Abstract
Chaperones TAPBPR and tapasin associate with class-I major histocompatibility complexes (MHC-I) to promote optimization (editing) of peptide cargo. Here, we use solution NMR to investigate the mechanism of peptide exchange. We identify TAPBPR-induced conformational changes on conserved MHC-I molecular surfaces, consistent with our independently determined X-ray structure of the complex. Dynamics present in the empty MHC-I are stabilized by TAPBPR, and become progressively dampened with increasing peptide occupancy. Incoming peptides are recognized according to the global stability of the final pMHC-I product, and anneal in a native-like conformation to be edited by TAPBPR. Our results demonstrate an inverse relationship between MHC-I peptide occupancy and TAPBPR binding affinity, where the lifetime and structural features of transiently bound peptides controls the regulation of a conformational switch, located near the TAPBPR binding site, which triggers TAPBPR release. These results suggest a similar mechanism for the function of tapasin in the peptide-loading complex.
Graphical abstract

INTRODUCTION
A protective immune response against endogenous antigens generated from infectious agents or tumors is elicited through interactions between αβ T cell receptors (TCRs) on CD8+ cytotoxic T cells and peptide-loaded major histocompatibility complex class I (pMHC-I) molecules displayed on the surface of antigen-presenting cells 1. Properly conformed pMHC-I is assembled by an intracellular pathway in which loading of the MHC-I nascent chain with high-affinity peptides dictates complex stability, cell surface lifetime, and immunogenicity 2. Peptide loading and editing is orchestrated by molecular chaperones tapasin and TAPBPR (TAP-binding protein related), which function as peptide exchange catalysts that influence the repertoire of MHC-I displayed antigens at the cell surface 3–7. These chaperones have been linked to human disease where altered expression results in deregulation of the peptide presentation pathway and dampening of T cell-mediated immune responses in the context of cancer, infections, and autoimmune diseases 8–12. A number of studies have gleaned key insights into the peptide editing functions of tapasin (a key component of the peptide-loading complex) and TAPBPR 13–15. While sharing a common function as MHC-I peptide editors, recent data suggest tapasin and TAPBPR perform discrete roles in the cell 15. This is supported by the finding that TAPBPR is unable to compensate for tapasin in situ 16,17, and elicits distinct effects on the repertoire of displayed peptides 5, possibly due to differences in MHC-I allelic dependencies between the two chaperones 13. Furthermore, while tapasin is associated with the transporter associated with antigen processing (TAP) in the endoplasmic reticulum (ER), TAPBPR does not interact with either TAP or ERp57 16 and lacks an ER retention motif, thus its localization is not restricted to the ER. As a result, TAPBPR is also found in the Golgi, and can chaperone MHC-I molecules farther along the trafficking pathway 16. Recently, TAPBPR has been suggested to perform an additional quality control in the peptide loading pathway by assisting in MHC-I re-glycosylation and consequent recycling of improperly conformed molecules to the ER 18. Despite these important insights, the molecular basis of the peptide-editing function of TAPBPR (and tapasin) remains incompletely characterized.
Recently, two X-ray structures of peptide-deficient complexes between TAPBPR and the murine MHC-I molecules H2-Dd and H2-Db were determined at resolutions of 3.4 Å and 3.3 Å, respectively 19,20. These studies propose a direct role for a ~3 Å widening of the MHC-I groove at the conserved α2-1 helix, induced by TAPBPR, in promoting the release of non-optimal peptides. Complementary biophysical data and molecular dynamics (MD) simulations have suggested a conformational switch role for the α2-1 helix “latch” during peptide exchange, both with and without chaperones 13,21,22. Furthermore, a 5.8 Å cryo-electron microscopy (cryoEM) model of the tapasin/MHC-I assembly 23 showed a consistent chaperone/MHC-I recognition mechanism. However, important mechanistic details concerning recognition of incoming peptides by the chaperoned groove are difficult to visualize crystallographically, or by cryoEM. Recent advances in solution nuclear magnetic resonance (NMR) spectroscopy have enabled the characterization of high molecular weight protein assemblies of considerable size and dynamic complexity 24. Here, we utilize these methods to explore the interaction of TAPBPR with MHC-I molecules of varying peptide occupancies, and provide new insights towards understanding how TAPBPR “edits” the peptide repertoire. Our results suggest a peptide exchange mechanism in which a transient pMHC-I/TAPBPR intermediate governs the selection of high-affinity peptides. Based on a range of biophysical data, we propose a model for chaperone release through negative allosteric coupling between non-overlapping chaperone and peptide binding sites along both ends of the MHC-I groove. Insights derived from our data provide a molecular blueprint for engineering novel molecular chaperones with desired specificities to promote peptide exchange both in vitro and in vivo.
RESULTS
Dynamics of pMHC-I in solution
To examine dynamic MHC-I regions and their putative role in peptide exchange13,21, we used solution NMR to characterize flexibility in a 45 kDa MHC-I molecule comprised of a murine H2-Dd heavy chain refolded with human light chain (hβ2m)25 and a high-affinity peptide (P18-I10, RGPGRAFVTI) derived from the HIV gp160 envelope protein 6. We first recorded 15N TROSY R1 and R2 relaxation rates, in conjunction with 15N-{1H} NOE values 26. We find several regions of increased mobility on H2-Dd, in particular the β1-β2 loop, β3-β4 loop, α3 domain and C-terminus, which show decreased 15N-{1H} NOE, R2 values and increased R1 values (Fig. 1A). We additionally acquired 1H-13C multiple-quantum Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments, in which the resonances of methyl groups undergoing conformational exchange in the μs-ms timescale exhibit changes in their effective relaxation rate (R2,eff) as a function of the frequency of the refocusing pulse (νCPMG) 27. In total, conformational exchange is observed for 14 AILV methyl groups on H2-Dd and 8 on hβ2m (Fig. 1B, C). For H2-Dd, sites undergoing dynamics are found on the pleated β-sheet of the groove, at the interface with hβ2m, the C-terminal end of the α2-1 helix, and within the α3 domain (Fig. 1B, D). For hβ2m, dynamic sites are localized at the interface with H2-Dd, suggesting the presence of restricted inter-domain motions in the pMHC-I (Fig. 1C, D). H2-Dd groove sites undergoing conformational exchange are localized rather than spanning the entire groove, suggesting sampling of alternative local conformations, rather than exchange with a peptide-free form (Fig. 1D, E). Notably, we find that MHC-I regions of increased flexibility correlate with residues required for TAPBPR association (Supplementary Fig. 1), supporting a direct role for dynamics in driving recognition of peptide-editing chaperones 13,21.
Figure 1. Heavy and light chain dynamics of free pMHC-I.
(A) Backbone dynamics using 15N probes revealed by measurement of 15N-{1H} NOE, R2 (s−1) and R1 (s−1) values recorded at 25°C at a 1H field of 600 MHz on H2-Dd-labeled P18-I10/H2-Dd/hβ2m. A diagram of the H2-Dd secondary structure is provided for reference. Gray boxes highlight dynamic regions of interest. (B) and (C) Representative relaxation dispersion curves highlighting methyl groups undergoing chemical exchange (Rex > 1 s−1) in 1H-13C multiple-quantum CPMG experiments recorded at 25°C and a 1H field of 600 MHz. NMR resonances with measured Rex > 1 s−1 are H2-Dd residues V12, V28, L95, V103, L110, L126, A135, A136, A150, L180, L215, L230, L270 and L272, as well as hβ2m residues I1, V27, L39, V49, L54, L64, V82 and V85. (D) Residues with NMR resonances exhibiting dynamics are indicated on the structure of P18-I10/H2-Dd/β2m (PDB ID 3ECB) with select residues from (A, B, C) labeled, except for C-terminal residues K276/E277, which are not present in the X-ray structure. (E) The location of all assigned AILV methyl probes is mapped on the structure of P18-I10/H2-Dd/β2m (PDB 3ECB).
TAPBPR recognition of a peptide-loaded MHC-I molecule
To evaluate the propensity for TAPBPR to bind H2-Dd molecules with distinct peptide occupancies, we used size exclusion chromatography (SEC) 6, to find that TAPBPR forms a high-affinity complex with either peptide-deficient H2-Dd (Supplementary Fig. 2), partially loaded RGPGC/H2-Dd S73C or fully-loaded P18-I10/H2-Dd (Supplementary Fig. 3A–C). Subsequent characterization by NMR reveals a tight complex between pMHC-I (Supplementary Fig. 4) and TAPBPR, in which chemical exchange between the free and bound states is slow on the chemical shift timescale. We observe two types of effects on the H2-Dd and hβ2m backbone amide and AILV methyl resonances upon TAPBPR binding: i) chemical shift deviations (CSDs), indicating a change in the local magnetic environment, and ii) conformational exchange-induced line broadening leading to reduced peak intensity ratios (I/I0), suggesting changes in μs-ms timescale dynamics (Supplementary Fig. 5, Supplementary Fig. 6 and Supplementary Fig. 7). Mapping effects onto the pMHC-I structure reveals affected H2-Dd sites on the α1 helix, α2 helix, the pleated β-sheet on the floor of the groove and on the α3 domain at the CD8 binding site (Supplementary Fig. 5A, B and Supplementary Fig. 6A, B and Supplementary Fig. 7A, C). For hβ2m, we identify a continuous molecular surface located at the interface with the H2-Dd α3 domain (Supplementary Fig. 5C, D, Supplementary Fig. 6C, D and Supplementary Fig. 7B, D).
