Abstract
The functional roles of the (His)17 region and an insert region in the eukaryotic nitrile hydratase (NHase, EC 4.2.1.84) from Monosiga brevicollis (MbNHase), were examined. Two deletion mutants, MbNHaseΔ238−257 and MbNHaseΔ219−272, were prepared in which the (His)17 sequence and the entire insert region were removed. Each of these MbNHase enzymes provided an α2β2 heterotetramer, identical to that observed for prokaryotic NHases and contains their full complement of cobalt ions. Deletion of the (His)17 motif provides an MbNHase enzyme that is ~55% as active as the WT enzyme when expressed in the absence of the Co-type activator (ε) protein from Pseudonocardia thermophila JCM 3095 (PtNHaseact) but ~28% more active when expressed in the presence of PtNHaseact. MbNHaseΔ219−272 exhibits ~55% and ~89% of WT activity, respectively, when expressed in the absence or presence of PtNHaseact. Proteolytic cleavage of MbNHase provides an α2β2 heterotetramer that is modestly more active compared to WT MbNHase (kcat = 163 ± 4 vs 131 ± 3 s−1). Combination of these data establish that neither the (His)17 nor the insert region are required for metallocentre assembly and maturation, suggesting that Co-type eukaryotic NHases utilize a different mechanism for metal ion incorporation and post-translational activation compared to prokaryotic NHases.
Keywords: Nitrile hydratase, enzyme kinetics, cobalt, protein biosynthesis, metal transport, metal insertion
Introduction
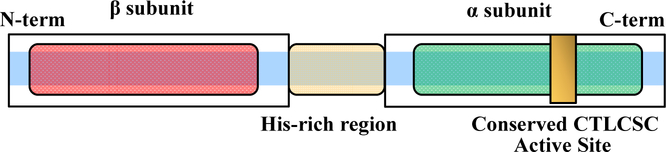
Nitrile hydratases (NHases, EC 4.2.1.84) are metalloenzymes that contain either a non-heme Fe(III) ion (Fe-type) or a non-corrin Co(III) ion (Co-type) in their active site [1, 2]. NHases catalyze the hydration of nitriles to their corresponding higher value amides under mild conditions (room temperature and physiological pH) and have attracted substantial interest as biocatalysts in preparative organic chemistry and bioremediation processes [3–8]. NHases have historically been only found in prokaryotes; however, multiple eukaryotic organisms were shown to contain genes that potentially encode NHase enzymes [9, 10]. We recently cloned and over-expressed the candidate gene from the eukaryotic organism Monosiga brevicollis in E. coli and characterized a fully functional Co-type NHase gene product, with fused α- and β-subunits linked by a (His)17 containing region (MbNHase) (Figure 1) [11]. Size-exclusion chromatography indicated that MbNHase is an (αβ)2 homodimer in solution, analogous to the α2β2 heterotetrameric architecture of prokaryotic NHases, of which numerous X-ray crystal structures exist [1, 2, 12, 13].
Figure 1.
Scheme showing the arrangement of the NHase β- and α-subunits in eukaryotes.
Several open reading frames (ORFs) have been identified just downstream from the structural α- and β-subunit genes in prokaryotic NHases, and one of these genes has been proposed to function as an activator (ε) protein [14–16]. The prevailing dogma is that both Co- and Fe-type NHase enzymes require the co-expression of an (ε) protein to be fully metallated, post-translationally modified, and fully functional [14–16]. No such (ε) protein has been identified for eukaryotic NHases, such as MbNHase [9, 10]. It is tempting to speculate that the insert region within MbNHase, which contains a (His)17 motif, plays the role of the (ε) protein. Histidine rich regions are present in some cobalamin (vitamin B12) biosynthetic pathway proteins, e.g., the chelatase CbiX enzyme from Bacillus megaterium and CobW from Pseudomonas denitrificans [17]. Histidine rich regions are also found in accessory proteins involved in metallocentre assembly of nickel hydrogenases and ureases, such as HypB from Bradyrhizobium japonicum and Rhizobium leguminosarum, SlyD from Escherichia coli and Helicobacter pylori, UreE from Klebsiella aerogenes, and Hpn and Hpn-like proteins from Helicobacter pylori [18].
To investigate metallocentre assembly in MbNHase, we created three different altered MbNHase enzymes. First, we obtained a proteolytically cleaved MbNHase enzyme in which the α-and β-subunits were separated providing an enzyme that structurally mimics prokaryotic NHase enzymes. Second, a mutant MbNHase enzyme, in which the entire (His)17 motif was removed (MbNHaseΔ238−257), was constructed and expressed in the presence and absence of the prototypical prokaryotic Co-type (ε) activator protein from Pseudonocardia thermophila JCM 3095 (PtNHaseact). Finally, a mutant was constructed in which the entire insert region was deleted (MbNHaseΔ219−272), leaving only the classical prokaryotic NHase α-and β-subunit analogs; this mutant was also expressed in the presence and absence PtNHaseact. Functional interrogation of these species provides important insight into the role of the insert region in MbNHase and how eukaryotic NHase enzymes are metallated and post-translationally modified.
Materials and Methods
Materials.
