Abstract
BACKGROUND
Global changes in gene expression underlying circuit and behavioral dysregulation associated with cocaine addiction remain incompletely understood. Here, we show how a history of cocaine self-administration (SA) “re-programs” transcriptome-wide responses throughout the brain’s reward circuitry at baseline and in response to context and/or cocaine re-exposure after prolonged withdrawal (WD).
METHODS
We assigned male mice to one of six groups: saline/cocaine SA + 24 hr WD; or saline/cocaine SA + 30 d WD + an acute saline/cocaine challenge within the previous drug-paired context. RNA-sequencing was conducted on six interconnected brain reward regions. Using pattern analysis of gene expression and factor analysis of behavior, we identified genes that are strongly associated with addiction-related behaviors and uniquely altered by a history of cocaine SA. We then identified potential upstream regulators of these genes.
RESULTS
We focused on three Patterns of gene expression that reflect responses to: a) acute cocaine, b) context re-exposure, and c) drug + context re-exposure. These Patterns revealed region-specific regulation of gene expression. Further analysis revealed that each of these gene expression Patterns correlated with an “Addiction Index”—a composite score of several addiction-like behaviors during cocaine SA—in a region-specific manner. CREB and nuclear receptor families were identified as key upstream regulators of genes associated with such behaviors.
CONCLUSIONS
This comprehensive picture of transcriptome-wide regulation in the brain’s reward circuitry by cocaine SA and prolonged WD provides new insight into the molecular basis of cocaine addiction, which will guide future studies of the key molecular pathways involved.
Keywords: RNA-sequencing, gene expression, nucleus accumbens, prefrontal cortex, basolateral amygdala, dorsal striatum, ventral hippocampus
INTRODUCTION
Addiction arises from genetic and environmental factors, which determine individual responses to initial and repeated drug exposure at the molecular, cellular, and circuit levels (1). A key feature of addiction is the ability for drug or drug-associated cues to trigger relapse, even after periods of prolonged abstinence (2). It is hypothesized that susceptibility to relapse depends on long-term neuroadaptations within the brain’s reward circuitry (3–5).
Behavioral responses to cocaine self-administration (SA) after withdrawal (WD) and re-exposure to drug or contextual cues are well characterized in rodent models. However, the underlying molecular mechanisms remain elusive. Most studies investigating transcriptional changes associated with long-term WD followed by cocaine/context re-exposure have focused on candidate genes within one or two brain regions. These studies have found that long-term WD from cocaine SA is associated with changes in growth factors and their signaling cascades (6–9), neurotransmitter and neuropeptide systems (10, 11), and immediate early genes (10, 12).
The few studies investigating transcriptome-wide changes after short-term WD from cocaine SA (13, 14), or long-term WD but without re-exposure (15), focused primarily on nucleus accumbens (NAc), ventral tegmental area (VTA) (14), or prefrontal cortex (PFC) (13, 15). No study has characterized transcriptome-wide changes across multiple interconnected brain reward regions. Furthermore, no transcriptomic study has compared multiple stages of WD plus drug/context re-exposure, while leveraging individual variability to identify genes transcriptome-wide associated with addiction-related behaviors.
Here, we performed RNA-sequencing on six reward-related brain regions in mice with a history of saline or cocaine SA. We profiled the transcriptome in these regions after short- and long-term WD with drug/context re-exposure. We hypothesized that a history of cocaine SA “reprograms” the transcriptome, resulting in “priming” or “desensitization” of molecular targets upon re-exposure to drug-related context ± cocaine.
METHODS AND MATERIALS
See supplemental information for detailed methods.
Experimental animals
Male C57BL/6J mice were obtained from The Jackson Laboratory. All experiments were conducted in accordance with guidelines of the Institutional Animal Care and Use Committee at Mount Sinai.
RNA-Sequencing
Brain regions were dissected rapidly and frozen on dry ice. RNA extraction, library preparation, and RNA-seq were conducted as described (16–18). Multiple targets were validated by qPCR using TaqMan assays (Figure S1; ThermoFisher, Foster City, CA).
Statistics and Bioinformatics
Behavior
Behaviors were analyzed using ANOVA or Kruskall-Wallis tests depending on homozygosity of variance. All analyses were conducted using SPSS Statistical Software, V24 (IBM, Armonk, NY).