To quantitatively characterize binding under conditions where peptide dissociation from the complex is minimal, we titrated TAPBPR into a 100 μM pMHC-I sample (labeled at the heavy chain) and performed a rigorous NMR line shape analysis for the resonances of nine methyl probes spanning both TAPBPR binding sites (Fig. 2A–D). The two regions are simultaneously and cooperatively engaged by TAPBPR with globally fitted Kd and koff values of 32 μM and 2.9 s−1, respectively (Supplementary Fig. 5E, F and Supplementary Fig. 6E, F, Supplementary Fig. 10). Notwithstanding our observation of a substantial interaction between TAPBPR and H2-Dd loaded with a high-affinity peptide, recognition of peptide-loaded molecules shows a strong MHC allelic dependency. No TAPBPR binding could be detected in experiments using three additional peptide-loaded class I HLA (human leukocyte antigen) molecules, either by SEC (HLA-B*15:01, HLA-B*27:02) or by NMR (HLA-A01:01), suggesting that if there is such an interaction it must be extremely transient (koff ≫ 3 s−1). (Supplementary Fig. 8, Supplementary Fig. 9).
Figure 2. NMR characterization of the 87 kDa pMHC-I/TAPBPR complex.
Representative selection from 2D 1H-13C HMQC spectra of P18-I10/H2-Dd/hβ2m 13C AILV methyl labeled at (A) H2-Dd or (B) hβ2m for unbound (red) and TAPBPR-bound (blue) states recorded at 25°C at a 1H field of 800 MHz. (C) Residues with affected methyl resonances from (A) and (B) are mapped on the X-ray structure (PDB ID 3ECB), except for I92 of hβ2m which is located on the surface opposing H2-Dd. (D) NMR line shape analysis of I124 and I142, performed in TITAN (Online Methods), upon titration of TAPBPR on H2-Dd labeled pMHC-I. See Supplementary Fig. 8A, B. (E) and (F) effects of TAPBPR binding to pMHC-I from solution NMR (using both amide and methyl probes) mapped on the X-ray structure of H2-Dd S73C/β2m/TAPBPR complex (PDB ID 5WER). MHC-I: (surface representation, H2-Dd green; β2m cyan); TAPBPR (cartoon representation, gray). The N-terminal domain (TN) and C-terminal domain (TC) of TAPBPR are noted. Residues with resonances affected in NMR experiments are shown in salmon (H2-Dd) and orange (hβ2m). (G) Comparison of MHC-I/TAPBPR contacts within 4 Å in the X-ray structure focusing on the H2-Dd α2-1 helix, adjacent to the F-pocket. H2-Dd residues that are in direct contact in the X-ray structure and affected in NMR experiments are colored salmon.
Finally, whether N-linked glycosylation of conserved Asn residues on the MHC-I modulates chaperone association (outside of a protein localization to the PLC) remains unclear 18. Towards further exploration of a potential structural basis for such an interaction we modeled the Glc1Man9GlcNAc2 glycan which is recognized by calreticulin 28, using the putative N-glycan acceptor sites 29 on H2-Dd or H2-Db in the two X-ray MHC-I/TAPBPR structures. Although we find no clear overlap between the glycans and TAPBPR by structural modeling (Supplementary Fig. 11), this important aspect of MHC-I recognition by chaperones remains to be addressed in future experiments.
Comparison of solution NMR mapping with X-ray structures
While NMR experiments were underway, the X-ray structures of the H2-Dd S73C/β2m/TAPBPR 20 and H2-Db/β2m/TAPBPR 19 complexes were solved. In both our NMR experiments and in the X-ray structures, the H2-Dd α2-1 helix and the CD8 recognition loop of the α3 domain are in direct contact with the TAPBPR TN (N/IgV) and TC (IgC) domains, respectively (Fig. 2E–G). The TAPBPR TC domain is nestled between the H2-Dd α3 domain and β2m, an unanticipated result, which is supported by our NMR experiments (Fig. 2E–F). Detailed inspection of MHC-I/TAPBPR contacts within 4 Å in the X-ray structure shows excellent agreement with MHC-I residues undergoing significant chemical shift perturbations in solution (Fig. 2G). Thus, independent characterization of the MHC-I/TAPBPR complex crystallographically and by NMR reveals a multifaceted interaction of TAPBPR with both peptide-deficient as well as peptide-loaded MHC molecules.
Modulation of dynamics in MHC-I/TAPBPR complexes
Electron density corresponding to peptide atoms is completely missing from the available MHC-I/chaperone structures 19,20,23. To elucidate how peptides influence groove dynamics, we compared the spectra of either peptide-deficient, partially loaded or fully loaded MHC-I/TAPBPR (Fig. 3A). While in the fully loaded complex most methyls in the H2-Dd groove give rise to well-resolved NMR peaks, in the other complexes several resonances are broadened due to conformational exchange at the intermediate (μs-ms) timescale (Fig. 3A, B, left). Notably, when the groove is partially occupied by a disulfide-conjugated pentamer peptide, resonances corresponding to residues in the A-, B-, C- and D-pockets become consistently less broadened, relative to the unoccupied state, while resonances corresponding to residues in the F-pocket remain exchange-broadened (Fig. 3A, B, middle). This ‘rescuing’ of attenuated NMR peaks becomes even more pronounced in a complex that is loaded with the full-length P18-I10 peptide, specifically for resonances corresponding to F-pocket residues (Fig. 3A, B, right). Pairwise comparisons of methyl chemical shift deviations, computed relative to the free pMHC-I reference spectrum, show close agreement between MHC-I/TAPBPR complexes containing different peptides (Supplementary Fig. 12). A difference can be observed at residue I124 (Supplementary Fig. 12, right), which we attribute to modulation of dynamics in the α2-1 helical segment when the adjacent F-pocket is unoccupied (RGPGC) or occupied (P18-I10) by peptide (Fig. 3B). Analysis of Cα distances between representative residues in MD simulations (Online Methods) reveals that the peptide-deficient groove undergoes a larger range of motions, relative to the fully loaded MHC-I molecule and the chaperoned peptide-deficient groove, an effect which becomes more pronounced at the C-, E- and F-pockets, near the α2-1 helix (Fig. 3C). Taken together, our results suggest that both TAPBPR and incoming peptides dampen dynamics in the MHC-I groove, culminating in a conformational change within the α2-1 helix.
Figure 3. Modulation of dynamics in MHC-I/TAPBPR complexes.
(A) Representative selection of 2D 1H-13C HMQC spectra recorded on: peptide-deficient H2-Dd/hβ2m/TAPBPR (left), RGPGC/H2-Dd S73C/hβ2m/TAPBPR (middle), and P18-I10/H2-Dd/hβ2m/TAPBPR (right). Dotted salmon boxes highlight examples of NMR peaks for the comparison shown in (B). An overlay of reference spectra of unbound RGPGC/H2-Dd S73C/hβ2m (middle) and P18-I10/H2-Dd/hβ2m (right) are shown in gray for comparison with the TAPBPR complex. See related Supplementary Fig. 12. (B) Comparison of residues with attenuated NMR resonances mapped on the X-ray structure of the MHC-I/TAPBPR complex (PDB ID 5WER), shown for three different groove occupancies with modeled peptide conformations. H2-Dd green; peptide magenta; TAPBPR gray. (C) All-atom molecular dynamics simulations (Online Methods) of peptide-loaded MHC-I, peptide-deficient MHC-I and peptide-deficient MHC-I/TAPBPR complexes. The diagram shows color-coded variance (Å2) of Cα-Cα atom distances computed between pairs of residues spanning the H2-Dd groove over 50 ns blocks of the simulation. Larger variance (red/white) indicates increased dynamics. The residue pairs are noted, which are highlighted on the P18-I10/H2-Dd X-ray structure with noted peptide binding pockets (right, PDB ID 3ECB).
NMR reveals a peptide-bound intermediate during exchange
To probe conformational intermediates along the TAPBPR-mediated peptide exchange pathway, we titrated P18-I10 peptide into a peptide-deficient MHC-I/TAPBPR complex. Initially, specific NMR resonances of residues in the H2-Dd groove are attenuated in the absence of peptide, while those of residues in the α3 domain remain observable (Supplementary Fig. 13A, blue, B–D). Upon the addition of sub-stoichiometric amounts of P18-I10, several exchange-broadened NMR resonances begin to appear in either similar (V76) or unique (I124, I142, A153, A158) chemical shift positions, relative to the reference spectrum of the native P18-I10/H2-Dd/hβ2m molecule (Supplementary Fig. 13A, purple). This suggests the formation of a peptide-bound intermediate state. As peptide is incrementally added to achieve a 1:1 molar ratio, the new resonances observed for several methyls (I124, I142, V231) are shifted and attenuated, while a set of resonances corresponding to the chemical shifts of the native pMHC-I reference can be simultaneously observed (Supplementary Fig. 13A, orange and green). The “transient” methyl resonances likely arise from an averaging of multiple conformations with distinct chemical shift environments. Their remission into the intermediate exchange NMR regime with increasing peptide concentration and consequent broadening is attributed to a phenomenon previously described as “transient narrowing” 30. A complementary LC-MS analysis unambiguously confirms that the “transient” resonances correlate with the formation of an intermediate state of the MHC-I/TAPBPR complex with peptide captured in the MHC-I groove. Finally, titration of 6-fold molar excess of P18-I10 peptide leads to recovery of the reference spectrum, as expected for the formation of the native free pMHC-I species in equilibrium with the TAPBPR-bound form (Supplementary Fig. 13A, red). The same trend is observed when P18-I10 is titrated onto RGPGC/H2-Dd S73C/hβ2m/TAPBPR complex, where the addition of the mild reducing agent tris(2-carboxyethyl) phosphine (TCEP) allows for complete annealing of P18-I10 and subsequent TAPBPR dissociation (Supplementary Fig. 14).
