Abstract
Abnormal airway smooth muscle cell (ASMC) proliferation and migration contribute significantly to increased ASM mass associated with asthma. MicroRNA (miR)-638 is a primate-specific miRNA that plays important roles in development, DNA damage repair, hematopoiesis, and tumorigenesis. Although it is highly expressed in ASMCs, its function in ASM remodeling remains unknown. In the present study, we found that in response to various mitogenic stimuli, including Platelet-derived growth factor (PDGF)-BB, Transforming Growth Factor Beta 1 (TGF-β1), and Fetal Bovine Serum (FBS), the expression of miR-638, as determined by quantitative real-time polymerase chain reaction (qRT-PCR), was significantly downregulated in the proliferative human ASMCs. Both gain- and loss-of-function studies were performed to study the role of miR-638 in ASMC proliferation and migration. We found that adenovirus-mediated miR-638 overexpression markedly inhibits ASMC proliferation and migration, while ablation of miR-638 by anti-miR-638 markedly increases cell proliferation and migration, as determined by WST-8 proliferation and scratch wound assays. Dual-luciferase reporter assay, qRT-PCR, and immunoblot analysis were used to investigate the effects of miR-638 on the expression of the downstream target genes in ASMCs. Our results demonstrated that miR-638 overexpression significantly reduced the expression of downstream target cyclin D1 and NOR1, both of which have been shown to be essential for cell proliferation and migration. Together, our study provides the first in vitro evidence highlighting the anti-proliferative and anti-migratory roles of miR-638 in human ASMC remodeling and suggests that targeted overexpression of miR-638 in ASMCs may provide a novel therapeutic strategy for preventing ASM hyperplasia associated with asthma.
Keywords: microRNA-638, airway smooth muscle, proliferation, migration, cyclin D1, asthma
1. Introduction
Asthma is a chronic inflammatory disease characterized by airway pathological remodeling, as manifested by abnormal smooth muscle hypertrophy, hyperplasia, basement membrane thickening, and subepithelial fibrosis in asthmatic patients (Gosens and Grainge, 2015) (Kudo et al., 2013). Airway smooth muscle cells (ASMCs) play important roles in the lung, and excessive proliferation and migration of ASMCs directly contribute to airway remodeling (Kudo et al., 2013; Makinde et al., 2007). Airway remodeling occurs as an airway repair response to the sustained injury as a result of inflammation. Disruption in this repair process often leads to airway remodeling. A crucial structural change in airway remodeling is increased ASMC proliferation and migration, which contribute significantly to cellular hyperplasia, and a subsequently increased airway smooth muscle mass (Johnson, 2001). At this point, the molecular mechanism(s) underlying the ASMC proliferation and migration is not completely understood.
microRNAs (miRNAs) are small (~22 nt) noncoding RNAs that regulate gene expression through binding to the 3′ untranslated region (UTR) of mRNA transcripts to induce mRNA degradation or translational inhibition of target mRNAs (He and Hannon, 2004) (Ameres and Zamore, 2013). Thus far, >1000 human miRNAs have been identified and they are thought to regulate up to 60% of gene transcripts, hence, having broad implications in the regulation of a wide range of biological activities, including development, cellular differentiation, and the pathogenesis of various diseases. Recently, multiple lines of evidence suggest that miRNAs play pivotal roles in the control of ASM phenotypes that are critically involved in asthma pathogenesis (Deshpande et al., 2015). Thus far, several miRNAs, including miR-10a, miR-23b, miR-143-3p, miR-21, and miR-203, have been implicated in the regulation of ASM cell proliferation and/or migration (Hu et al., 2014) (Cheng et al., 2016; Liu et al., 2015) (Chen et al., 2017a). Further identification of novel miRNAs involved in the regulation of ASMC proliferation and migration will help us develop novel therapeutic strategies for the treatment of asthma.
miR-638 is a primate-specific miRNA that plays important roles in development, DNA damage repair, hematopoiesis, and leukemogenesis (Li et al., 2013) (Tan et al., 2014) (He et al., 2016; Lu et al., 2015). In particular, the role of miR-638 in cancer development has recently received a significant attention. It has been shown that the expression of miR-638 is downregulated in various types of cancers, including gastric cancer, cervical cancer, and colon cancer, and overexpression of miR-638 inhibits cancer growth and metastasis, indicating miR-638 functions as a tumor suppressor (Parasramka et al., 2013) (Lu et al., 2015; Ma et al., 2014; Zhao et al., 2014). Although miR-638 is highly expressed in airway smooth muscle cells (Li et al., 2013), little is known about its role in regulating ASMC functions. In the present, we investigated whether miR-638 is involved in regulating ASMC proliferation and migration, if so, to determine the molecular mechanism(s) involved.
2. MATERIALS AND METHODS
2.1 Cell Culture
Human airway smooth muscle cells (ASMCs) were cultured with Dulbecco’s modified Eagles medium/Ham’s F12 (DMEM/F12; Gibco, Rockville, MD) containing 10% fetal bovine serum (HI FBS; Gibco, Grand Island, NY), 100 unit/mL penicillin and 100 μg/mL streptomycin (penicillin streptomycin solution, Manassas, VA,). HEK-293AD and HEK-293T cells were cultured in Dulbecco’s modified Eagles medium (DMEM; Corning, Manassas, VA) supplemented with 10% fetal bovine serum, 100 unit/mL penicillin and 100 μg/mL streptomycin. Human ASMCs between passage 4 and 6 were used for all experiments.
2.2 ASMC Proliferation Assay in Vitro
Human ASMC proliferation was determined by cell counting and cell proliferation kit WST-8 (Cayman Chemical, Ann Arbor, MI). For cell counting, cells were detached by trypsinization and resuspend in PBS (Gibco, Island, NY), then counted under a microscope. For WST-8 proliferation assay, 100 μl of ASMCs suspension were seeded into 96-well plates with or without PDGF-BB (20 ng/mL) (Peprotech, Rocky Hill, NJ), TGF-β1 (20 ng/mL) (Peprotech, Rocky Hill, NJ) treatment, and the plates were incubated for 48 h in a CO2 incubator at 37°C. Then 10 μl of the working solution containing WST-8 and 1-methoxy phenazine methosulfate (0.5 mM and 20 μM, respectively, as the final concentration) was added into each well and incubated for an additional 1 hour, the optical density was read at 450 nm with a microplate reader (BioTek Instruments, Inc., Winooski, VT).
2.3 Construction of Adenovirus
The adenovirus expressing miR-638 (Ad-miR-638) was made using RAPAd® Universal Adenoviral Expression System (Cell BioLab Inc. San Diego, CA) as we described previously (Li et al., 2013). PacAd5 9.2-100 Ad backbone vector was co-transfected with the pacAd5 K-NpA shuttle vector containing the miR-638 sequence into HEK-293AD Cells using FuGene 6 Transfection Reagent (Roche, Indianapolis, IN). Adenoviruses LacZ (Ad-LacZ) and NOR1 (Ad-NOR1) were made using AdMax system (Microbix Biosystems Inc., Toronto, Canada) essentially as we described previously. The viruses were propagated on HEK-293AD Cells and purified using CsCl2 banding followed by dialysis against 20 mmol/L Tris-buffered saline with 2% glycerol. Titering was performed with HEK-293AD cells using Adeno-X™ Rapid Titer Kit (Clontech Laboratories, Inc., Mountain View, CA) according to the manufacturer’s protocol.