Transcriptomic Analysis
Pairwise differential expression comparisons were performed as reported (16, 17) using Voom-Limma (19); a significance threshold of fold change>1.15 and nominal p<0.05 were applied.
Factor Analysis and Linear Modeling
Factor analysis was used to reduce the dimensions of interdependent behavioral variables. The transformed behavioral data were then used as continuous covariates to predict gene expression in linear models. A composite “Addiction Index” (AI) of 3 factors (Figure 4; Supplemental Figures S3,S4) most closely associated with SA behaviors was calculated (Supplemental Methods). Regression analysis was conducted using Voom-Limma to determine AI associations with gene expression (19).
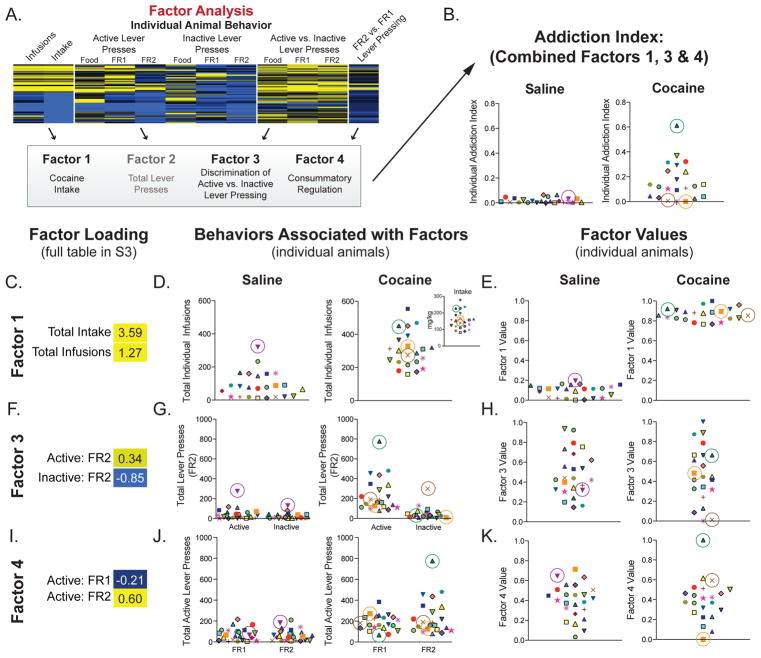
Figure 4. Generation of an “addiction index” for individual animals.
(A–B) Exploratory factor analysis on multiple behavioral endpoints reduced multi-dimensional behavioral data to 8 “factors.” A composite score, or “addiction index (AI),” of those factors most strongly associated with behaviors reflective of an addicted-like phenotype was generated using the individual transformed data for Factors 1, 3, & 4. (C–K) Data for individual animals for each behavior and each factor are presented. Each animal is represented by the same unique shape and color. (C, F, I) Factor loading, or associations, of Factors 1, 3, & 4 with SA behaviors (yellow = positive; blue = negative) are presented. (D, G, J) Individual data presented for the behaviors associated with each factor. (D) Factor 1 associated with intake and infusions; (G) Factor 3 is positively associated with active lever and negatively associated with inactive lever under an FR2 schedule; (J) Factor 4 is positively associated with FR2 lever presses and negatively associated with lever pressing on an FR1 schedule. (E, H, K) Individual transformed data for Factors 1 (E), 3 (H) and 4 (K). The product of these values was calculated to generate an AI for each individual. An animal must display high performance on all three factors (▲) to have a high AI. By contrast, if an animal performs poorly on one of the behaviors (× or ■) their AI is lower.
All other bioinformatic analyses were conducted as reported (16–18, 20, 21).