In addition, we compared binding of a sub-optimal peptide (photoP18-I10) to the peptide-deficient MHC-I/TAPBPR. PhotoP18-I10 utilizes the same N- and C-terminal anchors as the native P18-I10, but shows a reduced thermal stability in the pMHC-I (Supplementary Table 1), presumably due to steric hindrance of the bulky 2-nitrophenyl group and β-amino acid backbone relative to the native valine sidechain at position 8 6,31 (Supplementary Fig. 15). In contrast to P18-I10, photoP18-I10 does not promote TAPBPR dissociation from the MHC-I, even at large molar excess (Supplementary Fig. 15). Taken together, our NMR data suggest that the peptide-deficient MHC-I groove in complex with TAPBPR is sampling multiple conformations, relevant for both peptide recognition and TAPBPR release. Binding of peptide to the empty groove results in the formation of a distinct TAPBPR-bound intermediate state, consistent with our NMR results using purified complexes (Fig. 2A–C).
Negative allosteric coupling between peptide and TAPBPR
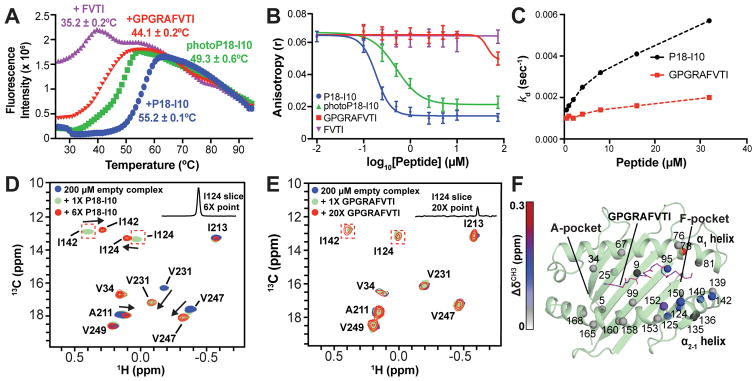
To identify peptide features that promote chaperone dissociation, we first screened a range of N- and C-terminal truncations for their efficiency at rescuing the thermal stability of free peptide-deficient MHC-I molecules prepared using conditional ligands 6,20,31. Examining binding of N- and C-terminal deletions of the native ligand P18-I10, we find that a single N-terminal deletion (ΔR1 - GPGRAFVTI) can modestly rescue H2-Dd thermal stability with a resulting Tm of ~44°C (Supplementary Table 1, Fig. 4A). The reduced stability is attributed to the loss of Arg1 contacts with Tyr7, Tyr171 and Glu163 in the A and B-pocket of H2-Dd, resulting in a reduced affinity (Supplementary Fig. 16A–C and Supplementary Table 2). We next used a fluorescence anisotropy (FA) assay to test binding of different peptides to a chaperoned MHC-I 5,6,32,33. We offered graded concentrations of peptide-deficient MHC-I/TAPBPR complex to 0.75 nM of a fluorescently labeled peptide (TAMRA-P18-I10), which is readily captured in the MHC-I groove. (Supplementary Fig. 17A). We find that an excess of 10 μM P18-I10 completely outcompetes TAMRA-P18-I10 for binding to empty groove (Supplementary Fig. 17B). Finally, we compared P18-I10, photoP18-I10, GPGRAFVTI, using a similar competition assay. Our results, in terms of the apparent dissociation constants of 0.09, 0.38 and 130 μM, suggest that peptide/MHC-I interactions spanning the entire groove are critical for TAPBPR-mediated peptide exchange (Fig. 4B). In both the DSF an FA assays, a negative control peptide, FVTI, shows minimal stability and exchange, as expected. Our FA results are also consistent with surface plasmon resonance (SPR) experiments, where addition of P18-I10 leads to a significant increase in the dissociation rate of TAPBPR from the peptide-occupied groove, relative to a peptide-deficient molecule (koff < 0.002 s−1). Addition of the sub-optimal GPGRAFVTI peptide leads to a more modest effect (Fig. 4C, Supplementary Fig. 17C, D).
Figure 4. Recognition of different peptide probes by the MHC-I/TAPBPR complex.
(A) DSF of UV-irradiated photoP18-I10/H2-Dd/hβ2m incubated with 20-fold molar excess of P18-I10 (RGPGRAFVTI), GPGRAFVTI or FVTI. Non-UV-irradiated photoP18-I10/H2-Dd/hβ2m is shown for reference. See related Supplementary Table 1. (B) FA (r) of 0.75 nM TAMRA-P18-I10 as a function competing unlabeled peptide P18-I10, GPGRAFVTI, photoP18-I10 or FVTI after incubation with 250 nM peptide-deficient H2-Dd/hβ2m/TAPBPR for 100 minutes. Fitted K2′ values are 0.186 μM (P18-I10), 0.501 μM (photoP18-I10), and 86.2 μM (GPGRAFVTI), which are close to values determined from a global fit (Supplementary Fig. 17A, B and Supplementary Fig. 18). K2′ for FVTI could not be determined. (C) SPR showing the TAPBPR dissociation phase from MHC-I as a function of increasing peptide concentration, assuming a single exponential decay model. See related Supplementary Fig. 17C, D. (D) and (E) 2D 1H-13C HMQC spectra of H2-Dd labeled peptide-deficient H2-Dd/hβ2m/TAPBPR complexes titrated with different peptides. Dotted red boxes highlight intermediate state peaks for I124 and I142 with bound peptide. 1H slices from the resonance of I124 in the bound state are inlayed for reference. See related Supplementary Fig. 17E. (F) Chemical shift deviation (CSD, ΔδCH3, ppm) of H2-Dd groove methyls (within 3.5 Å of the peptide) between GPGRAFVTI/H2-Dd/hβ2m/TAPBPR complex and free P18-I10/H2-Dd/hβ2m. Color coded CSD effects are plotted on the structure of H2-Dd (PDB ID 3ECB), with modeled GPGRAFVTI peptide. The methyl resonances of residues V9, A135 and A136 (shown in black) are missing due to conformational exchange-induced line broadening in the TAPBPR complex.
To further explore how peptides of different lengths may trigger TAPBPR release from the complex, we performed a series of NMR titrations. For P18-I10, we detect the formation of an intermediate state at a 1:1 molar ratio, highlighted by the appearance of methyl resonances corresponding to unique Ile124 and Ile142 conformations (Fig. 4D, green, red dotted boxes), while six-fold molar excess of peptide results in detectable formation of the free pMHC-I (Fig. 4D, red). Similarly, a titration of GPGRAFVTI shows the intermediate resonances, albeit with significantly attenuated intensities relative to P18-I10, as shown by a comparison of 1H slices, (Fig. 4E, green). Accordingly, formation of the final pMHC-I is not detectable by our NMR methyl probes using up to 20-fold molar excess of GPGRAFVTI (Fig. 4E, red). A combined NMR, SPR and FA analysis allows for a thorough description of the TAPBPR peptide-exchange cycle (Supplementary Fig. 18). According to our model, stable capture of a high-affinity peptide in the MHC groove can lead to as much as a three-order of magnitude increase in the dissociation rate of TAPBPR, from 0.002 s−1 to 2.9 s−1, as observed for P18-I10 (Supplementary Table 3). Notably, different peptides show a range of off-rates from the chaperoned MHC groove, from approx. 0.1 s−1 for P18-I10, to >2 s−1 for the N-terminal deletion mutant.
Finally, to examine specific structural adaptations that occur upon peptide annealing to the chaperoned groove, and their role in peptide exchange, we compared methyl NMR spectra of the “stalled” complex containing GPGRAFVTI relative to the free P18-I10/H2-Dd. Despite the missing N-terminal Arg from the GPGRAFVTI peptide, we measure small CSDs (< 0.1 ppm) for NMR peaks corresponding to residues in the A- and B-pockets, in contrast to the significant CSDs (0.1 to 0.3 ppm) for peaks corresponding to a continuous surface along the N-terminal α2-1 helical segment (residues 138–152) (Fig. 4F). This suggests that the conformation of the chaperoned MHC-I groove in the “stalled” complex is similar to the native molecule, with the exception of residues in the D-, E- and F- pockets. This finding is consistent with the sampling of multiple α2-1 helix conformations in fast exchange while the peptide is bound to the MHC-I/TAPBPR complex, where the ensemble average is skewed towards the “open” state observed both in the X-ray structure of TAPBPR-bound MHC-I and in our MD simulations (Supplementary Fig. 16C, D, F, red arrows). Taken together, our results support the view that the chaperoned MHC-I groove selectively captures incoming peptides and specific structural adaptations are required for TAPBPR dissociation and formation of the final pMHC-I product (Fig. 4A, Supplementary Table 1). TAPBPR release from the complex is driven by negative allosteric coupling between the peptide binding sites at both ends of the MHC-I groove, the TAPBPR binding site near the α2-1 helix and the anti-parallel pleated β-sheet underpinning the groove (Fig. 4F).
