The non-biological nucleic acid 2’-deoxy-2’-fluoro-arabinonucleic acid (2’F-ANA) may be of use because of its higher chemical stability than DNA in terms of resistance to hydrolysis and nuclease degradation. In order to investigate the charge transfer characteristics of 2’F-ANA, of relevance to applications in nucleic acid-based biosensors and chip technologies, we compare the electronic couplings for hole transfer between stacked nucleobase pairs in DNA and 2’F-ANA by carrying out density fuctional theory (DFT) calculations on geometries taken from molecular dynamics simulations. We find similar averages and distribution widths of the base-pair couplings in the two systems. On the basis of this result, 2’F-ANA is expected to have charge transfer properties similar to those of DNA, while offering the advantage of enhanced chemical stability. As such, 2’F-ANA may serve as a possible alternative to DNA for use in a broad range of nanobiotechnological applications. Furthermore, we show that the (experimentally observed) enhanced chemical stability resulting from the backbone modifications does not cause reduced fluctuations of the base-pair electronic couplings around the values found for “ideal” B-DNA (with standard step parameter values). Our study also supports the use of a DFT implementation, with the M11 functional, of the wave function overlap method to compute effective electronic couplings in nucleic acid systems.
DNA’s structural, biological, chemical and electronic properties make it of great interest in a wide range of fields, including nanobioelectronics,1, 2 biological sensing,3–5 redox protein signaling,6, 7 and biomedicine.8, 9 Therefore, research aims to understand and to optimize DNA charge transport,10–12 exploring its charge conduction mechanisms as a function of structure and sequence.13–20 Fast DNA charge transport can be achieved through transient coherence (flickering resonance21) or ‘deep-hole transfer’.22
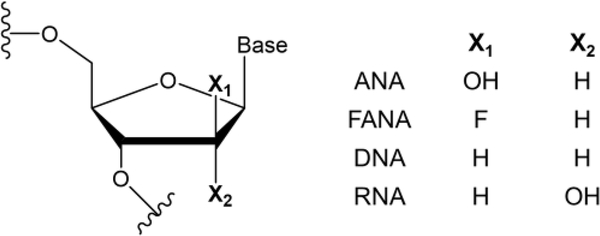
In contrast to the rich DNA literature, very little is known about hole transport through xeno-nucleic acids (XNA), although these chemically modified nucleic acids are known to also be capable of carrying out heredity and evolution functions (since genetic information can be stored in them and retrieved from them)23 while having greater resistance to nuclease degradation compared to DNA/RNA. The XNA investigated in this study, 2’F-ANA (Fig. 1), has notable chemical properties including: (i) high resistance to acid hydrolysis;24, 25 (ii) a wider operating pH range than DNA and RNA;24 (iii) the ability to form duplexes with DNA/RNA,26, 27 which can enable its use for the directed evolution of biopolymers; and (iv) the ability to induce the enzymatic degradation of target mRNA.28 These properties make 2’F-ANA an attractive vehicle for oral absorption of oligonucleotides29, 30 and intracellular delivery (with relevance to cancer treatments, as exemplified by the gymnotic silencing of the androgen receptor in prostate cancer cells by 2’F-ANA31) and could enable increased durability in diagnostic applications.24 For biosensing and chip technologies that rely on charge-transfer based signal transduction through a nucleic acid,32–34 2’F-ANA probes can potentially be an improved alternative to DNA probes. In fact, probes using 2’F-ANA can be washed with acid or base to remove the analytes without compromising the structural integrity of the 2’F-ANA array. Furthermore, unlike DNA-inspired systems such as the peptide nucleic acids (PNAs) (in which a neutral backbone can destabilize hole states localized on the nucleobases that support charge migration35), 2’F-ANA not only retains the negatively charged phosphate backbone but also contains a highly electronegative fluorine on the ribose moiety (Fig. 1) that increases the backbone charge compared to DNA.
