Abstract
Microtubules form from longitudinally and laterally assembling tubulin α/β-dimers. The assembly induces strain in tubulin, resulting in cycles of microtubule catastrophe and regrowth. This so-called dynamic instability is governed by GTP hydrolysis that renders the microtubule lattice unstable, but it is unclear how. We used the human microtubule nucleating and stabilising neuronal protein doublecortin and high-resolution cryo-EM to capture tubulin’s elusive hydrolysis intermediate GDP.Pi state, alongside the pre-hydrolysis analogue GMPCPP state, and the post-hydrolysis GDP state with and without an anti-cancer drug Taxol®. GTP hydrolysis to GDP.Pi, followed by Pi release, constitute distinct structural transitions, causing unevenly distributed compressions of tubulin dimers, thereby tightening longitudinal and loosening lateral inter-dimer contacts. We conclude that microtubule catastrophe is triggered because the lateral contacts can no longer counteract the strain energy stored in the lattice, while reinforcement of the longitudinal contacts may support generation of force.
Introduction
Microtubules (MTs) are key players in cell division, migration and signalling and are thus targets for herbicides, fungicides and in the treatment of many human diseases, most notably cancer. Their cylindrical lattice is built through longitudinal (head-to-tail) and lateral (side-by-side) supramolecular assembly of the tubulin α/β-heterodimer. In solution this heterodimer favours a ‘bent’ conformation, but the lattice geometry imposes and temporarily stabilizes a tense ‘straight’ conformation1–3 (Fig. 1a). Such spring-like conformational strain lies at the heart of MT dynamic instability and is vital for MT function4,5. The crucial switch between MT growth and shrinkage – known as ‘catastrophe’ - is driven by the β-subunit GTPase cycle, but its molecular basis has been unclear. The long-standing question is how the strain energy stored in the lattice is released to depolymerise MTs and do work in the cell.
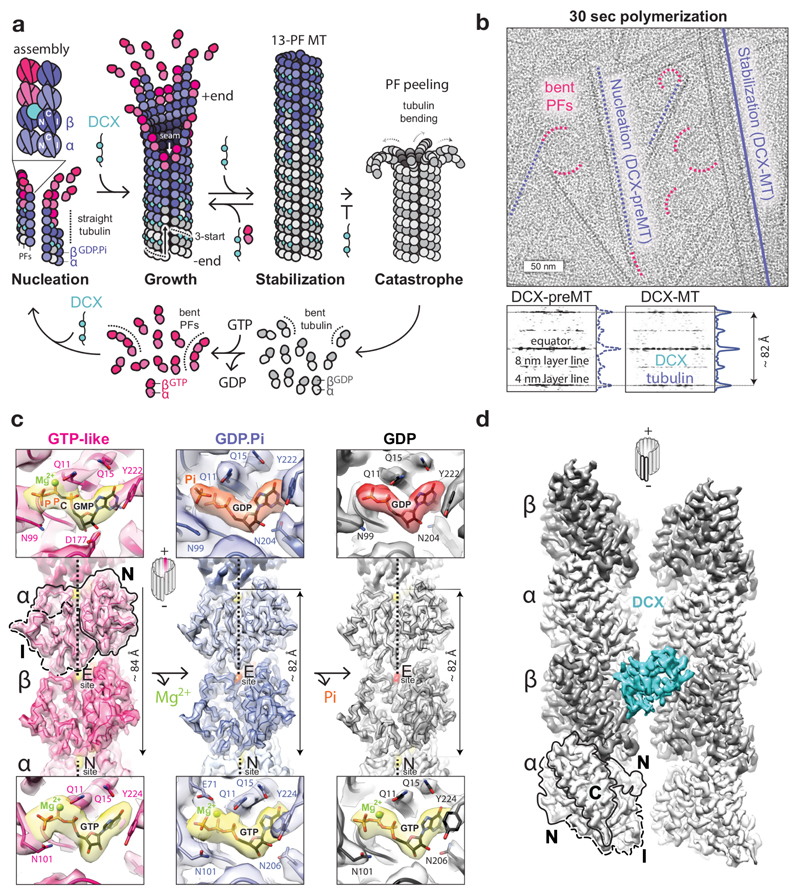
Figure 1. Tubulin GTPase cycle and MT structures stabilised by DCX.
a, DCX binds between 4 tubulin dimers, nucleating and stabilizing 13-PF 3-start MTs. In its absence, MTs undergo catastrophe once GTP hydrolysis and Pi release outpace MT growth. Then tubulin bends, whole protofilaments (PFs) peel away and depolymerise, and the cycle resumes. b, Top, representative cryo-micrograph showing the earliest tubulin assemblies (bent PFs), DCX-MT precursors (DCX-preMTs) and a mature 13-PF DCX-MT. Bottom, averaged power spectra and the layer line profile plots of DCX-MTs (n=16) and preMTs (n=9) showing compacted lattice spacing. c, Lumenal views of the cryo-EM reconstructions of MTs in GMPCPP, GDP-Pi and GDP states with fitted backbone traces of the refined atomic models of tubulin and close-up views of nucleotide densities (GTP and GMPCPP, yellow; GDP.Pi, orange; GDP, red) with selected residue side chains (coloured by heteroatom) around them at the exchangeable (E) and non-exchangeable (N) sites. N and I tubulin domains are outlined for one subunit. d, Front view of DCX binding site. N, I and C tubulin domains are outlined for one subunit.
α- and β-tubulin subunits are structurally very similar. Each comprises three functional domains6,7 determining MT polarity: 1) a GTP-binding N-terminal (N) domain, located towards the faster growing MT +end, 2) an intermediate (I) domain, facing the MT –end, and 3) a C-terminal (C) domain protruding from the MT wall (Fig. 1a). Tight ‘head-to-tail’ dimerisation of α/β-tubulin buries the GTP-binding site of the αN domain at its interface with the βI domain7, making it a non-exchangeable site (N-site), always occupied by GTP. The corresponding region in the β-subunit is exchangeable (E-site), and when occupied with GTP, it mediates analogous, but metastable ‘head-to-tail’ association with αI domain of another dimer, forming protofilaments (PFs). Individual PFs are compatible with the relaxed (energetically favoured) ‘bent’ tubulin conformation, but the lateral association of tubulin via specific interlocking of N and I domains gradually straightens tubulin dimers as well as the PFs formed by them2,8 (Fig. 1a). Thus, the mature MT geometry imposes and temporarily stabilises the straight tubulin conformation1–3,5, and the MT wall integrity relies on N and I domain involvement in both longitudinal and lateral lattice contacts (Fig. 1a). In vitro, MTs polymerise with a range of PF numbers, but in cells most MTs have a 13-PF architecture. This architecture necessitates that at one site called the seam, α- and β-tubulin form heterotypic lateral contacts, breaking the otherwise helical symmetry of an MT cylinder. Thus 13-PF MTs are pseudo-helical.
