Abstract
An association study was conducted to investigate the relation between 14 variants of glucose transporter 1 gene (SLC2A1) and the risk of type 2 diabetes (T2DM) leading to nephropathy. We also performed a meta-analysis of 11 studies investigating association between diabetic nephropathy (DN) and SLC2A1 variants. The cohort included 197 cases (T2DM with nephropathy), 155 diseased controls (T2DM without nephropathy) and 246 healthy controls. The association of variants with disease progression was tested using generalized odds ratio (ORG). The risk of type 2 diabetes leading to nephropathy was estimated by the OR of additive and co-dominant models. The mode of inheritance was assessed using the degree of dominance index (h-index). We synthesized results of 11 studies examining association between 5 SLC2A1 variants and DN. ORG was used to assess the association between variants and DN using random effects models. Significant results were derived for co-dominant model of rs12407920 [OR = 2.01 (1.17–3.45)], rs841847 [OR = 1.73 (1.17–2.56)] and rs841853 [OR = 1.74 (1.18–2.55)] and for additive model of rs3729548 [OR = 0.52 (0.29–0.90)]. The mode of inheritance for rs12407920, rs841847 and rs841853 was ‘dominance of each minor allele’ and for rs3729548 ‘non-dominance’. Frequency of one haplotype (C-G-G-A-T-C-C-T-G-T-C-C-A-G) differed significantly between cases and healthy controls [p = .014]. Regarding meta-analysis, rs841853 contributed to an increased risk of DN [(ORG = 1.43 (1.09–1.88); ORG = 1.58 (1.01–2.48)] between diseased controls versus cases and healthy controls versus cases, respectively. Further studies confirm the association of rs12407920, rs841847, rs841853, as well as rs3729548 and the risk of T2DM leading to nephropathy.
Keywords: Diabetes mellitus, diabetic nephropathy, glucose transporter 1 (GLUT1), genetic variants of SLC2A1
Introduction
Diabetic nephropathy is a major microvascular complication of diabetes mellitus [1]. It is the most frequent primary cause of end-stage renal disease and is characterized by a progressive clinical course, ultimately leading to death [2]. The main risk factor for developing diabetic nephropathy or any microvascular complication in diabetes is the poor glycemic control; though patients with good glycemic control may develop nephropathy [1]. This fact and a proven significant familial clustering of diabetic nephropathy [3–5] clearly implicate that specific genetically defined predisposition is involved in the pathogenesis of nephropathy in diabetes. However, the genes conferring susceptibility have not been identified yet [6–11].
The glucose transporter 1 (GLUT1), also called SLC2A1, i.e., the member 1 of the solute carriers family 2, is a most important representative of the facilitative glucose transporters. Its expression in the glomerular mesangial cell membrane is rate-limiting for intracellular glucose flux and utilization [12,13]. In mesangial cells, elevated levels of intracellular glucose, e.g., resulting from diabetes mellitus, activates cellular pathways involved in cellular growth and in the accumulation of extracellular matrix [14–16]. Exactly, these alterations are central in the pathogenesis of diabetic nephropathy. Mesangial cells over-expressing GLUT1 after gene transduction in vitro acquire a diabetic phenotype with accumulation of extracellular matrix even in the absence of enhanced glucose levels in the medium [13,17]. Furthermore, transgenic mice over-expressing GLUT1 on kidney glomerular cells develop diabetic nephropathy, despite normoglycemia [18]. Thus, it appears that the availability of GLUT1 transporters, rather than extracellular glucose concentrations per se, regulates mesangial cell glucose metabolic flux. In support of this contention, reduction of GLUT1 expression in mesangial cells [19] and in transgenic diabetic (db) mice [12,20] protects against diabetic complications, despite high glucose concentration. From these data, it becomes clear that GLUT1 on the cell membrane of glomerular cells may possess a central regulatory role for the development of diabetic nephropathy.
In the present candidate-gene study, we tested the hypothesis of association between 14 variants of SLC2A1 (rs12407920 C/T, rs2297976 G/T, rs710221 G/A, rs2086856 A/G, rs12130264 C/T, rs841847 C/T, rs841853 C/A, rs3729548 C/T, rs841855 G/A, rs3768029 C/T, rs12071418 C/G, rs3820549 C/G, rs3820546 G/A, rs11537641 G/A) and the progression of type 2 diabetes (i.e., from healthy status to diabetes without nephropathy and then, to diabetes leading to nephropathy). Thereafter, we tested the association between the SLC2A1 variants and the risk of diabetes leading to nephropathy. The former hypothesis was tested by the generalized linear odds ratio (ORG) [21,22]. The latter hypothesis was tested using the ORG as a genetic model-free approach and also by means of the additive and co-dominant inheritance models [21–23]. In addition, the mode of inheritance was estimated based on the degree of dominance index (h-index) [24,25]. Finally, an analysis of haplotypes was conducted.
To further investigate the contribution of SLC2A1 polymorphisms in the development and progression of DN, we performed a meta-analysis of all variants across SLC2A1 that had been examined in genetic association studies up-to-date. The variants included in meta-analysis were rs841853, rs1385129, rs841847, rs841848 and rs710218, out of which variants rs841853 and rs841847 were also genotyped in present case-control study.
Methods
Association study
Subjects
Ethics Committee of the University Hospital of Larissa, University of Thessaly, School of Medicine approved the study protocol. The study was conducted in the University Hospital of Larissa and all participants signed an informed consent before enrolment. All participants were recruited from patients attending the outpatient wards of Nephrology, Internal Medicine and Ophthalmology at the University Hospital of Larissa between 2009 and 2011. They were all Caucasians of Greek origin and during the study, they resided in the same region in central Greece (Thessaly).
The study cohort consisted of cases with type 2 diabetes and nephropathy, diseased controls (type 2 diabetes without nephropathy) and healthy controls. Diseased controls were matched to cases by age. Diagnosis of type 2 diabetes was based on the American Diabetes Association criteria of 2003 [26]. Type 2 diabetes with nephropathy was diagnosed on the basis of an overt albuminuria, urinary albumin excretion >300 mg/24 h (>200 µg/min; representing persistent albuminuria) with or without elevated serum creatinine levels (serum creatinine >1.3 mg/dl), determined in at least two separate occasions three months apart from one another, and in the absence of clinical or radiological evidence of non-diabetic renal disease. Infection was excluded by previous urine dipstick tests. Moderately increased albuminuria, formerly called microalbuminuria, i.e., urinary albumin excretion 30–300 mg/24 h (20–200 μg/min), was not categorized as diabetic nephropathy. Existence of arterial hypertension or cardiovascular disease and the glycosylated hemoglobin (HbA1c %) were registered. Blood samples for biochemical measurements and DNA extraction was taken from each individual.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using a salting out method. Based on Hapmap data for CEU population (Release 27, Phase II + III, Feb09, on NCBI B36 assembly, dbSNP b126) tag single nucleotide polymorphisms (SNPs) across SLC2A1 (spanning from 42925375 to 42959176, overall 33.802Kbp, on chromosome 1p34.2) were selected using the tagger algorithm (http://www.broadinstitute.org/mpg/tagger/) with a pair-wise approach, an r2 cut-off of ≥0.8 and a minor allele frequency >0.05. A total of 14 tag SNPs in three distinct gene regions were retrieved (rs12407920 C/T, rs2297976 G/T, rs710221 G/A, rs2086856 A/G, rs12130264 C/T, rs841847 C/T, rs841853 C/A, rs3729548 C/T, rs841855 G/A, rs3768029 C/T, rs12071418 C/G, rs3820549 C/G, rs3820546 G/A, rs11537641 G/A). Concretely, the tag SNPs were located in the intron 1 between exons 1 and 2 (rs12407920, rs2297976, rs710221, rs2086856) and in the intron 2 between exons 2–3 (rs12130264, rs841847, rs841853, rs3729548, rs841855, rs3768029, rs12071418, rs3820549, rs3820546) as well as in the exon 7 (rs11537641).
Genotyping was performed with TaqMan allele specific discrimination assays on an ABI PRISM® 7900 Sequence Detection System (Foster City, CA, USA) and analyzed with SDS software (Applied Biosystems, Foster City, USA). At least 10% of the samples were selected randomly for repeated genotyping, as an internal control. Genotyping was performed by laboratory personnel blinded to clinical status.
Data analysis
The data for continuous variables were expressed as mean value and standard deviation [mean ± SD] and data for categorical variables as count (or ratio) and percentage [n (%)]. The normality of continuous variables was tested by the Kolmogorov–Smirnov test. Pair-wise comparisons of continuous variables were performed with the t-test or the Mann–Whitney U test for unpaired data, as appropriate. The frequencies of categorical variables were compared by means of the χ2 test or the Fisher’s exact test.
The association between genotype distribution and disease progression (i.e., disease progression to diabetic nephropathy) was examined using the generalized linear odds ratio (ORG) [21,22]. The ORG expresses the probability of a subject being more diseased relative to the probability of being less diseased, given that diseased subjects have higher mutational load. Explicitly, the ORG shows how many cases/healthy-controls pairs exist in the study for which the cases have larger mutational load relative to the number of pairs for which the healthy controls have the larger mutational load; alternatively, ORG indicates whether the mutational load of a variant is implicating in disease susceptibility [21,22]. The association between genotype distribution and the disease status (i.e., healthy controls, diseased controls and cases) was additionally tested using the χ2 test.
For the investigation of the susceptibility to type 2 diabetes leading to nephropathy the co-dominant and additive inheritance models of cases were compared to healthy controls using univariate logistic regression. The magnitude of associations was expressed in terms of odds ratios (ORs) unadjusted and adjusted for age and gender with the corresponding 95% confidence interval (CI). These two inheritance models were selected since they are orthogonal [23,24]. From the respective ORs, we calculated the degree of dominance index (h-index) as an estimate for the mode of inheritance [24,25].
In healthy controls, deviation of the genotype distribution from the Hardy–Weinberg equilibrium (HWE) and existence of linkage disequilibrium (LD) between polymorphisms were evaluated using exact tests according to Weir [27,28]. A result was considered to be statistically significant when p < .05.
Genotype distribution and the respective unadjusted and adjusted ORs were calculated using IBM® SPSS® Statistics Version 21 (IBM Corp.©, Release 21.0.0.1, 2012, NY, USA). HWE and LD were tested using the Genetic Data Analysis (GDA) software [27,29]. The haplotype frequencies were estimated and compared by SHEsis [30,31]. ORG was calculated using ORGGASMA (http://biomath.med.uth.gr) [22].
