Abstract
The low frequency of HIV-1 recombinants within entire viral populations in both individual patients and in culture-based infection models impedes investigation of the underlying factors contributing either to the occurrence of recombinants or the survival of recombinants once they are formed. So far, most of the related studies have no consideration of recombinants’ functionality. Here we established a Functional Recombinant Production (FRP) system to produce pure and functional HIV-1 intersubtype Env recombinants, and utilized 454 pyrosequencing to investigate the distribution of over 4,000 functional and non-functional recombination breakpoints from either the FRP system or dual infection cultures. The results revealed that most of the breakpoints converged in gp41 (62%) and C1 (25.3%) domains of gp120, which has strong correlation with the similarity between the two recombining sequences. Yet, the breakpoints also appeared in C2 (5.2%) and C5 (4.6%) domains not correlated with the recombining sequence similarity. Interestingly, none of the intersubtype gp120 recombinants recombined between C1 and gp41 regions either from the FRP system or from the dual infection culture, and very few from the HIV epidemic, were functional. The present study suggests that the selection of functional Env recombinants is one of the reasons for the predominance of C1 and gp41 Env recombinants in the HIV epidemic, and provides an in vitro model to mimic selection of replication-competent HIV-1 intersubtype recombination in dual or superinfected patients.
Keywords: Human immunodeficiency virus type 1, Intersubtype recombination, Functional envelope recombinant
Graphical Abstract
Introduction
Recombination between two genetically distinct isolates of the same retrovirus species was first described in the 1970s [1,2]. Co-infection of a susceptible cell by two HIV-1 strains can produce heterodiploid HIV-1 particles [2]. Upon subsequent de novo infection by this heterodiploid virus, reverse transcriptase (RT) can jump between the two genomes during proviral DNA synthesis and create a chimeric HIV-1 genome [3]. During the course of infection, recombination is a major mechanism driving intrapatient HIV-1 evolution accomplished by shuffling reverse transcription derived point mutations between different HIV-1 genomes. HIV-1 recombination can also result in major antigenic shifts following dual/superinfections in individuals with different HIV-1 isolates and as such, is also referred to as a primitive form of sexual reproduction [4,5]. In some instances, recombination following dual/superinfections can rescue lethally mutated genomes generated from the high mutation rate [6-8] or can result in rapid host adaptation, escape from the elicited immune responses, and/or facilitate resistance to antiretroviral drugs consequently leading to faster disease progression in infected individuals [4]. Thus, intersubtype recombination is a major driving force for HIV evolution and is changing the global epidemiology of HIV-1 with circulating recombinant forms (CRFs) dominating regional epidemics and a number of unique recombinant forms (URFs) reported each year [9-18].
The HIV-1 env gene encodes the viral Env glycoprotein which mediates virus entry into a host cell [19]. Because the Env glycoprotein is exposed on the viral surface [20-22], the virus has to constantly change its Env sequence through mutation and/or recombination to avoid recognition by the host’s immune system. High rates of virus production and cell co-infections, continuous nucleotide substitutions, and subsequent inter-genomic recombination are ultimately responsible for tens of thousands of unique, replicating HIV-1 clones within an intrapatient HIV-1 population [23,24]. Recombination in the env gene can combine compensatory mutations to improve replicative fitness with mutations of reduced infectivity but necessary for e.g. immune evasion, a switch of co-receptor usage, or development of resistance to HIV entry inhibitors [25,26]. How HIV-1 recombines its env gene leading to these large genetic/antigenic shifts remains poorly understood.
Through intra- and HIV-1 intersubtype recombination, we recently showed that structural/functional relationships in the Env glycoprotein are not defined by the linear amino acid sequence, and as a result, escape from humoral or cell-mediated immune response may not be easily predicted [27]. The conserved domain 1 (C1) and C5/gp41 of Env, separated by ~337 amino acids, have co-evolved together based on their close structural/functional relations, whereas linear arrangement of V1/V2-C2-V3-C3-V4-C4-V5 (V1-to-V5) Env show reduced linkage and co-evolution [27]. Thus, intersubtype recombination within the V1-to-V5 region typically results in non-functional Env glycoprotein and “dead” virus [28]. However, even double recombination within the V1-to-V5 region between HIV-1 subtype strains can produce replication-competent virus if the C1 and C5/gp41 sequences are retained from the same HIV-1 strain. Our understanding of survival and propagation of HIV-1 intersubtype recombinants within susceptible cells, within infected individuals, and throughout the global population requires studies on (1) the mechanical process of retroviral reverse transcriptase jumping between the RNA templates, and (2) the role of HIV-1 sequence diversity in breakpoint selection, in relation to function, and the beneficial outcome of these processes on HIV-1 survival.
Many studies have characterized mechanisms and factors underlying HIV-1 intersubtype recombination, and how this impacts the env gene. For example, increased recombination frequency at 5’ and 3’ ends of the env gene, regions corresponding to the gp120 C1 and gp41, respectively, may be related to nucleotide and amino acid conservation [28-34]. However, with the exception of the study by Simon-Loriere et al. [28], functionality of the Env glycoproteins derived from in vitro recombination events has not been thoroughly investigated. Host cell entry by the Env glycoproteins is maintained despite high genetic variability in this gene (>20% amino acid diversity). However, the majority of intersubtype recombination events still result in non-functional glycoproteins due to breakpoints resulting in frameshifts, changes in codons causing deleterious point mutations, disruptions in the splice donors/acceptor sites (of overlapping Tat/Rev regulatory genes), and/or incompatibility of chimeric Env domains, as well as damage of genome secondary structure [35-37]. Until the advent of next generation sequencing (NGS), we could not access the complete frequency and sites of intersubtype recombination in HIV-1 (or any other retrovirus). For this study, we successfully established a unique Functional Recombinant Production (FRP) system to generate only functional HIV-1 intersubtype Env recombinants. By combining our novel FRP system with our in-house developed NGS assay, we were able to systemically investigate the distribution of functional and nonfunctional HIV-1 intersubtype Env recombinants from the FRP system and from an in vitro dual-infection system. Collectively this enabled us to reveal novel information into the possible mechanism behind why certain HIV-1 Env recombinants could pre-dominate in the HIV-1 epidemic.
Results
Construction of an HIV-1 Env Functional Recombinant Production (FRP) system producing functional HIV-1 intersubtype Env recombinants
We have previously utilized a dual infection method with or without HIV-1 isolate-specific siRNAs to generate HIV-1 intersubtype Env recombinants, but found significant limitations using that technique. With the dual infection system in the absence of siRNAs, less than 5% of the replicating virus were HIV-1 intersubtype recombinants and the parental viruses dominated the virus population [31]. HIV-isolate specific siRNA effectively inhibited parental viruses in a dual infection and enriched for HIV-1 intersubtype env recombinants. Over 70% of the siRNA-resistant virus had breakpoints in the env gene and between the siRNA target sites. However, siRNA inhibition was not complete and replication of parental viruses led to the rapid emergence of siRNA-resistant HIV-1 isolates with mutations in the siRNA target sequence [32]. In the present study, we established an HIV-1 Functional Recombinant Production (FRP) system to produce, enrich and screen for only functional HIV-1 intersubtype Env recombinants.
In order to select for the recombined HIV-1 with breakpoints in the env gene, we introduced two env genes from different HIV-1 isolates into two defective HIV-1 genome-containing vectors. In our previous study [38], we established an HIV-1 production system (pREC_nfl_HIV-1 and pCMV_cplt) which involved the production of two subgenomic RNA (sgRNA) segments that could complement each other to complete the reverse transcription of wild type proviral HIV-1 DNA. The sgRNAs were produced by co-transfections with the two complementary DNA vectors, one (pREC_nfl_NL4-3) of which transcribes a near full length (nfl) HIV-1 genome lacking a 5’LTR. The resultingnfl sgRNA and spliced mRNAs are also translated to produce the full HIV-1 proteome. The second complementary vector, pCMV_cplt produces a 5’LTR sgRNA and partial gag sequence, lacks the 3’LTR, the Gag AUG, pol, and env sequences. Again, the combination of both sgRNAs in a virus particle can support complete revere transcription leading to wild type HIV-1 propagation.