As described above, tubulin strain is inherent to building of an MT wall and is like a time bomb waiting for the trigger for disruption. This trigger involves GTP hydrolysis at the inter-dimer interface: the β-subunit’s GTP is hydrolysed due to recruitment of the α-subunit’s catalytic E254 residue7 during MT polymerisation. The equivalent residue within the intra-dimer interface is a non-catalytic βK254, hence the N-site GTP is never hydrolysed. E-site GTP hydrolysis somehow renders the MT wall unstable. Recent high-resolution cryo-EM structures using GTP analogues show9,10, in line with earlier studies11,12, that hydrolysis is accompanied by a longitudinal inter-dimer lattice compaction (~2 Å), but it is elusive how this rearrangement is linked to MT catastrophe. How is the strain energy stored in the lattice released to depolymerise MTs? The vital structure of the GDP.Pi intermediate has remained unknown, although GTPγS has been thought to mimic it10,13–15.
The imbalance between subunit strain energy and lattice binding energy leading to catastrophe would be predicted to result from perturbation of lattice contacts during and/or after lattice compaction. To detect these likely subtle (in the range of ~2 Å) changes in lattice contacts it is necessary to compare high-resolution MT structures representing sequential steps in the GTPase cycle in a uniform MT architecture. This has not been achieved to date. We used doublecortin (DCX), a unique MT-associated protein (MAP) and a perfect tool for this work, as it nucleates and stabilises a physiological 13-PF MT architecture by binding between 4 tubulin dimers in all nucleotide states (Fig. 1a, d) without perturbing known GTPase-linked transitions16,17. Crucially, DCX’s robust MT nucleation activity allowed rapid (~30 sec) sample preparation, such that we could capture the earliest stages of DCX-MT polymerisation (Fig. 1b), including the long-sought GDP.Pi transition state. By focusing on only the 13-PF MT architecture in the presence of only one MT binding partner – in contrast to previous work9,10 – our structures shed new light on the structural transitions that drive dynamic instability.
Results
Determination of 13-PF structures of DCX-MTs in different nucleotide states
To elucidate the mechanism of MT catastrophe, we analysed conformational changes in tubulin through its GTPase cycle with single particle cryo-EM, and assessed the impact of these changes on longitudinal and lateral lattice contacts. Overall, we determined 13-PF structures of 4 nucleotide states of DCX-MTs: 1) a GTP-like lattice (4.4 Å resolution) stabilised by GMPCPP, a slowly hydrolysable GTP analogue18; 2) a GDP.Pi lattice (4.2 Å) and 3) a control GTPγS lattice (4.4 Å), both of which show clear density equivalent to the γ-P of GTP, but no density for the magnesium ion10; and 4) a GDP lattice (3.8 Å) (Fig. 1c, Table 1, Supplementary Figs 1-3 and Supplementary Table 1). We calculated the lattice spacing and refined atomic models of tubulin for each reconstruction (Fig. 1c, Table 1 and Supplementary Table 1). The pre-hydrolysis GMPCPP lattice has an extended axial repeat (83.74 ± 0.20 Å), whereas the GDP.Pi, GTPγS and GDP lattices are compacted (81.74 ± 0.06, 82.10 ± 0.07 and 81.90 ± 0.06 Å, respectively). The compaction of the lattice together with the absence of magnesium density support the interpretation that we have captured the post-hydrolysis, GDP.Pi state. Even 10-20 dimer-long multi-PF MT precursor assemblies (DCX-preMTs) exhibit a compacted lattice (Fig. 1b), which emphasizes the speed of GTP hydrolysis and that the MT wall does not need to be complete for it to occur, at least with DCX bound at inter-dimer vertices (Fig. 1d). The overall tubulin conformation in GTPγS-DCX-MTs is in fact more similar to that in GDP-DCX-MTs than in GDP.Pi-DCX-MTs (Supplementary Fig. 3) despite the presence of γ-P/γ-S (Supplementary Fig. 2). This explains the slow MT growth observed with GTPγS and highlights the unusual properties of this nucleotide analogue14.
Table 1. Cryo-EM data collection, refinement and validation statistics.
| GMPCPP-DCX-MT (EMDB-3961) (PDB 6EVW) | GDP.Pi-DCX-MT (EMDB-3962) (PDB 6EVX) | GTPγS-DCX-MT (EMDB-3963) (PDB 6EVY) | GDP-DCX-MT (EMDB-3964) (PDB 6EVZ) | GDP-DCX-Taxol®-MT (EMDB-3965) (PDB 6EW0) | |
|---|---|---|---|---|---|
| Data collection and processing | |||||
| Magnification | 35,971 | 35,971 | 35,971 | 35,971 | 35,971 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 25 | 25 | 25 | 25 | 25 |
| Defocus range (μm) | -0.4 to -2.5 | -0.4 to -2.5 | -0.4 to -2.5 | -0.4 to -2.5 | -0.4 to -2.5 |
| Pixel size (Å) | 1.39 | 1.39 | 1.39 | 1.39 | 1.39 |
| Symmetry imposeda | 12-fold | 12-fold | 12-fold | 12-fold | 12-fold |
| Initial particle images (no.) | 11,552 | 7,727 | 9,395 | 32,256 | 18,651 |
| Final particle images (no.) | 3,244 | 6,591 | 6,037 | 30,434 | 17,626 |
| Map resolution (Å) | 4.4 | 4.2 | 4.4 | 3.8 | 3.8 |
| FSC thresholdb | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 4.1 – 5.8 | 4.0 – 5.8 | 4.0 – 5.8 | 3.2 – 4.8 | 3.2 – 4.8 |
| Refinement | |||||
| Initial model used (PDB code) | 6EVZ | 6EVZ | 6EVZ | 3JAR | 6EVZ |
| Model resolution (Å) | 4.5 | 4.2 | 4.4 | 3.8 | 3.8 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | 200 – 4.5 | 200 – 4.2 | 200 – 4.4 | 200 – 3.8 | 200 – 3.8 |
| Map sharpening B factor (Å2) | -137 | -167 | -156 | -163 | -99 |
| Model composition | |||||
| Non-hydrogen atoms | 40,536 | 40,896 | 40,890 | 40,866 | 41,238 |
| Protein residues | 5,112 | 5,166 | 5,166 | 5,166 | 5,166 |
| Ligands | 24 | 24 | 18 | 18 | 24 |
| B factors (Å2) | |||||
| Protein | 110 | 101 | 93 | 58 | 110 |
| Ligand | 143 | 86 | 82 | 64 | 103 |
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 |
| Bond angles (°) | 0.71 | 0.65 | 0.72 | 0.67 | 0.76 |
| Validation | |||||
| MolProbity score | 1.66 | 1.46 | 1.41 | 1.32 | 1.55 |
| Clashscore | 5.74 | 3.73 | 3.32 | 2.58 | 4.39 |
| Poor rotamers (%) | 0 | 0 | 0 | 0 | 0 |
| Ramachandran plot | |||||
| Favored (%) | 94.98 | 95.69 | 95.26 | 95.98 | 95.79 |
| Allowed (%) | 5.02 | 4.31 | 4.74 | 4.02 | 4.21 |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 |
DCX does not bind at the seam of the 13-PF MTs, thus pseudo-helical symmetry was applied across 12 PFs between which DCX binds.
