Abstract
Mechanical force sensation is fundamental to a wide breadth of biology from the classic senses of touch, pain, hearing, and balance to less conspicuous sensations of proprioception, blood pressure, and osmolarity and basic aspects of cell growth, differentiation, and development. These diverse and essential systems use force-gated (or mechanosensitive) ion channels that convert mechanical stimuli into cellular electrical signals. TRAAK, TREK1, and TREK2 are K+-selective ion channels of the two-pore domain K+ (K2P) family that are mechanosensitive: they are gated open by increasing membrane tension. TRAAK and TREK channels are expressed in are thought to play roles in somatosensory and other mechanosensory processes in neuronal and non-neuronal tissues. Here, we present protocols for three assays to study mechanical activation of these channels in cell membranes: (1) cell swelling, (2) cell poking, and (3) patched membrane stretching. Patched membrane stretching is also applicable to the study of mechanosensitive K2P channel activity in a cell-free system and a procedure for proteoliposome reconstitution and patching is also presented. These approaches have been used to show that TRAAK and TREK channels are membrane tension-gated mechanosensors are readily applicable to the study of other mechanosensitive ion channels.
Keywords: K2P ion channel, TREK1, TREK2, TRAAK, mechanosensitive ion channel, membrane tension gating, patch clamp, cell poking, cell swelling, patch inflation, proteoliposome reconstitution
1. Introduction
Mechanosensitive ion channels transduce mechanical forces into electrical currents. They underlie force sensation in hearing, balance, and touch sensation and an increasingly appreciated repertoire of less conspicuous force sensations. In fact, mechanosensitive ion channel activity is almost ubiquitous in cells, although the biological function of mechanosensitive currents in many contexts is unknown[1–5]. Cationic depolarizing, anionic depolarizing, and K+-selective hyperpolarizing mechanosensitive currents have been recorded due to the activity of different underlying ion channels. The K+-selective mechanosensitive currents are of two types: one is carried by a subset of voltage-gated K+-selective (Kv) channels[6, 7] and the second is carried by a subset of two pore domain K+-selective (K2P) channels[8, 9]. The three related mechanosensitive K2P channels TRAAK, TREK1, and TREK2 are intrinsically and remarkably mechanosensitive: they are activated by membrane tension generated by mechanical stimuli over essentially the entire range of their accessible open probability[10, 11].
While there are many possible ways to study ion channel mechanosensitivity, here we present three robust and complementary assays we routinely use for studying the mechanical activation of TRAAK and TREK K2P channels: cell swelling, cell poking, and patched membrane stretching. All three assays are applicable to recording channel activity in cells. Patched membrane stretching is also applicable to studying mechanical activation of K2Ps in a fully reconstituted system and a procedure for purification and reconstitution of mechanosensitive K2Ps into proteoliposomes for this purpose is also presented.
Cell swelling involves changing the osmolarity of the solution surrounding cells. Hypo-osmotic bath solution causes water influx into cells and the resulting volume change generates stress on the plasma membrane. Depending on the physiology of the cell under study, different cascades of downstream events can follow, but the initial swelling is thought to generate tension in the plasma membrane that can open mechanosensitive ion channels including TRAAK and TREK channels. Their activation can be assessed by electrophysiology. The advantages of this technique include simplicity and adaptability to most recording setups and experimental preparations. There are two major disadvantages of this technique. First, although solution composition can be controlled precisely, the assay is not perfectly quantitative because the mechanical response of different cells to the same osmotic stimulus can vary. Second, swelling is a slow process (typically on the order of seconds depending on the solution exchange system) compared to channel activation (which can occur on milli- to micro-second time scale). As a result, a number of intracellular signaling events are initiated in parallel with the increase in membrane tension. It is difficult to then distinguish whether changes in channel activity are due to tension changes or intracellular signaling events. Channel activation from hypo-osmotic shock thus may or may not be due to membrane tension sensitivity.
Cell poking involves mechanically stimulating an individual cell by depressing a blunt probe against its plasma membrane. The probe is mounted on a stacked piezo actuator to control displacement and the cell is simultaneously recorded with an electrode through a distal region of the membrane [12, 13]. Compared to swelling, poking stimuli can be delivered more rapidly (milliseconds). This permits kinetic aspects of channel activation to be reliably studied. Solution changes are not necessary and cells can be studied in a more physiological context. However, it is still a challenge to quantify mechanical activation that results from the membrane tension created by poking. While the position of the probe can be precisely controlled, the tension created by probe depression of the same distance will vary between cells as a result of their different volumes and morphologies and poking different parts of the same cell often generates different responses. Despite these caveats, cell poking is in many ways the best approach for studying mechanical activation of channels in a cellular context.
Patched membrane stretching involves applying steps of positive or negative pressure through the recording pipette to a patch of membrane under study. The membrane is stretched by the pressure change to create tension in the bilayer. Using a high-speed pressure clamp allows for rapid (milliseconds) and precise changes in the applied pressure[14]. There are several advantages to this approach compared to cell swelling and poking. First, it permits quantification of membrane tension. According to the Young-Laplace relationship, tension in the membrane is related to its radius of curvature and the pressure difference across the bilayer. These can be measured and tension calculated during patched membrane stretching [15]. Despite some caveats to this approach including a relatively high (~ 0.5–4 mN / m) and non-uniform tension in the patched membrane[16, 17], it has been instrumental in characterizing mechanosensitive channels [18–22]. Second, membrane stretching is applicable to patches from cells in any configuration (inside-out, outside-out, or cell-attached) and to patches from proteoliposomes containing purified channels reconstituted into fully defined lipid membranes[23]. The ability to study channel mechanosensitivity in the absence of all other cellular components is invaluable for understanding mechanisms of channel mechanosensitivity: it is the only rigorous way to exclude roles of other macromolecules in sensing and transmitting force to a mechanosensitive channel[24]. Without a reductionist approach, whether mechanical force opens the channel through tension in the surrounding membrane or by other means cannot be unequivocally determined. Reconstitution furthermore permits the chemical and physical properties of the surrounding lipid bilayer to be systematically varied so that their effects on channel activity can be determined.
Below we present procedures for studying mechanosensitivity of K2P ion channels using these complementary approaches: expression of mechanosensitive K2P channels in (1) cultured cells for study with cell swelling, cell poking, and patched membrane stretching assays and (2) in Pichia pastoris for purification, reconstitution into proteoliposomes, and study with patched membrane stretching.
2. Materials
2.1. Expression of mechanosensitive K2Ps in cultured cells
Host cells: HEK293T, CHO-K1, or SF9 cells (see Notes 1 & 2).