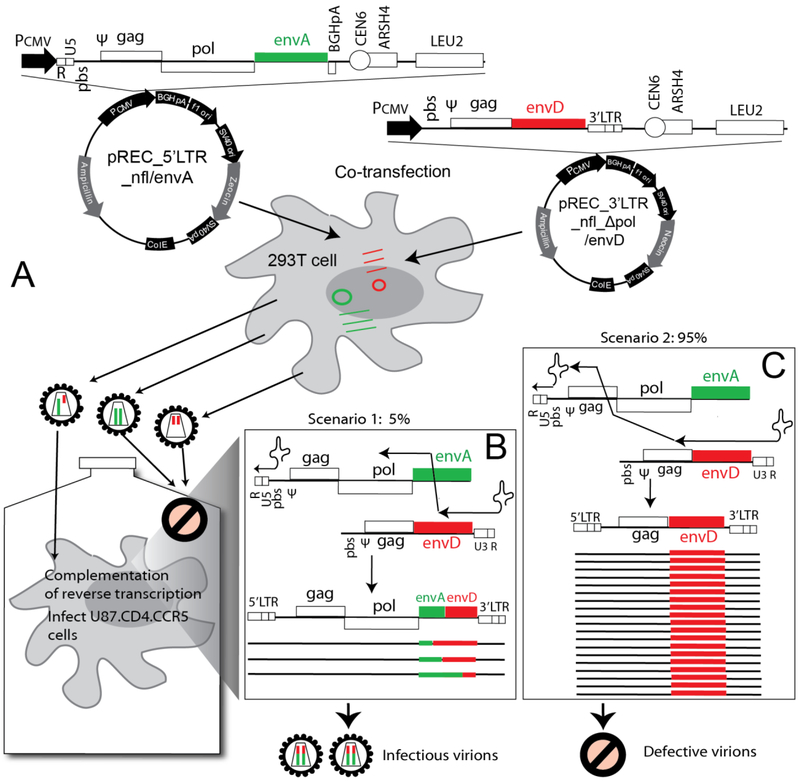
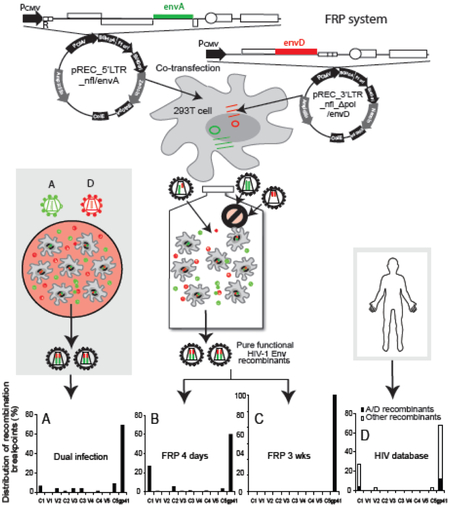
For this study, we designed a similar system, but using a newly constructed pREC_5’LTR_nfl vector which contained a near full length HIV-1 genome and a SV40 poly(A) sequence in replacement of the 3’LTR (Fig. 1A). This new 5’LTR vector now has stabilized HIV-1 mRNA species for the translation of the full HIV proteome. As described in Material and Methods, we then modified the pREC_nfl_3’LTR_HIV by deleting the pol coding region to generate the vector pREC_nfl_Δpol _3’LTR. When transfected with either plasmid alone, HEK-293T cells did not yield replication-competent virus in the supernatant (data not shown). When the two vectors were co-transfected, virus particles were produced harboring homo- or heterodimers of the two sgRNAs. We observed equivalent packaging of both sgRNAs due to the presence of the Ψ and GRPE sequences in both [39]. Upon de novo infection, the homodimeric viruses cannot complete reverse transcription and replicate. Reverse transcription in the heterodimeric virus can produce wild type, integration-competent proviral DNA with two LTRs but only if the (−) strand DNA elongating on the nfl_Δpol_3’LTR jumps to the 5’LTR_nfl within the env gene (Fig. 1B). It is important to note that our results show that the (−) strand DNAs copied on the nfl_Δpol_3’LTR sgRNA results in dead end, defective proviral DNAs lacking the 5’LTR, thus preventing integration and lacking intact Gag or Env ORFs (and deleted Pol) preventing new viral protein production. Based on previous studies [38], we predict that 10~20% of (−) strand DNA will jump to the 5’LTR_nfl sgRNA template within the env or gag genes. A template switch in gag would produce a proviral DNA with both LTRs but lack the pol gene (Fig. 1C). Thus, the lack of integrase would again prevent integration. A template switch within the env gene would result in a fully intact proviral DNA capable of host cell integration, a template for all HIV-1 mRNA species, a production of the full proteome, and new virus progeny capable of propagation (Fig. 1B). The ultimate proof for the FRP system was the emergence of replicating virus in U87.CD4.CCR5 or CXCR4 cells when infecting with the supernatant of HEK-293T cells co-transfected with the two NL4-3-based vectors.
Fig. 1.
Schematic for HIV-1 Env Functional Recombination Production (FRP) system. (A) pREC_5’LTR_nfl_envA (aka 5’LTR_envA) is the major plasmid containing all genes except the 3’LTR sequence, and expresses all of the viral proteins. The pREC__nfl_Δpol_3’LTR_envD (aka 3’LTR_envD) is a helper plasmid containing a truncated HIV-1 genome devoid of the 5’LTR and pol sequences. The HIV-1 strain NL4-3 env genes in the two plasmids have been replaced by a subtype A and D env sequences respectively. (B) and (C) Illustration of how intersubtype recombination occurs using the HIV-1 Env FRP system. The two defective HIV-1 genomes, if co-packaged, can complement each other to initiate and complete reverse transcription, however infectious complete genomes will only result when recombination occurs within the env region (B), while recombination in gag region will result in defective viral genome and noninfectious viruses (C).
In order to utilize the FRP system to generate HIV-1 intersubtype Env recombinants, yeast-based cloning was employed to replace the isogenic NL4-3 env genes in these two vectors with the env genes of HIV-1 isolates from different HIV-1 subtype classification, i.e. A91, A115, and D109. To test if this recombination system with different env genes produces infectious virus, we co-transfected into HEK-293T cells with the pREC_5’LTR_nfl_envA91 (aka 5’LTR_envA91) and pREC_nfl_Δpol_3’LTR_envD109 (aka 3’LTR_envD109) plasmids and recovered cell-free supernatant at 48 h with RT activity suggesting virus production. Again, the 5’LTR_envA91 plasmid contained the near full length HIV-1 genome (except the 3’LTR) driven by CMV promotor, thus could transcribe all of the HIV-1 mRNA species and produce the full HIV-1 proteome for new virus production. The cell-free HEK-293T transfection supernatant with RT activity was used to infect HIV-1 susceptible U87.CD4.CCR5 cells (Fig.1A). The de novo virus production in the U87.CD4.CCR5 cultures was detectable by RT assay by day 12 post-infection, which progressively increased and peaked after 14 days post-infection (Fig. 2). These findings suggested (1) that heterodiploid viruses were produced in the 293T cells, (2) that upon de novo infection of U87 cells, the heterodiploid virus supported reverse transcription of proviral DNA, (3) that this proviral DNA must have a recombination site producing a chimeric env gene, and finally (4) that this chimeric proviral DNA led to infectious HIV-1 propagation. To confirm points (3) and (4), the viral RNA was extracted from the supernatant of U87 cell cultures at peak viremia and then subject to RT-PCR amplification to obtain the env genes for clonal sequencing. All of 54 env clones from this U87 supernatant were A91/D109 recombinants with recombination breakpoints located primarily in the gp41 coding sequence.
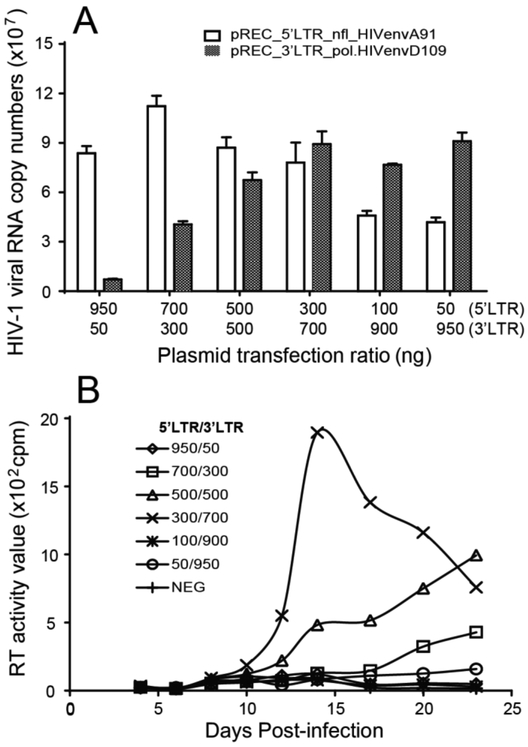
Fig. 2.
Optimization of transfection ratios of the two recombination vectors. Different ratio of the two vectors 5’LTR_envA and 3’LTR_envD were co-transfected into HEK-293T cells and the transfection supernatant was used to infect U87.CD4.CCR5 cells. Infection and virus propagation was monitored by real-time RT-PCR (A) and RT Assay (B).
Optimization of the HIV-1 Env FRP system
Success of this recombination system depends on the production of progeny viruses with heterodimeric genomes, suggesting equal packaging of the 5’LTR_env and 3’LTR_env sgRNAs into new virus particles. Equal packaging of both sgRNAs would ensure that half the progeny virus is heterodimeric based on the Hardy-Weinberg equilibrium. As described above, only the heterodimeric virus can generate a recombination event in the env gene which can then produce a replication-competent virus. To optimize heterodimeric virus production, recombination, and virus propagation, we transfected HEK-293T cells with different ratios of the 5’LTR_envA91 and 3’LTR_envD109 vectors (Fig. 2). RT-quantitative real time PCR (qPCR) was then performed on the viral RNA extracted from the cell-free supernatants to measure both the 5’LTR_envA91 and 3’LTR_envD109 sgRNAs using U5 and U3-specific primer/probes [38]. Transfection of 300ng: 700ng (3:7 ratio in mass, i.e. 1:3 ratio in DNA copy number based on the size of the two plasmids) of 5’LTR_envA91 (~17kb) : 3’LTR_envD109 (~13.3kb) respectively produced equal levels of both sgRNAs in the cell cytoplasm (data not shown) and in the virus of cell-free supernatant (Fig.2A). Virus produced from the HEK-293T cells transfected with the different plasmid ratios was then used to infect the U87.CD4.CCR5 cells. The 1:3 transfection ratio of 5’LTR_envA91 and 3’LTR_envD109 yielded the most virus propagation in the U87.CD4.CCR5 cells (Fig. 2B). Thus, a 1:3 transfection ratio of 5’LTR_env and 3’LTR_env was used in subsequent experiments.
With the current system, de novo replication in U87.CD4.CCR5 cells relies on the HIV-1 Env glycoprotein expressed from the 5’LTR_env vector. Several lentiviral vector studies suggest that co-expression of Vesicular Stomatitis Virus glycoprotein (VSV-G) will increase virus entry into target cells and increase virus transduction/infection. Thus, we performed plasmids 5’LTR_envA91 and 3’LTR_envD109 co-transfected with or without the helper plasmid pMDE-VSV.G, which expresses VSV-G in HEK-293T cells. Supernatants normalized for RT activity were then used to infect U87.CD4.CCR5 cells. Repeated experiments showed that the addition of VSV-G resulted in earlier detection of virus production and peak viremia in U87.CD4.CCR5 cell culture by 2-3 days (Fig. 3A). Since this system with VSV-G requires a triple transfection and a foreign component, and has minimal advantage for the production of HIV-1 intersubtype recombinants, all experiments were performed in the absence of VSV-G. Furthermore, the use of VSV-G could result in de novo infection/recombination within HEK-293T cells and may also reduce the infectious titers derived from these triple transfections. In contrast, the virus produced from the dual 5’LTR_env + 3’LTR_env transfection could not infect the HEK-293T cells of the same culture due to lack of CCR5 and CD4.