Acrylonitrile, 2-Amino-2-hydroxymethyl-propane-1,3-diol (Tris-HCl), and 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) were obtained from Sigma-Aldrich. Oligonucleotides and genes were obtained from Integrated DNA Technologies, Inc. All other reagents were purchased commercially and were the highest purity available.
Expression and Purification of MbNHase.
Protein sequences for the α- and β-subunit genes of the putative WT MbNHase were obtained from ORF 37534 (UniProt ID A9V2C1.1) of M. brevicollis and the predicted gene was synthesized by Integrated DNA Technologies, Inc. with optimized E. coli codon usage. This gene was cloned into the kanamycin resistant pET21a+ (EMD Biosciences) expression vector to create the plasmid pSMM0αβ, as previously reported [11].
A single colony was used to inoculate separate starter cultures of 50 mL LB Miller media containing the appropriate antibiotics (pET28a+: 50 mg/mL kanamycin) and allowed to grow at 37 °C with constant shaking overnight. These cultures were used to inoculate 6 L of LB Miller media containing the appropriate antibiotics and allowed to grow at 37 °C with constant shaking until an optical density of ~0.8–1.0 at 600 nm was reached. The cultures were cooled on ice to 20 °C and induced with 0.1 mM Isopropyl-β-D-1-thiogalactopyranoside (IPTG), supplemented with 0.25 mM of CoCl2, and shaken for an additional 16 hours at 20 °C.
Cells were pelleted by centrifugation at 6,370 × g for 10 minutes at 4 °C in a Beckman Coulter Avanti JA-10 rotor and resuspended in buffer A (50 mM sodium phosphate buffer, pH 7.5, containing 300 mM NaCl, 5% glycerol, and 10 mM imidazole) at a ratio of 5 mL per gram of cells. Cells were lysed by ultrasonication (Misonix Sonicator 3000) for 4 min (alternating 30s on and 45s off) at 21 W. Cell lysate was separated from cell debris by centrifugation in a JA-20 rotor at 31,000 × g at 4 °C for 20 min. Cell lysate was purified using immobilized metal affinity chromatography (IMAC; 100 mg protein/5mL column) on a GE ÄKTA Fast Protein Liquid Chromatography (FPLC) system at 4 °C. The column was washed with 50 mL of buffer A followed by 50 mL of buffer A containing additional imidazole (35 mM). The protein was eluted using a linear gradient (0–100%) of buffer B (50 mM NaH2PO4 pH 7.5, 300 mM NaCl, 10% glycerol, 525 mM imidazole) at a flow rate of 1 mL/min resulting in MbNHase being eluted between 150 to 240 mM imidazole.
Fractions containing WT MbNHase were pooled and concentrated to ~1 mL using an Amicon Ultra-15 (Millipore) and loaded onto a 16/60 Superdex 200 prep grade (GE Healthcare) polishing column using buffer C (50 mM HEPES and 300 mM NaCl at pH 8.0). Pure WT MbNHase was concentrated using an Amicon Ultra-15 (Millipore) and analyzed by SDS-PAGE with a 12.5% polyacrylamide SPRINT NEXT GEL™ (Amresco). Gels were stained with Gel Code Blue (Thermo-Fisher Scientific). The protein concentration of purified WT MbNHase was determined by measuring the absorbance at 280 nm on a Shimadzu UV-2450 spectrophotometer. The calculated molecular weight for the WT MbNHase homodimer is 111,207 g/mol with an extinction coefficient of 143,700 cm−1 M−1. The molecular weight is in good agreement with SDS-PAGE data [11].
Expression and Purification of MbNHase Mutants.
The genes encoding for the MbNHase α- and β-subunits, where the entire (His)17 motif was removed (MbNHaseΔ238−257), were synthesized by Integrated DNA Technologies, Inc. with optimized E. coli codon usage and cloned into the kanamycin resistant pET28a+ (EMD Biosciences) plasmid. The α- and β-subunit genes were designed for co-expression by overlapping the “TGA” stop codon of the β-subunit with the “ATG” start codon of the α subunit (Figure SI-1), like the expression of PtNHase [16]. In addition, the α- and β-subunit genes where the entire insert region was deleted (MbNHaseΔ219−272), were obtained by polymerase chain reaction (PCR) using the primers (Sense primer: CAACCATGGGTACCG AGCAGGCGGCGGTG; Anti-sense primer: CATAAGCTTTTAGTGGTGGTGGTGATGATGATCAACACGCGGCA) for the α-subunit and (Sense primer: ACG CATATG ATGCACCTGT TCA CCTACGACCTGCA; Anti-sense primer: AGTCCTCGAGTTACGCTTG CGGCGGGTTGCTA) for the β-subunit. The α-subunit gene was sub-cloned between NcoI and HindIII sites, while the β-subunit gene was sub-cloned between NdeI and XhoI sites within the pCOLADuet-1 expression vector (Novagen). The sequences were confirmed by Functional Biosciences. The plasmids containing the MbNHaseΔ238−257 and MbNHaseΔ219−272 deletion mutant genes were transformed into BL21 magic cells for the soluble expression of the MbNHaseΔ238−257 and MbNHaseΔ219−272 enzymes. Expression and purification of these mutant enzymes were carried out in an identical manner to that described above for WT MbNHase [11].
Expression and Purification of WT MbNHase and the Mutants in the Absence or Presence of PtNHaseact.
