Abstract
Endometrial carcinoma is one of the most common tumours in developed countries. In addition to the active role of genetic factors, epigenetic changes also have an important effect. The present study analysed the methylation status of kruppel like factor 4 (KLF4) and heparan sulfate-glucosamine 3-sulfotransferase 2 (HS3ST2) genes in three endometrial tissue types for carcinoma prediction. The sample comprised 91 women with histologically-confirmed endometrial carcinoma (64.16±9.64 years old), 36 women with hyperplasia (53.39±9.64 years old) and 45 with no signs or symptoms of malignancy (48.53±11.11 years old). The CpG dinucleotide methylation levels were examined by quantitative pyrosequencing, and the discrimination accuracy of the model was calculated using the Random Forest classification algorithm of the area under the ROC curve (AUC). The mean values of KLF4 and HS3ST2 methylation indices were 23.83±11.39 and 8.52±2.57 in the control samples; 30.40±8.52 and 33.76±20.66 in hyperplasia and 34.72±10.79 and 34.49±18.39 in the cancerous tissues. Multinomial logistic regression indicated that the HS3ST2 CpG1 methylation status is a predictor of hyperplasia (P<0.05) and that the KLF4 CpG2 dinucleotide can predict carcinoma formation (P<0.001). The AUC value of 0.95 indicates high discrimination accuracy of the CpG nucleotides methylation status model between the controls and the two other diagnoses. The results of the present study establish the likelihood that aberrations in KLF4 and HS3ST2 gene methylation levels are important in the development of endometrial hyperplasia and carcinoma, with hyperplasia an intermediate step between healthy and tumour tissues.
Keywords: CpG sites, quantitative methylation, pyrosequencing, Random Forest classification algorithm, area under the ROC curve
Introduction
Although endometrial carcinoma (EC) is one of the most frequent malignant gynaecological diseases in developed countries, its occurrence in developing countries is lower (1). EC in North American and European female populations accounts for almost 6% of all cancer cases and 3% of all cases of cancer-associated mortality, and the incidence varies between 19 and 25/100,000 women (2). Although ~10% of diagnosed EC is hereditary (3), the remaining 90% is sporadic; with EC typical in older, postmenopausal women. Statistics also show that 15% of women are diagnosed before the age of 50 years, and 5% before the age of 40 years (4).
The formation of EC is associated with the presence of polycystic ovary syndrome (5), obesity (6), nulliparity (7), hyperinsulinaemia (8) and excessive exposure to oestrogen manifested in earlier menarche (9). The therapeutic use of tamoxifen in women with, or at high risk of, breast cancer is also potentiality implicated in increased EC incidence (10) and there are further inter-connections, including in vitro-fertilisation treatment and higher frequencies of miscarriages and abortions (11).
Endometrial hyperplasia is another important risk factor in EC as it can develop into adenocarcinoma. There are four types of endometrial hyperplasia: Simple hyperplasia (progressing to cancer in 1% of cases), complex hyperplasia (progressing to cancer in 3%), simple atypical hyperplasia (progressing to cancer in 8%), and complex atypical hyperplasia (progressing to cancer in 29%) (12).
Endometrial malignancies are traditionally classified under types I and II, dependent on hyperplasia formation (13). Type I distinguishes 70–80% of tumours and these are characterised as oestrogen-dependent and preceded by hyperplasia formation. They are low grade, diploid and highly differentiated and more common in obese women. The remaining 20–30% is classified as Type II, which is considered oestrogen-independent and associated with atrophy and aneuploidy. These are high-grade, poorly differentiated and linked to higher metastatic risk and resultant poor prognosis (14).
Types of EC are differentiated by histopathological characteristics into endometrioid carcinoma (up to 75%, typically associated with type I tumours), serous carcinoma, carcinosarcoma and clear-cell carcinoma (15). Although this classification remains the main diagnostic tool, studies have highlighted the importance of incorporating genetic profile and risk determination models (16–18). Molecular approaches may also describe EC biological characteristics and features more accurately, distinguish between subtypes, and substantially improve predictive and treatment approaches (16,18–22) as each EC subtype has a distinct mutation profile (17,18,23).
In addition to genetic factors, several epigenetic mechanisms are involved in endometrial carcinogenesis (24,25). Previous analyses have predominantly focused on methylation changes in the promoter regions of genes involved in oestrogen metabolism, the DNA mismatch repair system and signalling pathways, including human mutL homolog 1 (26,27), cyclin-dependent kinase inhibitor 2A (27), estrogen receptor 1 (28), progesterone receptor-B (29), phosphatase and tensin homolog (30), Ras association domain family member 1A (31), O-6-methylguanine-DNA methyltransferase (32) and adenomatous polyposis coli (33,34).
For the purpose of the present study, the kruppel like factor 4 (KLF4) and heparan sulfate-glucosamine 3-sulfotransferase 2 (HS3ST2) cancer-related genes were analysed. KLF4 is part of the Kruppel-like gene family with ‘zinc-finger’ transcription factor. The main role of this gene is to maintain cell cycle integrity (35) and thus influence the growth, differentiation, proliferation and programmed apoptosis of somatic cells (36). KLF4 inhibits cell proliferation as a control protein via the activation of p21, which normally inhibits cyclin-dependent kinases (37). It also acts as a mediator in arresting the cell cycle following recognition of damage in the G1/S phase and eventually at the G2/M checkpoint, with this process being mediated by p53 activity (35,38). The methylation levels of KLF4 are generally lower in certain types of cancer, including oesophageal (39), pancreatic (40), lung (41), brain (42) and gastric cancer (43).
By contrast, the HS3ST2 gene encodes the heparansulfate 3-O-sulfotransferase 2 enzyme, which is a key component in heparansulfate (HS) fine structure biosynthesis involved in multiple biologic activities (44). Each enzyme in this cascade has a tissue-specific role and serves as a substrate for the subsequent reaction. Therefore, change in even one enzyme, including heparansulfate 3-O-sulfotransferase 2, leads to the diverse HS structure (45) involved in several types of cancer (44,46).
The present case-control study is unique in that it involves comparison of KLF4 and HS3ST2 methylation status in EC, hyperplasia and normal endometrial tissue; by the investigation of other clinical and histopathological data roles in methylation status, and by the quantification of predictor diagnostic accuracy by the area under the ROC curve (AUC).
Materials and methods
Patients and clinical pathological characteristics
The sample group comprised 172 Caucasian women hospitalised at the Department of Obstetrics and Gynaecology at Martin University Hospital (Martin, Slovakia) between 2011 and 2017. Tissue was analysed from 91 patients with EC, 36 with hyperplasia and 45 with normal endometrial tissue. Each tissue underwent standard histopathological analyses at the Department of pathology at Martin University Hospital. This provided histological type, degree of differentiation (G), and parameters of tumour-node-metastasis classification (Table I). Information on personal and gynaecological anamnesis was obtained during medical examination. This comprised body mass index (BMI), hypertension, diabetes mellitus, age at menarche, parity, abnormal uterine bleeding and abortion, and smoking habit.
Table I.
