Abstract
Inflammasomes are multimeric protein complexes that promote inflammation (through specific cleavage and production of bioactive IL-1β and IL-18) and pyroptotic cell death. The central role of inflammasomes in combating infection and maintaining homeostasis has been studied extensively. Although inflammasome-mediated inflammation and cell death are vital to limit pathogenic insults and to promote wound healing/tissue regeneration, unchecked/uncontrolled inflammation, and cell death can cause cytokine storm, tissue damage, autoinflammatory and autoimmune diseases, and even death in the afflicted individuals. NLRP3 is one of the major cytosolic sensors that assemble an inflammasome. Given the adverse consequences of uncontrolled inflammasome activation, our immune system has developed tiered mechanisms to inhibit NLRP3 inflammasome activation. In this review, we highlight and discuss recent advances and our current understanding of mechanisms by which NLRP3 inflammasome can be negatively regulated.
Keywords: ASC, caspase-1, inflammasome, negative regulation, NLRP3
1|. INTRODUCTION
Our immune system is comprised of germ-line encoded pathogen recognition receptors (PRR) that sense microbial invasion or tissue injury. PRR sensing of pathogen-associated molecular patterns (PAMP) or damage-associated molecular patterns (DAMP) initiate transcrip tional programming to induce inflammation that includes production of cytokines and chemokines. However, inflammation is a double-edged sword that needs to be tightly controlled: although critical for the control of pathogens and repair responses, excessive inflammation can be destructive.
TLR located on the cell membrane and endosomes were the first class of PRR to be identified and characterized. TLR guard the extra-cellular milieu for pathogens and tissue damage. Cytosolic sensors—including nod-like receptors (NLR), absent in melanoma-2 (AIM2)-like receptors (ALR), and retinoic acid inducible gene I-like receptors—protect the intracellular compartments of the cells from pathogens that escape sensing by the membrane-bound PRR. Upon recognition of PAMP and DAMP, both TLR and cytosolic PRR initiate a cascade of signaling pathways resulting in transcriptional regulation of several genes that are critical in controlling infection and maintaining cellular home-ostasis. In addition, some members of NLR and ALR can also assemble a multimeric protein complex called inflammasome.
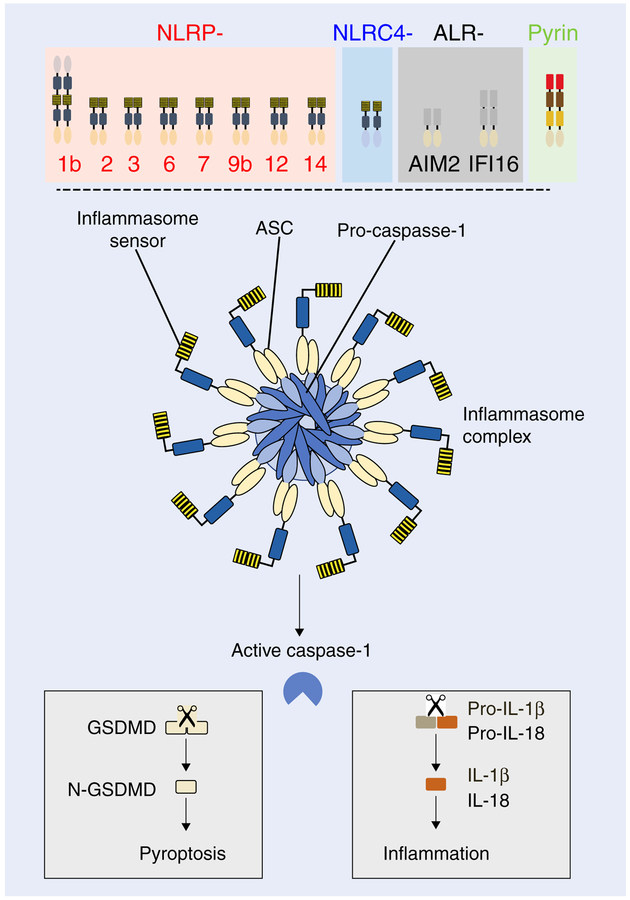
Upon sensing a danger signal, an inflammasome sensor recruits an adaptor molecule apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC), and a cysteine protease caspase-1, to form an inflammasome complex (Fig. 1).1,2 This multimeric complex has recently been shown to consist of 10–12 NLR molecules.3–5 Within this complex, caspase-1 undergoes autoproteolysis and becomes activated. Active caspase-1 has 2 major functions within the cells—first, it processes pro-IL-1β and pro-IL-18 into their bioactive forms to promote inflammation; second, it cleaves gasdermin D (GSDMD) to release N-terminal fragment of GSDMD (N-GSDMD), which promotes inflammatory cell death known as pyroptosis.6–9 Several cytosolic sensors have been described to form an inflammasome complex. To date, the 5 well-characterized inflammasomes include NLRP1b,1 NLRP3,10–13 NLRC4,14 AIM2,15–17 and Pyrin.18,19 Other reported sensors that can also assemble inflammasomes include NLRP2,20 NLRP6,21,22 NLRP7,23 NLRP9b,24 NLRP12,25 NLRP14,26 and IFI16.27
FIGURE 1. Cytosolic sensors that assemble inflammasome complex and the consequence of inflammasome activation.
Members of NLR family (NLRP1b, NLRP2, NLRP3, NLRP6, NLRP7, NLRP9b, NLRP12, NLRP14, NLRC4), HIN200 family (AIM2, IFI16), and Pyrin can assemble an inflammasome complex. Upon activation, these inflammasome sensors recruit adaptor ASC and caspase-1 to form the inflammasome complex. Inflammasome complex formation results in activation of caspase-1. Active caspase-1 cleaves pro-IL-1β and pro-IL-18 to release their bioactive forms to induce inflammation, and cleaves gasdermin D (GSDMD) to release N-terminal fragment of gasdermin D (N-GSDMD). N-GSDMD forms pores on the cell membrane to induce inflammatory cell death known as pyroptosis
In this review, we will focus on NLRP3 and discuss the recent literature that describes mechanisms to negatively regulate the NLRP3 inflammasome.
2|. NLRP3 INFLAMMASOME
NLRP3 is one of the well-studied NLR that assembles an inflamma-some. Unlike other cytosolic sensors, NLRP3 is activated in response to a wide array of stimuli including bacteria, viruses, fungi, protozoa, self-induced damage signals such as uric acid crystals and ATP, and environmental stimulants such as alum and silica. Thus, it is believed that NLRP3 recognizes a common secondary signal that is induced by these wide arrays of ligands. Whether a single, common secondary signal is recognized by NLRP3 to assemble an inflamma-some is still an area of intense debate; however, several studies have shown potassium (K+) efflux,28 calcium (Ca2+) mobilization,29 mitochondrial damage and reactive oxygen species (ROS) production,30 cardiolipin externalization,31 and Fas-associated death domain protein (FADD)/caspase-832 as important downstream events that are required for activation and assembly of the NLRP3 inflammasome.33 It is important to note that these downstream mechanisms are not mutually exclusive. More recently, downstream of K+ efflux, NIMA-related kinase 7 was identified as the immediate upstream kinase that directly interacted with the leucine-rich repeat (LRR)-domain of NLRP3 to regulate its inflammasome activity in murine bone marrow-derived macrophages (BMDM), bone marrow-derived dendritic cells (BMDC) and peritoneal macrophages.34–36
Importance of the NLRP3 inflammasome is highlighted by a plethora of studies that show a central role for this sensor in eliminating pathogens and maintaining cellular homeostasis.37 Conversely, aberrant activation of the NLRP3 inflammasome has been shown to cause metabolic disorders and autoinflammatory diseases such as gout, type II diabetes, cancer, familial cold autoinflammatory syndrome, Muckle-Wells syndrome, and neonatal-onset multisystem inflammatory disease.10,37–42 Here, we discuss mechanisms by which the NLRP3 inflammasomes are kept in check to limit excessive inflammation and disease.