The gene encoding the Co-type activator (ε) protein from Pseudonocardia thermophila JCM 3095 (PtNHaseact) was synthesized by Integrated DNA Technologies, Inc. with optimized E. coli codon usage and cloned into the kanamycin resistant pET21a+ (EMD Biosciences) expression vector to create the pSPTact plasmid. The pSPTact plasmid was co-expressed with the previously reported pSMMαβ plasmid encoding recombinant MbNHase [11] that had been freshly transformed into BL21(DE3) (Stratagene) competent cells. In addition, the pSPTact plasmid was co-expressed with the plasmids containing the MbNHaseΔ238−257 and MbNHaseΔ219−272 deletion mutant genes in BL21 magic cells. Expression and purification of WT MbNHase, MbNHaseΔ238−257 and MbNHaseΔ219−272 co-expressed with PtNHaseact was carried out in an identical manner to that described above for WT MbNHase [11].
Kinetic Analysis.
The enzymatic activity of WT MbNHase and each variant was determined using acrylonitrile as the substrate (acrylamide; Δε225 = 2.9 mM−1 cm−1). The rate of nitrile hydration was determined by continuously monitoring the formation of acrylamide at 225 nm using a Shimadzu UV-2450 spectrophotometer equipped with a TCC-240A temperature controlled cell holder [19]. A typical 1 mL reaction consisted of 50 mM Tris-HCl buffer pH 7.0 at 25 °C and various concentrations of acrylonitrile (0 to 450 mM). One unit (U) of MbNHase activity is defined as the formation of 1 μmol of acrylamide per minute. To obtain the kinetic parameters, Vmax and Km, the initial velocities from at least three independent measurements were fitted to the Michaelis-Menten equation using OriginPro 9.0 (OriginLab, Northampton, MA). Kinetic data for each MbNHase variant was determined more than three times for multiple purifications, all of which provided consistent results.
Metal Analysis.
The metal content of WT MbNHase and each variant, expressed in the presence and absence of CoCl2, was determined by inductively-coupled plasma mass spectrometry (ICP-MS). For comparison purposes, the Co-type NHase from Pseudonocardia thermophila JCM 3095 (PtNHase) was expressed and purified as previously described [20], and the metal content determined along with a buffer control that contained no protein. All protein samples were pretreated with 1 M urea and digested with concentrated nitric acid (0.863 mL) followed by heating at 70 °C for 1 h, allowed to cool to room temperature, and then diluted to a final concentration of 5% nitric acid. Samples were submitted for analysis at the Water Quality Center in the College of Engineering at Marquette University (Milwaukee, WI, USA).
Electronic Absorption and Electron Paramagnetic Resonance Spectra.
Electronic absorption spectra were recorded on a Shimadzu UV-2450 spectrophotometer equipped with a TCC-240A temperature-controlled cell holder. Spectra for WT MbNHase and each variant as well as PtNHase were obtained at 25 °C in a 1 cm quartz cuvette in 50 mM HEPES buffer containing 300 mM NaCl, pH 8.0. X-band EPR spectra for WT MbNHase were recorded at 4 K, 0.1 mW in 50 mM HEPES buffer containing 300 mM NaCl, pH 8.0 on a Bruker EMXplus spectrometer equipped with an ER4116DM (~ 9.6 GHz) resonator, an Oxford Instruments ESR900 helium flow cryostat, and Oxford Instruments ITC503 temperature controller.
Results and Discussion
Proteolytic Cleavage of WT MbNHase.
Active, mature MbNHase was obtained when expressed in the absence of an (ε) protein or the E. coli GroES/EL molecular chaperones [11][13], whereas prokaryotic NHase activation appears to absolutely require an activator protein [14–16]. These findings beg the question: “how is MbNHase functionally expressed?” We hypothesized that the insert region, which contains a His17 section, (Figure 1) plays a key role in metallocentre assembly and therefore, created three altered MbNHase enzymes targeting this insert region.