RESULTS
Cocaine Self-Administration Behavior
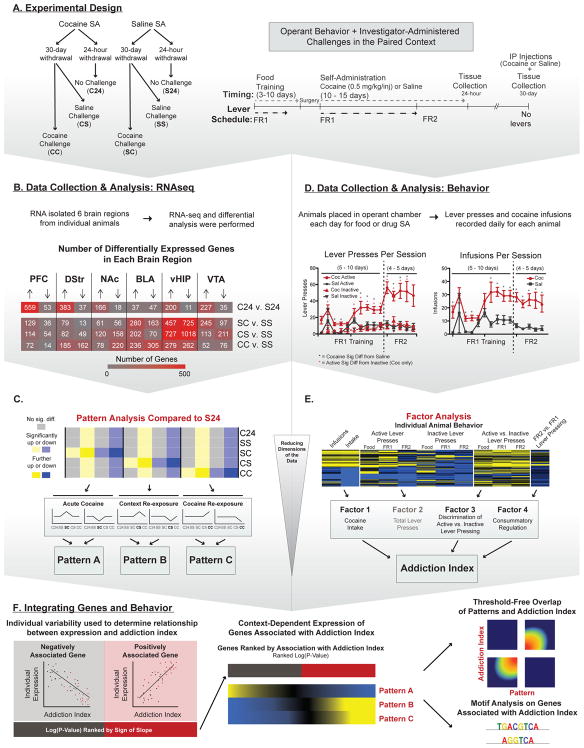
Figure 1 provides an outline of experimental procedures, which are explained in detail in Supplemental Methods. To determine how a history of cocaine SA influences circuit-wide transcriptomes, RNA-seq was performed on PFC, dorsal striatum (DStr), NAc, basolateral amygdala (BLA), ventral hippocampus (vHIP), and VTA, obtained from the following six groups of male mice (Figure 1A): saline SA + 24 hr WD (S24, n=5–8); cocaine SA + 24 hr WD (C24, n=5–8); saline SA + 30 d WD + saline re-exposure (SS, n=5–8); saline SA + 30 d WD + cocaine exposure (SC, n=5–8); cocaine SA + 30 d WD + saline exposure (CS, n=3–7); and cocaine SA + 30 d WD + cocaine re-exposure (CC, n=5–7). Supplemental Methods provides a complete breakdown of sample size by brain region.
Figure 1. Outline of Experimental Approach and Bioinformatic Analyses.
(A) Experimental design and summary of groups. Mice were food trained followed by 5–10 d of FR1 scheduling and 4–5 d of FR2. One group was euthanized 24 h after their last SA session while another cohort of animals were group housed in their home cage for 30 d. After WD, animals were given an injection of saline or cocaine and re-exposed to their original SA chamber for 1 h and euthanized immediately. (B) Data collection and RNA-seq data analysis. RNA-seq was performed on micro-dissections of 6 reward-associated brain regions. Differential expression analysis was performed to identify DEGs compared to their control group (S24 or SS). Number of DEGs per brain region are indicated (Red = greatest; Gray = least). (C) In an effort to identify genes that were uniquely altered by cocaine re-exposure we used pattern analysis and compared all groups to the same baseline (S24). Three patterns were investigated: Pattern A: genes uniquely altered by an initial dose of cocaine (SC; 1 h post-injection); Pattern B: genes uniquely altered by re-exposure to cocaine-paired context (CS); and Pattern C: genes uniquely altered by cocaine re-exposure (CC; 1 h post-injection). (D) Data collection and analysis of cocaine SA behavioral data. Because all animals (saline included), underwent varying numbers of SA trials at FR1, behavioral data was aligned to the day each animal transitioned onto an FR2 schedule (i.e., the last day on FR1). Therefore, data for days 5 - 10 of FR1, but not 1 -4, includes a majority of the animals in the study. In self-administering animals, cocaine (red) acted as a reinforcer as shown by increased active lever (solid line) vs. inactive lever (dotted line) responding on day 3 of FR1 (indicated by *). This did not occur for saline animals (black). Cocaine SA animals began pressing the active lever significantly more than saline (indicated by *) beginning on day 6 of FR1, which continued throughout FR2. Cocaine SA animals (red) received more infusions than their saline counterparts (black) and maintained the same number of infusions after switching to an FR2 schedule, indicating that cocaine was reinforcing lever pressing in these mice. (E) We generated an “addiction index” using exploratory factor analysis to reduce the multi-dimensional behavioral data to “factors” associated with components of cocaine SA behavior. We then combined the 3 factors most strongly associated with an addicted-like phenotype to differentiate between individual animals with high performance across multiple behavioral endpoints. (F) Integration of genes and behaviors to identify transcripts important for the addicted-like phenotype. Enrichment testing reveals transcripts regulated across multiple brain regions. In silico analysis of potential upstream regulators of the enriched genes. Rank-rank hypergeometric overlap used to determine if gene expression Patterns are associated with the addiction index within a brain region. Behavioral data were analyzed using Kruskal Wallis followed by Mann-Whitney Nonparametric Test; *p<0.05; **p<0.01; data are presented as mean ± SEM.