3|. NEGATIVE REGULATION OF NLRP3 INFLAMMASOME ACTIVATION
To understand how the NLRP3 inflammasome is negatively regulated, it is equally important to know what molecules/pathways are required for its activation. Activation of the NLRP3 inflammasome requires 2 signals—a priming signal through PRR that activates the NFκB and MAPK signaling pathway to up-regulate Nlrp3 expression, and an activating signal that promotes NLRP3 oligomerization to assemble the inflammasome complex.43 In this review, we will take a top-down approach and discuss how the NLRP3 inflammasome can be negatively regulated. The negative regulation of NFκB signaling relating to the NLRP3 inflammasome has been recently reviewed elsewhere,44 and thus, we will focus on direct negative regulation of Nlrp3 mRNA expression (posttranscriptional regulation) and NLRP3 protein levels (posttranslational regulation), and the NLRP3 inflammasome complex assembly.
3.1|. Posttranscriptional regulation of NLRP3 mRNA
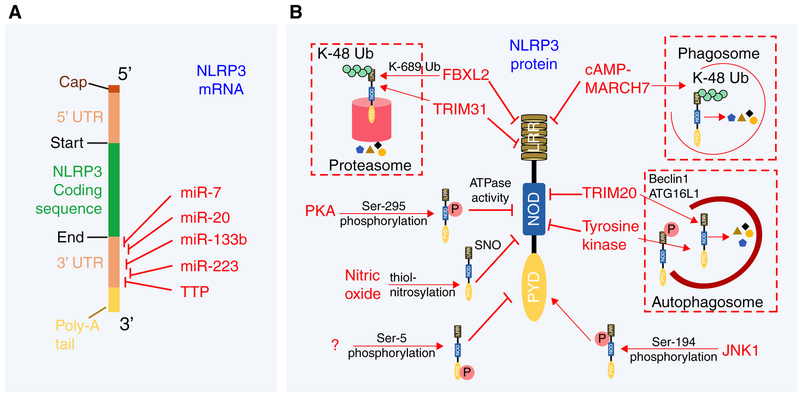
One of the major functions of a priming signal is to induce the expression of NLRP3 mRNA and accumulate NLRP3 protein for optimal activation of the inflammasome. Indeed, acute activation of BMDM and BMDC through TLR-2 agonist PAM3CSK4 or TLR-4 agonist LPS for up to 4 h dramatically increases both mRNA and protein levels of NLRP3.45 However, chronic TLR-2 and TLR-4 stimulation of BMDM for more than 12 h induces IL-10, a critical regulatory cytokine that negatively regulates protein levels of NLRP3.46 Thus, the NLRP3 protein that is available for activation of the inflammasome depends on expression and stability of the NLRP3 mRNA and the translated protein. All these facets could be potentially targeted to control NLRP3 inflammasome activation (Fig. 2A).
FIGURE 2. Posttranscriptional and posttranslational regulation ofNLRP3.
(A) Posttranscriptional regulation of NLRP3 mRNA. Miro-RNA (miR) and tristetraprolin (TTP) negatively regulate NLRP3 mRNA expression by binding to their complementary sequence in 3′-UTR regions. (B) Post-translational regulation of NLRP3 by phosphorylation, ubiquitination, and thiol-nitrosylation events. Phosphorylation of NLRP3 can inhibit ATPase activity, prevent oligomerization or promote its degradation. Thiol-nitrosylation of NLRP3 inhibits inflammasome activity via unknown mechanism
Micro RNAs (miR) are small noncoding RNAs that regulate gene expression by specifically binding and silencing target mRNA species. 3′ untranslated region (UTR) of NLRP3 mRNA contains miR-223 target sequence that is highly conserved among several mammalian species including humans, mice, rhesus monkeys, and dogs.47,48 Myeloid-specific miR-223 was demonstrated to bind to the 3′UTR of human NLRP3 mRNA and negatively regulate its expression in HEK293T cells.47,48 Indeed, the expression of miR-223 is inversely proportional to the level of NLRP3 observed in the different myeloid cell types. Although miR-223 expression decreases as monocytes differentiate into macrophages, NLRP3 mRNA expression increases as the cells progress from monocytes to macrophages.48 Likewise, miR-223 expression is much lower in dendritic cells (DC) compared with macrophages, and this correlates to higher NLRP3 mRNA expression in DC compared with macrophages.47 These studies have been further bolstered by a recent report that showed that neutrophils from mice lacking the conserved 3′UTR binding site for miR-223 in Nlrp3 mRNA (Nlrp3m223del mice) have increased NLRP3 protein levels and increased inflammasome activation as assessed by IL-1β production during LPS + nigericin stimulation in vitro.49,50 In a mouse model of dextran sodium sulfate (DSS)-induced colitis, Nlrp3m223del mice developed severe disease as demonstrated by increased weight loss, shortened colon length and total colon histology scores, similar to that observed in miR-223-deficient mice.49,50 Several other miRs have also been implicated in regulating Nlrp3 mRNA expression in various inflammatory settings. Studies with HEK293T cells transfected with miR-133b and a luciferase reporter conjugated to 3′UTR of murine Nlrp3 mRNA suggested that miR-133b binds to the 3′UTR of murine Nlrp3 mRNA to negatively regulate Nlrp3 mRNA expression.51 Over-expression of miR-133b in HEK293T cells reduced the protein levels of NLRP3, whereas inhibition of miR-133b increased the protein levels of HEK293T cells in vitro.51 Importantly, miR-133b inhibited NLRP3 inflammasome activation and ameliorated allergic inflammation in a mouse model of allergic rhinitis.51 Luciferase reporter assay in HEK293T cells demonstrated that miR-20b negatively regulated NLRP3 mRNA by binding to its 3′UTR.52 Furthermore, treatment of Mycobacterium tuberculosis-infected mice with miR-20b mimic inhibited NLRP3 protein induction and alleviated lung inflammation in mice.52 Activation of the NLRP3 inflammasome promoted Parkinson’s disease in a mouse model, which was ameliorated by injection of miR-7mimics.53 miR-7 directly bound 3′UTR of Nlrp3 mRNA in a luciferase reporter assay.53 Furthermore, overexpression of miR-7 in BV2 cells (immortalized murine microglial cells) reduced NLRP3 protein levels in a dose-dependent manner and abrogated inflammasome activation.53 More recently, RNA binding protein tristetraprolin (TTP) was shown to bind to the adenylate-uridylate (AU)-rich region in the 3′UTR of NLRP3 mRNA and repress NLRP3 3′UTR luciferase reporters in HEK293T cells. Importantly, siRNA-mediated knockdown of TTP in THP1 cells (human monocyte cell lines) increased NLRP3 protein levels and subsequently augmented inflammasome activation.54 Thus, TTP directly binds and regulates NLRP3 mRNA expression to repress NLRP3 inflammasome activity. Interestingly, most of these negative regulators described in here do not regulate other inflammasomes (Fig. 2A).
3.2|. Negative regulation of NLRP3 by posttranslational modifications
NLRP3 proteins are constitutively expressed in macrophages and DC. Priming through TLR increases NLRP3 protein levels required for optimal NLRP3 inflammasome activation. Upon stimulation with a ligand, NLRP3 translocates to the mitochondria and oligomerizes; both necessary steps for eventual NLRP3 inflammasome activation. How can these necessary steps be regulated by modulating NLRP3 protein to inhibit the activation of the NLRP3 inflammasome? We will discuss recent reports on posttranslational modification of NLRP3, and inhibition of NLRP3 trafficking, as mechanisms to negatively regulate NLRP3 inflammasome activation (Fig. 2B).