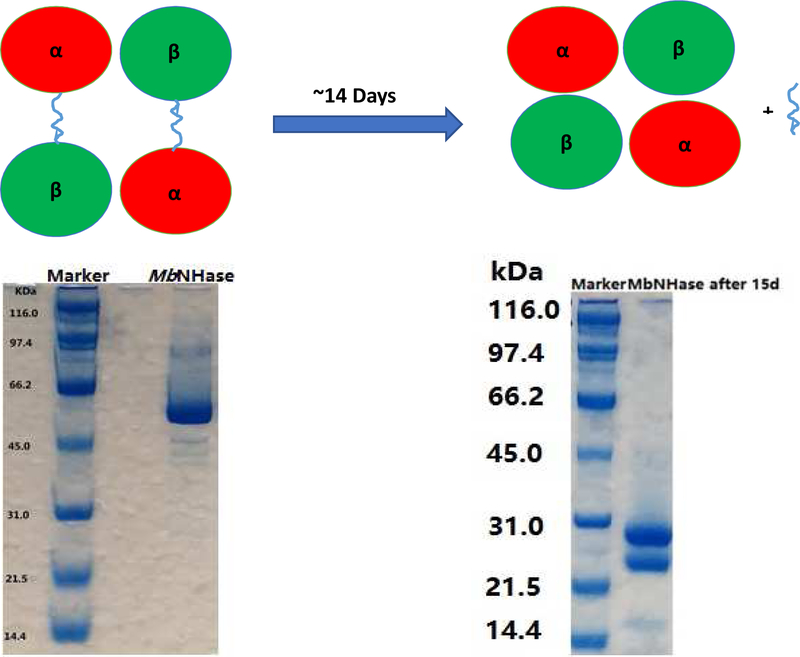
Purified WT MbNHase exhibits a single band at ~55 kDa on SDS-PAGE (Figure 2) and a kcat value of 131 ± 3 s−1 (Km = 83 ± 10 mM) in 50 mM Tris-HCl buffer pH 7.0 at 25 °C using acrylonitrile as the substrate, values that are indistinguishable from those reported previously (Table 1) [11]. Size-exclusion chromatography revealed that WT MbNHase exists primarily as an (αβ)2 homodimer in solution, analogous to the α2β2 heterotetramer architecture observed for prokaryotic NHases. However, storage of WT MbNHase at 4 °C in 50 mM Tris-HCl buffer, pH 7.0 for approximately two weeks, provided an MbNHase enzyme with a modestly increased kcat value of 163 ± 4 s−1 but a similar Km value of 93 ± 15 mM, using acrylonitrile as the substrate (Table 1). SDS-PAGE analysis of aged MbNHase samples revealed two polypeptides of ~24 and ~26 kDa (Figure 2), while size exclusion chromatography suggested an α2β2 heterotetramer, identical to that observed for prokaryotic NHases. These data suggested that WT MbNHase is cleaved, likely by trace amounts of proteases. The observed increase in kcat for the proteolytically cleaved MbNHase enzyme, represents an ~20% increase in rate over WT MbNHase. The increased rate was very reproducible for batch-to-batch preparations, providing the calculated error of ± 4 s−1. The observed increase in activity is perhaps due to an easing of conformational stress induced by the insert region in and around the active site, although structural characterization will be required to confirm this.
Figure 2.
The SDS-PAGE of (a) WT MbNHase and (b) the proteolytically cleaved MbNHase enzyme.
Table 1.
Kinetic constants for the wild-type and mutant MbNHasesa
| kcat (s−1) | Km (mM) | kcat/Km (s−1mM−1) | Metal Content | |
|---|---|---|---|---|
| Wild-typeb | 131 ± 3 | 83 ± 10 | 1.6 | 1.8 ± 0.1 |
| Cleaved | 163 ± 4 | 93 ± 15 | 1.8 | 1.9 ± 0.1 |
| Δ238–257 | 71 ± 4 | 104 ± 17 | 0.7 | 1.8 ± 0.1 |
| Δ238–257 + PtNHaseact | 166 ± 5 | 125 ± 17 | 1.3 | 2.3 ± 0.2 |
| Δ219–272 | 75 ± 8 | 73 ± 14 | 1.0 | 1.7 ± 0.1 |
| Δ219–272 + PtNHaseact | 117 ± 10 | 149 ± 23 | 0.8 | 1.7 ± 0.1 |
Acrylonitrile was used as the substrate.
Reference [11].
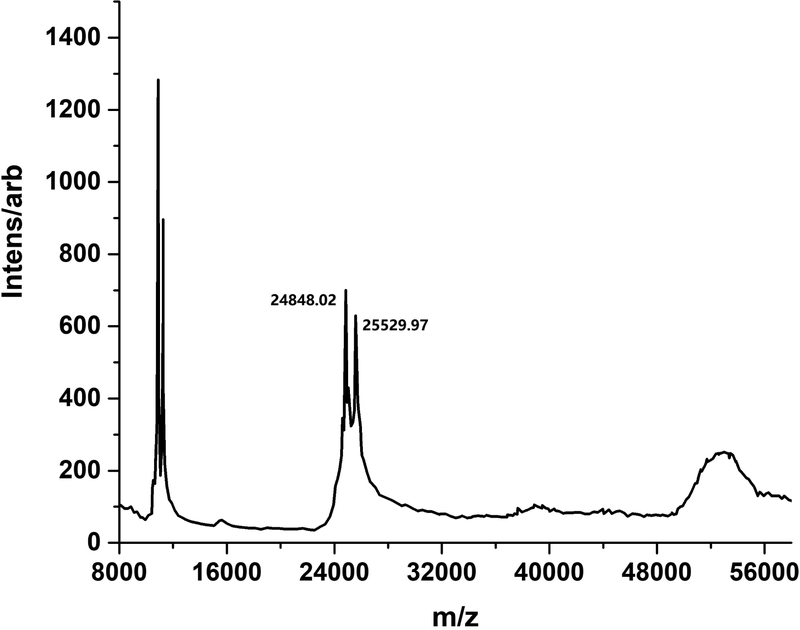
Confirmation of an α2β2 heterotetramer after proteolytic cleavage of WT MbNHase was obtained by MALDI-TOF mass spectroscopy (Figure 3). Two masses were clearly observed at 24,848 Da and 25,530 Da, values that are consistent with SDS-PAGE estimates (Figure 2) and correspond to the α- and β-subunits of MbNHase based on sequence comparison with the prototypical Co-type PtNHase enzyme (Figure 4). The two MALDI-TOF MS peaks were of similar intensities, indicating a ~1:1 α:β ratio. Interestingly, the observed molecular masses from MALDI-TOF MS suggests that a 5,226 Da peptide is lost after proteolytic cleavage, comparable to the size of the complete insert region of WT MbNHase. Attempts to isolate the cleaved fragment by gel-filtration were unsuccessful suggesting that the fragment is likely cleaved into small peptides by protease contaminants. Cleavage of MbNHase could be prevented by an additional gel-filtration purification step performed directly after IMAC; however, the addition of metal ion inhibitors such as EDTA or 1,10-phenanthroline had no effect on MbNHase cleavage. Addition of protease inhibitor cocktails, such as AEBSF, resulted in precipitation of MbNHase. Taken together, these data indicate that the single polypeptide of freshly isolated WT MbNHase, which contains fused α-and β-subunits linked by an insert region, is cleaved into separate α- and β-subunits upon ageing, likely by trace amounts of proteases, resulting in a modestly more active α2β2 heterotetrameric form of MbNHase.
