and ATR FTIR as well as electron microscopy.
Molecular weight distribution analysis of C. trachomatis Serovars D, E, F, and J nMOMP isolated from adherent HeLa cells
MALS in combination with refractive index (RI) and UV measurements was used in this study to determine the apparent molecular weight of nMOMP as well as the number of detergent molecules in the protein‐detergent micelles. MALS and RI detectors were directly connected to a size exclusion chromatography system to perform the analysis. nMOMP purifications obtained from adherent HeLa 229 cells infected with C. trachomatis Serovars D, E, F, or J were compared with C. muridarum nMOMP which has been reported to exist as a trimer in Zwittergent 3–14 detergent micelles.37, 39 The SEC UV chromatogram of C. muridarum nMOMP showed one major peak that accounted for approximately 95% of the area under curve. A small shoulder of earlier eluting aggregates was observed. The apparent molecular weight of the C. muridarum nMOMP zwittergent micelle was 196.8 kDa with nMOMP and zwittergent contributing 103 kDa and 93.8 kDa, respectively. The protein molecular weight of 103 kDa was observed consistently across the main peak [Fig. 1(A)] and calculated to an average of 2.6 nMOMP monomers enclosed in the protein detergent micelle, likely reflecting a mixture of nMOMP dimer and trimer detergent micelles. The mass contribution of the zwitterionic detergent Zwittergent 3–14 corresponded to an average of ~260 zwittergent molecules per micelle. Earlier eluting aggregates approached a protein‐detergent molecular weight of 400 kDa with nMOMP and Zwittergent detergent contributing ~ 200 kDa each.
Figure 1.
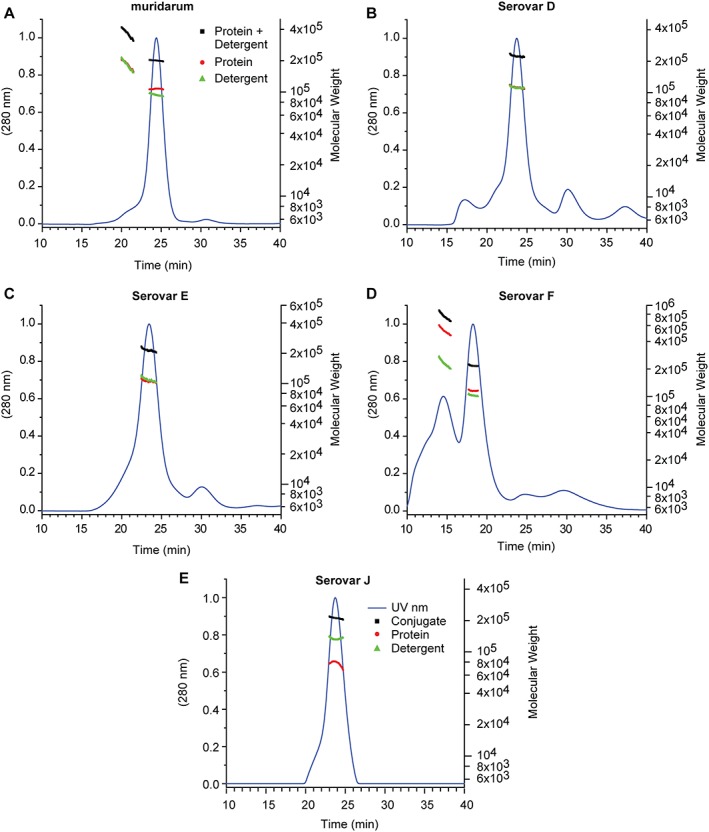
SEC MALS of nMOMP serovars expressed in adherent HeLa cells. UV absorption and molecular weight as a function of elution time. (A) Chlamydia muridarum. (B–E) Chlamydia trachomatis Serovars D, E, F*, and J. * The flow rate for Serovar F (Panel D) was higher (see Materials and Methods).
The SEC UV chromatogram of C. trachomatis Serovar D nMOMP showed a dominant peak that corresponded to nMOMP dimer and trimer. The average number of nMOMP monomers in the protein detergent micelle was 2.8. Similar to the C. muridarum nMOMP micelle, the micelle of C. trachomatis Serovar D nMOMP contained in average ~300 detergent molecules. A small shoulder of higher molecular weight aggregates was observed at an earlier retention time. Additional minor peaks at earlier and later elution times were observed but did not contain protein [Fig. 1(B)].
Similarly, SEC UV‐chromatograms with main peaks of micelles containing nMOMP dimer or trimer were observed for C. trachomatis Serovars E, F, and J nMOMP. The average number of nMOMP monomer per micelle was 2.5, 2.9, and 1.9, respectively (Fig. 1 and Table 1). The corresponding numbers of detergent molecules per nMOMP micelle were 304, 278, and 367. The C. trachomatis Serovar F nMOMP preparation showed, unlike the other serovars, a significant amount of aggregation.
Table 1.
Molecular weight determination by SEC‐MALS
| A. nMOMP serovars extracted from adherent HeLa cells | |||||
|---|---|---|---|---|---|
| Serovar | Protein M w (kDa) | Protein N | Detergent M w (kDa) | Detergent N | N Det/N Prot |
| Muridarum | 103.0 | 2.59 | 93.7 | 257.8 | 99.6 |
| D | 112.2 | 2.80 | 111.4 | 306.4 | 109.6 |
| E | 101.1 | 2.52 | 110.5 | 303.8 | 120.5 |
| F | 115.0 | 2.85 | 101.4 | 278 | 97.8 |
| J | 77.0 | 1.90 | 133.6 | 367.5 | 193.4 |
| B. Chlamydia trachomatis Serovar E nMOMP extracted from various suspension cells | |||||
|---|---|---|---|---|---|
| Cell line | Protein M w (kDa) | Protein N | Detergent M w (kDa) | Detergent N | N Det/N Prot |
| CAP | 54.8 | 1.36 | 63.2 | 173.8 | 127.4 |
| CLDK | 51.7 | 1.29 | 66.1 | 181.8 | 141.2 |
| CHO | 90.9 | 2.27 | 68.9 | 189.4 | 83.4 |
| HeLa | 92.2 | 2.30 | 118.8 | 326.7 | 142.2 |
Molecular weight determination of nMOMP obtained by SEC MALS measurements. The apparent protein molecular weight and the contribution of detergent molecules in the protein – detergent micelle were calculated for (A) C. muridarum and C. trachomatis Serovars D, E, F, and J nMOMP obtained from adherent HeLa cells. (B) Chlamydia trachomatis Serovar E nMOMP obtained from infected suspension cell lines.
Molecular weight distribution analysis of C. trachomatis Serovar E nMOMP isolated from suspension cell lines
SEC MALS was also applied to characterize the molecular weight distribution of C. trachomatis Serovar E nMOMP isolated from various Chlamydia‐infected suspension cell lines. The SEC UV chromatogram of nMOMP isolated from Chlamydia‐infected CAP suspension cells (human primary amniocytes, or CEVEC Amniocyte Production cells) showed three distinct peaks. The first peak eluted at a retention time of 16.5 min and contained residual co‐purifying DNA (with a maximum of the UV spectrum at 260 nm). The second peak with a retention time of 26 min contained nMOMP micelles. The average molecular weight of the micelles was 118 kDa. nMOMP protein contributed 54.8 kDa and the detergent contributed 63.2 kDa to the protein detergent micelle. The average number of nMOMP monomer units in CAP suspension cell derived protein micelles was 1.4. The micelles were formed by 175 zwittergent detergent molecules in average. The third peak with a retention time of 30 min did not contain protein [Fig. 2(A)].
Figure 2.
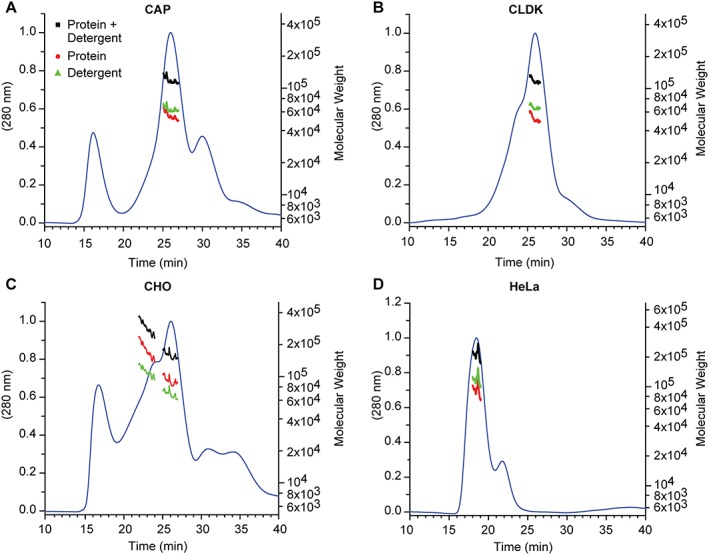
SEC MALS of Chlamydia trachomatis Serovar E nMOMP expressed in various suspension cell lines. UV absorption and molecular weight as a function of elution time. (A) CAP, (B) CLDK, (C) CHO, and (D) HeLa* *. The flow rate for HeLa (Panel D) was higher (see Materials and Methods).
The SEC UV chromatogram of nMOMP isolated from Chlamydia infected CLDK (Cutter Laboratories Dog Kidney) suspension cells showed one dominant peak of nMOMP micelles with an earlier eluting shoulder of nMOMP aggregate micelles. The apparent molecular weight of nMOMP micelles at the apex of the dominant peak was 118 kDa. Deconvolution of protein and detergent contributions showed that nMOMP protein and detergent accounted for 51.7 kDa and 66.1 kDa, respectively. The average number of nMOMP polypeptide chains in the nMOMP micelle was 1.3 and the average number of Zwittergent molecules per micelle was 180 [Fig. 2(B)].
Similar to nMOMP isolated from chlamydia infected CAP cells, co‐purifying DNA was observed in the preparation of nMOMP isolated from Chinese hamster ovary (CHO) suspension cells. The retention time of co‐purifying DNA in the SEC was 16.5 min. The apex of the main peak in the SEC UV chromatogram was centered at 26 min and contained nMOMP micelles. The average molecular weight of the nMOMP micelles was 160 kDa. nMOMP contributed 91 kDa to the molecular weight of the micelle, which corresponds to an average of 2.3 nMOMP monomers embedded in the micelle. The contribution of detergent to the mass of the nMOMP micelle was 68.9 kDa, which calculates to 189 detergent molecules per micelle. Equivalent to the analyses of nMOMP obtained from chlamydial particles in CAP and CLDK suspension cells, the chromatogram of nMOMP obtained from CHO suspension cells showed a noticeable shoulder to the left of the main peak. The shoulder contained higher molecular weight micelles of nMOMP aggregates. The aggregates were composed of approximately 6 monomer units and had a retention time of 22 min [Fig. 2(C)].