Histopathological characteristics of the endometrial cancer and hyperplasia groups.
| Characteristic | n | % |
|---|---|---|
| Endometrial cancer (n=91) | ||
| Endometroid | 78 | 85.7 |
| Endometroid with squamous differentiation | 9 | 9.9 |
| Othera | 4 | 4.4 |
| Stage (pT) | ||
| T0 | 1 | 1.1 |
| T1a | 35 | 38.4 |
| T1b | 35 | 38.4 |
| T2 | 11 | 12.2 |
| T3a | 6 | 6.6 |
| T3b | 3 | 3.3 |
| Lymph node metastasis (pN) | ||
| N0 | 34 | 37.0 |
| N1 | 6 | 6.5 |
| Nx | 51 | 56.5 |
| Histological grade | ||
| G1 | 19 | 20.9 |
| G2 | 49 | 53.8 |
| G3 | 23 | 25.3 |
| Hyperplasia (n=36) | ||
| Simplex hyperplasia | 21 | 58.3 |
| Simplex hyperplasia with atypia | 5 | 13.9 |
| Complex hyperplasia with atypia | 10 | 27.8 |
Endometroid with mucinous differentiation; clear cell carcinoma; undifferentiated endometrial sarcoma; serous adenocarcinoma.
DNA isolation and bisulfide modification
Tissue samples were stabilised in RNAlater solution immediately following sectioning and frozen at −20°C. DNA was then extracted by the column method (DNeasy Blood and Tissue Kit®, Qiagen GmbH, Hilden Germany). The qualitative parameters of the isolated DNA were assessed by 1.5% agarose gel electrophoresis and the DNA concentration was measured using a Nanodrop® device. Only samples with sufficient concentration of at least 100 ng/µl were considered for further processing. Genomic DNA (1 µg) was used for the bisulfite conversion performed using the Epitect bisulfite kit® (Qiagen GmbH): 1 µg of DNA dissolved in nuclease-free water was mixed with 85 µl of bisulfite mix and 35 µl of DNA protect buffer and amplified. The concentration of bisulfite modified samples was measured spectrophotometrically and samples were frozen to −20°C.
Methylation analyses
The methylation levels of three CpG sites in the HS3ST2 gene and six CpG sites in the KLF4 gene were analysed by pyrosequencing (Pyromark Q96 ID device). This is a quantitative, precise real-time sequencing methodology. The visible light emitted in the final step of the enzymatic cascade was scanned using a CCD camera; with the rate of light emission retaining continuous proportion with the number of incorporated nucleotides.
Pyro-sequencing has PCR amplification and sequencing phases; DNA amplification required 25 µl total volume (Pyromark PCR Kit® Qiagen GmbH) containing 2X pyromark PCR master mix, 10X coral load concentrate, 1 µl 25 mM MgCl2, 5X Q solution, 0.24 µM primer mix, RNase free water and bisulfide-modified DNA). The PCR reaction steps were as follows: Activation of polymerase (95°C, 15 min); 45 cycles of: Denaturation (94°C, 15 sec), annealing (56°C, 30 sec), extension (72°C, 30 sec) and final extension at 72°C for 10 min. The ampli-cons were then assessed by 1.5% agarose gel electrophoresis.
The PCR product (20 µl) was mixed with streptavidin-coated sepharose beads (GE Healthcare Life Sciences, Chalfont, UK), binding buffer and nuclease free water in a total volume of 80 µl. The 5′-biotiniled strand for sequencing was immobilised, transferred to 0.4 M sequencing primer and binding buffer solution (Qiagen GmbH) and incubated for 2 min at 80°C. The samples were analysed by Pyromark Q96 ID and interpreted by Pyromark Q96 software v. 2.5.8 (Qiagen GmbH) via calculation of the C/T ratio and the peak-high of each CpG site. The samples were analysed in duplicate, and controls comprised commercial methylated and unmethylated DNAs (diluted to series of 100, 75, 50, 25 and 0%). Commercially available Pyromark CpG assays® (Qiagen GmbH) provided methylation analyses of the following regulation sequences: KLF4 5′-CCCGACATACTGACGTGCTGGCGGGCCACGCGCGA-3′; HS3ST2 5′-TTGGCGAGATGTCGAGAGCGGGGGGA-3′.
Statistical analysis
The methylation levels were visualised by swarmplots (47) (Figs. 1 and 2). The data was not Gaussian, so robust one-way analysis of variance (ANOVA) (48) was used instead of simple ANOVA to determine the equality of the CpG methylation population median levels across the diagnostic groups. Rejection of the ANOVA hypothesis was followed by Tukey’s HSD post hoc test. Spearman’s correlation coefficient was used to quantify the strength of the linear association between quantitative variables.
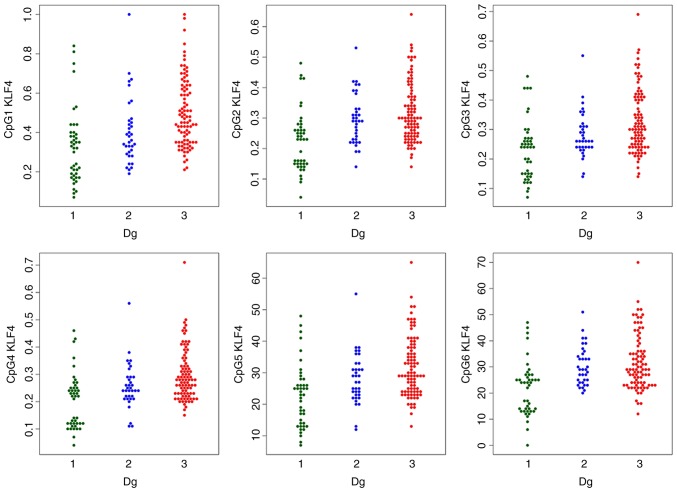
Figure 1.
Swarmplots of the methylation status of KLF4 CpG sites by diagnosis. 1, control; 2, hyperplasia; 3, endometrial cancer. Dg, diagnostic group; KLF4, kruppel like factor 4.
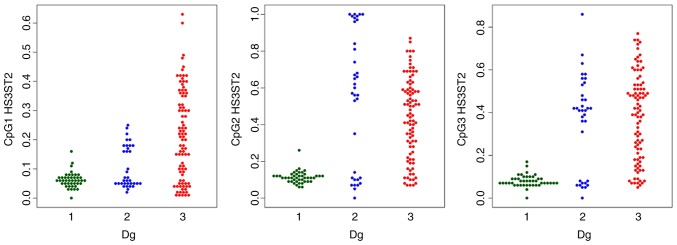
Figure 2.
Swarmplots of the methylation status of HS3ST2 CpG sites by diagnosis. 1, control; 2, hyperplasia; 3, endometrial cancer. Dg, diagnostic group; HS3ST2, heparan sulfate-glucosamine 3-sulfotransferase 2.
The methylation index (MI) was calculated as the mean-percent-methylation across all gene CpG sites (three CpG sites of the HS3ST2 gene and six CpG sites of the KLF4 gene). The promoter methylation status was theoretically classified as unmethylated (0–9%), methylated (10–29%) and highly methylated (30–100%) (49,50). Multinomial logistic regression then modelled dependence of the diagnosis on age, BMI, menarche, parity, CpGs and smoking. The model identified CpGs which are statistically significant predictors of diagnosis, while controlling the effect of other predictors.
The Random Forest algorithm assessed the diagnostic accuracy of the predictors. The subset of important predictors was identified by the minimum tree depth criterion in the nested cross-validation feature selection (51). The diagnostic accuracy was quantified by the ROC curve and summarised by the AUC. Finally, the Younden criterion identified the optimal sensitivity and specificity. Categorical variable independence was established using a χ2 test. The analyses were performed in R ver. 3.2.1. (52) and IBM SPSS ver. 21.
Results
Gynaecological anamnesis and risk factors
The statistically different characteristics in the EC, hyperplasia and control groups are listed in Table II. The statistics were also age-adjusted to eliminate age-effect on the variables examined.
