Abstract
Probiotic nutrition is frequently claimed to improve human health. In particular, live probiotic bacteria obtained with food are believed to reduce pathogen colonization and thus, susceptibility to infection. However, the underlying mechanisms remain poorly understood. Here, we report that the consumption of probiotic Bacillus bacteria comprehensively abolishes colonization with the dangerous pathogen, Staphylococcus aureus. We discovered that the widespread fengycin class of Bacillus lipopeptides achieves colonization resistance by inhibiting the S. aureus Agr quorum-sensing signaling system. Our study presents a detailed molecular mechanism underlining the importance of probiotic nutrition in reducing infectious disease. Notably, we provide human evidence supporting the biological significance of probiotic bacterial interference and show for the first time that such interference can be achieved by blocking a pathogen’s signaling system. Furthermore, our findings suggest a probiotic-based method for S. aureus decolonization and new ways to fight S. aureus infections.
There is increasing appreciation of the key role that the intestinal microbiome plays in preventing the colonization and overgrowth of pathogens1,2. The mechanisms that have been implicated in this beneficial function of probiotic bacteria are mostly indirect and include modulation of the immune system, enhancement of the intestinal epithelial barrier, or competition with pathogens for nutrients2–5. Whether there is direct interference between probiotic and pathogenic bacteria is less clear. Some probiotic strains produce bacteriocins, which can kill phylogenetically related pathogenic bacteria2, and it has been shown that a bacteriocin-producing Escherichia coli strain inhibited colonization with related pathogenic bacteria in the inflamed gut of mice6. However, no evidence has been obtained to indicate that such mechanisms matter or are widespread in humans. Furthermore, it is not known whether there are ways of direct probiotic bacterial interference that are not mediated by bacteriocins.
The genus Bacillus comprises different species of soil bacteria that form endospores with the ability to survive harsh environmental conditions, such as the high temperatures encountered during cooking procedures. Bacillus spores are commonly ingested with vegetables7. They can subsequently germinate to form metabolically active, vegetative cells8, which can temporarily colonize the intestinal tract9. Given the dependence on dietary customs, the concentration of Bacillus spores in human feces is highly variable. It has been reported to be ~ 105 CFU/g in average, occasionally reaching up to 108 CFU/g7. Several probiotic formulae contain Bacillus species10, which are believed to reduce pathogen colonization by mechanisms that - except for a described immune-stimulatory effect on epithelial cells11 - remain poorly defined.
Staphylococcus aureus is a widespread and dangerous human pathogen that can cause a variety of diseases, ranging from moderately severe skin infections to fatal pneumonia and sepsis12. Treatment of S. aureus infections is severely complicated by antibiotic resistance13, such as in methicillin-resistant S. aureus (MRSA), and there is no working S. aureus vaccine14. Therefore, alternative strategies to combat S. aureus infections are eagerly sought15. Because S. aureus infections commonly originate from previous asymptomatic colonization16,17, decolonization has recently gained considerable attention as a possible means to fight S. aureus infections in a preventive manner18. While the nares have traditionally been considered the primary S. aureus colonization site19, there is increasing evidence indicating that the intestinal tract is also commonly colonized by S. aureus20–22 and forms an important reservoir for S. aureus infectious disease outbreaks23,24. Several studies have reported levels of S. aureus in the feces of human adults of ~ 103 - 104 CFU/g25–27. Possibly, intestinal S. aureus colonization explains the failure of previous topical decolonization efforts aimed solely at the nose16,22,28.
Here, we hypothesized that the composition of the human gut microbiota affects intestinal colonization with S. aureus. To evaluate that hypothesis, we collected fecal samples from 200 healthy individuals from rural populations in Thailand (Fig. 1a). This exemplary population was selected to rule out, as much as possible, food sterilization and antibiotic presence as used frequently in highly developed urban areas, which potentially could diminish the abundance of probiotic bacteria in the food and the intestinal tract of the participating subjects. Our analysis revealed a comprehensive Bacillus-mediated S. aureus exclusion effect in the human population. By demonstrating that quorum-sensing is indispensable for S. aureus intestinal colonization and discovering that secreted Bacillus fengycin lipopeptides function as quorum-sensing blockers to achieve complete eradication of intestinal S. aureus, we provide evidence strongly suggesting that this pathogen exclusion effect in humans is due to a widespread and efficient probiotic-mediated mechanism that inhibits pathogen quorum-sensing signaling.
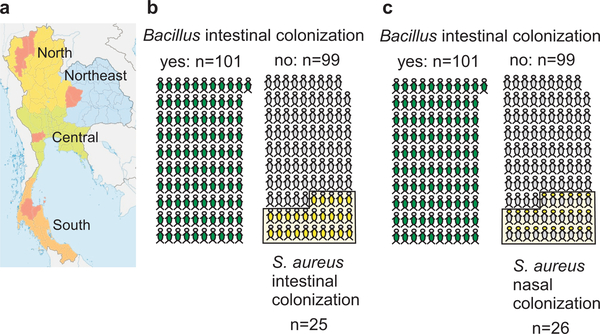
Figure 1 |. S. aureus colonization exclusion by dietary Bacillus in a human population.
a, Areas (in red) from which fecal samples were collected in rural populations and analyzed for presence of Bacillus and S. aureus. b,c, Intestinal (b) and nasal (c) colonization with S. aureus (yellow) in individuals that showed (green) or did not show (grey) intestinal colonization with Bacillus.
S. aureus exclusion by Bacillus
S. aureus intestinal carriage as determined by growth from fecal samples was found in 25/200 (12.5%) of human subjects. Nasal carriage was similar in frequency (26/200; 13%), a result that is in accordance with previous findings showing a correlation between nasal and intestinal colonization22. These rates are considerably lower than those commonly found in adult populations during cross-sectional culture-based surveys that were mainly performed in hospital-admitted individuals in urbanized areas (on average, ~ 20% for intestinal and ~ 40% for nasal carriage)16,21,22.
To examine the hypothesis that bacterial interactions in the gut determine intestinal S. aureus colonization, we first analyzed the composition of the gut microbiome by 16S rRNA sequencing. However, we did not detect significant differences in the composition of the microbiome between S. aureus carriers and non-carriers (Extended Data Fig. 1).
In contrast, we found a striking correlation between the presence of Bacillus bacteria and absence of S. aureus. Bacillus species (mostly B. subtilis, Extended Data Fig. 1) were found in 101/200 (50.5%) of subject samples. S. aureus was never detected in fecal samples when Bacillus species were present (p<0.0001, Fisher’s exact test) (Fig. 1b). Furthermore, this pathogen exclusion effect was not limited to the site of interaction, the gut, but extended to S. aureus colonization in a general fashion. While Bacillus was generally absent from nasal samples, S. aureus nasal colonization was never detected in the presence of intestinal Bacillus (p<0.0001, Fisher’s exact test) (Fig. 1c). Interestingly, the levels of S. aureus colonization we found in non-Bacillus-colonized individuals from rural Thailand approximately match those reported, using similar culture-based assays, in urbanized Western areas. These findings indicate a widespread mechanism exerted by Bacillus species that comprehensively inhibits colonization with S. aureus. Moreover, they suggest that S. aureus colonization is increased in urban populations due to the lack of a probiotic, Bacillus-containing diet. Intriguingly, they also imply that the intestinal site plays a previously underappreciated role in determining general S. aureus colonization, a notion in accordance with findings attributing a key role to fecal transmission in MRSA re-colonization28.
When we analyzed data from previous 16S rRNA-sequencing-based microbiome studies, we found strongly variant results and no correlation between the absence of S. aureus and the presence of B. subtilis: Studies that reported considerable B. subtilis or S. aureus numbers (samples with colonization by either species > 10%) did not reveal exclusion phenomena (average 14.89 ± 15.69% colonization by both species) (Extended Data Fig. 2). While we did not find a correlation, we feel this may be due to the fact that such sequencing-based analyses are set up to detect high-order taxonomic shifts rather than specific differences on the species or genus level.
Quorum-sensing and colonization
Our results showing no significant high-order taxonomic differences in the microbiome composition between S. aureus carriers and non-carriers exclude an indirect effect of Bacillus on the microbiome composition. Rather, we hypothesized that the Bacillus isolates produce a substance that directly and specifically inhibits S. aureus intestinal colonization. We first analyzed whether there is a growth-inhibitory effect of the Bacillus isolates on S. aureus. However, only a minor growth inhibition occurred in 6 out of 105 isolates (maximally 1 mm inhibition zone when analyzed using an agar diffusion test with five-times concentrated culture filtrate). This fails to explain the observed complete correlation between the presence of Bacillus and absence of S. aureus, and rules out a bacteriocin-mediated phenomenon.
It is poorly understood which factors are important for S. aureus intestinal colonization. Only one mouse study implicated wall teichoic acids and the surface protein, clumping factor A (ClfA)29. Prompted by our previous finding that ClfA is positively regulated by the Agr quorum-sensing system30, we hypothesized that the Bacillus isolates secrete a substance that interferes with quorum-sensing signaling. Quorum-sensing is responsible for sensing the density of the bacterial population (“quorum”) and controlling the concomitant alteration of cell physiology31. As quorum-sensing signals and sensors differ between different types of bacteria31, an underlying quorum-quenching mechanism would explain the specificity of the inhibitory effect that we detected.
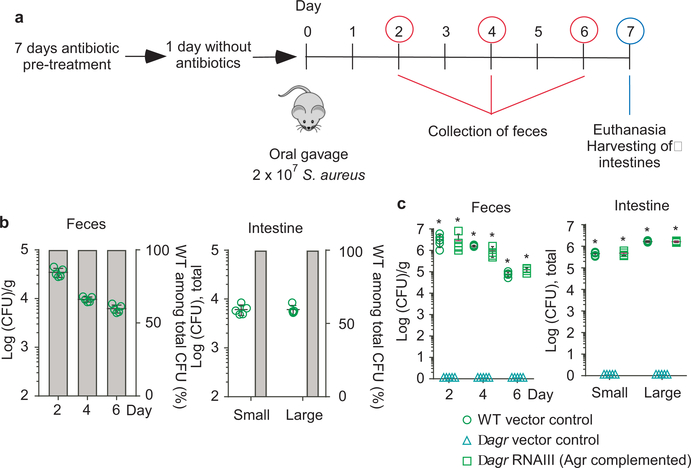
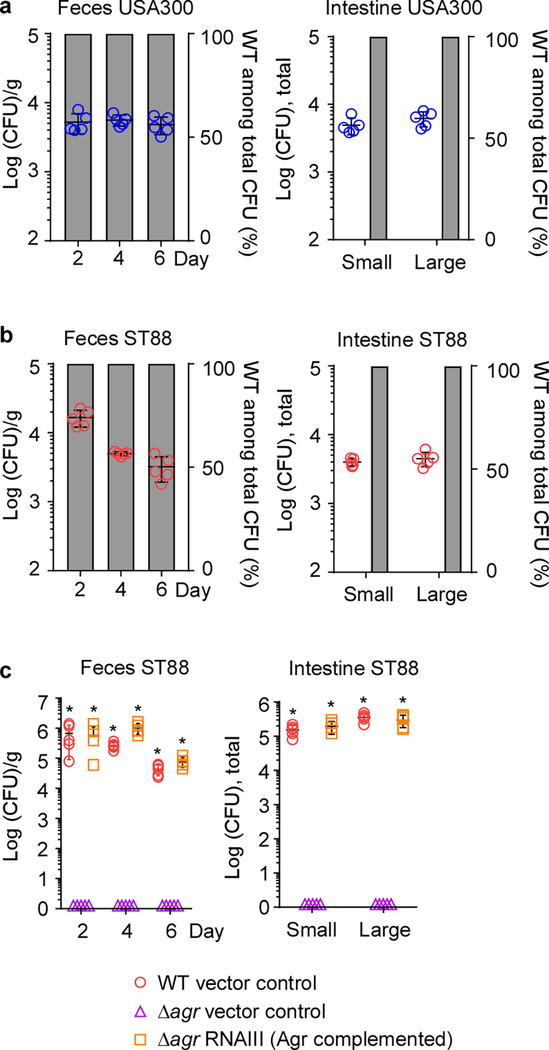
Because the role of quorum-sensing in S. aureus intestinal colonization is unknown, we first used a mouse model of S. aureus intestinal colonization to test whether Agr quorum-sensing plays a role (Fig. 2a). In all mouse models in our study, we included (i) a human fecal isolate belonging to a sequence type (ST) frequently detected in the obtained fecal isolates (ST2196), according to multi-locus sequence typing (MLST) we performed (Supplementary Tab. 1), (ii) a mouse infection isolate (ST88)32, and (iii) a human infection isolate of the highly virulent MRSA type USA30033. In competition experiments with equal amounts of wild-type and isogenic agr mutant strains, only wild-type S. aureus were detected in the feces and colonized the large and small intestines at the end of the experiment (competition index ≥ 100) (Fig. 2b, Extended Data Fig. 2 a,b). Furthermore, in a non-competitive experimental setup, only those bacteria expressing the intracellular Agr effector RNAIII34 achieved colonization, while agr-negative control strains never did (Fig. 2c, Extended Data Fig. 2c). These data show that in addition to its well-known role in infection30,35, the Agr quorum-sensing system is absolutely indispensable for intestinal colonization.