2.4 Oligonucleotide Transfection and Adenovirus Infection in Cultured ASMCs
miR-638 mimic (QIAGEN Foster City, CA) was used for miR-638 overexpression at a final concentration of 30 nM; Anti-miR-638 (QIAGEN Foster City, CA) was used for miR-638 knockdown at a final concentration of 100 nM. Negative control miR (QIAGEN, Foster City, CA) was used as a control for miR-638 mimic and Anti-miR-638 transfection experiments. Transfections were performed using HiPerFect transfection reagent (QIAGEN, Valencia, CA) at 24 h after seeding the cells into the wells following the manufacturer’s protocol. For adenovirus-mediated miR-638 overexpression, Ad-miR-638 was added into the culture medium at 100 multiplicity of infection (MOI) or at indicated MOIs. For NOR1 overexpression, cells were infected with Ad-NOR1 at 100 MOI. Ad-LacZ was used as an adenovirus control. 24 h or 48 h after transduction, ASMCs were harvested for RNA and protein isolation, respectively.
2.5 Quantitative RT-PCR (qRT-PCR) of miRNA and mRNA expression
miRNAs were isolated with miRNeasy Mini Kit (QIAGEN, Hilden, Germany) and the quantitative real-time polymerase chain reaction (qRT-PCR) was performed using All-in-one miRNA qRT-PCR Detection Kit (GeneCopoeia, Inc., Rockville, MD). mRNAs were isolated with TRIzol LS Reagent (Ambion, carsland CA). Complementary DNA (cDNA) was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Vilnius, Lithuania). qRT-PCR analyses were conducted to quantitate the relative mRNA expression using SYBR Master mix (Applied Biosystems, Austin, TX). U6 was used as an internal control for miRNA template normalization, and 18S was used for other template normalizations. The reactions were set up following the manufacturer’s protocol. After the reactions, the cycle threshold (Ct) data were determined using default threshold settings, and the mean Ct was determined from the duplicate PCRs. The target PCR Ct values were normalized by subtracting the U6 or 18S ΔCt value, which provided the Ct value. A comparative ΔCt method was used to compare each condition with the controls, and the values are expressed as 2−ΔCt. The relative DNA copy numbers were determined as described previously (Li et al., 2013).
2.6 Western Blotting
Cells were washed with PBS, and then lysed with RIPA Buffer (25 mM Tris•HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitors. After rocking at 4 °C for 60 mins, insoluble materials were removed by centrifugation. Equal amounts of protein were subjected to SDS-PAGE. Standard western blot was conducted using p-ERK1/2 (1:1000 diluted; Cell Signaling, cat. 4377S), T-ERK1/2 (1:1000 diluted; Cell Signaling, cat. 4696S), Cyclin D1 (1:1000 diluted; Cell Signaling, cat. 2978S), α-Tubulin (1:1000 diluted; Abcam, cat. ab176560), PCNA (1:500 diluted; Santa Cruz, cat. sc-56), NOR1 (1:500 diluted; Santa Cruz, cat. sc-292902) Actin (1:1000 diluted, Santa Cruz, cat. sc-8432) antibodies.
2.7 ASMC Migration Assays
Human ASMCs were seeded into 6-well plates and transduced with adenovirus or transfected with miRs, and then starved with 0.5% FBS medium for 48 h. After starvation, a linear wound was scratched in the center of the cell monolayer using sterile 200 μl pipette tip, and then washed 3 times with PBS to remove the cell debris. Migration of ASMCs into the wound was determined at different time points (0 and 24 h) in the presence and absence of PDGF-BB (20 ng/mL) stimulation. All images were captured by an EVOS FL Auto Cell Imaging System inverted microscope (Life Technologies, Carlsbad, CA). Migrated cells were quantitated using ImageJ 1.50b software program.
2.8 Flow Cytometry
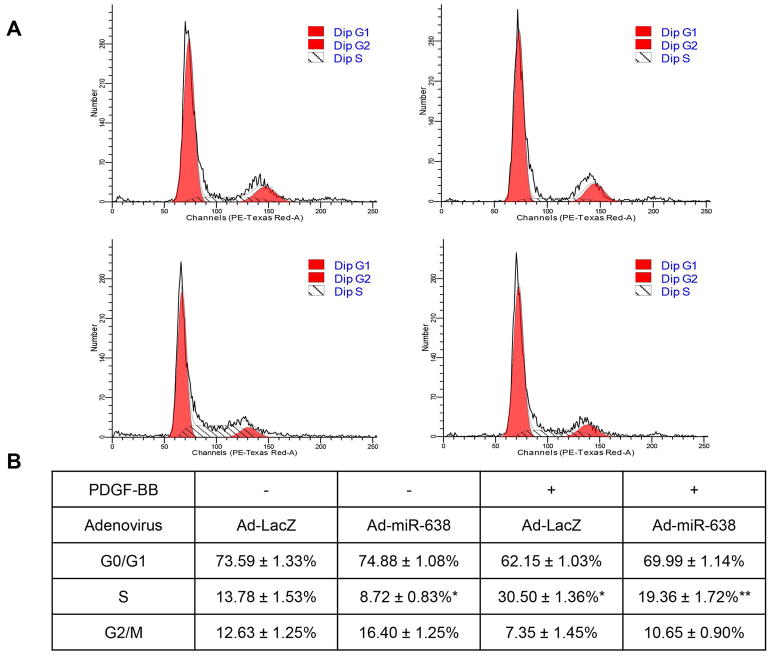
Human ASMCs were seeded into P-60 dishes and then infected with Ad-miR-638 or Ad-LacZ. 48 h after starvation with 0.5% FBS medium, cells were treated with or without PDGF-BB (20 ng/mL). Cells were then collected and fixed with 66% ethanol at 4°C for overnight, stained with propidium iodide + RNaseA (Abcam, Cambridge, UK) at 37 °C for 30 min. DNA content was analyzed using BD LSR-II flow cytometer, G0/1, S, and M/G2 phase were analyzed by ModFit LT 3.1 software.
2.9 Luciferase Assay
Luciferase assay was performed using the Dual-Luciferase reporter assay system (Gold Bio, St. Louis, MO) as previously described (Li et al., 2013). To construct the report vector, the 3′-untranslational region (UTR) sequence of cyclin D1 gene was cloned into the empty pLightSwitch-3′-UTR RenSP-luciferase reporter vector (SwitchGear Genomics, Menlo Park, CA). For the luciferase activity determination, HEK-293T cells were seeded into 24-well plates and co-transfected with luciferase reporter vector together with either miR-638 mimic, Anti-miR-638, or non-targeting control miRNA, following the transfection procedures provided by QIAGEN. 48 h after transfection, cells were lysed for luciferase activity measurement using a luminometer (BioTek Instruments Inc., Winooski, VT). To construct the cyclin D1-3′UTR-MUT vector, the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) was used to induce mutations in the 3′-UTR region of cyclin D1-3′UTR-WT vector as shown in Figure 6A.
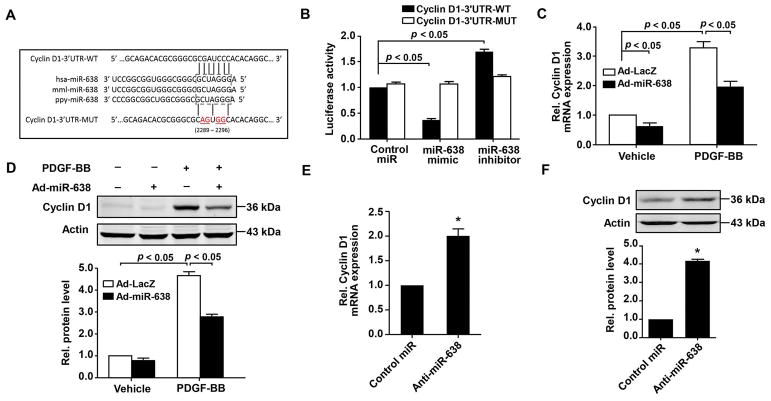
Figure 6.