Gene Up- and Downregulation as a Function of History of Cocaine SA and Drug Re-Exposure
Previous work demonstrates that repeated, non-contingent cocaine injections cause gene “priming” or “desensitization” in NAc upon cocaine re-exposure after prolonged WD (22, 23). We therefore used RNA-seq to investigate this phenomenon genome-wide and analyze transcriptomic changes throughout the reward circuitry in response to drug re-exposure after cocaine SA. Baseline transcriptional effects of cocaine SA were established by differential gene expression profiling in each brain region. Figure 1B shows pairwise comparisons of each cocaine treatment group with their saline controls (C24 vs. S24; SC, CS and CC vs. SS) and numbers of differentially expressed genes (DEGs; p<0.05 and fold change>15%) in each brain region (Supplemental Table S1).
To focus on genes that were uniquely altered following context/drug re-exposure after WD, we compared all groups to the same baseline (S24); Figures 1C, 2A; detailed description of pattern identification in Supplemental Methods). Figures 2B–D show heatmaps of DEG patterns within each brain region for all comparisons (C24, SS, SC, CS, and CC vs. S24). This approach revealed two key findings: 1) most DEGs change in the same direction across all re-exposure paradigms (SS – CC); and 2) the magnitude of change for these transcripts was significantly different depending on the animals’ history of cocaine SA and re-exposure (Figure 2B–D).
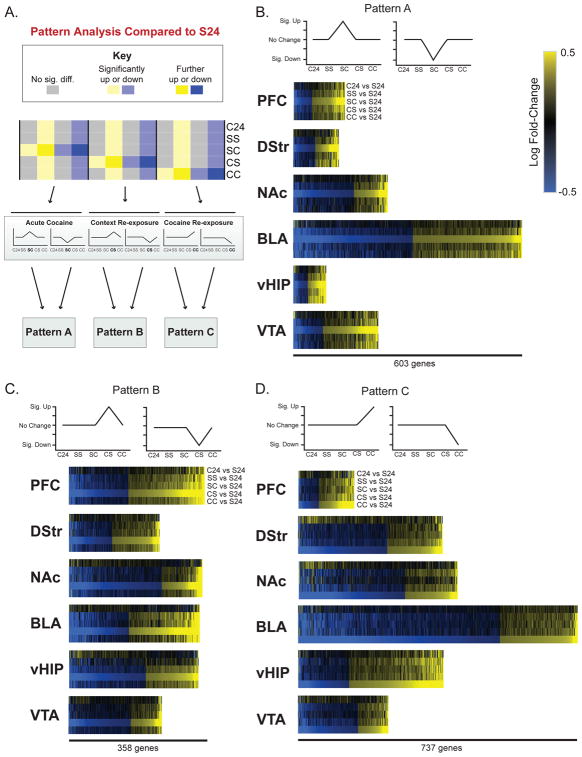
Figure 2. Gene expression Patterns associated with cocaine exposure.
(A) To reduce the dimensions of our RNA-seq data and identify genes that were uniquely changed by a specific exposure paradigm, we used pattern analysis to categorize genes into Patterns of expression when compared to the same S24 baseline. Categorization of genes affected uniquely by: (B) an initial dose of cocaine (Pattern A); (C) re-exposure to the cocaine-paired context after 30 d WD from cocaine SA (Pattern B); (D) re-exposure to cocaine in the cocaine-paired context after 30 d WD from cocaine SA (Pattern C). Heatmaps show that, for all brain regions, expression of genes categorized in each Pattern is, by definition, most pronounced in the comparison that represents that Pattern (e.g., Pattern A most pronounced in SC vs S24 when compared to other groups).
We focused on three patterns associated with drug use: first-ever exposure to cocaine (SC; Pattern A; Figure 2B), re-exposure to cocaine-paired context (CS, Pattern B, Figure 2C), and re-exposure to cocaine-paired context + cocaine (CC, Pattern C, Figure 2D). Each Pattern includes genes that were both differentially expressed from S24 (p<0.05; fold change>15%) and distinct from all other groups. Supplemental Table S2 provides complete gene lists for each pattern. Figure 3A–C shows the number of up- and downregulated DEGs in each Pattern, with a cell type analysis of DEGs shown in Supplemental Table S7.