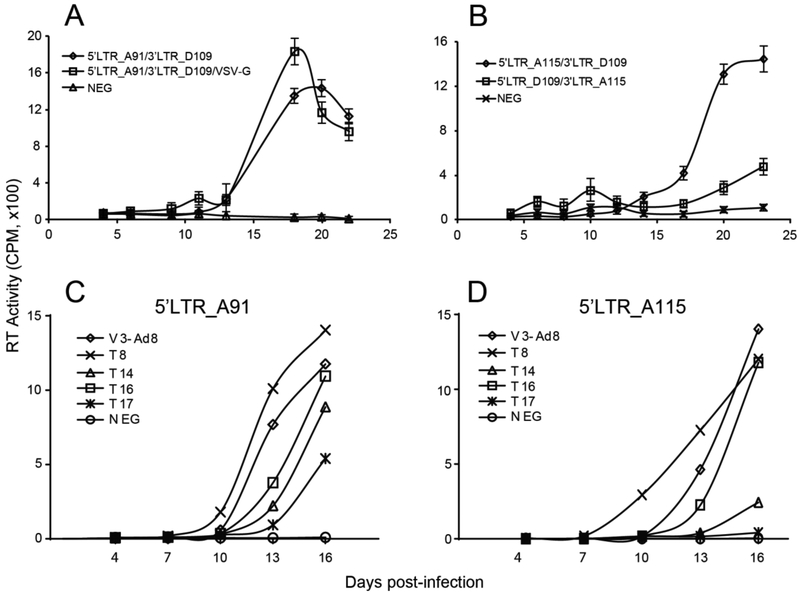
Fig. 3.
Production of infectious HIV-1 viruses through the HIV-1 Env FRP system. (A) 5’LTR_A91 and 3’LTR_D109 were co-transfected with or without VSV-G expressing plasmid. (B) Virus replication resulting from the HIV-1 Env FRP system with other set of subtype A (A115) and subtype D (D109). 5’LTR_A91 (C) or 5’LTR_A115 (D) was co-transfected with 3’LTR vector containing diverse HIV-1 envs for production of infectious HIV-1 intersubype Env recombinants. The error bars in panels C and D are approximately 3-15% of the mean.
To test the applicability of this recombination system with other env sequences, we co-transfected other different pairs: 5’LTR_envA115 + 3’LTR_envD109 and 5’LTR_envD109 + 3’LTR_envA115 into HEK-293T cells, and harvested the supernatant which was then used to infect U87.CD4.CCR5 cells. As described above, each pair of vectors led to the production and propagation of infectious viruses (Fig.3B). Clonal sequencing analyses (through TOPO-XL PCR cloning vector) of env fragments derived from viral RNA of the FRP system 3 weeks post-infection revealed that all HIV-1 env clones were A/D chimeras with breakpoints mapping to the gp41 region (Fig.4). To further test the system, we cloned five env sequences of T8 (subtype A1), V3.Ad8 (subtype B), T14 (A1U), T16 (CRF02-AG) and T17 (CRF02_AG) into pREC_nfl_Δpol_3’LTR_Δenv/URA3. The produced plasmids were then used to recombine with envA91 or envAl 15 in the plasmid pREC_5’LTR_HIV. Results showed that all of the five envs were able to produce replication-competent virus with A91 envelope and four envs (except T17) produced replication-competent virus with envA115 (Fig. 3C, D).
Fig. 4.
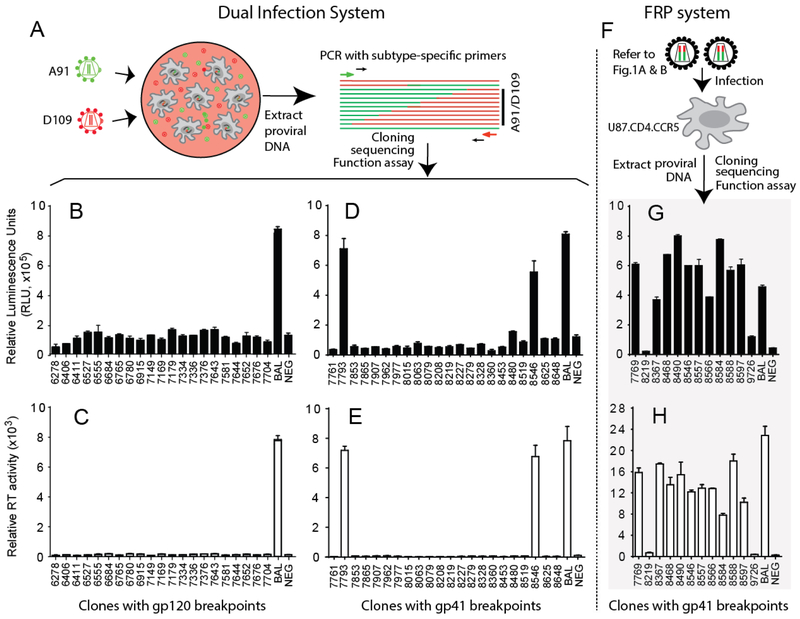
Detection of functional HIV-1 Env recombinants derived from dual infection and FRP systems. Schematics of generation of HIV-1 intersubtype recombinants from dual infection (A) and FRP (F) systems. Detection of functional HIV-1 EnvA/D recombinants with breakpoints in gp120 or gp41 from the dual infection culture (B, D) or from the HIV-1 Env FRP system (G) by Veritrop assay. Detection of virus production of chimeric HIV-1 clones containing different recombinants with breakpoints in gp120 or gp41 from dual infection culture (C, E) or FRP culture (H) through RT assay.
Most of the recombinants from FRP but not from dual infection system were functional
In our previous studies, the majority of the HIV-1 recombinants generated using either the dual infection or the siRNA enrichment method produced replication defective virus with non-functional Env glycoproteins. This new HIV-1 Env FRP system would generate pure HIV-1 Env recombinants within a wild type and replication-competent HIV backbone and without the presence of extraneous markers as required for selection of retroviral recombinants in other systems [30,31]. This FRP system provides a unique tool for investigating the functional breakpoints in HIV Env recombinants and is the template for designing systems for functional recombination in any HIV-1 gene product/region.
During the first round of virus production, the FRP system would mostly produce defective viral particles as described in Fig.1, but with continual propagation, only the HIV-1 viruses with functional intersubtype Env recombinants would survive. In contrast, with our dual infection system and in the absence of selection, the parental viruses dominate the culture with only a low production of HIV-1 intersubtype recombinants. In the dual infection system, frequency of HIV-1 intersubtype recombinants never increases beyond 5% in culture which is largely due to the generation of defective or less fit recombinants that cannot compete with the parental HIV-1 isolates. As described earlier, all of the A91/D109 and A115/D109 Env recombinants generated by the FRP system had breakpoints clustered in the gp41 coding region of Env (Fig.4G, H, and Fig. 5A, B, and see below for details). In contrast, when dual infecting with A91+D109 and A115+D109 viruses the recombination breakpoints were scattered throughout the env gene (Fig.4B-E, and Fig. 5C, D).
Fig. 5.
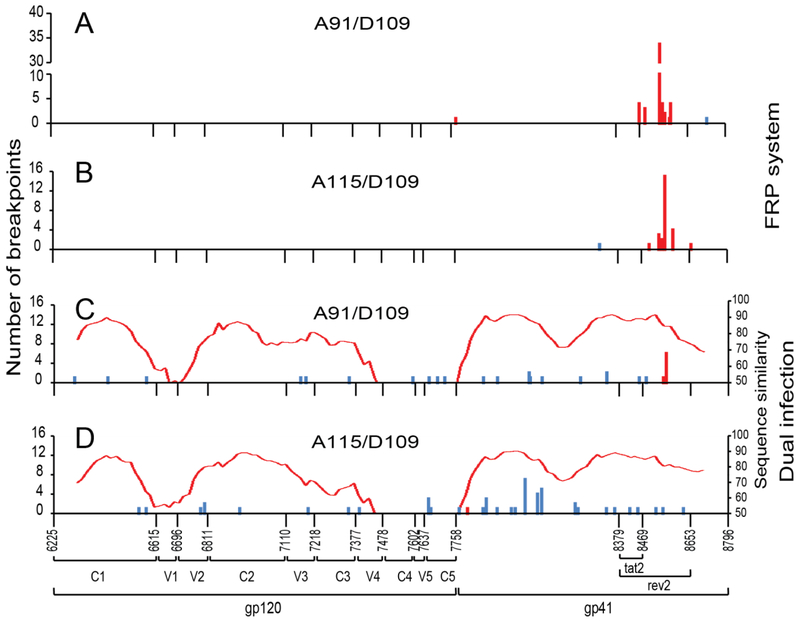
Distribution of recombination breakpoints in EnvA/D recombinants derived from FRP and dual infection culture by clonal sequencing. The location and frequency of recombination breakpoints at each nucleotide of the entire HIV-1 env region were shown from the HIV-1 Env FRP system with A91/D109 (A) and A115/D109 (B) sets, and from the dual infection system with A91/D109 (C) and A115/D109 (D) sets. Red bars stand for verified functional Env recombinants. The graphs also show the sequence similarity (right y-axis) between the two recombining envs (C, D). The red bars stand for verified functional recombinants while the blue bars stand for non-functional recombinants.
Following cloning, sequencing, and mapping of the intersubtype Env recombinants, we sub-cloned 12 unique recombinant envs from the FRP system and 43 recombinant envs (with intact ORFs) from dual infection system into a HIV-1 NL4-3 backbone and screened for Env expression and function using our cell-to-cell fusion assay, Veritrop. When derived from dual infections, only 2 of the 43 (4.6%) intersubtype A/D recombinant envs expressed the Env glycoproteins to mediate cell fusion through interactions with CD4 and CCR5 (Fig. 4B and D). Both of the two functional A/D Env recombinants had breakpoints in the gp41 coding region. When derived from the FRP system, 10 of 12 (83%) A/D Env recombinants were functional for cell-to-cell fusion and all had breakpoints in the gp41 coding region (Fig.4G). The only viruses capable of replication/propagation in U87.CD.CCR5 cells were those harboring the 3 and 10 A/D env genes (from dual infection and FRP systems, respectively) but not the other 42 A/D env (Fig. 4C, E, and H). It is interesting to note that the breakpoint at 8546 nt (HXB2 numbering) in the A91/D109 recombinants from both the FRP and dual infection systems resulted in functional Env and replication-competent virus.