Table II.
Mean values of age, BMI and menarche in the three study groups.
| Factor | Control (n=45) | Hyperplasia (n=36) | Cancer (n=91) | χ2/P-value |
|---|---|---|---|---|
| Age (years) | ||||
| Mean ± SD | 48.53±11.11 | 53.39±9.64 | 64.16±9.64 | P<0.001 |
| Median | 47.00 | 52.00 | 65.00 | |
| Menarche | ||||
| Mean ± SD | 13.22±2.01 | 13.17±1.87 | 13.21±1.42 | P=0.375 |
| Median | 13.00 | 14.00 | 13.00 | |
| BMI | ||||
| Mean ± SD | 28.09±4.82 | 32.05±5.07 | 35.57±3.81 | P<0.001 |
| Median | 28.41 | 32.16 | 35.60 | |
| Menopausal status, n (%) | ||||
| Pre | 17 (65.4) | 4 (28.6) | 2 (4.0) | χ2=42.22 |
| Peri | 4 (15.4) | 3 (21.4) | 3 (6.0) | P<0.001 |
| Post | 5 (19.2) | 7 (50.0) | 45 (90.0) | |
| Parity, n (%) | ||||
| Yes | 43 (95.6) | 36 (100.0) | 80 (87.9) | χ2=6.240 |
| No | 2 (4.4) | 0 (0.0) | 11 (12.1) | P=0.044 |
| Bleeding, n (%) | ||||
| Yes | 10 (38.5) | 11 (78.6) | 39 (78.0) | χ2=13.09 |
| No | 16 (61.5) | 3 (21.4) | 11 (22.0) | P=0.001 |
| Hypertension, n (%) | ||||
| Yes | 9 (34.6) | 7 (50.0) | 38 (76.0) | χ2=12.90 |
| No | 17 (65.4) | 7 (50.0) | 12 (24.0) | P=0.002 |
| Diabetes, n (%) | ||||
| Yes | 3 (11.5) | 3 (24.1) | 19 (38.0) | χ2=6.303 |
| No | 23 (88.5) | 11 (78.6) | 31 (62.0) | P=0.043 |
| Smoking, n (%) | ||||
| Yes | 9 (20.0) | 9 (25.0) | 8 (8.8) | χ2=6.415 |
| No | 36 (80.0) | 27 (75.0) | 83 (91.2) | P=0.040 |
BMI, body mass index; SD, standard deviation.
The following statistically significant differences were observed in gynaecological anamneses and reproductive characteristics: 90% of women with cancer were postmenopausal and 65.4% of the control group were premenopausal; 84.6% of women without children had cancer and the remaining 15.4% nulliparous women were in the control group, thus indicating the importance of nulliparity in EC. The statistics regarding abnormal bleeding were dominated by women from the cancer group (65.0%), followed by 18.3% of women with hyperplasia and 16.7% of controls. Menarche age was the only variable to have no significant impact on diagnostic typology in the study (P>0.05).
Analyses of other risk factors, including BMI, DM and hypertension, indicated higher cardiovascular risk, in addition to EC, as these metabolic parameters are also important cardiovascular risk variables. Women with cancer dominated the hypertension category (70.4% of cancer patients, 13.0% with hyperplasia and 16.7% of controls, P=0.002) and the diabetes mellitus category (76.0% prevailing in cancer sufferers, 12.0% in the hyperplasia group and 12.0% in the control group, P=0.043). The mean BMI values of classified women with cancer in class II on the obesity scale indicated severe obesity, and the women with hyperplasia were in class I obesity. The BMI values were not influenced by age (P=0.735), however, GLM analysis revealed they were influenced by diagnosis (P<0.001). Smoking results showed that 8.8% of women with cancer smoked; plus 25% with hyperplasia and 20% of controls (P=0.040).
Methylation levels
The MI of the two genes was statistically significantly different in the three study groups; with increasing tendency towards the EC group (Table III). The detailed comparison of groups using robust one-way ANOVA with Tukey’s HSD test confirmed a statistically significant difference in the HS3ST2 and KLF4 genes between median methylations levels of normal tissues, vs. hyperplasia, normal tissues, vs. cancer, and hyperplasia, vs. cancer. The only exception was HS3ST2 gene hyperplasia and cancer coincidence, where no significant difference was observed (P=0.847).
Table III.
Mean methylation values and medians of the MI and each CpG site in KLF4 and HS3ST2 genes according to diagnosis.
| Site | Normal (n=43) | Hyperplasia (n=35) | Cancer (n=91) | P-value |
|---|---|---|---|---|
| MI KLF4a | 23.83±11.39 | 30.40±8.52 | 34.72±10.79 | |
| 25.33 | 29.50 | 31.83 | 0.001 | |
| CpG1 | 32.65±18.77 | 40.49±17.07 | 50.22±17.42 | |
| 32.00 | 36.00 | 47.00 | <0.001 | |
| CpG2 | 22.51±9.96 | 29.69±8.29 | 31.91±9.80 | |
| 23.00 | 29.00 | 30.00 | <0.001 | |
| CpG3 | 23.05±10.05 | 28.14±7.69 | 32.76±10.68 | |
| 24.00 | 26.00 | 30.00 | <0.001 | |
| CpG4 | 20.30±10.01 | 25.69±8.23 | 29.78±9.31 | |
| 22.00 | 24.00 | 28.00 | <0.001 | |
| CpG5 | 22.79±10.01 | 28.09±8.05 | 31.85±9.52 | |
| 23.00 | 27.00 | 29.00 | <0.001 | |
| CpG6 | 21.70±10.32 | 30.34±7.35 | 31.80±10.47 | |
| 24.00 | 29.00 | 29.00 | <0.001 | |
| MI HS3ST2a | 8.52±2.57 | 33.76±20.66 | 34.49±18.39 | |
| 8.33 | 36.67 | 35.00 | <0.001 | |
| CpG1 | 6.21±2.63 | 10.43±7.11 | 21.59±15.35 | |
| 6.00 | 6.00 | 21.00 | <0.001 | |
| CpG2 | 11.33±3.26 | 54.77±36.14 | 43.88±21.81 | |
| 11.00 | 60.00 | 45.00 | <0.001 | |
| CpG3 | 8.02±2.76 | 36.09±21.96 | 38.00±20.26 | |
| 7.00 | 41.00 | 40.00 | <0.001 |
MI was calculated as mean methylation value of analysed CpG sites; values are expressed as the mean ± standard deviation and median. MI, methylation index; KLF4, kruppel like factor 4; HS3ST2, heparan sulfate-glucosamine 3-sulfotransferase 2.
The comparison of each CpG site methylation highlighted statistically significant differences between the cancer, hyperplasia and control samples in the KLF4 gene CpG1, CpG3, CpG4 and CpG5 sites, and the HS3ST2 CpG1 site. Differences in KLF4 gene CpG2 and CpG6 and HS3ST2 CpG2 and CpG3 were observed only in the control, vs. hyperplasia and control, vs. cancer groups, but not in the hyperplasia, vs. cancer group (Figs. 1 and 2).
The comparison of methylation in the EC histological subtypes and the hyperplasia subtypes in the two genes revealed no statistically different median methylation levels at P<0.05; nor were there differences in each CpG site or MIs. The HS3ST2 CpG1 site methylation levels were different between tumour stage status (pT) (0.027), however, no increasing or decreasing tendency in stage severity was detected. Similarly lacking any tendency, significant differences were established in lymph node metastasis status (pN) in the KLF CpG1 site (P=0.045), in the grading of all HS3ST2 CpG sites (CpG1 P=0.001, CpG2 P=0.008, CpG3 P=0.043) and in the HS3ST2 MI (P=0.004) (data not shown).