Figure 3.
MALDI-TOF Mass Spectra of proteolytically cleaved MbNHase revealing two peaks at 24.8 and 25.5 kDa corresponding to independent α- and β-subunits.
Figure 4.
Sequence alignment of MbNHase and PtNHase shows that the α-subunits share 23% identity while the β-subunits exhibit 32% identity (Orange: active site; green: insert region; yellow: histidine rich region).
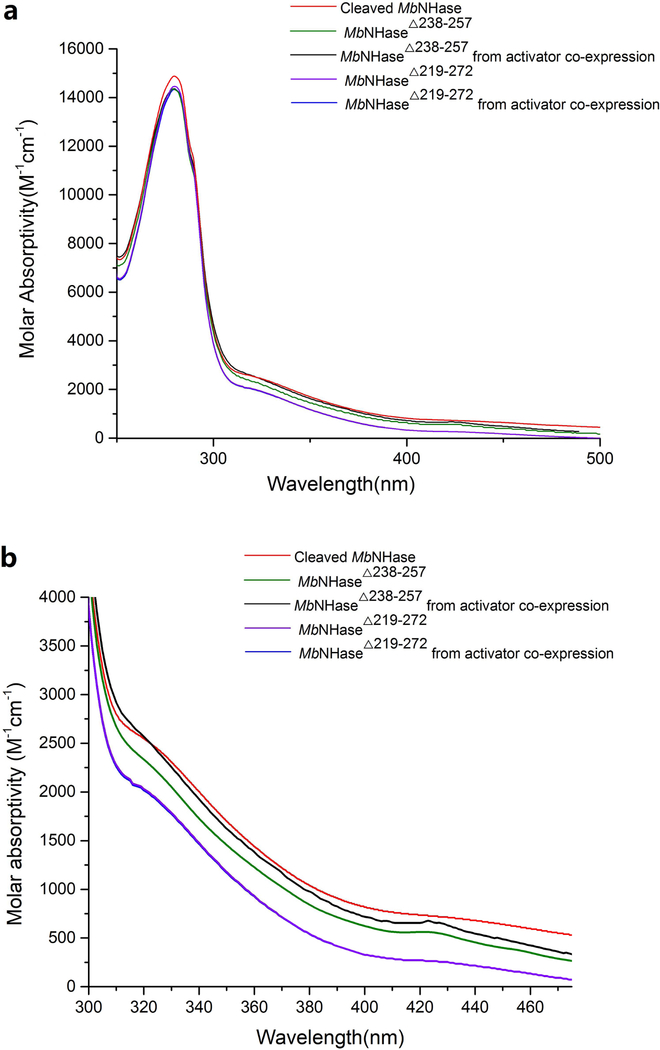
A combination of UV-Vis and EPR spectroscopy coupled with metal analyses was used to determine that the proteolytically cleaved MbNHase enzyme contained its full complement of Co(III). ICP-MS data indicated that the proteolytically cleaved MbNHase enzyme contained 1.9 ± 0.1 equivalents of cobalt per α2β2 heterotetramer, indistinguishable from WT MbNHase (Table 1) with no other metal ions detected above the background level of <10 ppb [11]. Proteolytically cleaved MbNHase exhibited the typical amber color of Co-type NHase enzymes in 50 mM HEPES buffer containing 300 mM NaCl, pH 8.0 (Figure 5) [1], and the UV-Vis spectrum reveals the characteristic S → Co(III) ligand-to-metal-charge-transfer (LMCT) band at 315 nm (ε = 2,500 M−1 cm−1) (Figure 5), which is blue-shifted by ~10 nm compared to WT MbNHase [11]. EPR spectra show no detectable Co(II) signals consistent with the presence of low-spin Co(III), which is diamagnetic (Figure SI-2). These data indicate that proteolytically cleaved MbNHase, retains its full complement of Co(III) and that neither the insert region nor the (His)17 motif is required for catalysis in the proteolytically cleaved MbNHase enzyme.
Figure 5.
UV-Vis spectra of various MbNHase constructs between (a)280 and 500 nm; (b) 300 and 480 nm in 50 mM HEPES buffer containing 300 mM NaCl at pH 8.0. (b)
Examination of the functional role of the (His)17 insert.