3.2.1|. Posttranslational modifications to negatively regulate the NLRP3 inflammasome
Proteins are tightly regulated by posttranslational modifications such as phosphorylation, glycosation, ubiquitination, sumoylation, neddylation, s-nitrosylation, methylation, and acetylation. Several of these modifications are also involved in the negative regulation of the NLRP3 protein; these have been reviewed in detail by Yang et al.55 recently. Using N1–8 macrophage cell lines (stably expressing C-terminal FLAG tagged NLRP3), Juliana et al.56 showed that NLRP3 proteins are heavily ubiquitinated in basal condition. Acute LPS priming for 10 min was sufficient to induce deubiquitination of NLRP3, a critical step for its activation.56 The deubiquitination of NLRP3 following priming event is not specific to LPS/TLR4 because similar results were also observed during acute PAM3CSK4 (TLR-2 agonist), poly I:C (TLR-3 agonist), R837 (TLR-7 agonist), and CpG (TLR-9 agonist) stimulations of murine BMDM.57,58 Inhibition of this deubiquitination step by deubiquitinating enzyme inhibitors PR-619 or WP1130 during LPS+ATP stimulation of N1–8 macrophages completely abolished caspase-1 activation.56 NLRP3 inflammasome activation in this acute TLR stimulation setting was independent of transcriptional events, but it required kinase activity of IL 1 receptor associated kinase 1 (IRAK1).57,58 A subsequent study identified deubiquitinating enzyme BRCA1/BRCA2-containing complex subunit 3 (BRCC3), which promoted NLRP3 deubiquitination to promote NLRP3 activation.59 How kinase activity of IRAK1 fits in this puzzle is unknown and needs further investigation. It is possible that IRAK1-mediated phosphorylation is a prerequisite step for BRCC3 to promote NLRP3 deubiquitination. Regardless, these studies indirectly suggested that ubiquitination of NLRP3 negatively regulates its activation. Recent studies have uncovered possible ubiquitin ligases that negatively regulate NLRP3 activation. Yan et al.60 showed that dopamine signals through the dopamine receptor D1 (DRD1) to inhibit monosodium urate (MSU), nigericin, ATP, and alum-induced NLRP3 inflammasome activation in LPS-primed murine BMDM. IL-1β and IL-18 cytokines produced during neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced neuroinflammation, LPS-induced shock, and MSU-induced peritonitis were significantly increased in Drd1-deficient mice, suggesting a protective role for dopamine signaling in regulating NLRP3 inflammasome activation in vivo.60 In LPS-primed murine BMDM, dopamine-DRD1 signaling induces cAMP expression; cAMP then binds NLRP3 and promotes K48-linked ubiquitination through the E3 ubiquitin ligase membrane-associated RING finger protein 7 (MARCH7).60 Interestingly, the ubiquitinated NLRP3 is degraded in a phagosome rather than a proteasome. More detailed biochemical studies are needed to identify specific lysine residues on NLRP3 that are ubiquitinated by MARCH7. Another report identified that E3 ubiquitin ligase F-box L2 (FBXL2) ubiquitinates NLRP3 to negatively regulate its expression in an in vitro ubiquitination tube assay.61 In vitro overexpression studies showed that FBXL2 directly interacts with NLRP3 but not with the W73A mutant of NLRP3.61 Thus, Tryptophan-73 residue of NLRP3 is critical for FBXL2 and NLRP3 interaction. In vitro ubiquitination assay further showed that K689R mutant of NLRP3 was poly-ubiquitinated to a lesser extent compared with wild-type (WT) NLRP3 by FBXL2.61 Thus, FBXL2 ubiquitinates NLRP3 on the Lysine-689 residue to promote proteasomal degradation.61 Of note, FBXL2 and NLRP3 studies were done in an in vitro system and whether Fbxl2-deficiency induces aberrant NLRP3 inflammasome activation in vivo is not known. Tripartite motif protein (TRIM) family members have been shown to negatively modulate NLRP3 protein expression. TRIM20 promotes NLRP3 degradation through unc-51-like autophagy activating kinase 1 (ULK1), Beclin 1, and ATG16L1-mediated autophagy.62 TRIM20 bound with both exogenously expressed and endogenous NLRP3, ULK1, Beclin1, and ATG16L1 in HEK293T cells.62 Mechanistically, in IFN-γ stimulated cells, TRIM20 interacts with NLRP3 via its SPRY domain and further recruits ULK1, Beclin 1, and ATG16L1 to facilitate autophagic degradation of NLRP3.62 TRIM31, a TRIM family protein with E3 ubiquitin ligase activity, negatively regulates ATP, nigericin, and alum-induced NLRP3 inflammasome activation in LPS-primed mouse peritoneal macrophages.63 TRIM31 directly interacted with NLRP3 as demonstrated by in vitro binding assays (in vitro transcribed/translated TRIM31 and NLRP3 in a tube interacted with each other) and promoted Lys-48-linked polyubiquitination and proteasomal degradation of NLRP3 in HEK293T and mouse peritoneal macrophages.63 As a result, compared with WT controls, Trim31-deficient mice were highly susceptible to alum-induced peritonitis and DSS-induced colitis, both NLRP3-dependent diseases.63 These studies highlight the crucial role of E3 ubiquitin ligases in posttranslational regulation of NLRP3.
Several studies have reported phosphorylation of NLRP3 as a potential mechanism that negatively regulates inflammasome activation. NLRP3 dephosphorylation by protein tyrosine phosphatase, non-receptor Type 22 (PTPN22) was reported to be critical for activation of the NLRP3 inflammasomes and induction of DSS-induced colitis in mice, suggesting phosphorylation as the limiting step for NLRP3 inhibition.64 Indeed, phosphorylated NLRP3 was shown to be sequestered in the autophagosomes in LPS + MSU-treated BMDC, and that the phosphorylated NLRP3 recovered from autophagsome enriched fractions were significantly increased in Drd1-deficient BMDC.65 PTPN22 interacted with NLRP3 upon MDP, MSU, and ATP-induced NLRP3 inflammasome activation in LPS-primed THP1, MM6, BMDC and PBMC, and dephosphorylated Tyrosine-861 on NLRP3.64 Although these studies did not elucidate the kinases that phosphorylate Tyrosine-861, it is clear that the kinase has to be a tyro-sine kinase.64 One of the likely candidates includes spleen tyrosine kinase, which has been implicated in the regulation of the NLRP3 inflammasome.66 Another study, by Mortimer et al., demonstrated that PGE2 at nanomolar concentrations can effectively inhibit ATP, nigericin, MSU, and silica-induced NLRP3 inflammasome activation in LPS-primed BMDM.67 Similarly, stimulation of PGE2 receptor E-prostanoid 4 (EP4) using EP4 agonist CAY10598 inhibited activation of the NLRP3 inflammasomes in LPS-primed BMDM.67 Moreover, PGE2-mediated inhibition of LPS + nigericin-induced caspase-1 and IL-1β activation was not observed in BMDM with siRNA-mediated deletion of protein kinase A (PKA), suggesting PGE2-EP4 needs PKA to inhibit NLRP3 inflammasome.67 In vitro PKA kinase assays demonstrated that PKA directly phosphorylates NLRP3 proteins purified from HEK293T cells at Ser-295 residue.67 Phosphorylation of NLRP3 Ser-295 turns off NLRP3 ATPase activity and inhibits inflammasome assembly.67 Mutation in the human NLRP3 corresponding to Ser-295 have been linked with autoinflammatory disorders, highlighting the importance of Ser-295 residue in negative regulation of NLRP3 inflammasomes.67 Phosphorylation of NLRP3 Pyrin domain at Ser-5 residue has also been reported to inhibit inflammasome activation.68 Immortalized Nlrp3-deficient macrophages reconstituted with WT NLRP3, but not S5E or S5D mutant, activated caspase-1 and produced IL-1β in response to LPS and nigericin.68 Although the Serine kinase that phosphorylates the Ser-5 residue remains unknown, Phosphatase 2A (PP2A) was identified as the phosphatase that dephosphorylates Ser-5 to regulate NLRP3 activity.68 Thus, immortalized macrophages with siRNA-mediated knockdown of PP2A produced significantly reduced levels of IL-1β in response to LPS and nigericin.68 Stutz et al.68 proposed that NLRP3 is phosphorylated at Ser-5 in an unprimed state, which limits Pyrin-Pyrin homotypic interaction and NLRP3 oligomerization to inhibit inflammasome activation.
It is important to note that phosphorylation of NLRP3 is not always associated with negative regulation. Indeed, a recent study by Song et al.69 has demonstrated NLRP3 phosphorylation as a critical priming step for inflammasome activation. The critical residue required for activation of NLRP3 inflammasome was identified to be Ser-198 in human NLRP3 and Ser-194 residue in mouse NLRP3.69 Thus, LPS-primed BMDM from mouse harboring a S194A mutant version of NLRP3 protein (Nlrp3S194A knock in mouse) failed to activate NLRP3 inflammasome in response to ATP, nigericin, MSU and poly I:C stimulations.69 Not surprisingly, Nlrp3S194A mice were highly resistant to 10 mg/kg LPS injections.69 In vitro kinase assays using murine NLRP3 and recombinant JNK1 demonstrated that JNK1 directly phosphorylated NLRP3.69 JNK1-mediated phosphorylation of NLRP3 at Ser-194 promoted NLRP3 self-association and deubiquitination, steps necessary for inflammasome activation.56–58,69 Identifying novel phosphatases that dephosphorylate Ser-194 on NLRP3 during inflammasome activation have potential of regulating aberrant NLRP3 inflammasome activation and treating NLRP3-associated inflammatory diseases.
