Abstract
LHFPL5, the gene for DFNB67, underlies autosomal recessive nonsyndromic hearing impairment. We identified seven Pakistani families that mapped to 6p21.31, which includes the LHFPL5 gene. Sanger sequencing of LHFPL5 using DNA samples from hearing impaired and unaffected members of these seven families identified four variants. Among the identified variants, two were novel: one missense c.452G>T (p.Gly151Val) and one splice site variant (c.*16+1G>A) were each identified in two families. Two known variants: c.250delC (p.Leu84*) and c.380A>G (p.Tyr127Cys) were also observed in two families and a single family, respectively. Nucleotides c.452G and c.*16+1G and amino acid residue p.Gly151 are under strong evolutionary conservation. In silico bioinformatics analyses predicted these variants to be damaging. The splice site variant (c.*16+1G>A) is predicted to affect pre-mRNA splicing and a loss of the 5’ donor splice site in the 3’untranslated region (3’ UTR). Further analysis supports the activation of a cryptic splice site approximately 357bp downstream, leading to an extended 3’ UTR with additional regulatory motifs. In conclusion, we identified two novel variants in LHFPL5, including a unique 3’UTR splice site variant that is predicted to impact pre-mRNA splicing and regulation through an extended 3’UTR.
Keywords: non-syndromic autosomal recessive hearing impairment, DFNB67, LHFPL5, 3’-untranslated region
Background
Hearing impairment (HI) is the most common sensory deficit in the world; one to two per 1000 children are born with congenital HI [1]. Over fifty percent of these cases are due to a genetic cause, most commonly with autosomal recessive inheritance. To date, ~70 genes have been identified for autosomal recessive (AR) nonsyndromic (NS) HI (Hereditary Hearing Loss Homepage). The DFNB67 locus was mapped to 6p21.1-p22.3 and afterwards Homo sapiens lipoma HMGIC fusion partner-like 5 (LHFPL5; MIM 609427), also known as Tetraspan membrane protein of hair cell stereocilia (TMHS), was identified as the causal gene for this locus [2]. Nine pathogenic variants in LHFPL5 (c.1A>G, c.89dupG, c.246delC, c.250delC, c.258_260delCTC, c.380A>G, c.494C>T, c.518T>A, c.649delG) have been reported in ARNSHI families without vestibular dysfunction from Pakistan, India, Turkey, Palestine, Algeria, Iran and Tunisia [(2–7)]. In the mouse, a missense variant c.482G>T (p.Cys161Phe) in the Tmhs gene of hurry-scurry mice was reported to cause deafness and vestibular dysfunction [8]. The protein encoded by LHFPL5 is transiently expressed in hair cell stereocilia bundles from E16.5 to P3 and is presumed to organize a transient cytoskeleton-cell membrane interaction necessary for proper hair cell bundle morphogenesis that is critical for auditory function [2].
In this study, seven Pakistani ARNSHI families were mapped to the DFNB67 region using genome wide linkage analyses. Sequencing of LHFPL5 revealed three families with previously reported pathogenic variants: two families with the c.250delC variant and one family with a c.380A>G variant. Novel variants were observed in four families: Two with missense variant c.452G>T (p.Gly151Val) and two with a splice site variant c.*16+1G>A located in the 3’-untranslated region (3’ UTR). Variants in LHFPL5 causing HI highlight the important roles of hair cell stereocilia and their ability to transmit auditory signals from external stimuli within the inner ear.