Correlation analysis
Correlation analysis of the entire sample confirmed the association between mean HS3ST2 and KLF4 gene methylation values and age (r=0.316, P<0.001; r=0.317, P<0.001), BMI (r=0.386, P<0.001; r=0.191, P=0.013) and diagnosis (r=0.496, P<0.001; r=0.387, P<0.001). The CpG sites in each gene correlated significantly with each other; with correlation coefficients varying between 0.677 and 0.934 for the HS3ST2 gene and between 0.850 and 0.944 for the KLF4 gene. This indicated a correlation in single CpG site methylation status (Table IV).
Table IV.
Correlation analyses between CpG sites of KLF4 and HS3ST2 gene.
|
KLF4
|
||||||
|---|---|---|---|---|---|---|
| CpG1 C | pG2 C | pG3 | CpG4 C | pG5 C | pG6 | |
| HS3ST2 | ||||||
| CpG1 | r=0.914 | r=0.927 | r=0.868 | r=0.916 | r=0.889 | |
| CpG2 | r=0.677 | r=0.933 | r=0.850 | r=0.923 | r=0.914 | |
| CpG3 | r=0.760 | r=0.934 | r=0.850 | r=0.944 | r=0.904 | |
| CpG4 | – | – | r=0.900 | r=0.873 | ||
| CpG5 | – | – | – | – | r=0.938 | |
r, Spearman’s correlation coefficient; P-value was <0.001 in all cases. KLF4, kruppel like factor 4; HS3ST2, heparan sulfate-glucosamine 3-sulfotransferase 2.
Multinomial logistic regression
The P-values of the predictors in Table V highlight the importance of age, BMI and the KLF4 CpG2 site as cancer predictors, and HS3ST2 CpG1 methylation was a significant factor in hyperplasia prediction.
Table V.
Multinomial logistic regression P-value coefficients of predictors in hyperplasia and cancer.
| Predictor | Hyperplasia (P-value) |
Endometrial cancer (P-value) |
|---|---|---|
| KLF4 | ||
| CpG1 | 0.464 | 0.106 |
| CpG2 | 0.263 | <0.001 |
| CpG3 | 0.101 | 0.470 |
| CpG4 | 0.846 | 0.892 |
| CpG5 | 0.821 | 0.926 |
| CpG6 | 0.058 | 0.164 |
| HS3ST2 | ||
| CpG1 | 0.044 | 0.247 |
| CpG2 | 0.434 | 0.806 |
| CpG3 | 0.225 | 0.149 |
| Age | 0.932 | 0.015 |
| Menarche | 0.812 | 0.964 |
| BMI | 0.057 | 0.001 |
| Smoking | 0.365 | 0.897 |
| Parity | 0.302 | 0.787 |
| Abort/UPT | 0.600 | 0.402 |
KLF4, kruppel like factor 4; HS3ST2, heparan sulfate-glucosamine 3-sulfotransferase 2; BMI, body mass index.
Selection of the diagnostic predictors and discrimination accuracy
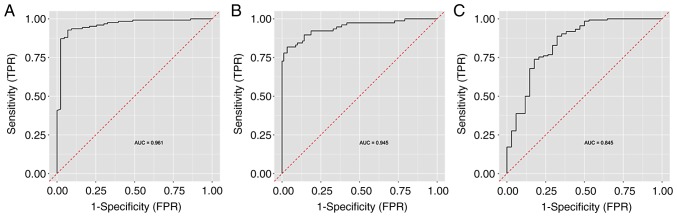
The Random Forest classification algorithm plot schematically visualised and ordered variables by their importance in diagnostic prediction (Fig. 3). On considering CpG methylation status together with clinical variables, the BMI was identified as the most important predictor, followed by CpG1 HS3ST2 dinucleotide methylation status, age and HS3ST2 CpG3 and CpG2 dinucleotides. Other parameters, including smoking, menarche and parity, had no significant impact on diagnostic prediction and were therefore omitted from the final model. The Random Forest algorithm with nested cross validation determined the predictive performance of this model with the selected important variables. The highest AUC value of 0.961 was attained in the discrimination of controls from the other groups, followed by the discrimination between the EC group and other groups (AUC 0.945). The lowest AUC (0.845) was determined in the discrimination of women with hyperplasia from the other groups (Fig. 4).
Figure 3.
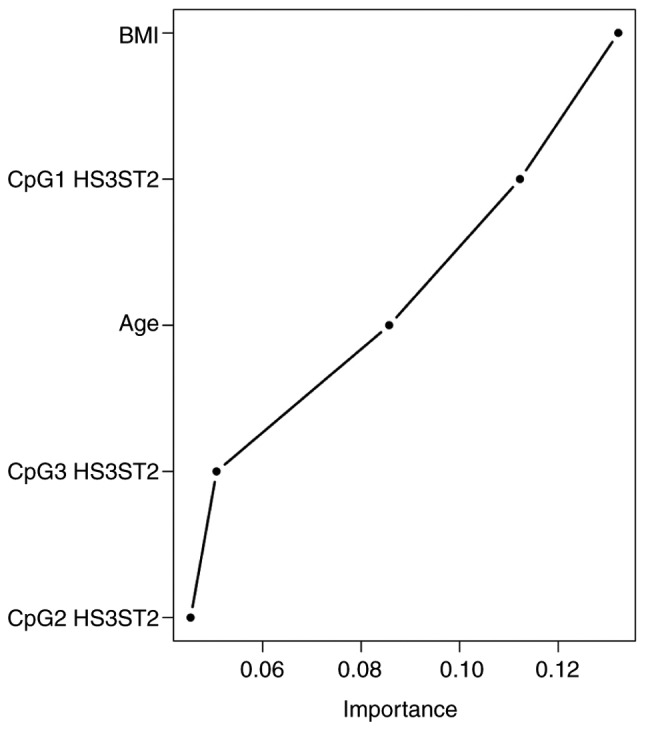
Importance plot with selected variables in diagnostic prediction. Importance increases to the right. HS3ST2, heparan sulfate-glucosamine 3-sulfotransferase 2; BMI, body mass index.
Figure 4.
ROC with AUC describing the predictive performance of the model. (A) Control, vs. cancer and hyperplasia; (B) endometrial cancer, vs. hyperplasia and control; (C) hyperplasia, vs. cancer and control. AUC, area under the ROC curve.
The model produced exclusively from the CpG methylation status determined that the HS3ST2 CpG sites were the most important predictors; followed by CpG1, CpG2 and CpG4 in the KLF4 gene. The 0.95 AUC value indicated the perfect discrimination accuracy of the CpG model between normal tissue and other diagnoses.