Several studies have demonstrated a role for nitric oxide in negatively regulating NLRP3 inflammasome activation. IFN-β signaling inhibited nigericin-induced NLRP3 inflammasome activation in PAM3CSK4-primed murine peritoneal macrophages in a nitric oxide-dependent manner.70 In support of a regulatory role for nitric oxide, treatment of LPS-primed peritoneal macrophages or THP1 macrophages with SNAP (a nitric oxide donor) attenuated NLRP3 inflammasome activation in response to ATP, nigericin, and MSU.70,71 Chronic LPS stimulation of peritoneal macrophages for 12 h induced nitric oxide, which inhibited NLRP3 inflammasome activation in response to nigericin in vitro.70 Similarly, IFN-γ pretreatment of murine BMDM inhibited NLRP3 inflammasome activation during Mycobacterium tuberculosis infection which required nitric oxide.72 Mechanistically, nitric oxide promotes S-nitrosylation of the NLRP3 protein, and this modification inhibits the ability of NLRP3 to assemble the inflammasome complex.70,72 S-nitrosylation of NLRP3 does not result in protein degradation or destabilization, as the NLRP3 protein remained stable during SNAP treatment of PAM3CSK4-primed peritoneal macrophages.70 Follow-up studies are needed to identify the exact cysteine residues that are S-nitrosylated and the molecular mechanisms (e.g., physical interference, deactivation of NLRP3) by which S-nitrosylation of NLRP3 inhibits its function.
3.3|. Regulation of processes that activate and assemble the NLRP3 inflammasome
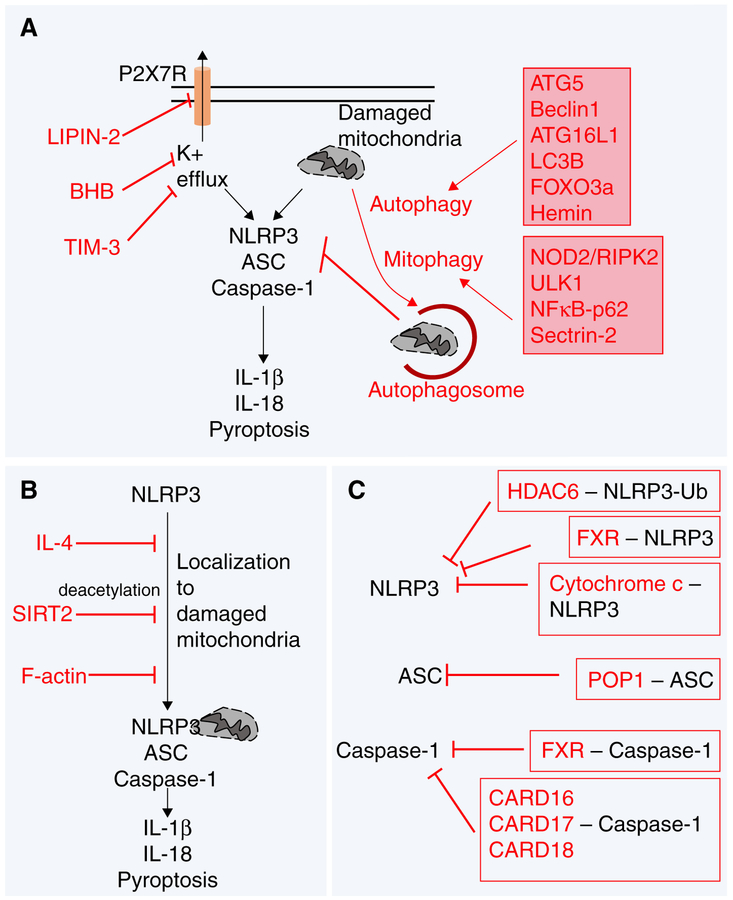
Upon receiving the priming signal and optimal production of NLRP3, the inflammasome is ready to be assembled. As discussed earlier, several common proximal events have been proposed to activate and assemble the NLRP3 inflammasome. The 2 major mechanisms that are well-established in the literature include (1) K+ efflux, and (2) mitochondrial damage-associated changes such as production of ROS and other host factors. As can be envisaged, mechanisms that restrict these activating cues can specifically inhibit the activation of NLRP3 inflammasome (Fig. 3A).
FIGURE 3. Mechanisms inhibiting NLRP3 assembly.
(A) Inhibition of K+ efflux and mitochondrial dysfunction as mechanisms to negatively regulate NLRP3 activation. (B) Factors that inhibit localization of NLRP3 to the mitochondria can negatively regulate inflammasome activation. (C) Proteins that sequester the components of NLRP3 inflammasome prevent its activation
3.3.1|. Preventing K+ efflux to inhibit the NLRP3 inflammasome
Petrillia et al.73 first observed that the addition of potassium chlo-ride (KCl) (130 mM concentration) to prevent K+ efflux inhibited activation of the NLRP3 inflammasome in LPS-primed mouse peritoneal macrophages as well as in THP1 cells by various stimuli including ATP, nigericin, MSU, and Escherichia coli. Thus, lowering the concentrations of cytosolic K+ was sufficient to activate the NLRP3 inflammasome. Interestingly, the low levels of K+ induced NLRP3 inflammasome assembly in a cell-free system; that is, THP1 cell lysates prepared in 10 mM KCl spontaneously activated caspase-1.73 Although these data suggest a direct role for K+ in promoting inflammasome assembly, further studies are needed to elucidate the precise molecular mechanisms. Given that K+ efflux seems to be the common trigger for activation of NLRP3 inflammasome in response to both bacterial toxins and particulate matter,28,74 inhibiting K+ efflux can negatively regulate NLRP3 inflammasome in various inflammatory conditions. The ketone metabolite β-hydroxybutyrate (BHB), which is elevated during starvation, caloric restriction, high-intensity exercise, and low-carb ketogenic diet,75,76 specifically inhibits NLRP3 inflammasome in murine BMDM and CD14+ primary human monocytes.77 Mechanistically, BHB prevents K+ efflux to reduce NLRP3 inflamma-some activation.77 Although the precise molecular details of how BHB prevents K+ efflux remain unknown, BHB-mediated NLRP3 inflammasome inhibition seems to be independent of starvation-associated mechanisms such as autophagy.77 Mutation in LIPIN2, a member of the LIPIN family of proteins, triggers the NLRP3-associated inflammatory disorder called Majeed syndrome.78 Primary human macrophages or murine BMDM with siRNA-mediated knockdown of LIPIN2 produced significantly elevated levels of IL-1β in response to LPS and ATP, a classical activator of the NLRP3 inflammasome.79 When compared with control cells, siRNA knockdown of LIPIN2 in RAW264.7 macrophages significantly increased K+ efflux following 2 mM ATP treatment.79 Thus, LIPIN regulates K+ efflux, a critical step for NLRP3 inflamma-some activation.28,73,74 Utilizing Lipin2-deficient mice, Lorden et al.79 showed in peritoneal macrophages that LIPIN-2 negatively regulates ATP-induced opening of P2X7 receptor channel to inhibit K+ efflux. More recently, transmembrane immunoglobulin and mucin domain (TIM)-3, a molecule involved in negative regulation of T cell activation and phagocytosis in macrophages, was demonstrated to negatively regulate LPS+ATP-induced NLRP3 inflammasome activation in murine peritoneal and J774 macrophages.80 Blocking TIM-3-signaling with soluble TIM-3 Ig protein for 1 h in J774 macrophages reduced intracellular K+ levels suggesting an inhibitory role for TIM-3 in K+ efflux.80 Further investigation is needed to elucidate precise mechanisms associated with TIM-3-mediated inhibition of NLRP3 activity.