Cyclin D1 is a direct target of miR-638 in human ASMCs. A,cyclin D1 is a direct target gene of miR-638, as predicted by TargetScan database. Schematic representation of the miR-638 putative binding site in the 3′-UTR of the human cyclin D1 gene and alignment of predicted wild-type and mutant cyclin D1 3′-UTR sequence at the binding site. Four mutated nucleotides are underlined. B, cyclin D1-3′UTR-WT or cyclin D1-3′UTR-MUT vector was co-transfected into HEK-293T cells, with either miR-638 mimic, anti-miR-638 or Non-targeting control miR. 48 h after transfection, renilla luciferase activities were measured (n=6). C and D, human ASMCs were pre-infected with either Ad-miR-638 or Ad-LacZ (100 MOI) for 24 h and then starved for 48 h, cyclin D1 mRNA was measured by qRT-PCR (n=4) at 24 h after stimulated with or without PDGF-BB (20 ng/mL). Cyclin D1 protein expression was detected by western blot (n=4). E and F, human ASMCs were pre-transfected with either anti-miR-638 or control miR (100 nM) for 24 h and then starved for 48 h. Relative levels of cyclin D1 mRNA and protein were detected at 24 h by qRT-PCR and western blot, respectively. n=4.
2.10 Statistical Analysis
All experiments were performed at least three times. All statistical analysis was performed using SPSS statistical analysis program (Version 17.0), and graphs were generated using GraphPad Prism software (Version 5.0.1). All data were presented as mean ± S.D.. For relative data analysis, the mean value of control group is defined as 1 or 100%. Differences between two groups were compared with the unpaired Student’s t-test. Differences between multiple groups were compared with one-way analysis of variance. A p value <0.05 was considered to be significant.
3. Results
3.1 Expression of miR-638 is decreased in proliferative ASMCs
To elucidate the role of miR-638 in airway smooth muscle biology, qRT-PCR was performed to determine the expression of miR-638 in both quiescent and proliferative human ASMCs in response to PDGF-BB, which is a potent stimulator for ASMC remodeling (Hirota et al., 2011; Walker et al., 1998). As shown in Figure 1A and B, stimulation of ASMCs with PDGF-BB substantially attenuated the expression of miR-638 in both time and dose dependent manners. PDGF-BB maximally inhibited miR-638 expression by approximately 75 % at 12 h after PDGF-BB stimulation (Figure 1A). The IC50 of PDGF-BB-induced inhibition on miR-638 expression is about 7 ng/mL (Figure 1B). To further determine whether the down-regulation of miR-638 expression is specific to PDGF-BB, human ASMCs were treated with other stimulators, such as 10% FBS, PGE2, ET-1, and TGF-β. Notably, we found that in addition to PDGF-BB, both 10% FBS and TGF-β1 markedly inhibited the expression of miR-638 in human ASMCs (Figure 1C and D), while Prostaglandin E2 (PGE2) and endothelin-1 (ET-1), which are involved in ASMC inflammartion (Liu et al., 2012) (Lin et al., 2015a), had no effect on miR-638 expression. Together, these results suggest that miR-638 may play an important role in ASMC remodeling.
Figure 1.
Expression of miR-638 is downregulated in proliferative ASMCs. Human ASMCs were seeded into 6-well plates and cultured in complete medium for 24 h at 40~60% confluence. Starvation was perform with 0.5% FBS medium and then treated with PDGF-BB (20 ng/mL) or 10% FBS. A, PDGF-BB induced a time-dependent decrease of miR-638 expression in ASMCs as determined by qRT-PCR (n=4). *p < 0.05 compared with the time at 0 h group. B, PDGF-BB induced a decreased expression of miR-638 in a dose-dependent manner at 24 h in ASMCs as determined by qRT-PCR (n=4). *p < 0.05 compared with the dose of 0 ng/mL group. C, FBS decreased expression of miR-638 in human ASMCs. Starved ASMCs were treated with 10% FBS for the indicated times and expression of miR-638 was determined by qRT-PCR (n=4). *p < 0.05 compared with the time at 0 h group. D, human ASMCs were treated with various mitogens for 24 h and expression of miR-638 was determined by qRT-PCR (n=4). miR-638 expression was significantly inhibited by PDGF-BB (20 ng/mL) and TGF-β1 (20 ng/mL), but not by other stimuli, such as PGE2 (20 ng/mL) and ET-1 (20 ng/mL). *p < 0.05 compared with vehicle.
To further determine the molecular signaling pathways involved in PDGF-BB-induced miR-638 downregulation, human ASMCs were pre-treated with inhibitors of various kinases involved in PDGF-BB-induced cell signaling in ASMCs. As shown in Figure 2A, treatment of ASMCs with MAPK kinase (MEK) 1/2 inhibitor PD98059 (50 μM) almost completely abolished the PDGF-BB-induced downregulation of miR-638 expression, whereas other kinases’ inhibitors, such as p38 inhibitor SB203580 (20 μM), PI3 kinase inhibitor LY294002 (50 μM), and JNK inhibitor SP600125 (20 μM), barely affected the inhibitory effects of PDGF-BB on miR-638 expression in human ASMCs. Indeed, treatment of cells with PD98059 (50 μM) completely blocked PDGF-BB-induced ERK1/2 phosphorylation in ASMCs, as shown in Figure 2B. Together, these results suggest that the MEK1/2 dependent signaling pathway is responsible for the PDGF-BB-induced down-regulation of miR-638 expression in proliferative human ASMCs.
Figure 2.
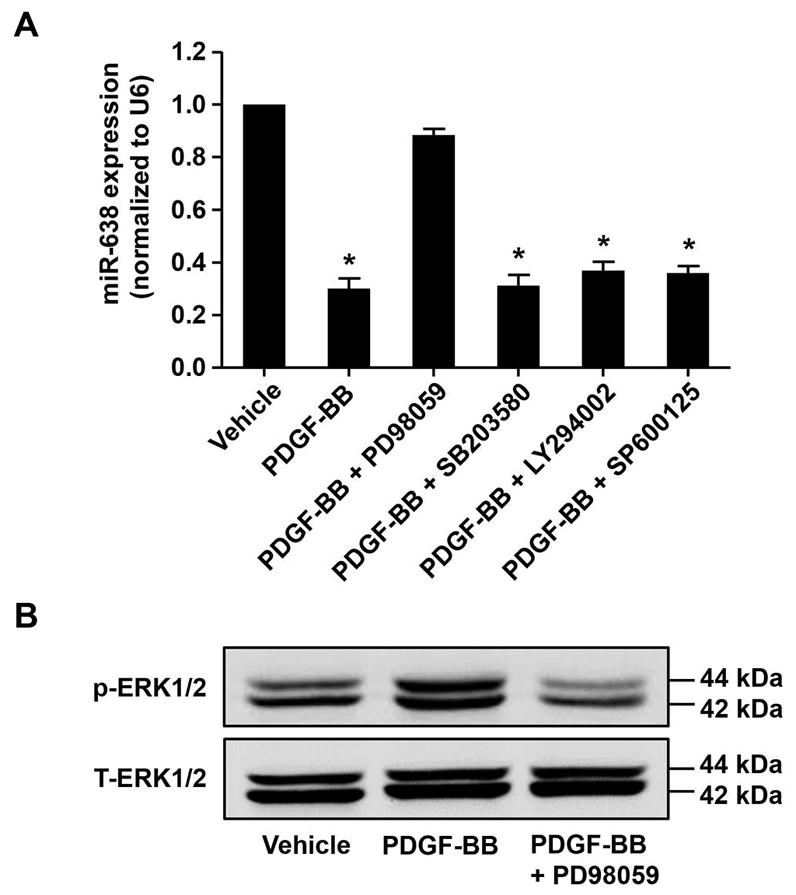
PDGF-BB-induced downregulation of miR-638 is MEK1/2 dependent. Starved human ASMCs were pre-treated with protein kinases’ inhibitors, including MEK1/2 inhibitor PD98059 (50 μM), p38 inhibitor SB203580 (20 μM), JNK inhibitor SP600125 (20 μM), and PI3K inhibitor LY294002 (50 μM) for 1 h and then stimulated with vehicle or PDGF-BB (20 ng/mL) for 24 h in (A) and 30 min in (B). A, the expression of miR-638 in ASMCs was determined by qRT-PCR (n=4). B, the phosphorylation and total expression of ERK1/2 were determined by western blot (n=4). *p < 0.05 compared with miR-638 expression in vehicle control group.