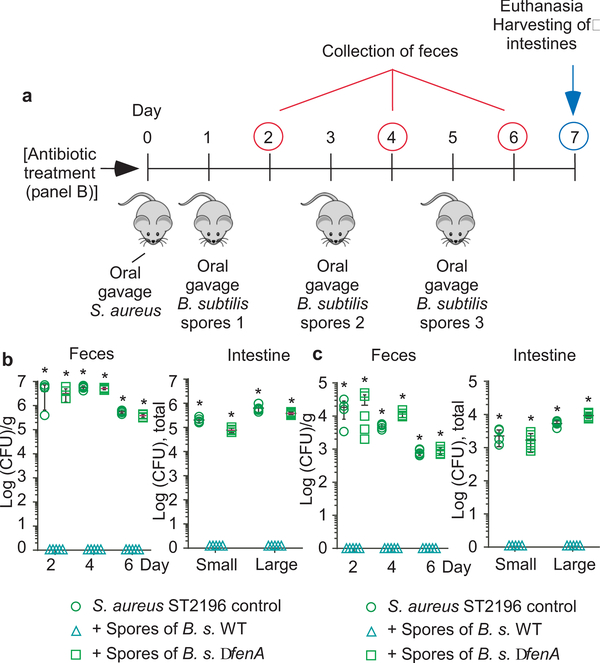
Figure 2 |. Quorum-sensing dependence of S. aureus intestinal colonization.
a, Experimental setup of the murine intestinal colonization model. Mice received by oral gavage 100 μl containing 108 CFU/ml of wild-type (WT) S. aureus strain ST2196 F12 and another 100 μl of 108 CFU/ml of the corresponding isogenic agr mutant (n=5/group, competitive experiment shown in b), or 200 μl containing 108 CFU/ml wild-type, isogenic agr mutant, or Agr (RNAIII)-complemented agr mutant (n=5/group, non-competitive experiment shown in c). CFU in the feces were determined two, four, and six days after infection. At the end of the experiment (day seven), CFU in the small and large intestines were determined. b, Competitive experiment. Total obtained CFU are shown as dot plots, also showing the means ±SD. Bars show the percentage of wild-type among total determined CFU, of which 100 were analyzed for tetracycline resistance that is present only in the agr mutant. No agr mutants were detected in any experiment; thus, all bars show 100%. Given that 100 isolates were tested, the competitive index wild-type/agr mutant in all cases is ≥ 100. c, Non-competitive experiment with genetically complemented strains. Wild-type and isogenic agr mutant strains all harbored the pKXΔ16 control plasmid; Agr-complemented strains harbored pKXΔRNAIII, constitutively expressing RNAIII, which is the intracellular effector of Agr. During the experiment, mice received 200 μg/ml kanamycin in the drinking water to maintain plasmids. Statistical analysis was performed using Poisson regression versus values obtained with the agr mutant strains. *, p<0.0001. Error bars shown the means ±SD. Note no bacteria were found in the feces or intestines of any mouse receiving S. aureus Δagr with vector control. The corresponding zero values are plotted on the x-axis of the logarithmic scale. See Extended Data Fig. 2 for the corresponding data using strains USA300 LAC and ST88 JSNZ.
Fengycin quorum-quenchers
Having established that the Agr quorum-sensing regulatory system is essential for S. aureus intestinal colonization, we analyzed whether culture filtrates of the Bacillus isolates collected from human feces can inhibit Agr. To that end, we used an S. aureus reporter strain, in which we had transferred the luminescence-conferring luxABCDE operon into the genome under control of the Agr P3 promoter34, which controls production of RNAIII. Remarkably, culture filtrates from all 105 isolates reduced Agr activity in that assay by at least 80% (Fig. 3a, Extended Data Tab. 1). No growth effects were observed, substantiating that growth inhibition does not underlie the inhibitory phenotype. Furthermore, culture filtrate from a reference B. subtilis strain suppressed the production of key Agr-regulated virulence factors (phenol-soluble modulins, α-toxin, and Panton-Valentine leukocidin) (Fig. 3b,c, Supplementary Fig. 1 ). These results indicated that the inhibitory effect of the Bacillus isolates on S. aureus colonization is due to a secreted substance that inhibits Agr signaling.
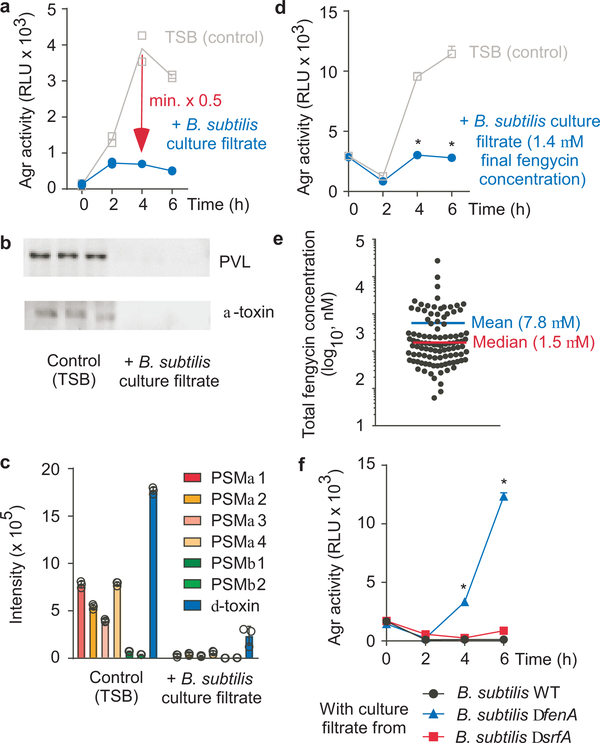
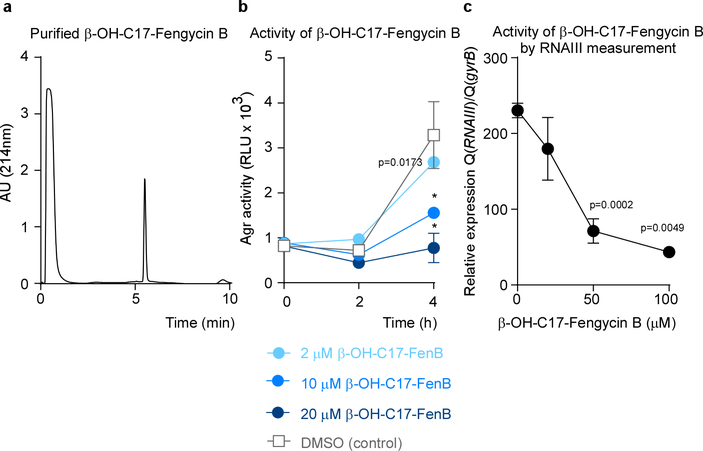
Figure 3 |. S. aureus quorum-sensing inhibition by Bacillus fengycin lipopeptides.
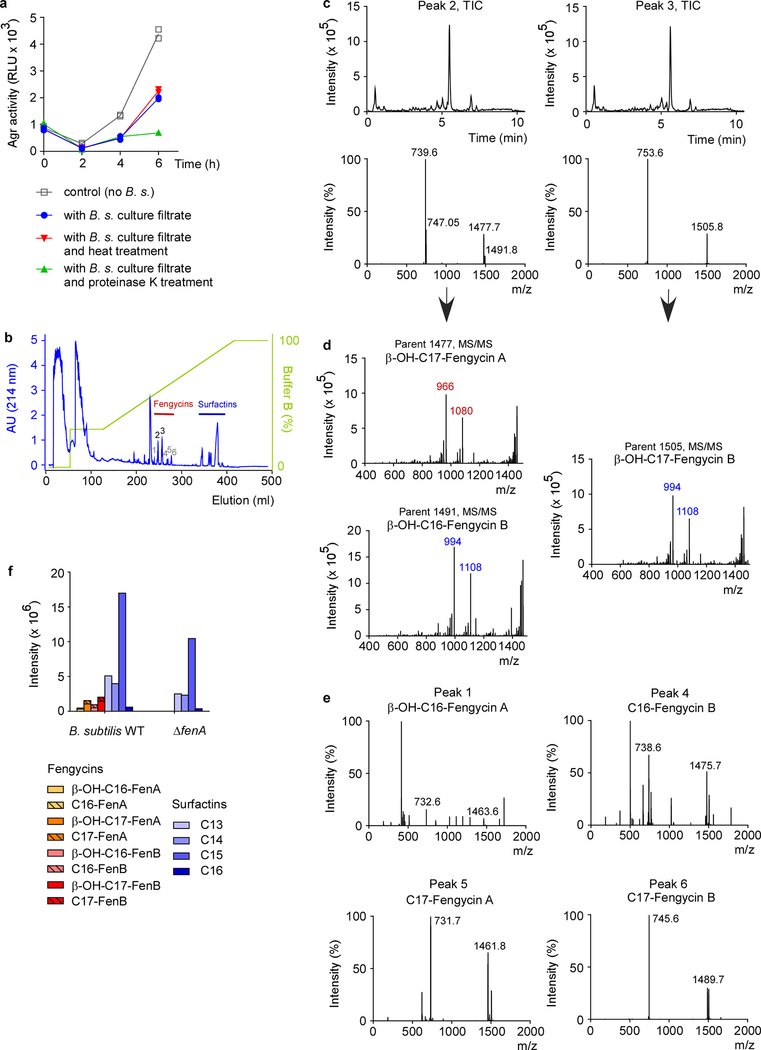
a, Example of an Agr inhibition experiment. The Bacillus isolate was considered inhibitory if luminescence after 4 h growth of S. aureus was ≤ 0.5 times that of the control value. RLU, relative light units. The experiment was performed with n=2 biologically independent samples. The lines connect the means. b, Inhibition of expression of PVL and α-toxin, using culture filtrate of the B. subtilis reference strain. Western blot analysis of n=3 biologically independent samples was performed with S. aureus culture filtrates grown for 4 h. See Supplementary Fig. 1 for the entire blots. c, Inhibition of expression of PSM toxins using culture filtrate of the B. subtilis standard strain. PSM expression was determined by RP-HPLC/ESI-MS after 4 h of growth. d, Test for inhibitory capacity of Bacillus culture filtrate applied to a final concentration representing the median concentration of total fengycin in the tested 106 Bacillus isolates. *, p<0.0001 (2-way ANOVA with Tukey’s post-test versus control). e, Total fengycin concentrations in stationary-phase culture filtrates of the 106 Bacillus isolates (see Extended Data Table 1 for details). f, Agr-inhibiting activities of B. subtilis wild-type (WT) in comparison to ΔfenA (fengycin-deficient) and ΔsrfA (surfactin-deficient) strains. *, p<0.0001 (2-way ANOVA with Tukey’s post-test versus WT). c,d,f, The experiments were performed with n=3 biologically independent samples. Error bars shown the means ±SD.