Gold standard resolution estimation according to Chen et al35
GTP hydrolysis in β-tubulin leads to uneven compression of α-tubulin
To characterise the origin of the lattice compaction in our reconstructions, we compared the different MT states by superposition on the β-subunit, which itself does not show large conformational changes10. This comparison reveals conformational rearrangements in the nucleotide-binding βT5 loop that likely cause changes in the E-site hydrogen bond network (βN domain) in the GDP.Pi state relative to the GMPCPP state (transition 1) (Fig. 2a, b and Videos 1, 2). The βT5 loop interfaces with the adjoining tubulin subunit via αS9 strand of the αI domain’s central β-sheet. This β-sheet is sandwiched between two inter-dimer anchor points: A1) a hydrophobic contact between the C-terminal end of helix αH8 plus αH8-S7 loop and helix βH11’10; and A2) a hydrogen bond between K336 side chain at the C-terminus of helix αH10 and K174 backbone carbonyl in βT5 loop (Fig. 2a-c). Thus, the conformational change in the βT5 loop leads to a ~1 Å shift of the αI domain towards the β-subunit. The αN domain, connected to the αI domain via αS6-S7 strands, undergoes a more prominent displacement (up to ~2.2 Å) towards both the MT -end and lumen, together with a part of the C domain associated with it. This results in an uneven compression of the whole α-subunit, globally manifested as lattice compaction. After Pi release (transition 2), the α-subunit slightly twists (by a further ~1 Å; Fig. 2a and Video 1), further narrowing the gap between the dimers.
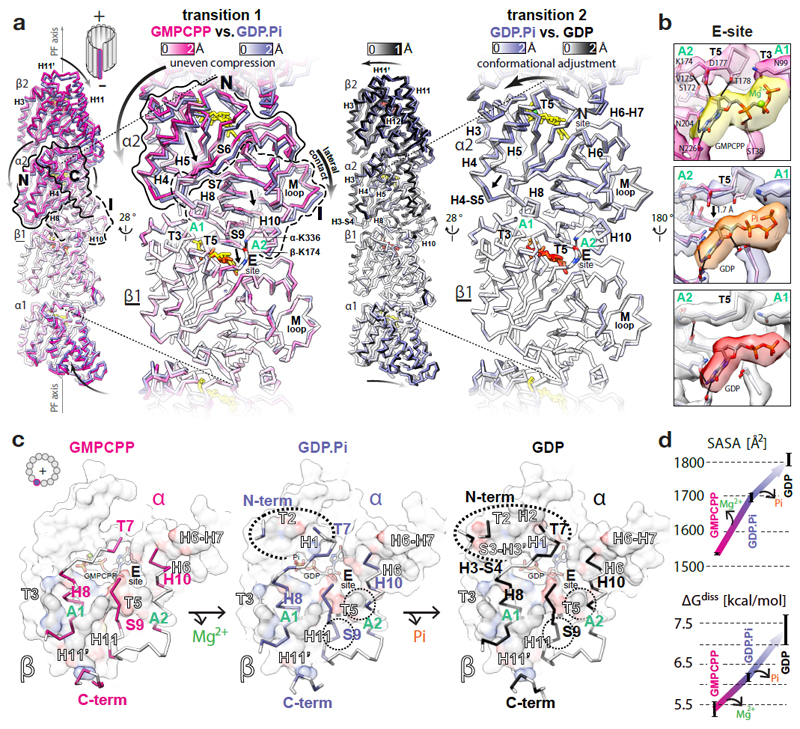
Figure 2. GTPase-dependent structural transitions strengthen the longitudinal MT lattice contacts.
a, Backbone front view and angled close-up cut-away view comparisons of different MT nucleotide states based on superposition on the β1-subunit (underlined); coloured by the degree of displacement, heteroatom or as follows: GMPCPP, red; GDP.Pi, orange; GDP, red; Mg2+, green. Tubulin domains are outlined and their overall relative movements around A1 and A2 anchor points are indicated with straight arrows. Curved arrows indicate how these transitions influence tubulin in the context of MT lattice. b, Conformational change of the T5 loop and the likely rearrangement of local hydrogen bonds (solid lines) shown with experimental densities and atomic models coloured as in a. c, +End view on the longitudinal inter-dimer interface; the relevant α-tubulin regions (colour-labelled) are shown as backbone traces superposed on top of the β-tubulin model-derived atomic surfaces of the whole β-subunits (white), with interfacing regions (black-and-white labels) coloured by element (C, grey; N, blue; O, red); nucleotides are shown as sticks; dotted ovals mark the interface expansion at each step. d, Plotted PDBePISA (www.ebi.ac.uk/pdbe/pisa/) calculations of the solvent-accessible surface areas (SASA) and the dissociation energies (ΔGdiss) of the longitudinal inter-dimer interfaces in different nucleotide states. The error bars represent 3 averaged inter-dimer interfaces generated with parallel refinements of non-crystallographic symmetry (NCS) mates in 6-dimer MT wall models (see Methods).
Uneven compression of α-tubulin reinforces longitudinal lattice contacts
Transitions 1 and 2 strengthen the longitudinal inter-dimer interface (Fig. 2c, d, Supplementary Fig. 4 and Video 3). While the core of the interface is conserved among all states, transition 1 results in additional interactions between αN-terminus and βT2 loop, αT7 loop and helix βH1, and more extended interaction of strand αS9 with helix βH11 (Fig. 2c), burying ~165 Å2 (~10%) more of the solvent accessible surface area (SASA) and increasing the predicted amount of energy needed to dissociate the complex (ΔGdiss) by ~0.8 kcal/mol (~15%) (Fig. 2d). Transition 2 further increases the interface area by ~110 Å2 and ΔGdiss by ~1.1 kcal/mol due to additional interactions of αH3-S4 loop with βS3-H3’ loop and αT7 loop with helices βH1 and βH2 (Fig. 2c, d).
