ABSTRACT
The discovery that extracellular vesicles (EVs) can transfer functional extracellular RNAs (exRNAs) between cells opened new avenues into the study of EVs in health and disease. Growing interest in EV RNAs and other forms of exRNA has given rise to research programmes including but not limited to the Extracellular RNA Communication Consortium (ERCC) of the US National Institutes of Health. In 2017, the International Society for Extracellular Vesicles (ISEV) administered a survey focusing on EVs and exRNA to canvass-related views and perceived needs of the EV research community. Here, we report the results of this survey. Overall, respondents emphasized opportunities for technical developments, unraveling of molecular mechanisms and standardization of methodologies to increase understanding of the important roles of exRNAs in the broader context of EV science. In conclusion, although exRNA biology is a relatively recent emphasis in the EV field, it has driven considerable interest and resource commitment. The ISEV community looks forward to continuing developments in the science of exRNA and EVs, but without excluding other important molecular constituents of EVs.
KEYWORDS: Extracellular Vesicles, extracellular RNA, Exosomes, microvesicles, ectosomes, oncosomes, extracellular vesicle function
Introduction
Studies of extracellular vesicles (EVs) and RNA were united in the mid- to late-2000s by findings that RNA molecules were incorporated into EVs and thereby transferred from cell to cell [1-5]. Communication via extracellular RNA (exRNA) was soon accepted as a novel set of mechanisms to explain the increasingly appreciated paracrine effects of EVs. Meanwhile, RNA profiling technologies advanced quickly, allowing comprehensive characterization from the small amounts of material often obtained from extracellular samples [6,7]. Coincidence or not, EV research outputs including patents and publications grew rapidly following the initial reports of EVs and exRNA [8]. Indeed, the first scientific workshop of the International Society for Extracellular Vesicles (ISEV) was dedicated to the topic of RNA and EVs [9-11]. Since then, ISEV has maintained attention to this topic [12]. In light of the importance of RNA in EV research—but also considering numerous unanswered questions about RNA EV loading, topology, transfer mechanisms, functionality and more—ISEV administered a worldwide survey on EVs and exRNA in 2017. The goal of this survey was to measure the pulse of the community on the state of EV and exRNA science.
Methods
An online survey was prepared with input from the ISEV Board of Officers and other community members, including program officers of the US National Institutes of Health (see Appendix 1 for the full list of questions). In addition to contact information (optional), 10 scientific questions were posed. Responses were mandatory for seven questions and optional for three. Five questions were multiple-choice or ranking, while the others were free-response text. Several questions also had “other” text box options. The survey was announced to the ISEV email list and through social media outlets (Facebook and Twitter) and remained open for 15 days. Two email reminders were sent to the ISEV email list prior to the deadline. At the close of the survey, data were collected in a spreadsheet and analyzed.
Results and Discussion
Numbers and geographical distribution
192 responses were received, including 188 unique responses based on IP addresses. Approximately 14% of respondents worked in the Asia Pacific region, 51% were in Europe or Africa and 35% were located in the Americas (Figure 1).
Figure 1.
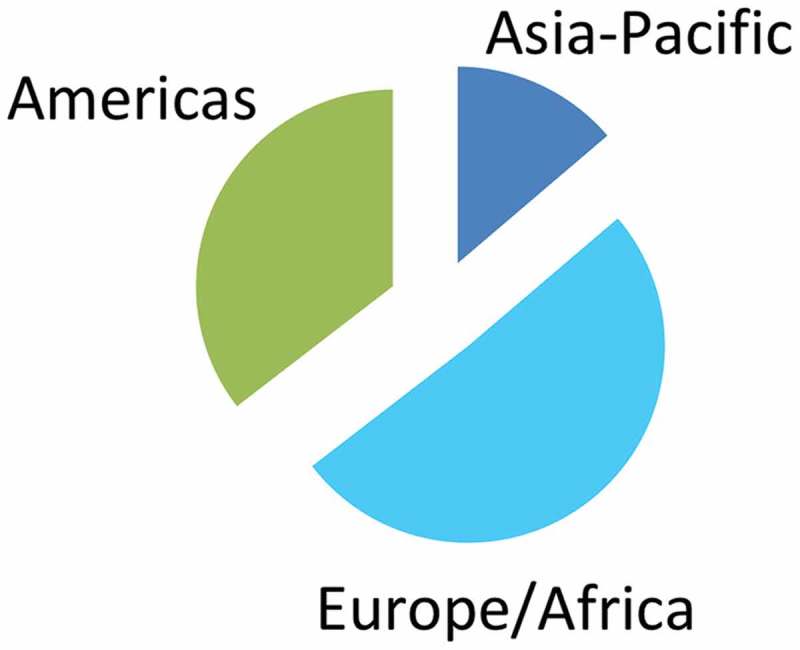
Geographical distribution of respondents by ISEV geographical chapter. The pie chart represents percentage from each chapter: Asia-Pacific, Europe/Africa and the Americas.