To characterize the Agr-inhibitory substance, we performed experiments with culture filtrate of the reference B. subtilis strain. The substance in question was thermostable and resistant to protease digestion (Extended Data Fig. 3 a). In reversed-phase high performance chromatography (RP-HPLC) (Extended Data Fig. 3b), significant Agr-inhibiting activity was associated with two peaks, which we then analyzed by RP-HPLC/electrospray ionization mass spectrometry (ESI-MS) (Extended Data Fig. 3c). This analysis, together with the elution behavior and published literature36, allowed us to identify the substances as members of the fengycin cyclic lipopeptide family. Because fengycins can differ in specific amino acids in addition to the length of the attached fatty acid, which usually is β-hydroxylated (β-OH), and different Bacillus strains produce somewhat different fengycin species37, we used further tandem mass spectrometric fragmentation analysis (MS/MS) to identify the specific fengycins present in the two active peaks (Extended Data Fig. 3d). Fengycins in the first peak were identified as β-OH-C17-fengycin A and β-OH-C16-fengycin B. The second peak consisted of one fengycin species, β-OH-C17-fengycin B. Smaller, adjacent peaks also contained fengycin species according to RP-HPLC/ESI-MS analysis, which we tentatively identified as β-OH-C17-fengycin A and the de-hydroxylated versions of the identified three major fengycins (Extended Data Fig. 3e). For further analyses, higher amounts of β-OH-C17-fengycin B were purified to homogeneity from culture filtrate and dose-dependent Agr-inhibiting activity of this pure substance was verified (Extended Data Fig. 4).
Using RP-HPLC/ESI-MS analysis, we found fengycin production in all isolates, substantiating the general character of the inhibitory interaction (Extended Data Tab. 1). While the production pattern of different fengycins varied in the analyzed isolates, in many of them β-OH-C17-fengycin B was the most strongly produced type. Notably, almost complete inhibition of Agr was detected at a concentration of ~ 1.4 μM total fengycin (Fig. 3d). This corresponds to the median concentration of total fengycin produced by stationary phase-cultures of the Bacillus isolates (1.5 μM) (Fig. 3e).
To provide definitive evidence that fengycin production underlies the Agr-inhibiting capacity of Bacillus, we produced an isogenic mutant in the reference B. subtilis strain of the fenA gene, which is essential for fengycin production38. RP-HPLC/ESI-MS showed specific absence of fengycins in that mutant strain, while surfactins, the predominant Bacillus lipopeptides, were still present (Extended Data Fig. 3f). Culture filtrate of the fenA mutant strain was devoid of Agr-inhibiting activity in contrast to that of the isogenic wild-type strain (Fig. 3f). We also measured an isogenic surfactin-negative mutant strain, which showed Agr-inhibiting activity similar to the wild-type strain (Fig. 3f). These results confirmed that fengycin production is the source of the observed Agr inhibition.
Mechanism of fengycin inhibition
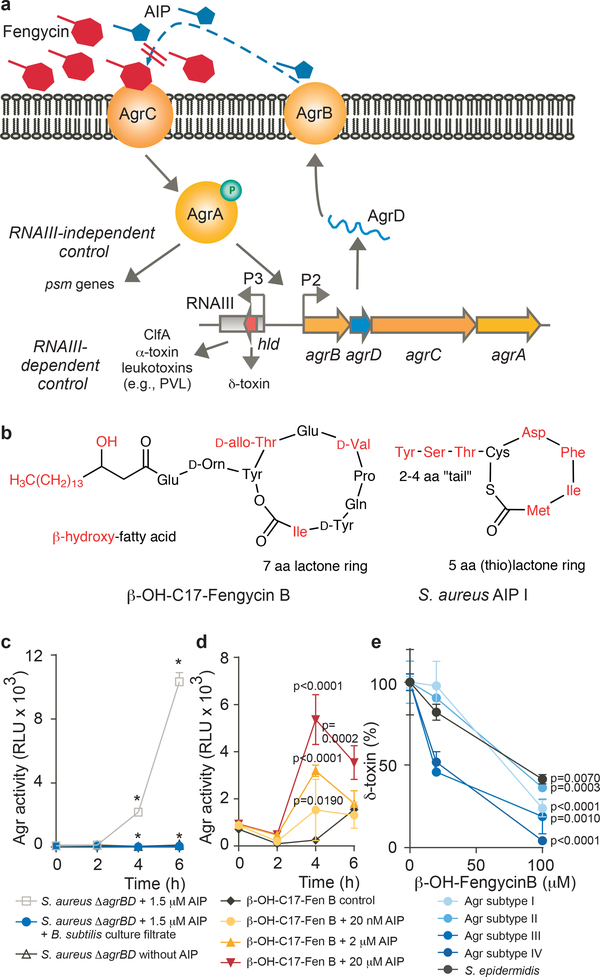
In the S. aureus Agr quorum-sensing regulatory circuit, the secreted Agr autoinducing-peptide (AIP) interacts with an extracellular domain of AgrC, the histidine kinase part of a two-component signal transduction system, to signal the cell density status (Fig. 4a)39. Different Agr subgroups of S. aureus, as well as different staphylococcal species, produce cyclic hepta- to nonapeptide AIPs35. AIPs from different subgroups or species frequently inhibit Agr signal transduction by competitive inhibition at the AgrC binding site39–41. Given that fengycins as cyclic lipopeptides show structural similarity to AIPs (Fig. 4b), it appears likely that fengycins compete with the natural AIP for AgrC binding. The only other theoretically possible site of interference from the extracellular space would be the membrane-located AIP production/secretion enzyme AgrB. Using an S. aureus agrBD deletion strain and stimulation of AgrC by synthetic AIP, which led to complete Agr inhibition, we ruled out that the target of Agr inhibition by Bacillus is AgrB (Fig. 4c). In further support of a mechanism that works by competition with AIP for binding to the AgrC receptor, fengycin inhibition could be reversed in a dose-dependent fashion by addition of AIP (Fig. 4d). Finally, we determined the AIP concentration in early stationary growth phase (6–8 h) to be ~ 1 μM (Extended Data Fig. 5a), which is approximately equal to the concentration of fengycin for which we found complete Agr inhibition (Fig. 3d). These findings indicate that fengycins inhibit Agr signal transduction by efficient competitive inhibition as structural analogues of AIPs.
Figure 4 |. Competitive inhibition of S. aureus AIP activity by fengycins.
a, Model of competitive Agr inhibition by fengycins. The agrBDCA operon, whose expression is driven by the P2 promoter, encodes the AgrD precursor of the autoinducing peptide (AIP), which is modified and secreted by AgrB. AIP binds to membrane-located AgrC, which upon auto-phosphorylation triggers phosphorylation and activation of the DNA-binding protein, AgrA. In addition to stimulating transcription from the P2 promoter (auto-induction), AgrA drives expression of RNAIII, which in turn regulates expression of target genes. RNAIII also encodes the δ-toxin. Furthermore, AgrA drives PSM expression in an RNAIII-independent fashion. b, Structural similarity of fengycins with AIPs. The structures of β-OH-C17-Fengycin B and AIP-I are shown as examples. In red are structures/amino acids that may differ in different subtypes. c, Fengycins work by inhibition of AgrC. Shown is the Agr inhibition by fengycin-containing Bacillus culture filtrate using an agrBD-deleted strain in which AgrC was stimulated by exogenously added AIP. * p<0.0001 [2-way ANOVA with Tukey’s post-test: Values obtained in ΔagrBD/AIP versus ΔagrBD/control (no AIP) and ΔagrBD/AIP/culture filtrate versus ΔagrBD/AIP]. d, Competitive titration of fengycin-mediated Agr inhibition by increasing amounts of AIP as assayed by the Agr luminescence assay. RLU, relative light units. Statistical analysis is by 2-way ANOVA with Tukey’s post-test versus control. e, Inhibition of Agr in different Agr subtype S. aureus and S. epidermidis (strain 1457) by β-OH-C17-Fengycin B as measured by relative expression of δ-toxin via RP/HPLC-MS. Statistical analysis is by 2-way ANOVA with Tukey’s post-test versus intensity values obtained without addition of fengycin. Values were calculated as percentage relative to intensity values obtained without addition of fengycin, due to different δ-toxin expression levels in the different strains. c-e, The experiments were performed with n=3 biologically independent samples. Error bars shown the means ±SD.
The fact that AgrC-AIP interaction differs by Agr subtype raises the question whether fengycins have general ability to inhibit Agr. We found that purified β-OH-C-17 fengycin B inhibited Agr in members of all S. aureus Agr subtypes, as well as in S. epidermidis (Fig. 4e). Furthermore, the S. aureus strains used in the mouse experiments belong to different Agr subtypes (USA300, type I, ST88, type III, ST2196, type I). These results indicate that fengycins have broad-spectrum Agr-inhibiting activity.
Bacillus spores eradicate S. aureus
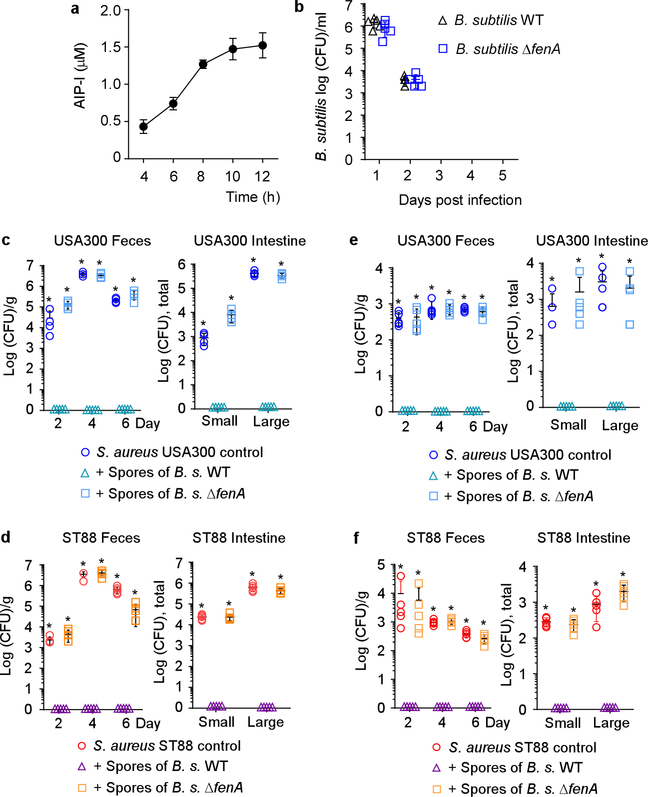
To validate our findings in vivo and demonstrate the specific role of fengycins in the inhibition of S. aureus intestinal colonization, we compared the impact of the B. subtilis wild-type reference strain and its isogenic fenA mutant on S. aureus colonization in a mouse intestinal colonization model. We first performed a control experiment analyzing the colonization kinetics of B. subtilis when given as spores, which corresponds to the form Bacillus would be taken up with food or probiotic formulae (Extended Data Fig. 5b). We observed transient colonization that strongly declined within two days. Importantly, colonization by the fenA mutant was not different from that by the B. subtilis wild-type strain, ruling out that fengycin production per se impacts B. subtilis colonization.