Fig. 1.
Molecular Structures of 2’F-ANA and DNA, compared with ANA (arabinonucleic acid) and with RNA.
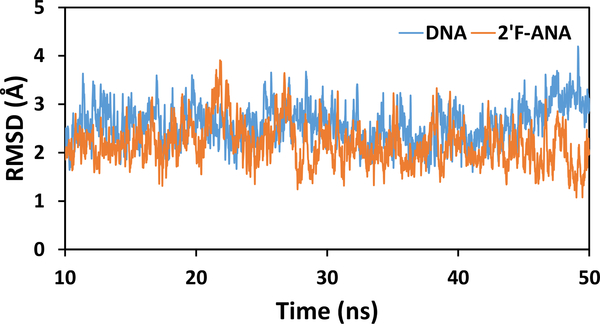
If charge transport in 2’F-ANA is at least as fast as in DNA, the 2’F-ANA’s enhanced structural properties favor its use in the nucleic acid applications mentioned above. Here, we compare the charge transfer properties of 2’F-ANA and DNA in terms of the effective36 electronic couplings between nucleobase pairs as a function of the nuclear conformation. DNA and 2’F-ANA were modeled using the Dickerson-Drew dodecamers (that has a B-DNA structure) (Fig. 2), using the structures with PDB codes 4C6437 and 2LSC,38 respectively. To obtain the 2’F-ANA duplex from the 2’F-ANA/ANA structure in the 2LSC PDB file, the U bases of the ANA strand were modified to T bases and the sugar moieties were replaced with the corresponding ones in 2’F-ANA, as detailed in the Supporting Information. The two systems were solvated with TIP3P water39 and simulated for 50 ns using classical molecular dynamics (MD), with the AMBER force field ff99 (see Supporting Information). In the 10–50 ns time window, the RMSDs of DNA and of 2’F-ANA fluctuate randomly around average values of 2.6 Å and 2.2 Å, with ranges of 1.6–4.2 Å and 1.1–3.9 Å, respectively (Fig. 3). The ranges of RMSD values suggest similar structural stability for 2’F-ANA and DNA, despite their different chemical stability. We selected MD snapshots for electronic coupling analysis each ns in the 10–50 ns time window of the MD production run.
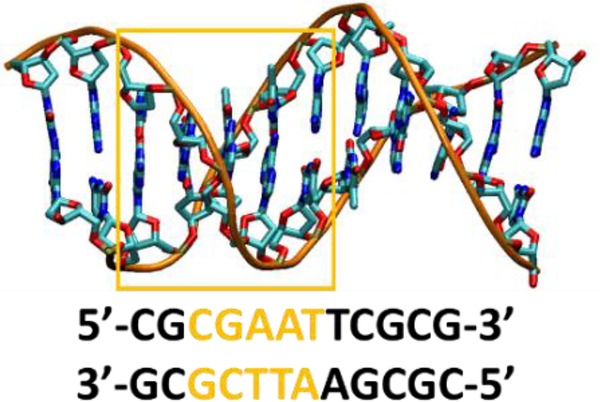
Fig. 2.
Structure and base pair sequence of the Dickerson-Drew dodecamer (PDB 4C6437). The base pairs used for VIF calculations are highlighted in yellow.
Fig. 3.
RMSDs (excluding H atoms) along the MD production runs for DNA (blue) and 2’F-ANA (orange).
The (effective) electronic coupling VIF (here, I and F denote the initial and final localized electronic states, respectively) is a crucial determinant of the charge-transfer rate constant.40, 41 In the nonadiabatic limit, at high temperatures (e.g., at room or physiological temperatures), and for slow modulation of the coupling by structural fluctuations,42, 43 the mean square electronic coupling is used to compute the rate. The variance of VIF is used to calculate the coherence parameter44 . C is a descriptor of coupling fluctuations. C = 1 occurs for rigid systems (σ = 0), while C ≅ 0 for very flexible systems with strong dependence of the coupling on the atomic conformation.