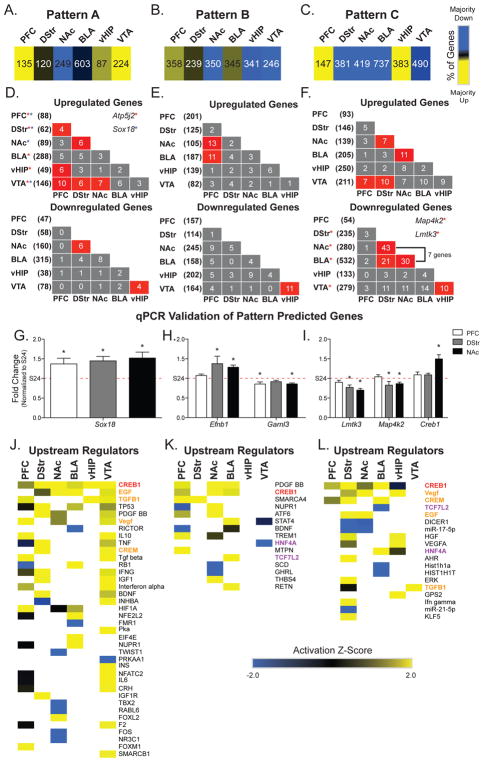
Figure 3. Gene expression patterns associated with cocaine exposure reveal circuit-wide transcriptional changes and upstream regulators.
(A–C) Number and percentage of genes up- and downregulated (yellow=>60% up; blue=>60% down) in each brain region for each of the three Patterns defined in Figure 2. (D–F) Overlap across brain regions of upregulated (top) and downregulated (bottom) genes, color-coded for significance. Total number of regulated genes in each region is shown in parentheses. Examples of transcripts up- or downregulated across more than two brain regions are listed in the insets. (G–I) Patterns were validated using qPCR on technical replicates. Patterns were validated for 8 transcripts across 3 brain regions. Representative transcripts from each pattern are presented. Fold-changes of at least 15% in the RNA-seq data were validated using qPCR across all patterns analyzed, supporting use of this fold-change in all analyses. (J–L) Upstream regulator analysis was conducted across brain regions for each Pattern. Five upstream regulators were consistently predicted to regulate genes across brain regions: CREB1 (highlighted in red) is a predicted upstream regulator of all Patterns. Regulators overlapping between Patterns A and C are highlighted in orange and are likely indicative of those important for regulating the response to acute cocaine exposure independent of a history of cocaine SA. Regulators overlapping between Patterns B and C are highlighted in purple and are likely indicative of those important for regulating the response to a cocaine-paired context after a history of cocaine SA. Activation Z-Scores in heatmaps: positive (yellow) = overrepresentation of targets activated by regulator; negative (blue) = overrepresentation of targets repressed by regulator; no direction (black) = no significant enrichment of activated versus repressed targets; white = not a predicted upstream regulator. *p<0.05; **p<0.01; * * = transcripts overlap across multiple brain regions.
One challenge in devising treatments for addiction is that many genes show different, sometimes opposite, regulation across brain regions. It was therefore of interest to identify specific transcripts that show similar directional changes across brain regions. Fisher’s exact tests (FETs) to compare overlap of DEGs associated with Patterns A–C (Figure 3E–F; Supplemental Table S3) revealed significant overlap of upregulated genes across brain regions in Patterns A–C and identified 2 transcripts that are upregulated across a majority of brain regions in Pattern A (Atp5j2 and Sox18). In Pattern C, overlap of 7 downregulated genes occurred in DStr, NAc and BLA, 2 of which were also downregulated in VTA (Lmtk3 and Map4k2). All genes with fold-change >15% from each Pattern were validated by qPCR in 3 brain regions (Figure 3G–I; Supplemental Figure S1). Therefore, we used fold-change cutoff of 15% for all comparisons.
Predicted Upstream Regulators Have Unique Gene Targets Based on Cocaine SA History and Re-Exposure Across Brain Regions
We hypothesized that these Pattern-associated genes might have common upstream regulators across brain regions, which could serve as potential targets for therapeutic intervention. Exploration of upstream regulators was conducted using Ingenuity Pathway Analysis (IPA; Qiagen Fredrick, MD) for each brain region and each Pattern. Comparison analysis was conducted to identify upstream regulators shared across brain regions (Figure 3J–L). Only those upstream regulators with an activation z-score>2 and p-value<0.01 in at least one brain region were included.