Distribution of recombination breakpoints in replication-competent HIV-1 intersubtype Env recombinants from the FRP system
Since the FRP system was designed to screen and propagate pure and functional Env recombinant viruses, we could determine the distribution of recombination breakpoints in these functional Env recombinants across the entire env gene. We plotted the frequency of breakpoints obtained from FRP cultures through clonal sequencing at each env nucleotide position (loci of HXB2:6225-8797) on bar graphs (Fig.5A, and B). Among the 54 and 27 recombinant env clonal sequences originating from the pairs of A91/D109 and A115/D109 respectively through the FRP technique, we found 9 and 7 unique breakpoints respectively, and only one recombinant from each pair was non-functional. However, none of these breakpoints were located in the gp120 region, instead all breakpoints from both pairs were found in the gp41 region mostly between nt8215–nt8726, i.e. tat/rev exon 2 region. We observed a major recombination hotspot nt8546 in envA91/envD109 set and nt8557 in envA115/envD109 set (Fig.5A, and B).
One of possible reasons for the absence of breakpoints in the gp120 region from our FRP system is that there were very few or no recombination events in this particular region in the system. Our previous studies have revealed that the recombination is related to the nucleotide sequence similarity between the two recombining strains amongst several other factors [30,31]. To exclude the possibility that the gp120 sequence similarity was not adequate for homologous recombination between envA91/envD109 or envA115/envD109 pairs, we constructed sequence similarity plots using SimPlot software [40] and found that the sequence similarity between envA91, envA115 and envD109 ranged up to 65-75% within the gp120 region (Fig. 5C and D) which should be enough to support effective recombination. We then analyzed the location of Env breakpoints derived from in vitro dual infection of U87.CD4.CCR5 cells with primary HIV-1 isolates A91/D109 or A115/D109 using a standard protocol [41]. Although the sequence at the gp41 coding region was still a hotspot for recombination, there were also substantial breakpoints distributed along the entire gp120 region in env recombinant clones from A91/D109 (10 out of 28, 36%) and A115/D109 (13 out of 48, 27%) sets (Fig. 5C, D). These findings show that recombination in these two env sets is feasible within the gp120 region.
Distribution of functional and non-functional recombination breakpoints in the HIV-1 env region
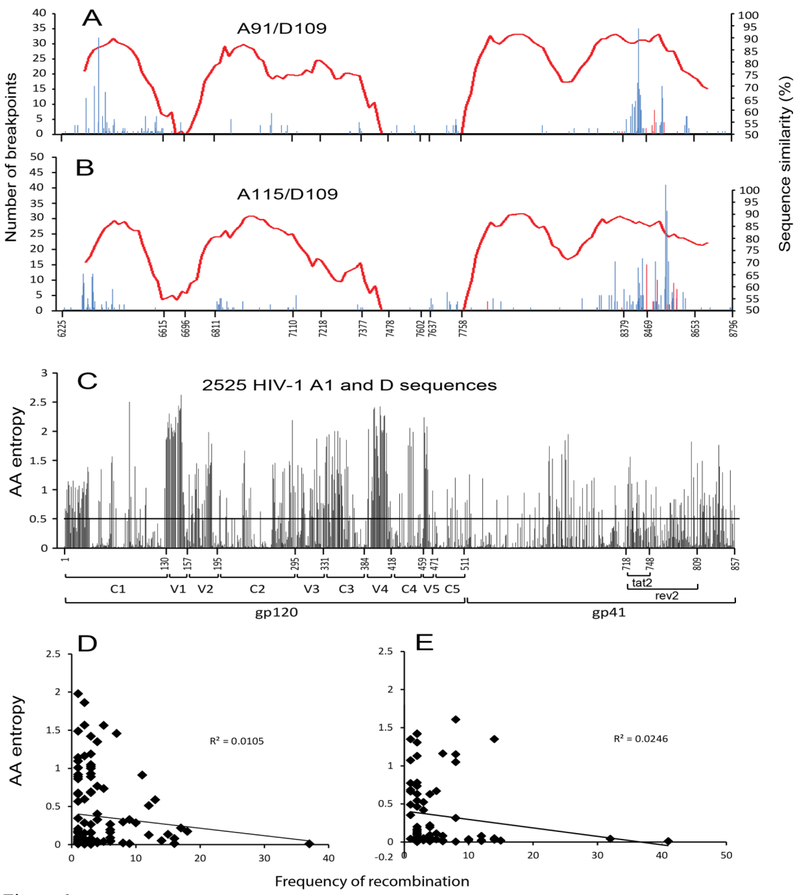
The presence of breakpoints in the HIV-1 gp120 coding region from the dual infection system, but not in the HIV-1 gp120 from the FRP system, suggested that breakpoints in gp120 region may be non-functional, and that with the FRP system, the only viruses to propagate after the first round of infection would be replication competent HIV-1 intersubtype Env recombinants. To detect both functional and non-functional recombinants from the FRP system, the DNA extracted from the U87.CD.CCR5 cells at 4 days and 3 weeks post-infection was PCR amplified using conserved env primers and subtype A and D specific primers for the 5’ and 3’ ends of the env gene. As expected, the full length D109 env was the major sequence in the PCR products using conserved primers at 4 days post-infection but absent in the Week 3 culture. When using the subtype A and D specific primers, we PCR amplified DNA products, which were then subject to amplicon-based next generation deep sequencing (NGS) for the day 4 samples. Due to the limited diversity of recombination breakpoints, we performed clonal sequencing on the intersubtype env amplicons derived at 3 weeks from the FRP system. Over 4000 A/D env sequences were obtained through NGS and then analyzed using the Recombination Identifier Program (RIP 3.0) [42] (http://www.hiv.lanl.gov/) (Fig.6). At day 4, the distribution of breakpoints in A91/D109 and A115/D109 env pairs were similar with the dual infection system and with FRP systems (Fig.6 and Fig. 7A, B). Recombination hotspots clustered in the C1 domain of gp120 (25.3%) and in the gp41 region (62%), especially in tat/rev exon 2. As expected, the majority of breakpoints in other gp120 regions mapped to the conserved domains (C1 25.3%, C2 5.2%, C5 4.6%, C3 0.7%, C4 0.5%) rather than the variable domains (V1 0.44%, V2 0.1%, V3 0.77%, V4 0.27%, V5 0.05%). The NGS results revealed that the FRP system produced over 30% of intersubtype Env recombinant viruses with breakpoints in the gp120 region at four days post-infection, indicating that the system was not defective in generating gp120 recombinants (Fig.6 and 7). In contrast, by week 3, all the A/D recombinants from the FRP system had breakpoints that mapped to the gp41 coding region and produced Env that were functional for HIV-1 entry into host cells which could also be found in the Day 4 FRP culture (red bars in Fig.6A and B).
Fig. 6.
Distribution of recombination breakpoints from early infection culture of FRP detected by 454 pyrosequencing. The proviruses from two HIV-1 env sets (A) A91/D109, and (B) A115/D109 paired by the FRP system were subjected to 454 pyrosequencing. The graphs show the location and frequency (left y-axis) of recombination breakpoints at each nucleotide of the entire HIV-1 env, and the sequence similarity (right y-axis) between the two recombining envs. Red bars stand for verified functional Env recombinants. (C) Shannon entropy analysis: The vertical axis represents entropy scores, while the horizontal axis shows the position of amino acids in the env gene. (D) and (E) show the correlation of amino acid sequence entropy and recombination frequency of A91/D109 and A115/D109, respectively.
Fig. 7.
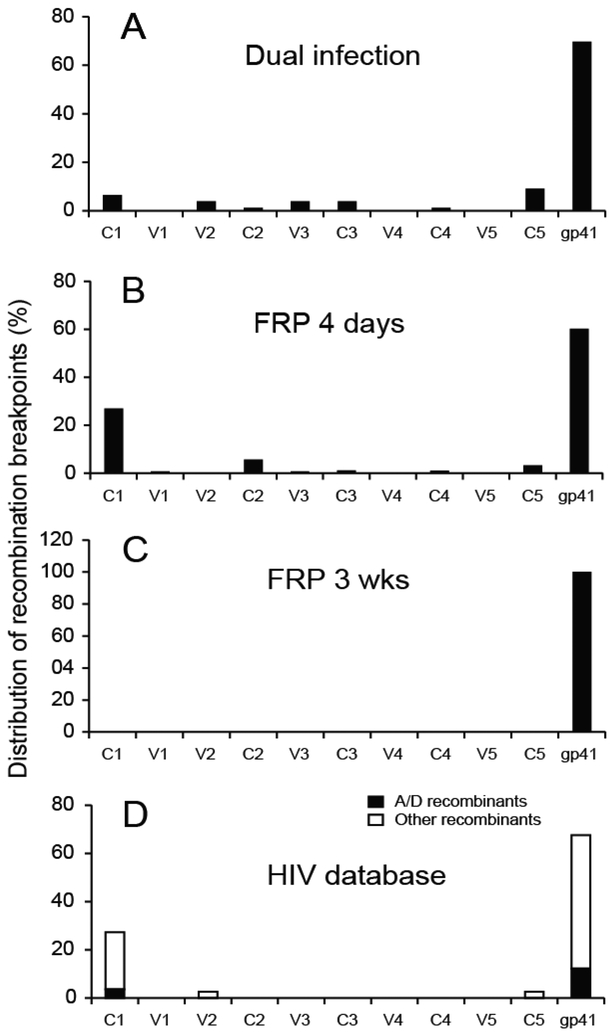
Distribution of recombination breakpoints in dual infection (A), 4 days (B) and 3 weeks (C) post-infection in the FRP system with A91/D109 and A115/D109 pairs. (D) Distribution of recombination breakpoints in HIV sequence database-derived HIV-1 Env recombinants with a single recombination breakpoint.
Analysis of sequence entropy and its correlation with frequency of recombination shows that The higher entropy seems to correspond to a lower frequency of recombination between subtype A and D isolates in V1, V2, V4, and V5 regions, and the lower entropy is associated with a higher frequency of recombination in C1 and tat2/rev2 regions (Fig. 6C). However, correlation analysis showed that there is no significant correlation between the amino acid entropy score and the frequency of recombination from either pair A91/D109 (R2=0.0105, Fig. 6D) or pair A115/D109 (R2=0.0246, Fig. 6E).
As described in the Material and Methods, to eliminate the possibility of PCR-induced env recombinants, we used Phusion High-Fidelity DNA Polymerase which has greater processivity than other Taq polymerases according to the product description, which would presumably reduce template switching through rapid DNA synthesis. We also set up control PCR amplifications with the same protocols as those employed in the dual infection, but with the DNA vector templates. Subsequent NGS revealed that only 1 recombinant was identified in the 912 sequences analyzed from the A91 + D109 and A115 + D109 control template amplifications. To rule out the possibility of recombination occurring during the transfection of cells between plasmid DNAs, we also analyzed the recombination events in the viruses directly from the transfection supernatant by RT-PCR amplification and NGS sequencing of env genes, and did not find any recombination events in those viruses.