Growth media and reagents. HEK293T cell media: DMEM with 10% Fetal bovine serum (FBS), 1% Glutamax, and 1% non-essential amino acids. CHO-K1 cell media: DMEM:F12 with 10% FBS, and 1% non-essential amino acids. Trypsin, Dulbecco’s phosphate-buffered saline (DPBS), 35 mm plastic tissue culture-treated dishes, and poly-D-lysine (PDL) coated coverslips.
Transfection media and reagents: Fugene HD, Optimem (see Note 3).
Mechanosensitive K2P cloned into a suitable mammalian cell expression vector (e.g. human TRAAK in pCEH [21]).
2.2. Expression of mechanosensitive K2Ps in Pichia pastoris
2.2.1. Transformation of Pichia pastoris
Host cells: Pichia pastoris strain (e.g. SMD1163) stored as glycerol stock.
Mechanosensitive K2P cloned into suitable Pichia expression vector to generate a protease cleavable, EGFP and His-tagged fusion protein (e.g. human TRAAK-EGFP-10xHis in pPICZ [21])
Solutions for transformation: 3 M Sodium Acetate pH 5.2, 100% Ethanol, 70% Ethanol, ice-cold 1 M Sorbitol (0.22 μm filtered), 1 M HEPES-KOH pH 8.0, 1 M DTT, YPD (1% w/v yeast extract, 2% peptone, 2% dextrose, see Note 4), 100 mg/mL Zeocin in H2O.
Selection plates: YPD with 1 M sorbitol, 2% w/v agar, and 0.5 mg/mL Zeocin (final concentrations).
Equipment for transformation: electroporator, electroporation cuvettes, 250 mL baffled flask, glass plating beads.
2.2.2. Growth and expression of Pichia pastoris
Media components: 10X YNB (per liter: 100 g (NH4)2SO4 and 34 g yeast nitrogen base without amino acids and without ammonium sulfate), 10X Glycerol (10% v/v), 10X Potassium Phosphate (KPi) pH 6 (per liter: 52.26 g K2HPO4 and 95.26 g KH2PO4), 250X Biotin (0.1 mg/mL in H2O), Methanol (see Note 5).
Prepared media: Glycerol growth media (BMGY: 1X YNB, 1X Glycerol, 1X Biotin, 1X KPi pH 6) and Methanol expression media (BMMY: 1X YNB, 1X Biotin, 1X KPi pH 6, 0.5% v/v Methanol).
Materials to freeze cells: Liquid nitrogen, silicone kitchen spatulas, metal strainer, plastic bags.
2.3. Purification of mechanosensitive K2Ps from Pichia
2.3.1. Materials common to large and small scale procedures
Cryogenic mixer mill and liquid nitrogen.
Lysis Buffer (see Note 6, all final concentrations): 50 mM Tris-HCl pH 8.0, 150 mM KCl, 1 mM EDTA pH 8.0, DNAse (50 μg/mL), protease inhibitors (Leupeptin (1 μg/ml), Pepstatin A (1 μg/ml), Soy Trypsin Inhibitor (10 μg/ml), Aprotinin (1 μg/ml), Benzamidine (1 mM), PMSF (1 mM), see Note 7).
Dodecyl maltoside (DDM) (see Note 8).
Size exclusion chromatography (SEC) buffer (all final concentrations): 20 mM Tris-HCl pH 8.0, 150 mM KCl, 1 mM EDTA, 1 mM DDM.
Liquid chromatography instrument and Superose 200 10/300 size exclusion column (see Note 9).
2.3.2. Materials for small scale lysis and extraction testing
Eppendorf tube adapter for mixer mill.
0.1–0.25 mm glass beads.
8X DDM: 320 mM DDM in Lysis Buffer (163 mg DDM added to 900 μL Lysis Buffer).
2.3.3. Materials for large scale lysis and purification
50 mL grinding jars, 25 mm diameter ball bearings, O-rings, Styrofoam box for chilling grinding jars in liquid nitrogen.
Fritted glass column, peristaltic pump, column flow adapter, UV monitor, flatbed recorder.
Cobalt immobilized metal chromatography (IMAC) resin (e.g. Talon).
Lysis Buffer with 40 mM DDM.
PreScission protease.
IMAC buffers (volumes are for a purification from ~25 grams Pichia cells): TK (40 mL, 50 mM Tris-HCl pH 8.0, 150 mM KCl), TKD (10 mL, TK + 6 mM DDM), TKDI10 (50 mL, TKD + 10 mM imidazole), TKDI30 (25 mL, TKD + 30 mM imidazole), TKDI300 (25 mL, TKD + 300 mM imidazole).
2.4. Reconstitution of mechanosensitive K2Ps into proteoliposomes.
Lipids (e.g. L-α-phosphatidylcholine from soybean or pure lipids).
Chloroform, Pentane.
Argon gas stream supplied through a glass Pasteur pipette.
De/Rehydration (DR) Buffer (100 mL/sample, 200 mM KCl, 5 mM HEPES-KOH pH to 7.2, 0.2 μm filtered).
Vacuum chamber with Drierite desiccant.
Purified mechanosensitive K2P.
Bio-Beads SM-2 absorbent washed according to manufacturer’s instructions followed by one wash into DR buffer.
Ultracentrifuge.
35 mm glass bottom petri dishes.
2.5. Electrophysiology
2.5.1. Materials common to all techniques
Electrophysiology rig: inverted microscope (see Note 10) with two high-resolution micromanipulators (<1 μm step size) on an air table to minimize vibration, amplifier with head stage, digitizer, computer, and computer interface.
Electrode preparation: bleach, 1.5 mL Eppendorf tubes, silver wire.
Pipettes: pipette puller, 10 cm Borosilicate glass with filament O.D.: 1.5 mm, I.D.: 1.17 mm, microforge, 10X Eyepiece with protractor, 2 mL syringes, 0.22 μm filters and fine injection needles or microfil tips for filling.
2.5.2. Cell swelling
Pipette solution: 10 mM HEPES, 150 mM KCl, 3 mM MgCl2, 5 mM EGTA, pH 7.2 (adjusted with KOH). Filter to 0.22 μm.
Bath solution, iso-osmotic: 10 mM HEPES, 100 mM NaCl, 15 mM KCl, 3 mM MgCl2, 1 mM CaCl2, 60 mM Sucrose or Sorbitol. pH 7.3 (adjusted with NaOH), 315mOsm/kg.
Bath solution, hypo-osmotic: 10 mM HEPES, 100 mM NaCl, 15 mM KCl, 3 mM MgCl2, 1 mM CaCl2, 60 mM Sucrose or Sorbitol. pH 7.3 (adjusted with NaOH), 250mOsm/kg.
Grounding reservoir solution: 150 mM KCl.
Diamond-shape perfusion chamber compatible with coverslips with a small accessory reservoir for the grounding electrode (see Note 11).