Methods
Subjects
This study was approved by the Institutional Review Boards of Quaid-i-Azam University and Baylor College of Medicine and Affiliated Hospitals. Informed consent was obtained from each family member participating in the study. Known and novel pathogenic variants in LHFPL5, which underlies ARNSHI were identified in seven consanguineous Pakistani families (Figure 1). These families are from different ethnic groups: Families 4072A, 4072B, 4298, and 4464 are from the Punjab province and speak Punjabi; Family 4275 is from the Punjab province, but speaks Saraiki; Family 4506 is from the Khyber Pakhtunkhwa province and speaks Pashto; and Family 4194 is from Balochistan and speak Balochi. Clinical histories were recorded to rule out non-genetic causes of HI, such as maternal or perinatal infections, administration of ototoxic medications, or trauma and syndromic forms of HI. Physical exams, including tandem gait and Romberg tests, were performed to evaluate for gross vestibular deficits. Pure tone air conduction audiometric testing at 250-8,000Hz was performed on hearing-impaired family members.
Figure 1.
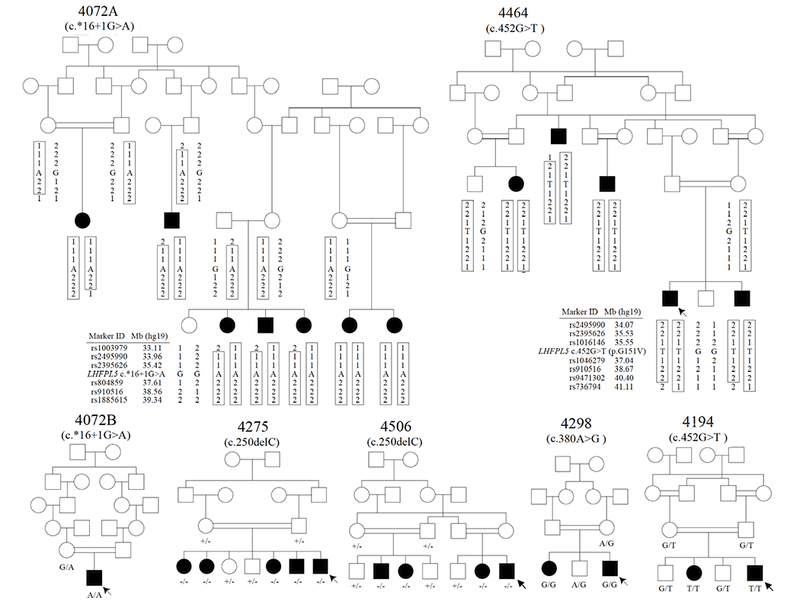
Pedigree drawings of the seven ARNSHI families with LHFPL5 variants. Families 4275 and 4506 segregate the known variant c.250delC, the −/− signifies that the family member is homozygous for the c.250delC variant and +/− indicates individuals that are heterozygous c.250delC variant carrier. Family 4298 segregates the known variant c.380A>G. Families 4194 and 4464 segregate the novel variant c.452G>T and families 4072A and 4072B segregate another novel variant c.*16+1G>A. It was reported that families 4072A and 4072B are distantly related but the exact relationship is unknown. Filled symbols represent individuals with HI and clear symbols hearing individuals. The six individuals with arrows indicate their audiograms are displayed in Figure 2. Haplotypes are presented for the two families 4072A and 4464, which have novel variants. A boxed haplotype carries the pathogenic variant. For the other five families, the corresponding nucleotide substitutions are presented below each sequenced individual.
Genotyping and Linkage analyses
Venous blood was obtained from both hearing and hearing-impaired members of the seven families (Figure 1). DNA extraction was performed following a phenol-chloroform protocol. The coding region of GJB2 was screened as well as two variants which are common causes of ARNHI in Pakistan: c.482 +1986_88delTGA in HGF and c.272A>G ( p.Phe91Ser) in CIB2. DNA samples underwent whole genome genotyping using the Illumina Human Linkage-panels containing ~6,000 SNP marker loci at the Center for Inherited Disease Research (CIDR).