The chaperoned groove recognizes peptides in their native conformation
We next titrated 13C labeled peptides onto unlabeled, peptide-deficient MHC-I/TAPBPR complexes to explore peptide annealing under conditions where TAPBPR dissociation from the MHC-I is negligible (according to our previously determined Kd of 32 μM). We find that 13C methyl and aromatic resonances of P18-I10 show large CSDs when the free peptide is compared to MHC-I and MHC-I/TAPBPR bound states (Fig. 5A, B). P18-I10 binds in slow exchange, where a unique set of peaks corresponding to formation of the pMHC-I/TAPBPR complex can be readily observed. TAPBPR binding only leads to minor additional changes, suggesting that chaperones do not alter the conformation of the MHC-I bound peptide (Fig. 5A–C). As expected, we observe significantly reduced binding for 13C-labelled GPGRAFVTI peptide, albeit still at the slow exchange NMR regime (Supplementary Fig. 19A, B), as highlighted by a peak intensity ratio analysis for all peptide methyl probes obtained at the 1:1 titration point (Fig. 5D). A detailed NMR line shape analysis suggests binding to be at least 100-fold weaker than P18-I10 (>900 μM vs 7.2 μM, respectively, Fig. 5E and Supplementary Fig. 19D). The slow exchange NMR titration behavior observed for GPGRAFVTI, despite its modest affinity, suggests that peptide binding necessitates kon-limiting structural adaptations in an “induced fit” mechanism 34. The resonances of the bound states are consistent between P18-I10 and GPGRAFVTI, showing that the two peptides adopt a similar conformation in the intermediate complex with TAPBPR, relative to the reference pMHC-I structure (Supplementary Fig. 19, Supplementary Fig. 20, Supplementary Table 4).
Figure 5. TAPBPR-mediated loading of isotopically labeled peptides.
(A) and (B) 2D 1H-13C HMQC spectra of 13C labeled P18-I10 (RGPGRAFVTI) in the free (green), H2-Dd/hβ2m bound (red) and H2-Dd/hβ2m/TAPBPR bound (blue) states for (A) methyl and (B) aromatic probes. See related Supplementary Fig. 18. Asterisks (*) denote alterative conformations observed for V8 methyls. (C) A correlation plot of CSDs (ΔδCH, ppm) for free versus MHC-I bound and free versus MHC-I/TAPBPR bound 13C labeled P18-I10. (D) Normalized peak intensity ratios for the H2-Dd/hβ2m/TAPBPR and H2-Dd/hβ2m bound states (IpMHC-I/TAPBPR/IpMHC-I) for P18-I10 (blue) and GPGRAFVTI (teal). (E) NMR line shape analysis from the titration of unlabeled peptide-deficient H2-Dd/hβ2m/TAPBPR complex with 13C isotopically labeled P18-I10. The 1D 13C spectral slices for the Cβ of Ala6 at complex:peptide molar ratios of 0.3 (black), 0.45 (red), 0.92 (green), and 1.34 (blue). Error bar represent noise RMSD in each spectrum. Curves without error bars are the best fit of data with the two-state model. The spectral traces were shifted vertically for visual clarity and are shown at different scales for each 1D trace. Estimated Kd and koff values are 7.2 ± 4.1 μM and < 80 s−1 for P18-I10. See related Supplementary Fig. 19D.
DISCUSSION
Despite tapasin and TAPBPR having been discovered more than a decade ago, only recently have structural and mechanistic details of their peptide editing function begun to emerge 13,15. Here, we use solution NMR to directly evaluate the role of structure and dynamics in mediating peptide exchange. In agreement with the recent X-ray structures 19,20, conserved surfaces on the H2-Dd heavy chain, including the pleated β-sheet floor of the groove, the α2-1 helix, and the CD8 recognition loop in the α3 domain are utilized to form a high-affinity interaction with TAPBPR (Fig. 2E–G). Our data further highlight effects on the α1 helix, opposite from the TAPBPR binding site (Supplementary Fig. 5E, F and Supplementary Fig. 6E, F), which further supports a widening of the MHC-I groove at the F-pocket (Supplementary Fig. 16C–F). The widening is promoted by a “Tyr84 swing”, where the conserved heavy chain residue Y84 is flipped and hydrogen bonds with TAPBPR E102 19,20. In the absence of TAPBPR, Y84 plays a role in closing of the α2-1 helix “latch” by associating with the C-terminus of bound peptides in the F-pocket 35. This phenomenon is similar to a “W43 swing”, mediating class II MHC interaction with the DM chaperone 36. Together, our independent studies of the MHC-I/TAPBPR complex by crystallography and solution NMR allow us to elucidate a detailed mechanism of peptide exchange.
Conformational plasticity within the groove of class I and II MHC molecules has been linked to peptide exchange 21,22. NMR has provided direct measurements of groove dynamics in peptide-deficient molecules 37, as well as domain mobility along the β2m interface 38. NMR relaxation dispersion experiments have revealed dynamics of the α2-1 helix and pleated β-sheet of peptide-loaded molecules, such as HLA-B*35:01 39, while hydrogen/deuterium exchange points to allotype-specific differences in dynamics between peptide-loaded and peptide-deficient MHC-I molecules 40. Our relaxation dispersion measurements of pMHC-I highlight motions at the α2-1 helix and pleated β-sheet floor of H2-Dd (near the C-, E- and F-pockets), and structural plasticity at the hβ2m/heavy chain interface (Fig. 1D), consistent with previous NMR results for HLA-B*27 38,41 and molecular dynamics simulations of HLA-B*44 42. Intriguingly, regions of mobility in the pMHC-I correlate with surfaces required for TAPBPR binding (Supplementary Fig. 1), suggesting a direct role of dynamics in driving chaperone recognition. In particular, the Cβ methyl of A150, which is located on a linker region between the short α2-1 and longer α2-2 helices, undergoes chemical exchange at the ~2 msec timescale, which could arise from a switching between distinct ϕ/ψ dihedral angles impacting the orientation of the α2-1 helix (Fig. 1B). In our MD simulations, sampling of the “open” conformation is observed for a high-affinity P18-I10/H2-Dd complex, but becomes more pronounced for a peptide-deficient molecule (Fig. 3C and Supplementary Fig. 16E, F), consistently with similar simulation results reported for other MHC-I alleles 43. Therefore, in conjunction with previous reports our data are consistent with a conformational selection mechanism driving the recognition of different pMHC-I molecules by TAPBPR.
Our NMR data provide evidence that peptide-deficient MHC-I/TAPBPR complexes are stable, properly conformed molecules in solution (Fig. 3A), in agreement with previous studies describing the stabilization of chaperoned MHC-I molecules in a peptide-receptive conformation 5,6. According to our MD simulations, TAPBPR stabilizes the peptide-deficient groove by dampening dynamics within the C-, E- and F-pockets relative to an empty, unchaperoned MHC-I groove (Fig. 3C and Supplementary Fig. 16E, F), as previously suggested by simulations of modeled MHC-I/tapasin complexes 44. Taken together, our results suggest a functional role of chaperones as “stand-ins” for the peptide in modulating MHC-I groove dynamics, allowing for enhanced kinetic and thermal stability 45.
The available structures of MHC-I/chaperone complexes entirely lack electron density corresponding to peptide 19,20,23. Here, we identify a peptide-bound MHC-I/TAPBPR intermediate state, and characterize structural adaptations in the MHC-I upon peptide annealing to the complex. Dynamics in the MHC-I groove are progressively reduced as a function of increasing peptide occupancy (Fig. 3A, B). We further uncover a link between crucial contacts in the A- and B-pockets with recognition and modulation of dynamics within the chaperoned MHC-I groove, and subsequent TAPBPR dissociation (Fig. 4E and Fig 5F). Our data provide evidence for negative allosteric coupling between the A- and F-pockets of the MHC-I groove, with the TAPBPR binding site near the α2-1 helix and the pleated β-sheet underlying the groove. Here, the TAPBPR-induced widening of the MHC-I groove, limits peptide binding to the F-pocket (Fig. 4F and Supplementary Fig. 16C, D, F). Consequently, while TAPBPR is bound to the MHC-I, initial peptide annealing is dominated by interactions within the A- and B-pockets, which remain properly conformed in the chaperoned groove. This “indirect” peptide sensing model utilized by chaperones is supported by the significantly reduced binding to peptide-deficient H2-Dd/TAPBPR observed for GPGRAFVTI lacking critical A- and B-pocket contacts. Notably, a similar peptide-editing mechanism has been observed for the DM chaperone, which induces structural rearrangements at the P1 pocket of empty MHC-II molecules to promote the rapid and specific selection of high-affinity peptides 36. A putative role for direct peptide sensing by TAPBPR via the ‘scoop’ loop spanning residues 22–26 (absent from tapasin), for which electron density is present in the H2-Db/TAPBPR but not in the H2-Dd/TAPBPR X-ray structure 19,20, has been hypothesized, but such effects remain to be rigorously tested in future experiments.