3.2 miR-638 is involved in ASMC proliferation
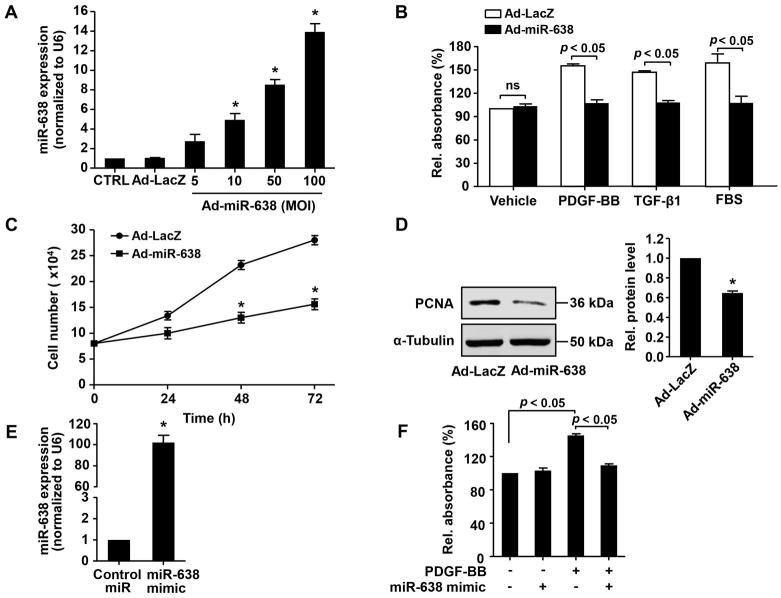
To substantiate the functional significance of miR-638 in ASM biology, we first performed a gain-of-function study by using adenovirus-mediated overexpression of miR-638 in human ASMCs. As shown in Figure 3A, Ad-miR-638 transduction markedly increased the expression levels of miR-638 in human ASMCs, as determined by qRT-PCR. Accordingly, both PDGF-BB and TGF-β-induced ASMC proliferation, as determined by WST-8 proliferation assay and cell counting (Figure 3B and C), were markedly inhibited in Ad-miR-638 transduced cells, while the airway smooth cell viability was barely affected. Moreover, miR-638 overexpression markedly inhibited PDGF-BB-induced expression of PCNA, a well-known marker of cell proliferation (Figure 3D). Likewise, transfection of ASMCs with miR-638 mimic led to a marked increase of miR-638 expression and a subsequent inhibition of PDGF-BB-induced ASMC proliferation (Figure 3E and F).
Figure 3.
miR-638 overexpression inhibits ASMC proliferation. A, human ASMCs were treated with vehicle control (CTRL), Ad-LacZ, or Ad-miR-638, at indicated MOI for 48 h and expression of miR-638 was then determined by qRT-PCR (n=4). *p < 0.05 compared with Ad-LacZ group. B and C, human ASMCs were transduced with either Ad-miR-638 or Ad-LacZ (100 MOI), 24 h after transduction, cells were starved for 48 h and stimulated with either vehicle, PDGF-BB (20 ng/mL), TGF-β1 (20 ng/mL), or 10% FBS. Cell proliferation was determined by WST-8 proliferation assay at 48 h in (B) and cell counting in (C) at indicated time points (n=6). *p < 0.05 compared with Ad-LacZ in PDGF-BB group at 24 h, 48 h, or 72 h. D, cultured human ASMCs were transduced with either Ad-LacZ or Ad-miR-638 (100 MOI). 48 h after transduction, expression of PCNA was determined by western blot (n=4). *p < 0.05 compared with Ad-LacZ group. E, human ASMCs were transfected with either miR-638 mimic or control miR (30 nM). 24 h after transfection, the expression of miR-638 was determined by qRT-PCR (n=4). *p < 0.05 compared with control miR group. F, human ASMCs were transfected with either miR-638 mimic or control miR (30 nM). 24 h after transfection, cells were starved for 48 h and then stimulated with or without PDGF-BB (20 ng/mL) for 48 h. Cell proliferation was determined by WST-8 proliferation assay (n=6).
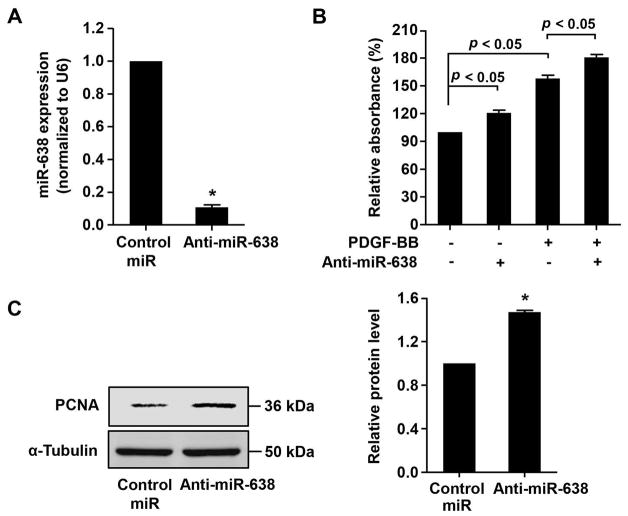
To further substantiate the role of miR-638 in ASMC proliferation, we performed a loss-of-function study. As shown in Figure 4A, transfection of human ASMCs with Anti-miR-638 led to a 90% inhibition of miR-638 expression, as compared with the transfection of control miR. Furthermore, we found that anti-miR-638 significantly increased human ASMC proliferation, under both baseline and PDGF-BB-stimulated conditions (Figure 4B). Together, these results suggest that miR-638 is an essential inhibitor for ASMC proliferation.
Figure 4.
miR-638 knockdown enhances ASMC proliferation. A, human ASMCs were transfected with either anti-miR-638 or control miR (100 nM). 72 h after transfection, miR-638 expression was determined by qRT-PCR (n=4). *p < 0.05 compared with control miR group. B, human ASMCs were transfected with either Anti-miR-638 or control miR (100 nM). 24 h after transfection, cells were starved for 48 h then stimulated with or without PDGF-BB (20 ng/mL) for 48 h, relative proliferation was determined by WST-8 proliferation assay (n=6). C, cultured human ASMCs were transfected with either anti-miR-638 or control miR (100 nM). 48 h after transduction, expression of PCNA was determined by western blot (n=4). *p < 0.05 compared with control miR group.
3.3 miR-638 is involved in ASMC migration
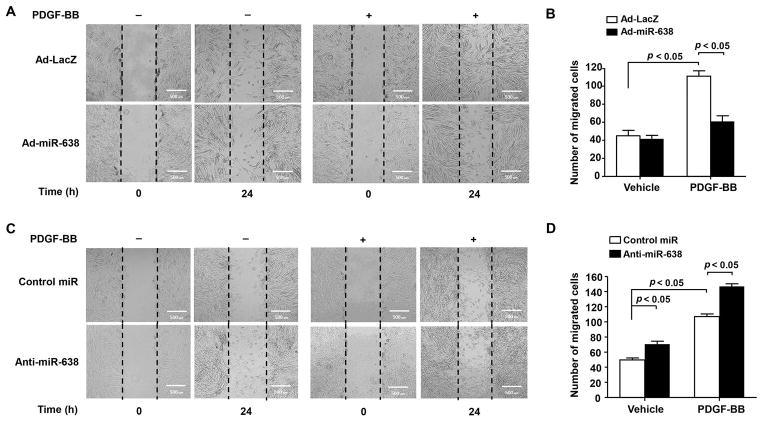
ASMC migration has been shown to play an important role in ASM remodeling (Aso et al., 2013) (Harada et al., 2015). To determine whether miR-638 plays a role in ASMC migration, both gain- and loss-of-function approaches were performed. As showed in Figure 5A and B, Ad-miR-638-mediated overexpression of miR-638 significantly attenuated both basal and PDGF-BB-induced ASMC migration, as determined by scratch wound assay. Likewise, Inhibition of miR-638 expression by its inhibitor markedly enhanced both basal and PDGF-BB-induced ASMC migration (Figure 5C and D). Together, these data demonstrated that miR-638 regulates not only ASMC proliferation, but also migration under both baseline and PDGF-BB-stimulated conditions.