Gravity-fed perfusion: 60 mL syringes, luer valve assortment kit and tubing, 18 or 20G Needles, tubing.
Salt bridges: 2–5% (w/v) agar in 1 M KCl, 10 cm Borosilicate glass with filament O.D.: 1.5 mm, I.D.: 1.17 mm, microforge.
2.5.3. Cell Poking
Pipette solution: 10 mM HEPES, 150 mM KCl, 3 mM MgCl2, 5 mM EGTA, pH 7.2 (adjusted with KOH), 0.22 μm filtered.
Bath solution: 10 mM HEPES, 15 mM KCl, 135 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, pH 7.3 (adjusted with NaOH), 0.22 μm filtered.
Piezo-driven actuator driven by a controller/amplifier controlled through the electrophysiology software.
Glass probes: pipette puller, 10 cm Borosilicate glass with filament O.D.: 1.5 mm, I.D.: 1.17 mm, microforge.
2.5.4. Patched membrane stretching
High Speed Pressure Clamp (HSPC), pump, and tubing.
Proteoliposome Bath Solution (see Note 12): 5 mM HEPES, 200 mM KCl, 40 mM MgCl2, pH 7.2 (adjust with KOH), 0.22 μm filtered.
Proteoliposome Pipette Solution: 5 mM HEPES, 20 mM KCl, 180 mM NaCl, pH 7.2 (adjust with NaOH), 0.22 μm filtered.
High magnification, high NA DIC objective (e.g. 100X 1.4 NA oil immersion) and image analysis software if calculating membrane tension from patch radius of curvature.
3. Methods
3.1. Expression of mechanosensitive K2Ps in cultured cells
HEK 293 or CHO cells should be kept using standard sterile technique in an appropriate cell culture environment. Cells should be routinely passaged before reaching 100% confluency.
Plate cells in 35 mm plastic dishes (for poking) or poly-D-lysine-coated coverslips (for cell swelling or best optics) 3 days before the experiment day at 0.2–0.5 × 106 cells per dish.
One day later, transfect cells using Fugene HD following recommended protocol (see Note 13). Cells will be optimal for recording 36–72 hours after transfection (see Note 14).
3.2. Expression of mechanosensitive K2Ps in Pichia pastoris
3.2.1. Transformation of Pichia pastoris
Purify pPICZ Pichia plasmid DNA with cloned mechanosensitive K2P.
Linearize 7.5 μg plasmid DNA with 2 μL PmeI at 37°C for 1.5 hours in the appropriate buffer in 50 μL total volume (see Note 15).
Ethanol-precipitate the DNA: Add 5 μL 3 M Sodium Acetate pH 5.2 and 225 μL 100% Ethanol and precipitate at −80°C for at least 1 hour. Centrifuge ≥ 13000 × g at 4°C for 20 minutes. Decant the supernatant carefully and wash the small clear pellet at least once with 1 mL 70% Ethanol. Dry the DNA at room temperature for at least 1 hour. Dissolve the dried DNA pellet in 10 μL 0.22 μm filtered H2O (see Note 16).
Inoculate 40 mL YPD in a 250 mL baffled flask with 600 μl of a SMD1163 Pichia pastoris glycerol stock.
Grow overnight (~18 hours) at 30°C with shaking at 250 RPM to an OD600 ~10.
Transfer to a 50 mL tube and centrifuge 4000 × g at 4°C for 5 min. Resuspend cell pellet with gentle vortexing in 30 mL YPD and 6 mL 1 M HEPES-KOH pH 8.0.
Add 1 mL sterile 1 M DTT and incubate at 30°C for ≥ 15 minutes.
Centrifuge 4000 × g at 4°C for 5 minutes and wash pellet twice with 50 mL ice cold 1 M Sorbitol.
Resuspend cell pellet with gentle vortexing in 300 μL ice cold 1 M Sorbitol. This suspension of competent Pichia cells should be stored on ice and is suitable for use for several hours.
Add 40 μL competent Pichia cells to the linearized DNA resuspended in H2O and mix.
Transfer cells and DNA to the bottom corner of pre-chilled electroporation cuvette. Gently tap to remove bubbles and small droplets bridging the electrodes (see Note 17).
Incubate on ice for ≥ 5 minutes.
Electroporate cells with voltage set to 2 kV. The decay time constant should be ~5 ms.
Add 1.0 mL ice cold 1 M Sorbitol, transfer to a culture tube and incubate at 30°C ≥ 1 hour.
Add 0.5 mL YPD and incubate at 30°C ≥ 1 hour with shaking at 250 RPM.
Plate 300 μL on YPDS plates with 500 μg/ml Zeocin.
Grow plates at 30°C for 3–4 days or until colonies are visible.
3.2.2. Growth and expression of Pichia pastoris
Because growth and expression conditions that maximize the yield of purified mechanosensitive K2Ps can vary based on the exact construct, strain, and growth environment, a procedure for optimization using a small-scale lysis and extraction test followed by fluorescence size exclusion chromatography is presented. Large-scale purification can then be performed using the optimal conditions.
3.2.2.1. Small scale growth
Inoculate 2.6 mL of BMGY + 0.5 mg/mL Zeocin in a culture tube with a single colony from a YPDS plate and grow at 30°C with shaking at 250 RPM overnight or until OD600 ~10–20.
Prepare glycerol stock by adding 600 μL cells to 300 μl 50% v/v sterile glycerol.
Centrifuge 4000 × g for 5 minutes, resuspend cells in 2 mL BMMY + 0.5 mg/mL Zeocin, and grow at 27°C with shaking at 250 RPM for 24 hours (see Note 18).
Centrifuge 4000 × g for 5 minutes, remove supernatant, and snap-freeze cells in liquid nitrogen. Store cell pellets at −80°C.
3.2.2.2. Large scale growth
Prepare starter culture: Inoculate 10 mL BMGY + 0.5 mg/mL Zeocin in a 50 mL baffled flask with ~100 μL of frozen glycerol stock and grow at 30°C with shaking at 250 RPM overnight (OD600 ≥ 10).
Prepare large-scale culture: Inoculate 1 L BMGY + 25 μg/mL Zeocin in a 2.8 L baffled flask with 10 mL starter culture and grow at 30°C with shaking at 250 RPM until OD600 ~20 (see Note 18).
Transfer culture to 1 L centrifuge bottle and centrifuge 8000 × g at 4°C for 10 minutes. Resuspend cells in 1 L BMMY + 25 μg/mL Zeocin, transfer to a clean 2.8 L baffled flask, and grow at 27°C with shaking at 250 RPM for 24 hours (see Note 19).
