Abstract
The role of lipid metabolism in epithelial stem cell (SC) function and carcinogenesis is poorly understood. The transcription factor Runx1 is known to regulate proliferation in mouse epithelial hair follicle (HF) SCs in vivo and in several mouse and human epithelial cancers. We found a novel sub-set of in vivo Runx1 HFSC target genes related to lipid metabolism and demonstrated changes in distinct classes of lipids driven by Runx1. Inhibition of lipid-enzymes Scd1 and Soat1 activity synergistically reduces proliferation of mouse skin epithelial cells and of human skin and oral squamous cell carcinoma cultured lines. Varying Runx1 levels induces changes in skin monounsaturated fatty acids (e.g. oleate, a product of Scd1) as shown by our lipidome analysis. Furthermore, varying Runx1 levels, the inhibition of Scd1, or the addition of Scd1-product oleate, individually affects the plasma membrane organization (or fluidity) in mouse keratinocytes. These factors also affect the strength of signal transduction through the membranes for Wnt, a pathway that promotes epithelial (cancer) cell proliferation and HFSC activation. Our working model is that HFSC factor Runx1 modulates the fatty acid production, which affects membrane organization, facilitating signal transduction for rapid proliferation of normal and cancer epithelial cells.
Graphical abstract
Introduction
Lipid metabolism regulates a variety of critical cell biological functions, including structural cell components, signaling, and energy resources (1). Lipids can be either synthesized de novo (via cell-intrinsic or endogenous metabolism) or imported from extracellular sources, such as diet or adipose reserves (2). Diet (e.g. high-fat diet), is known to affect the activity of tissue stem cells (SCs), in the nervous system (3, 4) and the intestine (5, 6). Essential fatty acids are only available from diet, and their metabolites can affect SC proliferation and differentiation (7).
Endogenous lipid metabolism may be important in SCs to render them independent of diet (8–10). For instance, distinct classes of glycolipids form specialized microdomains on the plasma membranes and are expressed preferentially by embryonic, neural and hematopoietic SCs (8). Furthermore, endogenous fatty acid synthesis regulates cellular reprograming and SC pluripotency (11). Genes necessary for fatty acid metabolism and lipid biosynthesis are up-regulated in adult neural SCs (NSCs) relative to more differentiated neuroblast, and de novo lipogenesis drives the proliferation of NSCs and downstream progenitors (2). This likely occurs by providing the building blocks necessary for membrane synthesis and to meet the energy demands of proliferative cells (2), but they may also be important in cell signaling (12, 13). It has been proposed that lipid metabolism may function as a bioenergetic rheostat of self-renewal versus differentiation to ensure ongoing neurogenesis (14, 15). Fatty acid oxidation is essential for hematopoietic SC fate decisions (16, 17) and for muscle stem cell fate, downstream of the Lkb1 kinase (18).
In addition to normal development and adult homeostasis, cell-intrinsic lipid metabolism plays an important role in various diseases such as stroke, diabetes, inflammation, and most importantly cancer (19–22). It is well established that de novo synthesis of lipids, including fatty acids, occurs in numerous cancers (23). The evidence to date suggests that the same classical pathways can regulate the behavior of both normal SCs and their related cancers (24). In this study, we asked if there is a cell-intrinsic rewiring of lipid-metabolic programs in epithelial cells through a known stem cell and cancer factor, Runx1, to ensure proper cell proliferation and survival.
We use genetic targeting of hair follicle stem cells (HFSCs) in mouse tissue, cultured epithelial skin cells (keratinocytes) and several human epithelial cancer cell lines and mouse tumors as model systems. The temporary lower HF region (bulb) periodically dies out and redevelops from the bulge region, where the HFSCs reside, in a regenerative process of homeostasis known as hair cycle (25, 26).
Previously, our laboratory uncovered the hematopoietic SC transcription factor Runx1 (27) as an important HFSC regulator during adult and embryonic skin homeostasis (27, 28). We implicated Runx1 in adult HFSC timely activation from quiescence and normal hair cycle progression (29) and in rates of self-renewal (25, 30). Runx1 is up-regulated in quiescent HFSCs migrating outside the niche (bulge), and promotes a reversible early progenitor (hair germ) cell-state prone for subsequent rapid activation and proliferation (29). We found Runx1 absolutely essential for keratinocyte proliferation and survival in cell culture and for skin and oral epithelial tumorigenesis in mice (30, 31). Furthermore, we found that several human oral and skin squamous cell carcinoma cell lines required Runx1 for their growth and survival, and several types of human epithelial tumors showed elevated levels of Runx1 (31). We have uncovered Stat3 (31), Wnt signaling (27), p21 and p15 (32) as signaling pathways mediating in part the role of Runx1 in proliferation and carcinogenesis. Wnt signaling is an important determinant of fate for different tissue SCs and cancers and promotes epithelial cell proliferation (33); it is also crucial for hair germ activation during hair cycle (34). Nuclear accumulation of β-catenin, which is the downstream effector of Wnt, turns on the target genes and promotes the activation of HFSCs and hair regeneration (35).
To systematically investigate how Runx1 is accomplishing its role in epithelial cell proliferation and survival, we characterized the global gene expression changes induced by Runx1 elevation in quiescent HFSCs in vivo (29). Among thousands of genes that changed mRNA expression upon Runx1 induction, we found a distinct GEO set of elevated genes with known functions in lipid metabolism (Table 1). Here we test the potential implication of some of these target genes in mediating Runx1-driven epithelial cell proliferation and survival in both normal tissue homeostasis and cancer.
Table 1.
Table depicting lipid-related genes increased by > 2-fold in response to Runx1 elevation in hair bulge in vivo, as shown by microarray analysis of sorted cells
| Lipid metabolism | Genes > 2 fold change in response to Runx1 Microarray of CD34+/α6+sorted HFSCs (Doxy PD19-20) (Lee, S.E. et al., Cell Rep, 2014. 6 (3):p. 499-513) |
|---|---|
| (1) Fatty Acid Metabolism | Slc16a1, Scd1, Scd3, Acox2, Acaa1b, Fa2h, Mgll, Cd36, Scd3, Tmem195, Prkar2b, Crat, Fads6, Far2, Ces1 |
| (2) Cholesterol metabolism | Nsdhl, Lpl, Cyp27a1, Soat1, Pparg, Cd36 |
| (3) Prostaglandins | Mgll, Pla2g2e, Pparg, Pla2g7, Edn1, Slco2a1, Pdpn, Avpr1a |
| (4) Phosphoinositides | Wipi1, Rasd2, Pitpnm1, Edn1, Plcb4, Snx5, lnpp1 |
| (5) Phosphatidylserine | Ptdss2 |
| (6) Phosphocholine | Pla2g2e, Lpcat3, Plcb4, Pla2g7 |
Material & Methods
Animal studies
Mouse work followed the Cornell University Institutional Animal Care and Use Committee guidelines. For Runx1 knockout (KO) studies we used K14-Cre or K14-CreERT2/Runx1Fl/FL (iKO) mice (31). For Runx1 deletion, mice were injected at PD17 intraperitoneally with 225µg Tamoxifen (Sigma) per gram of body weight dissolved in corn oil and sacrificed at PD20. Runx1Fl/Fl+TM and K14-CreERT2; Runx1Fl/Fl+oil littermates served as control. For overexpression studies, we used pTRE-mycRunx1 crossed with Tet-on K14-rTA mice previously generated (29). These mice were fed with doxycycline (doxy) chow (1,000 mg/kg) at PD19 to induce Runx1 expression and sacrificed at PD20, as described before (29). For hair-germ and bulge cell sorting we used previously generated K15-EGFP mice (36).
Immunofluorescence staining
Murine back skin or mouse and human tumors were embedded in Optimal Cutting Temperature (OCT) compound (Tissue Tek, Sakura) on dry ice and stored at −80°C. Frozen 10 µm skin sections were fixed with freshly made 4% paraformaldehyde for 10 minutes at room temperature (RT), blocked in blocking solution for 1– 2 hours, and incubated with primary antibodies for 24 hours at 4°C, as described before (29, 31). MOM Basic kit (Vector Laboratories) was used for mouse primary antibodies. After washing, we incubated for 1 hour with TxR, FITC, and Alexa-594 -conjugated secondary antibodies (Jackson Immuno Research), at a 1:500 dilution. TSA reagent (PerkinElmer) was applied (1:50 dilution) for 5 minutes on cell culture samples stained for Scd1 to enhance signal. After washing, the samples were counterstained with Hoechst and mounted in antifade solution onto glass slides for microscopy. HRP was used at 1:1000 dilution prior to application of TSA reagent. For Edu stainings, the Click-iT® Edu Alexa Flour® 594 Imaging kit was used (Thermo Fisher Scientific). Edu reagent (Thermo Fisher Scientific) was applied for 30 minutes at RT prior to blocking and application of other primary antibodies. Detailed list of antibodies with dilution is found in Supplementary Table 1.
Cell culture
The human oral squamous cell carcinoma lines SCC66 and SCC125 were obtained from Dr. Susanne Gollin (University of Pittsburg, Pittsburg, PA) (37) and grown in minimum essential media (MEM) as previously described (31). The human skin squamous cell carcinoma cell line SCC13 (Dr. Stuart Yuspa, NIH) was grown as described (38, 39). Murine primary keratinocytes were created and maintained in low-Ca2+ keratinocyte E medium as previously described (29, 31, 40). All cells were grown at 37°C with 5% CO2.
For chemical inhibitor experiments cells were grown to 70% confluency on coverslips and the media was replaced immediately prior to experimentation. Scd1 inhibitor (A939572) (41, 42) was purchased from Med Chem, concentration of 1µM was used in experimental studies. Soat inhibitor TMP-153 sc-200649 (43) was purchased from Santa Cruz Biotechnology. Soat inhibitor diluted in keratinocyte media were added to experimental wells up to 0.5 µM concentration for 24 hours and equivalent dilutions of ethanol were added to control wells. Cells were rinsed with phosphate buffer saline (PBS) prior to fixation in 1.6% paraformaldehyde for immunofluorescence staining. For the growth rescue experiment, keratinocytes isolated from the skin epithelium of Runx1 iKO newborn mice were treated with final working concentration of 2µM 4-hydroxy tamoxifen (4-OHT) dissolved in culture media and simultaneously added Oleate (Sigma) at the concentration of 12.5µM.
Membrane fluidity experiments
Measurements of membrane organization or fluidity were performed as described (44) with some modifications. Specifically, 2 million cells were suspended in 200 µL of 1X PBS and 1.8ml of Tyrodes/BSA buffer in 1 µM of Laurdan or Patman reagents (Molecular probe) for 15 minutes at 37°C with gentle shaking. Cells were subsequently washed and suspended in Tyrodes buffer. Measurements were performed at 25°C using an SLM steady state fluorimeter (# 8000C; SLM instruments, Urbana, IL) with excitation at 360 nm and emission spectra from 400 nm to 550 nm. Generalized polarization (Gp) was calculated as Gp = (F430 nm – F500 nm) / (F430 nm + F500 nm) after the background was subtracted.