Maturity of current understanding
Respondents were asked to rank the maturity of current understanding of five aspects of EV biology that could be seen roughly as sequential parts of an “EV life cycle”: biogenesis, cargo loading, transport/stability, uptake, functional cargo transfer. Ranking was from 1 (most knowledge) to 5 (least). The rank from most to least knowledge was biogenesis, transport/stability, uptake, cargo loading and cargo transfer. The distribution of votes (Figure 2) indicates diversity of opinion, but several results stand out. Biogenesis received by far the most “1” votes (high level of knowledge), nearly as many as all other categories combined; results also otherwise skewed towards the high end. Cargo transfer and cargo loading received the most and second-most “5” (least knowledge) votes, and these results were skewed towards low-knowledge scores. Ambivalence was apparent for the transport and uptake categories, with most votes in the middle of the range. These results suggest perceived unmet needs across all topics, but especially in cargo loading and cargo transfer.
Figure 2.
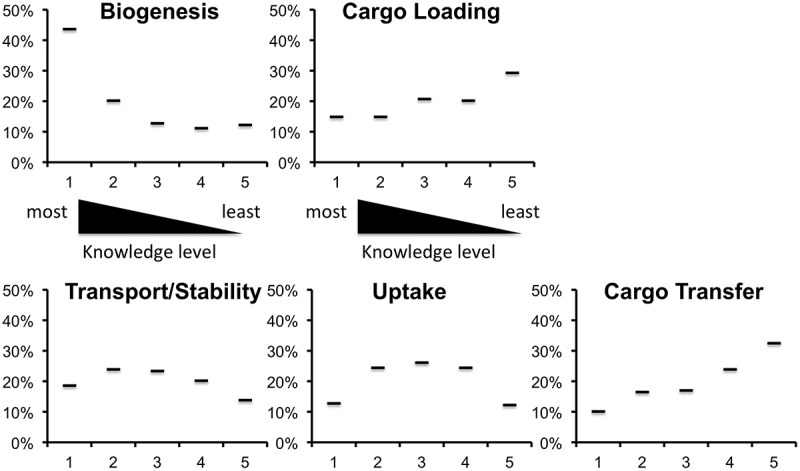
EV topics about which we know the “most” [1] to the “least” [5]. Respondents were required to rank the five topics from most [1] to least knowledge [2]. No rank could be used twice. Shown is the percentage of votes obtained by each. As depicted, biogenesis received the most votes for the best understood topic, while cargo loading and cargo transfer were thought to be least understood.
What is responsible for EV function?
Respondents next identified factors they felt were responsible for the reported effects of EVs: lipids, nucleic acids, proteins, metabolites, EVs as a whole (not any one constituent molecular category), or particles/molecules associated with the outside of EVs (but not part of the EVs themselves). Multiple selections were permitted, and an “other” box was included for comments. Only proteins were picked as the responsible factor by a majority of respondents (53%, Figure 3). However, not far behind were “EV as a whole” (47%) and nucleic acids (46%). Lipids, extra-EV particles and metabolites received fewer votes (<20% each). Several respondents suggested in “other” comments that the answer would depend on the experimental system, or that current knowledge was too limited to support a conclusion.
Figure 3.