The genotype data underwent quality control using MERLIN [9] to detect occurrences of double recombination events over short genetic distances which could be due to genotyping errors, and PEDCHECK [10] to identify Mendelian inconsistencies. Two-point and multipoint linkage analyses were performed using Superlink Online [11] while haplotype were constructed using Simwalk2 [12]. For linkage analysis an AR mode of inheritance with complete penetrance and disease allele frequency of 0.001 was used. Marker allele frequencies were estimated from observed genotypes and reconstructed genotypes of founders from Pakistani families that were genotyped at the same time. Genetic map positions of the marker loci were obtained through interpolation using the Rutgers combined linkage-physical map of the human genome (hg19) [13]. The linkage region was defined by the three-unit support intervals and regions of homozygosity.
DNA Sequencing
Primers for all four exons of LHFPL5 were created using Primer3 [14]. ExoSAP-IT (USB Corp., Cleveland, OH USA) was used to purify PCR-amplified products. Sequencing was performed on ABI 3730 DNA Analyzer using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc, Foster City, CA USA). DNA sequences were aligned and analyzed using Sequencher software v4.9 (GeneCodes Corp., Ann Arbor, MI USA).
Bioinformatics Analyses
Pathogenicity of the identified variants were investigated using Polyphen-2 [15], MutationTaster [16], SIFT [17], LRT [18], Mutation Assessor [19], FATHMM [20] and CADD [21]. Nucleotide conservation was predicted by GERP++ while amino acid residue conservation was investigated by importing similar non-human proteins found from UniProt [22] and aligning the protein sequences on ClustalW2 [23].The transmembrane helical structure was predicted on TMHMM 2.0 server [24].
The functional effects of donor splice site variant c.*16+1G>A were investigated using extensive bioinformatics approaches as RNA samples of hearing impaired individuals were not available. First, the effect of the variant and a possible cryptic splicing were predicted using three different softwares: NNsplice [25], HSF [26] and NetGene2 [27]. For the 3’-untranslated regions (UTR), based on natural and cryptic splice sites, RNA secondary structures and the Minimum Free Energies (MFE) were predicted on the Vienna RNA web-server [28]. Possible regulatory elements and miRNA binding sites were identified via the UTRscan [29] and PITA algorithm [30] respectively.
Results
Hearing impaired individuals from all families had no clinical history or physical examination findings that suggested that the HI is part of a syndrome. All hearing-impaired family members had prelingual bilateral profound HI. Air conduction audiometry showed bilateral hearing thresholds in the profound impairment range for all frequencies (Figure 2). There was no evidence of gross vestibular dysfunction based on the results of tandem gait and Romberg testing.
Figure 2.
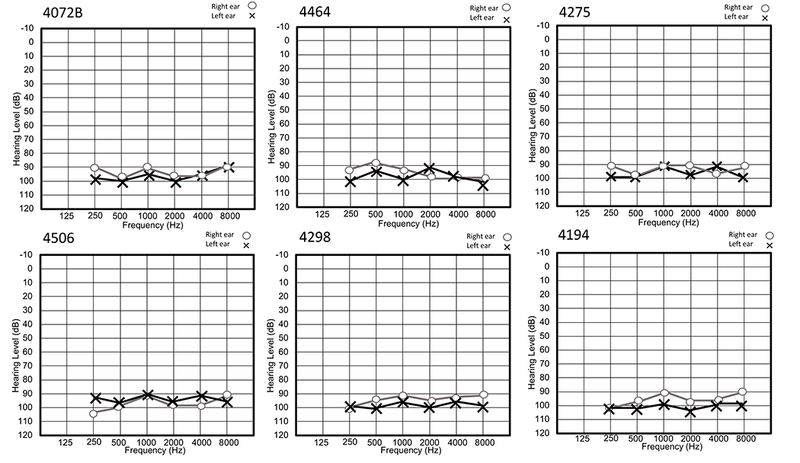
Air conduction thresholds of six hearing impaired individuals. Circles represent the right ear and crosses the left ear. All the tested subjects have bilateral and profound HI across all frequencies.