Meta-analysis
Identification and eligibility of relevant studies
In meta-analysis, the published genetic association studies (GAS) regarding variants of SLC2A1 gene were searched using HuGE Phenopedia (last update in January 2017), the NHGRI Catalog of Published Genome-Wide Association Studies (http://www.genome.gov/gwastudies/) regarding the disease term ‘diabetic nephropathies’ and PubMed with search terms such as ‘diabetic nephropathy’ AND ‘association’ AND (‘gene symbol’ OR ‘gene name’) (accessed on 18 January 2016). All included studies were published in English. We also persused articles from GWAS in HuGE Publit. Finally, relevant meta-analyses and references of the eligible articles were retrieved to identify articles not indexed in PubMed or HuGE Navigator.
The eligible studies should involve cases with persistent micro/macroalbuminuria with or without diabetic retinopathy, diseased controls with diabetes and normoalbuminuria or normal renal function and/or healthy controls. They should provide full genotypic data either genotype counts or allele frequencies and include human subjects. The diabetes could be either T1DM or T2DM.
We did not include in meta-analysis studies investigating disease progression, severity, phenotype modification, response to treatment or survival. Case reports, editorials, reviews and studies with other study designs, such as linkage studies, were also excluded. The eligibility of the articles was assessed independently by two investigators (MT, EZ), the results were compared and any disagreements were resolved by reaching a consensus.
Data extraction
From each article information regarding first author, year of publication, ethnicity, the PMID, type of diabetes, country and the phenotype was extracted. For cases and controls, we recorded their number, duration of diabetes, the selection criteria and the implementation of matching criteria. With regard to the genotypic data, we extracted, if available, the full genotype counts or allele frequencies.
Data synthesis and analysis
The association between genotype distribution and diabetic nephropathy was examined using the generalized linear odds ratio (ORG) [21,22]. The threshold for meta-analysis was the presence of 2 studies per variant. The associations are presented with generalized odds ratios with corresponding 95% confidence intervals using the random effect model. We tested for between-study heterogeneity with Cochran’s Q statistic (considered significant at p < .10) and assessed its extent with the I2 statistic, which is independent of the number of studies in the meta-analysis and takes values between 0 and 100%, with higher values denoting a greater degree of heterogeneity [32,33]. ORG was calculated using ORGGASMA (http://biomath.med.uth.gr) [22].
For each study, we examined if controls confronted with Hardy–Weinberg equilibrium (HWE) predicted genotypes using Fisher’s exact test. We also tested for ‘small-study effect’ with the Egger test [34].
We also performed subgroup analyses with regard to diabetes type (T1DM or T2DM) and ethnicity (Caucasians versus non-Caucasians), as well as a sensitivity analysis excluding the data of the present association study.
Results
Association analysis
Clinical profile of participants
The cohort consisted of 197 cases (patients with type 2 diabetes and nephropathy), 155 diseased controls (patients with type 2 diabetes without nephropathy) and 246 healthy controls. All participants were Caucasians of Greek origin. Table 1 shows the demographic and clinical characteristics. Among 197 cases with type 2 diabetes and nephropathy, 11 were under chronic renal replacement therapy. The distribution of age was as follows: above 60 years old were 165 cases (84%), 133 diseased controls (86%) and 223 healthy controls (91%). In 86% of the cases and 79% of diseased controls, the diabetes duration was more than 10 years.
Table 1.
Clinical profiles of the study-cohort.
| Parameters | Case-control study population groups (n = 498) |
||||||
|---|---|---|---|---|---|---|---|
| HC | DM | p Value* | DM-DN | DM + DN | p Value* | ||
| N | 246 | 352 | n.a. | 155 | 197 | n.a. | |
| Sex [m; n (%)] | 136 (55.3) | 181 (51.4) | .361 | 74 (47.7) | 107 (54.3) | .238 | |
| Age (years) | 71 ± 9.2 | 68 ± 8.9 | <.001 | 68 ± 9.1 | 69 ± 8.8 | .427 | |
| DM duration (years) | n.a. | 16.3 ± 8.0 | n.a. | 15.7 ± 8.3 | 16.8 ± 7.8 | .508 | |
| HbA1c (%) | n.d. | 7.35 ± 1.31 | n.a. | 7.20 ± 1.34 | 7.47 ± 1.29 | .019 | |
| Insulin treatment (%) | n.a. | 105 (29.8) | n.a. | 50 (32.3) | 55 (27.9) | .412 | |
| Hypertension (%) | 0 | 224 (63.6) | <.001 | 98 (63.2) | 126 (63.9) | .911 | |
| Cardiovascular disease (%) | 0 | 110 (31.3) | <.001 | 41 (26.5) | 69 (35.0) | .105 | |
| Creatinine (mg/dl) | 0.77 ± 0.15 | 1.46 ± 1.37 | <.001 | 0.90 ± 0.18 | 1.84 ± 1.67 | <.001 | |
| Urea (mg/dl) | 30 ± 7.9 | 59 ± 34 | <.001 | 42 ± 13.6 | 71 ± 38.3 | <.001 | |
| Albuminuria (mg/d) | n.d. | 470 ± 856 | n.a. | 43.9 ± 53.3 | 782 ± 1019 | <.001 | |
| Proteinuria (mg/d) | n.d. | 788 ± 1468 | n.a. | 105 ± 80.0 | 788 ± 1468 | <.001 | |
Clinical profiles of the study-cohort, consisting of 195 cases (i.e., T2DM-nephropathy; DM + DN), 157 diseased controls (i.e., T2DM without nephropathy; DM-DN) and 246 healthy controls (HC). Continuous data are given as mean and standard deviation [x ± SD] and categorical data as count and percentage [n (%)].
p Values were calculated by the Mann–Whitney U test for continuous variables or the χ2 test for categorical variables as appropriate.
Disease progression
The genotype distributions of the 14 variants in cases, diseased controls and healthy controls, and the respective ORG, are shown in Table 2. The healthy controls were conformed to HWE for all variants (p ≥ .05). There was a significant association between disease progression and genotype distribution of certain SLC2A1 variants (rs12407920 C/T, rs841847 C/T and rs841853 C/A). The model-free approach (ORG) produced significant results for these very variants, indicating that the risk of disease progression is related to the mutational load of these variants (i.e., diseased subjects have higher mutational load than healthy controls). In addition, the ORG for variant rs3729548 C/T was significant [ORG=0.78 (0.62–0.98)] indicating a protective role of allele T of rs3729548 for disease progression to nephropathy (Table 2).
Table 2.
Distribution of genotypes of SLC2A1 variants.
| Variant | Genotype | DM + DN N (%) | DM-DN N (%) | HC N (%) | *p Value | **ORG (95% CI) |
|---|---|---|---|---|---|---|
| rs12407920 | C C | 154 (79.4) | 124 (80.5) | 212 (88.0) | .044 | 1.55 (1.09–2.19) |
| C T | 38 (19.6) | 30 (19.5) | 26 (10.8) | |||
| T T | 2 (1.0) | 0 (0) | 3 (1.2) | |||
| rs2297976 | G G | 122 (63.9) | 96 (64.0) | 153 (63.2) | .973 | 0.99 (0.76–1.29) |
| G T | 62 (32.5) | 48 (32.0) | 82 (33.9) | |||
| T T | 7 (3.7) | 6 (4.0) | 7 (2.9) | |||
| rs710221 | G G | 66 (34.6) | 49 (33.3) | 77 (32.9) | .930 | 0.98 (0.77–1.24) |
| G A | 89 (46.6) | 74 (50.3) | 118 (50.4) | |||
| A A | 36 (18.8) | 24 (16.3) | 39 (16.7) | |||
| rs2086856 | A A | 86 (44.8) | 73 (48.7) | 111 (46.6) | .369 | 1.11 (0.87–1.41) |
| A G | 80 (41.7) | 64 (42.7) | 108 (45.4) | |||
| G G | 26 (13.5) | 13 (8.7) | 19 (8.0) | |||
| rs12130264 | C C | 176 (90.3) | 136 (89.5) | 209 (87.1) | .492 | 0.80 (0.52–1.23) |
| C T | 17 (8.7) | 16 (10.5) | 30 (12.5) | |||
| C C | 2 (1.0) | 0 (0.0) | 1 (0.4) | |||
| rs841847 | C C | 86 (44.8) | 71 (46.7) | 147 (60.2) | .015 | 1.49 (1.16–1.89) |
| C T | 87 (45.3) | 67 (44.1) | 79 (32.4) | |||
| T T | 19 (9.9) | 14 (9.2) | 18 (7.4) | |||
| rs841853 | C C | 70 (36.3) | 61 (41.2) | 126 (52.3) | .018 | 1.46 (1.15–1.86) |
| C A | 99 (51.3) | 69 (46.6) | 91 (37.8) | |||
| A A | 24 (12.4) | 18 (12.2) | 24 (10.0) | |||
| rs3729548 | C C | 71 (36.8) | 48 (32.9) | 72 (30.0) | .240 | 0.78 (0.62–0.98) |
| C T | 94 (48.7) | 69 (47.3) | 113 (47.1) | |||
| T T | 28 (14.5) | 29 (19.9) | 55 (22.9) | |||
| rs841855 | G G | 129 (68.3) | 113 (75.8) | 169 (70.4) | .509 | 1.05 (0.79–1.41) |
| G A | 55 (29.1) | 31 (20.8) | 65 (27.1) | |||
| A A | 5 (2.6) | 5 (3.4) | 6 (2.5) | |||
| rs3768029 | C C | 54 (28.6) | 32 (21.3) | 65 (26.6) | .213 | 0.86 (0.68–1.08) |
| C T | 100 (52.9) | 82 (54.7) | 115 (47.1) | |||
| T T | 35 (18.5) | 36 (24.0) | 64 (26.2) | |||
| rs12071418 | C C | 189 (97.9) | 147 (96.1) | 231 (96.7) | .583 | 0.76 (0.36–1.58) |
| C G | 4 (2.1) | 6 (3.9) | 8 (3.3) | |||
| G G | 0 (0.0) | 0 (0.0) | 0 (0.0) | |||
| rs3820549 | C C | 102 (53.1) | 86 (56.2) | 124 (50.8) | .842 | 0.94 (0.74–1.20) |
| C G | 72 (37.5) | 56 (36.6) | 98 (40.2) | |||
| G G | 18 (9.4) | 11 (7.2) | 22 (9.0) | |||
| rs3820546 | G G | 49 (25.8) | 42 (27.6) | 70 (29.0) | .362 | 0.96 (0.77–1.20) |
| G A | 107 (56.3) | 75 (49.3) | 113 (46.9) | |||
| A A | 34 (17.9) | 35 (23.0) | 58 (24.1) | |||
| rs11537641 | G G | 131 (67.9) | 107 (70.9) | 159 (66.0) | .484 | 0.91 (0.69–1.20) |
| G A | 58 (30.1) | 39 (25.8) | 70 (29.0) | |||
| A A | 4 (2.1) | 5 (3.3) | 12 (5.0) |
Distribution of genotypes of SLC2A1 variants among cases with T2DM-nephropathy (T2DM-nephropathy; DM + DN), diseased (T2DM without nephropathy; DM-DN) and healthy (HC) control subjects. *χ2p Values and **the generalized odds ratio (ORG) with respective 95% confidence intervals (95% CI) calculated for testing the association between genotype distribution of each variant and disease progression. ORG and p values are in bold in case of statistical significance.