Figure 5.
Overexpression of miR-638 inhibits ASMC migration. A, human ASMCs were seeded into 6-well plates and transduced with either Ad-miR-638 or Ad-LacZ (100 MOI). 24 h after transduction, cells were starved for 48 h, then scratched and stimulated with or without PDGF-BB (20 ng/mL), migration was monitored by cells within the wound area at 0 h and 24 h. Migrated cells were quantitated using ImageJ software program and the result was shown in (B). n=4. C, human ASMCs were seeded into 6-well plates and transfected with either anti-miR-638 or control miR (100 nM). 24 h after transfection, starvation was performed for 48 h, then cells were scratched and stimulated with or without PDGF-BB (20 ng/mL), migration was monitored by cells within the wound area at 0 h and 24 h. Migrated cells were quantitated using ImageJ software program as shown in (D). n=4.
3.4 Cyclin D1 is a direct target of miR-638 in human ASMCs
To understand the molecular mechanism(s) mediating miR-638-induced inhibition of ASMC proliferation, we searched the TargetScan database (Release 7.1: June 2016) and found that cyclin D1, a critical regulator for cell proliferation (Bonacci et al., 2006; Watanabe et al., 2009), is a potential target of miR-638, as shown in Figure 6A. To determine whether miR-638 directly binds to the 3′-UTR of cyclin D1 mRNA and impacts its expression, the 3′-UTR sequence of cyclin D1 mRNA containing the predicted miR-638 binding site was cloned into the pLightSwitch-3′-UTR RenSP-luciferase reporter vector. The reporter vector was then co-transfected with either miR-638 mimic or non-targeting control miR into HEK-293T cells. As shown in Figure 6B, the luciferase activity was inhibited by transfection of cells with miR-638 mimic, but not by the non-targeting control miR. Furthermore, transfection of anti-miR-638 increased the luciferase activity and mutation of miR-638 binding site abolished the miR-638-miediated inhibition on the luciferase activity (Figure 6B).
To further confirm whether cyclin D1 is a target gene of miR-638 in cultured ASMCs, both gain-of-function and loss-of-function approaches were performed. Consistent with previous reports (Bonacci et al., 2006), PDGF-BB stimulation significantly increased cyclin D1 expression in human ASMCs, which was substantially inhibited in Ad-miR-638-transduced cells, as determined by both qRT-PCR and western blot (Figure 6C and D). Conversely, transfection of ASMCs with anti-miR-638 significantly increased expression of cyclin D1 in ASMCs (Figure 6E and F). Together, these results suggest that miR-638 inhibits cell proliferation through targeted inhibition of cyclin D1 expression in ASMCs.
3.5 miR-638 inhibits cell cycle progression in ASMCs
Cyclin D1 is a crucial switcher of cell cycle for ASMCs (Watanabe et al., 2009). To determine whether miR-638 reduces cell proliferation through regulating cell cycle progression, we performed flow cytometry analysis in human ASMCs. As shown in Figure 7A and B, we found that transduction of Ad-miR-638 led to cell cycle arrest at G0/G1 phase and inhibition of cell cycle switch from G0/G1 phase to S phase, under both baseline and PDGF-BB-stimulated conditions.
Figure 7.
miR-638 inhibits ASMC cell cycle progression. A, human ASMCs were seeded into P-60 dishes at 40~60% confluence and transduced with either Ad-miR-638 or Ad-LacZ (100 MOI). 24 h after transduction, cells were starved for 48 h, then stimulated with or without PDGF-BB (20 ng/mL) for 48 h. Cells were fixed and stained with propidium iodide, cell cycle profiles were analyzed by flow cytometry. Data were analyzed as means ± standard deviation as shown in (B). n=4. *p < 0.05, compared with Ad-LacZ without PDGF-BB treatment. **p < 0.05, compared with Ad-LacZ with PDGF-BB treatment.
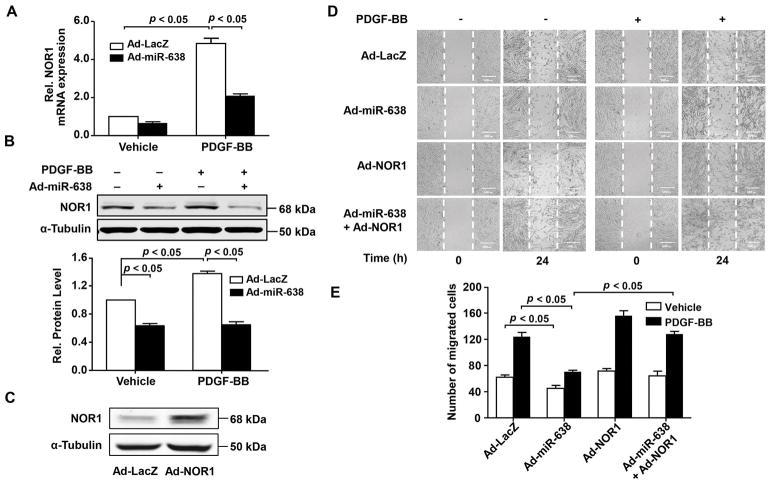
3.6 miR-638 inhibits ASMC migration through inhibiting NOR1 expression
Our previous data shows that orphan nuclear receptor NOR1 is one of the direct targets of miR-638 (Li et al., 2013). Several lines of evidence suggest that NOR1 is an essential regulator for migration in a wide range of cells (Chen et al., 2017b; You et al., 2009). Currently, little is known about the role of NOR1 in ASMC migration. As shown in Figure 8A and B, we found that PDGF-BB treatment markedly increased NOR1 expression in human ASMCs, which was inhibited by adenovirus-mediated miR-638 overexpression, as determined by both qRT-PCR and western blot, indicating that NOR1 is a potential target of miR-638 in ASMCs. To further determine the function of NOR1 in ASMC migration, we transduced human ASMCs with adenovirus bearing NOR1 cDNA (Ad-NOR1). 48 h after transduction of Ad-NOR1, NOR1 expression was increased significantly, as determined by western blot (Figure 8C). As shown in Figure 8D and E, transduction of Ad-NOR1 significantly increased ASMC migration, under both baseline and PDGF-BB-stimulated conditions. Importantly, miR-638-mediated inhibition of human ASMC migration was completely abolished in Ad-NOR1 transduced cells. Together, these results suggest that miR-638 inhibits ASMC migration, at least in part, through inhibiting NOR1 expression in ASMCs.
Figure 8.
NOR1 is critically implicated in ASMC migration. A, human ASMCs were transduced with either Ad-miR-638 or Ad-LacZ (100 MOI) for 24 h and then serum-deprived for 48 h. 3h after treated with PDGF-BB (20 ng/mL) or vehicle, NOR1 mRNA was measured by qRT-PCR (n=4). B, 6 h after treated with PDGF-BB (20 ng/mL) or vehicle, expression of NOR1 protein was detected by western blot (n=4). C, human ASMCs were infected with either Ad-NOR1 or Ad-LacZ (100 MOI) for 48 h, expression of NOR1 was detected by western blot (n=3). D, migration of human ASMCs was detected by scratch wound assay. Cells were seeded into 6-well plates and then transduced with adenovirus (100 MOI) as indicated. 24 h after transduction, cells were starved for 48 h, then scratched and stimulated with or without PDGF-BB (20 ng/mL). Migration was monitored by cell numbers within the wound area at 0 h and 24 h. Migrated cells were quantitated using ImageJ software program and quantitative results were shown in (E). n=4.