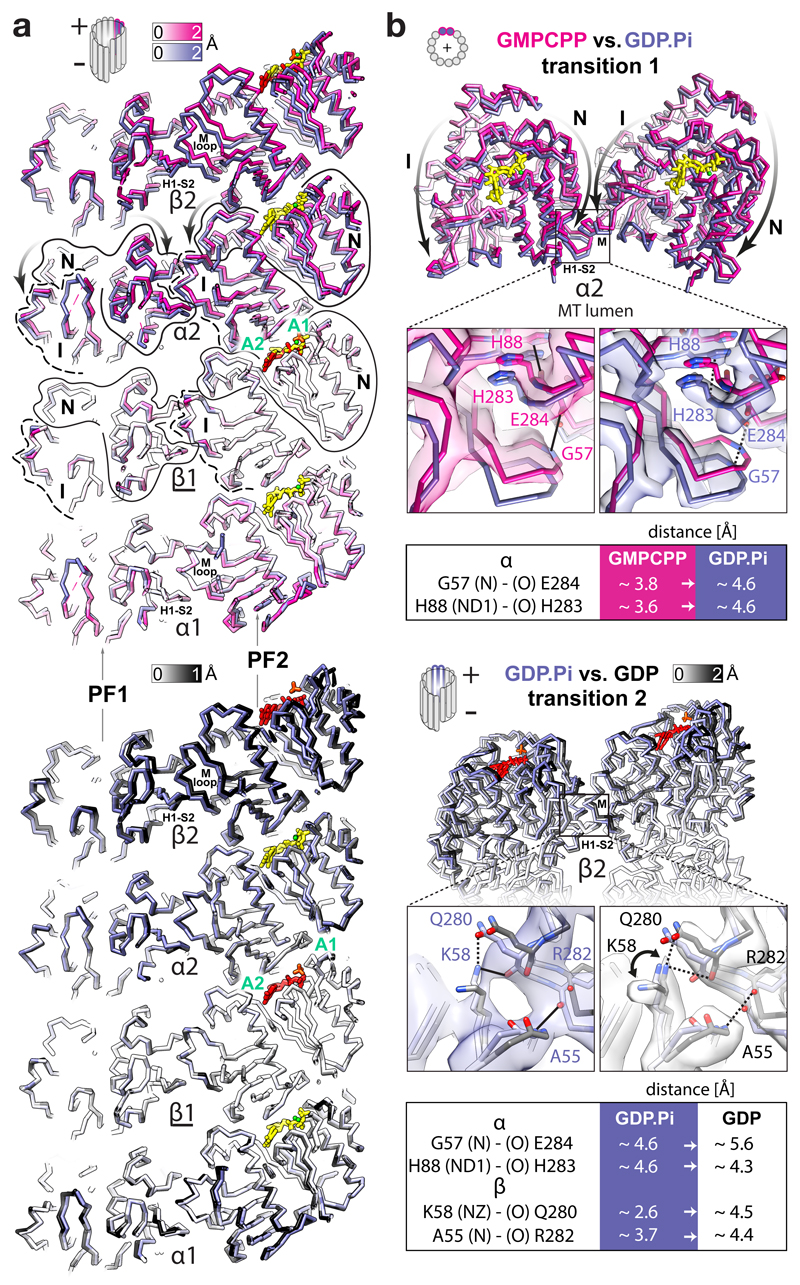
Uneven compression of α-tubulin weakens lateral lattice contacts
It seems paradoxical that the polymer becomes unstable while longitudinal contacts become tighter after lattice compaction. This draws attention to the lateral contacts. The N and I domains are connected in the lattice by both the longitudinal and the lateral interfaces, enabling transmission of structural information between PFs. Due to the uneven compression of the α-subunits during transition 1, the lattice compaction not only tightens the inter-dimer spaces, but also perturbs the lateral contacts; this is because the αN domains move further towards the MT –end and lumen than the laterally connected αI domains from the neighbouring PFs (Fig. 3 and Video 4). Thus, in our GTP state model, the conformation of the lateral contacts between the α-subunits are close enough (< 4 Å) to support multiple points of connectivity (e.g. Gly57-Glu284, His88-His283, His88-Glu284, Glu90-Lys280, Gln128-Gln285). After hydrolysis, two of these distances (the Gly57-Glu284 and His88-His283 pairs) increase > 4 Å as measured in our models of the GDP.Pi and GDP states. For example, αGly57-αGlu284 are ~3.8 Å apart in the GTP state model, ~4.6 Å apart in the GDP.Pi state model (Fig. 3b and Supplementary Fig. 5) and ~5.6 Å apart in the GDP state model. Transition 2 also results in noticeable perturbation of lateral contacts between the β-subunits. Out of several putative connections (< 4 Å) in our models of the GTP and GDP.Pi states (e.g. Ala55-Arg282, Lys58-Gln280, Gln83-Tyr281, Arg86-Tyr281 and Glu125-Lys336), separations of > 4 Å are observed between the βAla55- βArg282 and βLys58-βGln280 pairs in the GDP state model, while density corresponding to the side chain of βLys58 appears distinct in the GDP state reconstruction compared to the GDP.Pi state reconstruction, indicating that this residue is no longer involved in the lateral contact after Pi release (Fig. 3b, Supplementary Fig. 5 and Video 4).
Figure 3. FGTPase-dependent structural transitions weaken the lateral MT lattice contacts.
a, Backbone lumenal cut-away view at lateral contacts between two adjacent PFs in different nucleotide states, aligned and coloured as in Fig. 2a. Tubulin domains are outlined and their transitions around A1 and A2 anchor points are indicated with curved arrows. b, MT +end view of the α2-subunits undergoing the first transition (top) and a lumenal view of the β2-subunits undergoing the second transition; close-up views of selected connections (solid lines) compromised (dotted lines) during the transitions. Atomic models are coloured as in Fig. 2a or by heteroatom and are presented with experimental densities; tables summarise the likely affected lateral bonds according to measurements performed in the models. The interacting atoms are named (parentheses) according to the PDB convention.
DCX-MT lattice skew varies slightly with nucleotide state
The GTPase-dependent lattice transitions also manifest as changes in PF skew. The extended GMPCPP lattice shows a slightly right-handed skew (0.16 ± 0.08 °). Transition 1 straightens PFs (0.00 ± 0.08 °), and transition 2 restores some of the initial skew (0.08 ± 0.07 °) (Supplementary Fig. 6). The direct influence of DCX on this property is not known, but MTs bound by the end binding (EB) protein have a left-handed PF skew10, reaching 0.25 ± 0.01 ° in the GTPγS-EB-MT, whereas our GTPγS-DCX-MT is almost straight (-0.02 ± 0.08 °) (Supplementary Fig. 6 and Supplementary Table 1). Thus, PF skew is influenced by several variables, including the tubulin nucleotide state and bound MAPs, but it is not obvious from all these data that it is intrinsic to the mechanism of dynamic instability.