Cut off value
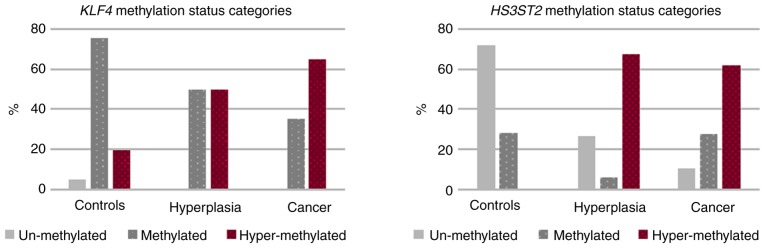
The methylation status in the three diagnostic categories is shown in Fig. 5. Unmethylated KLF4 gene status was detected only in normal tissue (4.9%). Although methylation and hypermethylation occurred at identical 50% frequency in hyperplasia, 35.2% of EC sufferers were identified as methylated and the remaining 64.8% were hypermethylated. Additional results were as follows: i) HS3ST2 gene analysis identified the high 71.8% unmethylated state in control tissue, but without hypermethylation; ii) there was high 64.9% hypermethylation in the hyperplasia group and 62.1% in the EC group. These differences were statistically significant (P<0.001).
Figure 5.
KLF4 and HS3ST2 methylation status categories according to diagnosis. KLF4, kruppel like factor 4; HS3ST2, heparan sulfate-glucosamine 3-sulfotransferase 2.
The Youden method calculated the cut-off value of class probability differentiating health status from the other two conditions. This was established at 0.5, with the diagnostic sensitivity and specificity of 88.8 and 60.5% for the KLF4 gene and 53.3 and 83.5% for the HS3ST2 gene. The corresponding cut-off for average methylation was 34.3 for KLF4 and 41.0 for HS3ST2. The Random Forest algorithm also assessed each pair of diagnostic groups, and the average methylation cut-offs remained the same in each gene, whereas the specificity and sensitivity varied. The best AUC values were obtained in the diagnosis status models distinguishing health and cancer in these genes; the KLF4 gene recorded an AUC of 0.751, 76.74% specificity and 73.52% sensitivity, and HS3ST2 returned an AUC of 0.789, 71.11% specificity and 86.81% sensitivity. It was not possible to differentiate between hyperplasia and cancer in the KLF4 gene due to its AUC of 0.488 (data not shown).
Discussion
It is evident that aberrant DNA methylation is a common factor in endometrial carcinogenesis. Decreased DNA methylation occurs early in carcinogenesis, and promoter hypermethylation leads to gene silencing and loss of gene expression. Therefore, carcinogenesis can be induced when the tumour suppressor gene or a critical gene involved in the cell cycle or in DNA repair is affected (53,54).
Aberrant methylation
The methylation of three CpG sites in the KLF4 gene and six CpG sites in the HS3ST2 gene were analysed in the present study. These are novel genes, and there have been few reports on their association with hyperplasia and EC (55–58). The functions of the protein products of these genes indicate the likelihood of increased methylation levels in cancer tissues, however, aberrant methylation also requires detection in types of hyperplasia that can evolve into EC. Consequently, there was increasing tendency of average gene methylation from normal endometrial tissue through hyperplasia to cancer (Table III).
The difference in methylation of each CpG and the MI in the two genes were statistically significant between normal tissue, hyperplasia and EC tissue. However, the difference between hyperplasia and EC tissues was less distinct as only one of the HS3ST2 gene CpG sites and four of the six KLF4 sites had statistically significant methylation levels at P<0.05. In addition, the comparison of hyperplasia and cancer MI revealed statistically significant difference only for the KLF4 gene (P=0.04). These results accentuate the importance of these genes in the genesis of cancer and hyperplasia, and they further support hyperplasia as a pre-cancerous tissue form (59,60). Similar results were recorded by Nieminen et al (61), who observed increasing methylation tendency from normal endometrial tissue through simple hyper-plasia to the complex type in 24 tumoursuppressor genes.
Although the methylation analyses of hyperplasia and carcinoma histological subtypes in the present study revealed no statistically significant differences in methylation levels, a difference was found between simple and complex hyperplasia. This inconsistency is likely due to unbalanced subtype incidence, as simple hyperplasia at 58.3% and endometroid adenocarcinoma at 85.7% formed the majority of the respective conditions, with the remaining subtypes registering at only minor frequencies. The small sample size in the present study limited statistical analyses to a certain extent; it was not possible to compare endometrial and hyperplasia sub-types, and the comparison of methylation mean values between hyperplasia and cancer was under-powered. Sample size determination analysis revealed the optimal sample size for HS3ST2 and KLF4 gene methylation is 11,268 and 80 individuals in each diagnostic group. In order to compare single CpG sites, the required number of individuals are as follows: HS3ST2 CpG1 19, CpG2 118 and CpG3 1,921, and KLF4 CpG1 50, CpG2 263, CpG3 64, CpG4 73, CpG5 87 and CpG6 603 in each diagnostic group. In the present study, only the analysis of HS3ST2 CpG1 site methylation between hyperplasia and cancer had adequate statistical power, and it was not possible to determine whether hypermethylated hyperplasia tissue is a prerequisite step in the carcinoma cascade. A higher sample size is also essential in terms of the detailed analysis of aberrant methylation in specific hyperplasia and cancerous subtypes. However, data from the preliminary analyses may be utilised by those who design investigations on the methylation status of these genes and include predominantly Caucasian subjects.
Methylation cut-offs
The literature surveys identified a lack of consistency in determining methylation cut-off values. A number of authors depend on three categories; unmethylated 0–9%, methylated 10–29% and highly methylated 30–100% (49,50), whereas others rely on two; unmethylated <15% and methylated >15% (62–64). In addition, several studies use ROC curve analysis to establish the optimal methylation threshold in discriminating diagnostic categories, cancer types, treatment decisions and outcomes, and patient survival (62,65,66). This highlights the importance of methylation status, which may be useful as a biomarker in cancer management.
In the present study, ROC analysis was performed using the Random Forest Algorithm with nested cross validation, as this provides realistic results. It was established that the model distinguishing control samples from hyperplasia and cancer provided the highest discriminatory ability at AUC=0.961, and that hyperplasia comparison with the other two conditions had the least discriminatory ability at AUC=0.845.
Pyrosequencing method
Several methods are available to analyse promoter region methylation status, and selection of the optimal method depends on the following: Relevant gene identification, gene analysis range, robustness, DNA quantity, the inclusion of bisulfite conversion and the availability of detection devices. Kurdyukov and Bullock (67) compared several methods and described their exploitation in practice. In the present study, pyrosequencing was selected as it is the standard technique in cancer research, detects small differences in methylation, is suitable for heterogeneous samples and provides quantitative results (67,68). However, it was not possible to determine whether it provides information on allele specificity or hemi-methylation, which may differentiate de novo methylation events from maintenance factors (69,70).
Risk factors
Risk factor analysis investigated the significant effects of metabolic factors, including BMI, hypertension and diabetes mellitus, in EC development. The high mean BMI values indicated severe obesity in women with cancer (35.57±3.81), compared with those in the control group (28.09±4,82, P<0.001). This risk factor is also often connected with hypertension, hypercholesterolaemia and diabetes mellitus. The high occurrence of hypertension and diabetes mellitus was noted in cancer patients (76.0 and 38.0%, respectively) and in hyperplasia (50.0 and 24.1%, respectively); therefore, future lipid profile analysis is worthwhile in determining the cluster effect of these factors and their combination in metabolic syndrome formation (71). These three conditions, high BMI, hypertension and diabetes mellitus, are also the main risk factors in cardiovascular disease (CVD) (72). Although cancer is considered second only to CVD in recently determined leading causes of mortality in Europe, America and Asia, current publications suggest that cancer, rather than CVD, is the most common cause of mortality (73–77). The results of the present study highlight the similarities and interactions between these diseases. The most common feature is inflammation as it contributes to both diseases and is specific in obesity, diabetes, hypertension and dyslipidaemia (74). Due to the high mortality rates of these diseases, it is paramount to unify preventive programmes to control and eliminate these risk factors and thus reduce risks of cancer and CVD.