Linkage analysis was performed on all families and resulting LOD Scores can be found in Table 1. Three of the seven ARNSHI families in this study had a significant LOD scores of ≥ 4.0 while the remaining families had suggestive evidence of linkage (LOD scores 1.7 – 2.7) to the 6p21.1-22.3 region which contains LHFPL5. Of the two families for which the novel variants were discovered, one family could establish linkage, i.e. c.*16+1G>Aa family 4072A LOD score=5.0 and p.Gly151Val family 4464 LOD score=5.4 (Table 1; Figure 1).
Table 1.
Bioinformatics analyses results of four pathogenic mutations found in seven Pakistani families
| Family | Maximum LOD | Chr6 coordinate | Variant | rs Id | GERP++ | Allele frequency |
PolyPhen-2 | SIFT | Mutation taster | LRT | Mutation assessor | FATHMM | CADD score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD | GME variome/BRAVOa | |||||||||||||
| 4072Ab | 5.0 | 35,787,241 | c.*16+1G>Ac | N.A | 3.49 | 0.00 | 0.00 | N.A.d | N.A. | Disease causing | N.A. | N.A. | N.A. | 23.3 |
| 4072Bb | 2.4 | 35,787,241 | c.*16+1G>Ac | N.A | 3.49 | 0.00 | 0.00 | N.A. | N.A. | Disease causing | N.A. | N.A. | N.A. | 23.3 |
| 4194 | 1.7 | 35,782,362 | c.452G>T (p.Gly151Val)c | rs762876554 | 5.53 | 8.1e-6 | 0.00 | Probably damaging | Damaging | Disease causing | D | Functional | Damaging | 28 |
| 4275 | 2.7 | 35,773,697 | c.250delC (p.Leu84*)e, f | N.A | 5.57 | 0.00 | 0.00 | N.A. | N.A. | Disease causing | N.A. | N.A. | N.A. | 25.8 |
| 4298 | 1.9 | 35,773,827 | c.380 A > G (p.Tyr127Cys)e, g | rs104893975 | 4.46 | 2.8e-5 | 0.00 | Probably damaging | Tolerated | Disease causing | D | Functional | Damaging | 24.8 |
| 4464 | 5.4 | 35,782,362 | c.452 G > T (p.Gly151Val)c | rs762876554 | 5.53 | 8.1e-6 | 0.00 | Probably damaging | Damaging | Disease causing | D | Functional | Damaging | 28 |
| 4506 | 4.0 | 35,773,697 | c.250delC (p.Leu84*)e, f | N.A | 5.57 | 0.00 | 0.00 | N.A. | N.A. | Disease causing | N.A. | N.A. | N.A. | 25.8 |
BRAVO is a database, which is powered by TOPMed Freeze 5 Public Subset. This dataset includes 463 million variants on 62,784 individuals
Families 4072A and 4072B reported to be related but the exact relationship is unknown
Mutations in bold indicate novel findings first reported in this study
Not applicable as those predictive software (except for MutationTaster) provide functional outcomes for missense variants
These mutations were previously reported in Pakistani and Indian families (Shabbir et al. 2006)
This mutation leads to NMD (nonsense-mediated decay) predicted by MutationTaster
rs104893975 is reported in dbSNP as a clinically associated pathogenic allele
LHFPL5 was selected for follow-up since for each family it is the only known HI gene within the linkage region. DNA samples from all available family members (Figure 1) underwent Sanger sequencing to determine if pathogenic variants lie within this gene. Known LHFPL5 variants segregated with HI in three of the seven families: c.250delC (p. Leu84*) for families 4275 and 4506 and c.380A>G (p. Try127Cys) for family 4298 (Figure 1). Families 4194 and 4464 had a novel missense variant c.452G>T (p. Gly151Val) in exon 2, and families 4072A and 4072B had a novel nucleotide substitution (G>A) at the 5’ donor splice site of exon 3 at c.*16+1 (Figure 3A and C) which segregated with HI. The known and novel variants were not observed in 200 and 600 Pakistani control chromosomes, respectively (Table 1). None of the variants were reported in the Greater Middle East (GME) Variome and Trans-omics for precision medicine (TOPMed) Bravo Database. Novel variant c.452G>T (p.Gly151Val) was observed in gnomAD exome data with a variant frequency 8.1 × 10−6 with two South Asians heterozygous individuals (MAF=6.5 × 10−5). Also in gnomAD exome sequence data known variant c.380A>G (p.Tyr127Cys; rs104893975) was observed with a MAF=2.8 × 10−5 with two heterozygous variants observed in South Asians (MAF 6.5×10−5) and five heterozygous variants observed in non-Finnish Europeans (MAF=4.5×10−5) and it is also reported in dbSNP and ClinVar as a clinically associated pathogenic variant (Table 1).