4. Discussion
Airway smooth muscle remodeling, including abnormal cell proliferation and migration, plays critical roles in the development of chronic respiratory diseases such as asthma. Under normal conditions, ASMCs remain in a quiescent and non-migratory state(Kudo et al., 2013; Makinde et al., 2007). Diverse stimuli, including growth factors, contractile agonists, inflammatory cytokines, and extracellular matrix proteins, have been shown to induce ASMC proliferation and migration, which contributes significantly to increased ASM hyperplasia associated with the development of severe asthma(Fixman et al., 2007). Recently, it is increasingly recognized that miRNAs play significant roles in regulating airway smooth muscle functions (Chiba and Misawa, 2010; Deshpande et al., 2015). Identification of novel miRNAs that are critically involved in ASMC remodeling is a critical step towards developing effective therapies for the treatment of asthma. In the present study, we identified miR-638 as a novel regulator in controlling airway smooth muscle cell proliferation and migration. We show that the expression of miR-638 is significantly downregulated in human ASMCs in response to multiple pathological stimuli, such as PDGF-BB, TGF-β1, and FBS. Overexpression of miR-638 significantly attenuates the ASMC proliferation and migration through targeting cyclin D1 and NOR1, which are critical regulators for ASMC remodeling. Furthermore, we found that the MEK1/2 pathway is responsible for the PDGF-BB-induced down-regulation of miR-638 expression in ASMCs, however, whether this pathway is involved in TGF-β1-and FBS-induced down-regulation of miR-638 expression remains to be further determined. In this regard, our study provides compelling evidence implicating miR-638 as a novel key regulator in human ASMC function.
miR-638 is expressed in human and non-human primates (Li et al., 2013; Ren et al., 2017). Recently, several published studies have focused on the function of miR-638 in cancer development. The expression of miR-638 has been found to be down-regulated in multiple types of cancers, including human gastric cancer, colorectal cancer, and acute leukemia (Lin et al., 2015b; Ren et al., 2017). Overexpression of miR-638 has been shown to inhibit cancer growth and metastasis, suggesting that it may function as a tumor suppressor (Lu et al., 2015; Shen et al., 2017). In addition, miR-638 has been recently implicated in the pathogenesis of lupus nephritis (Lu et al., 2012), and may function as a predictor of early virological response to interferon treatment in chronic hepatitis B patients (Kumar et al., 2015; Liu et al., 2010). A recent study has demonstrated that expression of miR-638 may regulate regional emphysema severity and gene expression networks associated with the oxidative stress response and aging in emphysematous lung tissue (Christenson et al., 2013). Although miR-638 is highly expressed in ASMCs, its role in ASMC function remains completely unknown. In the present study, we demonstrate that miR-638 is essential regulator for suppressing ASMC proliferation. Mechanistically, our study identified cyclin D1 as a direct target of miR-638 in human ASMCs. Cyclin D1 plays a central role in the regulation of cell proliferation and is required for cell cycle progression through the G1 phase (Bonacci et al., 2006). During the G1 phase, it is synthesized rapidly and accumulates in the nucleus, and then is degraded as the cell enters the S phase. Cyclin D1 binds cyclin dependent kinase 4 or 6 (CDK4/6) to form an active kinase that causes phosphorylation and inactivation of the retinoblastoma protein (Rb), which subsequently allows E2F transcription factors to promote the transcription of genes required for cell cycle progression (Garrido-Castro and Goel, 2017; Musgrove et al., 2011). Cyclin D1 has been the most widely studied cyclin in ASM biology. Cyclin D1 has been shown to be significantly increased in ASMCs in response to various mitogenic stimuli and inhibition of cyclin D1 can potently suppress ASMC proliferation, further highlighting the critical importance of cyclin D1 in ASMC proliferation. As cyclin D1 mutation and amplification are frequently observed in various tumors (Musgrove et al., 2011) (Palmero and Peters, 1996) and dysregulation of both cyclin D1 and miR-638 may contribute to tumorigenesis, identification of cyclin D1 as a direct target of miR-638 may help us understand the functional significance of miR-638 not only in ASM biology but also in tumorigenesis.
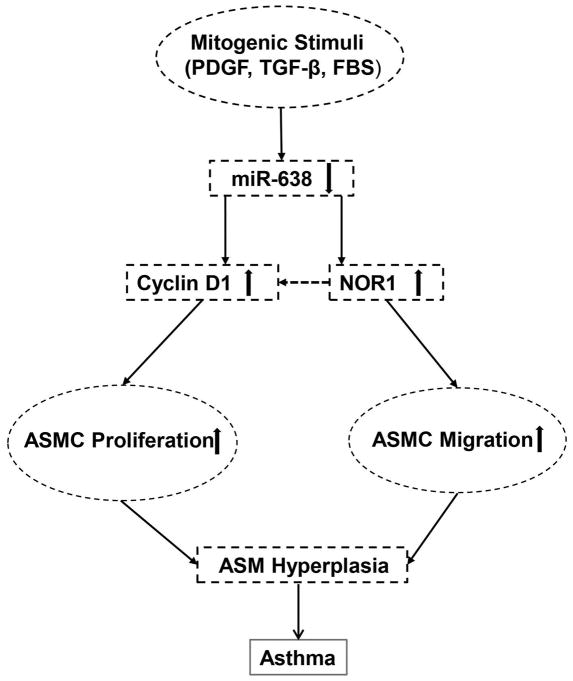
Orphan nuclear receptor NOR1 belongs to the nuclear receptor 4A (NR4A) family and the members of this family, including Nur77, Nurr1, and NOR1, participate in diverse biological functions through their genomic and non-genomic effects(Mohan et al., 2012). NOR1 expression is induced in various types of cells, by multiple stimuli, such as serum, PDGF-BB, ox-LDL, VEGF, and cytokines (Mohan et al., 2012; Nomiyama et al., 2006; Pei et al., 2005; You et al., 2009). Accumulating evidence suggests that NOR1 plays essential roles in cell proliferation and migration (Li et al., 2013; Wang et al., 2015). In Vascular SMCs, NOR1 is rapidly induced by mitogens including PDGF-BB and FBS (Nomiyama et al., 2006). Overexpression of NOR1 has been shown to exert pro-proliferative in vascular SMCs (Nomiyama et al., 2006). Knockdown of NOR1 expression with anti-sense oligonucleotides in cultured SMCs has been shown to reduce SMC proliferation and migration. In NOR1 knockout SMCs, PDGF-induced proliferation and migration is almost completely inhibited, which coincides with a strong reduction in cyclin D1 expression (Nomiyama et al., 2009). Since the expression and roles of NR4A family are context and tissue-specific (Rodriguez-Calvo et al., 2017), and the role of NOR1 in ASM biology has not been explored thus far, we attempted to investigate the expression and role of NOR1 in ASMC function. In the present study, we found that NOR1 is highly expressed in human ASMCs and its expression is markedly induced by PDGF-BB stimulation and inhibited by miR-638, which is consistent with our previous notion showing that miR-638 directly binds to the 3′-UTR of human NOR1 mRNA and down-regulates its expression in vascular SMCs (Li et al., 2013). Furthermore, we found that miR-638 only specifically downregulated the expression of NOR1, but had no effect on the expression of Nur77 and Nurr1 in human ASMCs (data not shown). Importantly, the inhibitory effects of miR-638 on ASMC migration is abolished by NOR1. Our results demonstrated that NOR1 is capable for promoting ASMC migration, further indicating that NOR1 is at least in part involved in the regulation of ASMC function by miR-638. Interestingly, in NOR1 knockout SMCs, the expression of cyclin D1 is strongly reduced. Indeed, sequence analysis of the cyclin D1 promoter identified a functional NGFI-B response element in human (−1061 bp) and in mouse (−2197 bp) (Nomiyama et al., 2006), which suggest that NOR1 may function as a transcriptional activator for cyclin D1 expression. Based on these observations, we attempt to speculate that miR-638 may regulate cyclin D1 expression through both directly binding the cyclin D1 3′-UTR and indirectly regulating NOR1 expression, thus exert potent anti-proliferative and migratory effects in human ASMCs (Figure 9). miR-638 exists only in human and non-human primates and it may have species-specific effect. Indeed, we found that the targeting sequences of miR-638 in the 3′-UTR of human cyclin D1 and NOR1 mRNAs are not conserved among different species, thus further testing its functional significance by using small animal models of asthma is not possible. Nevertheless, the clinical relevance of miR-638 in the pathogenesis of asthma warrants further investigation.