Transition from non-functional to functional breakpoints
By comparing the results from dual infection and FRP systems, we can now analyze intersubtype recombinants governed only by sequence conservation and mechanisms of template switching with the intersubtype recombinants (from the latter pool) that can only survive and propagated based on function and replicative fitness. In Fig.7A, with the dual infection method, we show a more random distribution of intersubtype A/D breakpoints across the env gene, with the highest recombination frequency in gp41 region (69.7%), as well as in all of the conserved domains (6.6% in C1, 1.3% in C2, 3.9% in C3, 1.3% in C4, and 9.2% in C5) and some of the variable regions (3.9% in V2 and 3.9% in V3) in gp120. With the FRP system at day 4, the majority of breakpoints of resulting intersubtype Env recombinants were located in the gp41 region (60.2%), with less proportion in the gp120 conserved domains (26.9% in C1, 5.7% in C2, 1.1% in C3, 1.0% in C4, and 3.2% in C5) (Fig.7B). By week 3, all the surviving intersubtype HIV recombinants had breakpoints within the gp41 coding region (Fig. 7C).
We further compared the distribution of breakpoints in our functional intersubtype A/D env genes derived from the FRP system with the intersubtype env sequences found in HIV-infected individuals (http://www.hiv.lanl.gov/). When analyzing the 77 in vivo intersubtype recombinants with a single breakpoint within the env coding region, we found that nt8215 to nt8595 gp41 region and the C1 domain were hot spots for recombination (67.5% and 27.2%, respectively), while only 5.6% were found in all other sequences of env (Fig.7D). Interestingly, when only looking at CRFs with a single EnvA/D recombination, there are 4 and 12 breakpoints in C1 and gp41 regions respectively, but no breakpoints between the two regions. Our previous dual infection system with or without siRNA selection did not generate HIV-1 intersubtype env recombination with a similar distribution of breakpoints as observed in HIV patients. However, similar hot spots and patterns for HIV-1 intersubtype recombination were observed in the env gene in the epidemic and derived from the in vitro FRP system. With both, only the functional intersubtype Env genes could survive.
Discussion
Our laboratory has studied intersubtype recombination in the HIV-1 env gene using a dual infection method in which two primary HIV-1 isolates are inoculated into the same susceptible cell culture at an equal multiplicity of infection (MOI) [30,31,41]. As we described previously and herein, this dual infection system is plagued by the limited number of functional breakpoints that arise, plus the dominance and outgrowth of the more fit parental strains [31,32]. To understand which intersubtype recombination breakpoints give rise to functional HIV-1 Env glycoproteins, we also developed an siRNA-based technique to select and enrich intersubtype Env recombinants within in vitro cultures [32]. During the first few days of culture, HIV-1 intersubtype recombinants with breakpoints between the siRNA target sites dominated virus production. However, the mutations in the parental HIV-1 quickly overcame the siRNA selection pressure, resulting in the emergence of the parental HIV-1 strains with siRNA-resistant mutations in the target sequences. These parental HIV-1 strains then outcompeted the earlier appearance of HIV-1 intersubtype recombinants. Therefore, neither dual infection with HIV-1 isolates of different subtypes nor the addition of HIV isolate-specific siRNA were effective as a model to compare the generation and evolution of functional HIV-1 intersubtype recombinants in the human population.
In the present study, an HIV-1 Env FRP system was designed to provide a new model for in vitro intersubtype recombination; the aim being to overcome the limitations of the previously described dual infection method and the siRNA selection technology. By introducing two different env genes into the two complementary vectors, the FRP system is able to generate and select for pure and functional intersubtype Env recombinants. In transfected HEK-293T cells, the two complementary plasmids pREC_5’LTR_nfl_envA (aka 5’LTR_envA) and pREC_nfl_Δpol_3’LTR_envD (aka 3’LTR_envD) produced a complete viral proteome from the former as well as two viral sgRNAs, 5’LTR_envA and 3’LTR_envD respectively. The two sgRNAs are randomly packaged into progeny virions according to Mendelian inheritance (Law of Segregation of genes and Law of Independent Assortment) to produce homodiploid (i.e. two same sgRNA) and heterodiploid (i.e. two different sgRNA) viruses. Homodiploid viruses with two 3’LTR_envD sgRNAs cannot initiate reverse transcription whereas two 5’LTR_envA sgRNAs cannot complete reverse transcription in susceptible cells (Fig.1), and thus, these homodiploid viruses are incapable of replication. In contrast, the heterodiploid viruses can replicate in susceptible U87.CD4.CCR5 cells but only through complementation and intersubtype recombination within the env genes of the two sgRNAs (Fig. 1B and C). We have previously described the fundamental principles of this heterodiploid virus producing replication-competent virus despite the lack of 5’ and 3’ LTR in each of the two complementary vectors [38].
From this FRP system, sequence analysis revealed that only chimeric viruses with an A/D env gene were present after ~2 weeks of virus propagation. In contrast, we detected virus production and amplification within 3-5 days with our previous dual infection system. In the latter case the recombination events occurring between the two viral RNAs generally resulted in nonfunctional viruses. It is important to stress that intersubtype recombination in our new system was not “forced” within the env gene. Based on previous estimates of recombination/breakpoints (~2-3% recombinants in entire viral population in 1000 nucleotide fragment) [30,31] and infection with 50% homodiploid virus, we estimate that ~97% of viruses in the initial round of U87 infection (using supernatants from 293T cell transfections) would result in non-recombinant viruses with dead end reverse transcripts, incapable of integration, mRNA transcription, and new HIV-1 protein translation. Thus, approximately 3% of the virus infecting U87 cells would recombine within the env gene and generate a chimeric A/D HIV-1 genome that may or may not produce a functional HIV-1 proteome. Only those HIV-1 genomes with a recombinant A/D env gene could integrate and produce the functional proteome for new virus progeny. These estimates would explain the slow emergence and dominance of replication-competent intersubtype recombinants, requiring ~2 weeks for detection instead of just 3-5 days with wild type virus. Please note that we originally planned to study the recombinants with multiple breakpoints as they are critical in causing rapid multiple drug resistance and immune evasion. However, HIV recombination occurs only in a cell simultaneously infected by two genetically distinct viruses and after the production of progeny viruses containing heterogeneous viral genomic RNAs. In our FRP system, the first round of viral infection produces diverse HIV-1 env recombinants requiring a relatively long culture period to grow out. We monitored virus production every two days and based on low production of recombinant virus, susceptible cell numbers in culture, and previous models on dual infection [43] we likely not reach titers sufficient to enable the co-infection of a cell with two different recombinants (but of course not impossible). Only when we achieved high MOI would recombinants of recombinants be produced with multiple breakpoints. Through clonal sequencing of the recombinants from Week 3 FRP culture, we only observed very few such recombinants that were insufficient to provide enough meaningful data to analyze the env recombinants with multiple breakpoints. Again, the detection of the infectious virus from the FRP system usually takes more than 12 days, while recombinants may appear in dual infected cultures within 2-3 days but the majority of these are non-infectious [27,32].
Several studies including ours have utilized in vitro systems to generate HIV-1 intersubtype recombinants but few have explored the function of these recombinant gene products. Simon-Loriere E. et al. examined the function of 39 A/B, A/C, A/G, C/B, C/G and G/B recombinant Env proteins derived from a single infection assay without selective pressure. They found that 14 of 24 intersubtype Env glycoproteins were functional at mediating host cell entry, whereas 14 of 15 with gp41 breakpoints were non-functional [28]. Recently, we have shown that intersubtype Env recombinants with breakpoints in gp120 are non-functional due in part to the close juxtaposition and intramolecular interactions of the C1, C5, and gp41 extracellular domain in the Env trimeric structure [27]. This C1/C5-gp41 extracellular domain interaction has led to co-evolution of these coding regions within HIV-1 subtypes. In that study, recombinants were constructed such that the C1/C5-gp41 were derived from a subtype B strain (i.e. NL4-3) but where the chimeric A/D gp120 region was placed between the subtype A C1 N-terminus and the subtype A C5/gp41 end. Despite those A/D recombinants being nonfunctional in the context of the complete chimeric A/D Env gene (subtype A C1 and subtype D C5/gp41), the maintenance of the C1/C5-gp41 extracellular domain from subtype B could maintain the core Env trimeric structure necessary for function while carrying the chimeric A/D gp120 sequences.
In the present study, the recombination breakpoints of over 4,000 different nonfunctional and functional HIV-1 intersubtype A/D Env recombinants from dual infection culture and the FRP system were mapped using a combination of clonal sequencing and pyrosequencing followed by analysis with the Recombination Identifier Program (RIP 3.0). We compared the distribution of A/D recombination sites in the dual infection system 1 week post-infection and in the FRP system 4 days and 3 weeks post-infection. As described in Fig. 4B-E and 5C, D, due to continuous recombination between the two parental viruses, the intersubytpe A/D recombination sites appeared to be randomly distributed throughout the env gene in the dual infection without a noticeable recombination site preference. This lack of recombination site preference could be attributed to the low number of recombinants analyzed within this study. Despite this, it should be noted that the clear majority of the resulting A/D Env recombinants (41 out of 43 tested) were nonfunctional for either virus replication or Env-mediated cell fusion. Within the FRP system, after just 4 days post-infection and before the intensive selection of functional recombinants, the distribution of intersubtype A/D recombination sites were also scattered throughout the env gene. This dissemination of recombination sites included most of variable regions (V1-V5) but with what appeared to be emerging hot spots in the gp120 C1 and gp41 regions (Fig. 6). Please note that, in the FRP system, the recombination between the two parental viral genomes only occurred in the initial round of viral replication. This is followed by the selection of functional recombinant viruses in the absence of parental viruses, which potentially explains the higher frequency of breakpoints in gp120 and gp41 regions in Day 4 FRP cultures than in the 1-week dual infection cultures. By week 3 within the FRP system, all of the recombination sites were mapped to the gp41 region, especially the second exon of rev (Fig. 5A, B). When comparing the recombinants with breakpoints in the gp41 region derived from Day 4 and Week 3 post-infection FRP cultures (Fig. 6A, B), there is an apparent decrease in number and frequency of observed breakpoints along with the viral growth. This is largely due to the screening of functional recombinants, as well as the competition amongst the individual recombinants. Nearly all of these intersubtype A/D env genes were functional for virus replication and host cell entry. Thus, we assume that the majority of recombinants detected 4 days post-infection are defective and incapable of replication while a few are replication competent. Therefore, these results suggest a transition from non-functional to functional A/D recombinants accompanied with a transition of recombination breakpoints from the entire env to gp41 region.