Figure 3.
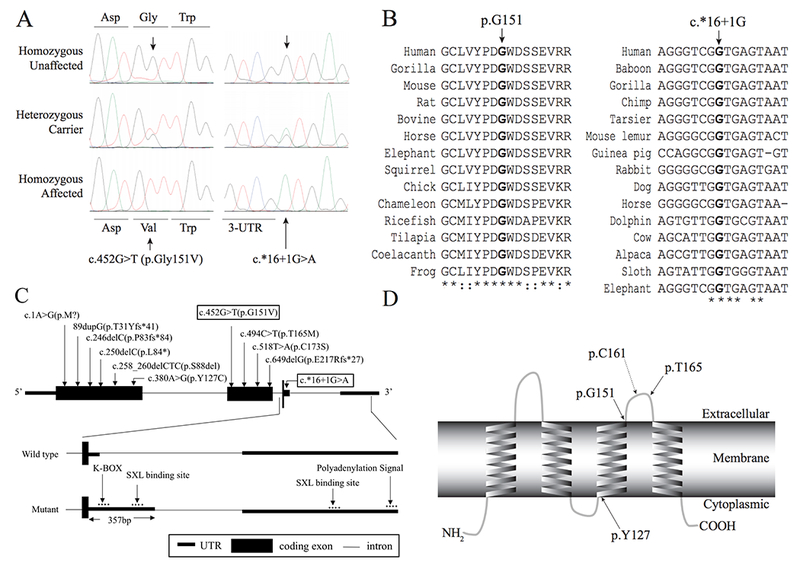
A) Chromatograms displaying the novel variants c.452G>T in families 4194 and 4464 and c.*16+1G>A in families 4072A and 4072B. B) ClustalW2 sequence alignment of amino acids across LHFPL5 proteins from various species with conserved amino acids indicated with an asterisk, while colons indicate conservation between groups with strongly similar properties. The glycine151 residue is indicated with an arrow, and is fully conserved across all species (Left Panel). DNA sequence alignment containing the guanine nucleotide c.*16+1, indicated with an arrow. The position is fully conserved across various species (Right Panel). C) Schematic presentation of the exon-intron structure with eleven pathogenic variants. The boxed variants indicate novel variants found in this study (Top panel). The wild type structure of exon3-intron3-exon4 of LHFPL5 (Middle panel). In the c.*16+1 mutant transcript, exon3 was extended by 357 bp due to the activation of a cryptic splice site. The regulatory element binding sites in 3’-UTR are indicated with an arrow (Bottom Panel). D) Predicted transmembrane helices in LHFPL5 (adapted from the result of TMHMM 2.0 analysis) and depiction of the amino acid positions of previously reported two missense variants and the novel variant found in this study. The dotted line arrow indicates the location of the missense variant found in hscy mice.
The novel missense variant c.452G>T (p.Gly151Val) has a CADD C-score of 28 and was predicted to be ‘Disease Causing’, ‘Damaging’, or ‘Functional’ by various bioinformatics prediction software (Table 1). The guanine nucleotide at c.452 has a GERP++ score of 5.53 indicating that it is under strong evolutionary constraint. Based on Clustalw2 alignment, the glycine residue at p.151 is fully conserved across 14 species ranging from frog to gorilla (Figure 3B). It is predicted the glycine residue (p.G151) is located on the second extracellular loop of the transmembrane protein (Figure 3D).