Topflash Wnt reporter activity assay
Transfection cocktail was made using TransIT-Keratinocyte reagents according to the manufacturer's instruction (Mirus, WI, USA). After 16 hours, media was refreshed with inhibitors, A939572: 1 µM, TMP-153 sc-200649 : 0.5 µM, Oleate: 12.5 µM (Sigma). After 16 hours of inhibitor pretreatment, cells were refreshed with media containing inhibitors with or without Wnt3a (50 ng/mL) (ab81484, Abcam, MA, USA). After 24 hours, we harvested cells with 50 µL protein lysis buffer (Promega, WI, USA), freeze-thawed 3 cycles, and determined TOP Flash activity by dual-luciferase™ reporter (DLR™) assay systems (Promega) using spectrophotometer (Glomax 20/20 Luminometer, Promega).
qRT-PCR and FACS Sorting
Total RNA was isolated from sorted HFSCs and HG cells using RNeasy Micro kit (Qiagen) and from cultured keratinocytes using RNeasy Mini kit (Qiagen). cDNA synthesis and qRT-PCR was performed as described earlier (45). Primers used are listed in Supplementary Table 2. Fluorescence activated cell sorting (FACS) using (BD FACS Aria) was done on single cell suspensions freshly isolated from K15-GFP+ skin and stained with CD34 and α6-integrin, as described (29) to isolate bulge and hair germ (HG) cells as HFSCs and early progenitor cells, respectively.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described before (29, 31), using Runx1 antibodies to test binding to the Scd1 and Soat1 promoter.
Lipid extraction from total skin for lipidome analysis
About 50 mg of mouse skin per sample was extracted by Bligh and Dyer extraction (46). For details check supplementary information. Fatty acid methyl esters (FAME) were analyzed by GC-FID and identified by GC MS/MS as described elsewhere (47). Response factors were calculated with an external standard mixture (GLC462; Nu-Chek Prep, Inc.) and applied to raw areas to correct for differential instrument responses. Percentages of each fatty acid were calculated by dividing the corrected area of individual fatty acid by the corrected area of total fatty acids for each lipid class (45).
Statistical Analysis
Unpaired t-tests were performed in Excel or GraphPad Prism, assuming equal variance among the groups, and P-values of ≤ 0.05 were considered significant.
Methods details are described in supplementary information.
Results
Lipid metabolism genes are changed in response to Runx1 expression in HFSCs and progenitor cells
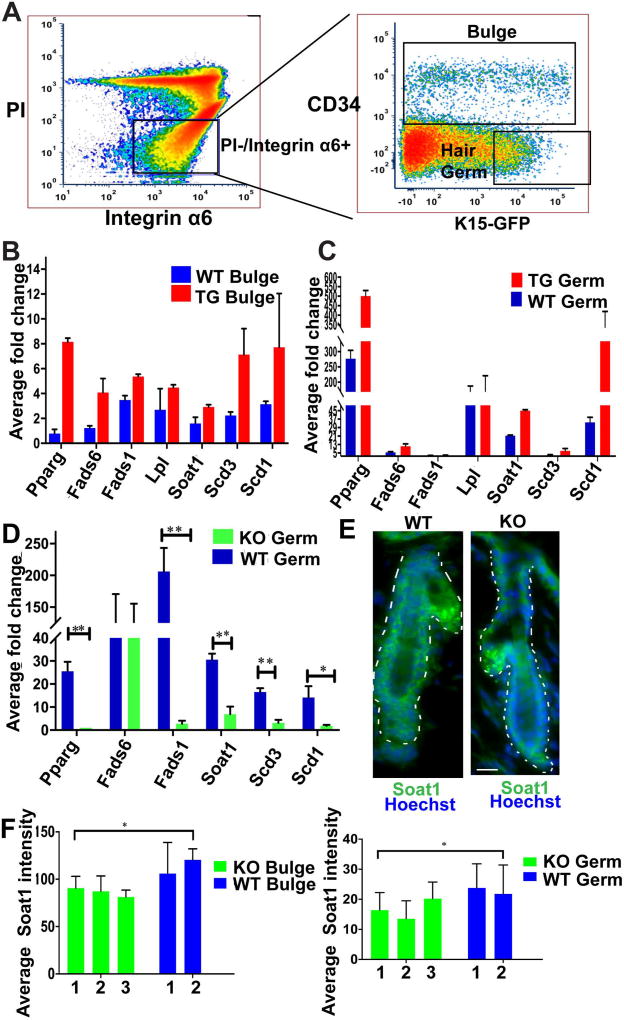
Previously, we identified potential Runx1 target genes from HFSC (bulge) and early progenitor (hair germ, HG) cells during the 1st telogen of mouse skin (29). For this, we employed pTRE-mycRunx1(29); K14-rTA transgenic (TG) mice with K15-EGFP (36) expression, which were doxy-fed for 1-day at PD19 to induce elevated Runx1 expression. Single-cell suspensions from mouse skin were then subjected to fluorescence activated cell sorting (FACS) to isolate bulge and HG cells respectively as CD34+/α6+ and CD34−/α6+/GFP high (29). Among the thousands of dis-regulated mRNAs obtained from Affymetrix microarrays of sorted cells, we identified a defined sub-set of 42 bulge up-regulated genes with known functions in lipid metabolism (Table 1). Using here the same experimental scheme we set out to verify the expression of some lipid-related genes by qRT-PCR (Fig. 1A–D). Runx1 TG bulge and HG sorted fractions (Fig. 1B–C) showed an increase in lipid related genes such as: Pparg, implicated in lipid homeostasis (48); Soat1, the rate limiting enzyme that converts cholesterol to cholesterol ester; Scd1 and Scd3 enzymes, which catalyze the conversion of saturated fatty acids (SFA) to D-9 monounsaturated fatty acids (MUFA); Fads6 and Fads1, which add double bonds on fatty acids. Lpl is involved in triacylglycerol hyrdrolysis (49) and its expression did not change.
Figure 1. Expression of lipid metabolism genes in mouse skin.
(A) Representative FACS dot plot shows sorting gates to isolate bulge (CD34+/α6 integrin+/GFP+/−) and hair germ (HG) early progenitor cells as (CD34−/α6+/GFP+), as previously characterized (1). The brightest 1/3 of the GFP+ cells were used, to avoid potential cross-contamination between bulge and hair germ populations (1).
(B) qRT-PCR from Runx1 inducible transgenic (TG) FACS sorted bulge cells shows an increase in expression of lipid metabolism genes as compare to WT littermates. (n=2 mice from each genotype).
(C) qRT-PCR from Runx1 inducible transgenic (TG) FACS sorted hair germ (HG) cells shows an increase in expression of lipid metabolism genes as compare to WT littermates. (n=2 mice from each genotype).
(D) qRT-PCR from Runx1 inducible knockout (iKO) FACS sorted HG cells shows a decrease in expression of lipid metabolism genes as compare to WT littermates. (n=3 mice from each genotype, P value throughout all figures that are <0.05 is represented as “*” on the graph and p value <0.005 represented as “**”.
(E) Immunofluorescence staining shows expression of Soat1 (green), Hoechst (blue) in HG and bulge is reduced in the Runx1 KO. Scale bar, 20 µm.
(F) Quantification of Soat1 expression in bulge and HG shows significant decrease upon Runx1 knockout. Numbers at bottom indicate individual mouse ID. (n=40 hair follicles per genotype).
Next, we tested the expression of these genes in bulge and HG cells sorted at PD20 from inducible knockout (iKO) Runx1fl/fl, K14-CreERT2; K15-EGFP mice and WT; K15-EGFP control littermates injected with tamoxifen at PD17. Some of the lipid-related genes, including Scd1 and Soat1 show significant decreased level upon Runx1 knockout (Fig. 1D).
Soat1 protein expression was also decreased in both bulge and HG in Runx1 KO skin by immunofluorescence staining of frozen skin sections followed by quantification (Fig. 1E and 1F). We also analyzed the expression of Scd1 using several commercial antibodies, and detected strong signal in the sebaceous gland, in line with previously reported mRNA by FISH (50). We surmise that Scd1 protein expression is lower in hair follicle cell types other than sebaceous glands, and hence difficult to detect, although the mRNA upregulation in response to Runx1 was easily detectable in the HG and bulge sorted cells.
Since Scd1 and Soat1 have previously been implicated in hair regulation and in several non-skin cancers, we asked whether they may also play a role in our reported Runx1-mediated control of cell proliferation in normal and cancer epithelial cells (29–31).
Runx1 dictate the expression of Scd1 and Soat1 in cultured keratinocytes
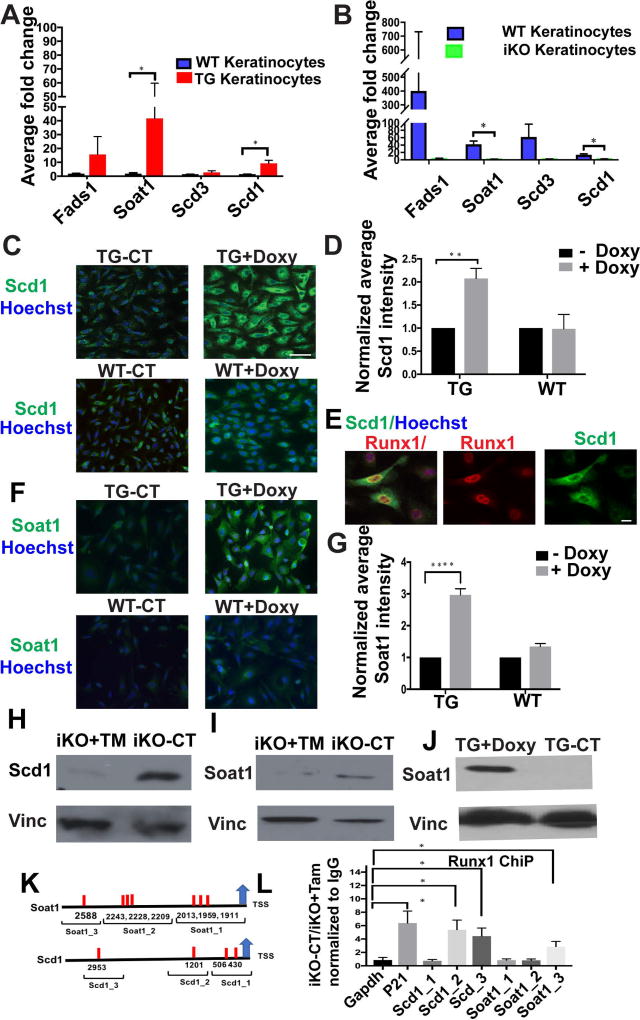
To further examine Runx1 regulation of potential downstream targets and to avoid compounding systemic effects in live tissues, we employed cultured keratinocytes genetically modified to inducibly manipulate Runx1 levels. Primary keratinocytes were isolated from the newborn skin epithelium of Runx1 TG and Runx1 iKO mice. Runx1 reduction is induced by administration of 4-OHT to the iKO cells for 48 hours (31), while Runx1 elevation is induced upon doxycycline treatment of TG cell lines (29) for 6 hours. First, we analyzed by qRT-PCR the mRNA expression of the several lipid enzymes we previously characterized in vivo. Significantly, the changes in expression levels in cell culture conditions (Fig. 2) were consistent with our in vivo data from Figure 1. In particular, upon Runx1 induction both Scd1 and Soat1 mRNAs were up-regulated in the TG cells and downregulated in the iKO, as expected (Fig. 2A, 2B). Furthermore, Runx1 elevation raised the Scd1 (Fig. 2C–E) and Soat1 protein levels (Fig. 2F–G, 2J).