Type 2 diabetes leading to nephropathy
Single-locus analysis
Table 3 shows the association results for type 2 diabetes leading to nephropathy. Analysis of the co-dominant and the additive inheritance model shown indicates that certain variants were associated with the risk of type 2 diabetes leading to nephropathy. Significant results were derived for the co-dominant inheritance model of the variants rs12407920 C/T [OR = 2.01 (1.17–3.45)], rs841847 C/T [OR = 1.73 (1.17–2.56)] and rs841853 C/A [OR = 1.74 (1.18–2.55)] as well as for the additive inheritance model of the variant rs3729548 C/T [OR = 0.52 (0.29–0.90)]. The mode of inheritance for the variants rs12407920 C/T, rs841847 C/T and rs841853 C/A was ‘dominance of each minor allele’and for the variant rs3729548 C/T was ‘non-dominance’. Therefore, for the variants rs12407920 C/T (h = 8.37), rs841847 C/T (h = 0.93) and rs841853 C/A (h = 0.94), the mode of inheritance is ‘dominance of each minor allele’, indicating that the homozygous for the minor allele has a greater risk of being diabetic with nephropathy than the homozygous for the frequent allele, and that the heterozygote has a risk of diabetes leading to nephropathy closer to the homozygote for the minor allele than to the midpoint between the two homozygotes. The mode of inheritance attributed to the variant rs3729548 C/T is ’non-dominance’ (h = 0.10), indicating that the heterozygote CT has a risk of being diseased that lies in the middle of the risk-protected CC and risk-exposed TT homozygous genotypes (Table 4).
Table 3.
Association between SLC2A1 gene variants and T2DM- nephropathy for the additive and co-dominant models.
| Variant | Genetic model | OR (95% CI) | *p-Value | ORadj. (95% CI) | #p-Value |
|---|---|---|---|---|---|
| rs12407920 C/T | Additive | 0.92 (0.15–5.56) | 0.926 | 0.69 (0.09-5.02) | .715 |
| Co-dominant | 2.01 (1.17–3.45) | 0.010 | 1.98 (1.14–3.43) | .015 | |
| rs2297976 G/T | Additive | 1.25 (0.43–3.67) | 0.679 | 1.17 (0.39–3.47) | .778 |
| Co-dominant | 0.94 (0.63–1.40) | 0.755 | 0.92 (0.61–1.38) | .677 | |
| rs710221 G/A | Additive | 1.08 (0.61–1,88) | 0.795 | 1.01 (0.56–1.81) | .987 |
| Co-dominant | 0.86 (0.58–1.26) | 0.432 | 0.85 (0.58–1.26) | .425 | |
| rs2086856 A/G | Additive | 1.77 (0.92–3.40) | 0.089 | 1.85 (0.94–3.65) | .075 |
| Co-dominant | 0.86 (0.59–1.26) | 0.441 | 0.85 (0.58–1.26) | .452 | |
| rs12130264 C/T | Additive | 2.37 (0.21–26,41) | 0.482 | 2.81 (0.25–31,79) | .403 |
| Co-dominant | 0.67 (0.36 1.25) | 0.209 | 0.72 (0.38–1.38) | .327 | |
| rs841847 C/T | Additive | 1.80 (0.90–3.62) | 0.097 | 1.77 (0.84–3.70) | .131 |
| Co-dominant | 1.73 (1.17–2.56) | 0.006 | 1.73 (1.16–2.58) | .007 | |
| rs841853 C/A | Additive | 1.80 (0.95–3.40) | 0.070 | 1.85 (0.94–3.61) | .073 |
| Co-dominant | 1.74 (1.18–2.55) | 0.005 | 1.75 (1.18–2.58) | .005 | |
| rs3729548 C/T | Additive | 0.52 (0.29–0.90) | 0.021 | 0.50 (0.28–0.92) | .025 |
| Co-dominant | 1.07 (0.73–1.56) | 0.737 | 1.06 (0.72–1.56) | .778 | |
| rs841855 G/A | Additive | 1.09 (0.33–3,66) | 0.887 | 1.04 (0.31–3.55) | .948 |
| Co-dominant | 1.10 (0.72–1.69) | 0.644 | 1.12 (0.73–1.73) | .608 | |
| rs3768029 C/T | Additive | 0.66 (0.38–1.14) | 0.135 | 0.65 (0.37–1,16) | .146 |
| Co-dominant | 1.26 (0.86–1.84) | 0.233 | 1.29 (0.87–1,90) | .202 | |
| rs12071418 C/G | Additive | **n.a. | **n.a. | **n.a. | **n.a. |
| Co-dominant | 0.61 (0.18–2.06) | 0.427 | 0.59 (0.17–2.01) | .396 | |
| rs3820549 C/G | Additive | 0.99 (0.51–1.95) | 0.988 | 0.99 (0.50–1.99) | .987 |
| Co-dominant | 0.89 (0.61–1,32) | 0.571 | 0.88 (0.59–1.30) | .517 | |
| rs3820546 G/A | Additive | 0.84 (0.48–1.46) | 0.534 | 0.85 (0.48–1,50) | .577 |
| Co-dominant | 1.46 (0.99–2.14) | 0.052 | 1.35 (0.92–2.00) | .126 | |
| rs11537641 G/A | Additive | 0.40 (0.13–1.28) | 0.125 | 0.36 (0.11–1,17) | .089 |
| Co-dominant | 1.05 (0.69–1.59) | 0.819 | 1.03 (0.68–1.58) | .874 |
Odds Ratio (OR) and the corresponding 95% Confidence Intervals (CI) for testing the association between the SLC2A1 gene variants and T2DM-nephropathy for the additive and co-dominant models. The ORs adjusted for age and sex calculated by univariate logistic regression are also shown. In all variants, the ORs are calculated considering the heterozygote as the risk genotype for the co-dominant model and the minor allele as the risk allele for the additive model.
χ2p Values.
Multivariate logistic regression p values.
Not applicable, because there are no G allele homozygotes. OR and p values are in bold in case of statistical significance.
Table 4.
The degree of dominance index (h-index).
| Variant | h-Index | Mode of inheritance |
|---|---|---|
| rs12407920 C/T | 8.37 | Dominance of allele T |
| rs841847 C/T | 0.93 | Dominance of allele T |
| rs841853 C/A | 0.94 | Dominance of allele A |
| rs3729548 C/T | 0.10 | None-dominance (additiveness) |
The degree of dominance index (h-index) as an estimate for the mode of inheritance, calculated on the basis of unadjusted odds ratio (OR) values and the respective mode of inheritance for all SLC2A1 variants with a significant association to type 2 diabetes leading to nephropathy, found for the additive and co-dominant inheritance models.
Linkage disequlibrium analysis
Table 5 shows the D′, r2 and p values for testing linkage disequilibrium (LD) between pairs of SCL2A1 variants for patients with diabetes leading to nephropathy (cases) and healthy controls. In cases 107 out of 182 (59%) and in healthy controls 120 out of 182 (66%) pairs of variants tested were in LD (p < .05). In 11 of the 14 SLC2A1 variants above 50% of variant pairs were in LD in both populations (cases and healthy controls). Especially, variant rs12071418 C/G was not in LD with any other variant in cases, whereas it was in LD with only 3 other variants (rs2297976 G/T, rs3820549 C/G, rs11537641 G/A) in controls. Moreover, omitting the rs12071418 C/G variant pairs, lead to 63% of pairs being in LD in cases and 69% in healthy controls, respectively.
Table 5.
Estimates (D′, r2 and p values) of linkage disequilibrium (LD) between pairs of SLC2A1 variants.