The donor splice site variant c.*16+1G>A was predicted to be disease causing by MutationTaster (Table 1) due to natural splice site disruption (Table 2). It is predicted to lead to a loss of the 5’ donor splice site in the 3’ UTR of LHFPL5, predicted by various bioinformatics tools (Table 2). In addition, the ADA and RF score for this variant are 0.99 and 0.93 respectively (> 0.60 is predicted to impact pre-mRNA splicing), shown by annotation of the dbscSNV database [31]. The guanine nucleotide at c.*16+1 has a GERP++ score of 3.49 indicating that the nucleotide is under strong evolutionary constraint (Figure 3B). A further in silico analysis using three splice site analysis tools (Table 2), shows that the c.*16+1G>A mutation is predicted to activate a cryptic splice site 357 bp toward the 3’ direction. The extended 3’-untranslated region (UTR) leads to a different RNA secondary structure with much less minimum free energy (Supplement Figure 1). A UTR analysis tool and microRNA target scanning software predicted that the extended 3’-UTR may include additional regulatory elements, ‘K-BOX’ and ‘SXL binding site’ (Figure 3C), and new microRNA binding sites (Supplement Table 1).
Table 2.
Splice site analyses results for the mutation, c.*16+1G>A, found in families 4072A and 4072B
| Prediction Software | Natural Donor Splice site a | Predicted effect of c.*16+1G>A | Cryptic Donor Splice site | 3’-UTR Length Variation in Cryptic Donor Splice site (bp) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Original Splice siteb | Chr6 Coordinate | Prediction Score c | 3′ UTR length (bp) | New Splice site b | Chr6 Coordinate | Prediction Score c | 3’ UTR length (bp) | |||
| NNsplice | GGTCGgtgagt | 35,787,241 | 0.98 | 1,110 | Natural Donor Splice site Broken d | TACAGgtgcgt | 35,787,598 | 0.99 | 1,467 | + 357 |
| HSF | GGTCGgtgagt | 35,787,241 | 92.1 | TACAGgtgcgt | 35,787,598 | 90.0 | + 357 | |||
| NetGene2 | GGTCGgtgagt | 35,787,241 | 0.99 | TACAGgtgcgt | 35,787,598 | 0.99 | + 357 | |||
Three splice site prediction software showed the highest donor splice site scores at the natural donor splice site position (NM_182548.3).
Nucleotides with capital letter indicate exon sequences and lowercase indicate intron sequences. Nucleotide in bold indicate the natural donor splice site (c.*16+1G) or the cryptic donor splice site.
Prediction score ranges 0-1, 0-100, 0-1 in NNsplice, HSF and NetGene2, respectively.
The three software predicted the mutation disrupts natural donor splice site and activate cryptic splice site.
Discussion
Variants underlying deafness and vestibular dysfunction in the Tmhs gene were identified in hscy mice [8]. This finding was followed by the identification of variants underlying HI in the human ortholog (LHFPL5) in families with HI without vestibular dysfunction that segregated DFNB67 [2–5]. Previously nine pathogenic variants were reported. LHFPL5 is predicted to be a tetra-span transmembrane protein with two extracellular loops (Figure 3D). The known variant c.250delC (p.Leu84*) observed in families 4275 and 4506 causes a frame-shift in exon 1, introducing a premature stop codon (Figure 3C), and the mRNA is eventually degraded by nonsense-mediated decay. The second known variant, c.380A>G (p. Tyr127Cys), seen in family 4298 replaces the tyrosine residue with a cysteine. This amino acid change occurring within the third transmembrane helix is predicted to result in the mis-localization of the protein (Figure 3D) [2].