Seven molecules (CREB1, EGF, TGFB1, CREM, VEGF, HNF4A, and TCF7L2) were predicted as upstream regulators in Pattern C and at least 1 other Pattern. Notably, CREB1 was a predicted upstream regulator across all 3 Patterns (highlighted in red, Figure 3J–L). CREB1 was the top upstream regulator in Patterns A and C and a predicted upstream regulator of genes in PFC, NAc, and BLA for Patterns A, B, and C (Figure 3J–L). CREB1 is activated by initial cocaine exposure and is critical for synaptic plasticity involved in cocaine reward (24, 25). Therefore, the prediction that CREB1 is an upstream regulator of genes responding to an acute dose of cocaine in all brain regions (Pattern A) validates our pattern identification methodology (Figure 3J). It should be noted that each gene list is unique for a Pattern within a brain region. Therefore, the finding that CREB1 is a predicted upstream regulator in all 3 Patterns in PFC, NAc, and BLA suggests that a history of cocaine SA with drug/context re-exposure results in different targets for CREB1 in these regions. TGFB1, CREM, EGF, and VEGF were predicted upstream regulators of patterns associated with an acute dose of cocaine, with or without a history of cocaine SA (Patterns A & C; highlighted in orange, Figure 3J & L). Finally, HNF4A, a nuclear receptor, and TCF7L2 were predicted upstream regulators in Patterns associated with cocaine SA + WD (Patterns B & C; highlighted in purple, Figure 3K & L). Molecular pathway analysis also identified biological processes associated with the three Patterns (Supplemental Figure S2).
Association of Gene Expression Regulation with Behavioral Features of Cocaine SA
We next studied whether individual differences in cocaine SA behavior contributed to the regulation of gene expression observed across brain regions and gene expression Patterns. We used exploratory factor analysis to reduce multidimensional behavioral data to factors associated with interrelated variables (Figures 1E, 4A; Supplemental Figure S3). We identified 3 factors that are associated with SA behaviors and reflect important components of addiction: Factor 1 – cocaine intake and infusion; Factor 3 – discrimination between active and inactive levers; and Factor 4 – consummatory regulation (altered intake between FR1 and FR2; Figures 1E and 4A).
To simplify these measures of addiction-related behaviors, we calculated a composite score, or “addiction index” (AI), for each animal (Figure 4B; Supplemental Methods). Individual data are presented for each factor (behavior: Figure 4D, G & J; factor values: Figure 4E, H & K). If an animal scored high on all 3 factors (e.g., ▲ in the cocaine SA group), it has a high AI. However, if an animal scored low on one factor (e.g., × does not discriminate between active and inactive levers and ■ does not increase lever pressing when moved to FR2) their AI is lower. Factor 2 was not included in the AI because it represents differences in total lever pressing (Supplemental Figure S4), a behavior more reflective of locomotor activity and not SA per se. Use of this factor analysis and calculated AI scores illustrates their utility in identifying key components of complex behavioral datasets and in discriminating between baseline individual differences in behavior and those driven specifically by cocaine SA.