Figure 2. Runx1 dictate the expression of Scd1 and Soat1 in cultured keratinocytes.
(A, B) qRT-PCR validation of mRNA levels for candidate genes in cultured Runx1 inducible knockout (iKO) following 48 hours of 4-OHT and transgenic (TG) keratinocytes following 6 hours of doxycycline relative to WT controls. (n=3 from each represented genotype). Significant p values (<0.05) are shown as *.
(C, F) Immunofluorescence staining of cultured keratinocytes shows expression of Scd1 or Soat1 (green) in response to doxycycline (doxy)-induced Runx1 (TG+Doxy), TG control with no doxy (TG-CT), and WT cells without doxy (WT-CT) ad with doxy (WT+Doxy). Scale bar, 20 µm.
(D, G) Quantification of average Scd1 or Soat1 staining after Runx1 induction (TG+Doxy) when compared with control (TG-CT) and (WT+ Doxy) when compared with no doxy control (WT-CT). (n=3 experiments, ~ 150–200 cells per experiment); normalized against control in each group, average intensity is arbitrary value. P value that are <0.05 is represented as “*” on the graph and P value <0.0001 represented as “****”.
(E) Immunofluorescence staining showing correlation of doxy-induced Runx1 elevation (red) with enhanced Scd1 expression (green). Nuclei (blue) were counterstained with Hoechst. Scale bar, 20 µm.
(H, I) Western blotting showing a decrease of Soat1 and Scd1 protein levels in cultured keratinocytes of Runx1 iKO after 48 hours of 4-OHT treatment, denoted as (TM). Vinculin (Vinc) works as loading control.
(J) Western blotting against vinculin (Vinc) or Soat1 antibodies showing an increase of Soat1 protein in Runx1 TG keratinocytes after 12 hours of doxy-induced Runx1 expression.
(K) Scheme representing ChIP probed regions to cover predicted Runx1 binding sites (red bar) the promoters of Scd1 and Soat1.
(L) ChIP of CT (iKO-CT) and Runx1 KO (iKO+Tam) keratinocytes using Runx1 antibody, P value for pair wise comparisons that are <0.05 is represented as “*” on the graph, (n= 3 independent experiments).
Conversely, consistent with mRNA expression patterns both Soat1 and Scd1 proteins were found down-regulated in the Runx1 iKO cells, 48 hours after addition of 4-OHT, as shown by Western blots (Fig. 2H, I). To further explore the molecular mechanism of gene regulation of Scd1 and Soat1, we examined Runx1 direct binding on the promoter of Scd1 and Soat1. We employed eukaryotic promoter database (https://epd.vital-it.ch/index.php) and TRANSFEC (http://gene-regulation.com/pub/databases.html) to determine binding sites for Runx1 in the promoter of Scd1 and Soat1; we experimentally tested 3 regions containing Runx1 binding sites in each of the promoters of Scd1 and Soat1 (Fig. 2K).
We performed ChIP as described before (25, 31) using K14-CreERT2;Runx1fl/fl cell lines in WT (iKO-CT) and Runx1 iKO (iKO+ Tam) cells. We used a region of the Gapdh promoter as a negative control (Fig. 2L), and previously known binding site to the P21 promoter (31) as a positive control (Fig. 2L). Our data showed a significant binding of Runx1 at site Scd1 1201, Scd1 2953 and Soat1 2958 as demonstrated by > 2.5 fold binding as compare to negative control Gapdh (Fig. 2L). Together, the data suggest that the transcription factor Runx1 controls the levels of mRNA and protein expression of Scd1 and Soat1 in mouse keratinocytes, and that this is likely through direct binding to its consensus DNA sites on the Scd1 and Soat1 promoters.
Scd1 & Soat1 are highly expressed in Squamous Cell Carcinomas
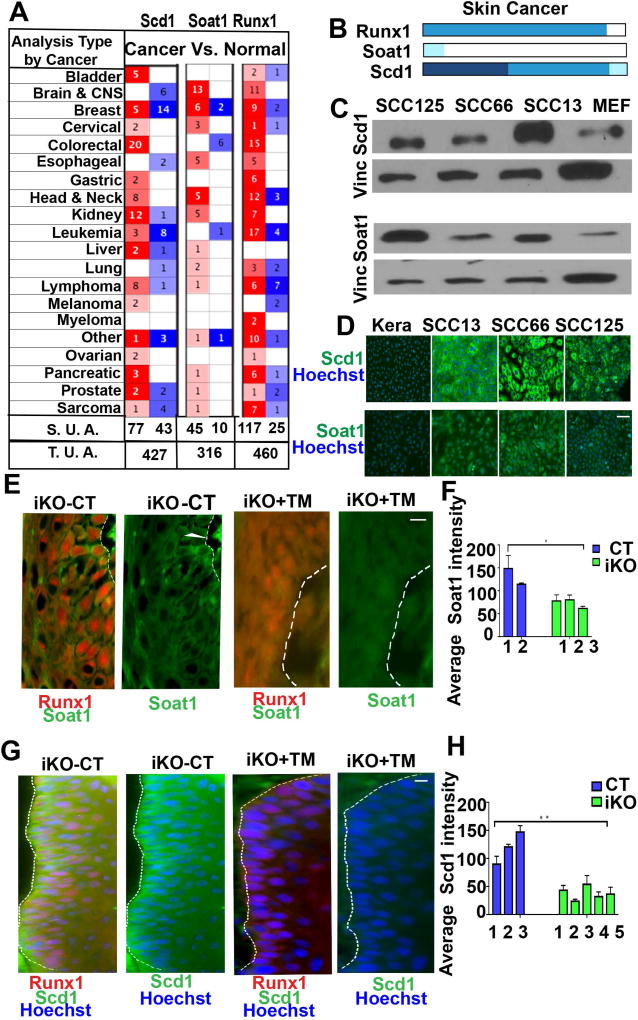
Cancer cells and tissue stem cells can use common pathways to regulate self-renewal and differentiation (24). We previously showed that the HFSC factor Runx1 is important for at least a subset of skin and oral (head and neck) squamous cell carcinoma (31), and others have implicated Runx1 in additional epithelial cancers and in leukemia (51). Scd1 and Soat1 have also been implicated in several cancers (41, 42, 52), but their potential role in squamous cell carcinomas has not been specifically analyzed yet. First, we broadly looked for Scd1 and Soat1 expression into a publically available database Oncomine™ (Compendia Bioscience, Ann Arbor, MI), which has gene expression data comparing primary tumors and normal tissues. We found that both Scd1 and Soat1 were among the top 10% of all overexpressed genes in head and neck as well as other types of tumors. This included epithelial tumors in which Runx1 is known to play a role, such as breast, cervical, and ovarian (Fig. 3A). Since Oncomine contained very few samples of squamous cell carcinomas of the skin, we next examined the Human Protein Atlas (53) (http://www.proteinatlas.org/) for Scd1, Soat1 and Runx1 protein expression in human patient tumor samples (Fig. 3B and Supplementary Fig. 1). Corroborating our previous findings (31), our collective analysis supports the notion that Runx1 is highly expressed in oral as well as skin carcinomas. We also find that Scd1 is moderately to highly expressed in both oral and skin squamous carcinoma samples. Finally, Soat1 is expressed at some level in few skin and oral carcinoma samples, but generally shows higher variability of expression in different tumor samples (Supplementary Fig. 1). Next, we examined the expression of Scd1 and Soat1 proteins by immunofluorescence staining and Western blots of human squamous cell carcinomas (SCC) cell lines, namely oral (SCC66, SCC125) (54), and skin SCC (SCC13) (38, 39). Previously we reported elevated Runx1 expression in all these human cancer lines (31) and here we show that they also express readily detectable levels of Scd1 and Soat1 (Fig. 3C, D; Supplementary Fig. 3). Previously we have shown that iKO of Runx1 during DMBA/TPA tumor initiation stage in mouse skin impairs tumor formation while iKO post-tumor formation results in tumor shrinkage (31). Suggestively, when we analyzed wild type and Runx1 iKO mouse skin papilloma tumors by immunofluorescence we found that Runx1 loss resulted in reduction of Scd1 and Soat1 expressions (Fig. 3E–H). Taken, together these data document expression of the Scd1 and Soat1 lipid enzymes in human epithelial cancer cells, including in cancer lines previously found to depend on Runx1 expression for proliferation (31).
Figure 3. Expression of Scd1 & Soat1 in Squamous Cell Carcinomas.
(A) Table showing the number of significant unique analyses (S. U. A.) and total unique analyses (T. U. A.) using Oncomine data sets curated for cancer versus normal tissue pointing to up (red) or down (blue) regulation of Runx1, Scd1, and Soat1 in a variety of cancers. Number in each cell corresponds to number of analysis that meets the threshold of top 10% of all overrepresented genes in tumors.
(B) Human Protein Atlas analyses showing expression profiles of Runx1, Soat1 and Scd1 in skin cancer which comprises tumors samples of squamous cell carcinoma and basal cell carcinoma. Runx1 is expressed in most skin cancer tumors, Soat1 shows low expression, while Scd1 shows moderate to high expression. Darker shades of blue correspond to higher expression.
(C) Western blotting of skin carcinoma (SCC13) and oral carcinomas (SCC66, SCC125) cell lines show elevated levels of Scd1 and Soat1 as compare to mouse embryonic feeders (MEFs) used as controls. Vinculin (Vinc) served as the loading control. Shown is one representative example of 3 independent experiments. (For quantification of data in C&D in repeat experiments see Supplementary Fig. 3)
(D) Immunofluorescence staining of skin squamous cell carcinoma (SCC13) and oral squamous cell carcinomas (SCC66, SCC125) showing Scd1 (green) and Soat1 (green). Note striking Scd1 and Soat1 up-regulation in SCCs as compare to wild type (WT) mouse keratinocytes. Scale bar, 20 µm.