The preparation of nMOMP obtained from Chlamydia infected HeLa suspension cells was homogeneous and contained nMOMP micelles with an average molecular weight of 211 kDa. The micelles eluted with a single symmetric peak and an average of 2.3 nMOMP monomers contributed 92.2 kDa to the mass of the micelles. The number of detergent molecules per micelles was 326, which was significantly higher than in micelles obtained from all other suspension cell line preparations. The higher number of detergent molecules per micelle may have prevented the formation of nMOMP aggregate micelles [Fig. 2(D)].
All nMOMP preparations from suspension cells with the exception of HeLa cells contained higher molecular weight species. These higher molecular weight species could be aggregates of nMOMP micelles, with a micelle composition identical to the composition observed in the main peak, or they could be high molecular weight species composed of nMOMP monomer aggregates that were embedded in a single large detergent micelle. It can be expected that in the latter case the ratio of nMOMP polypeptide chain molecules to detergent molecules would decrease with molecular weight, whereas in the first scenario, the ratio between nMOMP polypeptide chains (monomer units) and detergent molecules would remain the same. The measurements showed that the higher molecular weight species contained approximately 25% fewer detergent molecules when compared to the micelles detected in the main fraction, favoring the possibility that the higher molecular weight species were formed by aggregates of nMOMP monomer units which were embedded in detergent micelles.
Circular dichroism spectroscopy of C. trachomatis Serovars D, E, F, and J nMOMP isolated from adherent HeLa cells
CD spectroscopy measurements were performed to assess the secondary structure composition of C. trachomatis Serovars D, E, F, and J nMOMP micelles in zwittergent. The CD spectrum of C. muridarum nMOMP in Zwittergent has been published37 and a preparation of C. muridarum nMOMP obtained from adherent HeLa 229 cells was included in this analysis [Fig. 3(A)]. The CD spectrum of C. muridarum nMOMP showed a minimum at 216 nm which is indicative of β‐sheet contributing to the secondary structure. The presence of β‐sheet secondary structure is expected for a beta barrel protein. The molar ellipticity at 216 nm for C. muridarum nMOMP was −5245° cm2 dmol−1. The CD spectra of C. trachomatis Serovars D, E, F, and J nMOMP also displayed minima between 215 nm and 216 nm. The molar ellipticity at 216 nm for all C. trachomatis nMOMP Serovars D, E, F, and J was within a range of −6000 to −7500° cm2 dmol−1, which was slightly higher than the molar ellipticity observed for C. muridarum nMOMP. The shapes of the CD spectra of C. trachomatis Serovars D, E, F, and J nMOMP and C. muridarum nMOMP were qualitatively similar, with the spectrum of C. muridarum nMOMP being narrower toward higher wavelengths [Fig. 3(B–E)].
Figure 3.
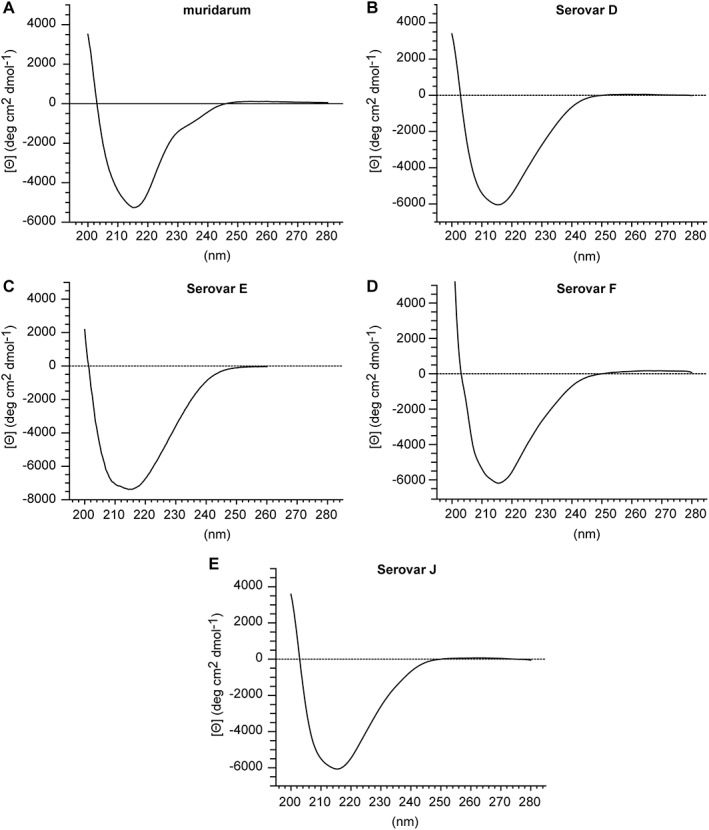
CD of nMOMP serovars expressed in adherent HeLa cells. Far‐UV CD spectra of native MOMP in 10 mM MES,150 mM NaCl, 0.1% Zwittergent 3–14 at pH 7.3. (A) Chlamydia muridarum. (B–E) Chlamydia trachomatis Serovars D, E, F, and J.
Deconvolution of the nMOMP CD spectra with the CDNN secondary structure prediction algorithm40 predicted the fraction of parallel and anti‐parallel β‐sheet secondary structure elements in C. trachomatis Serovars D, E, F, and J nMOMP to be in average 23.6% ± 0.7%. The fraction of β‐sheet elements of C. muridarum nMOMP was with 36.9% significantly higher. The algorithm of the CDNN neural network has been trained to distinguish parallel and anti‐parallel β‐sheet structures. Interestingly, the software predicted the β‐sheet orientation of C. muridarum nMOMP to be, as anticipated for beta barrel proteins, mostly anti‐parallel (31.1%), whereas the β‐sheet orientation of nMOMP of all C. trachomatis serovars was predicted to be equally parallel and anti‐parallel (11.8% ±0.4% each) (Table 2).
Table 2.
Secondary structure characterization by circular dichroism spectroscopy
| A. nMOMP serovars extracted from adherent HeLa cells | |||||||
|---|---|---|---|---|---|---|---|
| Serovar | Serovar D,E,F, and J | ||||||
| Muridarum (%) | D (%) | E (%) | F (%) | J (%) | Mean (%) | SD (%) | |
| Helix | 11.6 | 19.0 | 22.0 | 20.3 | 19.0 | 20.1 | 1.4 |
| Antiparallel | 31.1 | 12.1 | 11.5 | 11.5 | 12.1 | 11.8 | 0.3 |
| Parallel | 5.8 | 12.1 | 11.2 | 11.8 | 12.1 | 11.8 | 0.4 |
| Beta‐turn | 17.3 | 17.6 | 17.7 | 17.3 | 17.6 | 17.6 | 0.1 |
| Rndm. coil | 34.1 | 39.2 | 37.5 | 39.0 | 39.2 | 38.7 | 0.8 |
| All β‐sheet | 36.9 | 24.2 | 22.8 | 23.3 | 24.2 | 23.6 | 0.7 |
| B. Chlamydia trachomatis Serovar E nMOMP extracted from various suspension cells | ||||||
|---|---|---|---|---|---|---|
| Suspension cell line | ||||||
| CAP (%) | CLDK (%) | CHO (%) | HeLa (%) | Mean (%) | SD (%) | |
| Helix | 29.6 | 27.3 | 23.7 | 28.1 | 27.2 | 2.5 |
| Antiparallel | 11.7 | 12.1 | 13.8 | 11.7 | 12.3 | 1.0 |
| Parallel | 9.1 | 9.6 | 10.2 | 9.4 | 9.6 | 0.4 |
| Beta‐turn | 17.5 | 17.3 | 17.3 | 17.2 | 17.3 | 0.1 |
| Rndm. coil | 32.1 | 33.8 | 35.0 | 33.6 | 33.6 | 1.2 |
| All β‐sheet | 20.8 | 21.7 | 24.0 | 21.1 | 21.9 | 1.5 |
Secondary structure composition of nMOMP calculated by deconvolution of circular dichroism spectra. (A) Chlamydia muridarum and C. trachomatis Serovars D, E, F, and J nMOMP obtained from adherent HeLa cells. (B) Chlamydia trachomatis Serovar E nMOMP obtained from infected suspension cell lines.
Circular dichroism spectroscopy of C. trachomatis Serovar E nMOMP isolated from suspension cell lines
CD measurements of C. trachomatis Serovar E nMOMP isolated from various suspension cell lines were performed under conditions identical to the previous measurements. The minima of the CD spectra of C. trachomatis Serovar E nMOMP isolated from suspension cell lines were located between 208.5 nm and 210 nm. Compared to the CD spectra of C. trachomatis Serovars D, E, J, and F nMOMP obtained from adherent HeLa cells, the minima were shifted by 5 nm from 215 nm to a lower wavelength at 210 nm. The mean molar ellipticity at the minima of the CD spectra of C. trachomatis Serovar E nMOMP isolated from CAP, CHO, or HeLa suspension cell lines ranged from −8642° cm2 dmol−1 to −10,565° cm2 dmol−1 which was lower than the mean molar ellipticity of C. trachomatis Serovar E nMOMP isolated from adherent HeLa cells (−7367° cm2 dmol−1) and the molar ellipticity of C. trachomatis Serovar E nMOMP isolated from CLDK suspension cells (−7219° cm2 dmol−1).
The deconvolution of the CD spectra of C. trachomatis Serovar E nMOMP isolated from suspension cell lines predicted an average of 27.2% ± 2.5% α‐helix content. The β‐sheet content of nMOMP Serovar E isolated from suspension cell lines was 21.9% ± 1.5%, and β‐turn and random coil contributed 17.3% ± 0.1% and 33.6% ± 1.2%, respectively (Fig. 4 and Table 2).
Figure 4.