Feeding mice B. subtilis spores completely abrogated colonization of all tested S. aureus strains in the feces and intestines, in experimental setups with or without antibiotic pre-treatment to eliminate the pre-existing microbiota. (Fig. 5b,c, Extended Data Fig. 5c-f). In contrast, spores of the fenA mutant had no significant effect on colonization of any S. aureus test strain. As Bacillus intestinal colonization in humans has been shown to reach much higher levels than that by S. aureus7, a situation highly likely to be even more pronounced in the tested rural population, our mouse data obtained with S. aureus numbers approximately equaling or exceeding those of applied Bacillus spores suggest that fengycin-mediated quorum-sensing interference contributes to the S. aureus colonization exclusion we observed in humans.
Figure 5 |. Inhibition of S. aureus colonization by dietary fengycin-producing Bacillus spores in a mouse model.
a, Experimental setup. n=5 mice/group received 200 μl of 108 CFU/ml S. aureus strain ST2196 F12 by oral gavage. On the next and every following second day, they received 200 μl of 108 CFU/ml spores of the B. subtilis wild-type (WT) or its isogenic fenA mutant, also by oral gavage. CFU in the feces were determined two, four, and six days after infection. At the end of the experiment (day seven), CFU in the small and large intestines were determined. The experiment was performed with (b) or without (c) antibiotic pre-treatment. b,c Results. Statistical analysis was performed using Poisson regression versus values obtained with the B. subtilis WT spore samples. *, p<0.0001. Error bars shown the means ±SD. Note no S. aureus were found in the feces or intestines of any mouse challenged with any S. aureus strain receiving Bacillus wild-type spores. The corresponding zero values are plotted on the x-axis of the logarithmic scale. See Extended Data Fig. 5 for the corresponding data using strains USA300 LAC and ST88 JSNZ.
Conclusions
Scientific evidence to support the frequent claims that probiotic nutrients improve human health is scarce. This study provides evidence for a molecular mechanism of probiotic food bacteria-mediated direct interference that limits pathogen colonization. In particular, our data underscore the often-debated10,42 probiotic value of B. subtilis. Interestingly, we found the responsible agents to work by quorum-quenching, demonstrating that pathogen exclusion in the gut may work by inhibition of a pathogen signaling system. Furthermore, our findings emphasize the importance of quorum-sensing for pathogen colonization.
Our study suggests several highly valuable translational applications regarding alternative strategies to combat antibiotic-resistant S. aureus. First, the quorum-quenching fengycins, which previously had only been known for their anti-fungal activities43, may be used as quorum-sensing blockers for eagerly sought antivirulence-based efforts to treat staphylococcal infections15,44. Second, Bacillus-containing probiotics may be used for simple and safe S. aureus decolonization strategies. In that regard, it is particularly noteworthy that our human data indicate that probiotic Bacillus can comprehensively eradicate intestinal as well as nasal S. aureus colonization. Such a probiotic approach would have multiple advantages over the current standard topical strategy with antibiotics, which is aimed exclusively at decolonizing the nose45.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment, except for when noted.
Sample collection and bacterial screening
Nasal swabs and fecal samples were obtained from 200 Thai healthy volunteers from four different locations in southern, central, northeastern, and northern Thailand. One sterile nasal swab, a sample collection tube, a sterile container, and tissue paper were given to each participant. All participants provided informed written consent. The study was performed in compliance with all relevant ethical regulations and approved by the Siriraj Institutional Review Board (approval no. Si 733/2015). All participants were over 20 years old (age range: 20–87, median 57±14.5; 131 women and 69 men) and without history of intestinal disease. None had received any antibiotic treatment or stayed at a hospital within at least three months prior to the study. Nasal swabs and fecal samples were streaked on mannitol salt agar (MSA) and then incubated at 37 °C for 24 hours. Positive or negative S. aureus or Bacillus colonization could easily be distinguished by either strong growth on the entire plate, or absence of any colonies, respectively. At the time of this analysis, the purpose of the analysis was to obtain and archive colonizing S. aureus strains. As the hypothesis on Bacillus-S. aureus exclusion was only developed from the results of this analysis, the staff performing the analysis were blinded as for the exclusion hypothesis. Isolates were easily recognized as S. aureus or Bacillus by colony morphology and color; however, every isolate was confirmed for species identity using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (see below), and Bacillus species were also further distinguished by 16S rRNA sequencing (Extended Data Tab. 1). To that end, 16S rRNA genes were PCR-amplified using primers 27FB and 1492RAB46 and BLAST similarity analysis was used to identify the species. Subjects were considered as permanently colonized by S. aureus if two positive samples were obtained, tested in a 4-week interval. All individuals tested either negative or positive for S. aureus at both times. In total, 105 Bacillus isolates from 101 individuals were analyzed. In the samples from four individuals, two isolates each were taken due to apparent phenotypic differences.
Bacterial identification using MALDI-TOF MS
Isolates were inoculated onto sheep blood agar and incubated for 24 h at 37 °C. Bacterial colonies were applied onto a 96-spot target plate and allowed to dry at room temperature. Subsequently, 2 μl of MALDI matrix [a saturated solution of α-cyano-4-hydroxycinnamic acid (HCCA) in 50% acetonitrile and 2.5% trifluoroacetic acid] was applied onto the colonies and allowed to dry before testing. Then the target plate was loaded into the MALDI-TOF MS instrument (MicroFlex LT mass spectrometer, Bruker Daltonics). Spectra were analyzed using MALDI Biotyper automation control and the Bruker Biotyper 2.0 software and library (version 2.0, 3,740 entries; Bruker Daltonics). Identification score criteria used were those recommended by the manufacturer: a score of ≥ 2.000 indicated species-level identification, a score of 1.700 to 1.999 indicated identification to the genus level, and a score of <1.700 was interpreted as no identification. Isolates that failed to produce a score of <1.700 with direct colony or extraction methods were retested. S. aureus ATCC25923, E. coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 were used as controls.
Bacterial strains and growth conditions
The reference B. subtilis strain and parent of the fenA and srfA mutants used in this study was strain ZK3814 (genotype NCIB3610). The S. aureus strains used in all experiments (except the experiment analyzing different Agr-subtype S. aureus) were (i) the human fecal isolate F12 of ST2196 (Supplementary Tab. 1), (ii) strain JSNZ of ST88, a mouse isolate previously described as mouse-adapted32, (iii) strain LAC of pulsed-field type USA300, an MRSA lineage predominantly involved in community-associated infections, but now generally representing the major lineage responsible for S. aureus infections in the United States47.
Isogenic mutants in agr were previously described (for strain LAC)48 or produced in this study (for strains JSNZ and F12) by phage transduction of the agr deletion from strain RN6911. The agr system is entirely deleted in these strains, except for a 3’ part of RNAIII, which is not transcribed owing to the absence of the corresponding promoter. All mutants were verified by analytical PCR.
Due to the tetracycline resistance introduced in the agr deletion strains, kanamycin derivatives (pKXΔ) of the pTXΔ expression plasmid series were constructed and used for complementation of Agr. (This was not possible in strain LAC, which harbors multiple antibiotic resistance including to kanamycin). To that end, plasmid pKX1549 kindly provided by B. Krismer, University of Tübingen, was treated as described48 to delete the xylR promoter to make expression of any fragment cloned under control of the xyl promoter constitutive. To obtain plasmid pKXΔRNAIII, the RNAIII BamH1-MluI fragment was transferred from pTXΔRNAIII50. Plasmid pTXΔ16 is the corresponding empty control plasmid.
To construct the agrBD deletion mutant of strain LAC P3-lux, a 4.8 kb-PCR product from USA300 genomic DNA that included the agrBDCA operon, 1 kb upstream, and 1 kb downstream, was cloned into the SmaI site of pIMAY51 and inverse PCR was used to delete agrBD. Allelic exchange was then performed, and the chromosomal deletion was confirmed by PCR using one primer outside of the 1 kb homology arm, followed by sequencing the PCR product. See Supplementary Table 2 for oligonucleotides used.
To construct the tetracycline–resistant derivatives of S. aureus ST88 and ST2196, φ11 phage-mediated transduction was performed as described to transfer the tetracycline cassette in the donor strain (S. aureus RN4220 with integrated pLL29) to S. aureus ST88 and ST219652.
To construct the B. subtilis fengycin mutant strain, SPP1 phage-mediated transduction53 was performed to transfer the fenA deletion present in the donor strain (BKE18340 – a fenA(ppsA)::erm mutant in B. subtilis 168 obtained from the Bacillus Genetic Stock Center) to B. subtilis ZK3814. This was necessary as B. subtilis 168 bears a mutation in the sfp gene abolishing lipopeptide production.
Bacteria were generally grown in tryptic soy broth (TSB) with shaking unless otherwise indicated.
Typing of S. aureus isolates
S. aureus isolates were typed by multi-locus sequence typing (MLST) as previously described54. PCR amplicons of seven S. aureus housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were obtained from chromosomal DNA and their sequences compared with those available at the PubMLST database (https://pubmlst.org/saureus/). Not previously described alleles (arcC 520–521 and gmk 337) and STs (STs 4630 – 4638) were deposited to the website. The Agr subtype of S. aureus isolates was determined using a modified multiplex qRT-PCR protocol55. Two duplex qRT-PCR protocols using the respective described primer sets and two colored probes each were set up for Agr types I and II, and III and IV, respectively. Isolates whose Agr type could not be determined by that method were analyzed for the type of AIP production using HPLC/MS with the chromatography method also used for PSM detection (see below), integrating the three major m/z peaks for each AIP type.
Microbiome analysis
Genomic DNA from each fecal sample was extracted using a QIAamp DNA stool Minikit (Qiagen) according to the manufacturer’s instructions. The DNA was quantified using a Nanodrop spectrophotometer and 16S rRNA paired-end sequencing of the V4 region of 16S rRNA was performed by Illumina (San Diego, California) using an Illumina MiSeq system as described56.
For all obtained paired-end sequences, the abundance of Operational Taxonomic Units (OTU) and alpha and beta diversity were identified using Quantitative Insights Into Microbial Ecology (QIIME 1.9.1)57. This study used the Nephele (release 1.6) platform from the National Institute of Allergy and Infectious Diseases (NIAID) Office of Cyber Infrastructure and Computational Biology (OCICB) in Bethesda, Maryland. The sequences were assigned to OTUs with the QIIME’s uclust-based58 open-reference OTU picking protocol59 and the Greengenes 13_8 reference sequence set60 at 99% similarity. Alpha diversity was calculated using Chao1 and Shannon analyses61 and compared across groups using a nonparametric t test with 999 permutations.