The role of lateral contacts in MT integrity supported by Taxol® stabilisation mechanism
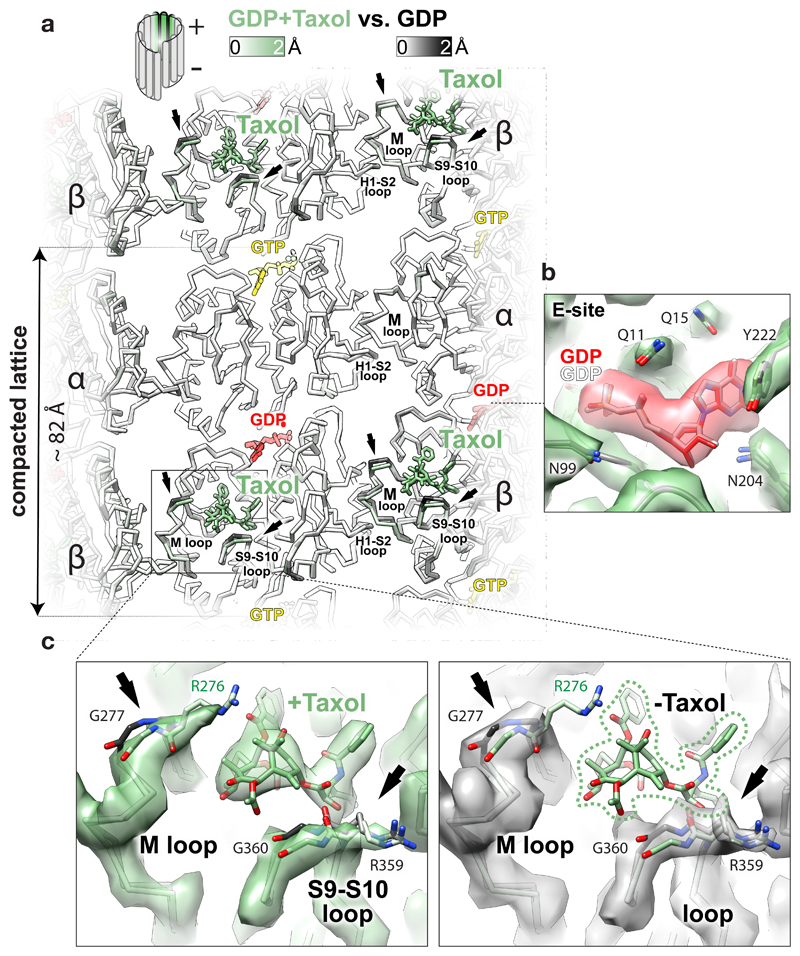
The blockbuster MT-stabilising drug Taxol® binds near the β-subunit lateral contacts6 and inhibits catastrophes. It has been suggested that it stabilises MTs by allosterically counteracting lattice compaction9. We investigated this idea using GDP-DCX-MTs and found that Taxol® binding to these pre-stabilised MTs does not reverse the GTPase-driven conformational changes. Rather, it locally displaces fragments of the βM and the βS9-S10 loops (≤ 1.7 Å) that it directly contacts (Fig. 4). Therefore, an alternative explanation of Taxol®’s MT-stabilising effect is that, while tubulin lateral contacts are loosened by GTP hydrolysis (Fig. 3b), Taxol® locks the βM lateral loops in the lattice-constrained conformation and thereby prevents PF peeling. Such stabilisation of the otherwise labile GDP-MTs may also explain why the drug renders MTs flexible19,20. Since it stabilises every other lateral contact along each PF (assuming saturation), Taxol® probably keeps the PFs only loosely associated within the GDP-MT lattice. Presumably as a consequence, we find MTs pre-stabilised with Taxol® to be poorly decorated with DCX in vitro (Supplementary Fig. 1b). The same is true for cellular MTs21, providing further evidence for looseness of the Taxol®-stabilised lattice, which DCX can override if it binds first.
Figure 4. Effect of Taxol® binding to GDP-DCX-MT.
a, Lumenal backbone view of GDP-DCX-MT structure with Taxol® superposed on the GDP-DCX-MT structure without Taxol®, coloured by the degree of displacement or as follows: Taxol®, green sticks; GTP, yellow sticks; GDP, red sticks. Regions showing the largest displacement are limited to loops directly contacting the drug (arrows). b, Unchanged βE-site before (grey ribbon and the nucleotide) and after (green ribbon and red/heteroatom coloured nucleotide) addition of Taxol®, shown in the density of the MT lattice containing Taxol®; selected residues are shown as sticks coloured by heteroatom. c, Close-up on Taxol® binding site in GDP-DCX-MT reconstruction with Taxol® (green map) and without Taxol® (grey map), focusing on local structural changes. The models are coloured as in a; the most displaced regions are shown as sticks where density is present, and pointed with arrows. Reconstruction without Taxol® has weaker density for R276 side chain (not visible at this density threshold), suggesting that Taxol® somewhat orders this residue in the Taxol®-bound MT. Taxol®’s close interaction with the βM loop – a main contributor to MT lateral contacts - explains how the lateral contacts are locked to promote MT lattice stability upon Taxol® binding.
Discussion
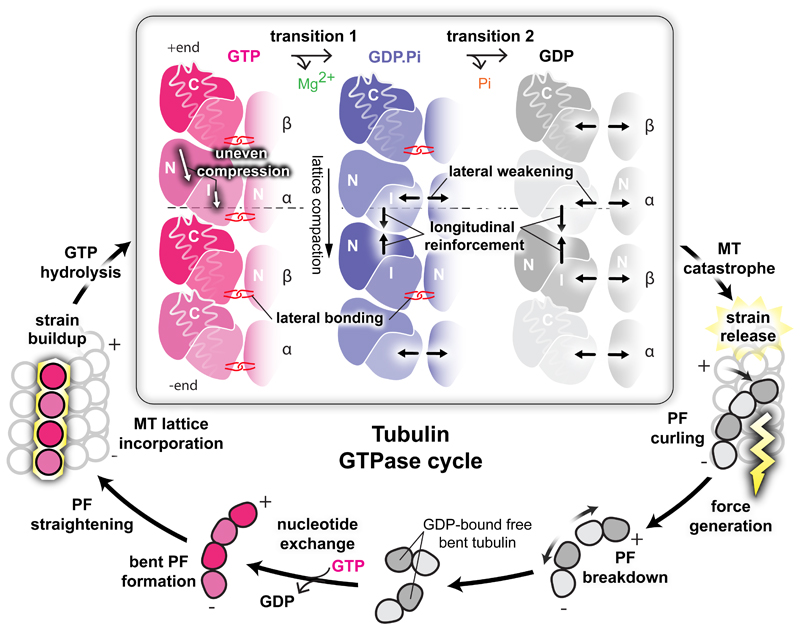
Our data suggest that a portion of GTP hydrolysis energy is absorbed by the lattice, triggering a 2-step loosening of its lateral contacts, first between the α-subunits (transition 1) and then between the β-subunits (transition 2), which coincides with strengthening of inter-dimer longitudinal interfaces (Fig. 5). This explains why whole PFs peel away from MTs after rapid induction of catastrophe1. Our data suggest that catastrophe happens because the lateral contacts become too weak to counteract the intrinsic strain of the straight tubulin dimers. The role of homotypic lateral contacts in dynamic instability has been discounted in previous studies9,10, but in comparing MTs stabilised by a single protein ligand (DCX) and in capturing the bona fide GDP.Pi state of the tubulin GTPase, we have identified their likely crucial role in contributing to MT dynamics. Very small differences in the energetics of lateral contact formation due to isoform-specific sequences could be an important mechanism for regulating MT dynamics in particular physiological settings22. As tubulin relaxes (bends), the disassembling PFs curl outward generating force3,23. The developing curvature in turn weakens the inter-dimer longitudinal contacts (Supplementary Fig. 7a-c), allowing complete PF breakdown. So, the fine-tuned MT dynamics seem to only require perturbation of a few connections per dimer to tip the balance towards MT catastrophe. In our GDP-DCX-MT, catastrophe is prevented by DCX stabilisation.