Although smoking presents an unequivocal negative risk factor, certain independent studies have suggested that it may be a protective mechanism against the development of EC (9,78). The present study did not confirm association between smoking and EC; although the number of smokers in the different diagnostic groups was significantly different (P=0.040), the identical smoking rate of 34.6% was recorded in the control group and hyperplasia group; with 30.8% incidence in the patients diagnosed with cancer.
In conclusion, to the best of our knowledge, the present study is the first report discriminating EC from hyperplasia and normal tissue using the AUC and to analyse KLF4 and HS3ST2 methylation cut-off points. The CpG methylation model revealed perfect discrimination accuracy between the control samples and other diagnoses. The AUC value was marginally higher when clinical variables, including BMI and age, were included. The aberrant CpG1 dinucleotide methylation level in the HS3ST2 gene regulation sequence was determined to be an important predictor in hyperplasia formation; similar to the KLF4 regulation sequence CpG2 dinucleotide effect in EC prediction.
The present study also confirmed the prominent role of BMI and other metabolic risk factors in EC formation. As these factors are important also in CVDs, this study sample is considered at high risk in the terms of morbidity and mortality rates for the two most common causes of mortality, CVD and cancer. Therefore, the implantation of effective and mutual preventive programs is required.
Acknowledgments
Not applicable.
Funding
This study was supported by Scientific Grant Agency VEGA (1/0199/17), Slovak Research and Development Agency (APVV-0224-12), ‘Biomedical Centre Martin’ project co-financed from EU sources (ITMS code: 26220220187) and by Comenius University Grants UK/22/2018 and UK/20/2018.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
PŽ, DB, DD and ZD conceived the study; DD, DB, ZD and VH performed laboratory analyses; MŇ, RF and TB were responsible for the collection of samples and clinicopathological data; ZD and MG performed statistical analyses, MG designed the figures; ZD, DB and DD designed and wrote the paper in consultation with PK, JH, EH and PŽ.
Ethics approval and consent to participate
The study was approved by The Regional Ethics Committee of the Jessenius Faculty of Medicine (code 169/2011, 1933/2016) and the research was performed in compliance with the Declaration of Helsinki. Informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Bansal N, Yendluri V, Wenham RM. The molecular biology of endometrial cancers and the implications for pathogenesis, classification, and targeted therapies. Cancer Contr. 2009;16:8–13. doi: 10.1177/107327480901600102. [DOI] [PubMed] [Google Scholar]
- 4.Sherman ME, Devesa SS. Analysis of racial differences in incidence, survival, and mortality for malignant tumors of the uterine corpus. Cancer. 2003;98:176–186. doi: 10.1002/cncr.11484. [DOI] [PubMed] [Google Scholar]
- 5.Hardiman P, Pillay OC, Atiomo W. Polycystic ovary syndrome and endometrial carcinoma. Lancet. 2003;361:1810–1812. doi: 10.1016/S0140-6736(03)13409-5. [DOI] [PubMed] [Google Scholar]
- 6.Schouten LJ, Goldbohm RA, van den Brandt PA. Anthropometry, physical activity, and endometrial cancer risk: Results from the Netherlands cohort study. J Natl Cancer Inst. 2004;96:1635–1638. doi: 10.1093/jnci/djh291. [DOI] [PubMed] [Google Scholar]
- 7.Kjaer SK, Mellemkjaer L, Brinton LA, Johansen C, Gridley G, Olsen JH. Tubal sterilization and risk of ovarian, endometrial and cervical cancer. A Danish population-based follow-up study of more than 65 000 sterilized women. Int J Epidemiol. 2004;33:596–602. doi: 10.1093/ije/dyh046. [DOI] [PubMed] [Google Scholar]
- 8.Troisi R, Potischman N, Hoover RN, Siiteri P, Brinton LA. Insulin and endometrial cancer. Am J Epidemiol. 1997;146:476–482. doi: 10.1093/oxfordjournals.aje.a009301. [DOI] [PubMed] [Google Scholar]
- 9.Gong TT, Wang YL, Ma XX. Age at menarche and endome-trial cancer risk: A dose-response meta-analysis of prospective studies. Sci Rep. 2015;5:14051. doi: 10.1038/srep14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. Endometrial cancer in tamoxifen-treated breast cancer patients: Findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86:527–537. doi: 10.1093/jnci/86.7.527. [DOI] [PubMed] [Google Scholar]
- 11.Ali AT. Reproductive factors and the risk of endometrial cancer. Int J Gynecol Cancer. 2014;24:384–393. doi: 10.1097/IGC.0000000000000075. [DOI] [PubMed] [Google Scholar]
- 12.Kurman RJ, Norris HJ. Evaluation of criteria for distinguishing atypical endometrial hyperplasia from well-differentiated carcinoma. Cancer. 1982;49:2547–2559. doi: 10.1002/1097-0142(19820615)49:12<2547::AID-CNCR2820491224>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 13.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 14.Felix AS, Weissfeld JL, Stone RA, Bowser R, Chivukula M, Edwards RP, Linkov F. Factors associated with type I and type II endometrial cancer. Cancer Causes Control. 2010;21:1851–1856. doi: 10.1007/s10552-010-9612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prat J. Prognostic parameters of endometrial carcinoma. Hum Pathol. 2004;35:649–662. doi: 10.1016/j.humpath.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Bendifallah S, Daraï E, Ballester M. Predictive modeling: A new paradigm for managing endometrial cancer. Ann Surg Oncol. 2016;23:975–988. doi: 10.1245/s10434-015-4924-2. [DOI] [PubMed] [Google Scholar]
- 17.McConechy MK, Ding J, Cheang MC, Wiegand K, Senz J, Tone A, Yang W, Prentice L, Tse K, Zeng T, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol. 2012;228:20–30. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talhouk A, McAlpine JN. New classification of endometrial cancers: The development and potential applications of genomic-based classification in research and clinical care. Gynecol Oncol Res Pract. 2016;3:14. doi: 10.1186/s40661-016-0035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karateke A, Tug N, Cam C, Selcuk S, Asoglu MR, Cakir S. Discrepancy of pre- and postoperative grades of patients with endometrial carcinoma. Eur J Gynaecol Oncol. 2011;32:283–285. [PubMed] [Google Scholar]
- 20.Sany O, Singh K, Jha S. Correlation between preoperative endometrial sampling and final endometrial cancer histology. Eur J Gynaecol Oncol. 2012;33:142–144. [PubMed] [Google Scholar]