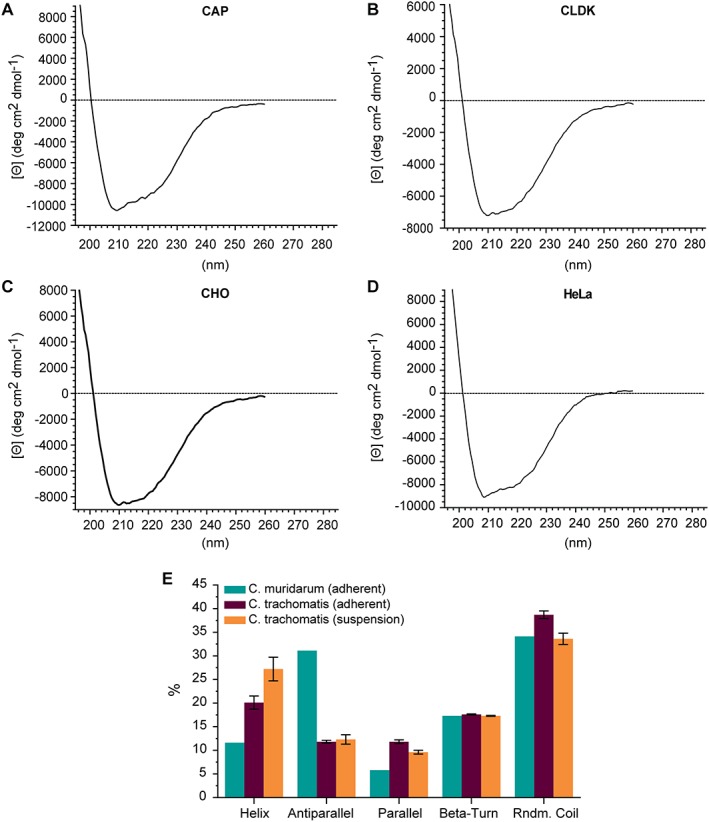
CD of Chlamydia trachomatis Serovar E nMOMP expressed in various suspension cell lines. Far‐UV CD spectra of native MOMP in 10 mM MES, 150 mM NaCl, 0.1% Zwittergent 3–14 at pH 7.3. (A) CAP, (B) CLDK, (C) CHO, (D) HeLa, and (E) Average of secondary structure composition of nMOMP calculated by spectral deconvolution using neuronal network analysis.
ATR FTIR spectroscopy of C. trachomatis nMOMP Serovars D, E, F, and J isolated from adherent HeLa cells
ATR FTIR is a method orthogonal to CD and is also used to assess the secondary structure of proteins. There is a direct correlation between IR amide I band frequencies and secondary structure components. The C=O stretching frequency in the amide I band is different for each secondary structure element. ATR FTIR was performed to further characterize the secondary structure of nMOMP. ATR FTIR spectra of nMOMP in Zwittergent micelles were acquired after applying the protein solution onto a ZnSe surface and generation of a hydrated protein film by drying the solution under a steady stream of nitrogen. Protein secondary structure elements such as α‐helix and β‐sheet exhibit distinct amide I infrared absorption bands in the amide I region between 1600 cm−1 and 1800 cm−1. The protein secondary structure absorption bands are not baseline resolved and require deconvolution before quantitative structural information can be obtained. The location of amide I bands was identified without bias by calculating the spectra's second derivative. The complex amide I infrared absorption spectrum was then reconstructed using iterative non‐linear least square regression analysis where Voigt shaped peaks were fixed at the location of the minima obtained from the second derivative analysis. ATR FTIR spectra of the amide I region of C. trachomatis Serovars D, E, F, and J nMOMP in Zwittergent, as well as their second derivative spectra, were highly similar [Fig. 5(B–E)]. Minima of the second derivative of the spectra were located at 1631 cm−1, 1657 cm−1, 1677 cm−1, and 1696 cm−1. Amide I bands close to 1631 cm−1 and 1696 cm−1 are indicative of β‐sheet secondary structure, while the amide I band at 1657 cm−1 corresponds to α‐helix structure and bands centered around 1677 cm−1 have been reported for β‐turn structures in proteins.41 Second derivative ATR FTIR spectra do not provide quantitative information for protein secondary structure, therefore, quantitative information was obtained by deconvolution of the FTIR spectra into Voigt‐shaped peaks. The peak area of individual amide I bands is correlated with secondary structure composition.41, 42, 43 The FTIR spectra were fitted with peaks centered at 1632 ± 3 cm−1 (β‐sheet), 1642 ± 3 cm−1 (random coil), 1657 ± 3 cm−1 (α‐helix), 1678 ± 4 cm−1 (β‐turn), and 1695 ± 4 cm−1 (β‐sheet). The second derivative of the spectra did not indicate a peak at 1642 cm−1 (random coil), but the addition of a peak at 1642 cm−1 to the deconvolution procedure greatly improved the quality of the fit and enabled the reconstruction of all nMOMP ATR FTIR spectra without significant residuals [Fig. S1(A–I)].
Figure 5.
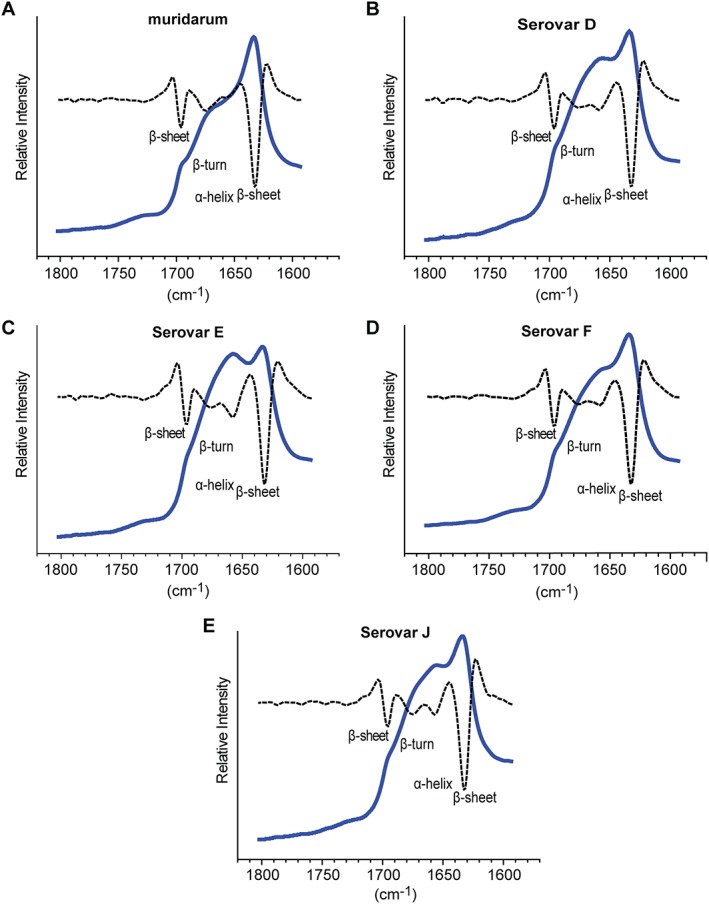
ATR FTIR of nMOMP serovars expressed in adherent HeLa cells. FTIR spectra (solid line) and second derivative (dotted line) of native MOMP. (A) Chlamydia muridarum. (B–E) Chlamydia trachomatis Serovars D, E, F, and J.
A spectral analysis of the ATR FTIR spectrum of C. muridarum nMOMP in Zwittergent was performed as a reference. The second derivative spectrum of C. muridarum nMOMP overlapped well with the second derivative spectra of all C. trachomatis nMOMP serovars, with the exception of the α‐helix band at 1657 cm−1 which was not well defined in the C. muridarum nMOMP spectrum (Fig. 5). Beta‐sheet amide I bands at 1631 cm−1 and 1696 cm−1 contributed to the spectra of C. trachomatis Serovars D, E, F, and J nMOMP 21.1% ± 0.3% and 4.8% ± 0.5%, respectively. The most abundant secondary structures of C. trachomatis Serovars D, E, F, and J nMOMP were random coil (28.8% ±4.0%) and α‐helix (27.6% ± 2.6%). The β‐turn band contributed an average 17.6% ± 1.1% to the spectra of C. trachomatis Serovars D, E, F, and J nMOMP. In comparison to C. trachomatis Serovars D, E, F, and J nMOMP, the α‐helix content of C. muridarum nMOMP was at 22.6% lower. For C. muridarum nMOMP, the contribution of random coil structure was 11.4% which was significantly lower than the 28.8% observed for C. trachomatis Serovars D, E, F, and J nMOMP. The β‐sheet amide I bands at 1631 cm−1 and 1696 cm−1 of C. muridarum nMOMP contribute 39.5% and 4.5%, respectively, which was almost 20% higher than the average β‐sheet signal intensity of C. trachomatis Serovars D, E, F, and J nMOMP. The contribution of β‐turn for C. muridarum nMOMP (22.6%) was similar to the contribution of β‐turn for C. trachomatis Serovars D, E, F, and J nMOMP [Fig. 6(E) and Table 3(A)].
Figure 6.
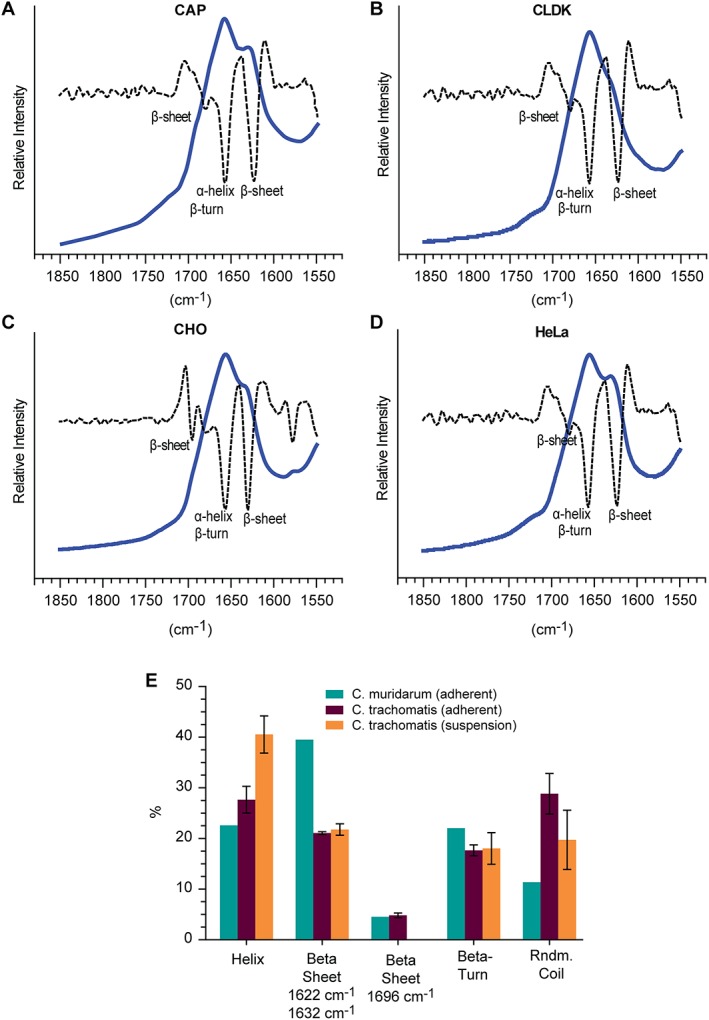
ATR FTIR of Chlamydia trachomatis Serovar E nMOMP expressed in various suspension cell lines. FTIR spectra (solid line) and second derivative (dotted line) of native MOMP. (A) CAP, (B) CLDK, (C) CHO, and (D) HeLa. (E) Average secondary structure composition of nMOMP calculated by spectral deconvolution and non‐linear least square regression analysis.