Figure 5. Mechanisms and implications of MT catastrophe and the tubulin GTPase cycle.
The specific area of this work (framed) is presented together with other stages of GTPase cycle discussed in the paper. Free GDP-bound tubulin exchanges E-site GDP to GTP and starts MT nucleation by assembling bent PFs, which gradually straighten as MT lattice assembly progresses. Completion of MT lattice leads to strain accumulation by imposing an unfavourable straight conformation on tubulin. GTP-hydrolysis enables release of this strain in stages, which lead to MT catastrophe: during the first transition, the α-subunits undergo uneven compression, weakening lateral, while tightening longitudinal lattice contacts. The second transition further tightens longitudinal contacts and weakens the lateral contacts between β-subunits. The strain overcomes the looser lateral contacts causing outward peeling of PFs and force generation (lightening). The developing curvature eventually breaks the longitudinal inter-dimer contacts, liberating tubulin to allow nucleotide exchange and resumption of the cycle.
These results explain why only GTP- or GMPCPP-tubulin spontaneously nucleate MTs, since only the extended lattice forms favourable lateral contacts, apparently balancing the energetic cost of tubulin straightening. To counteract tubulin bending, the longitudinal contact is also larger in the lattice (~1533 Å2) compared to X-ray structures of curved PFs (832-1026 Å2)24–28 (Supplementary Fig. 7c). Only this lattice-imposed straight conformation brings the catalytic αE254 residue sufficiently close to the E-site GTP to complete the GTPase machinery (Supplementary Fig. 7d).
The specific flexibility of Taxol®-MTs makes sense when the role of lateral contacts in dynamic instability is taken into account. Thus, where Taxol® is added to MTs after polymerisation has been allowed to proceed, our work suggests that the lateral contacts in these MTs would be loose due to the majority of tubulin being bound to GDP. Taxol® works by holding the β-tubulin lateral contacts in place, but the entirety of the lattice is presumably rather flexible, explaining the biophysical properties of these MTs29 and the documented knock-on effects on binding by microtubule regulators, including EBs and DCX21,30. Further, in the case of experiments where Taxol® is added at polymerisation initiation, it seems likely that its binding may alter or trap the GTPase-linked structural transitions in some more complex, time-dependent and structurally heterogeneous way. A previous study9 proposed that Taxol® prevents or reverses GTPase-dependent MT lattice compaction, and that MT stabilisation by the drug relies on preservation of the extended (GTP-like) lattice state. In our study and in another recent publication19, Taxol® binding was found to not induce lattice expansion. Similarly, other MT stabilising agents may be expected to also work by stabilizing lateral contacts in lattice-like conformations31.
The stochastic nature of GTP hydrolysis events in the lattice4 would be predicted to cause lateral contact mismatches between neighbouring pre- and post-hydrolysis dimers. These by themselves could cause tensions in the lattice, destabilising it and/or synchronising hydrolysis, presumably contributing to the multi-step nature of catastrophe32. The MT seam is an inbuilt source of post-hydrolysis lateral mismatch10, but not the only source of MT instability, because MTs do not simply unzip at the seam - rather their individual PFs depolymerise completely. Our analysis shows that the irreversible lattice transitions cause a global MT catastrophe, liberating tubulin to exchange its nucleotide and start the cycle again.
The intrinsic polarity of the tubulin dimers themselves2 and of the conformational changes we have described here (Figs 2, 3 and 5), likely contribute to the intrinsic but mechanistically poorly understood differences in MT dynamics at the MT + and -ends. Our data highlight the conformational plasticity of α-tubulin at the longitudinal tubulin interface. If this conformational plasticity is recapitulated at the MT -end, we predict that this would provide a previously unanticipated conformational barrier to stable addition of the incoming tubulin dimers. At the +end, in contrast, while we predict that α-tubulin retains its conformational plasticity in the incoming free tubulin dimers, the polymer-held β-tubulin would be predicted to provide a more stable platform/template for dimer docking, a stability that is reflected in faster +end growth.
With a multitude of MAPs, polymerases, depolymerases and modifying enzymes in vivo, MT dynamics are unlikely to be governed by GTP hydrolysis alone, but by a complex network of regulators. For example, recent work has indicated the importance of acetylation around the lateral contacts as cellular means to prolong MT longevity33,34; this reinforces our conclusions about their role in MT stability. Here we show that the majority of MTs - built from GDP.Pi or GDP tubulin - are structurally poised for immediate catastrophe and force generation. However, overriding cellular regulators can ensure that tubulin strain energy remains stored in the lattice until required, prompted by cell physiology.
Methods
Protein preparation
Human doublecortin isoform 2 (DCX, residues 1-360) was cloned into pNic28Bsa4 vector (Structural Genomics Consortium, Oxford, UK), which adds a tobacco etch virus (TEV) protease-cleavable N-terminal His tag to the protein. After expression in BL21 Star (DE3) E. coli cells (Invitrogen), the cells were sonicated in the lysis buffer (50 mM Na2HPO4 pH 7.2, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 2 mM DTT) supplemented with protease inhibitor cocktail (cOmplete Cocktail Tablet, Roche/Sigma Aldrich), and the lysates were clarified by centrifugation. Clear lysates were passed through nickel HisTrap HP column (GE Healthcare) and His-DCX was eluted with 10-250 mM imidazole gradient. To remove the His tag from the DCX protein, we used a His-tagged TEV protease expressed in-house and then removed both His-TEV and the cleaved tag by a passage through nickel beads (GE Healthcare). DCX was then captured on a HiTrap SP HP ion exchange column (GE Healthcare) equilibrated in BRB80 buffer (80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] pH 6.8, 1 mM EGTA [ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid], 1 mM MgCl2, 1 mM DTT [dithiotreitol]) and eluted with NaCl gradient (15-300 mM). DCX was then finally purified and desalted by gel filtration through Superdex 200 size exclusion column (GE Healthcare) equilibrated in the BRB80 buffer.