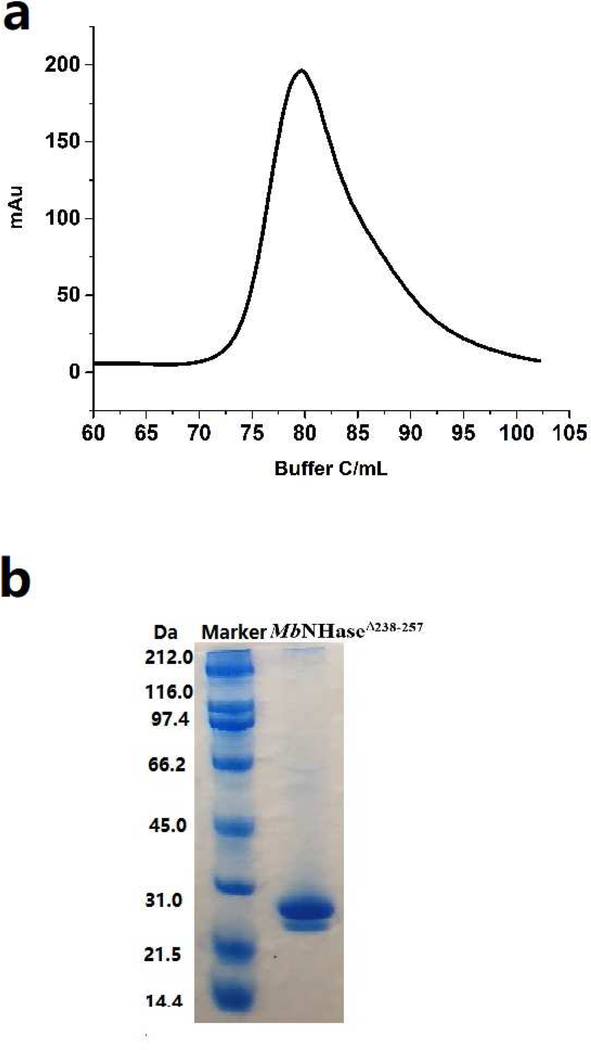
To investigate whether the (His)17 region plays a role in metal ion insertion and/or the posttranslational modification of the active site, the MbNHaseΔ238−257 (His)17 deletion mutant, was prepared. SDS-PAGE analysis revealed two bands at ~25.5 and ~28.5 KDa for MbNHaseΔ238−257 while size-exclusion chromatography indicated that MbNHaseΔ238−257 exists primarily as an α2β2 heterotetramer with a molecular weight of ~108 kDa, much like prokaryotic Co-type NHases (Figure 6). Kinetic analyses of the MbNHaseΔ238−257 (His)17 deletion mutant, expressed in the absence or presence of the prototypical Co-type activator (ε) protein, PtNHaseact, using acrylonitrile as the substrate, were performed in triplicate for multiple purifications in 50 mM Tris-HCl buffer, pH 7.0 at 25 °C providing k values of 71 ± 4 s−1 and 166 ± 5 s−1cat respectively. Km values for the MbNHaseΔ238−257 (His)17 deletion mutant expressed in the absence or presence of the PtNHaseact were 104 ± 17 mM and 125 ± 17 mM, respectively (Table 1). The MbNHaseΔ238−257 (His)17 deletion mutant expressed in the absence or presence of PtNHaseact contained 1.8 ± 0.1 and 2.3 ± 0.2 equivalents of cobalt per α2β2 heterotetramer, respectively, with no other metal ions detected above the background level of <10 ppb. The MbNHaseΔ238−257 (His)17 deletion mutant expressed in either the absence or presence of PtNHaseact in 50 mM HEPES buffer containing 300 mM NaCl, pH 8.0 exhibited the characteristic S → Co(III) LMCT band at ~320 nm (ε =2,200 M−1 cm−1) (Figure 5), nearly identical to WT MbNHase [11]. While the (His)17 region is not required for metal uptake or active site maturation, expression in the presence of the prototypical prokaryotic activator protein, PtNHaseact, does enhance the observed kcat value by ~60%, which is greater than would be expected from the slight increase (~20%) in metal content. Therefore, PtNHaseact does assist in the activation of the MbNHaseΔ238−257 (His)17 deletion mutant but is not required for post-translational modification of the active enzyme.
Figure 6.
Purification of the MbNHaseΔ238−268 mutant. a) size-exclusion column indicating a single peak corresponding to an α2β2 heterotetramer; b) SDS-PAGE gel page showing that the purified α and β-subunits are two independent proteins.
Investigation of the Insert region of MbNHase.