We calculated the effective electronic couplings with the formula (which is exact in the two-state approximation)45, 46
| (1) |
In eqn (1), a and b denote the coefficients of the ground state expansion on the diabatic (localized) electronic states, SIF is the overlap of such states and ΔEIF is their energy difference; ΔEv is the vertical energy difference between the adiabatic ground and first-excited states. The first expression in eqn 1 can also be used, with high accuracy,46 near the transition state (where ΔEIF ≪ 2VIF), while it has a removable discontinuity exactly at the transition state coordinates (which, however, are always a zero-measure subspace of the conformational space), where the first expression is easily reduced to the second one. As shown previously,46 eqn (1) enables CDFT states to be used as a suitable basis set to describe charge-transfer systems also in cases where the two CDFT states have a partial charge-transfer character47 that is reflected in a large overlap, as long as the two state approximation is valid (see refs 45, 46 and Supporting Information). The DFT implementation of eqn (1) is also robust with respect to basis set superposition errors.48
The quantities appearing in eqn (1) were obtained from DFT calculations. Constrained DFT49, 50 (as developed in refs. 51, 52 and implemented in the NWChem53 and QChem54 codes) was used to obtain the diabatic states. We used this theoretical approach on the selected MD structural snapshots. Since the Dickerson-Drew dodecamer has a palindromic sequence, we limited the coupling calculations to nearest-neighboring base pairs in the base set highlighted in Fig. 2. In the 5’-to-3’ direction, the base pairs are denoted CG-GC, GC-A1T, A1T-A2T, and A2T-TA. Our analysis first investigated the accuracy of different hybrid DFT approaches to calculate the GC-A1T and A1T-A2T couplings in the DNA MD snapshot at 10 ns. CASSCF and CAS-PT2 calculations (with an active space of 11 electrons in 12 orbitals) provided benchmark values55 in the range 36–46 meV for VG–A (to be approximately compared with the values in the first column of Table 1, which were calculated for the base-pair dimers, that is, in the presence of the complementary bases) and in the range of 2–4 meV for VA–A (cf. second column). Among the hybrid exchange-correlation (XC) functionals tested (Table 1), B3LYP56 and PBE057 (with 20% and 25% of Hartree-Fock (HF) exchange, respectively) give the largest VIF values because of excessive delocalization of the ground-state valence charge, as it was previously found with the double-zeta 6–31g* basis set48. Although the value of an electronic coupling can depend on the nuclear coordinates,48 the comparison with the benchmark coupling values and with the DFT values computed with XC functionals that have larger amounts of HF exchange (as well as a comparison with the ranges of values in Table S1) shows significant overestimation of the electronic couplings using B3LYP and PBE0. Although these XC functionals can produce appropriate optimized molecular geometries, their relatively small HF exchange components prevent accurate description of the ground-state tail on the acceptor nucleobase pair, which determines the value of the electronic coupling. The SOGGA11-X functional (with 40.15% HF exchange)58 produces smaller electronic couplings. Further improvement is achieved by using the range-separated functional CAM-B3LYP (19% and 65% of HF exchange in the short and long range, respectively59) and the Minnesota functionals M05–2X60 (56% of HF exchange), M06–2X61 (54% HF), M08-HX62 (52.23% HF), and M08-SO62 (56.79% HF). The coupling values obtained using these density functionals correlate with the amount of HF-type exchange. The relatively short distance between adjacent nucleobase pairs may explain the suboptimal performance of the CAM-B3LYP functional despite its range separation with a relatively large HF exchange component at long range. The ωB97M-V functional63 performs similarly to CAM-B3LYP for the GC-A1T coupling, while providing an A1T- A2T coupling much closer to the benchmark post-Hartree-Fock value compared to CAM-B3LYP. The BHLYP56, 64 (50% HF) and range-separated LRC-ωPBEh65 (20% and 100% of short and long range HF, respectively) XC functionals (which are suggested as the best performing XC functionals to describe charge transfer based on the error metric defined in ref. 66) lead to a further decrease in the electronic coupling values. However, the best performance (in comparison with the multi-configurational methods) is achieved with the M06-HF61 (100% HF exchange) and range-separated M1167 (42.8% short-range and 100% long-range HF exchange) XC functionals. M06-HF can underestimate the electronic couplings (as found recently by the combined use of constrained DFT and effective Hamiltonian approaches68), although its behavior in the long-range limit is not affected by electron self-interaction errors.