Interestingly, even though the sequence entropy and the frequency of recombination in some regions appeared to be negatively correlated, there is no significant such correlation when analyzing the entire env gene. This is reasonable based on our previous discoveries [30,31] which suggested two main factors determining the recombination site, being sequence similarity, which facilitates the jumping of the nascent DNA strand from one RNA template to the other during synthesis of the minus DNA stand, and the RNA secondary structure which may drive the preferential strand transfer and increased recombination frequency or vice versa.
Even though there are a few studies investigating the function of HIV-1 intersubtype Env recombinants, analysis of the circulating and unique recombinant forms (CRFs and URFs) in the HIV-1 epidemic are still able to reveal the possible mechanism for the selection of prevailing recombinants. Analysis of distribution of breakpoints in the functional intersubtype env sequences (i.e. CRFs and URFs) derived from HIV-infected individuals showed that nt8215 to nt8595 gp41 region and the C1 domain were hot spots for recombination to generate functional Env recombinants in vivo, but not the other regions within the env sequence. Similar hot spots for HIV-1 intersubtype recombination were observed in the env genes derived from the in vitro FRP system. With both, only the functional intersubtype Env genes could survive. When comparing the intersubtype Env A/D recombinants derived from our FRP system to those reported in patients with Env recombinants, especially Env A/D recombinants, we also found a paucity of recombination breakpoints in the gp120 coding region, which is consistent with our previous study [27] (Fig. 7). These results strongly suggest that the HIV-1 recombinants circulating in the epidemic are mainly selected by their functionality. Finally, we should point out that all of the Env recombinants analyzed in the present study have clade A sequences on their 5' ends and D sequences on their 3' ends, thus it is possible that a reciprocal system with D 3' and A 5' might look a bit different.
In the past 10 years, we have published several studies on the emergence and selection of intersubtype recombination using single-cycle assays and dual infection systems left untreated or inhibited by HIV isolate-specific siRNA [27,29-32,41,44]. These systems resulted mostly in nonfunctional Env recombinants or intersubtype HIV with poor replicative fitness, incapable of competing with the parental viruses. The newly developed FRP technique presented in this study represents a breakthrough in terms of investigation of functional HIV-1 intersubtype recombinants. The FRP system, through the use of yeast-based cloning vectors, has successfully generated a number of functional HIV-1 intersubtype Env recombinants, and provided an in vitro model to mimic selection of replication-competent HIV-1 intersubtype recombination in dual or superinfected patients. Furthermore, this FRP system is also adaptable to studying viral recombination between any two HIV-1 isolates of any subtypes, and we are now utilizing this technique to rapidly diversify and select for functional segments in the HIV-1 pol gene. Finally, the FRP system provides a new tool to obtain a functional and diversified pool of HIV-1 for subsequent studies evaluating viral resistance to antiretroviral drugs, immune selective pressure, and how host restriction factors influence HIV containment/escape. In conclusion, the FRP system employs only the natural HIV-1 process of strand switching during virus replication to produce intersubtype recombinants, does not involve the addition of any foreign genes (e.g. antibiotic resistance and fluorescent proteins), and is therefore a powerful new tool and an in vitro model for the study on HIV-1 intersubtype recombination.
Materials and methods
HIV-1 clones and primary isolates
HIV-1 subtype A isolates A91 and A115, and subtype D isolate D109 were collected from HIV patients in 1996 through a research collaboration between the Center for AIDS Research (CFAR) at Case Western Reserve University and the Joint Clinical Research Center (JCRC) in Uganda. The viruses were isolated and propagated by co-culturing HIV-1 patient-derived PBMCs with those from healthy donors. Prior to co-culture, PBMCs were pre-stimulated with phytohemagglutinin (PHA, 2ug/ml) and interleukin-2 (IL-2, 1ng/ml) in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) for 2-3 days culture. V3Ad8 is an HIV-1 clone containing Ad8 V3 sequence in NL4-3 backbone (a subtype B clone), T8 (Q168ENVa2, GenBank accession no.: AF407148) is a subtype A1 isolated from Kenya, T14 (33-7, GenBank accession no.: EU513186) is a 02A1U recombinant isolated from Cameroon, T16 (253-11, GenBank accession no.: EU513191) and T17 (250-4, GenBank accession no.: EU513189) are both CRF02 AG recombinants isolated from Cameroon. All these HIV-1 clones were obtained from the NIH AIDS Reagent Program. Tissue culture dose for 50% infectivity (TCID50) assays were performed to determine virus titer prior to subsequent experimentation. Titers were expressed as infectious units per milliliter.
Cell lines
U87.CD4.CXCR4 and U87.CD4.CCR5 cell lines were obtained from Dr. HongKui Deng and Dr. Dan R. Littman [45] through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, and were grown in DMEM supplemented with 15% FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), puromycin (1 μg/ml) and G418 sulfate (1 mg/ml). HEK-293T cells were obtained from the American Type Culture Collection (ATCC) and were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin. All cells were grown at 37°C in 5% CO2.
Dual infection
U87.CD4.CCR5 cells (1.2 ×105 per well in a 24-well plate) were inoculated with 0.1 multiplicity of infection (MOI) of each of the primary HIV-1 isolates pairs A91 and D109, or A115 and D109. Single virus infections were set up as controls alongside the dual infections in a 24-well plate. The viruses were propagated for 5 to 7 days, then both supernatant and cells were harvested. Proviral DNA was extracted using the PureLink Genomic DNA mini kit (Invitrogen) and was used as template for nested PCR to retrieve recombinant HIV-1 envs.
Conventional HIV-1 env cloning
To retrieve recombinant envs from the HIV-1 quasispecies in the dual infection cultures, nested PCR was performed with the external primers ENV A (sense, HXB2 numbering nt5954-nt5982, 5’-GGCTTAGGCATCTCCTATGGCAGGAAGAA) and ENV M (antisense, nt9068 to nt9098; 5’-TAGCCCTTCCAGTCCCCCCTTTTCTTTTA). The internal primers were specific for subtype A and D env recombinant amplification, i.e. VPU-A-1 (sense, nt6135-nt6155, 5’-TAGTAGGTATAGAATATAAGA) and Gp41-D-2 (antisense, nt8747-nt8769, 5’-GCCTAATTCTTCTAGGTATGTTG) for envA/D recombinants, and VPU-D-1 (sense, nt6135-nt6155, 5’-TAGTATTCATAGAGTGTAGAA) and Gp41-A-2 (antisense, nt8747-nt8769, 5’-GTCTTATTCGTCTGGGTATGTTA) for envD/A recombinants. PCR products were run on a 0.8% agarose gel, then the 2.7 kb fragments were purified using the QIAquick gel extraction kit (Qiagen) for subsequent cloning. Recombinant HIV-1 env PCR products were then cloned into the TOPO-XL PCR cloning vector (Invitrogen) and transformed into OneShot Top10 E. coli cells (Invitrogen) according to manufacturer’s instructions. Colony PCR was performed to screen for env positive clones which were used for plasmid extraction by using a plasmid miniprep kit (Qiagen), and the resultant DNA was sent for sequencing. Same procedure was performed for the proviral DNA from the FRP system 4 days postinfection but with different external primers (ENV-A and ENV-N) and internal primers (Vpu-1 and Env M) that were all located at conserved regions and were able to amplify from both subtype A and D env sequences.
Yeast-based HIV-1 env cloning
Saccharomyces Cerevisiae strain DY1457 (MATα-ade6-can1-his3-leu2-trp1-URA3) was obtained from the American Type Culture Collection (ATCC, MYA-906). Yeast was grown at 30°C in appropriate selection media, including Yeast Extract Peptone Dextrose (YEPD), complete supplement media (CSM)-Leu-URA3, CSM-Leu, and CMM-Leu + 5-fluoro-1,2,3,6-tetrahydro-2,6-dioxo-4-pyrimidine carboxylic acid (5-FOA) [38].
Cloning of HIV-1 env PCR products into the shuttle plasmid containing a neutral HIV-1 NL4-3 backbone, as well as into the recombination system plasmids, was done using the yeast homologous recombination-based method described previously [38]. Briefly, the prepared yeast was resuspended in 1 ml of fresh 1x LiAc/TE solution to induce competence. Transformation of yeast was performed using the lithium acetate method with a 1:3 ratio of the linearized plasmid and insert DNA. Yeast was then heat shocked at 42°C for 15 minutes and plated on proper selection agar plates. Plasmids were retrieved from yeast using a mixture of mechanical glass bead disruption and phenol-chloroform extraction, and transformed into STBL4 E.coli (Invitrogen). Plasmids containing various target recombinant HIV-1 env sequences were then extracted from STBl4 E. coli for further experimentation.