(E–H) Immunofluorescence staining (E, G) and quantifications (F, H) of mouse skin tumors show that Scd1 (green) and Soat1 (green) are highly expressed in WT tumors and are down-regulated in iKO tumors upon loss of Runx1. White line delineates epithelial cell clusters where Scd1 is expressed. Numbers represent tumors derived from different mice. Scale bar, 20 µm. P value that are <0.05 is represented as “*” and P value <0.005 represented as “**” on the graph.
Finally, we investigated whether Runx1, Scd1 and Soat1 expression may co-localize in human skin tumors (SCCs) by immunofluorescence staining of serial sections stained with either Runx1 and Scd1 or Runx1 and Soat1 (Supplementary Fig. 4, details discussed in Supplementary information). In conclusion, we were unable to find a positive correlation in Runx1 levels of expression with Scd1 and Soat1, suggesting that their potential inter-dependence is highly cell-type specific. This is not surprising since in normal tissue Scd1 is highly expressed in sebaceous glands, where Runx1 is absent. With the caveats of the serial sections imperfect overlap notwithstanding, these data suggest that rare cells distributed in specific patterns within the epithelial structures may express all three genes (Scd1, Soat, and Runx1). It is possible that these rare cells from primary tumor might be the ones that proliferate well in cell culture, thus explaining our results in the SCC cell lines.
Scd1 and Soat1 inhibition impairs epithelial normal and cancer cell proliferation
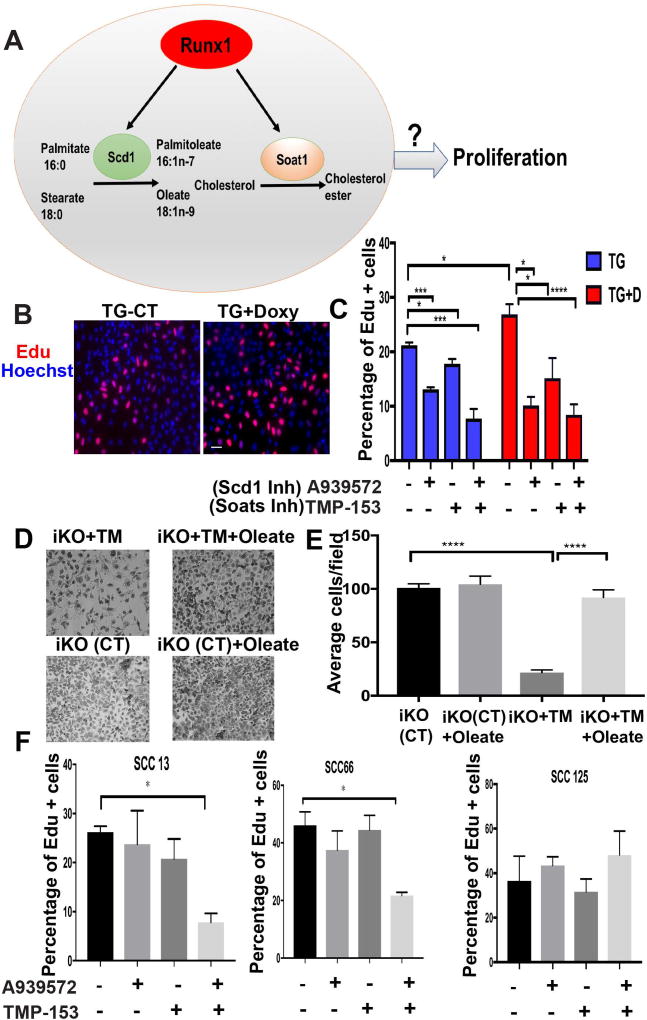
Previously we showed that Runx1 regulates proliferation of keratinocytes and of several human SCC lines (29) (31) including SCC-13, -66 and -125, employed in this study. Interestingly, lipid enzymes Scd1 and Soat1 augment tumor growth and proliferation of several cancer types and may be promising therapeutic targets via chemical inhibition (41, 55, 56). However, their possible involvement in SCC proliferation is unknown. Next we asked if chemical inhibition of Scd1 and Soat1 enzymatic activity may affect proliferation of mouse keratinocytes and human SCC cell lines (Fig. 4A). Primary keratinocytes isolated from mouse skin epithelium of Runx1 TG newborn mice were grown in the absence of doxycycline (TG-CT) or presence of doxycycline (TG+doxy) to elevate Runx1 expression. EdU added to culture medium 20 min. prior to fixation of labeled cells indicated S-phase, and revealed significant increased upon high Runx1 expression, as expected (29) (Fig. 4B). Next, we inhibited expression of Scd1 and Soat1 using well-established small molecule chemical inhibitors (A939572) (41, 42) (57–59) and TMP-153 (43, 60, 61), respectively. A939572 inhibits both mouse and human Scd1respectively (62). A939572 has no inhibitory activity to the co-enzymes of Scd1, cytochrome b5 and cytochrome b5 reductase, and interact directly and specifically with Scd1 (62). Furthermore, Scd1 shRNAs and A939572 have similar effects on renal carcinoma cell proliferation (41) attesting to the specificity of this inhibitor. TMP-153 is a potent, non-competitive Acetyl-coenzyme A acetyltransferase ACAT inhibitor of Soat1 and 2 enzymes (63), which drive rate limiting steps in forming cholesterol-esters (64). Since Soat 2 did not surface as a potential target in our microarray, we didn’t study it further. We found that A939572 and TMP-153 individually and synergistically reduce proliferation of both WT and Runx1 TG keratinocytes (Fig. 4B, 4C). This suggests that Scd1 and Soat1 may be needed for proliferation of cultured keratinocytes and that elevated Runx1 levels cannot rescue effects of Scd1 and Soat1 inhibition.
Figure 4. Scd1 and Soat1 activity affects epithelial cell proliferation.
(A) Scheme proposing Runx1 mediated pathway to regulate epithelial cell proliferation.
(B) Representatives images of Edu staining (red) to show effect of Runx1 on proliferation of keratinocytes. Runx1 is overexpressed after addition of doxycycline to TG cells (TG+Doxy); TG cells alone serves as a control (TG-CT).
(C) Quantification of percentage of proliferating cells with or without treatment of chemical inhibitors A939572 or TMP-153 to TG and TG+Doxy cells. (n=3, average 500 cells per sample were counted). P value that are <0.05 is represented as “*”, P value <0.005 represented as “**”, and P value <0.0001 represented as “**** ”.
(D) Representative images of Tamoxifen (TM)-induced Runx1 iKO keratinocytes with or without addition of Oleate, after 4 days of plating. (Also see Supplementary Figure 3) (n=2 different cell lines of represented genotype).
(E) Quantification of number of cells per field of Tamoxifen (TM)-induced Runx1 KO keratinocytes with or without addition of Oleate after 4 days of plating. P value that are <0.0001 represented as “****”.
(F) Quantification of percentage of proliferating cells (EdU+) with or without treatment of chemical inhibitors A939572 or TMP-153 to human SCC cells. Note that co-inhibiting Scd1 and Soat1 results in a synergistic decrease on Edu+ cell number of SCC 13 and SCC 66 cell lines. Significant P value that are <0.05 is represented as “*” on the graph.
Inhibition of Scd1 is known to result in depletion of its end-product oleate, which is needed for cell proliferation (65). Previously we reported Runx1 iKO cell proliferation defects (27), and here we attempted to rescue these defects by addition of oleate. Indeed, 4-OHT addition to the Runx1 iKO keratinocyte cell culture medium for 48 hours impaired cell proliferation relative to WT, and this was overcome by oleate (Fig. 4D, 4E & Supplementary Fig. 2). These data further suggest a role of Scd1 and its product oleate downstream of Runx1 in controling proliferation of normal keratinocyte.
Next, we tested whether inhibition of Scd1 and Soat1 also affects proliferation of human SCC cell lines. Interestingly, we found that unlike seen in normal keratinocytes (Fig. 4C) where either A939572 or TMP-153 alone was sufficient to significantly reduce proliferation, SCCs required both inhibitors to produce a significant effect, as shown by percentage of EdU+ cells. The combination of inhibitors resulted in an abrupt decrease in proliferation of SCC13 (skin) and SCC66 (oral), with no significant effect on SCC125 (oral) (Fig. 4F). The observed difference between normal and cancer cells growth in culture could indicate that cancer cells can better overcome growth obstacle inflicted by alteration of lipid metabolism. These data together suggest that Scd1 and Soat1 activity is important for epithelial cell proliferation in both normal and cancer conditions, potentially downstream of Runx1.
Lipids and membrane organization is altered in response to Runx1
Next, we asked if the mRNA changes in lipid-metabolism related genes downstream of Runx1 can be correlated with physiological changes in the lipid composition. GC-MS- based lipidome analysis from total skin of Runx1 TG, Runx1 iKO and their WT littermate controls shows that fatty acids in different classes of lipids were changed in response to Runx1 levels (Fig. 5A–C & Supplementary Table 3). Scd1 preferentially converts stearic acid (C18:0) to oleic acid (C18:1) and palmitic acid (C16:0) to palmitoleic acid (C 16:1). We observed a significant decrease in Scd1 activity upon Runx1 loss as shown by the ratio of its product/substrate (Fig. 5A) in the polar lipids category. This category of lipids is known to play important roles in signaling, and it was in line with our gene expression data and supporting its physiological relevance. Branched chain fatty acids (BCFA), which are synthesized in mammalian skin, appear as surface lipids in humans (66), mice (67), and fish (68). Our analysis show that BCFA appear at greater than over 10% in the cholesteryl ester (CE) fraction of whole skin samples (Supplmentary Table 3). Ratios of iso-16:0/16:0, iso-21:0/20:0 and iso-23:0/24:0 are significantly increased in the Runx1 knockout mice skin compared to controls (Fig. 5B). The CE lipid fraction also reveals significant decrease in Scd1 activity, as suggested by decrease in the ratio of oleate to stearic acid (Fig. 5C). Thus, our lipidome analysis of normal and epithelial-deficient Runx1 skin reveals changes in fatty acid composition that are consistent with a decrease in Scd1 activity.