Table 3.
Secondary structure characterization by ATR‐FTIR spectroscopy
| A. nMOMP serovars extracted from adherent HeLa cells. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Serovar | Serovar D, E, F, and J | |||||||
| cm−1 | Muridarum | D | E | F | J | Mean | SD | |
| 1642 | Rndm. coil | 11.4 | 23.4 | 28.3 | 31.8 | 31.8 | 28.8 | 4.0 |
| 1657 | Helix | 22.6 | 30.5 | 29.3 | 25.3 | 25.5 | 27.6 | 2.6 |
| 1632 | Beta sheet | 39.5 | 21.4 | 20.7 | 21.1 | 21.0 | 21.1 | 0.3 |
| 1696 | Beta sheet | 4.5 | 5.4 | 4.3 | 4.9 | 4.6 | 4.8 | 0.5 |
| 1678 | Beta‐turn | 22.0 | 19.2 | 17.4 | 16.9 | 17.0 | 17.6 | 1.1 |
| All β‐sheet | 44.0 | 26.8 | 25.1 | 25.9 | 25.6 | 25.9 | 0.7 | |
| B. Chlamydia trachomatis Serovar E nMOMP extracted from various suspension cells | |||||||
|---|---|---|---|---|---|---|---|
| Suspension cell line | |||||||
| cm−1 | CAPa | CLDK | CHO | HeLaa | Mean | SD | |
| 1642 | Rndm. coil | 18.9 | 18.2 | 13.9 | 27.8 | 19.7 | 5.9 |
| 1657 | Helix | 38.8 | 39.1 | 46.0 | 38.2 | 40.5 | 3.7 |
| 1632 | Beta sheet | 22.0 | 22.1 | 22.8 | 20.1 | 21.7 | 1.1 |
| 1696 | Beta sheet | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1678 | Beta‐turn | 20.2 | 20.6 | 17.3 | 13.9 | 18.0 | 3.1 |
| All β‐sheet | 22.0 | 22.1 | 22.8 | 20.1 | 21.7 | 1.1 | |
The location of the b‐sheet band shifted from 1632 cm−1 to 1625 cm−1.
Secondary structure composition of nMOMP calculated by deconvolution of ATR‐FTIR spectra. (A) Chlamydia muridarum and C. trachomatis Serovars D, E, F and J nMOMP obtained from adherent HeLa cells. (B) Chlamydia trachomatis Serovar E nMOMP obtained from infected suspension cell lines.
ATR FTIR spectroscopy of C. trachomatis Serovar E nMOMP isolated from suspension cell lines
ATR FTIR spectra of the amide I region of C. trachomatis Serovar E nMOMP obtained from suspension cell lines were of similar shape [Fig. 6(A–D)]. Peak fitting of the spectra showed that the relative contribution of β‐sheet was at 21.7% ± 1.1% identical for all C. trachomatis Serovar E nMOMP preparations obtained from suspension cell lines [Table 3(B)]. For nMOMP isolated from CAP and HeLa cell lines, the center of the β‐sheet bands shifted from 1632 cm−1 to 1625 cm−1. Beta‐sheet bands at 1696 cm−1 were not detected for C. trachomatis Serovar E nMOMP obtained from suspension cell lines. The secondary structure composition of nMOMP isolated from CAP and CLDK cell lines almost was identical [Table 3(B)]. Chlamydia trachomatis Serovar E nMOMP isolated from CHO cells showed in comparison to nMOMP Serovar E isolated from CAP and CLDK cell lines a 7% increase in α‐helix structure. This increase was accompanied by a 5% decrease in random coil conformation, decreasing from 19% to 14%. On the other hand, C. trachomatis Serovar E nMOMP isolated from HeLa cells showed a 9% increase in random coil conformation that was compensated by a 7% decrease in β‐turn structure in comparison to C. trachomatis nMOMP Serovar E isolated from CAP and CLDK cell lines [Table 3(B)].
CD thermal shift assay of C. muridarum nMOMP
We attempted to compare the thermal stability of C. trachomatis Serovars D, E, F, and J nMOMP as well as C. muridarum nMOMP by monitoring heat induced protein unfolding by CD spectroscopy.44 As shown in Fig. 7 the negative intensities of the C. muridarum nMOMP CD spectra between 200 nm and 260 nm gradually increased with increasing temperature, which is unusual. Thermal unfolding typically leads to loss of secondary structure and is consequently accompanied by a loss of negative dichroism. The enhanced CD spectrum of C. muridarum nMOMP heated to 90°C did not reverse 48 h after heating; showing that the heat induced structural conversion was irreversible. Cai et al. observed similar behavior for C. muridarum nMOMP36 and speculated that heating might lead to MOMP aggregates with increased intermolecular β structure. Chlamydia trachomatis Serovar E nMOMP showed in the CD thermal shift assay the same behavior as C. muridarum nMOMP (data not shown).
Figure 7.
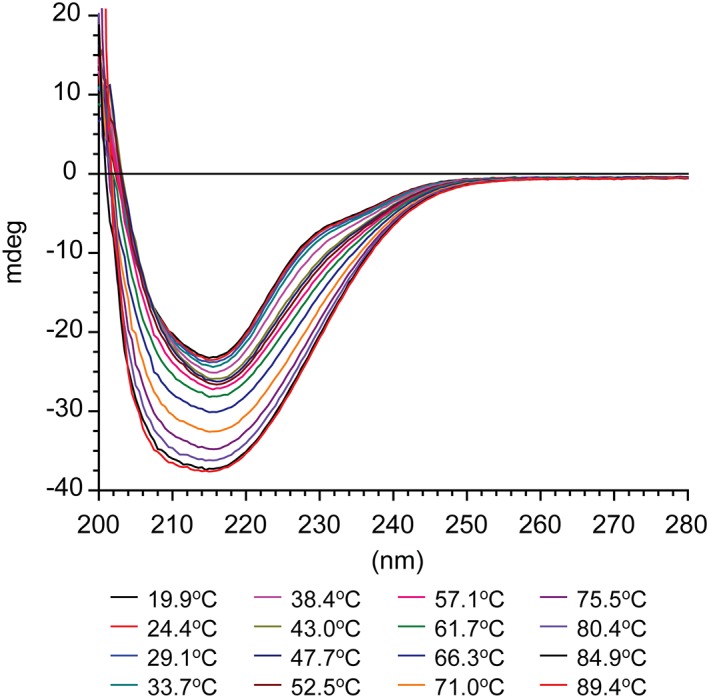
CD spectra of Chlamydia muridarum nMOMP at increasing temperatures. Far‐UV CD spectra of Chlamydia muridarum nMOMP in 10 mM MES, 150 mM NaCl, 0.1% Zwittergent 3–14 at pH 7.3 between 20°C and 90°C at ~5°C intervals.
Transmission electron microscopy
Negative stain transmission electron microscopy was used to visualize the morphological characteristics of C. muridarum nMOMP Zwittergent protein micelles. The grids contained large quantities of evenly distributed protein structures. The individual protein particles exhibited spherical forms measuring between 2.8 nm and 7.0 nm in diameter. The mean size of the protein particles was 5.8 nm [Fig. 8(A)]. The diameter of the 16‐strand beta barrel model of trimeric MOMP has a diameter of approximately 8 nm31 which is close to the biggest particle size observed by electron microscopy. The formation of soluble intermolecular aggregates of heat treated C. muridarum nMOMP was confirmed by electron microscopy. The electron micrograph of C. muridarum nMOMP after heating from 20°C to 90°C over the course of 2 ours showed elongated strands. Individual strands had a diameter between 3.2 nm and 7.4 nm and varied in length between 15.0 nm and 105.0 nm [Fig. 8(B)].
Figure 8.
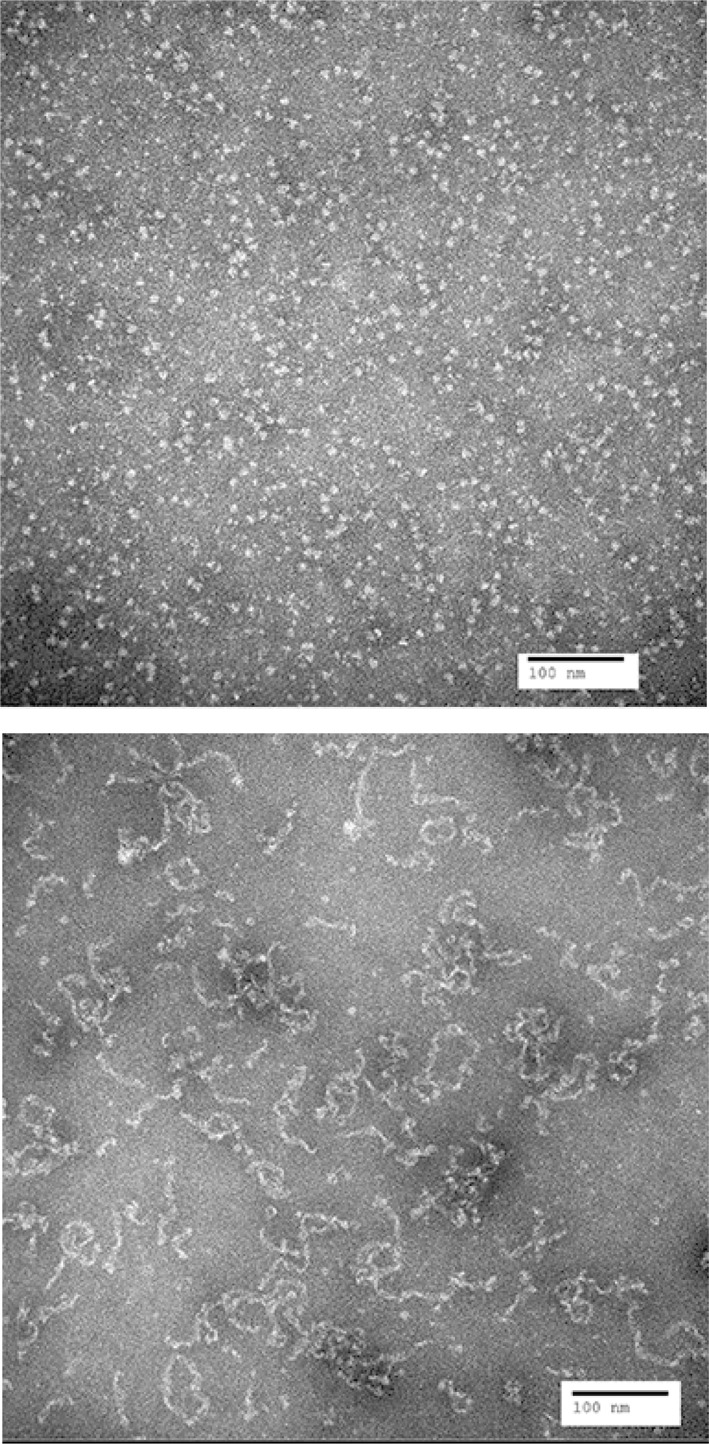
Electron micrograph of Chlamydia muridarum. Images were acquired at 150,000‐fold magnification: (A) not heated and (B) after heating to 90°C.