Our results outline a cycle of peptide-exchange governed by reversible transitions between ensembles of MHC-I states (Fig. 6A, Supplementary Fig 21A). Sampling of open and closed conformations at the α2-1 helix together with dynamics at the heavy chain/β2m interface regulates chaperone association and dissociation. Through subtle structural adaptations in the MHC-I groove, TAPBPR shifts the binding free energy landscape of the cellular peptide pool by several orders of magnitude, as illustrated by our results for the H2-Dd system (Fig. 6B). In the ER and Golgi, TAPBPR associates with MHC-I molecules to maintain them in a receptive conformation (Fig. 6C–F, Step 1). The α2-1 helix ‘latch’ remains in a dynamic equilibrium between open and closed conformations while bound to the chaperone (Fig. 6C–F, Step 1). TAPBPR performs its peptide editing function by promoting dissociation of low and moderate affinity peptides, thereby preferentially selecting peptides with higher affinities for the chaperoned groove. Low-affinity peptides are completely excluded from the MHC-I/TAPBPR complex (Fig. 6C, Steps 2 and 3). Moderate-affinity peptides interact transiently with the MHC-I/TAPBPR complex, but fail to promote TAPBPR release due to the lack of crucial A-, B-pocket contacts, which limits the lifetime within the chaperoned groove (fast koff) (Fig. 6D, Steps 2 and 3, Supplementary Fig. 21B). In contrast, high-affinity peptides are guided by initial contacts spanning both the A- and F-pockets to form a prolonged interaction within the groove, resulting in a closure of the α2-1 helix ‘latch’, which triggers TAPBPR release from the pMHC-I (Fig. 6E, step 3). Inside the ER, a competition between these processes occurs simultaneously, where high-affinity peptides are maintained within the chaperoned groove to become part of the displayed antigen repertoire. (Fig. 6F, step 3).
Figure 6. Molecular mechanism of chaperone-assisted peptide exchange.
(A) Thermodynamic cycle of TAPBPR-mediated MHC-I peptide exchange. Fitted equilibrium values determined from a combined NMR/FA analysis are shown (Supplementary Fig. 18). (B) Conceptual example of how TAPBPR may scrutinize the peptide repertoire by increasing free binding energy across the cellular pool of candidate peptides. (C–F) Peptide dependence on the dissociation of the MHC-I/TAPBPR complex. MHC-I molecules are stylized in a cartoon representation as previously used 13. Step 1: TAPBPR chaperoned MHC-I molecules are maintained in a peptide-receptive conformation and exhibit widespread dynamics (dotted arrows) within the groove. TAPBPR binding induces a widening of the MHC-I groove, specifically in the F-pocket near the α2-1 helix, which can sample open and closed conformations, serving as a “latch”. Step 2: Association of (C) low-affinity peptide (FVTI, orange), (D) moderate-affinity peptide (GPGRAFVTI or photoP18-I10, blue) and (E) high-affinity peptide (P18-I10, magenta) with the complex is dependent on specific interactions in the A- and F-pockets. Peptides exist in an ‘on’ and ‘off’ equilibrium (dotted arrows) with the chaperoned groove. Step 3: TAPBPR release requires i) sufficient lifetime of the peptide within the groove to modulate the α2-1 helix latch and ii) negative allosteric modulation between non-overlapping peptide binding sites in the MHC groove and TAPBPR binding sites. See related Supplementary Fig. 21. (F) Step 4: Following dissociation, TAPBPR is recycled and associates with nearby sub-optimally loaded or peptide-deficient MHC-I molecules.
In summary, we provide a detailed NMR characterization of dynamics of empty and peptide-loaded MHC-I/TAPBPR complexes and elucidate structural adaptions upon formation of a peptide-bound intermediate state in relation to peptide exchange. The shared similarities of structural and functional aspects between TAPBPR and tapasin implies that a similar exchange mechanism is utilized by tapasin in the peptide-loading complex, a hypothesis that can be readily tested. Future studies on other H2 and HLA alleles with model peptides will expand upon the role of conformational dynamics and allosteric communication in shaping the peptide editing functions of these highly efficient molecular machines.
ONLINE METHODS
Protein expression, refolding and purification
DNA encoding the luminal domain of class I MHC (MHC-I) heavy chains H2-Dd, HLA-A*01:01, HLA-B*15:01, HLA-B*27:02 and human β2m (hβ2m, light chain) were provided by Dale Long of the NIH Tetramer Core Facility and transformed into Escherichia coli BL21(DE3) (Novagen). Proteins were expressed in Luria-Broth and inclusion bodies were processed as previously described 46. For in vitro refolding, 10 mg of peptide and 200 mg mixtures of heavy chain:light chain at a 1:3 molar ratio were slowly diluted over 24 hours into refolding buffer (0.4 M Arginine-HCl, 2 mM EDTA, 4.9 mM reduced L-glutathione, 0.57 mM oxidized L-glutathione, 100 mM Tris pH 8.0) at 4°C while stirring. Refolding proceeded for four days at 4°C without stirring. Samples where photoP18-I10 was used were protected from light (except during UV-irradiation) with aluminum foil throughout purification and data acquiring procedures. Purification of pMHC-I complexes was performed by size exclusion chromatography (SEC) with a HiLoad 16/600 Superdex 75 pg column at 1 mL/min with running buffer (150 mM NaCl, 25mM Tris pH 8), followed by anion exchange chromatography with a mono Q 5/50 GL column at 1 mL/min using a 40 minute 0–100% gradient of buffer A (50 mM NaCl, 25mM Tris pH 8) and buffer B (1M NaCl, 25mM Tris pH 8). The luminal domain of TAPBPR was expressed as previously described using a Drosophila S2 cell expression system and purified in a similar manner to pMHC-I molecules 47. The C121S and S73C H2-Dd mutants were generated by site directed mutagenesis and confirmed with DNA sequencing. Refolding and purification of RGPGC/H2-Dd S73C/hβ2m was performed with all buffers contained 1 mM dipeptide (Gly-Leu) to improve stability throughout sample preparation. All proteins were exhaustively buffer exchanged into 100 mM NaCl, 20 mM sodium phosphate pH 7.2. Protein concentration was determined using A280 measurements on a NanoDrop with extinction coefficients estimated with the ExPASy ProtParam tool.
Peptides and mass spectrometry
Unlabeled peptides (≥ 98% purity) were prepared by chemical synthesis (Biopeptik Inc, Malvern, USA or GenScript, Piscataway, USA). The UV-labile RGPGRAFJ*TI, referred to as photoP18-I10, was derived from P18-I10 (RGPGRAFVTI) 6 and contains a modification with 3-amino-3-(2-nitrophenyl)-propionic acid (J*). The reducing agent labile RGPGC, also derived from P18-I10, contains a modification with a cysteine at position five and truncation of residues A6-I10. Peptide concentration was determined using measurements of A205 with a NanoDrop with extinction coefficients determined using the Protein Parameter Calculator by Anthis and Clore. Liquid chromatography-mass spectroscopy (LC-MS) was carried out with passage through a Higgins PROTO300 C4 column (5 μm, 100 mm x 2.1 mm) followed by electron ion spray mass spectroscopy performed on a Thermo Finnigan LC/MS/MS (LTQ). Analysis and deconvolution of LC-MS data was performed with MagTran.
Isotopically labeled peptides were prepared with DNA sequences corresponding to P18-10 (RGPGRAFVTI) or GPGRAFVTI as fusions with an N-terminal immunoglobulin-binding domain of streptococcal protein G (GB1) and a His6 tag subcloned in pET22b vector. For expression, Escherichia coli BL21(DE3) (Novagen) cells were transformed with plasmid and grown at 37°C in 1× M9 minimal media for U-[15N, 13C] labeling following standard protocols. Expression proceeded with the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 37°C, 200 r.p.m. for 5 hours. Cell extract was passed through a FF-HisTrap Column (GE Healthcare) and then His6-GB1-peptide construct was solid phase extracted using a Strata C-18T column (Phenomenex). The protein was resuspended in 50% (v/v) trifluoroacetic acid to perform cyanogen bromide (100-fold molar excess relative to the methionine molar concentration) cleavage overnight. Cleaved peptide was purified by reversed phase chromatography using an Aeris PEPTIDE XB-C18 column (3.6 μm, 150 x 4.6 mm, Phenomenex). Peptide identity and purity (≥ 98% purity) was confirmed by mass spectrometry.
Fluorescence Anisotropy
Fluorescence anisotropy (r, herein referred to as FA) was performed using a P18-I10 peptide labeled with TAMRA dye (KTAMRAGPGRAFVTI, herein called TAMRA-P18-I10) (Biopeptik Inc, Malvern, USA), which was modeled to not disrupt essential interaction with MHC-I residues compared to the native Arg1. Peptide-deficient H2-Dd/hβ2m/TAPBPR complexes for FA were prepared by UV-irradiation at 365 nm for 1 hour of photoP18-I10/H2-Dd/hβ2m/TAPBPR complexes as described below. For association experiments, graded concentrations (up to 800 nM) of peptide-deficient H2-Dd/hβ2m/TAPBPR was added to 0.75 nM TAMRA-P18-I10. For P18-I10 competition experiments, a mixture of 250 nM peptide-deficient H2-Dd/hβ2m/TAPBPR complex and 0.75 nM TAMRA-P18-I10 was offered graded concentrations of native P18-I10 ranging from 0 to 75 μM. Additional peptide competition assays were performed using the same parameters except by adding graded concentrations ranging from 0 to 75 μM of different peptides (GPGRAFVTI, photoP18-I10, FVTI). All FA experiments were performed in the dark to protect the TAMRA-P18-I10 and photoP18-I10 peptides from photolysis. In the competition experiments, the average of FA after incubation with 250 nM peptide-deficient H2-Dd/hβ2m/TAPBPR for 95–105 minutes at room temperature was plotted as a function of the log10 of excess peptide. Each experiment was performed at room temperature in a volume of 100 μL and loaded onto a black 96-well polystyrene assay plate (Costar 3915). FA data was recorded via a Perkin-Elmer Envision 2103 plate reader with excitation filter λex = 531 nm and emission filter λem = 595 nm. Each experiment was performed in triplicate and is representative of at least two independent experiments. Experimental values were subtracted from background FA values obtained from incubation of TAMRA-P18-I10 alone. All samples were prepared in matched buffer (100 mM NaCl, 20 mM sodium phosphate pH 7.2, 0.05% (v/v) tween-20). FA time courses were analyzed by global fitting to the 4-step using DynaFit.