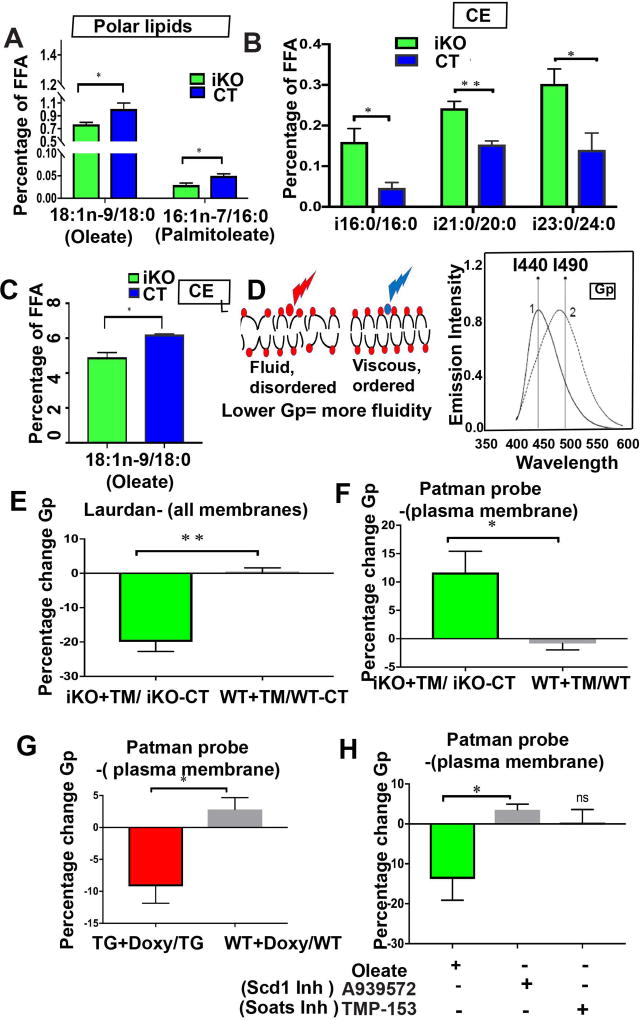
Figure 5. Runx1 levels impact lipid content and membrane organization.
(A–C) Data from lipidome analysis of total skin show changes upon Runx1 loss. P value that are <0.05 is represented as “*” and P value <0.005 represented as “**”: (A) ratios of Scd1 products (oleate and palmitoleate) and substrates (stearate and palmitate, respectively) change in the polar lipids category. (B) (C) Ratio of products and substrates in cholesterol ester (CE) lipid fraction with or without Runx1 loss. (B) Branched chain fatty acid iso (i)-16:0, i21, i23 increase significantly in response to Runx1 loss. (C) Oleate (Scd1 product) decreases in response to Runx1 loss. (n=3 CT mice and n=5 Runx1 iKO mice were used for lipidome analysis).
(D) Cartoon showing fluid or disordered state of membrane (left) due to unsaturated fatty acids versus viscous or ordered state of membrane (right). Laurdan is a membrane-intercalating fluorescent probe that shifts emission in the phospholipid bilayer of membranes from red (490 nm) in the membrane-disordered state to blue (440 nm) in the membrane-ordered state, as shown in the left cartoon and in the emission spectrum on the right. Fatty acid composition of membrane affects membrane organization or fluidity, which can be measured as a function of generalized polarization (Gp), using probes such as Laurdan and Patman.
(E) Membrane fluidity using Laurdan, which binds to all membrane, indicates decreased Gp value in the Runx1 iKO, which translates in more overall membrane fluidity. P value <0.005 represented as “**”:
(F–G) Membrane fluidity assays using Patman probe, which binds specifically to the plasma membrane. Note Runx1 elevation leads to decreased Gp (e.g. more membrane fluidity) and Runx1 knockout leads to increased Gp (e.g. less membrane fluidity). P value that are <0.05 is represented as “*” on the graph.
(H) Membrane fluidity assay using Patman probe indicates that fluidity can be altered by feeding cells with Oleate or A939572 but not TMP-153. (n>= 3 for E–G). P value that are <0.05 is represented as “*” on the graph.
Based on the classical models of lipid membrane assembly, the fatty acid composition of the lipid bilayer affects how membrane structure is organized, a characteristic sometimes referred to as membrane fluidity (69). Changes in unsaturated and branched fatty acids which we found to be changed in the polar lipids and CE fractions are expected to influences the phase transition behavior of membranes and the membrane fluidity (70). The double bonds present in unsaturated fatty acids (e.g. oleate) cause kinks in the long hydrocarbon tail resulting in poor packing within the membrane and hence a less ordered state (fluid or liquid phase). Conversely, saturated fatty acids pack more closely resulting in a more ordered state (viscous or crystalline phase) (71, 72). In our case, decreased Runx1 levels causes decrease Scd1 expression and activity (Fig. 5A), which in turn produces less oleate (an unsaturated fatty acid) in the polar lipid class (Fig. 5A), which contributes a major fraction to the plasma membrane thus likely resulting in less membrane fluidity.
In order to assess changes in lipid membrane organization with varying levels of Runx1 we first utilized the membrane specific fluorescent probe Laurdan, which binds to all lipid membranes; it is sensitive to the polarity of the environment and exhibits a 50 nm red shift in the emission spectrum over the gel to liquid –crystalline phase transition (Fig. 5D) (73). We found that upon Runx1 loss, there is a decrease in generalized polarization (GP), a mathematical variation consistent with an increase in membranes fluidity (74) (Fig. 5E). This observation was not consistent with our original expectation. However, Laurdan binds to all cellular membrane, not only the plasma membrane, and our lipidome analysis suggests complex changes in different classes of lipids not enriched in plasma membranes upon varying Runx1 levels (Fig. 5A–C). For instance, the changes in brached fatty acids may be responsible for this result. Nevertheless, these data further support the idea that Runx1 level affects the composition and the organization of lipids within the cellular membranes.
To specifically examine fluidity in the plasma membrane upon induced Runx1-level changes, we employed a specialized fluorescent dye named Patman probe (75). We found that upon Runx1 knockout there is an increase in GP, or plasma membranes are indeed less fluid (Fig. 5F), as expected from decreased Scd1 activity and low oleate production. Conversely, high Runx1 expression decreases GP, hence plasma membranes were more fluid (Fig. 5G). This was consistent with Runx1 induced higher activity of Scd1, and the over-production of an unsaturated fatty acid, oleate, as pointed by our gene expression and lipidome data (Fig 1B–C, 2A–D & 5A–C). Furthermore, we found that plasma membrane fluidity was decreased upon inhibiting Scd1 and increased upon addition of Scd1-product, oleate, as expected (Fig. 5H). In contrast, inhibition of Soat1 did not change the membrane fluidity (Fig. 5H). These data suggest that varying the expression of Runx1 changes plasma membrane organization and this is partly mediated by Scd1 and likely by the concentration of its product, oleate, within the lipid bilayer.
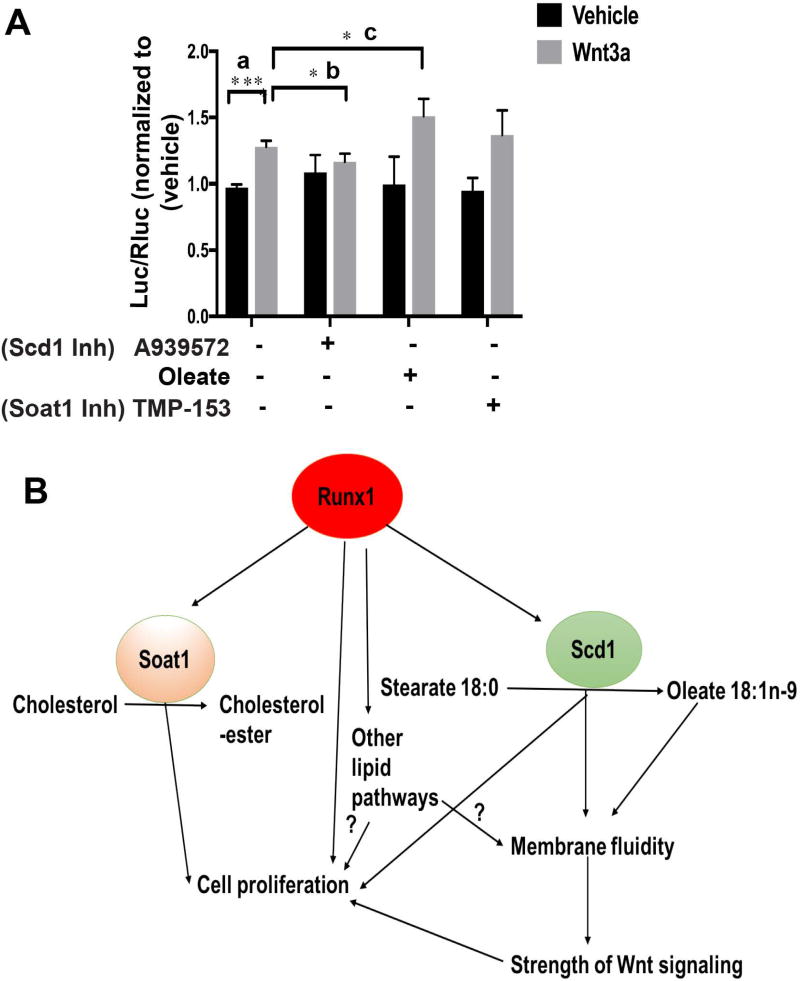
Membrane organization may in principle control different signaling outcomes (22, 76) although direct evidence for this model is scarce. Previously we linked Runx1 in the skin epithelium and cultured keratinocytes with increased Wnt signaling (28), a pathway important for epithelial cell proliferation (77), and for carcinogenesis (78). Thus, we next investigated a possible relationship between membrane fluidity and strength of Wnt signaling. We measured the strength of Wnt signaling using a TOPflash Wnt reporter assay (79) in the presence of oleate, and chemical inhibitors A939572 or TMP-153 (Fig. 6A). Upon adding exogenous Wnt3a to the medium as reported earlier TOPflash activity shows a significant increase (Fig. 6A, a). TOPflash activity relative to Wnt3a alone was significantly decreased by addition of A939572 (Fig. 6A, b) and increased by addition of oleate (Fig. 6A, c). This was in line with decreased and increased membrane fluidity changes, respectively (Fig. 5H). TMP-153 had no significant effect on TOPflash activity (Fig. 6A), consistent with no change in membrane fluidity (Fig. 5H). In summary, these data suggest that the strength of Wnt-induced signaling may be modulated by the activity of a lipid-modifying enzyme Scd1 and its product, oleate, and this positively correlates with membrane fluidity (Fig. 6B).
Figure 6. Scd1 activity affects Wnt signaling.
(A) Strength of Wnt signaling is measured by TOPflash assay, as described (2) upon treatment with inhibitors A939572, TMP-153 or Oleate for 16 hours followed by the addition of Wnt3a for 24 hours. (n=4 for each represented treatments). Y axis, Luc/Rluc. ratio of luciferase (Luc) units over Renilla luciferase (RLuc) in inhibitor-treated groups is normalized to vehicle. P value for pairwise comparisons a, b, c is represented on the graph. P value <0.0001 represented as “****” and P value that are <0.05 is represented as “*”.
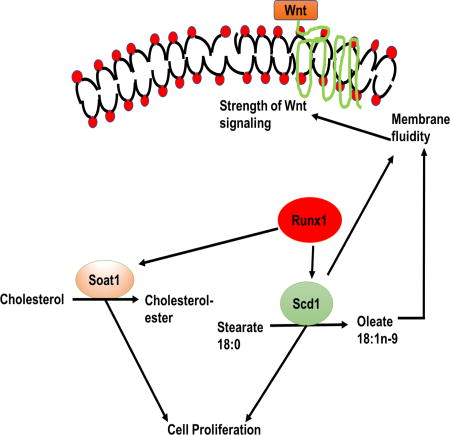
(B) Model: Runx1 modulates levels of Scd1 and Soat1 and its products oleate and cholesterol esters to regulate proliferation in part through membrane organization (or fluidity). Scd1 activity and oleate concentration in the plasma membrane phospholipids in turn alters Wnt activity resulting in higher responsiveness to signals and enhanced cell proliferation.