Table 1.
Effective electronic couplings between B-DNA nearest-neighboring base pairs calculated using the 6–311G** basis set and the listed hybrid XC functionals. All values are in meV.
| XC | ||
|---|---|---|
| B3LYP | 604.7 | 197.2 |
| PBE0 | 518.9 | 383.4 |
| SOGGA11-X | 272.4 | 25.1 |
| ωB97M-V | 150.4 | 7.6 |
| CAM-B3LYP | 147.6 | 33.1 |
| LRC-ωPBEh | 126.6 | 40.9 |
| BHLYP | 126.1 | 30.3 |
| M05–2X | 162.8 | 0.2 |
| M06–2X | 177.9 | 2.6 |
| M06-HF | 61.8 | 1.3 |
| M08-HX | 203.8 | 3.1 |
| M08-SO | 167.7 | 2.1 |
| M11 | 78.9 | 4.1 |
Based on the analysis above, we used the M11 functional to calculate the electronic couplings between nearest-neighboring base pairs in the selected MD snapshots of DNA and 2’F-ANA at an affordable computational cost. Nonetheless, based on the values in Table 1, the differences among the results using the ωB97M-V, BHLYP, M06-HF and M11 approximations could be appreciably reduced by averaging the electronic couplings over the MD snapshots, due to considerable cancellation of errors. The values of mean-square electronic coupling and coherence parameter for the selected base pair dimers are reported in Table 2. The individual coupling values in all selected MD snapshots are given in the Supporting Information.
Table 2.
Mean square electronic coupling ⟨VIF2⟩ (in meV2) and coherence parameter C for the indicated DNA and 2’F-ANA base pair dimers.
| base pair dimer |
DNA | 2’F-ANA | ||
|---|---|---|---|---|
| C | C | |||
| CG-GC | 188 | 0.52 | 211 | 0.60 |
| GC-A1T | 8118 | 0.56 | 1242 | 0.70 |
| A1T-A2T | 1681 | 0.65 | 2661 | 0.43 |
| A2T -TA | 1087 | 0.46 | 3941 | 0.25 |
The mean-square electronic couplings for the corresponding base-pair dimers in the two dodecamers are of the same orders of magnitude. The intra-strand GC-A1T coupling is stronger in DNA than in 2’F-ANA, while the converse holds for the inter-strand A2T-TA coupling. Nevertheless, the electronic couplings in 2’F-ANA are larger than the corresponding couplings in DNA for three of the four nucleobase stacks studied. This result indicates enhanced conductivity of 2’F-ANA, compared to DNA, for sequential hole hopping mechanisms, if similar free energy parameters are associated with the charge hopping steps in the two structures. The average values of the coherence parameters for the DNA and 2’F-ANA systems are 0.55 and 0.50, respectively, thus suggesting similar fluctuations of the electronic couplings across their nucleobase π-stacks. Further RMSD analysis (Supporting Information, Fig. S1) finds that the backbones of DNA and 2’F-ANA undergo structural fluctuations of similar magnitude, in reasonable agreement with previous findings.38 The nucleobase fluctuations span a slightly wider range in 2’F-ANA than in DNA and thus enable access to conformations with relatively large electronic coupling values (Figs. S2-S5).