Transfer culture to 1 L centrifuge bottle, centrifuge 8000 × g at 4°C for 10 minutes, and discard supernatant. Scoop out pellet with an ethanol rinsed silicone spatula and scrape into a metal strainer submerged in liquid nitrogen to snap-freeze the cells. Transfer frozen cells to a plastic bag, record cell mass, and store cell pellets at −80°C.
3.3. Purification of mechanosensitive K2Ps from Pichia pastoris
3.3.1. Small scale lysis and extraction testing
Resuspend cells from small-scale growth (or ~50 mg of frozen cells) in 750 μL of Lysis Buffer in a 1.5 mL tube.
Add ~0.75 mL glass beads.
Mill 3 cycles at 30 Hz for 1.5 minutes per cycle. Incubate on ice for 2 minutes between cycles.
Taking care to avoid glass beads, transfer supernatant to a new 1.5 mL tube on ice, wait for a minute to allow any transferred beads to settle, and transfer 490 μL of supernatant to second new 1.5 mL tube.
Add 70 μL 8X DDM to lysate, mix gently, and extract at 4°C with rocking for ≥ 1 hour.
Centrifuge ≥ 13000 × g at 4°C for 1 hour to pellet insoluble material.
Load 100 μL onto SEC column equilibrated in SEC buffer and monitor fluorescence using HPLC/FPLC to assess expression level and biochemical stability.
3.3.2. Large scale purification of mechanosensitive K2Ps
Fit two 50 mL grinding jars with ball bearing and o-rings, seal tightly, and submerge in liquid nitrogen in a styrofoam box to cool.
Transfer 12.5 grams of frozen cells into each chamber (see Note 20).
Mill 5 cycles at 30 Hz for 3 minutes per cycle. Submerge chambers in liquid nitrogen between cycles.
Use a spatula to transfer milled cell powder into stirring extraction buffer at 4°C, cover, and stir gently at 4°C for 3 hours. Check pH periodically (typically after 30 and 90 minutes) and adjust pH to 8 as needed with KOH.
Transfer lysate to centrifuge tubes and pellet insoluble material by centrifuging 30,000 × g at 4°C for 45 minutes.
Transfer 4 mL bed volume IMAC resin to a 15 mL falcon tube and wash sequentially with H2O, TK, and TKD buffers by mixing and centrifuging at 2000 × g at 4°C for 3 minutes.
Collect the soluble cell extract in a plastic beaker and add washed IMAC resin. Stir gently at 4°C for 3 hours.
Collect resin in a fritted glass column.
Wash peristaltic pump, tubing, and UV monitor with H2O and a small amount of detergent-containing buffer (TKDI10). Blank the UV monitor. Connect the column and chart recorder to the UV monitor and the peristaltic pump to the column adapter and a buffer inlet line.
Wash the IMAC resin sequentially with TKDI10 and TKDI30 buffers until absorbance at 280 nm falls to baseline (see Note 21).
Elute the channel from the IMAC resin with TKDI300 buffer. Collect the peak fraction in a 15 mL falcon tube on ice. Immediately after elution, add EDTA pH 8.0 to 1 mM final concentration.
Add PreScission protease at a 1:50–1:500 molar ratio and incubate overnight at 4°C with gentle inversion to cleave the fused EGFP-10xHis tag.
Wash a spin concentrator (15 mL, 50 kDa MWCO) in TKDI300 or SEC buffer by centrifuging 1500 × g at 4°C for 2 minutes.
Pellet any insoluble material in the elution fraction by centrifuging 5000 × g at 4°C for 5 minutes.
Concentrate the elution fraction containing mechanosensitive K2P by applying to the washed spin concentrator and sequentially centrifuging at 3000 × g at 4°C for 5 minutes and mixing until the volume is ≤ 500 μL.
Load onto SEC column equilibrated in SEC buffer and monitor absorbance at 280 nm using HPLC/FPLC. Fractionate mechanosensitive K2P containing peak that elutes at ~15 mL.
Assess purity of SEC fractions by SDS-PAGE and pool fractions with pure channel. Quantify concentration of purified channel and store at 4°C until use in reconstitution.
3.4. Reconstitution of mechanosensitive K2Ps into proteoliposomes.
3.4.1. Lipid preparation
Transfer 4 mg of lipid per sample to a clean 15 mL glass test tube flushed with argon and fitted with a plastic screw cap with Teflon plug. Add 2 mL chloroform and dissolve solid lipids by gently rotating or inverting tube.
Dry lipids to a thin film at the bottom of the tube under an argon stream inside of a chemical hood. Submerge tube in a beaker of water during evaporation to keep from cooling excessively. Pop any lipid bubbles at the bottom of the tube with the tip of the glass pipette and keep under argon stream 5 minutes after last visible traces of chloroform have evaporated to promote drying.
Add 2 mL pentane and dissolve lipids by gently rotating or inverting tube.
Repeat drying process under argon stream, transfer to tube vacuum chamber with desiccant, and remove all solvent under vacuum for at least 2 hours in the dark.
Remove from vacuum and layer dried lipids with argon.
Add DR buffer (1 mL per 10 mg lipid) and bath sonicate until solution is transparent to ensure complete formation of small liposomes.
3.4.2. Channel reconstitution
Prepare reconstitutions in 15 mL centrifuge tubes. Each reconstitution consists of 4 mg of prepared lipids (0.4 mL of 10 mg/mL solution), a variable amount of channel in SEC buffer (see Note 22), additional SEC buffer to ensure the final concentration of detergent remains above its critical micelle concentration, and DR buffer to make a final volume of 4 mL. Channel should be added last.
Layer with argon gas and rock at 4°C for ≥ 1 hour.
Add ~500 mg prepared Bio-Beads and rock at 4°C for ≥ 3 hours.
Transfer the supernatant to an ultracentrifuge tube. Avoid transferring the beads.
Centrifuge ≥ 130000 × g at 4°C for 1 hour.
Remove the supernatant, resuspend the pellet in 80 μL DR buffer, aliquot 20 μL samples into microcentrifuge tubes, snap-freeze in liquid nitrogen, and store at −80 °C until use.
3.5. Electrophysiology
3.5.1. Setup common to all techniques
Most electrophysiology rigs can be easily adjusted to perform these experiments. An inverted microscope most easily accommodates the patching electrode and poking probe above the cells. The patching electrode should be mounted on the headstage, which is attached to a micromanipulator positioned on one side of the sample.
Prepare electrodes: chlorinate the test and bath silver wire electrodes to replenish the active AgCl coating by immersing in fresh bleach in a 1.5 mL microcentrifuge tube for 10–20 minutes. Rinse with water before using. (see Note 23).
Prepare salt bridges (see Note 24): bend the shaft of a borosilicate glass tube under a flame to obtain a U-shaped cylinder and break to an appropriate size for bridging solutions in the recording chamber and grounding well. Fill the glass with agar by dipping the ends in 2% agar dissolved in 3 M KCl, dry bridges, and store in 3 M KCl at 4°C.