Discussion
In this study, we uncovered a novel molecular mechanism downstream of the stem cell and cancer factor Runx1, previously implicated in hematopietic and epithelial hair follicle stem cell function as well as in leukemia and epithelial squamous cell carcinoma (SCC) (80). We previously found that Runx1-expressing cells from the hair follicle bulge are at the origin of skin epithelial tumors (31). Recently Runx1 has been implicated in RNA biogenesis (81). While we have previously demonstrated Runx1 role in control of proliferation in part via p21 (32), Wnt (28) and Stat signaling (31), a full picture of its downstream mechanisms driven by its thousands of HFSC target genes (29), is still lacking. Here we bring evidence that among the Runx1 target genes are enzymes important in cell-intrinsic control of lipid metabolism. We show that Runx1 changes the overall lipid content in mouse skin in a manner that correlates with the known activity of specific target genes. In particular, we find that in mouse keratinocytes Runx1 controls the mRNA and protein level of lipid-metabolic enzymes Scd1 (and its product oleate) and Soat1 (and its product cholesterol-ester, CE) by directly binding to their promoters. Inhibiting the Scd1 and Soat1 enzymes has synergistic negative effects on cellular proliferation of mouse keratinocytes and of our previously demonstrated (31) Runx1-dependent human epithelial cancer cell lines from skin and oral epithelium. Addition of Scd1-product oleate rescues the Runx1-deficient keratinocyte proliferation defect, suggesting an important role of fatty acid synthesis downstream of Runx1 in control of epithelial cell proliferation and survival.
The Scd1-product oleate was decreased in the Runx1 knockout mice in the phospholipid fraction, which predominates in the plasma membrane. Because oleate is an unsaturated fatty acid its double bond would generate a kink in the hydrocarbon chain that may increase membrane fluidity, and possibly enhance membrane protein movement and signal transduction strength through the plasma membrane. Consistent with this model we show that Runx1 levels affect membrane fluidity in cultured keratinocytes, as does Scd1 activity and the concentration of its product, oleate. Significantly, we find a direct correlation of higher plasma membrane fluidity promoted by Runx1, Scd1 activity, and oleate, with the strength of Wnt signal transduction through the membrane. Wnt signaling is known to promote epithelial cell proliferation and carcinogenesis, HFSC activation and hair cycle progression (33, 34), and we previously showed that Runx1 promotes downstream Wnt signaling in mouse skin epithelium and keratinocytes (28).
Previously it was also shown that Scd1 deficiency impaires desaturation of de novo-synthesized of palmitoyl- to palmitoleoyl-CoA, which prohibits posttranslational O-palmitoleoylation of Wnt3a protein, essential for Wnt3a/β-catenin signaling in stem cell lineage decision in development of the epidermal barrier, hair growth cycle, and sebaceous glands (82). This mechanism might also be at play in regulating Wnt signaling downstream of Runx1 via Scd1 activity. Taken together our data suggest a working model in which Runx1 proliferation in epithelial cells is mediated in part via lipid metabolism, particularly through Scd1 and Soat1 (Fig. 6B). It is interesting to speculate that under the control of Runx1, Scd1 expression and oleate production in hair germ cells during quiescence in vivo may affect the plasma membrane fluidity and the O-palmitoleoylation of Wnt3a protein and thus promote Wnt activation and anagen onset. These possible mechanisms would explain in part the known Runx1 role in hair cycle progression. The composition of the plasma membrane, and possibly its fluidity, has been suggested to affect other signaling pathways, such as EGF and Notch signaling (83). Whereas Soat1 and Scd1 mutations have been associated with defects in sebaceous glands and with scarring alopecia, their potential roles in controlling hair cycle through directly promoting stem cell activation and proliferation have not been yet explored. Soat1 protein has been previously detected in the hair medulla and the sebaceous gland, and a mutation in this gene resulted in hair interior defects (84). We find Soat1 mRNA level and protein to be directly driven by Runx1 in the hair bulge and germ of the telogen hair follicle in adult mice. This expression pattern and the upstream regulation by Runx1, which is known to be essential for hair cycle and stem cell activation (25, 27, 29), together with the Soat1-inhibition effect on keratinocyte proliferation, may suggest a potential unexplored role of Soat1 in HFSCs to control hair cycle through stem cell activation. Scd1 loss, in turn, has been recently shown to result in the disruption of sebaceous gland and of the epidermal barrier function, and also in alopecia (82, 85). This phenotype was attributed to a metabolic syndrome and the loss of skin barrier function, while Scd1 role in the epithelial hair follicle stem cell biology remains undetermined. This is particularly appealing, since spontaneous mutations in the Scd1 gene in the Asebia mice cause all growth phases of the hair cycle, anagen, catagen, and telogen to last longer than those of the controls (86). Scd1 expression at high levels in the sebaceous gland (50) may have obscured its role in other cell types due to low expression that is difficult to detect with current antibodies. Our data in vivo detecting mRNA in bulge and hair germ and in keratinocytes (where both mRNA and protein are abundantly expressed), suggest that Scd1 may promote keratinocyte and hair germ cell activation and proliferation, a model that will need direct testing in future.
Our findings in human cancers are preliminary, but suggest a possible appealing model for future testing, in which the epithelial stem cell factor Runx1 regulates lipid metabolism in both normal and cancer epithelial cells. Many studies support the idea that cancer cells activate the cell intrinsic biosynthesis pathway for fatty acid metabolism to produce de novo lipogenesis (87, 88). Scd1 and Soat1 have been previously implicated in proliferation of other cancers including lung, prostate, and kidney (41, 55, 89) as well as glioblastoma and breast cancer (52, 56), respectively. Scd1 has been shown to decrease the ratio of saturated to unsaturated fatty acids during G1 phase of cell cycle, and to be essential for cancer cell growth and survival (88). We found that Scd1 is also expressed widely in epithelial human tumors as displayed our analyses of primary tumors of both Oncomine and the Human Protein Atlas databases. Strongly corroborating our Scd1 protein expression data in human SCCs cell lines and human SCC tumor samples also supported this notion. Soat1 showed a lower and variable expression throughout the human SCC tumors of the head and neck and SCC tumors of the skin suggesting broader heterogeneity of its expression in these cancers. Our immunostaining of serial primary human tumor sections indicates that only rare cells may express Runx1, Scd1 and Soat1 together. It is tempting to speculate that these are a postulated “cancer stem cell” population that also proliferates well in cell culture, explaining our results with the SCC lines. Very likely, pathways independent of Runx1 may also drive expression of Scd1 and Soat1 in a complex tumor microenvironment. We also found that Scd1 and Soat1 are down-regulated in mouse papillomas upon Runx1 loss in vivo. However, papilloma are early stages of tumorigenesis that eventually progress to SCC, it is also possible that Runx1 regulates Scd1 and Soat1 expression primarily during early stages of tumor initiation and growth. Clearly more work is needed to understand the relationship between Runx1 and lipid metabolism via Scd1 and Soat1 in human epithelial cancers.
Evidences from cancer preclinical and clinical trials suggest that chemical inhibition of lipid metabolism enzymes may be a potentially promissing approach to block cancer (90). TMP-153 inhibits the key enzymes for cholesterol esterification (i.e. Soat1 and Soat2) and aberrant cholesterol ester accumulation along with accumulation of oncogenic events promote prostate cancer aggressiveness (91). It is worth noting that our combination of A939572 and TMP-153 drugs to target Scd1 and Soat1 results in synergistic decrease in the proliferation of keratinocytes and of two of three human SCC cell lines. Unlike keratinocytes, which respond to either A939572 or TMP-153, human SCC cell line proliferation remains unaffected by each drug used individually, possibly because cancer cells have compensatory mechanisms to maintain their lipid production. Our data warrants further investigation of these drug combination for cancer therapy.
Although the role of lipids in the skin permeability barrier is well established (92, 93), there are no studies to date to investigate if lipids are crucial for HFSCs activation. Previously, lipid profiles in human patients with alopecia demonstrate abnormal lipid contents (94), suggesting that drugs targeting lipid metabolic enzymes may be beneficial in the treatment of alopecia. While we uncover here a small branch of lipid metabolism downstream of Runx1 via Scd1 and Soat1, many of our Runx1 targets with predicted lipid metabolism functions remain unexplored. In order to systematically understand the role of lipid metabolism in stem cell activation and thoroughly investigate the validity of our model (Fig. 6B), we need lipidome analysis from sorted hair germ and bulge stem cells at different hair cycle stages and in Runx1 mutants, and direct genetic targeting of Scd1, Soat1 and the other lipid enzymes in the HFSCs in vivo.
Conclusion
We propose a working model in which Runx1 induces changes in lipid metabolism genes along with thousand other downstream factors, to create a blueprint in the lineage primed early progenitor cells to prepare for subsequent rapid proliferation and differentiation. An intricate interplay may occur between lipid synthesis states and gene regulation through stem cell conserved transcription factors. When these factors promote stem cell activation and proliferation they may be highjacked by cancer cells originating in these stem cells to ensure their metabolic needs. This model can be tested in future not only in the hair follicle, but also in other systems where Runx1 plays a role, such as the hematopoietic stem cells and associated leukemia (80).
Supplementary Material
Representative images after staining with crystal violet of WT, 4-OHT-induced Runx1 iKO keratinocytes with or without addition of Oleate, after 4 days of plating in cell culture.
(A) (C) Quantification of immunofluorescence staining of squamous cell carcinoma (SCC) cells normalized to control non-cancer epithelial cells labeled as wild type. Note striking Scd1 and Soat1 up-regulation in SCCs as compare to control.
(B) (D) Quantification of western blotting of skin carcinoma (13) and oral carcinomas (66, 125) cell lines shows elevated levels of Scd1 and Soat1 as compared to non-cancer cell controls. Normalization in each experiment was to Vinculin (Vinc) and to non-cancer cell control.
P value that are <0.05 is represented as “*” on the graph and p value <0.005 represented as “**”, p value <0.0001 represented as “****”; Red * represents p value = 0.06.