Differential scanning fluorimetry
DSF experiments were performed on an Applied Biosystems ViiA 7 qPCR machine with excitation and emission wavelengths set to 470 nm and 569 nm with proteins in buffer of 100 mM NaCl, 20 mM sodium phosphate pH 7.2. Experiments were conducted in triplicate in MicroAmp Fast 96-well plates with 50 μL total volume containing final concentrations of 7 μM protein and 10× SYPRO orange dye (ThermoFisher). For UV-treated samples, proteins were UV-irradiated at 365 nm for 1 hour at room temperature and assayed following centrifugation at 13,000 rpm for 10 minutes to remove precipitates. For reducing agent treated samples, samples were incubated with 1 mM TCEP for 1 hour and assayed in the presence of TCEP, following centrifugation at 13,000 rpm for 10 minutes to remove precipitates. Rescue experiments were performed by incubation with 20-fold molar excess peptide throughout the duration of the experiment. Temperature was incrementally increased at a scan rate of 1°C/min between 25°C and 95°C. Data analysis and fitting was performed in GraphPad Prism v7.
Preparation of MHC-I/TAPBPR complexes and binding assays
To prepare peptide-deficient MHC-I/TAPBPR complexes, 200 μM photoP18-I10/H2-Dd/hβ2m and ~200 μM TAPBPR were mixed together (1:1 molar ratio) and incubated for 1 hour at room temperature. The mixture was UV-irradiated at 365 nm for 1 hour followed by centrifugation at 13,000 rpm for 10 minutes to remove precipitates. Pure 1:1 stoichiometric MHC-I/TAPBPR complexes were isolated by SEC with a Superdex 200 Increase 10/300 GL column at flow rate 0.5 mL/min in 100 mM NaCl, 20 mM sodium phosphate pH 7.2. For analytical SEC binding assays, 80 μM pMHC-I and 160 μM TAPBPR were mixed (1:2 molar ratio) and incubated at room temperature for 1 hour. Analytical SEC binding assays were performed using a Superdex 200 Increase 10/300 GL column at flow rate 0.5 mL/min in 100 mM NaCl, 20 mM sodium phosphate pH 7.2.
Surface plasmon resonance
SPR measurements were performed using a BiaCore T200 (GE Healthcare, Uppsala, Sweden) instrument at 25°C in 150 mM NaCl, 10 mM HEPES pH 7.3, 3 mM EDTA, 0.05% (v/v) tween-20. Approximately 300 resonance units (RU) of biotinylated TAPBPR was immobilized on a streptavidin-coated chip (GE Healthcare). H2-Dd S73C/β2m was then captured on the TAPBPR coated surface followed by a 120 second wash-out step with buffer. Graded concentrations of P18-I10 or GPGRAFVTI ranging from 0.5 to 32 μM were sequentially injected for 120 seconds and the dissociation of H2-Dd S73C/β2m was monitored. The surface was regenerated after each binding and dissociation cycle by injecting 100 μM P18-I10 to achieve complete dissociation.
Sample preparation for NMR
Samples for NMR were prepared as previously described 48. Protein expression was achieved by induction with 1 mM isopropyl-D-thiogalactoside (IPTG) at an OD600 of 0.6 followed by cell growth at 37°C for 5 hours at 200 r.p.m. Isotopically labeled pMHC-I was buffer exchanged into 100 mM NaCl, 20 mM sodium phosphate pH 7.2, 0.01% (v/v) NaN3 and 1 U Roche protease inhibitor cocktail in 90% H2O/10% D2O. MHC-I/TAPBPR complexes for NMR were prepared by mixing 200 μM isotopically labeled pMHC-I with 200 μM unlabeled TAPBPR at a 1:1 mole ratio in matched buffer (100 mM NaCl, 20 mM sodium phosphate pH 7.2) followed by isolation of pure 1:1 complex by SEC as described above. Peptide-deficient complexes were prepared as described above using isotopically labeled pMHC-I samples followed by isolation of pure 1:1 complex by SEC. Loss of peptide was confirmed by LC-MS.
NMR chemical shift assignments and dynamic measurements
Triple-labeled U-[15N, 13C, 2H] and AILV sidechain methyl (Ala 13Cβ, Ile 13Cδ1, Leu 13Cδ1/13Cδ2, Val 13Cγ1/13Cγ2) labeled (at either H2-Dd or hβ2m) pMHC-I molecules were prepared. We utilized a previously published multipronged assignment approach 48. Backbone amide resonances were assigned using a standard TROSY-based 3D HNCO, HNCA and HN(CA)CB experiments recorded at 600 MHz at 25°C. Acquisition times of 30 msec (15N), 20 msec (13CO) and 10/5 msec (13Cα/13Cβ) in the indirect dimension were used. Backbone amide assignments were validated by acquiring amide-amide NOEs using 3D HN-NHN and 3D N-NHN SOFAST NOESY experiments 49. AILV methyl resonances were assigned using a 3D HMCM[CG]CBCA methyl out-and-back experiment recorded on 1 mM pMHC-I samples at 800 MHz at 25°C. Acquisition parameters were 80, 80, 1,536 complex points in the 13Caliphatic, 13CM, 1HM dimensions corresponding to acquisition times of 4, 10 and 69 msec. A relaxation delay of 1.5 seconds was used with 40 scans/FID. Chemical shifts observed in the 3D HMCM[CG]CBCA experiment were compared to 13Cα and 13Cβ chemical shifts from 3D HNCA and 3D HN(CA)CB experiments. AILV methyl assignments were validated and stereo-specifically disambiguated by acquiring methyl-methyl NOEs with 3D HM-CMHM and 3D CM-CMHM SOFAST NOESY experiments 48. All 3D SOFAST NOESY experiments were acquired with standard parameters as previously described 47 on 1 mM pMHC-I samples with a relaxation delay (d1) of 0.2 sec and an NOE mixing time (d8) of 0.3 sec. To confirm the assignment of the TAPBPR bound pMHC-I states for the backbone amide and sidechain methyl resonances, additional 3D HM-CMHM and 3D HN-NHN SOFAST NOESY experiments were acquired on a 200 μM pMHC-I/TAPBPR complex and compared to similar NOE strips from the unbound pMHC-I reference. All NMR data was processed with NMRPipe 50 and analyzed using NMRFAM-SPARKY 51.
Backbone 15N amide relaxation rates were determined using TROSY-readout R1, R1ρ and 15N-{1H} heteronuclear NOE experiments at 25°C recorded at 1H field of 600 MHz on 1 mM perdeuterated pMHC-I samples. R2 rates were obtained from rotating frame R1ρ rates measured under a spin-lock field strength of 2 kHz. AILV methyl sidechain relaxation rates were determined using 1H-13C multiple quantum Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments on 1 mM perdeuterated pMHC-I samples recorded at 25°C at 600 MHz. Spectra were recorded in an interleaved manner with CPMG field strengths of 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 600, 700, 850 and 1000 Hz with a constant time delay of 40 msec. Peak intensities were converted to the R2, eff transverse decay rates with the equation R2, eff = 1/TCPMG × ln (I0/IνCPMG). CPMG curves were analyzed using GUARDD 52.
NMR chemical shift mapping and titrations
Chemical shift deviations (CSD, p.p.m.) were determined using the equations ΔδNH=[1/2(ΔδH2+ΔδN2/25)]1/2 for 15N amides and ΔδCH3=[1/2(ΔδH2+ΔδC2/4)]1/2 for 13C AILV methyls. The change in peak intensity was determined by calculating the ratio of Ibound/Iunbound for each assigned NMR resonance and normalized to the most intense NMR peak. For peptide titration experiments, NMR buffer (100 mM NaCl, 20 mM sodium phosphate pH 7.2, 10% D2O (v/v), 1U Roche protease inhibitor) matched samples of 200 μM peptide-deficient H2-Dd/hβ2m/TAPBPR or RGPGC/H2-Dd S73C/hβ2m/TAPBPR complexes (labeled at the heavy chain) were titrated with increasing concentrations of unlabeled peptide stock with 2D 1H-13C methyl SOFAST HMQC readouts recorded at 25°C at a 1H field strength of 800 MHz. Protein:peptide molar ratios were 1:0, 1:0.01, 1:0.03, 1:0.05, 1:0.1, 1:0.25, 1:0.4, 1:0.7, 1:1, 1:2, 1:3, 1:4, 1:5 and 1:6. At each point, 2D 1H-13C SOFAST HMQC spectra were acquired with 136 scans, 0.2 sec recycle delay (d1) and acquisition times of 30 msec in the 13C dimension. Titrations of 200 μM peptide-deficient H2-Dd/hβ2m/TAPBPR complex (labeled at the heavy chain) with unlabeled photoP18-I10, GPGRAFVTI and FVTI peptides into were performed under similar conditions using 1:0, 1: 0.25, 1:1, 1:6 and 1:20 protein:peptide molar ratios. All samples used in the titration containing photoP18-I10 were protected from light throughout data acquisition.