| Variant | LD | rs2297976 | rs710221 | rs2086856 | rs12130264 | rs841847 | rs841853 | rs3729548 | rs841855 | rs3768029 | rs12071418 | rs3820549 | rs3820546 | rs11537641 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12407920 | D′ | 0.99 (0.99) | 1.00 (0.99) | 1.00 (1.00) | 0.04 (0.59) | 0.31 (0.49) | 0.89 (0.83) | 0.85 (0.89) | 0.99 (0.99) | 0.04 (0.22) | 0.04 (1.00) | 0.19 (0.65) | 0.41 (0.57) | 0.69 (0.99) |
| r2 | 0.03 (0.02) | 0.09 (0.05) | 0.21 (0.15) | 0.00 (0.00) | 0.02 (0.06) | 0.16 (0.12) | 0.05 (0.05) | 0.03 (0.01) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.01) | 0.02 (0.02) | 0.01 (0.02) | |
| p | .03 (.09) | <.01 | <.01 | .18 (.29) | .1 (<.01) | <.01 | .12 (<.01) | .45 (.15) | .26 (.29) | 1.00 (.18) | .40 (.14) | .20 (.01) | .28 (.06) | |
| rs2297976 | D′ | – | 1.00 (1.00) | 0.99 (0.99) | 1.00 (0.99) | 0.76 (0.63) | 0.77 (0.64) | 0.87 (0.97) | 0.99 (0.53) | 0.90 (0.84) | 0.58 (0.80) | 0.65 (0.10) | 0.04 (0.24) | 0.17 (0.04) |
| r2 | – | 0.35 (0.33) | 0.13 (0.11) | 0.01 (0.02) | 0.30 (0.32) | 0.24 (0.25) | 0.12 (0.19) | 0.05 (0.01) | 0.17 (0.18) | 0.01 (0.04) | 0.04 (0.01) | 0.00 (0.01) | 0.00 (0.00) | |
| p | – | <.01 | <.01 | <.01 (.09) | <.01 | <.01 | <.01 | .26 (.80 | <.01 | .38 (.02) | .19 (.61) | .45 (.30) | .97 (.21) | |
| rs710221 | D′ | – | – | 0.38 (0.51) | 1.00 (0.99) | 0.18 (0.26) | 0.09 (0.21) | 0.88 (0.88) | 0.85 (0.93) | 0.76 (0.66) | 0.26 (0.99) | 0.31 (0.59) | 0.42 (0.43) | 0.89 (0.87) |
| r2 | – | – | 0.10 (0.16) | 0.04 (0.05) | 0.02 (0.03) | 0.00 (0.02) | 0.37 (0.49) | 0.21 (0.21) | 0.35 (0.32) | 0.00 (0.02) | 0.05 (0.19) | 0.11 (0.12) | 0.21 (0.25) | |
| p | – | – | <.01 | <.01 | .38 (.04) | .68 (.13) | <.01 | <.01 | <.01 | .73 (.09) | .02 (<.01) | <.01 | <.01 | |
| rs2086856 | D′ | – | – | – | 0.99 (0.99) | 0.47 (0.29) | 0.13 (0.05) | 0.81 (0.79) | 0.85 (0.85) | 0.43 (0.46) | 0.04 (0.63) | 0.40 (0.37) | 0.58 (0.65) | 0.70 (0.54) |
| r2 | – | – | – | 0.03 (0.03) | 0.05 (0.01) | 0.00 (0.00) | 0.21 (0.24) | 0.30 (0.31) | 0.08 (0.10) | 0.00 (0.00) | 0.12 (0.12) | 0.15 (0.18) | 0.19 (0.16) | |
| p | – | – | – | .04 (.28) | .02 (.03) | .48 (.47) | <.01 | <.01 | .01 (<.01) | .57 (.89) | <.01 | <.01 | <.01 | |
| rs12130264 | D′ | – | – | – | – | 0.16 (0.99) | 0.32 (0.81) | 0.72 (0.91) | 0.97 (0.99) | 0.47 (0.45) | 1.00 (0.99) | 0.99 (0.84) | 0.26 (0.72) | 0.96 (0.99) |
| r2 | – | – | – | – | 0.00 (0.02) | 0.00 (0.02) | 0.04 (0.07) | 0.01 (0.01) | 0.01 (0.01) | 0.00 (0.00) | 0.02 (0.02) | 0.01 (0.04) | 0.01 (0.02) | |
| p | – | – | – | – | .10 (.13) | .06 (.07) | <.01 | .28 (.65) | .12 (.09) | .17 (.86) | .10 (.03) | .01 (.02) | .22 (.23) | |
| rs841847 | D′ | – | – | – | – | – | 1.00 (1.00) | 1.00 (1.00) | 0.99 (0.99) | 0.93 (0.95) | 1.00 (0.96) | 0.33 (0.35) | 0.24 (0.40) | 0.81 (0.81) |
| r2 | – | – | – | – | – | 0.78 (0.74) | 0.30 (0.26) | 0.10 (0.06) | 0.34 (0.28) | 0.01 (0.01) | 0.02 (0.02) | 0.02 (0.04) | 0.06 (0.05) | |
| p | – | – | – | – | – | <.01 | <.01 | <.01 (.01) | <.01 | .25 (.37) | .23 (.18) | .21 (.00) | .02 (.00) | |
| rs841853 | D′ | – | – | – | – | – | – | 1.00 (1.00) | 0.99 (0.99) | 0.69 (0.63) | 0.99 (0.99) | 0.19 (0.13) | 0.29 (0.39) | 0.85 (0.80) |
| r2 | – | – | – | – | – | – | 0.39 (0.35) | 0.13 ((0.08) | 0.24 (0.16) | 0.01 (0.01) | 0.01 (0.00) | 0.04 (0.05) | 0.09 (0.06) | |
| p | – | – | – | – | – | – | <.01 | <.01 | <.01 | .36 (.38) | .29 (.46) | .06 (<.01) | <.01 | |
| rs3729548 | D′ | – | – | – | – | – | – | – | 1.00 (1.00) | 0.94 (0.84) | 0.04 (0.99) | 0.71 (0.75) | 0.63 (0.56) | 0.79 (0.89) |
| r2 | – | – | – | – | – | – | – | 0.13 (0.16) | 0.66 (0.61) | 0.00 (0.01) | 0.12 (0.19) | 0.29 (0.29 | 0.08 (0.17 | |
| p | – | – | – | – | – | – | – | <.01 | <.01 | .65 (.29) | <.01 | <.01 | <.01 | |
| rs841855 | D′ | – | – | – | – | – | – | – | – | 1.00 (1.00) | 0.59 (0.08) | 0.78 (0.94) | 0.72 (0.91) | 0.80 (0.85) |
| r2 | – | – | – | – | – | – | – | – | 0.17 (0.19) | 0.00 (0.00) | 0.34 (0.42) | 0.09 (0.14) | 0.60 (0.59) | |
| p | – | – | – | – | – | – | – | – | <.01 | 1.00 (.84) | <.01 | <.01 | <.01 | |
| rs3768029 | D′ | – | – | – | – | – | – | – | – | – | 0.91 (0.99) | 0.52 (0.57) | 0.57 (0.59) | 0.84 (0.88) |
| r2 | – | – | – | – | – | – | – | – | – | 0.01 (0.02) | 0.09 (0.13) | 0.30 (0.32) | 0.12 (0.18) | |
| p | – | – | – | – | – | – | – | – | – | .84 (.13) | <.01 | <.01 | <.01 | |
| rs12071418 | D′ | – | – | – | – | – | – | – | – | – | – | 0.99 (1.00) | 0.96 (0.99) | 0.23 (0.81) |
| r2 | – | – | – | – | – | – | – | – | – | – | 0.02 (0.04) | 0.01 (0.02) | <0.01 (0.05) | |
| p | – | – | – | – | – | – | – | – | – | – | .13 (.01) | .17 (.19) | .59 (.01) | |
| rs3820549 | D′ | – | – | – | – | – | – | – | – | – | – | – | 1.00 (1.00) | 0.93 (0.95) |
| r2 | – | – | – | – | – | – | – | – | – | – | – | 0.33 (0.37) | 0.45 (0.54) | |
| p | – | – | – | – | – | – | – | – | – | – | – | <.01 | <.01 | |
| rs3820546 | D′ | – | – | – | – | – | – | – | – | – | – | – | – | 1.00 (1.00) |
| r2 | – | – | – | – | – | – | – | – | – | – | – | – | 0.17 (0.22) | |
| p | – | – | – | – | – | – | – | – | – | – | – | – | <.01 |
Estimates (D, r2 and p values) of linkage disequilibrium (LD) between pairs of SLC2A1 variants (SNPs) in patients with type 2 diabetes leading to nephropathy (cases, DN) and in healthy controls (HC). LD estimates of HC are shown in brackets.
Analysis of haplotypes
The distribution of the estimated haplotype frequencies of the 14 SLC2A1 variants (rs12407920 C/T, rs2297976 G/T, rs710221 G/A, rs2086856 A/G, rs12130264 C/T, rs841847 C/T, rs841853 C/A, rs3729548 C/T, rs841855 G/A, rs3768029 C/T, rs12071418 C/G, rs3820549 C/G, rs3820546 G/A, rs11537641 G/A) for cases and healthy controls is presented in Table 6. The overall difference between cases and healthy controls was not significant (p = .132). In the analysis of the individual haplotypes, however, C-G-G-A-T-C-C-T-G-T-C-C-A-G derived significant results [p = .014; OR = 0.248 (0.075–0.817)]. This haplotype may confer protection for type 2 diabetes leading to nephropathy, as alleles, which were shown to increase the risk of diabetes leading to nephropathy (i.e., allele T of rs12407920 C/T, allele T of rs841847 C/T and allele A of rs841853 C/A), are all missing in the haplotype, whereas allele T of rs3729548 C/T, which seems to act protectively, is included.
Table 6.
Estimated haplotype frequencies for the 14 SLC2A1 variants.
| Estimated frequencies |
χ2 | Global | |||
|---|---|---|---|---|---|
| Haplotypes of SNPs 1–14* | DM + DN | HC | Odds ratio (95%CI) | p-Value | p-Value |
| C G A G C C C C A C C G G A | 0.127 | 0.121 | 1.009 (0.638–0.597) | .969 | .132 |
| C T A A C T A C G C C C A G | 0.074 | 0.067 | 1.063 (0.594–0.902) | .838 | |
| C G G A C C C T G T C C A G | 0.285 | 0.290 | 0.908 (0.637–0.292) | .591 | |
| C G G A T C C T G T C C A G | 0.010 | 0.038 | 0.248 (0.075–0.817) | .014 | |
| C T A A C T A C G C C C G G | 0.097 | 0.060 | 1.633 (0.927–0.875) | .087 | |
| C G G A C T A C G C C C G G | 0.031 | 0.015 | 2.067 (0.726–0.888) | .165 | |
| T G G G C T A C G C C C G G | 0.032 | 0.041 | 0.732 (0.330–0.627) | .443 | |
| C G G A C C C T G T C C G G | 0.039 | 0.048 | 0.773 (0.371–0.610) | .491 | |
| C G G A C T A C G C C G G G | 0.044 | 0.029 | 1.481 (0.667–0.288) | .332 | |
Estimated haplotype frequencies for the 14 SLC2A1 variants (SNPs 1–14: rs12407920 C/T, rs2297976 G/T, rs710221 G/A, rs2086856 A/G, rs12130264 C/T, rs841847 C/T, rs841853 C/A, rs3729548 C/T, rs841855 G/A, rs3768029 C/T, rs12071418 C/G, rs3820549 C/G, rs3820546 G/A, rs11537641 G/A) in cases (T2DM-nephropathy; DM + DN) and in healthy controls (HC). The p values for comparison between cases and HC of the frequencies of each haplotype and the global p values for testing the overall difference in haplotype frequencies are shown.