1,2, 3,4) Representative immunofluorescence staining of human skin squamous cell carcinoma sample from tumor H, A, C, E showing Scd1 (red) and Soat1 (green) & corresponding serial section immunofluorescence staining showing expression of Runx1 (green) & Scd1 (red), Blue (K14). Scale bar: 20µM. White dotted line represents enlarged view and white arrow shows region of colocalization. Scd1, Soat1 and Runx1 rarely colocalize in structures that are K14+made of a single layer of epithelial cells surrounding a void region (see Tumor A boxed areas) 5,6,7) Representative immunofluorescence staining of human skin squamous cell carcinoma samples from tumor E, D & A demonstrating various expression pattern of Scd1 (red) and Soat1(green), Dapi (blue). Scale bar: 20µM. Soat1 and Scd1 are colocalized in clusters of sebaceous gland like cells as indicated in Tumor E and in sebaceous gland in Tumor D; Tumor A show expression of Scd1 in a gradient manner opposite to basal layer.
(A), (B), (C) The Human Protein Atlas analyses showing expression profiles of Runx1, Soat1 and Scd1 in 17 major cancer types. Protein expression is derived from antibody-based protein profiling using immunohistochemistry. Runx1 is moderately expressed in most of the oral and skin squamous carcinomas tumor, Soat1 shows low expression, while Scd1 shows medium to high expression.
(D) Representative screen shot of Scd1 expression in individual SCC and BCC tumors from skin cancer patients. Out of 7 SCC tumor samples, 5 displays moderate Scd1 expression, 2 have high Scd1 expression. 3 out of 5 BCC tumor samples display high Scd1 expression, while 1 tumor have medium Scd1, while another BCC tumor shows low Scd1 expression.
Significance Statement.
Diet-derived lipids influence tissue stem cell activity, whereas recent evidence suggest that de novo endogenous lipogenesis may independently ensure their proliferation, activation and differentiation in a manner similar with that previously described in cancer cells. Lipid metabolism enzymes begin to emerge as potential therapeutic targets with multiple drugs developed that may inhibit tumor progression. Here, we uncover a potential intrinsic control of lipid metabolism via Scd1 and Soat1 by a known epithelial stem cell and cancer factor Runx1.
Acknowledgments
We thank Dr. C.J. Scheitz for creating the tumors used in Figure 3D–G experiments. We thank many colleagues for reagents and cell lines (see Experimental Procedures). The Cornell Cytometry core is supported in part by the Empire State Stem Cell Foundation (ESSCF), New York State-Department of Health (NYS-DOH), contract C026718. Funding was from New York Stem Cell Program (T028095) and National Institutes of Health (R56AR053201 and R01AR070157) to T.T. and Post-doctoral NYSTEM training award (DOH01-C30293GG-3450000) to P.J.
Footnotes
Disclosure of Potential Conflicts of Interest :
Authors declare no conflict of interests.
References
- 1.Ryan RO, van der Horst DJ. Lipid transport biochemistry and its role in energy production. Annu Rev Entomol. 2000;45:233–260. doi: 10.1146/annurev.ento.45.1.233. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton LK, Fernandes KJL. Neural stem cells and adult brain fatty acid metabolism: Lessons from the 3xTg model of Alzheimer's disease. Biol Cell. 2017 doi: 10.1111/boc.201700037. [DOI] [PubMed] [Google Scholar]
- 3.Sakayori N, Kimura R, Osumi N. Impact of lipid nutrition on neural stem/progenitor cells. Stem Cells Int. 2013;2013:973508. doi: 10.1155/2013/973508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Tang Y, Cai D. IKKbeta/NF-kappaB disrupts adult hypothalamic neural stem cells to mediate a neurodegenerative mechanism of dietary obesity and pre-diabetes. Nat Cell Biol. 2012;14:999–1012. doi: 10.1038/ncb2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyaz S, Mana MD, Roper J, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang JX, Wan JB, He C. Concise review: Regulation of stem cell proliferation and differentiation by essential fatty acids and their metabolites. Stem Cells. 2014;32:1092–1098. doi: 10.1002/stem.1620. [DOI] [PubMed] [Google Scholar]
- 8.Yu RK, Suzuki Y, Yanagisawa M. Membrane glycolipids in stem cells. FEBS Lett. 2010;584:1694–1699. doi: 10.1016/j.febslet.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodhi IJ, Wei X, Yin L, et al. Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab. 2015;21:51–64. doi: 10.1016/j.cmet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy E, Beaulieu JF, Delvin E, et al. Human crypt intestinal epithelial cells are capable of lipid production, apolipoprotein synthesis, and lipoprotein assembly. J Lipid Res. 2000;41:12–22. [PubMed] [Google Scholar]
- 11.Wang L, Zhang T, Wang L, et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017;36:1330–1347. doi: 10.15252/embj.201695417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GM C. The Cell:A Molecular Approach. 2000 [Google Scholar]
- 13.Ibarguren M, Lopez DJ, Escriba PV. The effect of natural and synthetic fatty acids on membrane structure, microdomain organization, cellular functions and human health. Biochim Biophys Acta. 2014;1838:1518–1528. doi: 10.1016/j.bbamem.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Knobloch M, Braun SM, Zurkirchen L, et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013;493:226–230. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton LK, Dufresne M, Joppe SE, et al. Aberrant Lipid Metabolism in the Forebrain Niche Suppresses Adult Neural Stem Cell Proliferation in an Animal Model of Alzheimer's Disease. Cell Stem Cell. 2015;17:397–411. doi: 10.1016/j.stem.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Yusuf RZ, Scadden DT. Fate through fat: lipid metabolism determines stem cell division outcome. Cell Metab. 2012;16:411–413. doi: 10.1016/j.cmet.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito K, Carracedo A, Weiss D, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan T, Xu Z, Liu J, et al. Lkb1 regulation of skeletal muscle development, metabolism and muscle progenitor cell homeostasis. J Cell Physiol. 2017;232:2653–2656. doi: 10.1002/jcp.25786. [DOI] [PubMed] [Google Scholar]
- 19.Lichtinger M, Ingram R, Hannah R, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 2012;31:4318–4333. doi: 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raz I, Eldor R, Cernea S, et al. Diabetes: insulin resistance and derangements in lipid metabolism. Cure through intervention in fat transport and storage. Diabetes Metab Res Rev. 2005;21:3–14. doi: 10.1002/dmrr.493. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Condello S, Thomes-Pepin J, et al. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell. 2017;20:303–314. e305. doi: 10.1016/j.stem.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao W, Prijic S, Urban BC, et al. Candidate Antimetastasis Drugs Suppress the Metastatic Capacity of Breast Cancer Cells by Reducing Membrane Fluidity. Cancer Res. 2016;76:2037–2049. doi: 10.1158/0008-5472.CAN-15-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray U, Roy SS. Aberrant lipid metabolism in cancer cells: the role of oncolipid-activated signaling. FEBS J. 2017 doi: 10.1111/febs.14281. [DOI] [PubMed] [Google Scholar]
- 24.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Tumbar T. Hairy tale of signaling in hair follicle development and cycling. Semin Cell Dev Biol. 2012;23:906–916. doi: 10.1016/j.semcdb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotsarelis G. Epithelial stem cells: a folliculocentric view. J Invest Dermatol. 2006;126:1459–1468. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- 27.Osorio KM, Lee SE, McDermitt DJ, et al. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development. 2008;135:1059–1068. doi: 10.1242/dev.012799. [DOI] [PubMed] [Google Scholar]
- 28.Osorio KM, Lilja KC, Tumbar T. Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. J Cell Biol. 2011;193:235–250. doi: 10.1083/jcb.201006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SE, Sada A, Zhang M, et al. High Runx1 levels promote a reversible, more-differentiated cell state in hair-follicle stem cells during quiescence. Cell Rep. 2014;6:499–513. doi: 10.1016/j.celrep.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoi CS, Lee SE, Lu SY, et al. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010;30:2518–2536. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheitz CJ, Lee TS, McDermitt DJ, et al. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–4139. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Hoi CS, Lilja KC, et al. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc Natl Acad Sci U S A. 2013;110:4634–4639. doi: 10.1073/pnas.1213015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 34.Greco V, Chen T, Rendl M, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lien WH, Polak L, Lin M, et al. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nat Cell Biol. 2014;16:179–190. doi: 10.1038/ncb2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris RJ, Liu Y, Marles L, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 37.White JS, Weissfeld JL, Ragin CC, et al. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007;43:701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rheinwald JG, Beckett MA. Defective terminal differentiation in culture as a consistent and selectable character of malignant human keratinocytes. Cell. 1980;22:629–632. doi: 10.1016/0092-8674(80)90373-6. [DOI] [PubMed] [Google Scholar]
- 39.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 40.Tumbar T, Guasch G, Greco V, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Roemeling CA, Marlow LA, Wei JJ, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19:2368–2380. doi: 10.1158/1078-0432.CCR-12-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noto A, Raffa S, De Vitis C, et al. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013;4:e947. doi: 10.1038/cddis.2013.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loizides-Mangold U, Clement S, Alfonso-Garcia A, et al. HCV 3a core protein increases lipid droplet cholesteryl ester content via a mechanism dependent on sphingolipid biosynthesis. PLoS One. 2014;9:e115309. doi: 10.1371/journal.pone.0115309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiziana Parasassi EKK, Bagatolli Luis, Gratton Enrico. Laurdan and Prodan as Polarity-Sensitive Fluorescent Membrane Probes. Journal of Fluorescence. 1998;8:365–373. [Google Scholar]
- 45.Wang AB, Zhang YV, Tumbar T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. EMBO J. 2017;36:61–78. doi: 10.15252/embj.201694572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 47.Ran-Ressler RR, Lawrence P, Brenna JT. Structural characterization of saturated branched chain fatty acid methyl esters by collisional dissociation of molecular ions generated by electron ionization. J Lipid Res. 2012;53:195–203. doi: 10.1194/jlr.D020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahli W. Peroxisome proliferator-activated receptors (PPARs): from metabolic control to epidermal wound healing. Swiss Med Wkly. 2002;132:83–91. doi: 10.4414/smw.2002.09939. [DOI] [PubMed] [Google Scholar]
- 49.Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med (Berl) 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y, Eilertsen KJ, Ge L, et al. Scd1 is expressed in sebaceous glands and is disrupted in the asebia mouse. Nat Genet. 1999;23:268–270. doi: 10.1038/15446. [DOI] [PubMed] [Google Scholar]