Growth inhibition analysis
Growth inhibition of S. aureus by Bacillus culture filtrates was tested with an agar diffusion assay. To that end, 10 μl of Bacillus culture filtrate of each isolate was spotted on sterile filter disks. The filters were let to dry and the procedure was repeated four times, after which filters were laid on agar plates containing S. aureus, resulting in the analysis of five-times concentrated culture filtrate.
Fengycin purification
To identify the Agr-inhibiting active substance, 10 ml culture filtrate of the B. subtilis reference strain grown for 48 h in TSB were applied to a Zorbax SB-C18 9.4 mm x 25 cm reversed-phase column (Agilent) using an AKTA Purifier 100 system (GE Healthcare). After washing with three column volumes with 100% buffer A [0.1% trifluoroacetic acid (TFA) in water] and five column volumes of 30% buffer B (0.1% TFA in acetonitrile), a 20-column volume gradient from 30 to 100% buffer B was applied. The column was run at a flow rate of 3 ml/min. Peak fractionation was performed using the absorbance at 214 nm and fractions were subjected to further analysis by RP-HPLC/ESI-MS and MS/MS and test for Agr inhibition (see below).
To purify larger amounts of the main active peak containing β-OH-C17-fengycin B, acetonitrile was added to 200 ml filtrate from cultures grown under the same conditions to a final concentration of 10%, precipitated material was removed by centrifugation for 10 min at 3700 x g using a Sorvall Legend RT centrifuge, and the obtained cleared supernatant was applied to a self-packed HR 16/10 column filled with Resource PHE (GE Healthcare) material (column volume, 17 ml). After sample application, the column was washed with 10 % buffer B for three column volumes and 25% buffer B for five column volumes, after which a gradient of 15 column volumes from 25 to 60% buffer B was applied. 10-ml fractions were collected and positive fractions (as determined by RP-HPLC/MS) were lyophilized. The lyophilisate was redissolved in 2 ml acetonitrile. Six ml of water was added and the precipitated material was removed by 5 min centrifugation in a tabletop centrifuge at maximum speed. The cleared supernatant was then further purified on a Zorbax SB-C18 9.4 mm x 25 cm reversed-phase column as described above.
PSM and lipopeptide detection by RP-HPLC/MS
PSMs were analyzed by RP-HPLC/ESI-MS using an Agilent 1260 Infinity chromatography system coupled to a 6120 Quadrupole LC/MS in principle as described62, but with a shorter column and a method that was adjusted accordingly. A 2.1 × 5 mm Perkin-Elmer SPP C8 (2.7 μm) guard column was used at a flow rate of 0.5 ml/min. After sample injection, the column was washed for 0.5 min with 90% buffer A /10% buffer B, then for 3 min with 25% buffer B. Then, an elution gradient was applied from 25% to 100% buffer B in 2.5 min, after which the column was subjected to 2.5 min of 100% buffer B to finalize elution.
Bacillus culture filtrates or (partially) purified lipopeptide (fengycins, surfactins) containing fractions were analyzed using the same column, system, and elution conditions. To quantify production of different fengycins, the two most abundant peaks, corresponding to double and triple charged ions, were used for the integration. Agilent Mass Hunter Quantitative Analysis Version B.07.00 was used for quantification.
Measurement of Agr activity
To determine the Agr-inhibiting activity of Bacillus culture filtrates or purified fractions, we measured luminescence emitted by an Agr P3 promoter-luxABCDE reporter fusion construct that was inserted into the genome of strain S. aureus LAC34. Strain LAC P3-luxABCDE was diluted 100-fold from a pre-culture grown overnight in TSB before distribution into a 96-well microtiter plate. To 100 μl of that dilution, 100 μl of sterilized culture filtrate sample was added, unless otherwise indicated. Plates were incubated at 37°C with shaking. Luminescence was measured with a GloMax Explorer luminometer (Promega) every 2 h for a total of 6 h. Inhibition was considered significant if the 4-h sample and control values differed by at least a factor of 2. Of note, the quorum-quenching effect exerted by the one-time initial dose of fengycin or fengycin-containing culture filtrates was transient and was overcome at later times by the increasing intrinsic AIP production. Agr-inhibiting activity of purified fengycin was also measured using quantitative real-time PCR of RNAIII as described previously30.
To determine the Agr-inhibiting activity on target strains other than LAC (Agr subtype I), the impact on the production of δ-toxin, whose gene is embedded in the Agr intracellular effector RNA, RNAIII, in most staphylococci, was used. Production of δ-toxin was measured by RP-HPLC/ESI-MS as described above. Strains LAC (Agr subtype I), A950085 (Agr subtype II), MW2 (Agr subtype III), and A970377 (Agr subtype IV) were used for testing the effect of β-OH-C17-fengycin B on S. aureus of different Agr subgroups. Strain 1457 was used for S. epidermidis. All strains were diluted 100-fold from a pre-culture grown in TSB. β-OH-C17-fengycin B dissolved in DMSO was added to each sample to a final concentration of 20 and 100 μM. All samples were incubated at 37 °C with shaking for 4 h. Samples were centrifuged and supernatant was collected for RP-HPLC/MS detection.
Analysis of PVL and α−toxin expression
S. aureus strain LAC was diluted 100-fold from a pre-culture grown in TSB and inoculated into 500 μl TSB. Two hundred and fifty μl of B. subtilis culture filtrate was added into the sample. Samples were incubated at 37 °C with shaking for 4 h. Samples were centrifuged in a tabletop centrifuge at maximum speed for 5 min; the supernatants were collected and loaded onto 12% SDS-PAGE gels, which were run at 160 V for 1 h. Proteins were transferred to nitrocellulose membranes using an iBlot Western blotting system. Membranes were incubated with Odyssey blocking buffer for 1 h at room temperature. Anti-α-toxin (polyclonal rabbit serum; Sigma S7531; dilution 1:5,000) or anti-LukF-PV (affinity-purified rabbit IgG specific for a peptide region of LukF-PV, produced by GenScript USA Inc. and kindly provided by F. DeLeo, NIAID; dilution 1:500) antibodies were added to the blocking buffer and membranes were incubated overnight at 4 °C. Then, membranes were washed five times with Tris-buffered saline containing 0.1% Tween-20, pH 7.4 and incubated with Cy5-labeled goat anti-rabbit IgG (diluted 1:10,000 in blocking buffer) in the dark for 1 h at room temperature. Membranes were washed five times with the washing buffer and scanned with a Typhoon TRIO+ Variable Mode Imager.
Preparation of Bacillus spores
B. subtilis wild-type or isogenic fengycin mutant strains were inoculated from a pre-culture (1:100) into 1 l of 2 x SG medium63 and allowed to sporulate for 96 hours. Cells were pelleted, washed with water, and resuspended in 20% metrizoic acid (Sigma). Five different concentrations (w/v) of metrizoic acid (60% - 20%) were added stepwise to a 50-ml centrifuge tube to obtain a density gradient. Then, a cell suspension was added to the top of the gradient, followed by centrifugation at 40,000 × g for 60 min at 4 °C64. Spores were found in the middle layers and were collected. They were washed with 10 ml water for three times. The total obtained number of viable spores per ml was determined by serial dilution, plating on TSA, and counting CFUs. The total number of heat-resistant spores per ml was determined by submerging the spores in water bath at 80 °C for 20 min followed by serial dilution and quantification of CFU/ml as described above.
Murine intestinal colonization model
In-vivo studies were approved by the Institutional Animal Care and Use Committee of the NIAID. Animal work was conducted adhering to the institution’s guidelines for animal use, and followed the guidelines and basic principles in the United States Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the Guide for the Care and Use of Laboratory Animals by certified staff in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International accredited facility.
All C57BL/6J mice were female and six to eight weeks of age at the time of use. In one setup, before S. aureus was given by oral gavage, mice were pre-treated to eradicate the pre-existing intestinal microbiota using an antibiotic mix consisting of ampicillin (1 g/l), metronidazole (1 g/l), neomycin trisulfate (1 g/l), and vancomycin (1 g/l), in the drinking water. The last day before gavage, antibiotics were omitted from the drinking water. No bacteria could be found in the feces or intestines of mice for 7 days after this treatment in a control experiment. In another setup, antibiotic pre-treatment was omitted. In all setups, S. aureus strains were grown to mid-exponential growth phase, washed, and resuspended in sterile PBS at 108 CFU/ml. Mice were inoculated by oral gavage with 200 μl of a 108 CFU/ml suspension of the indicated S. aureus strains, or 1:1 mixtures of wild-type and isogenic agr mutants to reach the same final concentration and volume. For the experiments with pKXΔ plasmid-containing strains, mice received kanamycin (0.2 g/l) in the drinking water during the experiment to maintain plasmids. For the B. subtilis spore competition experiment, oral gavage with 200 μl of spores of the Bacillus wild-type or its isogenic ΔfenA fengycin mutant (108 CFU/ml in sterile PBS) was performed on the day following the S. aureus gavage, and repeated every second day thereafter for a total of 3 times (days 2, 4, and 6). Intestinal colonization was evaluated by quantitative cultures of mice stool samples and samples from the small and large intestine. In detail, stool was collected and suspended to a final volume of 1 ml of PBS, diluted, and plated on TSB agar. Plates were incubated for 24 h at 37 °C, and colonies were enumerated. Moreover, after mice were euthanized seven days after infection, the small and large intestines were collected, resuspended each in 1 ml PBS, and homogenized. Serial dilutions of the homogenates were plated on TSB agar and incubated at 37 °C. Bacterial colonies were enumerated on the following day. In the experiments without antibiotic pre-treatment, extracts were plated on MSA plates containing 4 μg/ml oxacillin (for strain USA300 LAC) or 3 μg/ml tetracycline (for tetracycline-resistant derivatives of ST88 and ST2196), incubated for 48 h at 37 °C, and enumerated.
Statistics
Statistical analysis was performed using GraphPad Prism Version 6.05 using 1-way or 2-way ANOVA, or Fisher’s exact test, as appropriate, except for the experiments shown in Figures 2c, 5b,c, and Extended Data Figs. 2 b,cand 5c-f, for which Stata Release 15 and Poisson regression was used, due to the exclusive presence of 0 values in one group (no variance). In ANOVAs, Tukey post-tests were used, which correct for multiple comparisons using statistical hypothesis testing. All error bars show the mean and standard deviation (SD). All replicates are biological.
Data availability
Microbiome sequencing data are available at http://www.ncbi.nlm.nih.gov/bioproject/483343. All other data generated or analyzed during this study are included in this published article or in the supplementary information files.
Supplementary Material
Extended Data
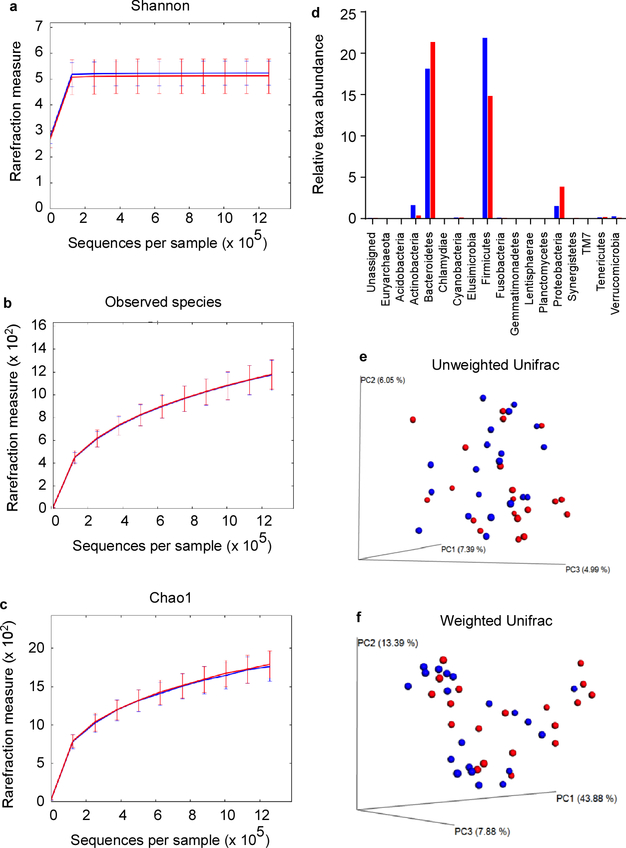
Extended Data Figure 1 |. Microbiome analysis of S. aureus carriers versus non-carriers.