Discussion
Over the past 20 years, MOMP has been identified as a promising vaccine target. Many animal studies have demonstrated that nMOMP has potential to be a major component of an efficacious subunit vaccine against chlamydial genital tract infections.6, 45, 46, 47 The effectiveness of nMOMP as a vaccine antigen has been linked to the preservation of its structural conformation following purification. It has been shown by Pal et al. that sonication which led to the reduction of the quaternary and possibly tertiary structure of C. muridarum nMOMP, diminished efficacy in a mouse protection study when compared to a MOMP preparation that was more likely to contain MOMP in a conformation that is close to the native state.39 Similarly, recombinant MOMP was less protective in mouse immunization studies performed with cationic liposomal adjuvants than MOMP directly isolated from the pathogen.48 This shows the importance of the ability to closely monitor structural quality attributes of MOMP preparations used for vaccination experiments.
We employed multiple spectroscopic methods to describe the structural properties of nMOMP obtained from C. trachomatis Serovars D, E, F, and J isolated from adherent HeLa cells as well as C. trachomatis Serovar E isolated form different suspension host cells and compared them to the structural properties of C. muridarum nMOMP.
SEC MALS analysis provides insights into the quaternary structure of protein detergent micelles and allows at the same time monitoring protein aggregation. The analysis showed that C. muridarum nMOMP micelles existed as a mixture of dimers and trimers, with a possible contribution of small amounts of monomer. The observed average molecular weight of 103 kDa calculated to an apparent oligomerization state of 2.6, which suggests that almost equal amounts of nMOMP exist in dimeric or trimeric conformations. This finding is consistent with the observations reported by Sun et al.37 who observed a mixture of monomers, dimers, and trimers in an SDS PAGE analysis of unheated, cross‐linked C. muridarum MOMP. The apparent oligomerization states for C. trachomatis Serovars D, E, and F nMOMP preparations were calculated to be 2.5–2.9 with the value we obtained for the C. muridarum nMOMP preparation being in this range. The average oligomerization state of the C. trachomatis Serovar J nMOMP preparation was significantly lower at 1.9, indicating that the majority of the nMOMP oligomerization state was dimeric. The 25% increase of detergent molecules that was required to solubilize Serovar J nMOMP is consistent with mostly nMOMP dimers being embedded by the detergent. All C. trachomatis Serovar E nMOMP preparations obtained from suspension cell lines had apparent oligomerization states between 1.4 and 2.3 which was lower than the apparent oligomerization state of C. trachomatis Serovar E nMOMP obtained from adherent HeLa cells (2.5). The cause for the lower apparent oligomerization state of Serovar E nMOMP isolated from suspension cell lines is unclear. It can be speculated that the cell line choice has an impact on the purification process, or that the relative quantities of nMOMP obtained from EBs and RBs are cell line dependent and impact the oligomerization state. Environmental factors of the host cell (redox potential, polarization) might also have an impact on the tertiary and quaternary structure of MOMP.
The molecular weight of detergent molecules constituting the nMOMP protein detergent micelles ranged from 63.2 kDa to 133.6 kDa. Geometric considerations require that more detergent molecules are needed to form a micelle that is formed around a MOMP trimer when compared to a micelle that surrounds a MOMP monomer. In addition, the number of detergent molecules per monomer is expected to decrease for monomeric to dimeric and trimeric MOMP micelles as lesser surface per monomer is solvent exposed. Overall our measurements were consistent with this expectation, but the spread in detergent molecules per monomer is considerable (Table 1 and Fig. S2). For example, the C. trachomatis Serovar E nMOMP preparations obtained from CHO and HeLa suspension cell lines were found to have an identical apparent oligomerization state of 2.3 but the number of micelle forming detergent molecules was 327 and 189, respectively, indicating that nMOMP micelles obtained from CHO cell preparations had an approximately 1.7‐fold higher detergent molecule content compared to the nMOMP micelles obtained from HeLa cell preparations. It seems likely that the tertiary and/or quaternary structure of nMOMP differs between the preparations. SEC MALS has theoretically enough resolution power to resolve MOMP monomers, dimers or trimers but it is not able to resolve protein oligomer micelles with a broad distribution in the number of micelle detergent molecules and hence returned non‐stoichiometric numbers for the oligomerization state of nMOMP. In addition, the measurement does not distinguish between large micelles that are formed around MOMP aggregates (aggregates being defined as more than three MOMP monomers) and the aggregation of micelles that are constituted of individual micelles around nMOMP dimers and trimers. The buffer pH can cause protein aggregation if it is close to the proteins isoelectric point. The calculated pI of all MOMP serovars is with 4.8 well below the buffer pH at 7.3 and 12 negatively charged side chains are calculated for MOMP at this pH.
To further characterize the structure of micelle embedded nMOMP, we performed CD and ATR FTIR spectroscopy. Both techniques provide quantitative information on protein secondary structure composition. CD spectra were recorded between 200 nm and 260 nm. Collection of CD data below 200 nm was precluded by light scattering and absorbance properties of Zwittergent detergent. The CD spectra of all MOMP preparations had minima near 215 nm, which is indicative of β‐sheet secondary protein structure49, 50 and consistent with MOMP being a β‐barrel protein. CD spectra of β‐barrel outer membrane proteins such as MBOMP from Entamoeba, OMP P66 from Borrelia burgdorferi and BamA from Treponema pallidum have been reported in the literature.51, 52, 53 The shape of the CD spectra of these β‐barrel proteins is diverse with minima varying between 210 nm and 220 nm. The CD spectrum we obtained for C. muridarum MOMP had a minimum at 215 nm and a shape that was consistent with the spectrum reported by Sun et al.37 We used the CDNN algorithm to deconvolute the CD spectra and estimate the secondary structure composition of all nMOMP preparations (Table 2).40 The major component of the C. muridarum nMOMP secondary structure was random coil (34.1%), and α‐helix contributed 11.6%. CDNN allows distinguishing between parallel and anti‐parallel β‐sheet secondary structure. Deconvolution of the CD spectrum of C. muridarum nMOMP estimated the anti‐parallel β‐sheet content to be 31.1%. The contribution of parallel β‐sheet was 5.8% and the contribution of β‐turn was 17.3%. These values are close to the report in the literature37 and an anti‐parallel β‐sheet conformation is consistent with MOMP adopting an outer membrane β‐barrel fold. The secondary structure predictions by CDNN for C. trachomatis Serovars D, E, F, and J nMOMP were very similar to each other, with standard deviations for the contribution of secondary structure elements between Serovars D, E, F, and J nMOMP being less than 1.5%. Interestingly, despite similar curve shapes and location of the minima, the deconvolution into secondary structure elements revealed differences between C. muridarum nMOMP and C. trachomatis nMOMP Serovars D, E, F, and J. The proportion of β‐sheet fold of C. trachomatis nMOMP Serovars D, E, F, and J obtained from adherent HeLa cells (23.6%) was significantly lower than for C. muridarum nMOMP (36.9%). Moreover, the CDNN algorithm assigned equal amounts of parallel and anti‐parallel β‐sheet fold to the C. trachomatis MOMP serovars. The high sequence similarity of >85% between C. muridarum MOMP and C. trachomatis MOMP [Table ST (1) and Fig. S(3)] makes it seem unlikely that the β‐sheet structure of C. muridarum nMOMP could form a drastically different fold than the β‐sheets structure of C. trachomatis nMOMP serovars.
One possible explanation is that the β‐barrel structure of C. trachomatis Serovars D, E, F, and J nMOMP is distorted and caused failure of the algorithm to correctly identify the β‐strand orientation. The contribution of β‐turn structures was essentially identical between C. trachomatis and C. muridarum nMOMP (17.6% vs. 17.3%). The smaller proportion of β‐sheet structure in C. trachomatis nMOMP was compensated by increased proportions of random coil (38.7% ± 0.8%) and α‐helix (20.1% ± 1.4%) structures. Based on the 16 strand β‐barrel model that has been proposed by Feher et al.,31 the 16 anti‐parallel strands require 113 amino acid residues, accounting for 36% of all amino acids. This proposed β‐strand percentage is in good agreement with the value that was observed by CD for C. muridarum nMOMP (36.9%) but is well above the value that was observed for C. trachomatis nMOMP (23.6%). The 16 strand β‐barrel model contains eight short β‐loops facing the cytosol which account for approximately 10% of the MOMP amino acid sequence. This is in agreement with the observed value of 17% from the CD measurement if additional outward facing, short β‐loops within the variable domains (VS1–4) are taken into consideration. The outward facing loops of the immunodominant variable domains account in the 16 strand β‐barrel model for ~50% of the MOMP amino acid sequence. The CD data suggest that the secondary structure composition of the variable domains is predominantly α‐helical and random coil.
The CD analysis of C. trachomatis Serovar E nMOMP obtained from suspension cell lines CAP, CLDK, CHO, and HeLa did not suggest host cell line dependent differences in the secondary structure composition of nMOMP. When compared to the analysis of C. trachomatis Serovars D, E, F, and J nMOMP obtained from adherent HeLa cells, the proportion of β‐strand conformation was at 20.8% decreased by 2.8% and the amount of α‐helix conformation increased by 7.8–27.2%. This suggests that certain areas of MOMP might be able to undergo a transformation to α‐helical conformation. Interestingly, the secondary structure prediction algorithms APSSP, CFPSS, GOR54, 55, 56 predict between 10% (APSSP) and 40% (CFPSS) of the Serovar E MOMP sequence to have α‐helix potential and the predicted α‐helix sequence locations are in conflict with β‐strand locations of the 16 stranded β‐barrel model (data not shown).