Construction of vectors for the HIV-1 Env Functional Recombinant Production (FRP) system
The FRP system is composed of two complementary vectors, pREC_5’LTR_nfl and pREC_nfl_Δpol_3’LTR (Fig. 1A). The previously constructed HIV-1 shuttle plasmid vector pREC_nfl_HIV-1 [38], which contains a near full length (nfl) genome but lacks the 5’ LTR region of the HIV-1 strain NL4-3, was the basis for construction of these two complementary plasmids for the HIV-1 Env FRP system. To create the pREC_5’LTR_nfl plasmid, the 3’LTR sequence of pREC_nfl_HIV_NL4-3 was deleted and a 5’LTR sequence was cloned back. To boost HIV-1 protein expression from this plasmid, the nef sequence was deleted and replaced with a bovine growth hormone poly(A) tail (BGHpA). To develop the pREC_nfl_Δpol_3’LTR plasmid, the NL4-3 sequence corresponding to HXB2 position nt2815–nt8797 was deleted from the original pREC_nfl_HIV_NL4-3 plasmid and replaced with URA3 by yeast homologous cloning. The amplification of URA3 sequence for this deletion was performed using primers Pol-SbfI-URA3-1 (sense, 5’-GATTTCTGGGAAGTTCAATTAGGAATACCACATCCTGCAGGGTTAAAACAGAAAAACCGCG GAGATTGTACTGAGAGTGCAC) and Env-URA3-2 (antisense, 5’-CTTATAGCAAAATCCTTTCCAAGCCCTGTCTTATTCTTCTAGCTGTGCGGTATTTCACACCG). The HIV-1 NL4-3 sequence corresponding to HXB2 position nt6202–nt8797 was then amplified using primers Pol-SbfI-envB (sense, 5’-GATTTCTGGGAAGTTCAATTAGGAATACCACATCCTGCAGGGTTAAAACAGAAAAACCGCG GAGATTGTACTGAGAGTGCACAGAAAGAGCAGAAGACAGTGGCAATGA) and Env-end (antisense, nt8797-nt8819, 5’-CTTTTTGACCACTTGCCACCCAT). The resultant PCR product was then used to replace the URA3 in the plasmid to create pREC_nfl_Δpol_3’LTR, a plasmid containing a defective HIV-1 genome without a 5’LTR and with a 3387 nucleotide deletion from HXB2 position nt2816 to nt6201 (including RT, IN, vif, vpr, vpu, tat1, and rev1). The final plasmids pREC_5’LTR_nfl_Δenv/URA3 and pREC_nfl_Δpol_3’LTR_Δenv/URA3 into which other primary HIV-1 env sequences can be cloned was created by replacing the env gene (nt6202-nt8797) with a URA3 sequence amplified with primers Env-B-1-URA3-1 (sense, 5’-TTAAGACAAAGAAAAATAGACAGGTTAATTGATAGACTAA) and Env-URA3-2 (antisense, 5’-CTTATAGCAAAATCCTTTCCAAGCCCTGTCTTATTCTTCTAGCCGCGGCTGTGCGGTATTTCA CACCG). The resulting two vectors can then be used to introduce env sequences from various HIV-1 strains to generate pREC_5’LTR_nfl/env and pREC_nfl_Δpol_3’LTR/env vectors for producing env recombinants (see Results and Fig.1B and 1C).
Veritrop assay
One million (1×106) U87.CD4.CCR5 cells were cultured overnight in 100 mm petri dishes (Day 1) and were transfected 24 hours later (Day 2) with pDM128fLuc (containing an HIV-1 LTR-driven luciferase gene) using FuGENE 6 transfection reagent (Roche) as previously described [46]. Also on day 1, 6.5×104 HEK-293T cells were plated in 24-well plates, and were transfected 24 hours later with recombinant env-containing plasmids pREC_nfl_HIV_envA/D. Approximately 6 hours after transfection, U87.CD4.CCR5 cells were plated into 24-well plate at 6.5×104 cells per well. On day 3, transfected HEK-293T cells were co-cultured with the transfected U87.CD4.CCR5 cells for 15 hours. The mixed cells were then lysed with 100 ul of Glo Lysis Buffer and combined with 50 μl of Bright Glo (Promega Biotech), and read on a Victor3 V (Perkin Elmer) luminometer to measure luciferase activity.
Virus production from the HIV-1 Env FRP system
To clone different envs into the recombination system, various HIV-1 envs (HXB2 numbering nt6203–nt8819) were amplified using primers VPU_6203_S (antisense, nt6203-nt6228, 5’-GAAAGAGCAGAAGACAGTGGCAA) and Env-end. Production of infectious HIV-1 viruses from the env chimeric genes in pREC_nfl_envA/D was performed using the complementation method according to Dudley et al. [38] since the cloned genomes lacked the 5’ LTR sequence. In brief, on day 1, 105 HEK-293T cells were plated, then 24 hours later (Day 2) 0.3 μg of each of the pREC_nfl_envA/D plasmids containing the various HIV-1 A/D recombinant envs were co-transfected with a complimenting pREC_5’LTR_pbs+Ψ. The next day (Day 3), 7×104 U87.CD4.CCR5 cells were plated in a 24-well plate and infected 24 hours later (Day 4) with the supernatant from transfected HEK-293T cells. After 24 hours (Day 5), the supernatant was removed and the infected U87.CD4.CCR5 cells were cultured for up to 2 weeks. Virus infection and propagation in U87.CD4.CCR5 cells was monitored by measuring reverse transcriptase (RT) activity in the culture supernatant every 2-3 days using a RT assay. Supernatant from virus-positive wells was harvested and frozen at −80°C for further studies.
RT Assay
Ten μl of cell culture supernatant was collected every other day to monitor virus replication by RT assay. The RT mixture comprised 1 μl of 10 mCi/mL α-32P dTTP (Perkin Elmer) in 1 ml of RT master mix [50 mM Tris-HCl (pH 7.8), 75 mM KCl, 2 mM dithiothreitol (DTT), 5 mM MgCl2, 5 μg/mL of poly(rA), 6.25 μg/mL oligo(dT), 0.5%(v/v) NP40], 25 μl of the RT mixture was added to each sample well and incubated at 37°C overnight. The next day, 10μl from each well was blotted onto a 96-well format DEAE filter mat (Perkin Elmer), dried and washed with 1X saline-sodium citrate (SSC) buffer on a shaking platform. The washed filter was dried on a 65°C heating block for 30 minutes and the α-32P radioactivity was quantified as counts per minute (CPM) on a Matrix 96-β-counter (Packard, Meriden, CT).
Clonal sequencing
We procured commercial sequencing services of ACGT, Inc. (Wheeling, Illinois). Full length env were sequenced using five universal primers Vpu-1 (sense, nt6108-nt6130, 5’-TAATAATAGCAATAGTTGTGTGG), E80 (sense, nt6862-nt6883, 5’-TTCCAATACACTATTGTGCTCC), EAD2 (antisense, nt8064-nt8084, 5’-CCAGAGATTTATTACTCCA), E15 (antisense, nt8424-nt8425; 5’-CTTGCTCTCCACCTTCTTCTTC), and ENV M. The retrieved sequences were joined correspondingly to form full length of individual env recombinants and aligned along with primary envs of isolates A91, D109, and A115 using BioEdit software. Aligned sequences were imported into SimPlot sequence similarity plotting software [40] and bootstrap plots were constructed with the recombinant sequence as the query and the two recombining primary envs as the references. Approximate recombination breakpoints were then determined according to HXB2 numbering using jpHMM at GOBICS tool available at http://www.hiv.lanl.gov.
454 sequencing
Proviral DNA derived from the HIV-1 Env FRP system infection experiments for two HIV-1 env sets A91/D109 and A115/D109 was extracted from infected U87.CD4.CCR5 cells using the PureLink Genomic DNA mini kit (Invitrogen). First-round PCR of proviral DNA (~200-500ng per reaction) was done using primers UNIV-VS1 (sense, 5’-AAACTTATGGGGATACTTGGG) and ENV-N (antisense, 5’-CTGCCAATCAGGGAAGTAGCCTTGTGT) with the following conditions: 94 °C 4 min, [94 °C 30 s, 55 °C 30 s, 72 °C 3 min] × 35 cycles, and 4 °C hold. The first-round PCR product (2ul) was then used as template for PCR of five −600 nt fragments covering the length of gp160 generated using fusion oligos. These oligos were comprised of 454 adaptor sequences (Roche Lib-A Primer A and Primer B) followed by a 10 nt multiplex identifier sequence (Roche MID 1-5) to permit sample pooling, and finally the following HIV-1 template specific oligos: set 1: E6203S (sense, nt6203-nt6225, 5’-GAAAGAGCAGAAGACAGTGGCAA) and E80R1 (antisense, nt6879-nt6858, 5’-CACAATAATGTATGGGAATTGG); set 2: ED31 (sense, nt6817-nt6845, 5’-CCTCAGCCATTACACAGGCCTGTCCAAAG) and E7413A (antisense, nt7413-nt7391, 5’-TCTCCCAAGTACTATTAAACAGT); set 3: E7356S (sense, nt7356-nt7379, 5’-TGTGGAGGGGAATTTTTCTACTGT) and E7963A (antisense, nt7963-nt7941, 5’-ACTCTTGCCTGGAGCTGCTTGAT); set 4: E7932F (sense, nt-7932-7951, 5’-GTCTGGGGCATCAAACAGCT) and E8572 (antisense, nt8572-nt8548, 5’-TCCACAATCCTCGCTGCAATTAAGA); and set 5: E8258F (sense, nt8232-nt8258, 5’-TGGAATTGGTTTGACATATCAAAGTGG) and E8911A (antisense, nt8911-nt8887, 5’-TTTCTAGGTCTCGAGATACTGCTCC). To eliminate the possible PCR-induced env recombinants, we used Phusion High-Fidelity DNA Polymerase (NEB Inc.) which can rapidly complete the DNA extension (15-30 seconds per Kb) and longer extension time (1min) than the request to prevent the possible template switching during amplification. Amplicons were purified using the Agencourt AMPure XP (Beckman Coulter) magnetic bead PCR purification system, and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies). Libraries were constructed using equimolar concentrations of each amplicon, and sequenced on a 454 GS Junior System (Roche Diagnostics) using the GS Junior Titanium emPCR Kit (Lib-A). The resulting reads were trimmed to exclude the MIDs and primer sequence, and low-quality reads were filtered using the GS Run Processor according to length and quality scores. The sequence reads were analyzed for recombinants by uploading them into the Recombination Identifier Program (RIP 3.0) (http://www.hiv.lanl.gov/) along with their parental reference sequences.
Analysis of sequence entropy and its correlation with frequency of recombination
To determine the correlation between the amino acid sequence entropy and the frequency of recombination, the Shannon entropy of 1652 subtype A and 873 subtype D Env amino acid sequences from the HIV database (https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html) was calculated using an online tool available at the Los Alamos National Laboratory (LANL) HIV Sequence Database: http://www.hiv.lanl.gov/content/sequence/ENTROPY/entropy_one.html.
Acquisition and analysis of HIV-1 env recombinant sequences from HIV database
Recombinant HIV-1 env sequences were downloaded from the HIV Sequence Database at http://www.hiv.lanl.gov/. The search options were selected as follows: “HIV-1”, “include recombinants”, and “any” sequences start from”6202” to “8797”. These search options returned 10,049 sequences. Sequences listing multiple subtypes in the “subtype” column of the search results were presumed to be intersubtype Env recombinants. 997 such sequences were screened, downloaded in “fasta file” format, and analyzed by the jpHMM tool to determine HXB2 breakpoints and env gene maps. The final 77 Env recombinants with a single breakpoint within env region were selected for further analysis.