Only haplotypes with a frequency of >0.03 in either cases (DM + DN) or controls (HC) are shown. OR and p values are in bold in case of statistical significance.
Meta-analysis
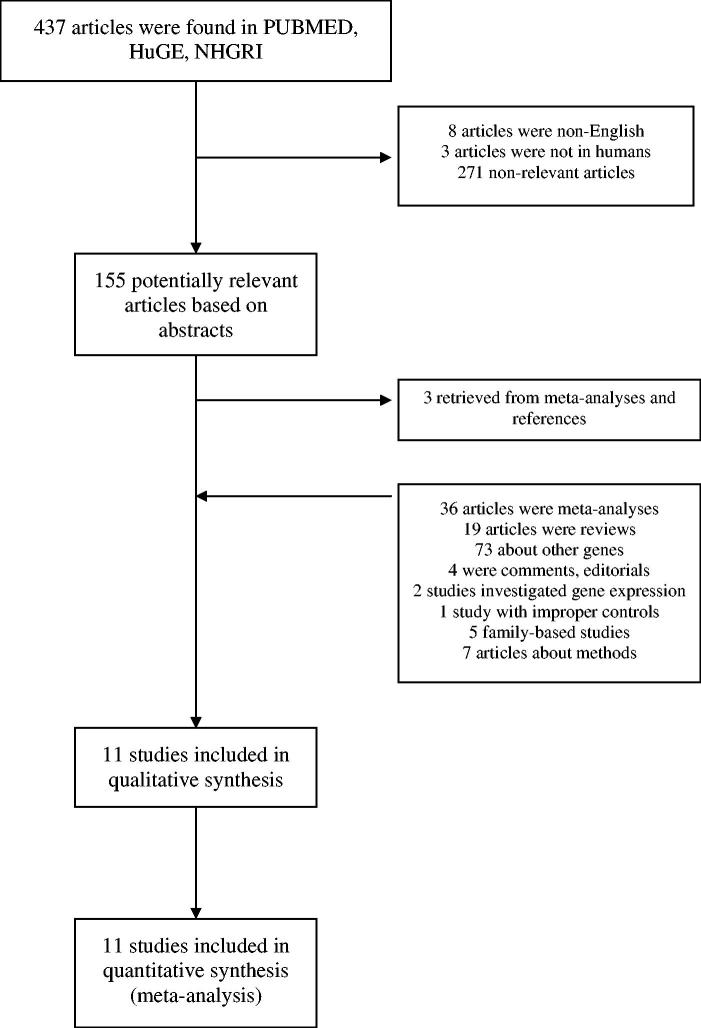
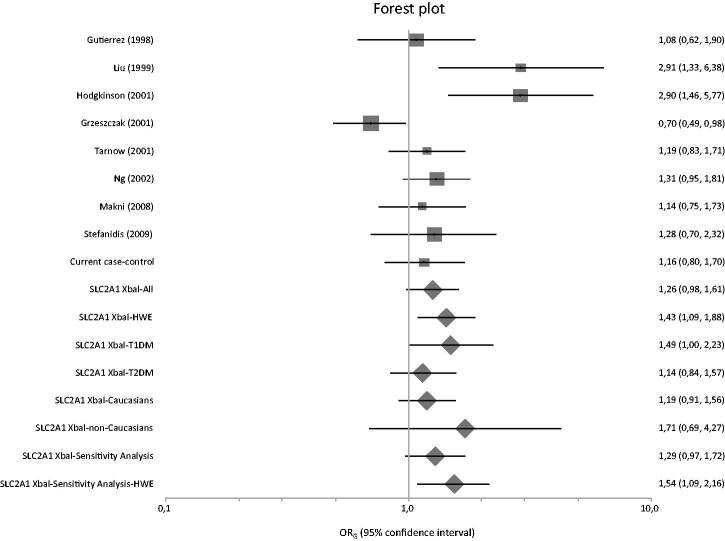
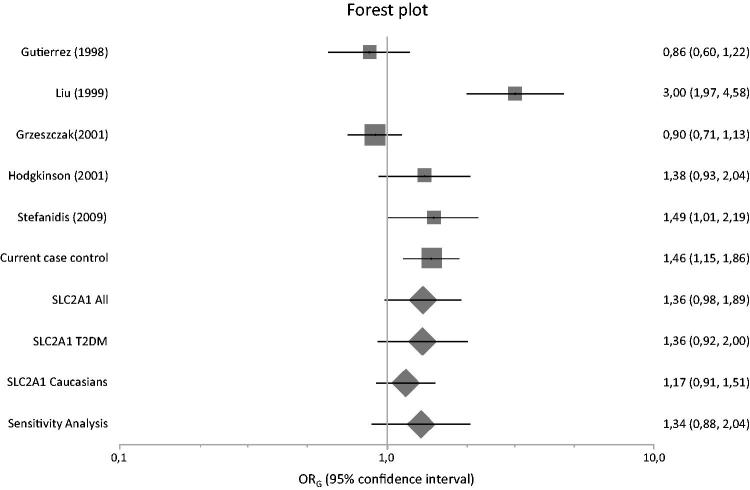
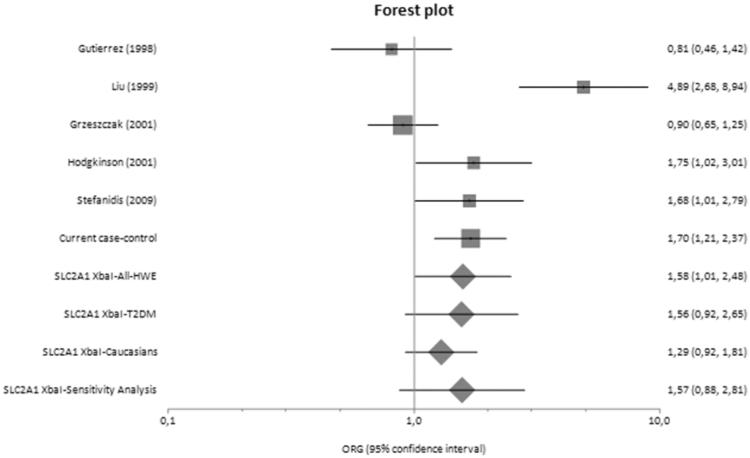
Figure 1 presents a flowchart of retrieved and excluded articles. The characteristics of each study are shown in Table 7. Across all available studies examining SLC2A1 variants, 8 genetic variants were studied (XbaI SNP, HaeIII SNP, Enhancer-1 SNP, Enhancer-2 SNP 1, Enhancer-2 SNP 2, Enhancer-3 SNP, HpyCH4V and rs3820589). Out of the aforementioned variants, only five variants examined in two studies or more and so meta-analyzed (XbaI SNP, HaeIII SNP, Enhancer-2 SNP 1, Enhancer-2 SNP 2 and HpyCH4V). Only XbaI SNP produced significant results in analysis using diseased controls versus cases and healthy controls versus cases giving a summary ORG of 1.428 (1.086, 1.877) and 1.581 (1.007, 2.482), respectively (Table 8). The studies comprised 1812 cases, 1763 diseased controls and 949 healthy controls and they were published between 1998 and 2015 [10,35–44]. Figures 2–4 are forest plot representations of variant rs841853.
Figure 1.
Flowchart showing how studies were selected for meta-analysis.
Table 7.
Characteristics of studies included in meta-analysis.
| Variant | References | Ethnicity | PMID | DM | Trait | N | Selection criteria | N | Selection criteria | N | Selection criteria | HWE HT | HWE DC | Analyses |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLC2A1 rs841853 | [10] | Caucasians | 19822956 | T2DM | DN | 92 | Pers. albuminuria | 56 | DM >15 y, pers. norm/ria matched for age, BMI | 92 | Non-diabetics matched for age, BMI | No | DC-C, HT-DC-C, HT-C | |
| [36] | Africans (Tunisia) | 18821326 | T2DM | DN | 126 | Pers. albuminuria, retinopathy | 273 | DM >10 y, norm/ria | No | DC-C | ||||
| [37] | Caucasians | 12086959 | T1DM | DN | 262 | Pers. macr/ria or d. ESRD | 230 | Pers. norm/ria , DM ≥15 y | DC-C | |||||
| [38] | Caucasians | 11231353 | T1DM | DN | 70 | Pers. proteinuria, retinopathy, DM ≥10 y | 44 | DM≥ 20 y, pers. norm/ria | 104 | Non-diabetics | DC-C, HT-DC-C, HT-C | |||
| [39] | Caucasians | 11477169 | T1DM | DN | 199 | Pers. macr/ria , retinopathy | 192 | Pers. norm/ria matched for gender, age, DM duration | DC-C | |||||
| [40] | E. Asians | 10231446 | T2DM | DN | 64 | Pers. albuminuria or proteinuria with or without impaired renal function | 45 | Only diabetics | 124 | Non-diabetics | DC-C, HT-DC-C, HT-C | |||
| [41] | Caucasians | 11168944 | T2DM | DN | 282 | Pers. micr/ria/proteinuria/CRF | 162 | Pers. norm/ria, DM ≥10 y | 194 | Non-diabetics | No | DC-C, HT-DC-C, HT-C | ||
| [42] | Caucasians | 9789717 | T2DM | DN | 60 | Pers. micro/macroalbuminuria | 100 | Pers. norm/ria matched for gender, age, BMI, DM duration and HbA1c and lipidic profile | 90 | Non-diabetics | DC-C, HT-DC-C, HT-C | |||
| SLC2A1 rs1385129 | [43] | Caucasians-Brazilians | 25701507 | T1DM | DN | 203 | Pers. micro/macroalbuminuria | 249 | Pers. norm/ria and serum Cr.<1.7 mg/dl | DC-C | ||||
| [44] | Asians | 26337659 | T2DM | DN | 126 | Pers. micr/ria | 150 | Pers. norm/ria | No | DC-C | ||||
| [36] | Africans (Tunisia) | 18821326 | T2DM | DN | 126 | Pers. albuminuria, retinopathy | 273 | DM >10 y, norm/ria | DC-C | |||||
| [37] | Caucasians | 12086959 | T1DM | DN | 262 | Pers. macr/ria or d. ESRD | 230 | Pers. norm/ria , DM duration ≥15 y | DC-C | |||||
| SLC2A1 rs841847 | [36] | Africans (Tunisia) | 18821326 | T2DM | DN | 126 | Pers. albuminuria, retinopathy | 273 | DM >10 y, norm/ria | DC-C | ||||
| [43] | Caucasians-Brazilians | 25701507 | T1DM | DN | 203 | Pers. micro/macroalbuminuria | 249 | Pers. norm/ria and serum Cr.<1.7 mg/dl | DC-C | |||||
| [37] | Caucasians | 12086959 | T1DM | DN | 262 | Pers. macr/ria or d. ESRD | 230 | Pers. norm/ria, DM ≥15 y | DC-C | |||||
| SLC2A1 rs841848 | [36] | Africans (Tunisia) | 18821326 | T2DM | DN | 126 | Pers. albuminuria, retinopathy | 273 | DM >10 y, norm/ria | DC-C | ||||
| [43] | Caucasians-Brazilians | 25701507 | T1DM | DN | 203 | Pers. micro/macroalbuminuria | 249 | Pers. norm/ria and s. Cr.<1.7 mg/dl | DC-C | |||||
| [37] | Caucasians | 12086959 | T1DM | DN | 262 | Pers. macr/ria or d. ESRD | 230 | Pers. norm/ria , DM ≥15 y | DC-C | |||||
| SLC2A1 rs710218 | [36] | Africans (Tunisia) | 18821326 | T2DM | DN | 126 | Pers. albuminuria, retinopathy | 273 | DM >10 y, norm/ria | No | DC-C | |||
| [38] | Caucasians | 15745834 | T1DM | DN | 131 | DM ≥10 y, pers. proteinuria, diabetic retinopathy | 72 | Uncomplicated, DM ≥20 y without retinopathy and proteinuria | 99 | Non-diabetics | DC-C | |||
| 35 | DM <10 y without retinopathy, proteinuria or overt neuropathy |
Table 8.