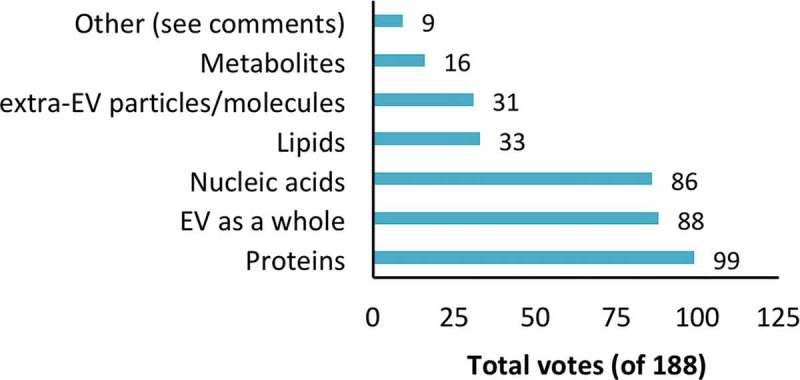
Factor(s) most responsible for observed effects of EVs. Respondents could select more than one answer. See supplementary material for selected comments.
Biomarker potential
As categories with biomarker potential, respondents again selected from the factors in the previous question. Nucleic acids and proteins were chosen by a majority of respondents (65% and 54%, respectively), while other factors were not highly rated (Figure 4). There were several “other” comments, one raising the point that the study of nucleic acids and protein is simply more advanced than that of other potential biomarkers.
Figure 4.
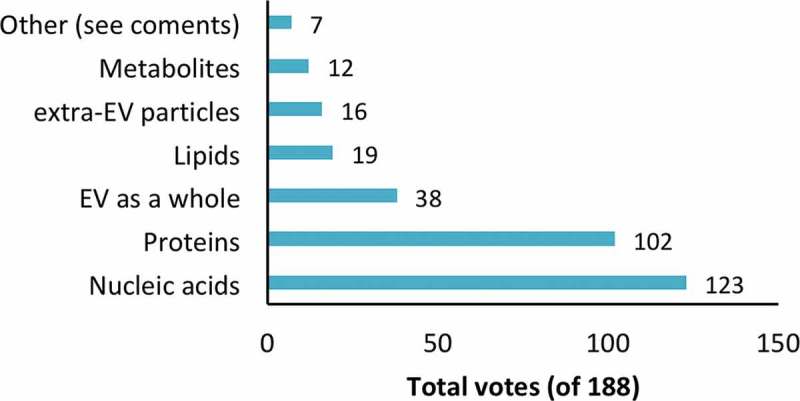
Factor(s) with the greatest biomarker potential. Respondents could select more than one answer. See supplementary material for selected comments.
Has there been a focus on RNA? If so, is it justified?
Most respondents agreed that an emphasis on RNA is currently evident in EV research. A majority (almost 62%) suggested that this perceived emphasis was justified, while 21% did not (Figure 5). Around 18% of respondents felt that there is not a specific emphasis on RNA in EV research. In numerous additional comments (Appendix 2), respondents observed that the emphasis on RNA was a recent development relative to the longer history of the EV field. Others stressed that proteins and lipids are also likely to contribute to EV functions, perhaps as much as or more than RNA. Respondents commonly favored a more holistic view of EV function, in which RNA, but also proteins, lipids and the EV package as a whole play functional roles. Some stated that certain forms of RNA, especially miRNA, had likely been emphasized due to stability, ease of detection or advances in detection methodologies, likening a focus on specific classes of molecules or types of RNA to “tunnel vision”. However, others remained enthused by the potential of RNA in EVs.
Figure 5.