Figure 9.
Proposed mechanism underlying miR-638-induced inhibition of airway smooth muscle cell proliferation and migration in asthmas. Pathological stimuli, such as PDGF-BB and TGF-β, downregulate the expression of miR-638, which in turn leads to an increased expression of cyclin D1 and orphan nuclear receptor NOR1 and a subsequent AMS hyperplasia in asthma.
In summary, the present study identified miR-638 as a novel regulator in human ASMCs by directly targeting both cyclin D1 and NOR1 expression. miR-638 expression was substantially down-regulated in proliferative human ASMCs and restoration of its expression markedly inhibited both ASMC proliferation and migration in response to PDGF-BB stimulation. In this regard, our study provides significant novel insight into the molecular mechanisms associated with ASMC remodeling, and suggests that targeted overexpression of miR-638 in ASMCs may present a potential novel therapeutic approach for inhibiting ASM hyperplasia associated with asthma pathogenesis.
TABLE 1.
Primers used in this study
| Name | Sequence (5′-3′) |
|---|---|
| Primers used in quantitative RT-PCR | |
| 18S rRNA-F | GTAACCCGTTGAACCCCATT |
| 18S rRNA-R | CCATCCAATCGGTAGTAGCG |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| hsa-miR-638 | AGGGAUCGCGGGCGGGUGGCGGCCU |
| cyclin D1-F | CCTCGGTGTCCTACTTCAAATG |
| cyclin D1-R | CACTTCTGTTCCTCGCAGAC |
| NOR1-F | ATAGTCTGAAAGGGAGGAGAGG |
| NOR1-R | ATCATGCAGATTGGAGGAGAAG |
|
| |
| Primers used in plasmid construction | |
|
| |
| Cyclin D1-3′-UTR-F | GAGATCTAGATTGCTGCTATTGGAGGAT |
| Cyclin D1-3′-UTR-R | GAGACTCGAGCCATAGCTACACGCAAAG |
| Cyclin D1-3′UTR-MUT-1F | GACACGCGGGCGCAGTCCCACACAGGCT |
| Cyclin D1-3′UTR-MUT-1R | AGCCTGTGTGGGACTGCGCCCGCGTGTC |
| Cyclin D1-3′UTR-MUT-2F | CGCGGGCGCAGTGGCACACAGGCTGG |
| Cyclin D1-3′UTR-MUT-2R | CCAGCCTGTGTGCCACTGCGCCCGCG |
Acknowledgments
This work was supported by the U.S. National Institutes of Health (R01HL103869 and R01GM123047), the AHA established investigator award (16EIA27710023), and grants from the Chinese Natural Science Foundation No. 81370418 to JS.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Aso H, Ito S, Mori A, Suganuma N, Morioka M, Takahara N, Kondo M, Hasegawa Y. Differential regulation of airway smooth muscle cell migration by E-prostanoid receptor subtypes. American journal of respiratory cell and molecular biology. 2013;48(3):322–329. doi: 10.1165/rcmb.2012-0158OC. [DOI] [PubMed] [Google Scholar]
- Bonacci JV, Schuliga M, Harris T, Stewart AG. Collagen impairs glucocorticoid actions in airway smooth muscle through integrin signalling. British journal of pharmacology. 2006;149(4):365–373. doi: 10.1038/sj.bjp.0706881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Shi J, Zhang W, Huang L, Lin X, Lv Z, Zhang W, Liang R, Jiang S. MiR-23b controls TGF-beta1 induced airway smooth muscle cell proliferation via direct targeting of Smad3. Pulm Pharmacol Ther. 2017a;42:33–42. doi: 10.1016/j.pupt.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Chen S, Zheng P, Wang W, Yi M, Chen P, Cai J, Li J, Peng Q, Ban Y, Zhou Y, Zeng Z, Li X, Xiong W, Li G, Xiang B. Abberent expression of NOR1 protein in tumor associated macrophages contributes to the development of DEN-induced hepatocellular carcinoma. Journal of cellular physiology. 2017b doi: 10.1002/jcp.26349. [DOI] [PubMed] [Google Scholar]
- Cheng W, Yan K, Xie LY, Chen F, Yu HC, Huang YX, Dang CX. MiR-143-3p controls TGF-beta1-induced cell proliferation and extracellular matrix production in airway smooth muscle via negative regulation of the nuclear factor of activated T cells 1. Mol Immunol. 2016;78:133–139. doi: 10.1016/j.molimm.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Misawa M. MicroRNAs and their therapeutic potential for human diseases: MiR-133a and bronchial smooth muscle hyperresponsiveness in asthma. Journal of pharmacological sciences. 2010;114(3):264–268. doi: 10.1254/jphs.10r10fm. [DOI] [PubMed] [Google Scholar]
- Christenson SA, Brandsma CA, Campbell JD, Knight DA, Pechkovsky DV, Hogg JC, Timens W, Postma DS, Lenburg M, Spira A. miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome medicine. 2013;5(12):114. doi: 10.1186/gm519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande DA, Dileepan M, Walseth TF, Subramanian S, Kannan MS. MicroRNA Regulation of Airway Inflammation and Airway Smooth Muscle Function: Relevance to Asthma. Drug Dev Res. 2015;76(6):286–295. doi: 10.1002/ddr.21267. [DOI] [PubMed] [Google Scholar]
- Fixman ED, Stewart A, Martin JG. Basic mechanisms of development of airway structural changes in asthma. The European respiratory journal. 2007;29(2):379–389. doi: 10.1183/09031936.00053506. [DOI] [PubMed] [Google Scholar]
- Garrido-Castro AC, Goel S. CDK4/6 Inhibition in Breast Cancer: Mechanisms of Response and Treatment Failure. Current breast cancer reports. 2017;9(1):26–33. doi: 10.1007/s12609-017-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosens R, Grainge C. Bronchoconstriction and airway biology: potential impact and therapeutic opportunities. Chest. 2015;147(3):798–803. doi: 10.1378/chest.14-1142. [DOI] [PubMed] [Google Scholar]
- Harada T, Yamasaki A, Chikumi H, Hashimoto K, Okazaki R, Takata M, Fukushima T, Watanabe M, Kurai J, Halayko AJ, Shimizu E. gamma-Tocotrienol reduces human airway smooth muscle cell proliferation and migration. Pulm Pharmacol Ther. 2015;32:45–52. doi: 10.1016/j.pupt.2015.04.003. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He M, Lin Y, Tang Y, Liu Y, Zhou W, Li C, Sun G, Guo M. miR-638 suppresses DNA damage repair by targeting SMC1A expression in terminally differentiated cells. Aging. 2016;8(7):1442–1456. doi: 10.18632/aging.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota JA, Ask K, Farkas L, Smith JA, Ellis R, Rodriguez-Lecompte JC, Kolb M, Inman MD. In vivo role of platelet-derived growth factor-BB in airway smooth muscle proliferation in mouse lung. American journal of respiratory cell and molecular biology. 2011;45(3):566–572. doi: 10.1165/rcmb.2010-0277OC. [DOI] [PubMed] [Google Scholar]
- Hu R, Pan W, Fedulov AV, Jester W, Jones MR, Weiss ST, Panettieri RA, Jr, Tantisira K, Lu Q. MicroRNA-10a controls airway smooth muscle cell proliferation via direct targeting of the PI3 kinase pathway. FASEB J. 2014;28(5):2347–2357. doi: 10.1096/fj.13-247247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR. Role of human airway smooth muscle in altered extracellular matrix production in asthma. Clinical and experimental pharmacology & physiology. 2001;28(3):233–236. doi: 10.1046/j.1440-1681.2001.03426.x. [DOI] [PubMed] [Google Scholar]