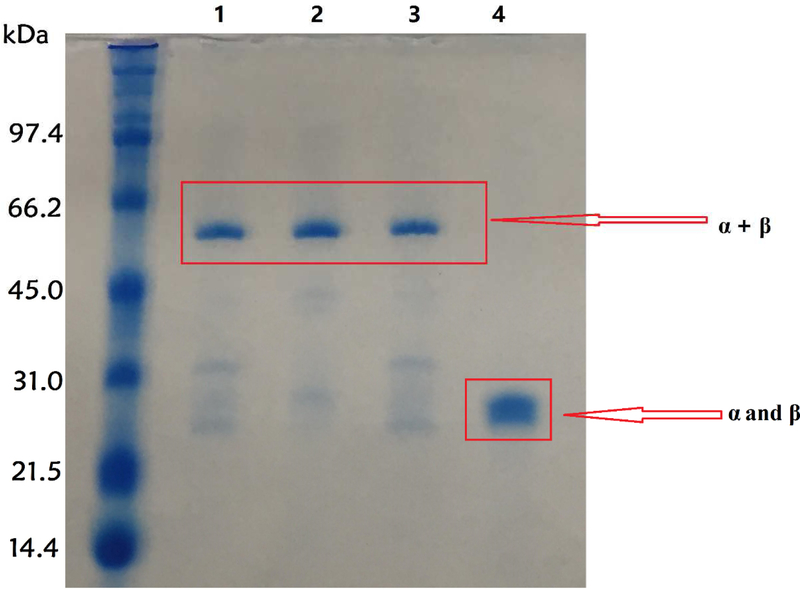
The MbNHaseΔ219−272 insert region deletion mutation provides an enzyme with the entire insert region between the α and β regions of WT MbNHase removed (Figure 1). Removal of this insert region essentially converts the eukaryotic MbNHase into a prokaryotic NHase analog, allowing the direct comparison of MbNHaseΔ219−272 and PtNHase. The MbNHaseΔ219−272 insert region deletion mutant expressed in the absence of PtNHaseact exhibited two bands on SDS-PAGE at ~25.5 and ~26.5 kDa. Size-exclusion chromatography indicated that MbNHaseΔ219−272 exists primarily as an α2β2 heterotetramer with a molecular weight of ~104 kDa, indistinguishable from proteolytically cleaved WT MbNHase and prokaryotic Co-type NHases such as PtNHase. Surprisingly, co-expression of MbNHaseΔ219−272 in the presence of PtNHaseact yielded a single band at ~52 kDa on SDS-PAGE which is consistent with an αβ complex. However, it could also be due to the formation of an α(ε)2 complex as PtNHaseact is ~14 kDa so an α(ε)2 complex would have a mass of ~53.5 kDa. Interestingly, the Co-type (ε) protein from Rhodococcus rhodochrous J1 was shown to form an α(ε)2 complex, which was proposed to bind Co(II) and insert it into apo-α2β2 NHase via a “self-subunit swapping” mechanism [21]. The Co-type (ε) protein was also proposed to facilitate oxidation of two active site Cys-residues. Attempts to separate the ~52 kDa proteins into their individual components were unsuccessful even in the presence of 8 M urea, 1 M dichlorodiphenyltrichloroethane (DDT) or SDS at 95 °C for 10 min (Figure 7).
Figure 7.
SDS-PAGE gel of MbNHaseΔ219–272. Column 1: MbNHaseΔ219−272 from activator co-expression; Column 2: MbNHaseΔ219−272 from PtNHase activator co-expression treated by 8mM Urea; Column 3: MbNHaseΔ219−272 from activator co-expression treated by 8M urea and 1M DDT; Column 4: MbNHaseD219−272 without activator.
Kinetic analysis of the MbNHaseΔ219−272 insert region deletion mutant expressed in the absence or presence of PtNHaseact in 50 mM Tris-HCl buffer, pH 7.0 at 25 °C provided kcat values of 75 ± 8 s−1 and 117 ± 10 s−1, respectively, using acrylonitrile as the substrate (Table 1). These kinetic data suggest that the MbNHaseΔ219−272 insert region deletion mutant expressedin the presence of PtNHaseact, is likely an αβ complex, even so, the Co-type NHase (ε) proteins are known to have significant sequence identity with the NHase β-subunit [22, 23], so α(ε)2 complex might be expected to be catalytically competent, but not likely more active than an α2β2 heterotetramer such as was observed for the MbNHaseD insert region deletion mutant expressed in the absence of PtNHaseact. The MbNHaseΔ219−272 insert region deletion mutant expressed in the absence or presence of PtNHaseact is not particularly stable in 50 mM Tris-HCl buffer, pH 7.0 at 25 °C and loses > 95% of its activity over the course of a few hours, indicating that the insert region plays a role in stabilizing the WT MbNHase enzyme. The MbNHaseΔ219−272 insert region deletion mutant contained 1.7 ± 0.1 equivalents of cobalt per α2β2 heterotetramer, irrespective of the co-expression of PtNHaseact, a value nearly identical to that observed for WT MbNHase (Table 1). UV-Vis spectra of the MbNHaseΔ219−272 insert region deletion mutant expressed in the absence or presence of PtNHaseact in 50 mM HEPES buffer containing 300 mM NaCl, pH 8.0 were indistinguishable from each other and WT MbNHase (Figure 5). Therefore, MbNHaseΔ238−257 binds its full complement of Co(III) ions without the assistance of an (ε) protein, indicating that the insert region is also not required for metal ion binding or active site maturation.