Prepare recording pipettes: Follow the pipette puller’s recommended settings to pull pipettes of 2.5–4.0 MΩ access resistance when filled with pipette solution. Lightly polish with a microforge. Fill pipettes with a microneedle attached to a 3 mL syringe with a 0.22 m filter immediately before use.
3.5.2. Cell Swelling
Prepare the gravity-assisted perfusion device: place two 60 mL syringes 30–40 cm above the microscope stage to serve as solution reservoirs. Connect their outlets through plastic tubing to a Y-shape connector and connect a needle at the end of the single output tube to deliver solution to the perfusion chamber. Place a roller or slider clamp on the single output tube and pinch clamp valves on the tubes of both solution reservoirs to control flow rate (see Note 25).
Prepare the solution collection system: Connect a second needle to a vacuum source through a fluid trap placed inside the Faraday cage. The tip of the collection needle will be placed on the side of the recoding chamber opposite to the perfusion delivery needle ~5 mm from the bottom of the chamber to keep the height of the solution constant without drying the dish.
Fill the solution reservoirs with iso- and hypo-osmotic bath solutions. Flush air from the tubes by opening all pinch clamps, turning the flow to high, and collecting the solution in a waste container. Once cleared of air bubbles, adjust the flow to 2–3 mL/minute and close pinch clamps. Fill tubing with iso-osmotic bath solution by flowing for 1–2 minutes.
Rinse cells with iso-osmotic bath solution and place one coverslip in the recording chamber in 0.5 mL iso-osmotic bath solution.
Place the chamber on the microscope stage. Fill the grounding reservoir with 150 mM KCl solution. Connect the grounding well to the perfusion chamber through the salt bridge. Place inlet and vacuum needles at opposing ends of the recording chamber.
Open the iso-osmotic solution reservoir first, and then the vacuum line, to establish the initial flow and liquid level.
Localize the cells on the microscope’s field of view using the 20X objective and adjust the microscope’s settings to achieve the best possible contrast.
Find a cell of interest and achieve a high resistance seal in whole cell mode.
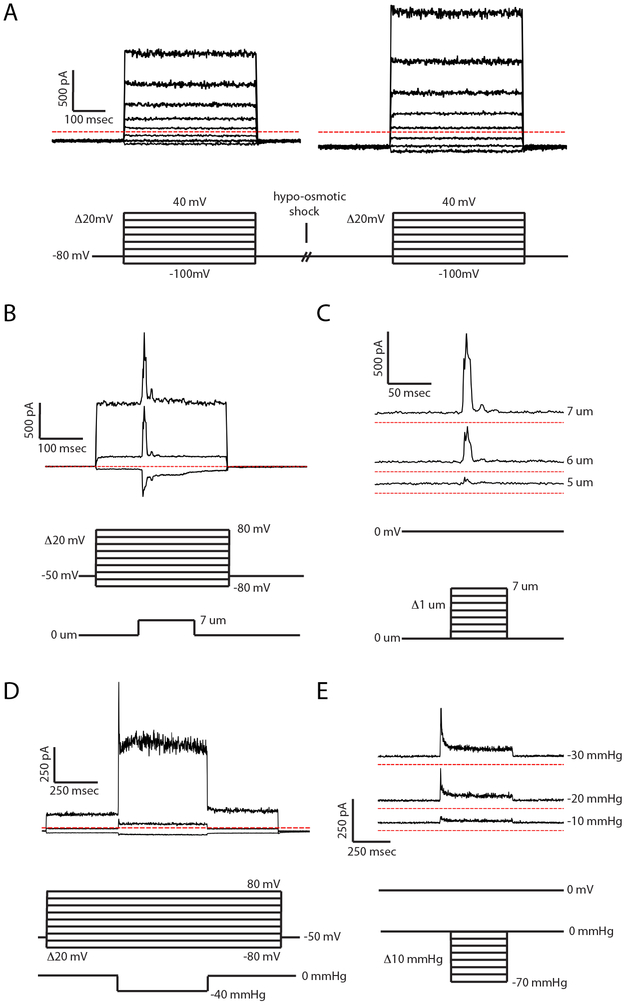
Record families of currents evoked by holding voltages varied from −100 mV to 100 mV in 20 mV steps until stable currents are obtained (Fig. 1A).
Swell cells by opening the clamp for the hypo-osmotic solution and closing the clamp for the iso-osmotic solution. Within a few seconds, the cell should be visibly swollen (see Note 26). Record families of currents at varying holding voltage using the same protocol. It is convenient to simultaneously record a movie of the process. The cell can be returned to the iso-osmotic solution to study reversibility of the process (see Notes 27 & 28).
Figure 1:
Protocols for electrical recording and mechanical stimulation for (A) cell swelling, (B,C) cell poking, and (D,E) patched membrane stretching (see Note 40). Example current recordings from human TRAAK expressed in cells are presented above the protocol diagrams and were obtained using the bath and pipette solutions described in the text. (A) Two voltage family protocols are separated by a solution exchange to induce hypo-osmotic swelling. Each voltage family consists of steps to test voltages ranging from −100 mV to +40 mV in 20 mV increments for 400 ms from a holding voltage of −80 mV. (B) A variable voltage, fixed probe depression family. The voltage family is analogous to (A) with the addition of a 150 ms probe depression of a set distance during each voltage step. Only currents from sweeps at −80 mV, 0 mV, and 80 mV are shown. (C) A fixed voltage, variable probe depression family. Sweeps are performed from a holding voltage of 0 mV. During each sweep, the probe is depressed for 100 ms. The depth of depression is increased by 1 μm in successive sweeps. Only currents from sweeps of 5, 6, and 7 μm depression are shown. (D) A variable voltage, fixed pressure application family. The voltage family is analogous to (A,C) with the addition of a 500 ms pressure pulse of a set value during each voltage step. (E) A fixed voltage, variable pressure family. Sweeps lasting one second each are performed from a holding voltage of 0 mV. During each sweep, pressure is applied for 250 ms. The amount of pressure applied is increased by 10 mmHg in successive sweeps. Only currents from −10, −20, and −30 mmHg are shown.
3.5.3. Cell Poking
Prepare poking apparatus: add a second micromanipulator opposite to the one holding the recording electrode and mount the poking apparatus onto it. The recording electrode and poking probe should each approach the sample ~45° from the horizontal surface.
Prepare poking probes: pull pipettes from borosilicate glass using a similar program as for patching. Immediately after pulling, use a microforge to seal the tips by holding the tip close to heat source until the opening appears rounded and closed. The tips should be smooth and ~2–3 μm in diameter.