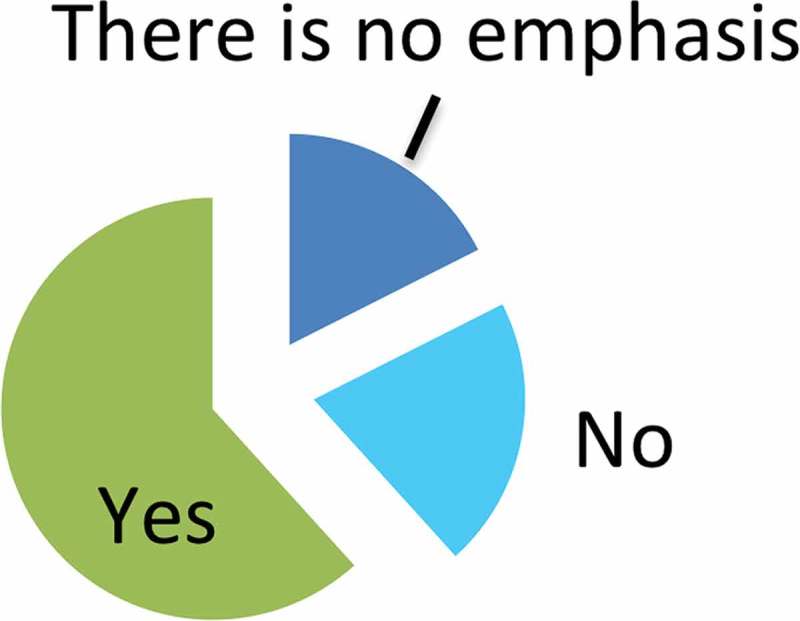
Is the recent emphasis on RNA justified? Only one answer was permitted. The pie chart represents percentage. Total numbers for each answer are given.
What are the current needs in the study of EVs and exRNA?
Four open response questions completed the survey. Respondents were asked about the major remaining challenges in EV/exRNA research and what is needed to overcome them; fundamental mechanisms of exRNA and EV biology that must be better understood for the field to flourish; and what standardized protocols or resources are needed. An open comment box was also available for thoughts not captured in the earlier parts of the survey. Since the responses to these questions overlapped partially, we have attempted to extract broad themes below, but most substantive answers are included in the supplementary materials.
A broader view of EVs and exRNA
Responses cautioned against placing exclusive emphasis on specific types of EVs or exRNA. The existence of multiple subtypes of EVs has become apparent, and RNA is incorporated not only into exosomes. Similarly, many exRNA molecules are not found in EVs; they are abundant in ribonucleoproteins and other non-EV particles. Like DNA, RNA may also associate with the outside of EVs, and not be exclusively luminal. As in the answers to previous questions, several respondents emphasized the diversity of small exRNA beyond miRNAs, to include tRNA fragments, Y RNAs, mRNAs and beyond small RNA to include longer RNAs. Molecular context was also underlined. For example, if RNAs are important, what about RNA binding proteins?
Function and stoichiometry: in vitro and in vivo
Numerous respondents contrasted in vitro and in vivo studies. In vitro findings are informative and suggestive but must ultimately be placed into the in vivo context. That is, to what extent are the numbers of EVs or exRNA molecules used to establish principles in vitro also relevant to whole organisms? Diverse in vivo models are of course needed to establish disease relevance and study mechanism. Respondents encouraged more development of models conducive to EV and exRNA studies.
Techniques to deal with small amounts, plus single EV and single molecule studies
The question of sensitivity is inherent in studies of particles such as EVs, most of which are a small fraction of the size of a single cell. Many survey respondents highlighted aspects of sensitivity, asking for better methods to increase EV or exRNA yield (without compromising purity) and improved techniques to obtain and analyze EV subsets. Several answers tied a lack of concordance of some profiling findings to challenges of low-abundance studies: throughout, researchers must establish the limits of detection of their techniques, then be mindful of these limits to avoid overinterpretation of below-the-threshold data. A repeated emphasis was on the need for single-EV capabilities, which respondents found to be vital for understanding EV subtypes and their functions. These studies will require ever-increased sensitivity and optimized techniques for reliable detection of single molecules or at least low numbers of surface antigens (flow-based technologies) and RNAs (e.g., digital PCR).