Novel variant c.452G>T (p.Gly151Val) replaces the glycine residue located on the second extracellular loop with a valine. This variant is in close proximity to the p.Cys161Phe hscy mouse variant [8] and a previously reported human pathogenic variant p.Thr165Met [4] (Figure 3D), implicating the functional significance of the second extracellular loop. Since LHFPL5 is presumed to organize a transient cytoskeleton-membrane interaction in the stereocilia of sensory hair cells, the variant p.Gly151Val may cause dysfunction or mis-localization of LHFPL5, leading to stereocilia pathology similar to that found in hscy mice [8].
The second novel variant c.*16+1G>A introduces a 5’ donor splice site disruption in exon 3 (Figure 3C), activating a cryptic splice site predicted to occur 357 bp downstream and extending the 3’ UTR. The change occurs in the 3’-untranslated region (UTR), to which regulatory molecules and microRNAs (miRNAs) can bind to regulate gene expression. The extended 3’-UTR sequence may affect the expression of LHFPL5 in different ways: 1) longer 3’-UTR sequences may introduce unstable RNA secondary structures, which eventually lead to translational repression; 2) additional regulatory elements - ‘K-BOX’, ‘SXL binding site’ (Figure 3C) - may negatively affect the gene expression; 3) new miRNA binding sites in the extended 3’-UTR sequences (Supplement table 1) may reduce the mRNA activity. Causal variants in 3’-UTR regions have not frequently been reported, especially variants affecting 3’UTR splicing and therefore affecting gene function [32]. This low identification rate could be attributed to the limited understanding of their functional impact.
miRNAs play very important roles in the auditory system and variants in miR-96 have been reported to cause deafness in humans [33,34] and mice [35]. Micro-RNAs are involved with hearing functions inhibiting target mRNAs by repressing translational activity and destabilizing RNA secondary structure [36]. Among the new miRNA binding sites found in the extended region, miR-5787 was reported to repress cellular growth targeting eukaryotic translation initiation factor 5 (elF5) in fibroblasts [37]. Interestingly, in Drosophila, ‘K-Box’ bound miRNAs (K-Box miRNAs) were reported to inhibit the Notch pathway [38], which is involved in cochlear development and deafness [39]. Thus, new miRNA binding sites and regulatory element such as ‘K-box’ in the longer 3’-UTR sequences may alter the expression of LHFPL5, which may result in pathogenic effects.
Variants in LHFPL5 are a relatively rare cause of ARNSHI, but knowledge of all variants contributing to the HI phenotype provides a valuable resource for diagnostic genetic testing. Moreover, the unusual splice site variant in 3’-UTR provides a deeper understanding of the functional roles of various 3’-UTR regulatory motifs in the etiology of human deafness.
Supplementary Material
Acknowledgements:
The authors would like to thank the family members who participated in the study. This work was funded by the Higher Education Commission, Islamabad, Pakistan and by the National Institutes of Health (NIH) – National Institute of Deafness and other Communication Disorders (DC03594 and DC011651). Genotyping services were provided by CIDR through a fully funded federal contract from the NIH to the Johns Hopkins University, Contract Number N01-HG-65403.
Footnotes
Competing interests: The authors declare no competing interests
Electronic Database Information
The following URLs were accessed for data in this article:
Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org
UCSC Genome Browser, http://genome.ucsc.edu
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
1000 genomes, http://www.1000genomes.org
http://gnomad.broadinstitute.org
Supplementary Information
The Supplementary information in this paper includes one figure (Supplementary figure 1) and one table (Supplementary table 1).