Highlights.
Successfully established a Functional Recombinant Production (FRP) system to generate pure and functional HIV-1 Env recombinants.
Established an in vitro model to mimic selection of replication-competent HIV-1 intersubtype recombination in dual or superinfected patients.
Selection of functional Env recombinants is one of the reasons for the predominance of certain HIV-1 recombinants in the epidemics.
Acknowledgements
This project is supported by NIAID, NIH, Grant R01 AI084816, the Fogarty International Center at NIH, Grant R21 AI079852, and CIHR grant HBF143165. We thank Dr. Kelly Summers at Western University for revising the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beemon K, Duesberg P, Vogt P: Evidence for crossing-over between avian tumor viruses based on analysis of viral RNAs. Proc Natl Acad Sci U S A 1974, 71: 4254–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogt PK: Genetically stable reassortment of markers during mixed infection with avian tumor viruses. Virology 1971, 46: 947–952. [DOI] [PubMed] [Google Scholar]
- 3.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA: Strand transfer events during HIV-1 reverse transcription. Virus Res 2008, 134: 19–38. [DOI] [PubMed] [Google Scholar]
- 4.Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U et al. : Recombination: Multiply infected spleen cells in HIV patients. Nature 2002, 418: 144. [DOI] [PubMed] [Google Scholar]
- 5.Wain-Hobson S, Renoux-Elbe C, Vartanian JP, Meyerhans A: Network analysis of human and simian immunodeficiency virus sequence sets reveals massive recombination resulting in shorter pathways. J Gen Virol 2003, 84: 885–895. [DOI] [PubMed] [Google Scholar]
- 6.Bull JJ, Sanjuan R, Wilke CO: Theory of lethal mutagenesis for viruses. J Virol 2007, 81: 2930–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quan Y, Liang C, Brenner BG, Wainberg MA: Multidrug-resistant variants of HIV type 1 (HIV-1) can exist in cells as defective quasispecies and be rescued by superinfection with other defective HIV-1 variants. J Infect Dis 2009, 200: 1479–1483. [DOI] [PubMed] [Google Scholar]
- 8.Summers J, Litwin S: Examining the theory of error catastrophe. J Virol 2006, 80: 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartolo I, Calado R, Borrego P, Leitner T, Taveira N: Rare HIV-1 Subtype J Genomes and a New H/U/CRF02_AG Recombinant Genome Suggests an Ancient Origin of HIV-1 in Angola. AIDS Res Hum Retroviruses 2016, 32: 822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskaleychik FF, Laga VY, Delgado E, Vega Y, Fernandez-Garcia A, Perez A et al. : [Rapid spread of the HIV-1 circular recombinant CRF02-AG in Russia and neighboring countries]. Vopr Virusol 2015, 60: 14–19. [PubMed] [Google Scholar]
- 11.Parczewski M, Leszczyszyn-Pynka M, Witak-Jedra M, Rymer W, Zalewska M, Gasiorowski J et al. : Distribution and time trends of HIV-1 variants in Poland: Characteristics of non-B clades and recombinant viruses. Infect Genet Evol 2016, 39: 232–240. [DOI] [PubMed] [Google Scholar]
- 12.Saeng-Aroon S, Loket R, Plipat T, Lumyai S, Chu PY, Sangkitporn S et al. : Circulation of HIV-1 Multiple Complexity Recombinant Forms Among Female Sex Workers Recently Infected with HIV-1 in Thailand. AIDS Res Hum Retroviruses 2016, 32: 694–701. [DOI] [PubMed] [Google Scholar]
- 13.Tongo M, Dorfman JR, Martin DP: High Degree of HIV-1 Group M (HIV-1M) Genetic Diversity within Circulating Recombinant Forms: Insight into the Early Events of HIV- 1M Evolution. J Virol 2016, 90: 2221–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuilleumier S, Bonhoeffer S: Contribution of recombination to the evolutionary history of HIV. Curr Opin HIV AIDS 2015, 10: 84–89. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Liang Y, Feng Y, Wang B, Li Y, Wu Z et al. : HIV-1 prevalence and subtype/recombinant distribution among travelers entering China from Vietnam at the HeKou port in the Yunnan province, China, between 2003 and 2012. J Med Virol 2015, 87: 1500–1509. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Wei H, Feng Y, Li J, Kalish ML, Lu H et al. : Genomic characterization of two novel HIV-1 second-generation recombinant forms among men who have sex with men in Beijing, China. AIDS Res Hum Retroviruses 2015, 31: 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei H, Xing H, Hsi JH, Jia M, Feng Y, Duan S et al. : The sexually driven epidemic in youths in China's southwestern border region was caused by dynamic emerging multiple recombinant HIV-1 strains. Sci Rep 2015, 5: 11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, Takebe Y, Wei H, He X, Hsi JH, Li Z et al. : Geographic origin and evolutionary history of China's two predominant HIV-1 circulating recombinant forms, CRF07_BC and CRF08_BC. Sci Rep 2016, 6: 19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Checkley MA, Luttge BG, Freed EO: HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 2011, 410: 582–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H et al. : Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med 1997, 3: 205–211. [DOI] [PubMed] [Google Scholar]
- 21.Frost SD, Wrin T, Smith DM, Kosakovsky Pond SL, Liu Y, Paxinos E et al. : Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A 2005, 102: 18514–18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rong R, Gnanakaran S, Decker JM, Bibollet-Ruche F, Taylor J, Sfakianos JN et al. : Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol 2007, 81: 5658–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neher RA, Leitner T: Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput Biol 2010, 6: e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shriner D, Rodrigo AG, Nickle DC, Mullins JI: Pervasive genomic recombination of HIV-1 in vivo. Genetics 2004, 167: 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mild M, Esbjornsson J, Fenyo EM, Medstrand P: Frequent intrapatient recombination between human immunodeficiency virus type 1 R5 and X4 envelopes: implications for coreceptor switch. J Virol 2007, 81: 3369–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratcliff AN, Shi W, Arts EJ: HIV-1 resistance to maraviroc conferred by a CD4 binding site mutation in the envelope glycoprotein gp120. J Virol 2013, 87: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagaya BS, Vega JF, Tian M, Nickel GC, Li Y, Krebs KC et al. : Functional bottlenecks for generation of HIV-1 intersubtype Env recombinants. Retrovirology 2015, 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon-Loriere E, Galetto R, Hamoudi M, Archer J, Lefeuvre P, Martin DP et al. : Molecular mechanisms of recombination restriction in the envelope gene of the human immunodeficiency virus. PLoS Pathog 2009, 5: e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archer J, Pinney JW, Fan J, Simon-Loriere E, Arts EJ, Negroni M et al. : Identifying the important HIV-1 recombination breakpoints. PLoS Comput Biol 2008, 4: e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baird HA, Galetto R, Gao Y, Simon-Loriere E, Abreha M, Archer J et al. : Sequence determinants of breakpoint location during HIV-1 intersubtype recombination. Nucleic Acids Res 2006, 34: 5203–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baird HA, Gao Y, Galetto R, Lalonde M, Anthony RM, Giacomoni V et al. : Influence of sequence identity and unique breakpoints on the frequency of intersubtype HIV-1 recombination. Retrovirology 2006, 3: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, Abreha M, Nelson KN, Baird H, Dudley DM, Abraha A et al. : Enrichment of intersubtype HIV-1 recombinants in a dual infection system using HIV-1 strain-specific siRNAs. Retrovirology 2011, 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCutchan FE, Hoelscher M, Tovanabutra S, Piyasirisilp S, Sanders-Buell E, Ramos G et al. : In-depth analysis of a heterosexually acquired human immunodeficiency virus type 1 superinfection: evolution, temporal fluctuation, and intercompartment dynamics from the seronegative window period through 30 months postinfection. J Virol 2005, 79: 11693–11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Powell RL, Lezeau L, Kinge T, Nyambi PN: Longitudinal quasispecies analysis of viral variants in HIV type 1 dually infected individuals highlights the importance of sequence identity in viral recombination. AIDS Res Hum Retroviruses 2010, 26: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galli A, Kearney M, Nikolaitchik OA, Yu S, Chin MP, Maldarelli F et al. : Patterns of Human Immunodeficiency Virus type 1 recombination ex vivo provide evidence for coadaptation of distant sites, resulting in purifying selection for intersubtype recombinants during replication. J Virol 2010, 84: 7651–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golden M, Muhire BM, Semegni Y, Martin DP: Patterns of recombination in HIV-1M are influenced by selection disfavouring the survival of recombinants with disrupted genomic RNA and protein structures. PLoS One 2014, 9: e100400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo J, Robertson DL, Lovell SC: Constraints from protein structure and intra-molecular coevolution influence the fitness of HIV-1 recombinants. Virology 2014, 454–455: 34–39. [DOI] [PubMed] [Google Scholar]
- 38.Dudley DM, Gao Y, Nelson KN, Henry KR, Nankya I, Gibson RM et al. : A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques 2009, 46: 458–467. [DOI] [PubMed] [Google Scholar]
- 39.Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y et al. : A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 2013, 13: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG et al. : Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999, 73: 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinones-Mateu ME, Ball SC, Marozsan AJ, Torre VS, Albright JL, Vanham G et al. : A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol 2000, 74: 9222–9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuiken CL, Foley B, Hahn B, Korber B, Marx PA, McCutchan F et al. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, New Mexico: HIV Sequence Compendium 2001 LA-UR 01–3860. 2001. Ref Type: Edited Book [Google Scholar]
- 43.Immonen T, Gibson R, Leitner T, Miller MA, Arts EJ, Somersalo E et al. : A hybrid stochastic-deterministic computational model accurately describes spatial dynamics and virus diffusion in HIV-1 growth competition assay. J Theor Biol 2012, 312: 120–132. [DOI] [PubMed] [Google Scholar]
- 44.Quinones-Mateu ME, Gao Y, Ball SC, Marozsan AJ, Abraha A, Arts EJ: In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J Virol 2002, 76: 9600–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjorndal A, Deng H, Jansson M, Fiore JR, Colognesi C, Karlsson A et al. : Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol 1997, 71: 7478–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber J, Vazquez AC, Winner D, Gibson RM, Rhea AM, Rose JD et al. : Sensitive cell-based assay for determination of human immunodeficiency virus type 1 coreceptor tropism. J Clin Microbiol 2013, 51: 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]