Rinse cells with bath solution and add 2 mL bath solution or enough volume to obtain a depth of ~0.5 cm in the recording chamber.
Localize the cells on the microscope’s field of view using the 20X objective and adjust the microscope’s settings to achieve the best possible contrast.
Place a poking probe in its holder and position its tip near the region of interest ~200 μm above the cells in a corner of the field of view. Avoid crashing the tip against the glass surface (see Note 29).
Find a cell of interest and achieve a high resistance seal in whole cell mode. Optimal cells for this experiment are relatively large (~15 μm in one dimension) and minimally flat against the surface. Choose a corner of the cell to establish the gigaseal in order to leave most of the cell surface open for mechanical stimulation.
Record families of currents evoked by voltage from −100 mV to 100 mV in 20 mV steps until stable currents are obtained. Ensure the cell is healthy, the seal is tight, the access resistance is not too high, and the cell displays the expected currents.
Choose a bulky area of the cell for poking. Bring the tip of poking probe to within 2 μm from the surface of the cell (see Note 30). Record families of currents at varying holding voltage with a fixed depth probe stimulation or families of currents at a fixed holding voltage with varying depth probe stimulations. It is convenient to simultaneously record a movie of the process (see Notes 31 & 32) (Fig. 1B,C).
3.5.4. Patched membrane stretching
Membrane patches from cells or from proteoliposomes can be stretched to activate embedded mechanosensitive K2Ps. Cell preparations are as described above. Considerations for bending pipettes to enable patch imaging and membrane tension calculation, preparation of proteoliposomes for patching, and setup of the recording rig for patched membrane stretching are described below.
3.5.4.1. Bending pipette tips
If desired, pipette tips can be bent in order to observe changes in the radius of curvature of the patched membrane so that resulting changes in membrane tension can be calculated.
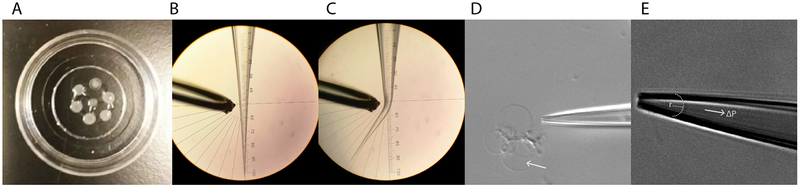
Place polished 2.5–4 MΩ tip in the microforge, center and align the pipette tip along the vertical axis of the eyepiece protractor, and orient the heating filament in the plane of the tip on the side of the eyepiece protractor (Fig 2B,C).
Heat the shank of the tip where the horizontal and vertical axes of the eyepiece protractor cross. The tip will bend towards the filament. Match the angle between the micromanipulator and the sample plane such that when the bent tips are loaded in the pipette holder, the tip shank is approximately parallel to the sample plane (see Note 33).
Figure 2:
Proteoliposome reconstitution and patched membrane stretching. (A) Photograph of dehydrated/rehydrated proteoliposomes in a 35 mm glass bottom dish prior to addition of bath solution. Seven, three-microliter drops of proteoliposomes were dehydrated prior to rehydration overnight with 20 μL DR buffer. (B) A pipette tip positioned next to the filament of a microforge (left) prior to and (right) after application of heat to bend the tip to a ~35° angle. (D) A bent pipette tip positioned adjacent to a group of unilamellar proteoliposomes to be patched. (E) A bent pipette tip with patched membrane. The radius of curvature of the membrane can be fit and used with the recorded pressure difference to calculate tension within the membrane.
3.5.4.2. Preparing proteoliposomes for patching
Thaw an aliquot of proteoliposomes and dispense in several drops on a 35 mm glass bottom dish.
Dry ≥ 3 hours in a vacuum chamber in the dark to dehydrate the proteoliposomes.
Rehydrate the proteoliposomes with 20 μL DR buffer. The dry lipids spots should not be touching. Add DR buffer such that each lipid cake is covered and drag the solution between the cakes to connect as many as possible. Place rehydrating proteoliposomes within a humid chamber at 4°C overnight (Fig. 2A).
Move the sample to the microscope stage and gently add ~5 mL of proteoliposome bath solution to the dish. Avoid letting solution rush over the proteoliposomes to limit floating debris that can clog pipettes and inhibit seal formation.
Localize a suitable unilamellar proteoliposome in the microscope’s field of view using the 20X or 40X objective and adjust the microscope’s settings to achieve the best possible contrast (Fig. 2D, see Note 34).
3.5.4.3. Patched membrane stretching and analysis
Prepare pressure clamp: connect a pressure clamp headstage to the port of the recording electrode holder with a short piece of tubing. Stable connections are important to minimize pipette drift (see Note 35). Connect the pressure clamp headstage to a vacuum and pressure pump and controller such that pressure can be controlled through the digitizer and recording software.
Load a pipette and achieve a high resistance seal in the appropriate configuration. Cell-attached, inside-out, or outside-out patches can be studied from cells. Proteoliposome patches form in the inside-out configuration (see Note 36 & 37).
Record families of currents evoked by voltage from −100 mV to 100 mV in 20 mV steps until stable currents are obtained and to ensure good seal formation.
Record families of currents at varying holding voltage with a fixed pressure pulse or families of currents at a fixed holding voltage with varying pressure stimulations (Fig. 1D,E see Note 38).
If using bent pipette tips, a movie can be simultaneously recorded. Before stimulation, move the pipette to a field of view free from background material. Switch to a high magnification objective (e.g. 100X DIC 1.4NA oil immersion) prior to imaging.
Tension (T) may be estimated in each frame by applying the Young-LaPlace equation, T= ΔPR/2, with R equal to the radius of curvature of the patched membrane and ΔP equal to the pressure difference across the membrane (Fig. 2E). Patched membrane radii of curvature can be fit manually in conventional image analysis software (e.g. ImageJ) or automatically with a custom script. Tension changes can then be correlated with the electrophysiological data (see Note 39).
Footnotes
We have found that common cell lines vary greatly in their expression of endogenous currents (both mechanosensitive and otherwise) and ease of transfection, patching, and growth. In our experience, CHO-K1 cells are particularly suitable for studying mechanosensitive channel activity because they are electrically quiet and have only small (or not measurable) mechanically activated currents. A disadvantage of CHO cells is that they tend to express transfected channels at lower current densities compared to other cells. HEK293T cells are easier to transfect, patch, and tend to grow in most conditions, but typically contain significant endogenous voltage-gated potassium channel activity and Piezo1-dependant mechanosensitive currents [25]. If HEK cells are to be used, endogenous Piezo1 activity can be distinguished from transfected K+ channels through the reversal potential of the currents evoked. Piezo1 currents reverse around 0 mV under the ten-fold potassium concentration gradient used throughout this chapter, while K+ currents reverse at EK = −59 mV.