While our current results show similar structural flexibility, and related similar electronic coupling fluctuations, for the DNA and 2’F-ANA duplexes, further theoretical studies need to investigate whether the more negative backbone of 2’F-ANA compared to DNA may lead to appreciable changes in the free energy parameters (reaction free energies and reorganization energies) associated with the hole transfer steps and to partial hole transfer to the backbone (namely, a partial transfer of electron charge to the positively charged bases).
From a methodological point of view, our study supports the use of eqn (1) in conjunction with the M11 Minnesota functional to compute electronic couplings in base-pair dimers (see Table 2). BHLYP and ωB97M-V are the next most accurate density functionals to compute nucleobase electronic couplings based on the results of this study (however, it was shown that the Becke half-and-half approximation can enable very accurate electronic coupling calculation by including diffuse functions in the basis set46).
The computational results of Fig. 3 and Table 2 indicate that the modification of the DNA backbone leading to 2’F-ANA (which yields chemical protection against the external environment) does not enhance the intrinsic structural stability, with an appreciably increased charge transport efficiency of the π-stacking pattern, as quantified in terms of nucleobase pair electronic couplings. Nonetheless, our results indicate that 2’F-ANA combines its greater chemical stability with similar or slightly improved hole conduction compared to DNA (as quantified in Table 2), thereby promoting 2’F-ANA as a potentially better candidate than DNA for applications in (nano)biotechnology.
Supplementary Material
Acknowledgements
We thank Dr. Carlos Gonzalez for his help with retrieving the 2’F-ANA force field parameters, and Prof. Head-Gordon group for useful discussion on the use of the ωB97M-V functional. We acknowledge use of the Duke Compute Cluster and ET Cluster at Duke University, and support of our research by the National Institutes of Health (Grant GM-48043).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available. See DOI: 10.1039/x0xx00000x
References
- 1.Korol R and Segal D, J. Phys. Chem. C, 2018, 122, 4206–4216. [Google Scholar]
- 2.Doye JPK, Ouldridge TE, Louis AA, Romano F, Sulc P, Matek C, Snodin BEK, Rovigatti L, Schreck JS, Harrison RM and Smith WPJ, Phys. Chem. Chem. Phys, 2013, 15, 20395–20414. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen KV, Holade Y and Minteer SD, ACS Catal., 2016, 6, 2603–2607. [Google Scholar]
- 4.Ricci F, Lai RY and Plaxco KW, Chem. Commun, 2007, DOI: 10.1039/B708882E, 3768–3770. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Yan Z, Sun Y, Ren J and Qu X, Chem. Commun, 2017, 53, 6215–6218. [DOI] [PubMed] [Google Scholar]