Rigor and reproducibility
One of the most pronounced themes of the survey responses was scientific rigor and reproducibility, including standardization. Respondents urged the application of rigorous and reproducible approaches to each stage of the EV/exRNA workflow, from pre-analytical factors/sample collection through EV and RNA isolation and bioinformatics analysis. The ISEV minimal information initiative (MISEV) was praised, but updates and additional efforts were recommended. These might include biofluids-specific SOPs; a range of standard reference materials that can be used to assess isolation efficiency or spike-ins that are appropriate for specific classes of EVs or RNAs; and normalization procedures specific to EV and exRNA studies (see also Table 1, selected suggested protocols, resources and approaches). Answers suggested that some published “EV” studies are not really about EVs at all, relying on commercial kits or substandard techniques that are not fit for purpose. At the end of the pipeline, development of new bioinformatics tools and curated databases was encouraged, with a goal of more facile and effective integrated data analysis.
Table 1.
Selected suggested protocols, resources and approaches.
| Biofluids-specific EV and exRNA isolation protocols, including those for plasma, serum, CSF, milk. |
| Methods for isolation and quantification of cell- or tissue-specific EVs from biofluids. |
| Methods for isolation and quantification of EV subtypes. |
| SOPs to help increase purity of EVs. |
| Guidelines for handling and studying small-volume samples. |
| Evidence-based EV storage methods. |
| Housekeeping genes for EV RNA normalization, perhaps different for different classes of RNA. |
| More efficient protocols for removal of extra-EV RNA: it is difficult to remove all “outer” RNA even with combined protease/RNase treatments. |
| Methods to load EVs with specific RNAs. |
| Better or more direct sensor systems to measure functions of EV/exRNA uptake. |
| EV flow (nano-flow) guidelines, including for single-EV assays. |
| A catalog of EV-relevant antibodies that have been used successfully. |
| Better molecular labelling methods. |
| Regular updates to existing protocols and guidelines, including MISEV. |
| Updated nomenclature. |
| Simple bioinformatics analysis pipelines. |
| Updates of existing EV databases to exclude studies that are likely contaminated or rank quality of evidence. |
| Open minds—avoid standardizing too early or too dogmatically. |
Synergy of better mechanistic understanding and better tools
Many responses referred to stages of the EV “life cycle” also covered in “Maturity of current understanding, above, suggesting that improved technologies are needed to develop better mechanistic understanding and vice versa. RNA loading into EVs was seen as an area with particular needs, since there is currently a good deal of uncertainty about specific versus random incorporation of cargo molecules into EVs. Greater understanding could lead to new technologies for incorporation of specific RNA molecules into EVs generally or into subtypes of EVs. Questions about EV transfer to recipient cells brought up the need for better ways to track individual EVs and their clearance; assess random versus non-random “trafficking” of EVs, perhaps based on surface markers; and means to track uptake or fusion. For RNA function, after an EV reaches the recipient cell, tools are required to visualize or otherwise track RNAs as they incorporate into functional machinery of the recipient cell (or are degraded). Overall, how does one prove that specific RNAs are responsible for a given effect? In addition to genome editing, the needed technologies include improved tools to load specific molecules into EVs (again), better label RNA inside cells and vesicles and delete or functionally disable RNAs after incorporation into EVs.
Let’s get together: from meetings to training and collaboration
Many respondents mentioned a need for more collaboration, funding, meetings and/or training. Large annual meetings were found to be important, but some felt that more regular local meetings are also needed. Funded consortia like the ERCC of the US National Institutes of Health were said to be useful but needed in other regions. Several respondents felt that newcomers to the field needed more assistance and encouragement and should not be excluded from networks. Some newcomers said that they had struggled to find collaborators or slots at training workshops, indicating a need for more training capacity. Indeed, in answer to a previous question (for which more than one answer was allowed), 66% of respondents suggested that more international collaboration was needed, while 62% asked for more national or regional collaboration. 38% indicated a need for both. Only 10% felt that the current levels of collaboration were appropriate (Figure 6).
Figure 6.