Cultured insect cells can be used as an alternative to mammalian cells. We have successfully used these techniques with Sf9 cells, which express low levels of endogenous channel activity and are amenable to patching.
We prefer Fugene for transfection as it is minimally toxic to cells.
Prepare by autoclaving yeast extract and peptone, cooling, and adding 0.22 μm filtered 20% dextrose.
YNB, glycerol, and KPi can be autoclaved and Biotin should be filter sterilized.
This general lysis buffer is appropriate for purification of human TRAAK and zebrafish TREK1 channels. Modifications may be necessary to promote biochemical stability of other channels.
Leupeptin, Soy Trypsin Inhibitor, Aprotinin, and Benzamidine (1 mM) are prepared as 1000X stocks in water, aliquoted, and stored at −20°C. Pepstatin A is prepared as a 1000X stock in 9:1 v/v Methanol:Acetic acid, aliquoted, and stored at −20°C. PMSF is prepared as a 100X stock in ethanol and stored at room temperature with molecular sieves to keep anhydrous. PMSF should be added immediately prior to addition of cells.
Detergents are stored under argon in a desiccator at −20°C. Prior to use, they are warmed to room temperature before opening to prevent condensation inside the bottle.
A single HPLC or FPLC equipped with an absorbance detector (280 nm), fluorescence detector (EGFP fluorescence; 488 nm excitation 508 nm emission), and fraction collector would be sufficient for both small and large scale procedures. We use a dedicated HPLC with an autosampler for small scale testing and FPLC for large scale purifications.
Optics should minimally be sufficient to discern features of mammalian cells. A 20X DIC objective is sufficient for most procedures, but 40X-100X DIC objectives are desirable.
In our experience, a diamond-shaped chamber gives the best result as it ensures laminar flow across the bath. The recording chamber should be able to easily accommodate coverslips and have a small volume (less than 500 μL). If the perfusion chamber does not include the reservoir for the grounding electrode, a de-capped 1.5 mL tube attached to one side of the chamber can be used.
If recording from patched membranes from cells, use the pipette and bath solutions in 2.5.3.
Other transfection reagents require higher cell confluency at the time of transfection.
Overgrown cells are not optimal for recording. HEK cells in particular will develop electrical connections between adjacent cells in close proximity, generating very large capacitive peaks and endogenous currents.
PmeI linearizes pPICZ within the 5’ AOX1 promotor region. Additional PmeI sites must not be present within the mechanosensitive K2P gene.
The presence of residual salts in the linearized DNA reduces the transformation efficiency.
Arcing during electroporation will result in a loud popping or cracking sound. Cells may still be plated, but the efficiency will be lower. The two causes of this are high concentrations of residual salts from insufficient washing of DNA and the presence of small bubbles or droplets bridging the cuvette plates.
We find reducing the concentration of zeocin at this stage to a level sufficient to inhibit bacterial growth is sufficient to prevent contamination and had no negative consequences on expression or yield.
Expression times can be varied and optimal expression assessed by small scale lysis and FSEC. Human TRAAK is optimally expressed ~48 hours post methanol induction.
In our experience, ~0.5 mg of human TRAAK or zebrafish TREK1 can be purified from 25 g Pichia cells which is approximately the amount recovered from a 1 L culture. Volumes are reported for purification from 25 g of cells.
The amount of contaminating proteins eluting in TKDI30 will vary and human TRAAK and zebrafish TREK1 will elute almost exclusively in TKDI300.
We have found that reconstitution ratios of 1:10 (w/w) channel:lipid give large (nA) currents in patches and ratios of 1:1000 result in a few channels per patch.
Electrodes should be routinely chlorinated to recoat with AgCl. Once every 1–3 days of active recording is typically acceptable.
The grounding electrode should be placed in a separate grounding well connected to the recording chamber with a salt bridge to avoid offset artifacts from the changes in liquid junction potential during the experiment. This is particularly important for swelling experiments where the bath is constantly perfused. We store prepared salt bridges for months at 4°C with no contamination, but sodium azide can be added to the storage solution to prevent microbial growth if desired.
This simple setup can be extended to deliver more solutions by using manifold valves.
Most cells subjected to hypo-osmotic shock will gradually swell and then recover to close to their original size upon switching back to iso-osmotic solution. However, in some cases we observe a membrane bleb form around the cell during swelling. This may be due to membrane detachment from the underlying cytoskeleton, appears to be irreversible, and accompanies cell death. We do not use recordings from cells in this state.
HEK and CHO cells are healthy to record from for 1–3 hours after being removed from the incubator. However, we do not record from a single cell for more than ~15 minutes as cells usually begin showing signs of compromised health after this time.
We replace the coverslip after each swelling trial because it is unclear whether multiple rounds of swelling are detrimental to cell health or result in changes to cellular physiology.
We use the same poking probe for successive experiments.
The probe can be brought down until it just touches the cell to establish the position at which contact occurs and then immediately brought back up by a known distance. It is important to start the recording without stimulation such that the poking probe is not touching the cell.
Occasionally the cell will show a surface indentation at the point of probe contact. In this case, we withdraw the poking probe and reposition it in another part of the cell.
We find that different parts of the cell respond with variable magnitude, suggesting that there is a positional aspect to the experiment. It is unclear whether this is due to nonuniform spatial distribution of channels and/or membrane tension upon poking. Different target regions of the cell can be evaluated to identify the position for recording maximal poking-evoked currents.
In practice, angles ~5 degrees shallower than the angle between the micromanipulator and the sample plane are preferred. This prevents tips from being discarded because they are over bent and cannot reach the sample.
Multilamellar proteoliposomes will appear thicker at their edges and will not form proper high resistance seals. Unilamellar proteoliposomes are typically located at the edges of lipid cakes, or more frequently, loosely tethered to veins of dried proteoliposomes that emerge from and connect the dried lipid cakes. Intact unilamellar proteoliposomes will quiver if the microscope stage is gently tapped.
The most common sources of pipette drift in our experience are: (1) over or under-tightened micropipette holder connections and (2) cable forces from the connection between the pipette holder and pressure clamp headstage. Incorporating a luer connection which is fixed to the microscope stage with wax between the pressure clamp headstage and pipette holder can minimize these cable forces and drift.
If patching proteoliposomes, apply positive 5–10 mmHg pressure to keep the pipette tip clean while approaching the target membrane. Upon contacting the proteoliposomes, dropping the pressure to between 0 and −10 mmHg typically results in a rapid GΩ seal in the inside-out configuration. Maintain zero or slightly positive pressure on the patch in between negative test pressure pulses to minimize creep during the experiment.