3.3.2|. Clearance of damaged mitochondria to limit NLRP3 inflammasome activation
One of the central mechanisms that promote NLRP3 inflammasome activation is mitochondrial dysfunction. The downstream mechanisms by which mitochondrial damage promotes NLRP3 inflammasome activation has been reviewed previously.18 Conversely, pathways that regulate clearance of damaged mitochondria can negatively regulate the NLRP3 inflammasome by limiting the proximal signals released from damaged mitochondria, which are necessary for NLRP3 activation. Autophagy is a major process involved in clearance of damaged mitochondria from the cells and thus could regulate NLRP3 inflammasome activation. In support of this hypothesis, siRNA-mediated deletion of autophagy-associated molecules ATG5 and Beclin1 in THP1 monocytes increased NLRP3 inflammasome activation by MSU crystals and nigericin in vitro.30 Similarly, deficiency of LC3B or Beclin1 in BMDM resulted in defective clearance of damaged mitochondria and increased release of mitochondrial (mt) DNA and mtROS in the cytosol, which promoted excessive NLRP3 inflammasome activation during LPS + ATP stimulation.81 Moreover, mice deficient in LC3B are highly susceptible to LPS shock and produce copious amounts of IL-1β and IL-18.81 Lc3b-deficient mice also produce highly increased levels of IL-1β and IL-18 in a cecal-ligation and puncture (CLP)-induced polymicrobial sepsis model.81 Further strengthening the role for autophagy in negatively regulating the NLRP3 inflamma-some, loss of ATG16L1 (protein essential for autophagy) in fetal liver macrophages promoted aberrant NLRP3 inflammasome activation during LPS, LPS + ATP, and LPS + MSU stimulations.82 Forkhead box O 3a (FOXO3a) overexpression in murine Kupffer cells promoted Bim expression, which restored autophagy flux and attenuated NLRP3 inflammasome activation during LPS and palmitic acid stimulation in vitro.83 Thus, FOXO3a promotes autophagy to negatively regulate the NLRP3 inflammasome. More recently, Hemin and its derivative cobalt, protoporphyrin, were reported to promote both autophagosome formation and autophagy flux.84 As a result Hemin inhibited NLRP3 inflammasome activation in both human and murine macrophages in vitro, and during MSU-induced peritonitis in mice in vivo.84
Similar to autophagy, several studies have reported a role for mitophagy in clearance of damaged mitochondria and regulation of NLRP3 inflammasome activity. Nucleotide-binding oligomerization domain containing 2 (NOD2) and receptor interacting protein kinase 2 (RIPK2) promote phosphorylation and activation of a mitophagy-associated molecule, ULK1, during influenza virus (mouse adapted PR8 H1N1 strain) infection of murine BMDC to clear damaged mitochondria, clear infection, and promote homeostasis.85 Thus, mice deficient in NOD2 and RIPK2 exhibit uncontrolled hyperactive inflammasome activation (as demonstrated by aberrant IL-18 production) and are susceptible to influenza virus infection when compared with controls.85 Given that PR8-infection induces the production of NLRP3 inflammasome-associated IL-1β and IL-18 cytokines,86,87 Lupfer et al.85 proposed that NOD2/RIPK2-signaling axis negatively regulates the NLRP3 inflammasome. Follow up studies by Lupfer et al.88 showed that Citrobacter rodenitum infection of Nod2 and Ripk2-deficient BMDM induced increased NLRP3 inflammasome activation as demonstrated by caspase-1 cleavage and IL-18 production. Zhong et al.89 further showed that NFκB signaling (which is required for priming of NLRP3) negatively regulates excessive inflammation by promoting p62-dependent clearance of damaged mitochondria. As such, LPS-primed BMDM deficient in p62 have dramatic accumulation of damaged mitochondria following ATP, alum, and MSU stimulation (classical activators of the NLRP3 inflammasomes), which promote aberrant caspase-1 activation, IL-1β production, and cell death.89 Thus, the NFκB-p62-mitophagy pathway serves as a cell-intrinsic mechanism to restrict host response by negatively regulating excessive NLRP3 inflammasome activation.89 sestrin2 (SESN2) facilitates mitophagy by promoting mitochondrial aggregation and K63-linked ubiquitination of damaged mitochondria, and by increasing the levels of ULK1 in murine BMDM during LPS + ATP stimulation.90 Thus, Sesn2-deficient BMDM display increased caspase-1 activation during LPS + ATP-induced activation of the NLRP3 inflammasome.90 As a result, Sesn2deficient mice are highly susceptible to LPS shock as demonstrated by rapid death of 100% of the Sesn2-deficient mice by 48 h following 12 mg/kg intraperitoneal LPS injection.90 Sesn2-deficient mice also produce increased levels of IL-1β and IL-18 during CLP-induced polymicrobial sepsis and significantly higher percentage of Sesn2-deficient mice succumb to CLP-induced septic death.90
Taken together, these studies highlight the importance of autophagy and mitophagy in clearance of damaged mitochondria and regulation of the NLRP3 inflammasomes (Fig. 3A).
3.3.3|. Inhibition of NLRP3 trafficking to the mitochondria
Upon activation, NLRP3 needs to translocate to the mitochondria to assemble the inflammasome complex.91 Thus, mechanisms that target the cytoskeleton molecules such as microtubule and actin can negatively regulate activation of the NLRP3 inflammasome (Fig. 3B). Recombinant IL-4 inhibited ATP and nigericin-induced caspase-1 activation and IL-1β maturation in THP1 cells and murine BMDM.92 IL-4 treatment markedly reduced ATP-induced localization of NLRP3 to the mitochondrial fraction in murine BMDM,92 a critical step needed for inflammasome assembly.91 IL-4 signaling reduced ATP-induced micro-tubule polymerization in LPS-primed murine BMDM, which could be a possible mechanism by which IL-4 limits NLRP3 enrichment into the mitochondria.92 NAD-dependent deacetylase, sirtuin2 (SIRT2), negatively regulates NLRP3 inflammasome activation by restricting NLRP3 trafficking to the mitochondria.93 Inhibition of SIRT2 activity by AGK2 (a SIRT2 inhibitor) in LPS-primed J774 macrophages increased IL-1β production after nigericin stimulation.93 Acetylated α-tubulin is required for Dynein-dependent localization of NLRP3 to the mitochondria.93 SIRT2 actively deacetylates α-tubulin to restrict NLRP3 trafficking to mitochondria and inhibit assembly of the inflammasome complex.93 Actin polymerization regulates various cellular processes that include motility, cytokinesis, and vesicular trafficking.94
Upon activation with ATP or nigericin, NLRP3 interacts with F-actin, which negatively regulates NLRP3 activity in PMA-primed THP1 cells.94 F-actin-mediated interaction and inhibition of NLRP3 required Flightless-I (Fli-I) and LRR Fli-I-interacting protein 2 (LRRFIP2).94
Concurrently, production of IL-1β and caspase-1 activation were significantly increased in PMA-primed THP1 monocytes or LPS-primed murine peritoneal macrophages upon shRNA-mediated deletion of Fli-I or LRRFIP2.94,95 Biochemical studies showed that LRRFIP2 and Fli-I facilitates NLRP3’s interaction with F-actin to inhibit NLRP3 inflammasome activity.94,95 Fli-I also sequesters caspase-1 and inhibits caspase-1 activation in this complex.95 Importantly, siRNA-mediated LRRFIP2 knockdown in mice increased alum-induced activation of caspase-1 and IL-1β in peritoneal exudate cells and exhibited severe peritonitis.95
3.4|. Sequestering NLRP3, ASC, or caspase-1 to inhibit inflammasome assembly
Assembly of the inflammasome requires NLRP3, ASC, and caspase-1 oligomerization. NLRP3 recruits ASC through Pyrin–Pyrin interaction and ASC subsequently recruits caspase-1 via CARD-CARD interaction to assemble the NLRP3 inflammasome complex. Molecules that can sequester NLRP3, ASC, and caspase-1 out of the complex can thus negatively regulate NLRP3 inflammasome activation (Fig. 3C). Histone deacetylase 6 (HDAC6) negatively regulate NLRP3 inflammasome activation by sequestering NLRP3.96 Murine BMDM with shRNA-mediated knockdown of HDAC6 demonstrated higher levels of caspase-1 activation and IL-1β production following LPS + ATP and LPS + nigericin stimulation.96 Interestingly, this negative regulation was independent of HDAC6’s deacetylase activity as demonstrated by failure of Tubastatin A (TSA—a selective HDAC6 deacytlase activity inhibitor) to rescue the increased caspase-1 and IL-1β cleavage in HDAC6-silenced BMDM.96 Given that the protein level of NLRP3 did not change following shRNA-mediated HDAC6 knockdown in BMDM, it was posited that HDAC6 sequesters NLRP3.96 In support, NLRP3 coimmunoprecipitated with HDAC6 when expressed together in HEK293T cells.96 Binder of Ubiquitin Zinc finger (Buz) domain of HDAC6 was required for HDAC6’s interaction with NLRP3, suggesting HDAC6 interaction with ubiquitinated NLRP3 negatively regulates the