Results from meta-analyses based on genotype counts.
| Diseased controls versus cases | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Variant | RS | Studies (n) | Cases/Controls (n) | RE ORG | 95% LL | 95% UL | I2(%) | PQ | PE |
| SLC2A1 | XbaI(+)>XbaI(−) | rs841853 | 9 | 1311/1226 | 1.26 | 0.98 | 1.61 | 62.31 | 0.01 | 0.04 |
| SLC2A1 | All in HWE | 6 | 811/735 | 1.43 | 1.09 | 1.88 | 50.90 | 0.07 | 0.08 | |
| Subgroup analyses | ||||||||||
| T1DM | XbaI(+)>XbaI(−) | rs841853 | 3 | 494/443 | 1.49 | 1.00 | 2.23 | 62.13 | 0.07 | 0.16 |
| T2DM | XbaI(+)>XbaI(−) | rs841853 | 6 | 817/783 | 1.14 | 0.84 | 1.57 | 60.86 | 0.03 | 0.05 |
| Caucasians | XbaI(+)>XbaI(−) | rs841853 | 7 | 1121/908 | 1.19 | 0.91 | 1.56 | 62.17 | 0.01 | 0.12 |
| Non-Caucasians | XbaI(+)>XbaI(−) | rs841853 | 2 | 190/318 | 1.71 | 0.69 | 4.27 | 76.70 | 0.04 | na |
| Sensitivity analysis | ||||||||||
| Excluding current case-control | XbaI(+)>XbaI(−) | rs841853 | 8 | 1118/1078 | 1.29 | 0.97 | 1.72 | 67.020 | 0 | 0.05 |
| All in HWE | 5 | 618/587 | 1.54 | 1.09 | 2.16 | 57.70 | 0.05 | 0.13 | ||
| SLC2A1 | HaeIII SNP | rs1385129 | 4 | 716/899 | 0.74 | 0.350 | 1.57 | 92.05 | 0 | 0.18 |
| All in HWE | 3 | 590/749 | 1.14 | 0.73 | 1.76 | 74.96 | 0.02 | 0.11 | ||
| SLC2A1 | Enh2 SNP1 G>A | rs841847 | 4 | 783/904 | 1.48 | 0.85 | 2.60 | 89.27 | 0 | 0.13 |
| SLC2A1 | All in HWE | 4 | ||||||||
| Subgroup analyses | ||||||||||
| T1DM | Enh2 SNP1 G>A | rs841847 | 2 | 465/479 | 1.10 | 0.87 | 1.40 | 0 | 0.92 | na |
| T2DM | Enh2 SNP1 G>A | rs841847 | 2 | 318/425 | 2.02 | 0.58 | 7.00 | 94.83 | 0 | na |
| Caucasians | Enh2 SNP1 G>A | rs841847 | 2 | 454/382 | 1.08 | 0.85 | 1.39 | 0 | 0.96 | na |
| Non-Caucasians | Enh2 SNP1 G>A | rs841847 | 2 | 329/522 | 2.06 | 0.62 | 6.86 | 94.81 | 0 | na |
| Sensitivity analysis | ||||||||||
| Excluding current case-control | Enh2 SNP1 G>A | rs841847 | 3 | 591/752 | 1.65 | 0.78 | 3.51 | 92.21 | 0 | 0.16 |
| All in HWE | 3 | |||||||||
| SLC2A1 | Enh2 SNP2 C>T | rs841848 | 3 | 588/750 | 1.18 | 0.95 | 1.45 | 0 | 0.62 | 0.35 |
| All in HWE | 3 | |||||||||
| Subgroup analyses | ||||||||||
| T1DM | Enh2 SNP2 C>T | rs841848 | 2 | 462/477 | 1.14 | 0.88 | 1.48 | 0 | 0.37 | na |
| T2DM | Enh2 SNP2 C>T | rs841848 | 1 | na | na | na | na | na | na | na |
| Caucasians | Enh2 SNP2 C>T | rs841848 | 2 | 462/477 | 1.14 | 0.88 | 1.48 | 0 | 0.37 | na |
| Non-Caucasians | Enh2 SNP2 C>T | rs841848 | 1 | na | na | na | na | na | na | na |
| Sensitivity analysis | ||||||||||
| Excluding current case-control | Enh2 SNP2 C>T | rs841848 | na | na | na | na | na | na | na | na |
| SLC2A1 | HpyCH4V | rs710218 | 2 | 257/380 | 3.87 | 0.61 | 24.38 | 96.94 | 0.00 | na |
| All in HWE | 2 | |||||||||
| Healthy controls versus diseased controls versus cases | ||||||||||
| SLC2A1 | XbaI(+)>XbaI(−) | rs841853 | 6 | 761/554/845 | 1.36 | 0.98 | 1.89 | 84.03 | 0 | 0.10 |
| SLC2A1 | All in HWE | 6 | ||||||||
| Subgroup analyses | ||||||||||
| T1DM | XbaI(+)>XbaI(−) | rs841853 | 1 | na | na | na | na | na | na | na |
| T2DM | XbaI(+)>XbaI(−) | rs841853 | 5 | 691/510/741 | 1.36 | 0.92 | 2.00 | 87.14 | 0 | 0.17 |
| Caucasians | XbaI(+)>XbaI(−) | rs841853 | 5 | 697/509/721 | 1.17 | 0.91 | 1.51 | 69.98 | 0.01 | 0.21 |
| Non-Caucasians | XbaI(+)>XbaI(−) | rs841853 | 1 | na | na | na | na | na | na | na |
| Sensitivity analysis | ||||||||||
| Excluding current case-control | XbaI(+)>XbaI(−) | rs841853 | 5 | 568/406/604 | 1.34 | 0.88 | 2.04 | 86.30 | 0 | 0.07 |
| All in HWE | 5 | |||||||||
| Healthy controls versus cases | ||||||||||
| SLC2A1 | XbaI(+)>XbaI(−) | rs841853 | 6 | 761/845 | 1.58 | 1.01 | 2.48 | 83.03 | 0 | 0.23 |
| All in HWE | 6 | |||||||||
| Subgroup analyses | ||||||||||
| T1DM | XbaI(+)>XbaI(−) | rs841853 | 1 | na | na | na | na | na | na | na |
| T2DM | XbaI(+)>XbaI(−) | rs841853 | 5 | 691/741 | 1.56 | 0.92 | 2.65 | 86.15 | 0 | 0.28 |
| Caucasians | XbaI(+)>XbaI(−) | rs841853 | 5 | 697/721 | 1.29 | 0.92 | 1.81 | 66.67 | 0.02 | 0.42 |
| Non-Caucasians | XbaI(+)>XbaI(−) | rs841853 | 1 | na | na | na | na | na | na | na |
| Sensitivity Analysis | XbaI(+)>XbaI(−) | rs841853 | 5 | 568/604 | 1.568 | 0.875 | 2.81 | 85.765 | 0 | 0.15 |
| Excluding current case-control | All in HWE | 5 | ||||||||
na: non-applicable.
Figure 2.
Forest plot presenting results of individual studies and pooled estimates from both main and subgroup meta-analyses between diseased controls versus cases.
Figure 3.
Forest plot presenting results of individual studies and pooled estimates from both main and subgroup meta-analyses between healthy controls versus diseased controls versus cases.
Figure 4.
Forest plot presenting results of individual studies and pooled estimates from both main and subgroup meta-analyses between healthy controls versus cases.
Regarding the subgroup analyses according to diabetes types and ethnicity, the relevant results were not significant. However, in sensitivity analysis, when the present association study was excluded, the patterns of results changed (Figure 2).
Discussion
The present study investigated whether SLC2A1 variants, certain 14 tag SNPs, are associated with the type 2 diabetes disease progression and with the development of type 2 diabetes mellitus leading to nephropathy and also provided the most comprehensive overview assessing for all genetic variants of SLC2A1 that have been examined in genetic association studies regarding diabetic nephropathy.
Upon examining the association between SLC2A1 variants and type 2 diabetes leading to nephropathy, we selected as a control population the healthy subjects and not the patients with diabetes type 2 without nephropathy since every participant of the latter population is always a candidate to become a future case with diabetic nephropathy. Moderately increased albuminuria was not categorized as diabetic nephropathy because the diagnosis of diabetic nephropathy cannot be based only on the presence of moderately increased albuminuria. Apart from diabetic nephropathy there are several other causes for moderately increased albuminuria in diabetic patients. In addition, patients with moderately increased albuminuria do not invariably develop nephropathy. The strict selection criteria in our study ensured a relative clear case definition. At the end, only a histological examination would ensure the diagnosis of diabetic nephropathy, however, kidney biopsies are not routinely carried out in diabetes. Among patients with diabetes and nephropathy, who underwent kidney biopsy, the prevalence of diabetic nephropathy was found to be about 73% [45].