References
- 1.Morton CC, Nance WE. Newborn Hearing Screening - A Silent Revolution. N Engl J Med Boston. 2006;354:2151–64. [DOI] [PubMed] [Google Scholar]
- 2.Shabbir MI, Ahmed ZM, Khan SY, Riazuddin S, Waryah AM, Khan SN, et al. Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J Med Genet. 2006;43:634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bensaïd M, Hmani-Aifa M, Hammami B, Tlili A, Hakim B, Charfeddine I, et al. DFNB66 and DFNB67 loci are non allelic and rarely contribute to autosomal recessive nonsyndromic hearing loss. Eur J Med Genet. 2011;54: e565–9. [DOI] [PubMed] [Google Scholar]
- 4.Kalay E, Li Y, Uzumcu A, Uyguner O, Collin RW, Caylan R, et al. Mutations in the lipoma HMGIC fusion partner-like 5 (LHFPL5) gene cause autosomal recessive nonsyndromic hearing loss. Hum Mutat Hoboken. 2006;27:633. [DOI] [PubMed] [Google Scholar]
- 5.Shahin H, Walsh T, Rayyan AA, Lee MK, Higgins J, Dickel D, et al. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet EJHG Leiden. 2010;18:407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ammar‐Khodja F, Bonnet C, Dahmani M, Ouhab S, Lefèvre GM, Ibrahim H, et al. Diversity of the causal genes in hearing impaired Algerian individuals identified by whole exome sequencing. Mol Genet Genomic Med. 3:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloan-Heggen CM, Babanejad M, Beheshtian M, Simpson AC, Booth KT, Ardalani F, et al. Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet. 2015;52:823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, Johnson KR. A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc Natl Acad Sci U S A. 2005;102:7894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 10.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D. Online System for Faster Multipoint Linkage Analysis via Parallel Execution on Thousands of Personal Computers. Am J Hum Genet. 200678:922–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37. [PMC free article] [PubMed] [Google Scholar]
- 13.Matise TC, Chen F, Chen W, Vega FMDL, Hansen M, He C, et al. A second-generation combined linkage–physical map of the human genome. Genome Res. 2007;17:1783–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol Clifton NJ. 2000;132:365–86. [DOI] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. [DOI] [PubMed] [Google Scholar]
- 17.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. [DOI] [PubMed] [Google Scholar]
- 18.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, et al. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum Mutat. 2013;34:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012;40:D71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. [DOI] [PubMed] [Google Scholar]
- 24.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J Mol Biol. 2001;305:567–80. [DOI] [PubMed] [Google Scholar]
- 25.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol J Comput Mol Cell Biol. 1997;4:311–23. [DOI] [PubMed] [Google Scholar]
- 26.Desmet F- O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. [DOI] [PubMed] [Google Scholar]
- 28.Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, et al. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2010;38:D75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007; 39:1278–84. [DOI] [PubMed] [Google Scholar]
- 31.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014; 42:13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin C, Jiang J, Wang W, Yao K. Identification of a MIP mutation that activates a cryptic acceptor splice site in the 3′ untranslated region. Mol Vis. 2010; 16:2253–8. [PMC free article] [PubMed] [Google Scholar]
- 33.Mencía Á, Modamio-Høybjør S, Redshaw N, Morín M, Mayo-Merino F, Olavarrieta L, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009; 41:609–13. [DOI] [PubMed] [Google Scholar]
- 34.Soldà G, Robusto M, Primignani P, Castorina P, Benzoni E, Cesarani A, et al. A novel mutation within the MIR96 gene causes non-syndromic inherited hearing loss in an Italian family by altering pre-miRNA processing. Hum Mol Genet. 2012; 21:577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009; 41:614–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudnicki A, Avraham KB. microRNAs: the art of silencing in the ear. EMBO Mol Med. 2012; 4:849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo H, Yoo JK, Lee J, Lee DR, Ko JJ, Oh SH, et al. The hsa-miR-5787 represses cellular growth by targeting eukaryotic translation initiation factor 5 (eIF5) in fibroblasts. Biochem Biophys Res Commun. 2011; 415:567–72. [DOI] [PubMed] [Google Scholar]
- 38.Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, Chou Y- T, et al. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development. 2012; 139:2821–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terrinoni A, Serra V, Bruno E, Strasser A, Valente E, Flores ER, et al. Role of p63 and the Notch pathway in cochlea development and sensorineural deafness. Proc Natl Acad Sci U S A. 2013;110:7300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