inflammasome activity.96 Given that NLRP3 is a cytosolic sensor, it is likely that the HDAC6 sequesters NLRP3 in the cytoplasm and not in the nucleus of the cell. However, recent studies have shown that NLRP3 is also present in the nucleus of T cells.97 Thus, it is also possible that HDAC6 may sequester NLRP3 in the nucleus. Farnesoid X receptor (FXR), a master regulator of bile acid homeostasis, was shown to negatively regulate bile acid-induced NLRP3 inflammasome activation.98 Bile acids such as deoxycholic acid and chenodeoxycholic acid activated caspase-1 and IL-1β in WT peritoneal macrophages, which were increased in Fxr-deficient peritoneal macrophages.98 Although the precise molecular mechanisms remain to be explored, FXR coimmunoprecipitated with NLRP3 and caspase-1 in THP1 cells both in unstimulated and LPS-stimulated conditions, suggesting FXR-mediated sequestration of NLRP3 and caspase-1 as a possible mechanism for tempering NLRP3 inflamma-some activation.98 Cytochrome c sequesters NLRP3 in the cytosol to inhibit inflammasome activation.99 Transducing cytochrome c into the cytoplasm of LPS-prime THP1 cells reduced caspase-1 activation and IL-1β cleavage following ATP stimulation.99 Conversely, siRNA-mediated silencing of cytochrome c in THP1 cells increased caspase-1 and IL-1β activation in response to LPS + ATP stimulation.99 ATP-induced activation of the NLRP3 inflammasome triggered the release of cytochrome c from inner mitochondrial membrane into the cytosol in LPS-primed THP1 cells.99 In LPS-primed THP1 cells, endogenous NLRP3 immunoprecipitated with cytochrome c following ATP stimulation.99 Furthermore, coexpression of cytochrome c with different NLRP3 constructs in HEK293T cells demonstrated that cytochrome c bound to the LRR domain of NLRP3.99 Cytochrome c prevents NLRP3’s association with cardiolipin on mitochondria to negatively regulate activation of the NLRP3 inflammasome.31,99 Furthermore, cytochrome c initiates the formation of apoptosome, a complex that promotes apoptosis,100,101 suggesting a potential crosstalk between apoptotic and pyroptotic cell death pathways. In concurrence, caspase-8 and FADD, both critical molecules that induce apoptosis, are required for activation of the NLRP3 inflammasome.32
Pyrin and CARD domains mediate protein–protein interaction via homotypic Pyrin–Pyrin or CARD–CARD interactions. This provides the potential for regulatory proteins to sequester Pyrin- or CARD-containing molecules to regulate their activities. Indeed, several Pyrin-only Proteins (POP) and CARD-only Proteins (COP) have been reported as negative regulators of inflammasome signaling; they have been reviewed recently.102,103 Interestingly, these POP and COP proteins have evolved rather recently, as these molecules are only found in primates and humans but are completely absent in mice.102 The POP family of proteins includes POP1, POP2, POP3, and POP4. POP1 has a higher degree of homology (64%) with ASC. POP1 sequesters ASC, thereby restricting the recruitment of ASC to suppress NLRP3 inflammasome activation.104,105 Although there is no ortholog of POP1 in mouse, transgenic expression of human POP1 protected mice from NLRP3 inflammasome-mediated inflammatory diseases.104 POP2, POP3, and POP4 have not yet been shown to interact with either NLRP3 or ASC in humans. Future studies will determine whether these POP proteins can regulate the NLRP3 inflammasomes in specific inflammatory settings.
CARD16,106,107 CARD17,108 and CARD18109 constitute the CARD-only proteins. The COP proteins CARD16, CARD17 and CARD18 are all located on chromosome 11q22 adjacent to caspase-1 and bear a high degree of homology to the CARD domain of human caspase-1.102 Thus, it was proposed that these COP proteins arose from caspase-1 through a recent gene duplication event.110 CARD16 also known as Pseudo-ICE was first identified in 2001 through a database screen of expressed sequence tags that resembled high degree of homology to human pro-caspase-1.106,107 CARD16, which shares 92% sequence homology to the CARD domain of human caspase-1, can self-associate in a homodimer, directly bind with caspase-1 CARD and inhibit IL-1β secretion by IFN-γ and LPS-stimulated THP1 cells.106,107 These interactions were specific as CARD16 did not bind other CARD containing proteins including caspase-2, caspase-4, caspase-9, APAF-1, Bcl10, cIAP1, cIAP2, and CRADD.106,107 CARD17, previously known as INCA (Inhibitory CARD), was identified in 2004 by in silico bioinformatics approach and share 81% sequence homology to the CARD domain of human pro-caspase-1. CARD17 directly binds with caspase-1 CARD and inhibit LPS-induced release of IL-1β and from THP1 cells.108,111 CARD17 interaction with caspase-1 CARD is specific as CARD17 does not interact with CARD domains of RIPK2 or caspase-2.108 A recent study further demonstrated that CARD17 interaction with caspase-1 inhibits its polymerization and filament formation,111 events necessary for inflammasome activation.112,113 CARD18 also known as ICEBERG shares 51% sequence homology with the CARD domain of human pro-caspase-1.109 CARD18 interacts with other CARD18 in a homotypic interaction and also with CARD of human pro-caspase-1, but not the CARD domains of caspase-7, caspase-8, caspase-9, or RIPK2.106,109,111 These data demonstrate specificity of CARD18 in binding caspase-1 CARD. When expressed in THP1 cells, CARD18 inhibited LPS-induced release of IL-1β [109]. Of note, a recent study with THP1 cells expressing doxycycline inducible CARD18 did not find any role for CARD18 in inhibiting IL-1β secretion upon LPS + nigericin stimulation, although caspase-1 activation was slightly reduced in CARD18 expressing THP1 cells.111 The precise mechanisms by which these COP inhibit caspase-1 activation remain to be elucidated, and why they interact with certain CARD proteins and not others (i.e., their specificity) are not completely understood.
4|. CONCLUSIONS
NLRP3 is a central global inflammasome sensor that activates and assembles inflammasome complex in response to a wide array of stimuli. Given the promiscuous nature of NLRP3, the negative regulation of NLRP3 is critical to prevent unwanted inflammation and tissue damage. In this review, we focused on the cellular mechanisms that are employed to keep NLRP3 inflammasome activation and inflammation in control. Positive and negative regulations are 2 faces of the same coin. Likewise, understanding positive regulation of the NLRP3 inflammasome is central to examining and identifying the basis of its negative regulation. In this review, we take a systematic, step-wise approach to explain and discuss potential ways the NLRP3 inflammasome can be negatively regulated. In brief, we identified 5 major points in the assembly and activation of the NLRP3 inflammasome that could be potentially targeted for its negative regulation: (1) Priming events that lead to activation of NFκB and MAP kinase and subsequent up-regulation of NLRP3 mRNA, (2) posttranscriptional regulation of NLRP3 mRNA, (3) posttranslational regulation of NLRP3 protein, (4) inhibition of proximal events that activate NLRP3, (5) sequestration of NLRP3, ASC, or caspase-1.
In addition to the cell-intrinsic factors that inhibit activation of the NLRP3 inflammasome discussed in this review, several microbe-associated factors can also negatively regulate the NLRP3 inflammasome.114 Despite the amount of papers published on NLRP3 activation, we still lack a complete understanding of negative regulatory pathways, and several important questions still remain unanswered. As discussed in this review, there are many layers of negative regulation to inhibit NLRP3 activation, yet aberrant activation of the NLRP3 inflammasome is commonly observed in various disease settings. Specifically, deficiency in just one negative regulatory pathway is enough to promote aberrant NLRP3 activation. What are the reasons for this failure to regulate aberrant NLRP3 activation? Are these diverse mechanisms of NLRP3 regulation not redundant? Are negative regulatory pathways specifically activated in response to specific stimuli? Importantly, most of these mechanistic studies are always performed and validated in macrophages and DC. Do the same rules of negative regulation also apply to other immune cells such as neutrophils, T cells, and B cells that play major roles in inflammatory disorders? Future efforts should be geared towards understanding the contribution of these negative regulators in translational settings. Undoubtedly, a clear understanding of the basic biology and precise cellular and molecular mechanisms of negative regulation of NLRP3 will aid in the development of future therapeutics that can effectively treat NLRP3-associated inflammatory diseases.