The titration of TAPBPR onto AILV methyl labeled (at heavy chain) P18-I10/H2-Dd/hβ2m was performed on a 100 μM sample in NMR buffer (100 mM NaCl, 20 mM sodium phosphate pH 7.2) at the following pMHC-I/TAPBPR ratios: 1:0, 1:0.08, 1:0.2, 1:0.36, 1:0.52, 1:0.68, 1:0.85, 1:1.16, 1:1.48, 1:1.80, and 1:2.28 with 2D 1H-13C SOFAST HMQC spectra as a readout. Data were processed with 4 Hz and 10 Hz Lorentzian line broadening in the direct and indirect dimensions, respectively and fit using a two-state model in TITAN 53 with bootstrap error analysis of 100 replicas. A total of nine NMR peaks were used for the global fitting procedure, including resonances corresponding to methyl groups of residues spanning the entire H2-Dd heavy chain (I124, A125, L126, A140, I142, L224, V231, A245, V247).
NMR experiments with isotopically labeled peptide
For structure determination of unbound P18-I10 in solution, a sample containing 1 mM P18-I10 (natural isotopic abundance) was used to record a 2D 1H-1H NOESY (300 msec mixing time, 75 msec acquisition time in the indirect dimension) spectrum was acquired at a 1H field of 800 MHz at 4°C in addition to 2D 1H-15N HSQC, 2D 1H-13C HSQC spectra, and 2D 1H-1H TOCSY spectrum (80 msec isotropic mixing time, 30 msec acquisition time in the indirect dimension) acquired at a 1H field of 600 MHz at 4°C. In all experiments the recycle delay was set to 1 sec. Automatic NOE assignments and structure calculations were performed in CYANA 2.1 followed by manual inspection and validation. The P18-I10 ensemble was generated from the 20 lowest energy structures (1.51 Å RMSD).
NMR spectra of P18-I10 or GPGRAFVTI in the unbound, MHC-I or MHC-I/TAPBPR bound states were obtained using 2D 1H-13C SOFAST HMQC experiments with selective excitation of either the methyl (13CM-SOFAST HMQC) or aromatic sidechain (13CAro-SOFAST HMQC) resonances as previously described 48. CSDs and peak intensity ratios analysis was performed as described above. For peptide titration experiments, 113.6 μM of unlabeled peptide-deficient H2-Dd/hβ2m/TAPBPR complex was titrated with 1.2, 5.7, 28.4, 79.8, 113.6, 227.2 and 340.8 μM of 13C labeled peptide. Each sample was prepared in matched NMR buffer and experiments were acquired with 2D 1H-13C SOFAST HMQC experiments using identical parameters with methyl probes as a readout. Experiments were performed at 25°C at a 1H field of 800 MHz.
NMR line shape fitting was performed in Integrative Data Analysis Platform (IDAP, available from Dr. Evgenii Kovrigin, http://lineshapekin.net/#IDAP) 54. Data were processed with an exponential window function with 4 Hz and 10 Hz Lorentzian line broadening in the direct and indirection dimensions, respectively. The 13C dimension from 2D 1H-13C SOFAST HMQC spectra was extracted, NMR peaks were normalized to unity area, and fit to ‘U’ model corresponding to a two-state binding mechanism with an adjustable constant vertical offset for each spectrum in MATLAB. Error bar represent noise root-mean-squared deviation in each spectrum. NMR line shape fitting allows for estimate for the upper and lower bounds for the Kd and Koff.
An apparent Kd of each peptide for MHC-I/TAPBPR complex was also determined by calculating the fraction peptide bound (fb - the intensity of the bound NMR peak divided by the sum of the intensities of the bound and free peaks). Titration points were fit to the equation:
in Mathematica 11 (Wolfram Research, Inc.). The final apparent Kd was determined by taking the average of each Kd value determined for NMR peaks corresponding to each AILV residue at each titration point (i.e. a global fit).
Molecular Dynamics simulations
All-atom molecular dynamics (MD) simulations in explicit solvent were carried out as previously described for MHC-I/tapasin complexes 55 in GROMACS version 5.1.4 using an AMBER99SB-ILDN protein forcefield and TPI3P water model. LINCS and SETTLE constraint algorithms were used to constrain peptide/protein and water molecules, respectively. An integration time step of 4 fsec was used with coordinates output every 10 psec. Short range interactions were treated with a Verlet cut-off scheme with 10 Å electrostatic and van der Walls cutoffs and long-range electrostatics were treated with the PME method with a grid spacing of 1.2 Å and cubic interpolation. Periodic dodecahedron boundaries were used. The thermodynamic ensemble was nPT where temperature was kept constant at 300K by a V-rescale modified thermostat with 0.1 psec time constant and pressure was kept constant at 1 bar pressure using an isotropic Berendsen barostat with 0.5 psec time constant and 4.5 × 10−5 bar−1 isothermal compressibility. X-ray structures served as inputs: empty H2-Dd S73C/β2m/TAPBPR (PDB ID 5WER), P18-I10/H2-Dd/β2m (PDB ID 3ECB) and empty H2-Dd/β2m (PDB ID 3ECB modified such that the atoms corresponding to P18-I10 were removed) with missing densities built in using Rosetta. The system was solvated to overall neutral charge and contained Na+ and Cl− ions to yield physiological concentration of 0.15 M. Final systems ranged from ~95,000 (empty MHC-I) to 196,000 (empty MHC-I/TAPBPR complex) atoms. Following 500 steps of steepest-descent energy minimization, initial velocities were generated at 65 K with linear heating up to 300 K over 2 nsec. Trajectories for each molecule were acquired for 500 nsec. Heat maps of the variance of distances between Cα atoms of pairs of residues spanning the groove were generated using the heatmapper plugin in VMD. Structures were extracted after the final 500 nsec simulation using VMD.
DATA AVAILABILITY
NMR assignments for the P18-I10 peptide, H2-Dd heavy chain and hβ2m light chain have been deposited into the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession numbers 30350, 27248 and 27249, respectively. The solution NMR structure of unbound P18-I10 peptide and X-ray structures of RGPGC/H2-Dd S73C/hβ2m and RGPGC/H2-Dd S73C/β2m/TAPBPR are deposited in the RCSB Protein Data Bank with accession codes PDB ID 6B9K, 5WES and 5WER, respectively.
Supplementary Material
Acknowledgments
The authors would like to acknowledge G. Morozov and A. Bax for helpful discussions, J. Ying and V. Tugarinov for assistance with recording NMR relaxation data, and C. Waudby for help with NMR line shape fitting in TITAN. E.L.K. was supported by the Regular Research Grant 2016 from Committee on Research (COR), Marquette University. This research was supported by the Intramural research program of the NIAID, NIH, a K-22 Career Development and an R35 Outstanding Investigator Award to N.G.S. through NIAID(AI2573-01) and NIGMS(1R35GM125034-01), respectively, and by the Office of the Director, NIH, under High End Instrumentation (HIE) Grant S10OD018455 which funded the 800 MHz NMR spectrometer at UCSC.