The microbiota of n=20 randomly selected S. aureus carriers (red) and n=20 non-carriers (blue) were analyzed in fecal samples. a-c, Rarefaction curves of 16S rRNA gene sequences. Error bars shown the means ±SD. a, Shannon index. b, Observed species against the number of sequences per sample. c, Chao1 index. d, Relative taxa abundance comparison between S. aureus carriers (red) and non-carriers (blue), e-f, beta diversity, represented by a Principal Coordinate Analysis (PCoA) plot based on unweighted UniFrac (e) and weighted UniFrac metrics (f) for samples from S. aureus carriers (red) and non-carriers (blue).
Extended Data Figure 2 |. Quorum-sensing dependence of S. aureus intestinal colonization.
Data from strains USA300 LAC and ST88 JSNZ. The experimental setup is the same as in Fig. 2 of the main manuscript: Mice received by oral gavage 100 μl containing 108 CFU/ml of wild-type (WT) S. aureus strains USA300 LAC or ST88 JSNZ and another 100 μl of 108 CFU/ml of the corresponding isogenic agr mutant (n=5/group, competitive experiment shown in a,b), or 200 μl containing 108 CFU/ml wild-type, isogenic agr mutant, or Agr (RNAIII)-complemented agr mutant (n=5/group, non-competitive experiment shown in c). CFU in the feces were determined two, four, and six days after infection. At the end of the experiment (day seven), CFU in the small and large intestines were determined. a,b Competitive experiment. Total obtained CFU are shown as dot plots, also showing the means ±SD. Bars show the percentage of wild-type among total determined CFU, of which 100 were analyzed for tetracycline resistance that is present only in the agr mutant. No agr mutants were detected in any experiment; thus, all bars show 100%. Given that 100 isolates were tested, the competitive index wild-type/agr mutant in all cases is ≥ 100. c, Non-competitive experiment with genetically complemented strains. Wild-type and isogenic agr mutant strains all harbored the pKXΔ16 control plasmid; Agr-complemented strains harbored pKXΔRNAIII, constitutively expressing RNAIII, which is the intracellular effector of Agr. During the experiment, mice received 200 μg/ml kanamycin in the drinking water to maintain plasmids. Statistical analysis was performed using Poisson regression versus values obtained with the agr mutant strains. *, p<0.0001. Error bars show the means ±SD. Note no bacteria were found in the feces or intestines of any mouse receiving S. aureus Δagr with vector control. The corresponding zero values are plotted on the x-axis of the logarithmic scale.
Extended Data Figure 3 |. Analysis of Agr-inhibitory substances.
a, Influence of heat and protease on Agr inhibition. B. subtilis (B. s.) culture filtrate was subjected to heat (95 °C, 20 min) or proteinase K digestion (50 μg/ml, 37 °C, 1 h) and the impact on inhibition of Agr activity was measured using the luminescence assay with the USA300 P3-luxABCDE reporter strain (see Fig. 3a). RLU, relative light units. The experiment was performed with n=2 independent biological samples. Lines connect the means. (The observed additional suppression of Agr activity in the proteinase K-treated sample at 6 h, as compared to the B.s. culture filtrate sample, is expected due to proteolytic inactivation of intrinsic AIP.) b, Preparative RP chromatography of B. subtilis culture filtrate to determine the Agr-inhibiting substance. The peaks labeled 2 and 3 showed significant Agr-inhibiting activities in the Agr activity assay and were identified as fengycins using subsequent RP-HPLC/ESI-MS and MS/MS analysis (see c,d). The peaks labeled 1 and 4–6 also contained fengycin species (see e). c, Fractions corresponding to the Agr-inhibitory peaks 2 and 3 from the preparative RP run (b) were subjected to RP-HPLC/ESI-MS. Top, total ion chromatograms (TICs) of the RP-HPLC/ESI-MS runs; bottom, ESI mass spectrogram of the major peaks. d, MS/MS analysis of the peak 2 and 3 fractions. Peaks that are characteristic for a given fengycin subtype (A or B in this case) are marked in color. e, Analysis of further fengycin-containing fractions. Peaks 1, 4, 5, and 6 from the preparative RP run (b) were also found to contain fengycin species as determined by subsequent RP-HPLC/ESI-MS analysis. Shown are the mass spectrograms of the major peaks of those runs and the tentative characterization for fengycin type. The preparative and analytical chromatography and HPLC/MS analyses (as shown in b and d) were repeated multiple (> 10) times for fengycin purification with similar results. MS/MS analyses were not repeated. f, Analysis of fengycin and surfactin lipopeptide expression of the B. subtilis wild-type strain and its isogenic ΔfenA mutant.
Extended Data Figure 4 |. Assessment of purity and functionality of purified β-OH-C17-Fengycin B.
a, RP-HPLC run. b, Agr inhibition at different concentrations in the luminescence assay. RLU, relative light units. Statistical analysis was by 2-way ANOVA with Tukey’s post-test. Comparisons shown are those versus DMSO control. c, Agr inhibition as measured by inhibition of expression of RNAIII by qRT-PCR. *, p<0.0001 (1-way ANOVA with Tukey’s post-test. Comparisons shown are those versus 0 μM value). b,c, The experiments were performed with n=3 independent biological samples. Error bars show the means ±SD.
Extended Data Figure 5 |. Inhibition of S. aureus colonization by dietary fengycin-producing Bacillus spores in a mouse model.
a, AIP concentration during S. aureus growth. Strain LAC (USA300) was grown in TSB and AIP concentrations were measured by HPLC/MS. Calibration was performed using synthetic AIP. The detection limit of this assay is ~ 0.3 μM. The experiment was performed with n=3 independent biological samples. Error bars show the means ±SD. b, B. subtilis colonization kinetics in the mouse intestinal colonization experiment. Mice (n=5) received 200 μl of a 108 CFU/ml suspension of B. subtilis wild-type or ΔfenA mutant spores by oral gavage and CFU in the feces were analyzed up to 5 days afterwards. Error bars show the means ±SD. c-f, Inhibition mouse model with strains USA300 LAC and ST88 JSNZ. The experimental setup was the same as shown in Fig. 5a. n=4 or 5 mice/group received 200 μl of 108 CFU/ml S. aureus strains USA300 LAC or ST88 JSNZ by oral gavage. On the next and every following second day, they received 200 μl of 108 CFU/ml spores of the B. subtilis wild-type (WT) or its isogenic fenA mutant, also by oral gavage. CFU in the feces were determined two, four, and six days after infection. At the end of the experiment (day seven), CFU in the small and large intestines were determined. The experiment was performed with (c,d) or without (e,f) antibiotic pre-treatment. Statistical analysis was performed using Poisson regression versus values obtained with the B. subtilis WT spore samples. *, p<0.0001. Error bars shown the means ±SD. Note no S. aureus were found in the feces or intestines of any mouse challenged with any S. aureus strain receiving Bacillus wild-type spores. The corresponding zero values are plotted on the x-axis of the logarithmic scale.
Extended Data Tab. 1 |.
Fengycin production and Agr inhibitory potency of Bacillus fecal isolates*
| Isolate Code |
Bacillus species† | β - OH-C16- FenA | C16-FenA | β - OH-C17- FenB | C17-FenB | β - OH-C17- FenA | C17-FenA | β - OH-C16- FenB | C16-FenB | % Agr inhibition‡ | Total Fengycin Concentration |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | licheniformis | 100 | 48 | 65 | 80 | 33 | 33 | 51 | 32 | 98 | 442 |
| 14 | subtilis | 82 | 27 | 104 | 138 | 52 | 111 | 186 | 0 | 97 | 700 |
| 15 | amyloliquefaciens | 7 | 212 | 212 | 83 | 152 | 56 | 274 | 81 | 92 | 1076 |
| 16 | sonorensis | 5833 | 437 | 984 | 288 | 2751 | 1248 | 1760 | 1090 | 98 | 14390 |
| 18 | subtilis | 107 | 0 | 58 | 118 | 0 | 0 | 53 | 23 | 97 | 359 |
| 19 | ticheniformis | 0 | 61 | 0 | 75 | 13 | 0 | 149 | 31 | 97 | 329 |
| 21 | sonorensis | 0 | 85 | 159 | 48 | 0 | 104 | 18 | 33 | 95 | 447 |
| 26 | megaterium | 47 | 0 | 112 | 8 | 33 | 134 | 48 | 23 | 98 | 405 |