ACKNOWLEDGMENTS
This work was supported by K22AI127836 and University of Iowa Startup funds to Dr. Prajwal Gurung.
Abbreviations:
- AIM2
absent in melanoma-2
- ALR
absent in melanoma-2-like receptors
- ASC
apoptosis-associated speck-like protein containing a CARD
- ATG
autophagy-related
- BHB
β-hydroxybutyrate
- BMDC
bone marrow-derived dendritic cells
- BMDM
bone marrow-derived macrophages
- BRCC3
BRCA1/BRCA2-containing complex subunit 3
- Ca2+
calcium
- CARD
caspase activation and recruitment domain
- CLP
cecal ligation and puncture
- COP
CARD-only proteins
- DAMP
damage-associated molecular patterns
- DC
dendritic cells
- DRD1
dopamine receptor D1
- DSS
dextran sodium sulfate
- EP4
E-prostanoid 4
- FADD
Fas-associated death domain protein
- FBXL2
F-box and leucine-rich repeat protein 2
- FliI
Flightless-I
- FOXO3a
Forkhead box O 3a
- FXR
Farnesoid X receptor
- GSDMD
gasdermin D
- HDAC6
histone deacetylase 6
- IFI16
interferon gamma inducible protein 16
- IRAK1
IL 1 receptor associated kinase 1
- KCl
potassium chloride
- LC3B
microtubule-associated protein light chain 3B
- LRR
leucine-rich repeat
- LRRFIP2
leucine-rich repeat Fli-I-interacting protein 2
- MARCH7
membrane-associated RING finger protein 7
- miR
micro RNA
- MSU
monosodium urate
- N-GSDMD
N-terminal fragment of GSDMD
- NLR
Nod-like receptors
- NLRC
Nod-like receptor family CARD domain containing protein
- NLRP
Nod-like receptor family pyrin domain containing protein
- NOD2
nucleotide-binding oligomerization domain containing protein 2
- PAMP
pathogen-associated molecular patterns
- PKA
protein kinase A
- POP
pyrin-only proteins
- K+
potassium
- PRR
pathogen recognition receptor
- PTPN22
protein tyrosine phosphatase, non-receptor Type 22
- RIPK2
receptor interacting protein kinase 2
- ROS
reactive oxygen species
- SESN2
sestrin2
- SIRT2
sirtuin2
- TIM
transmembrane immunoglobulin and mucin domain
- TRIM
tripartite motif
- TTP
tristetraprolin
- ULK1
Unc-51 like autophagy activating kinase 1
- UTR
untranslated region
- WT
wild-type
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. [DOI] [PubMed] [Google Scholar]
- 2.Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–21122. [DOI] [PubMed] [Google Scholar]
- 3.Hu Z, Zhou Q, Zhang C, et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 2015;350:399–404. [DOI] [PubMed] [Google Scholar]
- 4.Tenthorey JL, Haloupek N, Lopez-Blanco JR, et al. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science. 2017;358:888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Chen S, Ruan J, et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kesavardhana S, Kanneganti TD. Mechanisms governing inflamma-some activation, assembly and pyroptosis induction. Int Immunol. 2017;29:201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Zhang Z, Ruan J, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. [DOI] [PubMed] [Google Scholar]
- 9.Kayagaki N, Stowe IB, Lee BL, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526: 666–671. [DOI] [PubMed] [Google Scholar]
- 10.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. [DOI] [PubMed] [Google Scholar]
- 11.Mariathasan S, Weiss DS, Newton K, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440: 228–232. [DOI] [PubMed] [Google Scholar]
- 12.Kanneganti TD, Ozoren N, Body-Malapel M, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. [DOI] [PubMed] [Google Scholar]
- 13.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. [DOI] [PubMed] [Google Scholar]
- 14.Mariathasan S, Newton K, Monack DM, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. [DOI] [PubMed] [Google Scholar]
- 15.Hornung V, Ablasser A, Charrel-Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burckstummer T, Baumann C, Bluml S, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. [DOI] [PubMed] [Google Scholar]
- 18.Gurung P, Lukens JR, Kanneganti TD. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med. 2015;21:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Yang J, Gao W, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature.2014;513:237–241. [DOI] [PubMed] [Google Scholar]
- 20.Minkiewicz J, de Rivero Vaccari JP, Keane RW. Human astrocytes express a novel NLRP2 inflammasome. Glia. 2013;61:1113–1121. [DOI] [PubMed] [Google Scholar]
- 21.Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy M, Thaiss CA, Zeevi D, et al. Microbiota-Modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015;163:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khare S, Dorfleutner A, Bryan NB, et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36:464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu S, Ding S, Wang P, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546:667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vladimer GI, Weng D, Paquette SW, et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 2012;37:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abe T, Lee A, Sitharam R, Kesner J, Rabadan R, Shapira SD. Germ-Cell-Specific inflammasome component nlrp14 negatively regulates cytosolic nucleic acid sensing to promote fertilization. Immunity.2017;46:621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerur N, Veettil MV, Sharma-Walia N, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflamma-some activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee GS, Subramanian N, Kim AI, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. [DOI] [PubMed] [Google Scholar]
- 31.Iyer SS, He Q, Janczy JR, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurung P, Anand PK, Malireddi RK, et al. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J Immunol. 2014;192:1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y, Hara H, Nunez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi H, Wang Y, Li X, et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat Immunol. 2016;17:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid-Burgk JL, Chauhan D, Schmidt T, et al. A genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J Biol Chem. 2016;291: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y, Zeng MY, Yang D, Motro B, Nunez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 2016;530:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menu P, Vince JE. The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin Exp Immunol. 2011;166:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurung P, Kanneganti TD. Autoinflammatory skin disorders: the inflammasomme in focus. Trends Mol Med. 2016;22: 545–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masters SL, Dunne A, Subramanian SL, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1beta in type 2 diabetes. Nat Immunol. 2010;11: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandanmagsar B, Youm YH, Ravussin A, et al. The NLRP3 inflamma-some instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen H, Gris D, Lei Y, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12:408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daley D, Mani VR, Mohan N, et al. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J Exp Med. 2017;214:1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malik A, Kanneganti TD. Inflammasome activation and assembly at a glance. J Cell Sci. 2017;130:3955–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonina IS, Zhong Z, Karin M, Beyaert R. Limiting inflammation-the negative regulation of NF-kappaB and the NLRP3 inflammasome. Nat Immunol. 2017;18:861–869. [DOI] [PubMed] [Google Scholar]
- 45.Gurung P, Lamkanfi M, Kanneganti TD. Cutting edge: sHARPIN is required for optimal NLRP3 inflammasome activation. J Immunol. 2015;194:2064–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gurung P, Li B, Subbarao Malireddi RK, Lamkanfi M, Geiger TL, Kanneganti TD. Chronic TLR stimulation controls NLRP3 inflammasome activation through IL-10 mediated regulation of NLRP3 expression and caspase-8 activation. Sci Rep. 2015;5:14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol. 2012;189:4175–4181. [DOI] [PubMed] [Google Scholar]
- 48.Haneklaus M, Gerlic M, Kurowska-Stolarska M, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189:3795–3799. [DOI] [PubMed] [Google Scholar]
- 49.Neudecker V, Haneklaus M, Jensen O, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med. 2017;214:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanneganti TD. Inflammatory bowel disease and the NLRP3 inflammasome. N Engl J Med. 2017;377:694–696. [DOI] [PubMed] [Google Scholar]
- 51.Xiao L, Jiang L, Hu Q, Li Y. MicroRNA-133b ameliorates allergic inflammation and symptom in murine model of allergic rhinitis by targeting Nlrp3. Cell Physiol Biochem. 2017;42:901–912. [DOI] [PubMed] [Google Scholar]