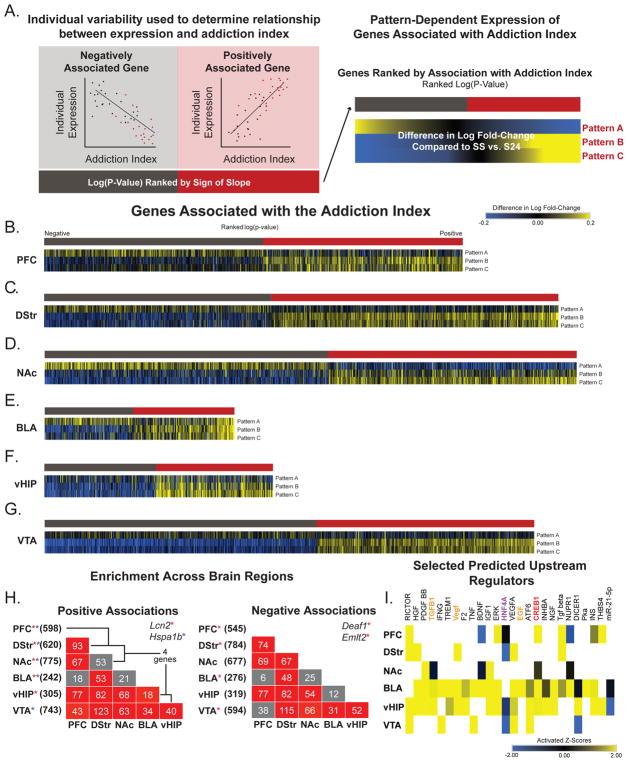
We used linear modeling to identify genes associated with AI scores (Figures 1F and 5A; Supplemental Table S4) to test the hypothesis that individual differences in SA behavior are associated with transcriptional regulation. We noted that the direction of expression changes in genes associated with AI scores were similar across all four 30 d WD groups (Supplemental Figure S5). Because we observed changes in magnitude but not direction in genes categorized as Patterns, we hypothesized a similar effect would be observed in genes associated with AI scores. We calculated magnitude change by subtracting the log fold-change in expression of SS vs. S24 from all other comparisons (SC, CS and CC vs. S24; Figure 5B). This allowed us to adjust for gene expression differences observed between the two saline control groups. For example, if a gene is further downregulated after cocaine re-exposure, it has a negative value (blue). However, if the downregulation is blunted in comparison to that of the SS controls, it has a positive value (yellow). Heatmaps of genes significantly associated with AI scores are displayed (p<0.05, |slope|>0.2) ranked by -log(p-value) and sign of slope (red=positive association; gray=negative association).
Figure 5. Genes associated with the AI are reprogrammed by cocaine SA to be responsive to drug or cocaine-paired context.
(A) Linear modeling was used to identify genes associated with the AI within each brain region. Only genes with a slope of at least ±15% and a nominal p<0.05 were investigated. Similar to the gene expression Patterns (Figure 2), we observed that directional changes in expression were similar across all re-exposure comparisons (SS, SC, CS & CC vs. S24). Genes that were negatively associated with AI (gray bar) were downregulated and genes positively associated with AI (red bar) were upregulated (Supplemental Table S4). (B–G) Heatmaps were transformed to indicate change in expression from SS controls. Blue = fold change in the negative direction from SS vs. S24 and yellow = fold change in the positive direction from SS vs. S24. Cocaine SA programs those transcripts associated with AI to be hyper-response to context either with or without drug. (H) Overlap of genes positively (left) or negatively (right) associated with AI across brain regions, color-coded for significance. Total number of genes in each brain region listed in parentheses and total number of genes overlapping between regions indicated in corresponding boxes. There is significant overlap of genes associated with the AI across most brain regions. (I) Upstream regulator analysis reveals similar putative transcriptional regulators in genes associated with AI as those associated with specific gene expression Patterns. Colors correspond to regulators overlapping in multiple Patterns (see Figure 3). Activation Z-Scores: positive (yellow) = overrepresentation of targets activated by regulator; negative (blue) = overrepresentation of targets repressed by regulator; no direction (black) = no significant enrichment of activated or repressed targets; white = not a predicted upstream regulator.
The heatmaps reveal that a history of cocaine SA (Patterns B & C) augments the transcriptional response observed in the SS groups of those genes positively and negatively associated with AI in all 6 brain regions (Figure 5C–H). The same is not true after an animal’s first dose of cocaine. Notably, in NAc, the transcriptional response of genes associated with AI is attenuated when compared to the SS group (Figure 5E). These data suggest that one dose of cocaine has little impact on genes associated with addiction-related behaviors.
We next used FETs to identify specific transcripts positively or negatively associated with AI across brain regions (Figure 5H). More transcripts overlapped across brain regions in our pair-wise comparisons than in the Patterns. Notably, genes encoding AP-1 transcription factors, including Fos, Fosb, and Fosl2 were associated with AI in the BLA, vHIP, and NAc. This is consistent with prior work implicating AP-1 as an important transcriptional mediator of drug action (25). Genes associated with AI were enriched for neuronal-specific transcripts in all regions (Supplemental Table S7). Six transcripts (Hspb1, Dnajc3, Mpdz, Tmem252, Lcn2, and Hspa1b) were positively associated across 5 brain regions. Notably, Lipocalin 2 (Lcn2) was associated with AI all regions except the VTA, where there was a trend (slope=1.84; p-value=0.07), suggesting that Lcn2 may be a potential novel therapeutic target for addiction.