- 6.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK and Barton JK, Proc. Natl. Acad. Sci. U.S.A, 2009, 106, 15237–15242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, Chazin WJ and Barton JK, Science, 2017, 355, eaag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousmail D, Amrein L, Fakhoury JJ, Fakih HH, Hsu JCC, Panasci L and Sleiman HF, Chem. Sci, 2017, 8, 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS and Mirkin CA, Science, 2006, 312, 1027–1030. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Xiang L, Zhang Y, Zhang P, Beratan DN, Li Y and Tao N, Nat. Chem, 2016, 8, 941. [DOI] [PubMed] [Google Scholar]
- 11.Berlin YA, Kurnikov IV, Beratan D, Ratner MA and Burin AL, in Long-Range Charge Transfer in DNA II, ed. Schuster GB, Springer Berlin Heidelberg, Berlin, Heidelberg, 2004, DOI: 10.1007/b94471, pp. 1–36. [DOI] [Google Scholar]
- 12.Venkatramani R, Keinan S, Balaeff A and Beratan DN, Coord. Chem. Rev, 2011, 255, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis FD, Liu X, Liu J, Miller SE, Hayes RT and Wasielewski MR, Nature, 2000, 406, 51. [DOI] [PubMed] [Google Scholar]
- 14.Lewis FD, Daublain P, Cohen B, Vura-Weis J, Shafirovich V and Wasielewski MR, J. Am. Chem. Soc, 2007, 129, 15130–15131. [DOI] [PubMed] [Google Scholar]
- 15.Lewis FD, Letsinger RL and Wasielewski MR, Acc. Chem. Res, 2001, 34, 159–170. [DOI] [PubMed] [Google Scholar]
- 16.Thazhathveetil AK, Trifonov A, Wasielewski MR and Lewis FD, J. Am. Chem. Soc, 2011, 133, 11485–11487. [DOI] [PubMed] [Google Scholar]
- 17.Kalosakas G and Spanou E, Phys. Chem. Chem. Phys, 2013, 15, 15339–15346. [DOI] [PubMed] [Google Scholar]
- 18.Meggers E, Michel-Beyerle ME and Giese B, J. Am. Chem. Soc, 1998, 120, 12950–12955. [Google Scholar]
- 19.Fujitsuka M and Majima T, Chem. Sci, 2017, 8, 1752–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujitsuka M and Majima T, Phys. Chem. Chem. Phys, 2012, 14, 11234–11244. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu C, Balaeff A, Skourtis SS and Beratan DN, Proc. Natl. Acad. Sci. U.S.A, 2014, 111, 10049–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renaud N, Harris MA, Singh APN, Berlin YA, Ratner MA, Wasielewski MR, Lewis FD and Grozema FC, Nat. Chem, 2016, 8, 1015. [DOI] [PubMed] [Google Scholar]
- 23.Pinheiro VB, Taylor AI, Cozens C, Abramov M, Renders M, Zhang S, Chaput JC, Wengel J, Peak-Chew S-Y, McLaughlin SH, Herdewijn P and Holliger P, Science, 2012, 336, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watts JK, Katolik A, Viladoms J and Damha MJ, Org. Biomol. Chem, 2009, 7, 1904–1910. [DOI] [PubMed] [Google Scholar]
- 25.Noronha AM, Wilds CJ, Lok C-N, Viazovkina K, Arion D, Parniak MA and Damha MJ, Biochemistry, 2000, 39, 7050–7062. [DOI] [PubMed] [Google Scholar]
- 26.Wilds CJ and Damha MJ, Nucleic Acids Res., 2000, 28, 3625–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damha MJ, Wilds CJ, Noronha A, Brukner I, Borkow G, Arion D and Parniak MA, J. Am. Chem. Soc, 1998, 120, 12976–12977. [Google Scholar]
- 28.Kalota A, Karabon L, Swider CR, Viazovkina E, Elzagheid M, Damha MJ and Gewirtz AM, Nucleic Acids Res, 2006, 34, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manallack DT, Prankerd RJ, Yuriev E, Oprea TI and Chalmers DK, Chem. Soc. Rev, 2013, 42, 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akhtar S, Hughes MD, Khan A, Bibby M, Hussain M, Nawaz Q, Double J and Sayyed P, Adv. Drug Deliv. Rev, 2000, 44, 3–21. [DOI] [PubMed] [Google Scholar]