- 51.Scheitz CJ, Tumbar T. New insights into the role of Runx1 in epithelial stem cell biology and pathology. J Cell Biochem. 2013;114:985–993. doi: 10.1002/jcb.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geng F, Cheng X, Wu X, et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin Cancer Res. 2016;22:5337–5348. doi: 10.1158/1078-0432.CCR-15-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 54.Yeh HY, Cheng SW, Lin YC, et al. Identifying significant genetic regulatory networks in the prostate cancer from microarray data based on transcription factor analysis and conditional independency. BMC Med Genomics. 2009;2:70. doi: 10.1186/1755-8794-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang J, Fan XX, He J, et al. SCD1 is associated with tumor promotion, late stage and poor survival in lung adenocarcinoma. Oncotarget. 2016;7:39970–39979. doi: 10.18632/oncotarget.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antalis CJ, Arnold T, Rasool T, et al. High ACAT1 expression in estrogen receptor negative basal-like breast cancer cells is associated with LDL-induced proliferation. Breast Cancer Res Treat. 2010;122:661–670. doi: 10.1007/s10549-009-0594-8. [DOI] [PubMed] [Google Scholar]
- 57.Bednarski T, Olichwier A, Opasinska A, et al. Stearoyl-CoA desaturase 1 deficiency reduces lipid accumulation in the heart by activating lipolysis independently of peroxisome proliferator-activated receptor alpha. Biochim Biophys Acta. 2016;1861:2029–2037. doi: 10.1016/j.bbalip.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Liu J, Cinar R, Xiong K, et al. Monounsaturated fatty acids generated via stearoyl CoA desaturase-1 are endogenous inhibitors of fatty acid amide hydrolase. Proc Natl Acad Sci U S A. 2013;110:18832–18837. doi: 10.1073/pnas.1309469110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paton CM, Ntambi JM. Loss of stearoyl-CoA desaturase activity leads to free cholesterol synthesis through increased Xbp-1 splicing. Am J Physiol Endocrinol Metab. 2010;299:E1066–1075. doi: 10.1152/ajpendo.00388.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bate C, Tayebi M, Williams A. Cholesterol esterification reduces the neurotoxicity of prions. Neuropharmacology. 2008;54:1247–1253. doi: 10.1016/j.neuropharm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Sugiyama Y, Odaka H, Itokawa S, et al. TMP-153, a novel ACAT inhibitor, lowers plasma cholesterol through its hepatic action in golden hamsters. Atherosclerosis. 1995;118:145–153. doi: 10.1016/0021-9150(95)05601-r. [DOI] [PubMed] [Google Scholar]
- 62.Xin Z, Zhao H, Serby MD, et al. Discovery of piperidine-aryl urea-based stearoyl-CoA desaturase 1 inhibitors. Bioorg Med Chem Lett. 2008;18:4298–4302. doi: 10.1016/j.bmcl.2008.06.088. [DOI] [PubMed] [Google Scholar]
- 63.Sugiyama Y, Ishikawa E, Odaka H, et al. TMP-153, a novel ACAT inhibitor, inhibits cholesterol absorption and lowers plasma cholesterol in rats and hamsters. Atherosclerosis. 1995;113:71–78. doi: 10.1016/0021-9150(94)05429-m. [DOI] [PubMed] [Google Scholar]
- 64.Rudel LL, Lee RG, Cockman TL. Acyl coenzyme A: cholesterol acyltransferase types 1 and 2: structure and function in atherosclerosis. Curr Opin Lipidol. 2001;12:121–127. doi: 10.1097/00041433-200104000-00005. [DOI] [PubMed] [Google Scholar]
- 65.Mason P, Liang B, Li L, et al. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS One. 2012;7:e33823. doi: 10.1371/journal.pone.0033823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicolaides N, Fu HC, Ansari MN, et al. The fatty acids of wax esters and sterol esters from vernix caseosa and from human skin surface lipid. Lipids. 1972;7:506–517. doi: 10.1007/BF02533016. [DOI] [PubMed] [Google Scholar]
- 67.Oku H, Shudo J, Mimura K, et al. Age-related changes in branched-chain fatty acid concentration of the skin surface lipid from hairless mouse. Comp Biochem Physiol B Biochem Mol Biol. 1996;114:103–106. doi: 10.1016/0305-0491(96)00009-0. [DOI] [PubMed] [Google Scholar]
- 68.Wang DH, Jackson JR, Twining C, et al. Saturated Branched Chain, Normal Odd-Carbon-Numbered, and n-3 (Omega-3) Polyunsaturated Fatty Acids in Freshwater Fish in the Northeastern United States. J Agric Food Chem. 2016 doi: 10.1021/acs.jafc.6b03491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marguet D, Lenne PF, Rigneault H, et al. Dynamics in the plasma membrane: how to combine fluidity and order. EMBO J. 2006;25:3446–3457. doi: 10.1038/sj.emboj.7601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiological reviews. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edidin M. Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol. 2003;4:414–418. doi: 10.1038/nrm1102. [DOI] [PubMed] [Google Scholar]
- 72.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez SA, Tricerri MA, Gratton E. Laurdan generalized polarization fluctuations measures membrane packing micro-heterogeneity in vivo. Proc Natl Acad Sci U S A. 2012;109:7314–7319. doi: 10.1073/pnas.1118288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris FM, Best KB, Bell JD. Use of laurdan fluorescence intensity and polarization to distinguish between changes in membrane fluidity and phospholipid order. Biochim Biophys Acta. 2002;1565:123–128. doi: 10.1016/s0005-2736(02)00514-x. [DOI] [PubMed] [Google Scholar]
- 75.Klymchenko AS, Kreder R. Fluorescent probes for lipid rafts: from model membranes to living cells. Chem Biol. 2014;21:97–113. doi: 10.1016/j.chembiol.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Srivastava K, Dash D. Altered membrane fluidity and signal transduction in the platelets from patients of thrombotic stroke. Mol Cell Biochem. 2001;224:143–149. doi: 10.1023/a:1011979801366. [DOI] [PubMed] [Google Scholar]
- 77.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 78.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 79.Sukhdeo K, Mani M, Zhang Y, et al. Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci U S A. 2007;104:7516–7521. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deltcheva E, Nimmo R. RUNX transcription factors at the interface of stem cells and cancer. Biochem J. 2017;474:1755–1768. doi: 10.1042/BCJ20160632. [DOI] [PubMed] [Google Scholar]
- 81.Cai X, Gao L, Teng L, et al. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell. 2015;17:165–177. doi: 10.1016/j.stem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stoffel W, Schmidt-Soltau I, Jenke B, et al. Hair Growth Cycle Is Arrested in SCD1 Deficiency by Impaired Wnt3a-Palmitoleoylation and Retrieved by the Artificial Lipid Barrier. J Invest Dermatol. 2017;137:1424–1433. doi: 10.1016/j.jid.2017.02.973. [DOI] [PubMed] [Google Scholar]
- 83.Weber U, Eroglu C, Mlodzik M. Phospholipid membrane composition affects EGF receptor and Notch signaling through effects on endocytosis during Drosophila development. Dev Cell. 2003;5:559–570. doi: 10.1016/s1534-5807(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 84.Wu B, Potter CS, Silva KA, et al. Mutations in sterol O-acyltransferase 1 (Soat1) result in hair interior defects in AKR/J mice. J Invest Dermatol. 2010;130:2666–2668. doi: 10.1038/jid.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sampath H, Flowers MT, Liu X, et al. Skin-specific deletion of stearoyl-CoA desaturase-1 alters skin lipid composition and protects mice from high fat diet-induced obesity. J Biol Chem. 2009;284:19961–19973. doi: 10.1074/jbc.M109.014225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sundberg JP, Boggess D, Sundberg BA, et al. Asebia-2J (Scd1(ab2J)): a new allele and a model for scarring alopecia. Am J Pathol. 2000;156:2067–2075. doi: 10.1016/S0002-9440(10)65078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mihaylova MM, Sabatini DM, Yilmaz OH. Dietary and metabolic control of stem cell function in physiology and cancer. Cell Stem Cell. 2014;14:292–305. doi: 10.1016/j.stem.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Igal RA. Roles of StearoylCoA Desaturase-1 in the Regulation of Cancer Cell Growth, Survival and Tumorigenesis. Cancers (Basel) 2011;3:2462–2477. doi: 10.3390/cancers3022462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim SJ, Choi H, Park SS, et al. Stearoyl CoA desaturase (SCD) facilitates proliferation of prostate cancer cells through enhancement of androgen receptor transactivation. Mol Cells. 2011;31:371–377. doi: 10.1007/s10059-011-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Q, Luo Q, Halim A, et al. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Yue S, Li J, Lee SY, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wertz PW. Lipids and barrier function of the skin. Acta Derm Venereol Suppl (Stockh) 2000;208:7–11. doi: 10.1080/000155500750042790. [DOI] [PubMed] [Google Scholar]
- 93.Bouwstra JA, Ponec M. The skin barrier in healthy and diseased state. Biochim Biophys Acta. 2006;1758:2080–2095. doi: 10.1016/j.bbamem.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 94.Kim MW, Shin IS, Yoon HS, et al. Lipid profile in patients with androgenetic alopecia: a meta-analysis. J Eur Acad Dermatol Venereol. 2017;31:942–951. doi: 10.1111/jdv.14000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative images after staining with crystal violet of WT, 4-OHT-induced Runx1 iKO keratinocytes with or without addition of Oleate, after 4 days of plating in cell culture.
(A) (C) Quantification of immunofluorescence staining of squamous cell carcinoma (SCC) cells normalized to control non-cancer epithelial cells labeled as wild type. Note striking Scd1 and Soat1 up-regulation in SCCs as compare to control.
(B) (D) Quantification of western blotting of skin carcinoma (13) and oral carcinomas (66, 125) cell lines shows elevated levels of Scd1 and Soat1 as compared to non-cancer cell controls. Normalization in each experiment was to Vinculin (Vinc) and to non-cancer cell control.
P value that are <0.05 is represented as “*” on the graph and p value <0.005 represented as “**”, p value <0.0001 represented as “****”; Red * represents p value = 0.06.
1,2, 3,4) Representative immunofluorescence staining of human skin squamous cell carcinoma sample from tumor H, A, C, E showing Scd1 (red) and Soat1 (green) & corresponding serial section immunofluorescence staining showing expression of Runx1 (green) & Scd1 (red), Blue (K14). Scale bar: 20µM. White dotted line represents enlarged view and white arrow shows region of colocalization. Scd1, Soat1 and Runx1 rarely colocalize in structures that are K14+made of a single layer of epithelial cells surrounding a void region (see Tumor A boxed areas) 5,6,7) Representative immunofluorescence staining of human skin squamous cell carcinoma samples from tumor E, D & A demonstrating various expression pattern of Scd1 (red) and Soat1(green), Dapi (blue). Scale bar: 20µM. Soat1 and Scd1 are colocalized in clusters of sebaceous gland like cells as indicated in Tumor E and in sebaceous gland in Tumor D; Tumor A show expression of Scd1 in a gradient manner opposite to basal layer.
(A), (B), (C) The Human Protein Atlas analyses showing expression profiles of Runx1, Soat1 and Scd1 in 17 major cancer types. Protein expression is derived from antibody-based protein profiling using immunohistochemistry. Runx1 is moderately expressed in most of the oral and skin squamous carcinomas tumor, Soat1 shows low expression, while Scd1 shows medium to high expression.
(D) Representative screen shot of Scd1 expression in individual SCC and BCC tumors from skin cancer patients. Out of 7 SCC tumor samples, 5 displays moderate Scd1 expression, 2 have high Scd1 expression. 3 out of 5 BCC tumor samples display high Scd1 expression, while 1 tumor have medium Scd1, while another BCC tumor shows low Scd1 expression.