Conclusion
Characterization of the eukaryotic NHase from Monosiga brevicollis constructs described herein, confirm that MbNHase does not require an NHase activator protein or the E. coli chaperone proteins GroEL/ES for metallocentre assembly, including metal ion insertion and active site post-translational maturation. In addition, these data indicate that the (His)17 region found within the insert that links the α- and β-subunits is not required for metal ion incorporation or active site maturation. The fact that the proteolytically cleaved WT MbNHase enzyme and the MbNHaseΔ238−257 (His)17 deletion mutant each exhibit a modest increase in activity compared to WT MbNHase, suggests that the insert region likely induces a structural strain on the active site that has a limiting effect on the catalytic rate of hydration. The lack of the need for either an intrinsic or an extrinsic activator polypeptide for MbNHase is in stark contrast to the absolute requirement for an activator for assembly and activation of the otherwise similar prokaryotic NHases. The pertinent and related outstanding questions are, “How can the MbNHase metallocentre self-assemble and self-activate?” and, “Why do the prokaryotic NHase metallocenters require activators for assembly and activation?” The genetically engineered functional constructs of MbNHase described herein represent an important new tool with which to further address these important questions using structural and spectroscopic methods.
Supplementary Material
Highlights.
Metallocentre assembly in the eukaryotic nitrile hydratase from Monosiga brevicollis (MbNHase).
WT MbNHase expresses as a single polypeptide with fused α- and β-subunits linked by a (His)17 and an insert region.
Two insert region mutant MbNHase enzymes were examined, MbNHaseΔ238−257 and MbNHaseΔ219–272.
Neither the (His)17 nor the entire insert region are required for metallocentre assembly and maturation.
Acknowledgments
This work was supported by National Science Foundation (CHE-1808711, RCH and BB; CHE-1532168 BB & RCH), the Todd Wehr Foundation, Bruker Biospin, and the National Institutes of Health/NIBIB National Biomedical EPR Center (P41-EB001980).
Abbreviations.
- NHase
nitrile hydratase
- ORF
open reading frame
- ICP-MS
inductively-coupled plasma mass spectrometry
- IMAC
immobilized metal affinity chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information. Supplementary information includes the gene design for the MbNHaseΔ238−257 mutant (Figure SI-1) and an EPR spectrum of WT MbNHase (Figure SI-2).
References
- 1.Kovacs JA (2004) Chem Rev 104:825–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrop TC and Mascharak PK (2004) Acc Chem Res 37:253–260 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi M, Nagasawa T and Yamada H (1992) Trends Biotechnol 10:402–408 [DOI] [PubMed] [Google Scholar]
- 4.Nagasawa T, Shimizu H and Yamada H (1993) Appl Microbiol Biotechnol 40 [DOI] [PubMed] [Google Scholar]
- 5.Nagasawa T and Yamada H (1995) Pure Appl Chem 67:1241–1256 [Google Scholar]
- 6.Yamada H and Kobayashi M (1996) Biosci Biotech Biochem 60:1391–1400 [DOI] [PubMed] [Google Scholar]
- 7.Prasad S and Bhalla TC (2010) Biotechnology Advances 28:725–741 [DOI] [PubMed] [Google Scholar]
- 8.Nagasawa T, Mathew CD, Mauger J and Yamada H (1988) Appl Environ Microbiol 54:1766–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marron AO, Akam M and Walker G (2012) PLoS ONE 7:e32867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foerstner KU, Doerks T, Muller J, Raes J and Bork P (2008) PLoS ONE 3:e3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez S, Yang X, Bennett B and Holz RC (2017) Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1865:107–112 [DOI] [PubMed] [Google Scholar]
- 12.Tsujimura M, Odaka M, Nakayama H, Dohmae N, Koshino H, Asami T, Hoshino M, Takio K, Yoshida S, Maeda M and Endo I (2003) J Am Chem Soc 125:11532–11538 [DOI] [PubMed] [Google Scholar]
- 13.Dey A, Chow M, Taniguchi K, Lugo-Mas P, Davin S, Maeda M, Kovacs JA, Odaka M, Hodgson KO, Hedman B and Solomon EI (2006) J Am Chem Soc 128:533 – 541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiyama M, Horinouchi S, Kobayashi M, Nagasawa T, Yamada H and Beppu T (1991) J Bacteriol 173:2465–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N. Hashimoto Y, M., Horinouchi S, Beppu T (1994) Bioscience, biotechnology, and biochemistry 58:1859–1869 [DOI] [PubMed] [Google Scholar]
- 16.Nojiri M, Yohda M, Odaka M, Matsushita Y, Tsujimura M, Yoshida T, Dohmae N, Takio K and Endo I (1999) Journal of Biochemistry 125:696–704 [DOI] [PubMed] [Google Scholar]
- 17.Mendel RR, Smith AG, Marquet A and Warren MJ (2007) Natural Product Reports 24:963–971 [DOI] [PubMed] [Google Scholar]
- 18.Higgins KA, Carr CE and Maroney MJ (2012) Biochemistry 51:7816–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens JM, Rao Saroja N, Jaouen M, Belghazi M, Schmitter J-M, Mansuy D, Artaud I and Sari M-A (2003) Protein Expression and Purification 29:70–76 [DOI] [PubMed] [Google Scholar]
- 20.Martinez S, Wu R, Sanishvili R, Liu D and Holz R (2014) Journal of the American Chemical Society 136:1186–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Hashimoto Y, Cui T, Washizawa Y, Mino H and Kobayashi M (2010) Biochemistry 49:9638–9648 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Hashimoto Y and Kobayashi M (2009) The Journal of Biological Chemistry 284:14930–14938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z, Hashimoto Y, Shiraki K and Kobayashi M (2008) Proceedings of the National Academy of Sciences 105:14849–14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.