- 21.Batista TP, Cavalcanti CL, Tejo AA, Bezerra AL. Accuracy of preoperative endometrial sampling diagnosis for predicting the final pathology grading in uterine endometrioid carcinoma. Eur J Surg Oncol. 2016;42:1367–1371. doi: 10.1016/j.ejso.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Han G, Sidhu D, Duggan MA, Arseneau J, Cesari M, Clement PB, Ewanowich CA, Kalloger SE, Köbel M. Reproducibility of histological cell type in high-grade endometrial carcinoma. Mod Pathol. 2013;26:1594–1604. doi: 10.1038/modpathol.2013.102. [DOI] [PubMed] [Google Scholar]
- 23.Banno K, Nogami Y, Kisu I, Yanokura M, Umene K, Masuda K, Kobayashi Y, Yamagami W, Susumu N, Aoki D. Candidate biomarkers for genetic and clinicopathological diagnosis of endometrial cancer. Int J Mol Sci. 2013;14:12123–12137. doi: 10.3390/ijms140612123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stampoliou A, Arapantoni-Dadioti P, Pavlakis K. Epigenetic mechanisms in endometrial cancer. J BUON. 2016;21:301–306. [PubMed] [Google Scholar]
- 25.Banno K, Yanokura M, Iida M, Masuda K, Aoki D. Carcinogenic mechanisms of endometrial cancer: Involvement of genetics and epigenetics. J Obstet Gynaecol Res. 2014;40:1957–1967. doi: 10.1111/jog.12442. [DOI] [PubMed] [Google Scholar]
- 26.Banno K, Yanokura M, Susumu N, Kawaguchi M, Hirao N, Hirasawa A, Tsukazaki K, Aoki D. Relationship of the aberrant DNA hypermethylation of cancer-related genes with carcinogenesis of endometrial cancer. Oncol Rep. 2006;16:1189–1196. [PubMed] [Google Scholar]
- 27.Guida M, Sanguedolce F, Bufo P, Di Spiezio Sardo A, Bifulco G, Nappi C, Pannone G. Aberrant DNA hypermethylation of hMLH-1 and CDKN2A/p16 genes in benign, premalignant and malignant endometrial lesions. Eur J Gynaecol Oncol. 2009;30:267–270. [PubMed] [Google Scholar]
- 28.Sasaki M, Kotcherguina L, Dharia A, Fujimoto S, Dahiya R. Cytosine-phosphoguanine methylation of estrogen receptors in endometrial cancer. Cancer Res. 2001;61:3262–3266. [PubMed] [Google Scholar]
- 29.Ren Y, Liu X, Ma D, Feng Y, Zhong N. Down-regulation of the progesterone receptor by the methylation of progesterone receptor gene in endometrial cancer cells. Cancer Genet Cytogenet. 2007;175:107–116. doi: 10.1016/j.cancergencyto.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Salvesen HB, Stefansson I, Kretzschmar EI, Gruber P, MacDonald ND, Ryan A, Jacobs IJ, Akslen LA, Das S. Significance of PTEN alterations in endometrial carcinoma: A population-based study of mutations, promoter methylation and PTEN protein expression. Int J Oncol. 2004;25:1615–1623. [PubMed] [Google Scholar]
- 31.Arafa M, Kridelka F, Mathias V, Vanbellinghen JF, Renard I, Foidart JM, Boniver J, Delvenne P. High frequency of RASSF1A and RARb2 gene promoter methylation in morphologically normal endometrium adjacent to endometrioid adenocarcinoma. Histopathology. 2008;53:525–532. doi: 10.1111/j.1365-2559.2008.03147.x. [DOI] [PubMed] [Google Scholar]
- 32.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 33.Ignatov A, Bischoff J, Ignatov T, Schwarzenau C, Krebs T, Kuester D, Costa SD, Roessner A, Semczuk A, Schneider-Stock R. APC promoter hypermethylation is an early event in endometrial tumorigenesis. Cancer Sci. 2010;101:321–327. doi: 10.1111/j.1349-7006.2009.01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno-Bueno G, Hardisson D, Sánchez C, Sarrió D, Cassia R, García-Rostán G, Prat J, Guo M, Herman JG, Matías-Guiu X, et al. Abnormalities of the APC/beta-catenin pathway in endometrial cancer. Oncogene. 2002;21:7981–7990. doi: 10.1038/sj.onc.1205924. [DOI] [PubMed] [Google Scholar]
- 35.Ghaleb AM, Yang VW. Krüppel-like factor 4 (KLF4): What we currently know. Gene. 2017;611:27–37. doi: 10.1016/j.gene.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Karim EA, Hagos EG, Ghaleb AM, Yu B, Yang VW. Krüppel-like factor 4 regulates genetic stability in mouse embryonic fibroblasts. Mol Cancer. 2013;12:89. doi: 10.1186/1476-4598-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagos EG, Ghaleb AM, Dalton WB, Bialkowska AB, Yang VW. Mouse embryonic fibroblasts null for the Krüppel-like factor 4 gene are genetically unstable. Oncogene. 2009;28:1197–1205. doi: 10.1038/onc.2008.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 40.Zammarchi F, Morelli M, Menicagli M, Di Cristofano C, Zavaglia K, Paolucci A, Campani D, Aretini P, Boggi U, Mosca F, et al. KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma. Am J Pathol. 2011;178:361–372. doi: 10.1016/j.ajpath.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T, Chen X, Zhang W, Liu J, Avdiushko R, Napier DL, Liu AX, Neltner JM, Wang C, Cohen D, Liu C. KLF4 regulates adult lung tumor-initiating cells and represses K-Rasmediated lung cancer. Cell Death Differ. 2016;23:207–215. doi: 10.1038/cdd.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray SK. The transcription regulator Krüppel-like factor 4 and its dual roles of oncogene in glioblastoma and tumor suppressor in neuroblastoma. For Immunopathol Dis Therap. 2016;7:127–139. doi: 10.1615/ForumImmunDisTher.2016017227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei D, Gong W, Kanai M, Schlunk C, Wang L, Yao JC, Wu TT, Huang S, Xie K. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–2754. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 44.Vijaya Kumar A, Salem Gassar E, Spillmann D, Stock C, Sen YP, Zhang T, Van Kuppevelt TH, Hülsewig C, Koszlowski EO, Pavao MS, et al. HS3ST2 modulates breast cancer cell invasiveness via MAP kinase- and Tcf4 (Tcf7l2)-dependent regulation of protease and cadherin expression. Int J Cancer. 2014;135:2579–2592. doi: 10.1002/ijc.28921. [DOI] [PubMed] [Google Scholar]
- 45.Suhovskih AV, Domanitskaya NV, Tsidulko AY, Prudnikova TY, Kashuba VI, Grigorieva EV. Tissue-specificity of heparan sulfate biosynthetic machinery in cancer. Cell Adhes Migr. 2015;9:452–459. doi: 10.1080/19336918.2015.1049801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikolova V, Koo CY, Ibrahim SA, Wang Z, Spillmann D, Dreier R, Kelsch R, Fischgräbe J, Smollich M, Rossi LH, et al. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis. 2009;30:397–407. doi: 10.1093/carcin/bgp001. [DOI] [PubMed] [Google Scholar]
- 47.Eklund A. Bee swarm: The Bee Swarm plot, an alternative to stripchart. R package version 0.2.3. 2016 https://CRAN.R-project.org/package=beeswarm.