A complementary secondary structure analysis was performed using ATR‐FTIR spectroscopy. Both techniques detected significantly higher β‐sheet structure in C. muridarum nMOMP when compared to C. trachomatis Serovars D, E, F, and J nMOMP isolated from adherent HeLa cells. By ATR FTIR C. muridarum nMOMP had 18.1% more β‐sheet structure than C. trachomatis Serovars D, E, F, and J nMOMP which corroborated the 13.3% increase observed by CD. Both spectroscopic methods assigned between 17% and 22% β‐turn structure to C. muridarum and C. trachomatis nMOMP. nMOMP secondary structure predictions for random coil and α‐helix agreed less between the different spectroscopic methods. The contributions of random coil calculated from the ATR‐FTIR spectra were consistently lower than the contributions determined by CD. For example, for C. muridarum nMOMP the proportion of random coil calculated by ATR‐FTIR was 11% which is lower than the 23% determined by CD. Similarly, the contribution of random coil for Serovar E nMOMP obtained from suspension cell lines was 20% by ATR‐FTIR and 34% by CD. On the other hand, the contributions of α‐helix calculated from ATR‐FTIR measurements were consistently higher than the contributions determined by CD. In case of C. trachomatis Serovar E nMOMP obtained from suspension cell lines, the contribution of α‐helix was 40.5% by ATR‐FTIR and 27.2% by CD. Interestingly, both spectroscopic methods observed the same trend for α‐helix content of the different preparations, with α‐helix content being the lowest for C. muridarum nMOMP, higher for C. trachomatis nMOMP obtained from adherent HeLa cells, and the highest for C. trachomatis nMOMP obtained from suspension cell lines. The contribution of α‐helix structure was 40.5% for C. trachomatis Serovar E nMOMP from suspension cell lines and 27.6% and 22.6% for C. trachomatis nMOMP Serovars D, E, F, and J and C. muridarum nMOMP. These differences in α‐helix composition were consistent with the CD measurements which observed the same trend (27.2%, 20.1%, and 11.6%).
It could be speculated that the nMOMP aggregates of C. trachomatis Serovar E that were observed by SEC MALS for samples obtained from suspension cell lines contain an increased amount of α‐helix. On the other hand, we did not observe a direct correlation between the fraction of aggregated protein in each sample and the secondary structure composition, which was consistent for all nMOMP preparations obtained from adherent or suspension cell lines. The maximum deviation of secondary structure composition was 2.5% for the CD and 5.9% for the ATR FTIR measurement (Tables 2 and 3).
It is also noteworthy that all ATR FTIR spectra of C. trachomatis Serovars D, E, F, and J nMOMP obtained from adherent HeLa cells displayed a β‐sheet band maximum centered at 1632 cm−1 (Fig. 5), whereas ATR FTIR spectra of C. trachomatis nMOMP Serovar E obtained from suspension cell lines CAP, CLDK, CHO, and HeLa displayed a maximum at the α‐helix band centered at 1657 cm−1 (Fig. 6). This difference became even more apparent in the second derivative analysis. Pronounced local minima at the α‐helix band at 1657 cm−1 and at the β‐sheet band were recorded in the FTIR analysis of C. trachomatis Serovar E nMOMP from suspension cell lines. The β‐sheet bands were centered either at 1632 cm−1 (CLDK and CHO) or at 1625 cm−1 (CAP and HeLa). FTIR bands at 1632 cm−1 and 1625 cm−1 have been both reported to represent β‐sheet structure. The secondary structural difference that is causing this shift is not known.41 Another qualitative difference between the ATR FTIR spectra of C. trachomatis nMOMP obtained from adherent HeLa cells and from suspension cell lines was that spectra obtained for C. trachomatis Serovar E nMOMP from suspension cell lines were missing the second β‐sheet band at 1696 cm−1. These differences provide an insight into the flexibility of the secondary and/or tertiary structure of MOMP. The results we obtained from CD and ATR‐FTIR analysis of MOMP were overall consistent and small differences might have been caused by the CD deconvolution algorithm being trained on soluble proteins and, therefore, not being optimized for bacterial membrane proteins.
Conclusion
Chlamydial MOMP (OmpA) is best characterized by its four immunodominant variable domains and is thought to fold as a pore forming outer membrane beta barrel protein. Folding of MOMP into its native structure is complicated by the presence of 8–10 cysteine residues. The disulfide bonding pattern of C. muridarum MOMP has been investigated by Wang et al.32 but is still not fully understood. The cysteine residues of MOMP are thought to not only form intramolecular bonds, but might also form intermolecular bonds stabilizing the MOMP trimer or to form covalent bonds between MOMP and the 60 kDa and 15 kDa cysteine‐rich outer membrane proteins. High‐resolution structure elucidation by NMR or X‐ray crystallography remains elusive but several models have been proposed.10, 31, 32, 33 All published models of C. trachomatis MOMP propose a 16 stranded anti‐parallel β‐strand conformation with the caveat that the location of the β‐strands in the MOMP primary sequence varies greatly between these models. Atanu et al. proposed a 14‐strand β‐barrel model for the highly homologous MOMP of C. pneumoniae (sequence consensus 76%).34 In addition, the PRED TMBB2 prediction algorithm predicts C. muridarum and C. trachomatis MOMP to fold into a 12 stranded β‐barrel.57 MOMP monomers are thought to associate into homo‐trimers. This work represents the first spectroscopic structural characterization of human serovars of C. trachomatis nMOMP. We assessed the structure of nMOMP solubilized in Zwittergent micelles from different C. trachomatis human serovars and compared it to the highly homolog genus C. muridarum using spectroscopic techniques. Our results for C. muridarum MOMP were consistent with published literature and in agreement with the model of a trimeric 16 stranded β‐barrel, even though the diameter of C. muridarum MOMP observed by electron microscopy is slightly lower than expected for the 16 stranded β‐barrel. For C. trachomatis MOMP the spectroscopic measurements suggest that the percentage of β‐sheet conformation might be insufficient to form 16 β‐strands. Our measurements are in agreement with a trimeric MOMP quaternary structure, but also indicate the presence of MOMP dimers and possibly monomers. The detergent micelles are not homogenous with a wide distribution of detergent molecules per micelle. It is possible that the stability of C. muridarum and C. trachomatis MOMP differs or that the native structure is differently maintained throughout the isolation procedure. In addition, we found that the spectroscopic properties, and therefore the structure of C. trachomatis MOMP, depended to some degree on whether nMOMP was isolated from adherent or suspension cells. It remains to be elucidated if this structural difference is caused by structural adjustment of MOMP in different biophysical environments or if it is an artifact of slight variations in the isolation procedures. The choice of the detergent class for MOMP micelle formation can be expected to play an important role in future studies of MOMP structure as well as the prevention of protein aggregation.58 The redox potential of the buffer system is another variable that might influence MOMP heterogeneity and aggregation and requires further systematic analysis. A flexible conformation of MOMP, with different structures in the EB and the RB life cycle, is conceivable and might add to the difficulty of isolating MOMP in a single conformation. A well founded understanding of the MOMP structure and its dynamics will be essential for the discernment of MOMP based Chlamydia vaccination studies.
Material and Methods
Materials
Chlamydia muridarum (strain Nigg); C. trachomatis mouse pneumonitis (MoPn) (obtained from Dr Bernard Arulanandam, University of Texas at San Antonio).
Chlamydia trachomatis Serovar D [UW‐3Cx] (ATCC # VR‐885) and C. trachomatis Serovar E [Bour] (obtained from Suchland lab at the University of Washington).
Chlamydia trachomatis Serovar F (ATCC #VR‐346), C. trachomatis Serovar J (ATCC #VR‐886).
Suspension HeLa 229 cells (ATCC# CCL‐2.1).
Suspension CHO‐K1 cells (ATCC).
Suspension CAP cells (CEVEC Pharmaceuticals GmbH).
Suspension CLDK (MSD; US9441207B2).
Methods
Cell growth and propagation of chlamydial bodies
HeLa 229 (ATCC # CCL‐2.1, Manassas, VA) cells were used for propagation of Chlamydia serovars. HeLa 229 cells were grown in Eagle's Minimal Essential Medium (EMEM, ATCC) supplemented with 10% heat‐inactivated fetal bovine serum (FBS; Hyclone, Logan, UT), 50 μg/mL vancomycin (Sigma, St. Louis, MO), and 10 μg/mL gentamicin (Invitrogen, Carlsbad, CA). Host cells were seeded into tissue culture flasks at a cell density of 5 × 105 cells/mL and incubated overnight at 37°C in 5% CO2 to achieve a confluent monolayer. Cell monolayers were treated with 45 μg/mL DEAE‐Dextran Hydrochloride (Sigma) in Hanks’ Balanced Salt Solution (HBSS; Invitrogen) for 10 min at 37°C with slow rocking and then washed with HBSS. Chlamydia trachomatis Serovar D [strain UW‐3/Cx] (ATCC # VR‐885), Serovar E [Bour] (ATCC #VR‐348B), Serovar F [UW‐6/Cx] (ATCC #VR‐346), Serovar J [UW‐36/Cx] (ATCC #VR‐886), or C. muridarum (obtained from Dr Bernard Arulanandam [U. Texas San Antonio]) diluted in sucrose‐phosphate‐glutamate (SPG; Boston BioProducts, Ashland, MA) buffer were overlaid onto the monolayer at an MOI of 1.5:1 and incubated for 2 h at 37°C with slow rocking. Cycloheximide (Sigma‐Aldrich) was added at 2 μg/mL to the flasks without removing the inoculum. Incubations continued at 37°C with no rocking for 44–48 h for all serovars, except Serovar E which was incubated for 68–72 h. The supernatant was removed and centrifuged at 15,900 rpm for 90 min. The Chlamydia‐infected HeLa cells were removed from the flask using sterile glass beads and the cell suspension was centrifuged at 12,000 rpm for 1 h. The resulting pellets were frozen at −80°C. Chlamydiae were propagated either in‐house or by Paragon BioServices, Inc. (Baltimore, MD).
Suspension‐adapted CHO‐K1 (ATCC), CAP (CEVEC Pharmaceuticals GmbH), CLDK (MSD), and HeLa 229 (ATCC) cells were maintained in CD CHO (Gibco), Protein Expression Media (Gibco), Veggie #8 (SAFC) or EX‐CELL Hela (SAFC), respectively, containing 2–6 mM l‐glutamine (Gibco). For production of nMOMP, cells were infected with C. trachomatis, Serovar E, strain BOUR. Prior to incubation with cells, infectious elementary body (EB) stocks were pre‐treated for 30 min with 10 μg/mL polyethylenimine (linear, 25 kDa, Polysciences) in SPG buffer in a total volume of 2.5 mL. During EB pre‐treatment, cells were washed with HBSS w/o calcium, magnesium, or phenol red (Hyclone) and were pelleted by centrifugation (300g for 10 min). Pre‐treated EBs were then diluted to 5 mL in SPG and used to resuspend the cell pellet. The cell pellet/EB mixture was transferred to a TPP TubeSpin Bioreactor and incubated at 37°C for approximately 2 h in a Kuhner shaker/incubator (200 rpm, 2 in. throw, 5% CO2, 85% relative humidity). Following this incubation, infected cells were transferred to a disposable non‐baffled ventilated 1 L polycarbonate shake flask (Corning) and diluted to a final volume of 250 mL with the appropriate cycloheximide‐supplemented production media. Cells were incubated at 37°C, 5% CO2, and 85% relative humidity, with shaking (200 rpm, 2 in. throw). Four days post‐infection, infected cells were pelleted and media was removed. Cell pellets were stored at −70°C until nMOMP purification. For each cell line, the multiplicity of infection was 1.5; however, the cell density at the time of infection, the production media and the cycloheximide concentration varied. For suspension‐adapted CHO‐K1 cells, 4.2 × 106 viable cells/mL were infected and grown in 0.25 μg/mL cycloheximide in CD CHO media supplemented with 2 mM l‐glutamine. CAP cells were infected at a density of 3.7 × 106 viable cells/mL and maintained in protein expression media containing 1 μg/mL cycloheximide and 4 mM l‐glutamine. CLDK cells were infected at a density of 2.6 viable cells/mL and grown in Veggie #8 containing 0.5 μg/mL cycloheximide. Lastly, suspension‐adapted HeLa 229 cells were infected at density of 4.5 × 106 viable cells/mL and grown in EX‐CELL media supplemented with 0.5 μg/mL cycloheximide and 6 mM l‐glutamine.