- Kudo M, Ishigatsubo Y, Aoki I. Pathology of asthma. Frontiers in microbiology. 2013;4:263. doi: 10.3389/fmicb.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Sharma Y, Bandi S, Gupta S. Endogenous antiviral microRNAs determine permissiveness for hepatitis B virus replication in cultured human fetal and adult hepatocytes. Journal of medical virology. 2015;87(7):1168–1183. doi: 10.1002/jmv.24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Liu Y, Yi B, Wang G, You X, Zhao X, Summer R, Qin Y, Sun J. MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc Res. 2013;99(1):185–193. doi: 10.1093/cvr/cvt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Lin WN, Hou WC, Hsiao LD, Yang CM. Endothelin-1 induces VCAM-1 expression-mediated inflammation via receptor tyrosine kinases and Elk/p300 in human tracheal smooth muscle cells. American journal of physiology Lung cellular and molecular physiology. 2015a;309(3):L211–225. doi: 10.1152/ajplung.00232.2014. [DOI] [PubMed] [Google Scholar]
- Lin Y, Li D, Liang Q, Liu S, Zuo X, Li L, Sun X, Li W, Guo M, Huang Z. miR-638 regulates differentiation and proliferation in leukemic cells by targeting cyclin-dependent kinase 2. J Biol Chem. 2015b;290(3):1818–1828. doi: 10.1074/jbc.M114.599191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Laidlaw TM, Feng C, Xing W, Shen S, Milne GL, Boyce JA. Prostaglandin E2 deficiency uncovers a dominant role for thromboxane A2 in house dust mite-induced allergic pulmonary inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):12692–12697. doi: 10.1073/pnas.1207816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398(1):57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yang K, Shi H, Xu J, Zhang D, Wu Y, Zhou S, Sun X. MiR-21 modulates human airway smooth muscle cell proliferation and migration in asthma through regulation of PTEN expression. Exp Lung Res. 2015;41(10):535–545. doi: 10.3109/01902148.2015.1090501. [DOI] [PubMed] [Google Scholar]
- Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow KM, Wang G, Li PK, Szeto CC. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology. 2012;17(4):346–351. doi: 10.1111/j.1440-1797.2012.01573.x. [DOI] [PubMed] [Google Scholar]
- Lu X, Chen L, Chen Y, Shao Q, Qin W. Bafilomycin A1 inhibits the growth and metastatic potential of the BEL-7402 liver cancer and HO-8910 ovarian cancer cell lines and induces alterations in their microRNA expression. Experimental and therapeutic medicine. 2015;10(5):1829–1834. doi: 10.3892/etm.2015.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Pan X, Fan P, He Y, Gu J, Wang W, Zhang T, Li Z, Luo X. Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Molecular cancer. 2014;13:118. doi: 10.1186/1476-4598-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinde T, Murphy RF, Agrawal DK. The regulatory role of TGF-beta in airway remodeling in asthma. Immunol Cell Biol. 2007;85(5):348–356. doi: 10.1038/sj.icb.7100044. [DOI] [PubMed] [Google Scholar]
- Mohan HM, Aherne CM, Rogers AC, Baird AW, Winter DC, Murphy EP. Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(12):3223–3228. doi: 10.1158/1078-0432.CCR-11-2953. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nature reviews Cancer. 2011;11(8):558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- Nomiyama T, Nakamachi T, Gizard F, Heywood EB, Jones KL, Ohkura N, Kawamori R, Conneely OM, Bruemmer D. The NR4A orphan nuclear receptor NOR1 is induced by platelet-derived growth factor and mediates vascular smooth muscle cell proliferation. J Biol Chem. 2006;281(44):33467–33476. doi: 10.1074/jbc.M603436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama T, Zhao Y, Gizard F, Findeisen HM, Heywood EB, Jones KL, Conneely OM, Bruemmer D. Deficiency of the NR4A neuron-derived orphan receptor-1 attenuates neointima formation after vascular injury. Circulation. 2009;119(4):577–586. doi: 10.1161/CIRCULATIONAHA.108.822056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero I, Peters G. Perturbation of cell cycle regulators in human cancer. Cancer surveys. 1996;27:351–367. [PubMed] [Google Scholar]
- Parasramka MA, Ali S, Banerjee S, Deryavoush T, Sarkar FH, Gupta S. Garcinol sensitizes human pancreatic adenocarcinoma cells to gemcitabine in association with microRNA signatures. Molecular nutrition & food research. 2013;57(2):235–248. doi: 10.1002/mnfr.201200297. [DOI] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280(32):29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- Ren Y, Chen Y, Liang X, Lu Y, Pan W, Yang M. MiRNA-638 promotes autophagy and malignant phenotypes of cancer cells via directly suppressing DACT3. Cancer letters. 2017;390:126–136. doi: 10.1016/j.canlet.2017.01.009. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Calvo R, Tajes M, Vazquez-Carrera M. The NR4A subfamily of nuclear receptors: potential new therapeutic targets for the treatment of inflammatory diseases. Expert opinion on therapeutic targets. 2017;21(3):291–304. doi: 10.1080/14728222.2017.1279146. [DOI] [PubMed] [Google Scholar]
- Shen Y, Chen H, Gao L, Zhang W, He J, Yang X, Qin L, Xue X, Guo Z. MiR-638 acts as a tumor suppressor gene in gastric cancer. Oncotarget. 2017;8(64):108170–108180. doi: 10.18632/oncotarget.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Peng J, Fu Y, An S, Rezaei K, Tabbara S, Teal CB, Man YG, Brem RF, Fu SW. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast cancer research : BCR. 2014;16(5):435. doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TR, Moore SM, Lawson MF, Panettieri RA, Jr, Chilvers ER. Platelet-derived growth factor-BB and thrombin activate phosphoinositide 3-kinase and protein kinase B: role in mediating airway smooth muscle proliferation. Molecular pharmacology. 1998;54(6):1007–1015. doi: 10.1124/mol.54.6.1007. [DOI] [PubMed] [Google Scholar]
- Wang CG, Lei W, Li C, Zeng DX, Huang JA. Neuron-derived orphan receptor 1 promoted human pulmonary artery smooth muscle cells proliferation. Exp Lung Res. 2015;41(4):208–215. doi: 10.3109/01902148.2014.993776. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Yamasaki A, Hashimoto K, Shigeoka Y, Chikumi H, Hasegawa Y, Sumikawa T, Takata M, Okazaki R, Watanabe M, Yokogawa T, Yamamura M, Hayabuchi T, Gerthoffer WT, Halayko AJ, Shimizu E. Expression of functional leukotriene B4 receptors on human airway smooth muscle cells. The Journal of allergy and clinical immunology. 2009;124(1):59–65. e51–53. doi: 10.1016/j.jaci.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You B, Jiang YY, Chen S, Yan G, Sun J. The orphan nuclear receptor Nur77 suppresses endothelial cell activation through induction of IkappaBalpha expression. Circulation research. 2009;104(6):742–749. doi: 10.1161/CIRCRESAHA.108.192286. [DOI] [PubMed] [Google Scholar]
- Zhao LY, Yao Y, Han J, Yang J, Wang XF, Tong DD, Song TS, Huang C, Shao Y. miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Digestive diseases and sciences. 2014;59(8):1743–1753. doi: 10.1007/s10620-014-3087-5. [DOI] [PubMed] [Google Scholar]