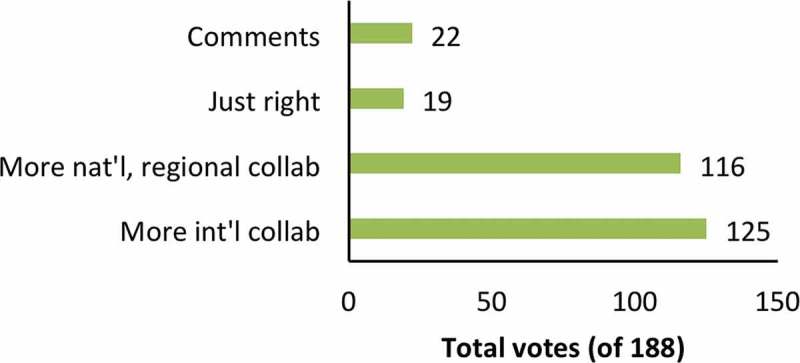
Collaboration needs. Respondents could select more than one answer. Abbreviations: national (nat’l), international (int’l). See supplementary material for selected comments.
Summary
Overall, the respondents to this ISEV survey on EVs and exRNA echoed the general enthusiasm about the role of RNAs in EV function. However, they also stated that much remains to be learned about the mechanisms of RNA loading, transfer and function in recipient cells. Furthermore, while the contribution of RNA to function was disputed by few, there was an evident desire to place RNA in the total EV context alongside other bioactive molecules. On the wish-list of the respondents were improved techniques, standardized protocols, enhanced exchange of knowledge and more resources to advance this exciting area of science.
Supplementary Material
Funding Statement
This work was supported in part by funding to the drafting authors from the US Department of Defense under W81XWH-16-1-0736 (to CS) and the US National Institutes of Health, National Institute on Drug Abuse, under DA040385 (to KWW).
Acknowledgements
The authors are grateful to all respondents to this ISEV EV and exRNA survey. The authors also thank the US National Institutes of Health Extracellular RNA Communication Consortium for soliciting related feedback on the state of the exRNA and EV fields, which contributed to the decision to administer this survey.
Disclosure statement
No potential conflict of interest was reported by the authors.
Authors’ contributions
KWW, AFH, SS, DDV, MW, EIB, CG and JL designed the survey. KWW prepared the online survey. CS and KWW analysed the results and wrote the manuscript. All authors reviewed and approved the final draft of the manuscript.
References
- 1.Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006May;20(5):847–6. [DOI] [PubMed] [Google Scholar]
- 2.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007June9;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 3.Aliotta JM, Sanchez-Guijo FM, Dooner GJ, et al. Alteration of marrow cell gene expression, protein production, and engraftment into lung by lung-derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells [Internet]. 2007September;25(9):2245–2256. DOI: 10.1634/stemcells.2007-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008December ;10(12):1470–1476. Internet Cited 2008 November18 . Internet: http://www.ncbi.nlm.nih.gov/pubmed/19011622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010. ;107(14):6328–6333. Internet Cited 2010 March23 Internet: http://www.ncbi.nlm.nih.gov/pubmed/20304794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolte-’T Hoen EN, Buermans HP, Waasdorp M, et al. 2012. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. Internet Internet Internet2012July24 Internet http://www.ncbi.nlm.nih.gov/pubmed/22821563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng L, Quek CYJ, Sun X, et al. The detection of microRNA associated with Alzheimer’s disease in biological fluids using next-generation sequencing technologies. Front Genet. Internet. 2013;4 Internet 2018 March18]: http://journal.frontiersin.org/article/10.3389/fgene.2013.00150/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S, Hochberg FH, Jones PS.. Extracellular vesicles: the growth as diagnostics and therapeutics; a survey. J Extracell Vesicles. Internet; 2018January26 ;7(1):1438720 cited 2018 July10 Internet: http://www.ncbi.nlm.nih.gov/pubmed/29511461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Théry C. ISEV RNA Workshop-New York City, October 1- 2,2012. J Extracell vesicles. Internet. 2012;1:19857 cited 2018 July10 Internet: http://www.ncbi.nlm.nih.gov/pubmed/26082070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell vesicles. 2013;2:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill AF, Pegtel DM, Lambertz U, et al. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell vesicles [Internet]. 2013;2 [cited 2016 September22] Available from: http://www.ncbi.nlm.nih.gov/pubmed/2437690922859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateescu B, Kowal EJK, van Balkom BWM, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - An ISEV position paper. J Extracell Vesicles. 2017;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