- 31.Souleimanian N, Deleavey GF, Soifer H, Wang S, Tiemann K, Damha MJ and Stein CA, Mol. Ther. Nucleic Acids, 2012, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horny MC, Lazerges M, Siaugue JM, Pallandre A, Rose D, Bedioui F, Deslouis C, Haghiri-Gosnet AM and Gamby J, Lab Chip, 2016, 16, 4373–4381. [DOI] [PubMed] [Google Scholar]
- 33.Wong ELS and Gooding JJ, Anal. Chem, 2006, 78, 2138–2144. [DOI] [PubMed] [Google Scholar]
- 34.Arnold AR, Grodick MA and Barton JK, Cell Chem. Biol, 2016, 23, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beratan DN and Waldeck DH, Nat. Chem, 2016, 8, 992. [DOI] [PubMed] [Google Scholar]
- 36.Farazdel A, Dupuis M, Clementi E and Aviram A, J. Am. Chem. Soc, 1990, 112, 4206–4214. [Google Scholar]
- 37.Lercher L, McDonough MA, El-Sagheer AH, Thalhammer A, Kriaucionis S, Brown T and Schofield CJ, Chem. Commun, 2014, 50, 1794–1796. [DOI] [PubMed] [Google Scholar]
- 38.Martin-Pintado N, Yahyaee-Anzahaee M, Campos-Olivas R, Noronha AM, Wilds CJ, Damha MJ and Gonzalez C, Nucleic Acids Res, 2012, 40, 9329–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW and Klein ML, J. Chem. Phys, 1983, 79, 926. [Google Scholar]
- 40.Marcus RA and Sutin N, Biochim. Biophys. Acta, 1985, 811, 265–322. [Google Scholar]
- 41.Nitzan A, Chemical dynamics in condensed phases, Oxford University Press, Oxford, 2007. [Google Scholar]
- 42.Troisi A, Nitzan A and Ratner MA, J. Chem. Phys, 2003, 119, 5782–5788. [Google Scholar]
- 43.Skourtis SS, Balabin IA, Kawatsu T and Beratan DN, Proc. Natl. Acad. Sci. U.S.A, 2005, 102, 3552–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balabin IA and Onuchic JN, Science, 2000, 290, 114–117. [DOI] [PubMed] [Google Scholar]
- 45.Migliore A, J. Chem. Phys, 2009, 131, 114113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migliore A, Chem J. Theory Comput., 2011, 7, 1712–1725. [DOI] [PubMed] [Google Scholar]
- 47.Mao Y, Ge Q, Horn PR and Head-Gordon M, J. Chem. Theory Comput, 2018, 14, 2401–2417. [DOI] [PubMed] [Google Scholar]
- 48.Migliore A, Corni S, Varsano D, Klein ML and Di Felice R, J Phys Chem B, 2009, 113, 9402–9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dederichs PH, Blügel S, Zeller R and Akai H, Phys. Rev. Lett, 1984, 53, 2512–2515. [DOI] [PubMed] [Google Scholar]
- 50.Wesolowski TA and Warshel A, J Phys Chem-Us, 1993, 97, 8050–8053. [Google Scholar]
- 51.Wu Q and Van Voorhis T, Phys. Rev. A, 2005, 72, 024502. [Google Scholar]
- 52.Wu Q and Van Voorhis T, J. Chem. Phys, 2006, 125, 164105. [DOI] [PubMed] [Google Scholar]
- 53.Valiev M, Bylaska EJ, Govind N, Kowalski K, Straatsma TP, Van Dam HJJ, Wang D, Nieplocha J, Apra E, Windus TL and de Jong WA, Comput. Phys. Commun, 2010, 181, 1477–1489. [Google Scholar]