- 48.Mair P, Schoenbrodt F. WRS2: Wilcox robust estimation and testing. R package version 0.9.2. 2017 [Google Scholar]
- 49.Brigliadori G, Foca F, Dall’Agata M, Rengucci C, Melegari E, Cerasoli S, Amadori D, Calistri D, Faedi M. Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. J Neurooncol. 2016;128:333–339. doi: 10.1007/s11060-016-2116-y. [DOI] [PubMed] [Google Scholar]
- 50.Gurrieri L, De Carlo E, Gerratana L, De Maglio G, Macerelli M, Pisa FE, Masiero E, Aprile G, Follador A, Puglisi F, et al. MGMT pyrosequencing-based cut-off methylation level and clinical outcome in patients with glioblastoma multiforme. Future Oncol. 2018;14:699–707. doi: 10.2217/fon-2017-0437. [DOI] [PubMed] [Google Scholar]
- 51.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841–860. doi: 10.1214/08-AOAS169. [DOI] [Google Scholar]
- 52.R Development Core Team . R: A language and environment for statistical computing. The R Foundation for Statistical Computing; Vienna: http://www.R-project.org, Accessed February 10, 2015. [Google Scholar]
- 53.Takeda T, Banno K, Yanokura M, Adachi M, Iijima M, Kunitomi H, Nakamura K, Iida M, Nogami Y, Umene K, et al. Methylation analysis of DNA mismatch repair genes using DNA derived from the peripheral blood of patients with endometrial cancer: Epimutation in endometrial carcinogenesis. Genes (Basel) 2016;7:E86. doi: 10.3390/genes7100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang SW, Li J, Podratz K, Dowdy S. Application of DNA methylation biomarkers for endometrial cancer management. Expert Rev Mol Diagn. 2008;8:607–616. doi: 10.1586/14737159.8.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CC, Wang HC, Liao YP, Chen YC, Weng YC, Yu MH, Lai HC. The feasibility of detecting endometrial and ovarian cancer using DNA methylation biomarkers in cervical scrapings. J Gynecol Oncol. 2018;29:e17. doi: 10.3802/jgo.2018.29.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai HC, Wang YC, Yu MH, Huang RL, Yuan CC, Chen KJ, Wu CC, Chiang KJ, Chao TK. DNA methylation as a biomarker for the detection of hidden carcinoma in endometrial atypical hyperplasia. Gynecol Oncol. 2014;135:552–559. doi: 10.1016/j.ygyno.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Wentzensen N, Bakkum-Gamez JN, Killian JK, Sampson J, Guido R, Glass A, Adams L, Luhn P, Brinton LA, Rush B, et al. Discovery and validation of methylation markers for endometrial cancer. Int J Cancer. 2014;135:1860–1868. doi: 10.1002/ijc.28843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, Xing X, Li J, Lowdon RF, Zhou Y, Lin N, Zhang B, Sundaram V, Chiappinelli KB, Hagemann IS, et al. Comparative DNA methylome analysis of endometrial carcinoma reveals complex and distinct deregulation of cancer promoters and enhancers. BMC Genomics. 2014;15:868. doi: 10.1186/1471-2164-15-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen YC, Tsao CM, Kuo CC, Yu MH, Lin YW, Yang CY, Li HJ, Yan MD, Wang TJ, Chou YC, Su HY. Quantitative DNA methylation analysis of selected genes in endometrial carcinogenesis. Taiwan J Obstet Gynecol. 2015;54:572–579. doi: 10.1016/j.tjog.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Trimble CL, Method M, Leitao M, Lu K, Ioffe O, Hampton M, Higgins R, Zaino R, Mutter GL, Society of Gynecologic Oncology Clinical Practice Committee Management of endo-metrial precancers. Obstet Gynecol. 2012;120:1160–1175. doi: 10.1097/aog.0b013e31826bb121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nieminen TT, Gylling A, Abdel-Rahman WM, Nuorva K, Aarnio M, Renkonen-Sinisalo L, Järvinen HJ, Mecklin JP, Bützow R, Peltomäki P. Molecular analysis of endometrial tumorigenesis: Importance of complex hyperplasia regardless of atypia. Clin Cancer Res. 2009;15:5772–5783. doi: 10.1158/1078-0432.CCR-09-0506. [DOI] [PubMed] [Google Scholar]
- 62.Yuan XL, Zhang Z, Li B, Gao N, Zhang H, Sangild PT, Li JQ. Genome-wide DNA methylation analysis of the porcine hypothalamus-pituitary-ovary axis. Sci Rep. 2017;7:4277. doi: 10.1038/s41598-017-04603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegel EM, Riggs BM, Delmas AL, Koch A, Hakam A, Brown KD. Quantitative DNA methylation analysis of candidate genes in cervical cancer. PLoS One. 2015;10:e0122495. doi: 10.1371/journal.pone.0122495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dvorakova E, Chmelarova M, Laco J, Palicka V, Spacek J. Methylation analysis of tumor suppressor genes in endometroid carcinoma of endometrium using MS-MLPA. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013;157:298–303. doi: 10.5507/bp.2013.035. [DOI] [PubMed] [Google Scholar]
- 65.Dong X, Hou Q, Chen Y, Wang X. Diagnostic value of the methylation of multiple gene promoters in serum in hepatitis B virus-related hepatocellular carcinoma. Dis Markers. 2017;2017;2929381 doi: 10.1155/2017/2929381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boers A, Wang R, van Leeuwen RW, Klip HG, de Bock GH, Hollema H, van Criekinge W, de Meyer T, Denil S, van der Zee AGJ, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin Epigenetics. 2016;8:29. doi: 10.1186/s13148-016-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurdyukov S, Bullock M. DNA methylation analysis: Choosing the right method. Biology (Basel) 2016;5:E3. doi: 10.3390/biology5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delaney C, Garg SK, Yung R. Analysis of DNA methylation by pyrosequencing. Methods Mol Biol. 2015;1343:249–264. doi: 10.1007/978-1-4939-2963-4_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patiño-Parrado I, Gómez-Jiménez Á, López-Sánchez N, Frade JM. Strand-specific CpG hemimethylation, a novel epigenetic modification functional for genomic imprinting. Nucleic Acids Res. 2017;45:8822–8834. doi: 10.1093/nar/gkx518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forné I, Pichler G, Hörl D, Fellinger K, et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res. 2015;25:911–929. doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, Gordon DJ, Krauss RM, Savage PJ, Smith SC, Jr, et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute scientific statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 72.Expert Panel on Detection, Evaluation Treatment of High Blood Cholesterol in Adults Executive summary of the third report of The National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 73.Wilson L, Bhatnagar P, Townsend N. Comparing trends in mortality from cardiovascular disease and cancer in the United Kingdom, 1983–2013 Joinpoint regression analysis. Popul Health Metr. 2017;15:23. doi: 10.1186/s12963-017-0141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koene RJ, Prizment AE, Blaes A, Konety SH. Shared risk factors in cardiovascular disease and cancer. Circulation. 2016;133:1104–1114. doi: 10.1161/CIRCULATIONAHA.115.020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weir HK, Anderson RN, Coleman King SM, Soman A, Thompson TD, Hong Y, Moller B, Leadbetter S. Heart disease and cancer deaths-trends and projections in the United States 1969–2020. Prev Chronic Dis. 2016;13:E157. doi: 10.5888/pcd13.160211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nichols M, Townsend N, Scarborough P, Rayner M. Trends in age-specific coronary heart disease mortality in the European Union over three decades: 1980–2009. Eur Heart J. 2013;34:3017–3027. doi: 10.1093/eurheartj/eht159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bosetti C, Bertuccio P, Malvezzi M, Levi F, Chatenoud L, Negri E, La Vecchia C. Cancer mortality in Europe, 2005–2009 and an overview of trends since 1980. Ann Oncol. 2013;24:2657–2671. doi: 10.1093/annonc/mdt301. [DOI] [PubMed] [Google Scholar]
- 78.Merritt MA, Tzoulaki I, Tworoger SS, De Vivo I, Hankinson SE, Fernandes J, Tsilidis KK, Weiderpass E, Tjønneland A, Petersen KE, et al. Investigation of dietary factors and endometrial cancer risk using a nutrient-wide association study approach in the EPIC and Nurses’ Health Study (NHS) and NHSII. Cancer Epidemiol Biomarkers Prev. 2015;24:466–471. doi: 10.1158/1055-9965.EPI-14-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.