Isolation and purification of major outer membrane protein
Chlamydia muridarum or C. trachomatis MOMP was purified using a modified methodology from Pal et al.59 In brief, MOMP was extracted from intact Chlamydia‐infected HeLa cells using sonication. Initial steps used 200 mM phosphate buffer, pH 5.5 with 2% CHAPS (Calbiochem, La Jolla, CA), 1 mM EDTA, and 100 mM DTT (Acros Organics, Fair Lawn, NJ) with 1× protease inhibitor cocktail. Subsequent steps used 200 mM phosphate buffer, pH 5.5 with 2% Zwittergent 3–14 (Calbiochem) with 100 mM DTT, 1 mM EDTA, and 1× protease inhibitor. MOMP was purified post‐extraction with hydroxyapatite chromatography (BioRad BioScale Mini Cartridge CHT Type II, 40 μm Media). The extraction product was loaded onto the hydorxyapatite cartridge in 20 mM phosphate buffer pH 5.5, 0.2% Zwittergent 3–14 and step eluted from the cartridge using 500 mM phosphate buffer, pH 5.5, 0.2% Zwittergent 3–14. The hydroxyapatite product was concentrated using tangential flow filtration (TFF), and then further purified by anion exchange chromatography (0.75 mL – 2 cm Pall Ceramic Hyper D 20066‐023, Pall, Port Washington, NY). The concentrated hydroxyapatite product was loaded with 20 mM MES (Sigma) pH 6.3, 0.1% Zwittergent Z‐3,14 as running buffer and subsequently step eluted from the column using 20 mM MES pH 6.3, 0.1% Zwittergent 3–14, 1 M NaCl (Sigma‐Aldrich). MOMP product was then dialyzed into 10 mM MES pH 7.3, 150 mM NaCl, 0.1% Zwittergent 3–14. The yield of MOMP isolated from adherent cell lines varied between 0.3 mg and 0.6 mg per 100 cm2 cell culture surface with a wet cell weigh of ~ 1 g. The yield of MOMP isolated from suspension cell lines varied between 4 mg and 10 mg per liter of cell culture. Product purities were validated by LC/MS2 and estimated to be ≥95% by label free quantitation. The nMOMP protein samples were stored at 4°C and analyzed within 1 week after purification.
Protein concentration determination by amino acid analysis
Vapor phase hydrolysis of the MOMP micelles was performed with constant‐boiling 6 N HCl at 110°C for 20–70 h using an ELDEX (Napa, CA) H/D workstation. Protein hydrolysates and amino acid standards were derivatized with AQC using the Waters AccQ Tag Ultra derivatization kit according to the manufacturer's directions. AQC reacts with primary and secondary amines to form stable, fluorescent derivatives that can be separated by RP‐HPLC.62 The fluorescent AQC derivatives were separated on an AccQ Tag Ultra C18, 1.7 μm, 2.1 × 100 mm column using a modified gradient program.
Size exclusion chromatography/multi‐angle laser light scattering
Separation and analysis of MOMP micelles was performed on an Agilent 1100 HPLC system consisting of a quaternary pump, a vacuum degasser, a thermostated autosampler and column compartment with a Superdex 200 30/100 GL column (GE Healthcare Life Sciences). The column temperature was kept at room temperature (20°C). The mobile phase (10 mM MES, pH 7.3, 150 mM NaCl, 0.1% Zwittergent 3–14) was pumped at 0.5 mL/min. A flow rate of 0.6 mL/min was used for samples indicated in the legend of Fig. 1. To reduce baseline noise in the light scattering detector, a 25 mm high‐pressure filter with 0.1 μm pores (Millipore) was used for in‐line filtration of the mobile phase. Detection was carried out using the Agilent HPLC DAD UV‐detector G1315B (280 nm) and Wyatt Technology's (Santa Barbara, CA) DAWN Heleos II light‐scattering and T‐Rex refractive index detectors. The refractive index increment value (dn/dc) of 0.185 mL/g for protein (literature) and 0.1485 mL/g for Zwittergent 3–14 (experimentally determined) were used for molecular weight calculations. Protein UV extinction coefficients were calculated based on the primary sequence using Vector NTI Advance 11 (Invitrogen). The Dawn Heleos II detector was calibrated with toluene according to the manufacturer's instructions. About 200 μg of protein sample was injected for each analysis. Determination of protein mass content and molecular weight was performed by the ASTRA 6.1 software (Wyatt Technology) using the conjugate analysis algorithm. More detail on the use of three detector systems for the analysis of protein complexes is reported elsewhere.60, 61
CD spectroscopy
CD measurements were performed with a Chirascan spectrophotometer (Applied Photophysics, Leatherhead, UK) in quartz cuvettes with 0.05 cm path length (Starna Cells, CA). Spectra were recorded from 200 nm to 260 nm or 280 nm at 20°C. The resolution was set to 1 nm and data acquisition was performed at a rate of five time points per second at a band width of 2 nm. Final spectra were obtained by averaging five consecutive scans and corrected for background by subtraction of spectra of protein‐free samples that were recorded under the same conditions. The MOMP concentration ranged from 0.8 to 1.5 mg/mL. Measurements were performed in 10 mM MES pH 7.3, 150 mM NaCl, 0.1% Zwittergent 3–14. Mean molar ellipiticities per residue were calculated based on the molar protein concentration and the amino acid composition. MOMP concentrations were determined by amino acid analysis. Normalized secondary structure compositions were predicted using the CDNN algorithm.40
ATR‐FTIR spectroscopy
ATR‐FTIR was performed using a Nicolet 6700 FTIR spectrometer (Thermo Scientific). The spectrometer was continuously purged with nitrogen gas to remove water vapor. The internal reflection element was a ZnSe crystal (Smart ITR plate). One μL of MOMP in 10 mM MES, 2% Zwittergent Z 3–14, 10 mM DTT, pH 7.3 at protein concentrations between 1 mg/mL and 2 mg/mL was applied to the ATR plate and dried under a stream of nitrogen. Infrared spectra were recorded from 1400 cm−1 to 2000 cm−1. The data are means of 254 scans at a resolution of 4 (data spacing 0.482 cm−1). Buffer spectra without protein were obtained using identical conditions and then subtracted from the protein spectrum. All processing procedures were carried out in the amide I region. The quality of the FTIR spectra was ensured by monitoring a straight baseline between 2000 cm−1 and 1750 cm−1. The second‐order derivative was obtained by the method of Savitsky‐Golay using 21 data points and third‐order polynomial (Omnic software V 8.3.103). Non‐linear curve fitting procedures to deconvolute the FTIR spectrum were performed using the manufacturer's software. Voigt shaped peak centers were fixed at the minima obtained from the second derivative analysis. The peak width was set as a floating parameter and not constrained. The fitting procedure was performed until a stable solution was achieved and the fit‐curve best overlaid with the measurement (Fig. S1).
Electron microscopy
Electron microscopy measurements were performed by Electron Microscopy BioServices (Frederick, MD) using a FEI Tecnai Spirit Twin Transmission Electron Microscope (FEI, OR) at high magnifications at 80 kV. Negative stain was performed by incubation for 60 s with 2% phosphotungstic acid. Excess stain was removed with filter paper.
Conflict of Interest
All co‐authors are current or former employees of Merck & Co and may own stock or hold stock options in the company.
Statement for broader audience
Chlamydial major outer membrane protein (MOMP) is one of the lead components for a human vaccine. In the absence of a well‐defined functional assay, a thorough biophysical structure characterization by various methods is a prerequisite to monitoring consistency of protein structure due to variation in growth and isolation procedures.