| 30 | subtilis | 0 | 0 | 63 | 28 | 109 | 77 | 87 | 43 | 96 | 407 |
| 31 | sonorensis | 23 | 0 | 52 | 0 | 157 | 15 | 0 | 0 | 97 | 246 |
| 32 | sonorensis | 23 | 118 | 145 | 0 | 0 | 0 | 74 | 59 | 94 | 418 |
| 33 | ticheniformis | 89 | 119 | 70 | 99 | 98 | 102 | 150 | 0 | 96 | 727 |
| 35 | ticheniformis | 0 | 0 | 98 | 0 | 149 | 72 | 28 | 132 | 96 | 479 |
| 36 | pumilus | 0 | 0 | 10 | 0 | 0 | 136 | 144 | 0 | 98 | 290 |
| 37 | subtilis | 116 | 232 | 67 | 0 | 15 | 45 | 0 | 114 | 96 | 589 |
| 38 | subtilis | 59 | 167 | 215 | 0 | 79 | 114 | 50 | 212 | 93 | 896 |
| 39 | ticheniformis | 150 | 117 | 249 | 0 | 241 | 271 | 226 | 230 | 96 | 1484 |
| 40 | subtilis | 753 | 174 | 1260 | 1124 | 1548 | 538 | 841 | 821 | 80 | 7058 |
| 41 | subtilis | 2298 | 860 | 1777 | 0 | 4524 | 816 | 1563 | 1957 | 91 | 13796 |
| 42 | sonorensis | 34 | 43 | 0 | 0 | 41 | 57 | 88 | 0 | 96 | 263 |
| 43 | sonorensis | 477 | 0 | 2488 | 816 | 1667 | 604 | 1216 | 858 | 96 | 8126 |
| 45 | sonorensis | 98 | 0 | 24 | 25 | 0 | 0 | 0 | 39 | 99 | 187 |
| 47 | pumilus | 342 | 14 | 237 | 0 | 0 | 108 | 105 | 0 | 97 | 806 |
| 48 | subtilis | 0 | 0 | 49 | 0 | 7 | 0 | 0 | 0 | 94 | 56 |
| 49 | subtilis | 5007 | 828 | 979 | 0 | 3147 | 0 | 2027 | 1210 | 97 | 13200 |
| 50 | subtilis | 429 | 0 | 933 | 0 | 1065 | 354 | 614 | 526 | 90 | 3922 |
| 51 | subtilis | 712 | 185 | 911 | 208 | 1187 | 665 | 863 | 586 | 88 | 5316 |
| 52 | subtilis | 9630 | 1571 | 923 | 0 | 2365 | 1775 | 4690 | 1099 | 95 | 22052 |
| 53 | ticheniformis | 0 | 0 | 0 | 0 | 0 | 0 | 58 | 25 | 95 | 83 |
| 55 | subtilis | 45 | 43 | 113 | 12 | 94 | 0 | 0 | 216 | 96 | 523 |
| 56 | sonorensis | 167 | 53 | 0 | 0 | 83 | 201 | 109 | 58 | 98 | 671 |
| 57 | subtilis | 127 | 104 | 166 | 301 | 61 | 96 | 98 | 246 | 96 | 1200 |
| 58 | sonorensis | 498 | 510 | 841 | 0 | 967 | 222 | 0 | 0 | 94 | 3039 |
| 59 | subtilis | 0 | 0 | 1008 | 0 | 0 | 297 | 715 | 488 | 82 | 2508 |
| 61 | subtilis | 124 | 84 | 21 | 0 | 160 | 39 | 42 | 335 | 97 | 805 |
| 62 | ticheniformis | 93 | 128 | 7 | 48 | 40 | 188 | 0 | 153 | 97 | 657 |
| 63 | amyloliquefaciens | 4153 | 380 | 975 | 0 | 2584 | 1047 | 1975 | 1084 | 95 | 12197 |
| 64 | sonorensis | 133 | 91 | 187 | 230 | 29 | 387 | 62 | 34 | 95 | 1154 |
| 65 | subtilis | 9 | 126 | 0 | 0 | 35 | 90 | 239 | 36 | 97 | 535 |
| 66 | sonorensis | 254 | 0 | 0 | 0 | 377 | 0 | 131 | 0 | 97 | 762 |
| 67 | subtilis | 215 | 84 | 200 | 0 | 0 | 0 | 155 | 168 | 97 | 821 |
| 68 | subtilis | 41 | 12 | 144 | 0 | 0 | 0 | 115 | 182 | 93 | 494 |
| 69 | subtilis | 266 | 27 | 236 | 290 | 132 | 0 | 54 | 195 | 98 | 1200 |
| 70 | subtilis | 54 | 55 | 390 | 0 | 196 | 0 | 187 | 112 | 94 | 994 |
| 71 | subtilis | 14881 | 4578 | 88106 | 34879 | 39939 | 16967 | 42502 | 23859 | 97 | 265710 |
| 74 | pumilus | 157 | 56 | 54 | 14 | 0 | 62 | 177 | 41 | 95 | 560 |
| 75 | ticheniformis | 281 | 22 | 40 | 0 | 0 | 292 | 125 | 204 | 95 | 964 |
| 76 | subtilis | 124 | 0 | 74 | 0 | 0 | 101 | 82 | 192 | 97 | 573 |
| 77 | sonorensis | 0 | 0 | 0 | 25 | 0 | 0 | 93 | 13 | 97 | 131 |
| 78 | amyloliquefaciens | 73 | 99 | 0 | 0 | 79 | 17 | 176 | 13 | 94 | 458 |
| 79 | amyloliquefaciens | 10 | 0 | 0 | 0 | 0 | 180 | 51 | 63 | 97 | 304 |
| 80 | subtilis | 1741 | 322 | 4222 | 1105 | 3327 | 910 | 2207 | 1529 | 91 | 15363 |
| 81 | subtilis | 1739 | 426 | 5073 | 1579 | 3371 | 998 | 3241 | 1933 | 87 | 18361 |
| 82 | subtilis | 1002 | 0 | 3413 | 921 | 1998 | 0 | 1710 | 992 | 91 | 10037 |
| 83 | ticheniformis | 356 | 0 | 536 | 4 | 83 | 201 | 107 | 52 | 97 | 1338 |
| 85 | subtilis | 52 | 0 | 49 | 0 | 84 | 161 | 0 | 132 | 95 | 479 |
| 87 | subtilis | 1327 | 312 | 3931 | 0 | 2763 | 667 | 2624 | 1167 | 98 | 12790 |
| 88 | pumilus | 101 | 0 | 215 | 156 | 313 | 367 | 276 | 216 | 96 | 1643 |
| 89 | subtilis | 105 | 59 | 266 | 0 | 0 | 0 | 51 | 38 | 93 | 519 |
| 91 | subtilis | 325 | 91 | 493 | 17 | 120 | 290 | 302 | 186 | 98 | 1825 |
| 92 | subtilis | 254 | 0 | 234 | 156 | 275 | 0 | 154 | 66 | 96 | 1140 |
| 93 | subtilis | 493 | 273 | 91 | 342 | 204 | 287 | 277 | 435 | 96 | 2402 |
| 94 | subtilis | 876 | 115 | 134 | 37 | 384 | 190 | 445 | 529 | 91 | 2712 |
| 95 | amyloliquefaciens | 351 | 175 | 157 | 146 | 110 | 225 | 196 | 197 | 97 | 1557 |
| 97 | subtilis | 1845 | 912 | 4714 | 1310 | 3686 | 1492 | 2484 | 1557 | 86 | 18000 |
| 98 | subtilis | 1367 | 804 | 3572 | 1512 | 2803 | 1511 | 2005 | 1626 | 93 | 15200 |
| 99 | subtilis | 77 | 375 | 705 | 0 | 28 | 170 | 115 | 77 | 91 | 1547 |
| 100 | amyloliquefaciens | 81 | 117 | 337 | 267 | 105 | 166 | 237 | 502 | 81 | 1811 |
| 103 | pumilus | 249 | 45 | 350 | 207 | 162 | 249 | 536 | 279 | 98 | 2077 |
| 104 | subtilis | 293 | 77 | 105 | 269 | 75 | 509 | 286 | 17 | 99 | 1632 |
| 106 | subtilis | 978 | 796 | 4415 | 3419 | 1874 | 1378 | 2478 | 1527 | 97 | 16866 |
| 107 | subtilis | 423 | 199 | 322 | 150 | 520 | 517 | 384 | 160 | 98 | 2675 |
| 108 | pumilus | 224 | 114 | 397 | 0 | 229 | 211 | 413 | 43 | 99 | 1631 |
| 110 | subtilis | 140 | 77 | 317 | 286 | 404 | 127 | 117 | 139 | 95 | 1607 |
| 111 | pumilus | 184 | 104 | 319 | 67 | 96 | 212 | 294 | 120 | 93 | 1395 |
| 112 | subtilis | 470 | 183 | 1412 | 637 | 1211 | 732 | 950 | 655 | 96 | 6251 |
| 113 | pumilus | 463 | 202 | 276 | 204 | 211 | 0 | 156 | 137 | 95 | 1650 |
| 115 | subtilis | 268 | 232 | 297 | 62 | 313 | 410 | 713 | 88 | 97 | 2382 |
| 116 | subtilis | 352 | 205 | 369 | 0 | 172 | 298 | 561 | 350 | 96 | 2306 |
| 117 | subtilis | 143 | 104 | 716 | 328 | 0 | 149 | 97 | 266 | 98 | 1803 |
| 118 | subtilis | 163 | 34 | 1788 | 0 | 0 | 155 | 306 | 42 | 93 | 2488 |
| 119 | amyloliquefaciens | 604 | 256 | 947 | 258 | 0 | 350 | 361 | 104 | 93 | 2880 |
| 121 | subtilis | 503 | 151 | 0 | 364 | 63 | 84 | 174 | 162 | 98 | 1502 |
| 122 | subtilis | 152 | 311 | 24 | 165 | 86 | 213 | 296 | 83 | 91 | 1329 |
| 123 | subtilis | 8801 | 3540 | 23045 | 10778 | 16465 | 6428 | 16077 | 10706 | 98 | 95839 |
| 124 | subtilis | 106 | 139 | 316 | 168 | 64 | 175 | 195 | 103 | 86 | 1266 |
| 125 | subtilis | 0 | 0 | 0 | 158 | 0 | 318 | 290 | 0 | 95 | 765 |
| 126 | subtilis | 288 | 157 | 211 | 110 | 428 | 421 | 185 | 112 | 87 | 1913 |
| 127 | subtilis | 478 | 193 | 103 | 435 | 240 | 303 | 551 | 132 | 96 | 2434 |
| 128 | subtilis | 177 | 156 | 228 | 118 | 96 | 276 | 426 | 48 | 97 | 1525 |
| 129 | pumilus | 249 | 37 | 0 | 162 | 0 | 224 | 0 | 144 | 97 | 816 |
| 130 | subtilis | 1267 | 0 | 4668 | 1266 | 2940 | 1168 | 2158 | 1863 | 88 | 15332 |
| 131 | subtilis | 4164 | 496 | 513 | 307 | 1226 | 671 | 1948 | 662 | 83 | 9986 |
| 132 | subtilis | 441 | 291 | 491 | 391 | 200 | 562 | 381 | 279 | 95 | 3036 |
| 134 | subtilis | 6647 | 750 | 1011 | 0 | 3143 | 1166 | 2829 | 953 | 97 | 16498 |
| 136 | subtilis | 751 | 233 | 2569 | 773 | 1813 | 890 | 1701 | 1010 | 89 | 9740 |
| 137 | subtilis | 1572 | 328 | 3297 | 1036 | 2221 | 627 | 2266 | 0 | 82 | 11347 |
| 138 | pumilus | 288 | 311 | 232 | 11 | 415 | 236 | 236 | 403 | 85 | 2133 |
| 139 | subtilis | 1898 | 709 | 7830 | 2453 | 5328 | 2073 | 3880 | 2210 | 98 | 26381 |
| 140 | sonorensis | 0 | 106 | 217 | 815 | 225 | 0 | 103 | 3987 | 91 | 5454 |
| 141 | thuringiensis | 258 | 26 | 325 | 230 | 0 | 264 | 0 | 0 | 87 | 1102 |
| 142 | pumilus | 422 | 159 | 262 | 0 | 402 | 335 | 639 | 251 | 87 | 2471 |
| 143 | subtilis | 250 | 37 | 210 | 16 | 64 | 68 | 293 | 89 | 83 | 1027 |
| 144 | sonorensis | 110 | 208 | 134 | 191 | 0 | 304 | 361 | 91 | 87 | 1399 |
| B. subtilis ZK3814 | 2563 | 1781 | 17078 | 5725 | 11963 | 4448 | 5444 | 4902 | 94 | 53904 |
The table shows intensity values of the integration of m/z peaks associated with the specific fengycin species as obtained by HPLC/MS. The two most abundant peaks, corresponding to double and triple charged ions, were used for the integration. Values are in nM, obtained by calibration using weighed and diluted aliquots of the Bacillus lipopeptide surfactin.
Bacillus species were determined by sequencing 16S RNA encoding DNA, as specified in Methods.
Percentage of Agr inhibition was determined by dividing the 4-h value in the luminescence assay (using 100 μl of culture filtrate) obtained for the sample by that obtained for the control and multiplying by 100.
Extended Data Tab. 2 |.