- 54.Shao Y, Gan Z, Epifanovsky E, Gilbert ATB, Wormit M, Kussmann J, Lange AW, Behn A, Deng J, Feng X, Ghosh D, Goldey M, Horn PR, Jacobson LD, Kaliman I, Khaliullin RZ, Kuś T, Landau A, Liu J, Proynov EI, Rhee YM, Richard RM, Rohrdanz MA, Steele RP, Sundstrom EJ, Woodcock HL, Zimmerman PM, Zuev D, Albrecht B, Alguire E, Austin B, Beran GJO, Bernard YA, Berquist E, Brandhorst K, Bravaya KB, Brown ST, Casanova D, Chang C-M, Chen Y, Chien SH, Closser KD, Crittenden DL, Diedenhofen M, DiStasio RA, Do H, Dutoi AD, Edgar RG, Fatehi S, Fusti-Molnar L, Ghysels A, Golubeva-Zadorozhnaya A, Gomes J, Hanson-Heine MWD, Harbach PHP, Hauser AW, Hohenstein EG, Holden ZC, Jagau T-C, Ji H, Kaduk B, Khistyaev K, Kim J, Kim J, King RA, Klunzinger P, Kosenkov D, Kowalczyk T, Krauter CM, Lao KU, Laurent AD, Lawler KV, Levchenko SV, Lin CY, Liu F, Livshits E, Lochan RC, Luenser A, Manohar P, Manzer SF, Mao S-P, Mardirossian N, Marenich AV, Maurer SA, Mayhall NJ, Neuscamman E, Oana CM, Olivares-Amaya R, O’Neill DP, Parkhill JA, Perrine TM, Peverati R, Prociuk A, Rehn DR, Rosta E, Russ NJ, Sharada SM, Sharma S, Small DW, Sodt A, Stein T, Stück D, Su Y-C, Thom AJW, Tsuchimochi T, Vanovschi V, Vogt L, Vydrov O, Wang T, Watson MA, Wenzel J, White A, Williams CF, Yang J, Yeganeh S, Yost SR, You Z-Q, Zhang IY, Zhang X, Zhao Y, Brooks BR, Chan GKL, Chipman DM, Cramer CJ, Goddard WA, Gordon MS, Hehre WJ, Klamt A, Schaefer HF, Schmidt MW, Sherrill CD, Truhlar DG, Warshel A, Xu X, Aspuru-Guzik A, Baer R, Bell AT, Besley NA, Chai J-D, Dreuw A, Dunietz BD, Furlani TR, Gwaltney SR, Hsu C-P, Jung Y, Kong J, Lambrecht DS, Liang W, Ochsenfeld C, Rassolov VA, Slipchenko LV, Subotnik JE, Van Voorhis T, Herbert JM, Krylov AI, Gill PMW and Head-Gordon M, Mol. Phys, 2015, 113, 184–215. [Google Scholar]
- 55.Blancafort L and Voityuk AA, J. Phys. Chem. A, 2006, 110, 6426–6432. [DOI] [PubMed] [Google Scholar]
- 56.Becke AD, J. Chem. Phys, 1993, 98, 5648–5652. [Google Scholar]
- 57.Adamo C and Barone V, J. Chem. Phys, 1999, 110, 6158–6170. [Google Scholar]
- 58.Peverati R and Truhlar DG, J. Chem. Phys, 2011, 135, 191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanai T, Tew DP and Handy NC, Chem. Phys. Lett, 2004, 393, 51–57. [Google Scholar]
- 60.Zhao Y, Schultz NE and Truhlar DG, J. Chem. Theory Comput, 2006, 2, 364–382. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y and Truhlar DG, Theor Chem Acc, 2008, 120, 215–241. [Google Scholar]
- 62.Zhao Y and Truhlar DG, J. Chem. Theory Comput, 2008, 4, 1849–1868. [DOI] [PubMed] [Google Scholar]
- 63.Mardirossian N and Head-Gordon M, J. Chem. Phys, 2016, 144, 214110. [DOI] [PubMed] [Google Scholar]
- 64.Becke AD, J. Chem. Phys, 1993, 98, 1372–1377. [Google Scholar]
- 65.Rohrdanz MA, Martins KM and Herbert JM, J. Chem. Phys, 2009, 130, 054112. [DOI] [PubMed] [Google Scholar]
- 66.Kim H, Goodson T and Zimmerman PM, J Phys Chem Lett, 2017, 8, 3242–3248. [DOI] [PubMed] [Google Scholar]
- 67.Peverati R and Truhlar DG, J. Phys. Chem. Lett, 2011, 2, 2810–2817. [Google Scholar]
- 68.Gillet N, Berstis L, Wu XJ, Gajdos F, Heck A, de la Lande A, Blumberger J and Elstner M, J. Chem. Theory Comput, 2016, 12, 4793–4805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.