Lyophilised bovine brain tubulin was purchased from Cytoskeleton and reconstituted to 100 μM concentration in BRB80 supplemented with either 1 mM GTP for dynamic MTs (GTP-tubulin) or one of the nucleotide analogues: GMPCPP [Guanosine-5'-[(α,β)-methyleno]triphosphate] (Jena Biosciences) (GMPCPP-tubulin) or GTPγS [Guanosine 5'-O-(3-thiotriphosphate)] (Roche) (GTPγS-tubulin).
Preparation of cryo-EM samples
To obtain GMPCPP-DCX-MTs with high GMPCPP occupancy, 30 μM GMPCPP-tubulin was cycled twice through: 30 min polymerization at 37 °C in BRB80 buffer containing 1 mM GMPCPP, pelleting and depolymerisation in cold buffer. In the third polymerisation round, 5 μM GMPCPP-tubulin was co-polymerised with 3.5 μM DCX under the same conditions. This polymerisation strategy was required to obtain a fully extended 13-PF MT lattice.
GTPγS does not nucleate MTs, hence it cannot be enriched in the lattice through tubulin polymerisation and depolymerisation cycles and its occupancy is not known. For GTPγS-DCX-MTs 5 μM GTPγS-tubulin was co-polymerised for 30 min with 3.5 μM DCX at 37 °C in BRB80 buffer containing 2 mM GTPγS.
For GDP-DCX-MTs 5 μM GTP-tubulin was co-polymerised for 30 min with 3.5 μM DCX at 37 °C in BRB80 buffer containing 1 mM GTP. For GDP-DCX-Taxol®-MTs 1 mM Taxol® was added after 30 min of polymerisation and the sample was incubated for further 30 min at 37 °C.
In all the above MT preparations, sub-stoichiometric concentration of DCX versus tubulin was used to nucleate MTs with a desired 13-PF architecture, while keeping MT bundling minimal. These MTs were applied directly to glow-discharged Lacey grids (Agar) and incubated for 30 sec at room temperature. Then the grids were briefly blotted and 50 μM DCX solution in BRB80 buffer was added to maximise MT decoration with DCX. The grids were then transferred to Vitrobot (FEI/Thermo Fisher Scientific) and incubated there for 1 min at 30 °C and 95 % humidity, before finally blotting and plunge freezing in liquid ethane.
For GDP.Pi-DCX-MTs 10 μM GTP-tubulin was mixed with 50 μM DCX in cold BRB80 buffer containing 1 mM GTP and immediately applied to a glow-discharged Lacey grid for rapid polymerisation directly on the grid inside the Vitrobot set to 30 °C and 95 % humidity. After 30 sec incubation the grid was blotted and vitrified as before.
Cryo-EM data collection
Micrographs were acquired on a 300 kV Polara microscope (FEI) combined with a K2 Summit camera (Gatan) operated in counting mode after energy filter with a 20 eV slit. The magnification at the specimen plane was 35,971x resulting in a pixel size of 1.39 Å. The dose rate was ~5 e-/pixel/sec, corresponding to ~2.6 e-/Å2/sec. The total dose on the specimen was ~23.4 e- collected over 9 sec exposures fractionated into 36 movie frames (0.25 sec/frame). We used SerialEM software (http://bio3d.colorado.edu/SerialEM/) to manually collect exposures with -0.4 to -2.5 μm defocus range.
Image processing and 3D reconstruction
We used MotionCor236 to globally and locally (25 tiles/image) align movie frames. Using EMAN 1 Boxer37 we picked MT segments from these drift-corrected image sums. Boxes of 652 x 652 pixel size spanned ~11 tubulin dimers and were cut along MTs with ~8 dimer overlap. These segments were subsequently treated as single particle input to Chuff38,39, a custom-designed multi-script processing pipeline using Spider40 and Frealign41. The initial seam finding alignment was done in Spider by projection matching to a synthetic 13-PF DCX-MT reference filtered to 30 Å. The contrast transfer function (CTF) parameters were estimated with CTFFIND342, and the CTF correction was performed during local refinement within Frealign, producing isotropic 3D reconstructions with pseudo-helical symmetry applied 12 times. Independently processed half maps were combined in Relion 1.443 and subjected to its standard post-processing routine, involving: (1) estimation of map resolution based on Fourier Shell Correlation (FSC) between the two half maps, (2) computation of the average B-factor based on Guinier plot using the EMBfactor program44, and (3) map sharpening using the computed B-factor value. The resolutions of the final maps were estimated using 0.143 FSC cut-off criterion and the absence of over-fitting was confirmed with high-resolution noise substitution test35 (Table 1, FSCtrue). Side chains of acidic residues are mostly missing, likely due to their exceptional susceptibility to radiation damage, unless they are stabilised by an interaction (e.g. D177 H-bonding with GMPCPP, Fig. 1c). Reconstructions using data collected with the first 4 e- did not recover these vulnerable side chains.
Atomic model refinement
We used a high-resolution cryo-EM 6 GDP-tubulin dimer model (PDB code: 3JAR10, devoid of EB3 chains) as a starting point for refinement in all of our cryo-EM density maps. Nucleotides were substituted as necessary with structures downloaded from the Grade Server (http://grade.globalphasing.org/cgi-bin/grade/server.cgi) and DCX density was masked away and excluded from the refinements by zoning maps around tubulin structures in UCSF Chimera45. Each isolated map was placed in a new unit cell with P1 space group. Ten macro-cycles of refinements in real space were carried out at each round using phenix.real_space_refine (http://phenix-online.org/) with default settings (Ramachandran plot, C-beta deviations, rotamer and secondary structure restraints). Non-crystallographic symmetry (NCS) group definitions were manually provided as constraints for the related tubulin chains. The program automatically determined weight between data and the restraints to achieve RMS deviations for covalent bonds not greater then 0.01, and for angles not greater than 1.0. Model geometry was evaluated by MolProbity46 after each round of refinement, and problematic regions in the models were manually corrected in Coot47. This process was repeated for every structure until satisfactory level of model:map agreement with excellent model stereochemistry were accomplished (Table 1).
Estimation of PF skew in different MT lattices
We calculated the average difference between the assigned ϕ-angles (rotation angle around MT axis) of the consecutive DCX-MT segments cut along individual MTs in each lattice state. These individual values of skew were then averaged over > 50 MTs per lattice state (Supplementary Fig. 6). To determine statistical significance between the means we used one-way analysis of statistical variance (ANOVA), since standard deviations across all datasets were not significantly different by both Brow-Forsythe and Bartlett’s tests, according to Prism 6 (graphpad.com). The degree of freedom within each group (residual) amounted to 355 and between the groups to 4, resulting in the F ratio of 68.67 and the significance level (P value) of < 0.0001.