The analysis showed that certain variants of SLC2A1 (rs12407920 C/T, rs841847 C/T, rs841853 C/A and rs3729548 C/T) are involved in disease progression. In addition, these variants are associated with the risk of diabetes leading to nephropathy: significant results were derived for the co-dominant model of the variants rs12407920 C/T [OR = 2.01 (1.17–3.45)], rs841847 C/T [OR = 1.73 (1.17–2.56)] and rs841853 C/A [OR = 1.74 (1.18–2.55)] as well as for the additive model of the variant rs3729548 C/T [OR = 0.52 (0.29–0.90)]. The mode of inheritance for the variants rs12407920 C/T, rs841847 C/T and rs841853 C/A was ‘dominance of each minor allele’ and for the variant rs3729548 C/T was ‘non-dominance’. The frequency of one haplotype (C-G-G-A-T-C-C-T-G-T-C-C-A-G) was significantly different on a comparison between cases and healthy controls [p = .014; OR = 0.248 (0.075–0.817)]. This haplotype may confer protection for type 2 diabetes leading to nephropathy, as all the alleles contributing to the risk of diabetes leading to nephropathy (i.e., allele T of rs12407920 C/T, allele T of rs841847 C/T and allele A of rs841853 C/A) are missing in the haplotype, whereas allele T of rs3729548 C/T, which seems to act protectively, is included.
In agreement to our findings, a previous systematic review and meta-analysis of nine genetic association studies in patients with either type 1 or type 2 diabetes found that certain genetic variants in SLC2A1 (rs1385129, rs841847, rs841848 and rs841853) enhance susceptibility to diabetic nephropathy [46]. The similarity of findings in type 1 and type 2 diabetes mellitus is not surprising. The pathogenesis of diabetic nephropathy is generally the same in all types of diabetes. It is principally related to hyperglycemia and to arterial hypertension. In this context, the putative pathogenic role of the cell-membrane glucose transporter GLUT1 is mainly depending on hyperglycemia, i.e., it is depending on diabetes control and not on diabetes type.
In our study the genetic association was performed in patients with type 2 diabetes of Greek (Caucasian) origin. Subgroup analyses for Caucasians, in the above systematic review, revealed association between diabetic nephropathy due to both type 1 and type 2 diabetes and SLC2A1 variants [46]. All four studies concerning type 1 diabetes mellitus in the analysis were including only Caucasian populations. There were two studies (out of five) concerning type 2 diabetes mellitus, which included non-Caucasian populations, one in Asians and one in Tunisians, and both showed a positive association between diabetic nephropathy and SLC2A1 variants. A more recent genetic association study in Brazilian patients with type 1 diabetes mellitus and inadequate blood glucose control showed that another variant of SLC2A1 (rs3820589) is associated with progression of nephropathy [42]. Along with our findings, these reports clearly implicate a modulating role for SLC2A1 variants in diabetic nephropathy.
However, any genetic association study on the risk for development of diabetic nephropathy in either type of diabetes mellitus might be readily confounded if the genetic factors under investigation are also predisposing to diabetes. This is especially true when healthy controls, i.e., controls without diabetes, are included. SLC2A1 may not be involved in the pathogenesis of type 1 diabetes but its involvement in type 2 diabetes is plausible according to functional criteria. Nevertheless, according to a study of two populations in the Pacific, the variant rs841853 of SLC2A1 was not predisposing to diabetes type 2. Concretely, the rs841853 alleles frequency was the same (high) in both populations, one Polynesian population with high prevalence for type 2 diabetes and one highland New Guinean population with a notable absence of type 2 diabetes [47]. In contrast, the meta-analysis by Cui et al. (2013) provides strong evidence for Asians and marginal evidence for Caucasians, that the rs841853 variant of SLC2A1 may confer increased susceptibility to type 2 diabetes mellitus [48]. In our study, these putative confounding effects might have affected results and the possibility of more conclusive inferences.
Single nucleotide polymorphisms of SLC2A1, involved in disease progression in our study, are all intron variants (rs12407920, rs841847, rs841853 and rs3729548). As intronic variants, they cannot possibly cause changes in the protein sequence and they are not associated to alterations of the SLC2A1 expression. For these reasons, although significant associations were detected in this study, their functional significance seems questionable and their relevance would need further experimental proof. These polymorphisms may not be causative, but linkage disequilibrium with other loci with an etiologic role in diabetic nephropathy cannot be excluded.
In addition, in our study, the sample size was relatively small. The association of diabetic nephropathy and 4 genetic variants in the GLUT1 gene is remarkable. However, the number of patients and controls is low for the inference of genetic association. This is a common phenomenon in candidate-gene association studies [49]. In general, in order to achieve a power >80% for identifying a modest genetic effect (odds ratio 1.2) of a polymorphism present in 10% of the individuals, a sample size of more than 10,000 subjects would be needed [49]. It is obvious that a single institution will never be able to provide a sufficient number of patients to predict association, if it really exists. Then, future collaborative studies may provide more power to detect significant associations by pooling of data. Finally, future meta-analysis of multiple studies may overcome the deficiency of small power and to provide more conclusive evidence for the implication of SLC2A1 in complications in diabetes [23]. However, the validity of the present findings should be replicated from other gene-candidate or genome-wide association studies (GWAS) [6,39,46,50,51].
Diabetic nephropathy is a complex disease with multifactorial etiology and it involves epistatic and gene-environment interactions, and therefore, single type of genetic studies, such as gene-candidate association studies, have a reduced likelihood to provide conclusive inferences. In addition to hypothesis-driven studies (i.e., the gene-candidate association studies), hypothesis-free studies such as GWAS [23,52,53], microarrays gene expression analyses [54,55] and whole genome linkage scans [56,57] may assist in providing more conclusive evidence regarding the significance of SLC2A1 as a marker in diabetes leading to complication. This can be achieved by examining the genomic convergence of these different types of studies [53]. Although GWAS represent a superior strategy for unraveling genetic complexity [52], the findings of gene-candidate association studies may be supportive in replicating existed evidence and in revealing genuine genetic effects that could merit prioritization in future studies. However, GWAS themselves lack replication and therefore, replication of their findings from different investigators and different methodologies (such as gene-candidate association studies) are essential to interpret the mass of associations likely to result from GWAS [23,56,57].
However, since the sample size of the present association study was relatively small, we performed a meta-analysis considering all published studies, which investigated the association between SLC2A1 variants and diabetic nephropathy. In total, eight SLC2A1 variants were investigated, out of which only five variants were examined in two studies or more and so considered in meta-analysis. Among these five variants, two were also genotyped in our case-control study (rs841853, rs841847). We recorded a significant association between XbaI polymorphism and diabetic nephropathy in analysis of diseased controls versus cases and healthy controls versus cases, after accounting between study heterogeneity using the random effects model. It is noteworthy to mention that meta-analysis when included, our case-control study changed the pattern of results in comparison with healthy controls versus cases, as a significant association was detected for XbaI polymorphism suggesting that this genetic variant maybe associated with diabetic nephropathy.
To the best of our knowledge, it is the most comprehensive meta-analysis regarding SLC2A1 variants, since it includes all polymorphisms with available data for meta-analysis and all available comparisons between cases, diseased controls and healthy controls. These three comparisons made the trait discrimination more feasible. An important issue in all genetic studies regarding diabetic nephropathy is the demarcation of genetic loci associated with diabetic nephropathy per se and not with the type of diabetes which caused the renal disease. One additional strength of our meta-analysis is the strict definition of cases, as only subjects with persistent moderately increased albuminuria were considered as cases.
However, our meta-analysis has also some limitations. A common issue in meta-analysis is the publication bias, as only published studies were included in meta-analysis. Furthermore, the search was restricted in studies published in English. We should also interpret with caution the results of the meta-analysis because the number of studies is small and the sample size of each study also small.
Conclusion
In conclusion, the present study presents the results of an association study, which investigated the relation between 14 tag variants across SLC2A1 and the risk of type 2 diabetes leading to nephropathy and it also reviews the current epidemiology findings regarding the contribution of SLC2A1 variants in diabetic nephropathy. The results suggest that SLC2A1 variants and haplotypes may be involved in the pathogenesis of diabetic nephropathy. However, additional studies and a genetic convergence analysis of different data sources are needed in order to merit prioritization in future studies producing more conclusive claims of the association between SLC2A1 and genetic susceptibility to diabetic nephropathy.