Proteoliposome patches containing mechanosensitive K2Ps will immediately show increases in K+ current upon pressure stimulation. Patches from cells, in contrast, often display only small increases in K+ current upon pressure stimulation immediately after formation or excision. The magnitude of the response increases over time to a stable regime and can be accelerated by “exercising” the patch with steps of applied pressure. This run-up phenomena is likely due to the gradual dissolution of cortical cytoskeleton that is initially intact in the patch membrane as inclusion of cytoskeleton disrupting drugs in the bath solution accelerates this process. We exercise patches from cells with steps of moderate pressure until a stable response is observed.
While pressure steps in either direction will activate mechanosensitive K2Ps in patches of any configuration, in our experience, cell-attached, inside-out, and proteoliposome patches are most mechanically stable when stimulated with negative pressure and outside-out patches are most stable when stimulated with positive pressure.
A trigger cable can be used to imitate camera acquisition at the onset of electrical recording to facilitate data synching. Alternatively, images and recordings may be synchronized using time stamps in the metadata.
Mechanosensitive potassium currents should display a reversal potential of close to the equilibrium potential for K+ which is approximately −59 mV in the ten-fold K+ concentrations used here. This feature can be used to distinguish mechanically evoked activation of potassium currents from background and leak currents. It is convenient to using a holding voltage of 0 mV in the constant voltage protocols because currents from non-selective mechanosensitive channels (and leak currents) will be zero at this voltage, while K+-selective currents from mechanosensitive K2Ps will be outward and non-zero.
References
- 1.Ranade SS, Syeda R, Patapoutian A (2015) Mechanically Activated Ion Channels. Neuron 87:1162–1179. doi: 10.1016/j.neuron.2015.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delmas P, Coste B (2013) Mechano-Gated Ion Channels in Sensory Systems. Cell 155:278–284. doi: 10.1016/j.cell.2013.09.026 [DOI] [PubMed] [Google Scholar]
- 3.Nilius B, Honore E (2012) Sensing pressure with ion channels. Trends Neurosci 35:477–486. doi: 10.1016/j.tins.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 4.Anishkin A, Loukin SH, Teng J, Kung C (2014) Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc Natl Acad Sci USA 111:7898–7905. doi: 10.1073/pnas.1313364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Lewis AH, Grandl J (2016) Touch, Tension, and Transduction – The Function and Regulation of Piezo Ion Channels. Trends Biochem Sci 1–15. doi: 10.1016/j.tibs.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao J, Padilla F, Dandonneau M, et al. (2013) Kv1.1 Channels Act as Mechanical Brake in the Senses of Touch and Pain. Neuron 77:899–914. doi: 10.1016/j.neuron.2012.12.035 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt D, del Mármol J, Mackinnon R (2012) Mechanistic basis for low threshold mechanosensitivity in voltage-dependent K+ channels. Proc Natl Acad Sci USA 109:10352–10357. doi: 10.1073/pnas.1204700109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enyedi P, Czirják G (2010) Molecular background of leak K+ currents: two-pore domain potassium channels. Physiol Rev 90:559–605. doi: 10.1152/physrev.00029.2009 [DOI] [PubMed] [Google Scholar]
- 9.Honore E (2007) The neuronal background K2P channels: focus on TREK1. Nat Rev Neurosci 8:251–261. doi: 10.1038/nrn2117 [DOI] [PubMed] [Google Scholar]
- 10.Brohawn SG (2015) How ion channels sense mechanical force: insights from mechanosensitive K2P channels TRAAK, TREK1, and TREK2. Ann N Y Acad Sci 1352:20–32. doi: 10.1111/nyas.12874 [DOI] [PubMed] [Google Scholar]
- 11.Brohawn SG, Campbell EB, Mackinnon R (2014) Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature 516:126–130. doi: 10.1038/nature14013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao J, Delmas P (2011) Recording of mechanosensitive currents using piezoelectrically driven mechanostimulator. Nat Protoc 6:979–989. doi: 10.1038/nprot.2011.343 [DOI] [PubMed] [Google Scholar]
- 13.Coste B, Mathur J, Schmidt M, et al. (2010) Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330:55–60. doi: 10.1126/science.1193270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Besch S, Suchyna T, Sachs F (2002) High-speed pressure clamp. Pflugers Arch 445:161–166. doi: 10.1007/s00424-002-0903-0 [DOI] [PubMed] [Google Scholar]
- 15.Sokabe M, Sachs F, Jing ZQ (1991) Quantitative video microscopy of patch clamped membranes stress, strain, capacitance, and stretch channel activation. Biophys J 59:722–728. doi: 10.1016/S0006-3495(91)82285-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sukharev SI, Sachs F (2012) Molecular force transduction by ion channels - diversity and unifying principles. J Cell Sci 125:3075–3083. doi: 10.1242/jcs.092353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opsahl LR, Webb WW (1994) Lipid-glass adhesion in giga-sealed patch-clamped membranes. Biophys J 66:75–79. doi: 10.1016/S0006-3495(94)80752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moe P, Blount P (2005) Assessment of Potential Stimuli for Mechano-Dependent Gating of MscL: Effects of Pressure, Tension, and Lipid Headgroups †. Biochemistry 44:12239–12244. doi: 10.1021/bi0509649 [DOI] [PubMed] [Google Scholar]
- 19.Lewis AH, Grandl J (2015) Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. eLife 4:143. doi: 10.7554/eLife.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukharev SI (2002) Purification of the Small Mechanosensitive Channel of Escherichia coli (MscS): the Subunit Structure, Conduction, and Gating Characteristicsin Liposomes. Biophys J 83:290–298. doi: 10.1016/S0006-3495(02)75169-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brohawn SG, Su Z, Mackinnon R (2014) Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc Natl Acad Sci USA 111:3614–3619. doi: 10.1073/pnas.1320768111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berrier C, Pozza A, de Lacroix de Lavalette A, et al. (2013) The purified mechanosensitive channel TREK-1 is directly sensitive to membrane tension. Journal of Biological Chemistry. doi: 10.1074/jbc.M113.478321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinac B, Rohde PR, Battle AR, et al. (2009) Studying Mechanosensitive Ion Channels Using Liposomes In: Lipid-Protein Interactions. Humana Press, Totowa, NJ, pp 31–53 [DOI] [PubMed] [Google Scholar]
- 24.Teng J, Loukin S, Anishkin A, Kung C (2014) The force-from-lipid (FFL) principle of mechanosensitivity, at large and in elements. Pflugers Arch. doi: 10.1007/s00424-014-1530-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubin AE, Murthy S, Lewis AH, et al. (2017) Endogenous Piezo1 Can Confound Mechanically Activated Channel Identification and Characterization. Neuron 94:266–270.e3. doi: 10.1016/j.neuron.2017.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]