- 52.Lou J, Wang Y, Zhang Z, Qiu W. MiR-20b inhibits mycobacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Exp Cell Res. 2017;358:120–128. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y, Lu M, Du RH, et al. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener. 2016;11:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haneklaus M, O’Neil JD, Clark AR, Masters SL, O’Neill LAJ. The RNA-binding protein Tristetraprolin (TTP) is a critical negative regulator of the NLRP3 inflammasome. J Biol Chem. 2017;292:6869–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang J, Liu Z, Xiao TS. Post-translational regulation of inflammasomes. Cell Mol Immunol. 2017;14:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juliana C, Fernandes-Alnemri T, Kang S, Farias A, Qin F, Alnemri ES. Non-transcriptional priming and deubiquitination regulate NLRP3 inflammasome activation. J Biol Chem. 2012;287: 36617–36622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin KM, Hu W, Troutman TD, et al. IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proc Natl Acad Sci USA. 2014;111:775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandes-Alnemri T, Kang S, Anderson C, Sagara J, Fitzgerald KA, Alnemri ES. Cutting edge: tLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. J Immunol. 2013;191: 3995–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–338. [DOI] [PubMed] [Google Scholar]
- 60.Yan Y, Jiang W, Liu L, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160:62–73. [DOI] [PubMed] [Google Scholar]
- 61.Han S, Lear TB, Jerome JA, et al. Lipopolysaccharide primes the nalp3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCFFBXL2 E3 Ligase. J Biol Chem. 2015;290: 18124–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura T, Jain A, Choi SW, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210:973–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song H, Liu B, Huai W, et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spalinger MR, Kasper S, Gottier C, et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest. 2016;126:4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spalinger MR, Lang S, Gottier C, et al. PTPN22 regulates NLRP3-mediated IL1B secretion in an autophagy dependent manner. Autophagy. 2017:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gross O, Poeck H, Bscheider M, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. [DOI] [PubMed] [Google Scholar]
- 67.Mortimer L, Moreau F, MacDonald JA, Chadee K. NLRP3 inflamma-some inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat Immunol. 2016;17:1176–1186. [DOI] [PubMed] [Google Scholar]
- 68.Stutz A, Kolbe CC, Stahl R, et al. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med. 2017;214:1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song N, Liu ZS, Xue W, et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol Cell 68 2017;185–197: e6. [DOI] [PubMed] [Google Scholar]
- 70.Hernandez-Cuellar E, Tsuchiya K, Hara H, et al. Cutting edge: nitric oxide inhibits the NLRP3 inflammasome. J Immunol. 2012;189: 5113–5117. [DOI] [PubMed] [Google Scholar]
- 71.Mao K, Chen S, Chen M, et al. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 2013;23:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mishra BB, Rathinam VA, Martens GW, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nat Immunol. 2013;14:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–1589. [DOI] [PubMed] [Google Scholar]
- 74.Kanneganti TD, Lamkanfi M. K(+) drops tilt the NLRP3 inflamma-some. Immunity. 2013;38:1085–1088. [DOI] [PubMed] [Google Scholar]
- 75.Cotter DG, Schugar RC, Crawford PA. Ketone body metabolism and cardiovascular disease. Am J Physiol Heart Circ Physiol. 2013;304:H1060–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends Endocrinol Metab. 2014;25:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Youm YH, Nguyen KY, Grant RW, et al. The ketone metabolite beta-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat Med. 2015;21:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ferguson PJ, Chen S, Tayeh MK, et al. Homozygous mutations in LPIN2 are responsible for the syndrome of chronic recurrent multifocal osteomyelitis and congenital dyserythropoietic anaemia (Majeed syndrome). J Med Genet. 2005;42:551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lorden G, Sanjuan-Garcia I, de Pablo N, et al. Lipin-2 regulates NLRP3 inflammasome by affecting P2X7 receptor activation. J Exp Med. 2017;214:511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, Shi Q, Dou S, et al. Negative regulation of Nod-like receptor protein 3 inflammasome activation by T cell Ig mucin-3 protects against peritonitis. Immunology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakahira K, Haspel JA, Rathinam VA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. [DOI] [PubMed] [Google Scholar]
- 83.Liu Y, Zhang W, Wu X, Gong J. Foxo3a-dependent Bim transcription protects mice from a high fat diet via inhibition of activation of the NLRP3 inflammasome by facilitating autophagy flux in Kupffer cells. Oncotarget. 2017;8:34258–34267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nurmi K, Kareinen I, Virkanen J, et al. Hemin and cobalt protoporphyrin inhibit nlrp3 inflammasome activation by enhancing autophagy: a novel mechanism of inflammasome regulation. J Innate Immun. 2017;9:65–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lupfer C, Thomas PG, Anand PK, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14:480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomas PG, Dash P, Aldridge JR, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza a virus via the regulation of caspase-1. Immunity. 2009;30: 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lupfer CR, Anand PK, Liu Z, et al. Reactive oxygen species regulate caspase-11 expression and activation of the non-canonical NLRP3 inflammasome during enteric pathogen infection. PLoS Pathog. 2014;10:e1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhong Z, Umemura A, Sanchez-Lopez E, et al. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim MJ, Bae SH, Ryu JC, et al. SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages. Autophagy. 2016;12:1272–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hwang I, Yang J, Hong S, et al. Non-transcriptional regulation of NLRP3 inflammasome signaling by IL-4. Immunol Cell Biol. 2015;93:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Misawa T, Takahama M, Kozaki T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14:454–460. [DOI] [PubMed] [Google Scholar]
- 94.Burger D, Fickentscher C, de Moerloose P, Brandt KJ. F-actin dampens NLRP3 inflammasome activity via Flightless-I and LRRFIP2. Sci Rep. 2016;6:29834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jin J, Yu Q, Han C, et al. LRRFIP2 negatively regulates NLRP3 inflammasome activation in macrophages by promoting flightless-I-mediated caspase-1 inhibition. Nat Commun. 2013;4:2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hwang I, Lee E, Jeon SA, Yu JW. Histone deacetylase 6 negatively regulates NLRP3 inflammasome activation. Biochem Biophys Res Commun. 2015;467:973–978. [DOI] [PubMed] [Google Scholar]
- 97.Bruchard M, Rebe C, Derangere V, et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol. 2015;16:859–870. [DOI] [PubMed] [Google Scholar]
- 98.Hao H, Cao L, Jiang C, et al. Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab. 2017;25:856–867:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi CS, Kehrl JH. Cytochrome c Negatively Regulates NLRP3 Inflammasomes. PLoS One. 2016;11:e0167636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102:17545–17550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. [DOI] [PubMed] [Google Scholar]
- 102.Matusiak M, Van Opdenbosch N, Lamkanfi M. CARD- and pyrin-only proteins regulating inflammasome activation and immunity. Immunol Rev. 2015;265:217–230. [DOI] [PubMed] [Google Scholar]
- 103.Dorfleutner A, Chu L, Stehlik C. Inhibiting the inflammasome: one domain at a time. Immunol Rev. 2015;265:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Almeida L, Khare S, Misharin AV, et al. The PYRIN domain-only protein pop1 inhibits inflammasome assembly and ameliorates inflammatory disease. Immunity. 2015;43:264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003;373:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1beta generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8:649–657. [DOI] [PubMed] [Google Scholar]
- 107.Lee SH, Stehlik C, Reed JC. Cop, a caspase recruitment domain-containing protein and inhibitor of caspase-1 activation processing. J Biol Chem. 2001;276:34495–34500. [DOI] [PubMed] [Google Scholar]
- 108.Lamkanfi M, Denecker G, Kalai M, et al. INCA, a novel human caspase recruitment domain protein that inhibits interleukin-1beta generation. J Biol Chem. 2004;279:51729–51738. [DOI] [PubMed] [Google Scholar]
- 109.Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM. ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell. 2000;103:99–111. [DOI] [PubMed] [Google Scholar]
- 110.Kersse K, Vanden Berghe T, Lamkanfi M, Vandenabeele P. A phylogenetic and functional overview of inflammatory caspases and caspase-1-related CARD-only proteins. Biochem Soc Trans. 2007;35: 1508–1511. [DOI] [PubMed] [Google Scholar]
- 111.Lu A, Li Y, Schmidt FI, et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat Struct Mol Biol. 2016;23:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cai X, Chen J, Xu H, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lu A, Magupalli VG, Ruan J, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rathinam VA, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nat Immunol. 2012;13:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]