Ethical approval
This study was approved by the Ethics Committee of the University Hospital of Larissa, University of Thessaly, School of Medicine. The study was conducted in the University Hospital of Larissa and all participants signed an informed consent before enrollment.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Rich SS. Genetics of diabetes and its complications. J Am Soc Nephrol. 2006;17:353–360. [DOI] [PubMed] [Google Scholar]
- 2.Gross JL, de Azevedo MJ, Silveiro SP. Diabetic nephropathy: diagnosis, prevention, and treatment. Diabetes Care. 2005;28:164–176. [DOI] [PubMed] [Google Scholar]
- 3.Strojek K, Grzeszczak W, Morawin E, et al. Nephropathy of type II diabetes: evidence for hereditary factors? Kidney Int. 1997;51:1602–1607. [DOI] [PubMed] [Google Scholar]
- 4.Quinn M, Angelico MC, Warram JH, et al. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 1996;39:940–945. [DOI] [PubMed] [Google Scholar]
- 5.Borch-Johnsen K, Norgaard K, Hommel E, et al. Is diabetic nephropathy an inherited complication? Kidney Int. 1992;41:719–722. [DOI] [PubMed] [Google Scholar]
- 6.Zintzaras E, Stefanidis I. Association between the GLUT1 gene polymorphism and the risk of diabetic nephropathy: a meta-analysis. J Hum Genet. 2005;50:84–91. [DOI] [PubMed] [Google Scholar]
- 7.Zintzaras E, Uhlig K, Koukoulis GN, et al. Methylenetetrahydrofolate reductase gene polymorphism as a risk factor for diabetic nephropathy: a meta-analysis. J Hum Genet. 2007;52:881–890. [DOI] [PubMed] [Google Scholar]
- 8.Tziastoudi M, Stefanidis I, Hadjigeorgiou GM, et al. A systematic review and meta-analysis of genetic association studies for the role of inflammation and the immune system in diabetic nephropathy. Clin Kidney J. 2017;10:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zintzaras E, Papathanasiou AA, Stefanidis I. Endothelial nitric oxide synthase gene polymorphisms and diabetic nephropathy: a HuGE review and meta-analysis. Genet Med. 2009;11:695–706. [DOI] [PubMed] [Google Scholar]
- 10.Stefanidis I, Kytoudis K, Papathanasiou AA, et al. XbaI GLUT1 gene polymorphism and the risk of type 2 diabetes with nephropathy. Dis Markers. 2009;27:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanidis I, Kreuer K, Dardiotis E, et al. Association between the interleukin-1β Gene (IL1B) C-511T polymorphism and the risk of diabetic nephropathy in type 2 diabetes: a candidate-gene association study. DNA Cell Biol. 2014;33:463–468. [DOI] [PubMed] [Google Scholar]
- 12.Heilig CW, Deb DK, Abdul A, et al. GLUT1 regulation of the pro-sclerotic mediators of diabetic nephropathy. Am J Nephrol. 2013;38:39–49. [DOI] [PubMed] [Google Scholar]
- 13.Heilig CW, Concepcion LA, Riser BL, et al. Overexpression of glucose transporters in rat mesangial cells cultured in a normal glucose milieu mimics the diabetic phenotype. J Clin Invest. 1995;96:1802–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigert C, Brodbeck K, Brosius FC III, et al. Evidence for a novel TGF-beta1-independent mechanism of fibronectin production in mesangial cells overexpressing glucose transporters. Diabetes. 2003;52:527–535. [DOI] [PubMed] [Google Scholar]
- 15.Larkins RG, Dunlop ME. The link between hyperglycaemia and diabetic nephropathy. Diabetologia 1992;35:499–504. [DOI] [PubMed] [Google Scholar]
- 16.Heilig CW, Brosius FC III, Cunningham C. Role for GLUT1 in diabetic glomerulosclerosis. Expert Rev Mol Med. 2006;8:1–18. [DOI] [PubMed] [Google Scholar]
- 17.Mogyorosi A, Ziyadeh FN. GLUT1 and TGF-beta: the link between hyperglycaemia and diabetic nephropathy. Nephrol Dial Transplant. 1999;14:2827–2829. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Heilig K, Saunders T, et al. Transgenic overexpression of GLUT1 in mouse glomeruli produces renal disease resembling diabetic glomerulosclerosis. Am J Physiol Renal Physiol. 2010;299:F99–F111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilig CW, Kreisberg JI, Freytag S, et al. Antisense GLUT-1 protects mesangial cells from glucose induction of GLUT-1 and fibronectin expression. Am J Physiol Renal Physiol. 2001;280:F657–F666. [DOI] [PubMed] [Google Scholar]
- 20.Heilig CW, Saunders T, Brosius FC III, et al. Glucose transporter-1-deficient mice exhibit impaired development and deformities that are similar to diabetic embryopathy. Proc Natl Acad Sci USA. 2003;100:15613–15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zintzaras E. The power of generalized odds ratio in assessing association in genetic studies with known mode of inheritance. J Appl Stat. 2012;39:2569–2581. [Google Scholar]
- 22.Zintzaras E. The generalized odds ratio as a measure of genetic risk effect in the analysis and meta-analysis of association studies. Stat Appl Genet Mol Biol. 2010;9:Article21. doi: 10.2202/1544-6115.1542. [DOI] [PubMed] [Google Scholar]
- 23.Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61:634–645. [DOI] [PubMed] [Google Scholar]
- 24.Zintzaras E, Santos M. Estimating the mode of inheritance in genetic association studies of qualitative traits based on the degree of dominance index. BMC Med Res Methodol. 2011;11:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zintzaras E, Santos M. Performance of MAX test and degree of dominance index in predicting the mode of inheritance. Stat Appl Genet Mol Biol. 2012;11:Article-6115. [DOI] [PubMed] [Google Scholar]
- 26.Genuth S, Alberti KG, Bennett P, et al. Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 2003;26:3160–3167. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez JL, Weir BS. A disequilibrium coefficient approach to Hardy-Weinberg testing. Biometrics. 1989;45:53–70. [PubMed] [Google Scholar]
- 28.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data Sunderland, Massachusetts: Sinauer Associates, 1996. [Google Scholar]
- 29.Genetic Data Analysis : Computer program for the analysis of allelic data. Version 1.0 (d16c). Free program distributed by the authors over the internet. 2001. [Google Scholar]
- 30.YONG Y, HE L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19:519–523. [DOI] [PubMed] [Google Scholar]
- 32.Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10:101. [Google Scholar]
- 33.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:315–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makni K, Jarraya F, Rebaï M, et al. Risk genotypes and haplotypes of the GLUT1 gene for type 2 diabetic nephropathy in the Tunisian population. Ann Hum Biol. 2008;35:490–498. [DOI] [PubMed] [Google Scholar]
- 36.Ng DPK, Canani L, Araki S, et al. Minor effect of GLUT1 polymorphisms on susceptibility to diabetic nephropathy in type 1 diabetes. Diabetes. 2002;51:2264–2269. [DOI] [PubMed] [Google Scholar]
- 37.Hodgkinson AD, Millward BA, Demaine AG. Polymorphisms of the glucose transporter (GLUT1) gene are associated with diabetic nephropathy. Kidney Int. 2001;59:985–989. [DOI] [PubMed] [Google Scholar]
- 38.Tarnow L, Grarup N, Hansen T, et al. Diabetic microvascular complications are not associated with two polymorphisms in the GLUT-1 and PC-1 genes regulating glucose metabolism in Caucasian type 1 diabetic patients. Nephrol Dial Transplant. 2001;16:1653–1656. [DOI] [PubMed] [Google Scholar]
- 39.Liu ZH, Guan TJ, Chen ZH, et al. Glucose transporter (GLUT1) allele (XbaI-) associated with nephropathy in non-insulin-dependent diabetes mellitus. Kidney Int. 1999;55:1843–1848. [DOI] [PubMed] [Google Scholar]
- 40.Grzeszczak W, Moczulski DK, Zychma M, et al. Role of GLUT1 gene in susceptibility to diabetic nephropathy in type 2 diabetes. Kidney Int. 2001;59:631–636. [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez C, Vendrell J, Pastor R, et al. GLUT1 gene polymorphism in non-insulin-dependent diabetes mellitus: genetic susceptibility relationship with cardiovascular risk factors and microangiopathic complications in a Mediterranean population. Diabetes Res Clin Pract. 1998;41:113–120. [DOI] [PubMed] [Google Scholar]
- 42.Marques T, Patente TA, Monteiro MB, et al. Association of single nucleotide polymorphisms in the gene encoding GLUT1 and diabetic nephropathy in Brazilian patients with type 1 diabetes mellitus. Clin Chim Acta. 2015;444:170–175. [DOI] [PubMed] [Google Scholar]
- 43.Amini S, Javanmardi M, Mokarizadeh A, et al. Association of HaeIII single nucleotide polymorphisms in the SLC2A1 gene with risk of diabetic nephropathy; evidence from Kurdish patients with type 2 diabetes mellitus. QJM: An International Journal of Medicine. 2016;109:399–404. [DOI] [PubMed] [Google Scholar]
- 44.Hodgkinson D, Page T, Millward B, et al. A novel polymorphism in the 5’ flanking region of the glucose transporter (GLUT1) gene is strongly associated with diabetic nephropathy in patients with Type 1 diabetes mellitus. J Diabetes Complications. 2005;19:65–69. [DOI] [PubMed] [Google Scholar]
- 45.Mazzucco G, Bertani T, Fortunato M, et al. Different patterns of renal damage in type 2 diabetes mellitus: a multicentric study on 393 biopsies. Am J Kidney Dis. 2002;39:713–720. [DOI] [PubMed] [Google Scholar]
- 46.Cui W, Du B, Zhou W, et al. Relationship between five GLUT1 gene single nucleotide polymorphisms and diabetic nephropathy: a systematic review and meta-analysis. Mol Biol Rep. 2012;39:8551–8558. [DOI] [PubMed] [Google Scholar]
- 47.Myles S, Hradetzky E, Engelken J, et al. Identification of a candidate genetic variant for the high prevalence of type II diabetes in Polynesians. Eur J Hum Genet. 2007;15:584–589. [DOI] [PubMed] [Google Scholar]
- 48.Du B, Liu S, Cui C, et al. Association between glucose transporter 1 rs841853 polymorphism and type 2 diabetes mellitus risk may be population specific (1rs8418532). J Diabetes. 2013;5:291–299. [DOI] [PubMed] [Google Scholar]
- 49.Zintzaras E, Lau J. Trends in meta-analysis of genetic association studies. J Hum Genet. 2008;53:1–9. [DOI] [PubMed] [Google Scholar]
- 50.Mooyaart AL, Valk EJ, van Es LA, et al. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia. 2011;54:544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu CC, Kao WL, Steffes MW, et al. Genetic variation of glucose transporter-1 (GLUT1) and albuminuria in 10,278 European Americans and African Americans: a case-control study in the Atherosclerosis Risk in Communities (ARIC) study. BMC Med Genet. 2011;12:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitsios GD, Zintzaras E. Genomic convergence of genome-wide investigations for complex traits. Ann Hum Genet. 2009;73:514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitsios GD, Zintzaras E. Genome-wide association studies: hypothesis-"free" or "engaged"? Transl Res. 2009;154:161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zintzaras E, Ioannidis JP. Meta-analysis for ranked discovery datasets: theoretical framework and empirical demonstration for microarrays. Comput Biol Chem. 2008;32:38–46. [DOI] [PubMed] [Google Scholar]
- 55.Zintzaras E, Ioannidis JP. METRADISC-XL: a program for meta-analysis of multidimensional ranked discovery oriented datasets including microarrays. Comput Methods Programs Biomed. 2012;108:1243–1246. [DOI] [PubMed] [Google Scholar]
- 56.Thomas DC. Are we ready for genome-wide association studies?. Cancer Epidemiol Biomarkers Prev. 2006;15:595–598. [DOI] [PubMed] [Google Scholar]
- 57.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]