Supporting information
Appendix S1: Supporting Information
References
- 1. Kuo CC, Jackson LA, Campbell LA, Grayston JT (1995) Chlamydia pneumoniae (TWAR). Clinic Microbiol Rev 8:451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collina F, De Chiara A, De Renzo A, De Rosa G, Botti G, Franco R (2012) Chlamydia psittaci in ocular adnexa MALT lymphoma: a possible role in lymphomagenesis and a different geographical distribution. Infect Agents Cancer 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mariotti SP (2004) New steps toward eliminating blinding trachoma. New Engl J Med 351:2004–2007. [DOI] [PubMed] [Google Scholar]
- 4. Brunham RC, Rey‐Ladino J (2005) Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 5:149–161. [DOI] [PubMed] [Google Scholar]
- 5. Liang S, Bulir D, Kaushic C, Mahony J (2017) Considerations for the rational design of a Chlamydia vaccine. Human Vacc Immunotherap 13:831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de la Maza LM, Zhong G, Brunham RC (2017) Update in Chlamydia trachomatis vaccinology. Clin Vaccine 20:e00543–e00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Budrys NM, Gong S, Rodgers AK, Wang J, Louden C, Shain R, Schenken RS, Zhong G (2012) Chlamydia trachomatis antigens recognized in women with tubal factor infertility, normal fertility, and acute infection. Obstet Gynecol 119:1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malhotra M, Sood S, Mukherjee A, Muralidhar S, Bala M (2013) Genital Chlamydia trachomatis: an update. Ind J Med Res 138:303–316. [PMC free article] [PubMed] [Google Scholar]
- 9. Manavi K (2006) A review on infection with Chlamydia trachomatis . Best Pract Res Clinic Obstet Gynaecol 20:941–951. [DOI] [PubMed] [Google Scholar]
- 10. Findlay HE, McClafferty H, Ashley RH (2005) Surface expression, single‐channel analysis and membrane topology of recombinant Chlamydia trachomatis Major Outer Membrane Protein. BMC Microbiol 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang SP, Kuo CC, Barnes RC, Stephens RS, Grayston JT (1985) Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J Infect Dis 152:791–800. [DOI] [PubMed] [Google Scholar]
- 12. Cruz‐Fisher MI, Cheng C, Sun G, Pal S, Teng A, Molina DM, Kayala MA, Vigil A, Baldi P, Felgner PL, Liang X, de la Maza LM (2010) Identification of immunodominant antigens by probing a whole Chlamydia trachomatis open reading frame proteome microarray using sera from immunized mice. Infect Immun 79:246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Molina DM, Pal S, Kayala MA, Teng A, Kim PJ, Baldi P, Felgner PL, Liang X, de la Maza LM (2010) Identification of immunodominant antigens of Chlamydia trachomatis using proteome microarrays. Vaccine 28:3014–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cambridge CD, Singh SR, Waffo AB, Fairley SJ, Dennis VA (2013) Formulation, characterization, and expression of a recombinant MOMP Chlamydia trachomatis DNA vaccine encapsulated in chitosan nanoparticles. Intl J Nanomed 8:1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang PF, Cai YQ, Chen J, Ye XX, Mao SS, Zhu SL, Xue XY, Chen S, Zhang LF (2017) Evaluation of tandem Chlamydia trachomatis MOMP multi‐epitopes vaccine in BALB/c mice model. Vaccine 35:3096–3103. [DOI] [PubMed] [Google Scholar]
- 16. Kalbina I, Wallin A, Lindh I, Engstrom P, Andersson S, Strid A (2011) A novel chimeric MOMP antigen expressed in Escherichia coli, Arabidopsis thaliana, and Daucus carota as a potential Chlamydia trachomatis vaccine candidate. Prot Express Purif 80:194–202. [DOI] [PubMed] [Google Scholar]
- 17. Schautteet K, Stuyven E, Beeckman DSA, Van Acker S, Carlon M, Chiers K, Cox E, Vanrompay D (2011) Protection of pigs against Chlamydia trachomatis challenge by administration of a MOMP‐based DNA vaccine in the vaginal mucosa. Vaccine 29:1399–1407. [DOI] [PubMed] [Google Scholar]
- 18. Tifrea DF, Pal S, Popot JL, Cocco MJ, de la Maza LM (2014) Increased immunoaccessibility of MOMP epitopes in a vaccine formulated with amphipols may account for the very robust protection elicited against a vaginal challenge with Chlamydia muridarum . J Immunol 192:5201–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Millman KL, Tavare S, Dean D (2001) Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar‐specific differences in tissue tropism, immune surveillance, and persistence of the organism. J Bacteriol 183:5997–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeffrey BM, Suchland RJ, Quinn KL, Davidson JR, Stamm WE, Rockey DD (2010) Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 78:2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moulder JW (1991) Interaction of chlamydiae and host cells in vitro. Microbiolog Rev 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barbour AG, Amano KI, Hackstadt T, Perry L, Caldwell HD (1982) Chlamydia‐trachomatis has penicillin‐binding proteins but not detectable muramic acid. J Bacteriol 151:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghuysen JM, Goffin C (1999) Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob Agents Chemother 43:2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yen TY, Pal S, de la Maza LM (2005) Characterization of the disulfide bonds and free cysteine residues of the Chlamydia trachomatis mouse pneumonitis major outer membrane protein. Biochemistry 44:6250–6256. [DOI] [PubMed] [Google Scholar]
- 25. Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, Valdivia RH (2011) Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol 82:1185–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X, Afrane M, Clemmer DE, Zhong G, Nelson DE (2010) Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J Bacteriol 192:2852–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Elwell C, Mirrashidi K, Engel J (2016) Chlamydia cell biology and pathogenesis. Nat Rev 14:385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wen Z, Boddicker MA, Kaufhold RM, Khandelwal P, Durr E, Qiu P, Lucas BJ, Nahas DD, Cook JC, Touch S, Skinner JM, Espeseth AS, Przysiecki CT, Zhang L (2016) Recombinant expression of Chlamydia trachomatis major outer membrane protein in E. coli outer membrane as a substrate for vaccine research. BMC Microbiol 16:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hughes ES, Shaw KM, Ashley RH (2001) Mutagenesis and functional reconstitution of chlamydial major outer membrane proteins: VS4 domains are not required for pore formation but modify channel function. Infect Immun 69:1671–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schulz GE (2002) The structure of bacterial outer membrane proteins. Biochim Biophys Acta 1565:308–317. [DOI] [PubMed] [Google Scholar]
- 31. Feher VA, Randall A, Baldi P, Bush RM, de la Maza LM, Amaro RE (2013) A 3‐dimensional trimeric beta‐barrel model for Chlamydia MOMP contains conserved and novel elements of Gram‐negative bacterial porins. PLoS One 8:e68934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX (2006) Identification of surface‐exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci 15:122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodriguez‐Maranon MJ, Bush RM, Peterson EM, Schirmer T, de la Maza LM (2002) Prediction of the membrane‐spanning beta‐strands of the major outer membrane protein of Chlamydia. Protein Sci 11:1854–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Atanu FO, Oviedo‐Orta E, Watson KA (2013) A novel transport mechanism for MOMP in Chlamydophila pneumoniae and its putative role in immune‐therapy. PLoS One 8:e61139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feinstein HE, Tifrea D, Sun GF, Popot JL, de la Maza LM, Cocco MJ (2014) Long‐term stability of a vaccine formulated with the amphipol‐trapped major outer membrane protein from Chlamydia trachomatis . J Memb Biol 247:1053–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cai S, He F, Samra HS, de la Maza LM, Bottazzi ME, Joshi SB, Middaugh CR (2009) Biophysical and stabilization studies of the Chlamydia trachomatis mouse pneumonitis major outer membrane protein. Mol Pharm 6:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun G, Pal S, Sarcon AK, Kim S, Sugawara E, Nikaido H, Cocco MJ, Peterson EM, de la Maza LM (2007) Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis . J Bacteriol 189:6222–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tifrea DF, Sun G, Pal S, Zardeneta G, Cocco MJ, Popot JL, de la Maza LM (2011) Amphipols stabilize the Chlamydia major outer membrane protein and enhance its protective ability as a vaccine. Vaccine 29:4623–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pal S, Theodor I, Peterson EM, de la Maza LM (2001) Immunization with the Chlamydia trachomatis mouse pneumonitis major outer membrane protein can elicit a protective immune response against a genital challenge. Infect Immun 69:6240–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bohm G, Muhr R, Jaenicke R (1992) Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng 5:191–195. [DOI] [PubMed] [Google Scholar]
- 41. Goormaghtigh E, Cabiaux V, Ruysschaert JM (1990) Secondary structure and dosage of soluble and membrane‐proteins by attenuated total reflection Fourier‐transform infrared‐spectroscopy on hydrated films. Eur J Biochem 193:409–420. [DOI] [PubMed] [Google Scholar]
- 42. Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S (2001) Tom40, the pore‐forming component of the protein‐conducting TOM channel in the outer membrane of mitochondria. J Cell Biol 153:1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kong J, Yu S (2007) Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim Biophys Sin 39:549–559. [DOI] [PubMed] [Google Scholar]
- 44. Greenfield NJ (2006) Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc 1:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pal S, Tatarenkova OV, de la Maza LM (2015) A vaccine formulated with the major outer membrane protein can protect C3H/HeN, a highly susceptible strain of mice, from a Chlamydia muridarum genital challenge. Immunology 146:432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pal S, Theodor I, Peterson EM, de la Maza LM (1997) Immunization with an acellular vaccine consisting of the outer membrane complex of Chlamydia trachomatis induces protection against a genital challenge. Infect Immun 65:3361–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kari L, Whitmire WM, Crane DD, Reveneau N, Carlson JH, Goheen MM, Peterson EM, Pal S, de la Maza LM, Caldwell HD (2009) Chlamydia trachomatis native major outer membrane protein induces partial protection in nonhuman primates: implication for a trachoma transmission‐blocking vaccine. J Immunol 182:8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pal S, Tifrea DF, Follmann F, Andersen P, de la Maza LM (2017) The cationic liposomal adjuvants CAF01 and CAF09 formulated with the major outer membrane protein elicit robust protection in mice against a Chlamydia muridarum respiratory challenge. Vaccine 35:1705–1711. [DOI] [PubMed] [Google Scholar]
- 49. Kelly SM, Jess TJ, Price NC (2005) How to study proteins by circular dichroism. Biochim Biophys Acta 1751:119–139. [DOI] [PubMed] [Google Scholar]
- 50. Greenfield NJ (1996) Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal Biochem 235:1–10. [DOI] [PubMed] [Google Scholar]
- 51. Santos HJ, Imai K, Makiuchi T, Tomii K, Horton P, Nozawa A, Ibrahim M, Tozawa Y, Nozaki T (2015) A novel mitosomal beta‐barrel outer membrane protein in Entamoeba. Sci Rep 5:8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kenedy MR, Luthra A, Anand A, Dunn JP, Radolf JD, Akins DR (2014) Structural modeling and physicochemical characterization provide evidence that P66 forms a beta‐barrel in the Borrelia burgdorferi outer membrane. J Bacteriol 196:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Desrosiers DC, Anand A, Luthra A, Dunham‐Ems SM, LeDoyt M, Cummings MA, Eshghi A, Cameron CE, Cruz AR, Salazar JC, Caimano MJ, Radolf JD (2011) TP0326, a Treponema pallidum beta‐barrel assembly machinery A (BamA) orthologue and rare outer membrane protein. Mol Microbiol 80:1496–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266:540–553. [DOI] [PubMed] [Google Scholar]
- 55. Murzin AG (2001) Progress in protein structure prediction. Nat Struct Biol 8:110–112. [DOI] [PubMed] [Google Scholar]
- 56. Chou PY, Fasman GD (1974) Prediction of protein conformation. Biochemistry 13:222–245. [DOI] [PubMed] [Google Scholar]
- 57. Tsirigos KD, Elofsson A, Bagos PG (2016) PRED‐TMBB2: improved topology prediction and detection of beta‐barrel outer membrane proteins. Bioinformatics 32:665–671. [DOI] [PubMed] [Google Scholar]
- 58. Seddon AM, Curnow P, Booth PJ (2004) Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta 1666:105–117. [DOI] [PubMed] [Google Scholar]
- 59. Pal S, Fielder TJ, Peterson EM, de la Maza LM (1994) Protection against infertility in a BALB/c mouse salpingitis model by intranasal immunization with the mouse pneumonitis biovar of Chlamydia trachomatis . Infect Immun 62:3354–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wen J, Arakawa T, Philo JS (1996) Size‐exclusion chromatography with on‐line light‐scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem 240:155–166. [DOI] [PubMed] [Google Scholar]
- 61. Kendrick BS, Kerwin BA, Chang BS, Philo JS (2001) Online size‐exclusion high‐performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein–protein or protein–ligand association states. Anal Biochem 299:136–146. [DOI] [PubMed] [Google Scholar]
- 62.Cohen SA (2003) Amino acid analysis using pre‐column derivatization with 6‐aminoquinolyl‐N‐hydroxysuccinimidyl carbamate. Analysis of hydrolyzed proteins and electroblotted samples. Methods Mol Biol 211:143–154. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