Estimation of DCX occupancy in different MT lattices
A Fourier-transformed image of any DCX-MT shows a ~1/4 nm layer line corresponding to ~4 nm tubulin subunit repeat and an ~1/8 nm layer line corresponding to ~8 nm DCX repeat. To estimate the relative DCX occupancy in each lattice state we averaged power spectra of all the DMX-MT segments in a given state using EMAN37 and calculated intensities of the 1/8 nm layer lines in relation to the 1/4 nm layer lines. To calculate the error of that ratio for each lattice state we divided each dataset into 3 approximately equal subsets and calculated the average ratio and its standard deviation (Supplementary Fig. 1b).
Figure and video preparation
Molecular visualisations in all figures and videos (Supplementary Information) were prepared using UCSF Chimera45.
Data availability
Our maps and coordinates were deposited in EMDB/PDB with the following accession codes: GMPCPP-DCX-MT, EMD-3961 and PDB ID 6EVW; GDP.Pi-DCX-MT, EMD-3962 and PDB ID 6EVX; GTPγS-DCX-MT, EMD-3963 and PDB ID 6EVY; GDP-DCX-MT, EMD-3964 and PDB ID 6EVZ; GDP-DCX-Taxol®-MT, EMD-3965 and PDB ID 6EW0.
Supplementary Material
Acknowledgments
S.W.M. and C.A.M. are supported by the Medical Research Council, U.K (MR/J000973/1 and MR/R000352/1). We thank A. Roberts and M. Steinmetz for invaluable discussions about this work.
Footnotes
Accession Codes
Our maps and coordinates were deposited in EMDB/PDB with the following accession codes: GMPCPP-DCX-MT, EMD-3961 and PDB ID 6EVW; GDP.Pi-DCX-MT, EMD-3962 and PDB ID 6EVX; GTPγS-DCX-MT, EMD-3963 and PDB ID 6EVY; GDP-DCX-MT, EMD-3964 and PDB ID 6EVZ; GDP-DCX-Taxol®-MT, EMD-3965 and PDB ID 6EW0.
Author Contributions
S.W.M. conceived experimental strategies, designed and carried out experiments and computations, processed data, interpreted results and wrote the manuscript; C.A.M. proposed and supervised research, interpreted results and wrote the manuscript.
Competing Financial Interest Statement
Competing interests. None to declare.
References
- 1.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci U S A. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driver JW, Geyer EA, Bailey ME, Rice LM, Asbury CL. Direct measurement of conformational strain energy in protofilaments curling outward from disassembling microtubule tips. eLife. 2017;6 doi: 10.7554/eLife.28433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 5.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 7.Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 8.Guesdon A, et al. EB1 interacts with outwardly curved and straight regions of the microtubule lattice. Nat Cell Biol. 2016;18:1102–1108. doi: 10.1038/ncb3412. [DOI] [PubMed] [Google Scholar]
- 9.Alushin GM, et al. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vale RD, Coppin CM, Malik F, Kull FJ, Milligan RA. Tubulin GTP hydrolysis influences the structure, mechanical properties, and kinesin-driven transport of microtubules. J Biol Chem. 1994;269:23769–23775. [PubMed] [Google Scholar]
- 12.Hyman AA, Chrétien D, Arnal I, Wade RH. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J Cell Biol. 1995;128:117–125. doi: 10.1083/jcb.128.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci U S A. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer SP, et al. EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr Biol CB. 2014;24:372–384. doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moores CA, et al. Mechanism of microtubule stabilization by doublecortin. Mol Cell. 2004;14:833–839. doi: 10.1016/j.molcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Fourniol FJ, et al. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 A resolution. J Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellogg EH, et al. Insights into the Distinct Mechanisms of Action of Taxane and Non-Taxane Microtubule Stabilizers from Cryo-EM Structures. J Mol Biol. 2017;429:633–646. doi: 10.1016/j.jmb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikumoto M, Kurachi M, Tosa V, Tashiro H. Flexural rigidity of individual microtubules measured by a buckling force with optical traps. Biophys J. 2006;90:1687–1696. doi: 10.1529/biophysj.104.055483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ettinger A, van Haren J, Ribeiro SA, Wittmann T. Doublecortin Is Excluded from Growing Microtubule Ends and Recognizes the GDP-Microtubule Lattice. Curr Biol CB. 2016;26:1549–1555. doi: 10.1016/j.cub.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vemu A, Atherton J, Spector JO, Moores CA, Roll-Mecak A. Tubulin isoform composition tunes microtubule dynamics. Mol Biol Cell. 2017;28:3564–3572. doi: 10.1091/mbc.E17-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zakharov P, et al. Molecular and Mechanical Causes of Microtubule Catastrophe and Aging. Biophys J. 2015;109:2574–2591. doi: 10.1016/j.bpj.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prota AE, et al. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules. Mol Pharmacol. 2016;89:233–242. doi: 10.1124/mol.115.100149. [DOI] [PubMed] [Google Scholar]
- 26.Prota AE, et al. Pironetin Binds Covalently to αCys316 and Perturbs a Major Loop and Helix of α-Tubulin to Inhibit Microtubule Formation. J Mol Biol. 2016;428:2981–2988. doi: 10.1016/j.jmb.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Doodhi H, et al. Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes. Curr Biol CB. 2016;26:1713–1721. doi: 10.1016/j.cub.2016.04.053. [DOI] [PubMed] [Google Scholar]
- 28.Nawrotek A, Knossow M, Gigant B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J Mol Biol. 2011;412:35–42. doi: 10.1016/j.jmb.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 29.Mickey B, Howard J. Rigidity of microtubules is increased by stabilizing agents. J Cell Biol. 1995;130:909–917. doi: 10.1083/jcb.130.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bechstedt S, Lu K, Brouhard GJ. Doublecortin recognizes the longitudinal curvature of the microtubule end and lattice. Curr Biol CB. 2014;24:2366–2375. doi: 10.1016/j.cub.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 31.Field JJ, Díaz JF, Miller JH. The Binding Sites of Microtubule-Stabilizing Agents. Chem Biol. 2013;20:301–315. doi: 10.1016/j.chembiol.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 33.Xu Z, et al. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science. 2017;356:328–332. doi: 10.1126/science.aai8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portran D, Schaedel L, Xu Z, Théry M, Nachury MV. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol. 2017;19:391–398. doi: 10.1038/ncb3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 38.Sindelar CV, Downing KH. The beginning of kinesin’s force-generating cycle visualized at 9-A resolution. J Cell Biol. 2007;177:377–385. doi: 10.1083/jcb.200612090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fourniol FJ, et al. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 A resolution. J Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 41.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 43.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 46.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our maps and coordinates were deposited in EMDB/PDB with the following accession codes: GMPCPP-DCX-MT, EMD-3961 and PDB ID 6EVW; GDP.Pi-DCX-MT, EMD-3962 and PDB ID 6EVX; GTPγS-DCX-MT, EMD-3963 and PDB ID 6EVY; GDP-DCX-MT, EMD-3964 and PDB ID 6EVZ; GDP-DCX-Taxol®-MT, EMD-3965 and PDB ID 6EW0.