Analysis of previous microbiome studies* for correlation between S. aureus and B. subtilis presence in the human intestinal tract
| Study ID | Study Name | Samples | Only B. subtilis | Only S. aureus | Both | Neither |
|---|---|---|---|---|---|---|
| ERP012803 | American Gut Project | 6635 | 1 (0.015%) | 304 (4.58%) | 0 | 6330 (95.4%) |
| ERP011001 | Human gut bacteria that rescue growth and metabolic defects transmitted by microbiota from undernourished children | 1732 | 408 (23.61%) | 70 (4.05%) | 71 (4.11%) | 1179 (68.23%) |
| ERP005437 | 16S sequencing of Malawian children | 1515 | 118 (7.79%) | 6 (0.4%) | 4 (0.26%) | 1387 (91.55%) |
| SRP049113 | Human gut microbiota from the ALADDIN study | 664 | 2 (0.30%) | 61 (9.19%) | 7(1.05%) | 594 (89.46%) |
| ERP019564 | Role of Gut Microbiota in Pathophysiology of Parkinson's Disease | 481 | 8(1.66%) | 7(1.45%) | 0 | 466 (96.88%) |
| SRP073172 | DNA from FIT can replace stool for microbiota-based colorectal | 408 | 63 (15.44%) | 71 (17.40%) | 99 (24.26%) | 175 (42.89%) |
| SRP068240 | Human feces metagenome 16s rDNA sequencing | 350 | 52 (14.85%) | 189 (54%) | 89 (25.43%) | 20 (5.71%) |
| SRP064846 | Homo sapiens fecal microbiome transplant | 271 | 20 (7.38%) | 47 (17.34%) | 6(2.21%) | 198 (73.06%) |
| SRP065497 | Human gut environment Targeted loci environmental | 270 | 54 (20%) | 8 (2.96%) | 19 (7.04%) | 189 (70%) |
| ERP021093 | Gut microbiome from patients obtained by 16s rRNA sequencing. | 268 | 88 (32.84%) | 14 (5.22%) | 57 (21.27%) | 109 (40.67%) |
| ERP010229 | Gut microbial succession follows acute secretory diarrhea in humans | 260 | 12 (4.62%) | 92 (35.38%) | 122 (46.92%) | 34 (13.08%) |
| ERP010458 | Gut microbiota of stroke patients differentiates from healthy controls | 233 | 3(1.29%) | 32 (13.73%) | 4(1.72%) | 194 (83.26%) |
Inclusion criteria: All studies found on the EBI Metagenomics website (https://www.ebi.ac.uk/metagenomics/) with > 200 participants (independent samples) using Illumina Miseq instruments were included in the analysis.
Analysis: Raw 16S rRNA sequencing data were pooled from the EBI Metagenomics website. Taxonomic assignment (TSV) files were used for analysis. The number of sequence reads was used to analyze how many samples contain S. aureus or B. subtilis. Samples with a read number of more than 0 were defined as colonized. When there were no reads, samples were designated as non-colonized.
Acknowledgements
The authors thank Roberto Kolter, Harvard Medical School, for providing the B. subtilis srfA mutant, David Dubnau, Rutgers University, for the SPP1 phage, Silvia Holtfreter, University of Greifswald and Wei Ping Zeng, Texas Tech University Health Sciences Center, for providing strain JSNZ/ST88, Bernhard Krismer, University of Tübingen, for plasmid pKX15, Frank DeLeo, NIAID, for anti-LukF-PV, and Nana A. Amissah for technical assistance. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH) (project ZIA AI000904–16, M.O.) and the Thailand Research Fund through the Royal Golden Jubilee PhD Program (grant no. PHD/0072/2557 to P.P. and P.K.). P.K. was also supported by the Faculty of Medicine Siriraj Hospital, Mahidol University, Grant Number (IO) R015833012, P.P. by the Graduate Partnership Program of the NIH, and S.W.D. by the Postdoctoral Research Associate Program of the National Institute of General Medical Sciences (1FI2GM11999101).
Footnotes
The authors declare that there are no conflicts of interest.
References
- 1.Guarner F & Malagelada JR Gut flora in health and disease. Lancet 361, 512–519 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Chen GY, Inohara N & Nunez G Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14, 685–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gourbeyre P, Denery S & Bodinier M Probiotics, prebiotics, and synbiotics: impact on the gut immune system and allergic reactions. J Leukoc Biol 89, 685–695 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Macpherson AJ & Harris NL Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 4, 478–485 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Bermudez-Brito M, Plaza-Diaz J, Munoz-Quezada S, Gomez-Llorente C & Gil A Probiotic mechanisms of action. Ann Nutr Metab 61, 160–174 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Sassone-Corsi M et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tam NK et al. The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol 188, 2692–2700 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casula G & Cutting SM Bacillus probiotics: spore germination in the gastrointestinal tract. Appl Environ Microbiol 68, 2344–2352 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duc Le H, Hong HA, Barbosa TM, Henriques AO & Cutting SM Characterization of Bacillus probiotics available for human use. Appl Environ Microbiol 70, 2161–2171 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong HA, Duc Le H & Cutting SM The use of bacterial spore formers as probiotics. FEMS Microbiol Rev 29, 813–835 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Fujiya M et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1, 299–308 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lowy FD Staphylococcus aureus infections. N Engl J Med 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Lowy FD Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111, 1265–1273 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Septimus EJ & Schweizer ML Decolonization in Prevention of Health Care-Associated Infections. Clin Microbiol Rev 29, 201–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickey SW, Cheung GYC & Otto M Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wertheim HF et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5, 751–762 (2005). [DOI] [PubMed] [Google Scholar]
- 17.von Eiff C, Becker K, Machka K, Stammer H & Peters G Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 344, 11–16 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Simor AE & Daneman N Staphylococcus aureus decolonization as a prevention strategy. Infect Dis Clin North Am 23, 133–151 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Williams RE Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev 27, 56–71 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mody L, Kauffman CA, Donabedian S, Zervos M & Bradley SF Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin Infect Dis 46, 1368–1373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eveillard M et al. Evaluation of a strategy of screening multiple anatomical sites for methicillin-resistant Staphylococcus aureus at admission to a teaching hospital. Infect Control Hosp Epidemiol 27, 181–184 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Acton DS, Plat-Sinnige MJ, van Wamel W, de Groot N & van Belkum A Intestinal carriage of Staphylococcus aureus: how does its frequency compare with that of nasal carriage and what is its clinical impact? Eur J Clin Microbiol Infect Dis 28, 115–127 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Senn L et al. The Stealthy Superbug: the Role of Asymptomatic Enteric Carriage in Maintaining a Long-Term Hospital Outbreak of ST228 Methicillin-Resistant Staphylococcus aureus. MBio 7, e02039–02015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squier C et al. Staphylococcus aureus rectal carriage and its association with infections in patients in a surgical intensive care unit and a liver transplant unit. Infect Control Hosp Epidemiol 23, 495–501 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Lindberg E et al. High rate of transfer of Staphylococcus aureus from parental skin to infant gut flora. J Clin Microbiol 42, 530–534 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhalla A, Aron DC & Donskey CJ Staphylococcus aureus intestinal colonization is associated with increased frequency of S. aureus on skin of hospitalized patients. BMC Infect Dis 7, 105 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray AJ, Pultz NJ, Bhalla A, Aron DC & Donskey CJ Coexistence of vancomycin-resistant enterococci and Staphylococcus aureus in the intestinal tracts of hospitalized patients. Clin Infect Dis 37, 875–881 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Klotz M, Zimmermann S, Opper S, Heeg K & Mutters R Possible risk for re-colonization with methicillin-resistant Staphylococcus aureus (MRSA) by faecal transmission. Int J Hyg Environ Health 208, 401–405 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Misawa Y et al. Staphylococcus aureus Colonization of the Mouse Gastrointestinal Tract Is Modulated by Wall Teichoic Acid, Capsule, and Surface Proteins. PLoS Pathog 11, e1005061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung GY, Wang R, Khan BA, Sturdevant DE & Otto M Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79, 1927–1935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller MB & Bassler BL Quorum sensing in bacteria. Annu Rev Microbiol 55, 165–199 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Holtfreter S et al. Characterization of a mouse-adapted Staphylococcus aureus strain. PLoS One 8, e71142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diep BA et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Dastgheyb SS et al. Role of Phenol-Soluble Modulins in Formation of Staphylococcus aureus Biofilms in Synovial Fluid. Infect Immun 83, 2966–2975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick RP & Geisinger E Quorum sensing in staphylococci. Annu Rev Genet 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Pathak KV, Keharia H, Gupta K, Thakur SS & Balaram P Lipopeptides from the banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. J Am Soc Mass Spectrom 23, 1716–1728 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Cochrane SA & Vederas JC Lipopeptides from Bacillus and Paenibacillus spp.: A Gold Mine of Antibiotic Candidates. Med Res Rev 36, 4–31 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Chang LK et al. Construction of Tn917ac1, a transposon useful for mutagenesis and cloning of Bacillus subtilis genes. Gene 150, 129–134 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Lyon GJ, Wright JS, Muir TW & Novick RP Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41, 10095–10104 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Ji G, Beavis R & Novick RP Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Otto M, Echner H, Voelter W & Gotz F Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun 69, 1957–1960 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brisson J HSO’s Part 2 – Is Bacillus subtilis Dangerous?, <http://fixyourgut.com/hso-probiotics-part-2-danger-supplementing-bacillus-subtilis/> (2014).
- 43.Vanittanakom N, Loeffler W, Koch U & Jung G Fengycin--a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29–3. J Antibiot (Tokyo) 39, 888–901 (1986). [DOI] [PubMed] [Google Scholar]
- 44.Khan BA, Yeh AJ, Cheung GY & Otto M Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin Investig Drugs 24, 689–704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poovelikunnel T, Gethin G & Humphreys H Mupirocin resistance: clinical implications and potential alternatives for the eradication of MRSA. J Antimicrob Chemother 70, 2681–2692 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Miranda CA, Martins OB & Clementino MM Species-level identification of Bacillus strains isolates from marine sediments by conventional biochemical, 16S rRNA gene sequencing and inter-tRNA gene sequence lengths analysis. Antonie Van Leeuwenhoek 93, 297–304 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Carrel M, Perencevich EN & David MZ USA300 Methicillin-Resistant Staphylococcus aureus, United States, 2000–2013. Emerg Infect Dis 21, 1973–1980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang R et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13, 1510–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Gauger T et al. Intracellular monitoring of target protein production in Staphylococcus aureus by peptide tag-induced reporter fluorescence. Microb Biotechnol 5, 129–134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Queck SY et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell 32, 150–158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monk IR, Shah IM, Xu M, Tan MW & Foster TJ Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luong TT & Lee CY Improved single-copy integration vectors for Staphylococcus aureus. J Microbiol Methods 70, 186–190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasbin RE & Young FE Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14, 1343–1348 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enright MC, Day NP, Davies CE, Peacock SJ & Spratt BG Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38, 1008–1015 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francois P et al. Rapid Staphylococcus aureus agr type determination by a novel multiplex real-time quantitative PCR assay. J Clin Microbiol 44, 1892–1895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caporaso JG et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Rideout JR et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McDonald D et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lozupone CA, Hamady M, Kelley ST & Knight R Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73, 1576–1585 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joo HS & Otto M The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods Mol Biol 1106, 93–100 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Nicholson WL & Setlow P in Molecular biological methods for Bacillus (eds Harwood CR & Cutting SM) 391–450 (John Wiley and Sons, 1990). [Google Scholar]
- 64.Fukushima T et al. Characterization of a polysaccharide deacetylase gene homologue (pdaB) on sporulation of Bacillus subtilis. J Biochem 136, 283–291 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microbiome sequencing data are available at http://www.ncbi.nlm.nih.gov/bioproject/483343. All other data generated or analyzed during this study are included in this published article or in the supplementary information files.