Footnotes
AUTHOR CONTRIBUTIONS
A.C.M., K.N., D.H.M. and N.G.S. designed the research, interpreted data and wrote the manuscript. K.N. performed SPR experiments. A.C.M performed differential scanning fluorimetry experiments. J.S.T. performed fluorescence anisotropy experiments with analysis performed by C.R.B. A.C.M., K.N. and M.B. generated constructs, performed protein expression and purification. A.C.M. and D.F.S. prepared and purified isotopically labeled peptides. A.C.M., V.K., D.F.S. and N.G.S. acquired and analyzed NMR data. A.C.M. and E.L.K. performed NMR line shape analysis. A.C.M. and D.F.S. performed and analyzed MD simulations. J.J. provided X-ray structures of the RGPGC/H2-Dd S73C/β2m and RGPGC/H2-Dd S73C/β2m/TAPBPR. K.N., J.J., and D.H.M. conceived and validated the disulfide-linked covalent constructs and their binding behavior.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.Rock KL, Reits E, Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 2007;8:873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- 4.Barnden MJ, Purcell AW, Gorman JJ, McCluskey J. Tapasin-mediated retention and optimization of peptide ligands during the assembly of class I molecules. J Immunol Baltim Md 1950. 2000;165:322–330. doi: 10.4049/jimmunol.165.1.322. [DOI] [PubMed] [Google Scholar]
- 5.Hermann C, et al. TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. eLife. 2015;4:e09617. doi: 10.7554/eLife.09617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morozov GI, et al. Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc Natl Acad Sci U S A. 2016;113:E1006–1015. doi: 10.1073/pnas.1519894113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul S, et al. HLA class I alleles are associated with peptide binding repertoires of different size, affinity and immunogenicity. J Immunol Baltim Md. 2013;1950:191. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shionoya Y, et al. Loss of tapasin in human lung and colon cancer cells and escape from tumor-associated antigen-specific CTL recognition. Oncoimmunology. 2017;6:e1274476. doi: 10.1080/2162402X.2016.1274476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen QR, Hu Y, Yan C, Buetow K, Meerzaman D. Systematic Genetic Analysis Identifies Cis-eQTL Target Genes Associated with Glioblastoma Patient Survival. PLOS ONE. 2014;9:e105393. doi: 10.1371/journal.pone.0105393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park B, et al. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity. 2004;20:71–85. doi: 10.1016/s1074-7613(03)00355-8. [DOI] [PubMed] [Google Scholar]
- 11.Montserrat V, Galocha B, Marcilla M, Vázquez M, López de Castro JA. HLA-B*2704, an allotype associated with ankylosing spondylitis, is critically dependent on transporter associated with antigen processing and relatively independent of tapasin and immunoproteasome for maturation, surface expression, and T cell recognition: relationship to B*2705 and B*2706. J Immunol Baltim Md 1950. 2006;177:7015–7023. doi: 10.4049/jimmunol.177.10.7015. [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, et al. Further Examination of the Candidate Genes in Chromosome 12p13 Locus for Late-Onset Alzheimer Disease. Neurogenetics. 2008;9:127–138. doi: 10.1007/s10048-008-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas C, Tampé R. Proofreading of Peptide-MHC Complexes through Dynamic Multivalent Interactions. Front Immunol. 2017;8:65. doi: 10.3389/fimmu.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Hateren A, Bailey A, Elliott T. Recent advances in Major Histocompatibility Complex (MHC) class I antigen presentation: Plastic MHC molecules and TAPBPR-mediated quality control. F1000Research. 2017;6:158. doi: 10.12688/f1000research.10474.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neerincx A, Boyle LH. Properties of the tapasin homologue TAPBPR. Curr Opin Immunol. 2017;46:97–102. doi: 10.1016/j.coi.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Boyle LH, et al. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci U S A. 2013;110:3465–3470. doi: 10.1073/pnas.1222342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann C, Strittmatter LM, Deane JE, Boyle LH. The binding of TAPBPR and Tapasin to MHC class I is mutually exclusive. J Immunol Baltim Md 1950. 2013;191:5743–5750. doi: 10.4049/jimmunol.1300929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neerincx A, et al. TAPBPR bridges UDP-glucose:glycoprotein glucosyltransferase 1 onto MHC class I to provide quality control in the antigen presentation pathway. eLife. 2017;6:e23049. doi: 10.7554/eLife.23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas C, Tampé R. Structure of the TAPBPR–MHC I complex defines the mechanism of peptide loading and editing. Science. 2017:eaao6001. doi: 10.1126/science.aao6001. [DOI] [PubMed] [Google Scholar]
- 20.Jiang J, et al. Crystal structure of a TAPBPR–MHC-I complex reveals the mechanism of peptide editing in antigen presentation. Science. 2017:eaao5154. doi: 10.1126/science.aao5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieczorek M, et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayres CM, Corcelli SA, Baker BM. Peptide and Peptide-Dependent Motions in MHC Proteins: Immunological Implications and Biophysical Underpinnings. Front Immunol. 2017;8:935. doi: 10.3389/fimmu.2017.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blees A, et al. Structure of the human MHC-I peptide-loading complex. Nature. 2017;551:525. doi: 10.1038/nature24627. [DOI] [PubMed] [Google Scholar]
- 24.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 25.Pedersen LO, et al. The interaction of beta 2-microglobulin (beta 2m) with mouse class I major histocompatibility antigens and its ability to support peptide binding. A comparison of human and mouse beta 2m. Eur J Immunol. 1995;25:1609–1616. doi: 10.1002/eji.1830250621. [DOI] [PubMed] [Google Scholar]
- 26.Lakomek NA, Ying J, Bax A. Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korzhnev DM, Kloiber K, Kay LE. Multiple-quantum relaxation dispersion NMR spectroscopy probing millisecond time-scale dynamics in proteins: theory and application. J Am Chem Soc. 2004;126:7320–7329. doi: 10.1021/ja049968b. [DOI] [PubMed] [Google Scholar]
- 28.Wearsch PA, et al. Major histocompatibility complex class I molecules expressed with monoglucosylated N-linked glycans bind calreticulin independently of their assembly status. J Biol Chem. 2004;279:25112–25121. doi: 10.1074/jbc.M401721200. [DOI] [PubMed] [Google Scholar]
- 29.Ryan SO, Cobb BA. Roles for major histocompatibility complex glycosylation in immune function. Semin Immunopathol. 2012;34:425–441. doi: 10.1007/s00281-012-0309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovrigin EL. NMR line shapes and multi-state binding equilibria. J Biomol NMR. 2012;53:257–270. doi: 10.1007/s10858-012-9636-3. [DOI] [PubMed] [Google Scholar]
- 31.Rodenko B, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat Protoc. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Hateren A, et al. A Mechanistic Basis for the Co-evolution of Chicken Tapasin and Major Histocompatibility Complex Class I (MHC I) Proteins. J Biol Chem. 2013;288:32797–32808. doi: 10.1074/jbc.M113.474031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latham MP, Zimmermann GR, Pardi A. NMR chemical exchange as a probe for ligand-binding kinetics in a theophylline-binding RNA aptamer. J Am Chem Soc. 2009;131:5052–5053. doi: 10.1021/ja900695m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hein Z, et al. Peptide-independent stabilization of MHC class I molecules breaches cellular quality control. J Cell Sci. 2014;127:2885–2897. doi: 10.1242/jcs.145334. [DOI] [PubMed] [Google Scholar]
- 36.Pos W, et al. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 2012;151:1557–1568. doi: 10.1016/j.cell.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurimoto E, et al. Structural and functional mosaic nature of MHC class I molecules in their peptide-free form. Mol Immunol. 2013;55:393–399. doi: 10.1016/j.molimm.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Beerbaum M, et al. NMR spectroscopy reveals unexpected structural variation at the protein-protein interface in MHC class I molecules. J Biomol NMR. 2013;57:167–178. doi: 10.1007/s10858-013-9777-z. [DOI] [PubMed] [Google Scholar]
- 39.Yanaka S, et al. Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex. J Biol Chem. 2014;289:24680–24690. doi: 10.1074/jbc.M114.566174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Hateren A, et al. Direct evidence for conformational dynamics in Major Histocompatibility Complex class I molecules. J Biol Chem. 2017 doi: 10.1074/jbc.M117.809624. jbc.M117.809624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hee CS, et al. Dynamics of free versus complexed β2-microglobulin and the evolution of interfaces in MHC class I molecules. Immunogenetics. 2013;65:157–172. doi: 10.1007/s00251-012-0667-4. [DOI] [PubMed] [Google Scholar]
- 42.Bailey A, et al. Selector function of MHC I molecules is determined by protein plasticity. Sci Rep. 2015;5:srep14928. doi: 10.1038/srep14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sieker F, Springer S, Zacharias M. Comparative molecular dynamics analysis of tapasin-dependent and -independent MHC class I alleles. Protein Sci Publ Protein Soc. 2007;16:299–308. doi: 10.1110/ps.062568407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisette O, Wingbermühle S, Tampé R, Schäfer LV. Molecular mechanism of peptide editing in the tapasin-MHC I complex. Sci Rep. 2016;6:19085. doi: 10.1038/srep19085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abualrous ET, et al. The Carboxy Terminus of the Ligand Peptide Determines the Stability of the MHC Class I Molecule H-2Kb: A Combined Molecular Dynamics and Experimental Study. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0135421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Natarajan K, Malchiodi EL, Margulies DH, Mariuzza RA. Three-dimensional structure of H-2Dd complexed with an immunodominant peptide from human immunodeficiency virus envelope glycoprotein 120. J Mol Biol. 1998;283:179–191. doi: 10.1006/jmbi.1998.2091. [DOI] [PubMed] [Google Scholar]
- 47.Morozov GI, et al. Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc Natl Acad Sci U S A. 2016;113:E1006–1015. doi: 10.1073/pnas.1519894113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natarajan K, et al. An allosteric site in the T-cell receptor Cβ domain plays a critical signalling role. Nat Commun. 2017;8:15260. doi: 10.1038/ncomms15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi P, Xia Y, Khanra N, Veglia G, Kalodimos CG. 15N and 13C- SOFAST-HMQC editing enhances 3D-NOESY sensitivity in highly deuterated, selectively [1H,13C]-labeled proteins. J Biomol NMR. 2016;66:259–271. doi: 10.1007/s10858-016-0074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinforma Oxf Engl. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kleckner IR, Foster MP. GUARDD: user-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J Biomol NMR. 2012;52:11–22. doi: 10.1007/s10858-011-9589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waudby CA, Ramos A, Cabrita LD, Christodoulou J. Two-Dimensional NMR Lineshape Analysis. Sci Rep. 2016;6:24826. doi: 10.1038/srep24826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovrigin EL. NMR line shapes and multi-state binding equilibria. J Biomol NMR. 2012;53:257–270. doi: 10.1007/s10858-012-9636-3. [DOI] [PubMed] [Google Scholar]
- 55.Fisette O, Wingbermühle S, Tampé R, Schäfer LV. Molecular mechanism of peptide editing in the tapasin-MHC I complex. Sci Rep. 2016;6:19085. doi: 10.1038/srep19085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR assignments for the P18-I10 peptide, H2-Dd heavy chain and hβ2m light chain have been deposited into the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession numbers 30350, 27248 and 27249, respectively. The solution NMR structure of unbound P18-I10 peptide and X-ray structures of RGPGC/H2-Dd S73C/hβ2m and RGPGC/H2-Dd S73C/β2m/TAPBPR are deposited in the RCSB Protein Data Bank with accession codes PDB ID 6B9K, 5WES and 5WER, respectively.