Upstream regulator analysis identified 192 molecules predicted to regulate genes associated with AI (Figure 5J; Supplemental Table S5). RICTOR was the top-predicted regulator in PFC, DStr, vHIP, and VTA, and CREB1 (highlighted in red) was a predicted upstream regulator of genes in PFC, NAc, BLA, and vHIP. Finally, HNF4A was a predicted upstream regulator in 4 out of 6 brain regions. Notably, CREB1 and HNF4A were both predicted in cocaine SA + WD Patterns (Patterns B and C).
Transcriptome-Wide Expression Profiles Dependent on a History of Cocaine SA and Re-Exposure Reflect Region-Specific Roles in Addiction-Related Behaviors
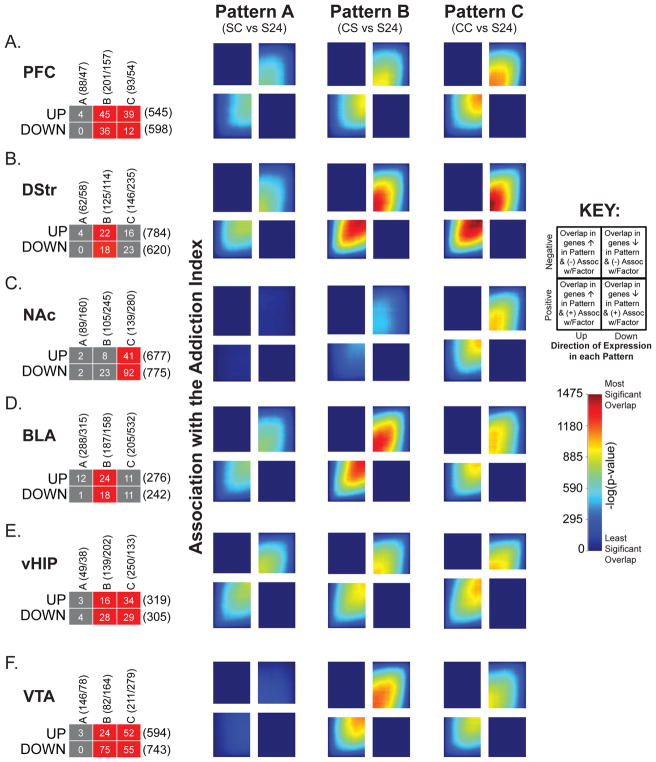
To determine if genes associated with AI overlap with genes changed in the condition defining each Pattern of gene expression, we used rank rank hypergeometric overlap (RRHO) analysis, which compares large datasets in a threshold-free manner (16, 21, 26, 27) (Figure 1F & 6). In each brain region, there was significant overlap of genes up- and downregulated in Patterns B and C —Patterns related to cocaine SA—and genes positively and negatively associated with AI, respectively. This finding is supported by FETs on filtered lists (left) showing significant overlap of up- and downregulated genes in Patterns B in all brain regions except NAc. In contrast, overlap between Pattern A—associated with initial, acute cocaine exposure—and AI was absent or far weaker. This is similar to SS vs. S24 comparisons (Supplemental Figure S7) in all brain regions except vHIP, where AI overlaps strongly with Pattern A (Figure 6E). Additionally, each region showed some Pattern-specific associations with the AI (Figure 6). Notably, NAc displayed strong associations with Pattern C (Figure 6C) only and BLA showed the strongest associations with Pattern B (Figure 6D).
Figure 6. Overlap of transcriptional profiles related to the AI and gene expression Patterns reveals which Pattern contributes most to AI.
A–F) Overlap of genes positively or negatively associated with AI and also up- or downregulated within each gene expression Pattern within the gene lists filtered for significance (Fisher’s exact test; left) or transcriptome-wide expression profiles (RRHO plots; right). Overlap of genes associated with AI are specific to brain regions. For example, significant overlap of up- and downregulated genes across Patterns B & C with AI are observed in PFC and VTA. vHIP. BLA and DStr are enriched in genes in Pattern B and NAc only shows enrichment of genes in Pattern C. RRHO plots to the right of each panel reveal significance of overlap between region-specific transcriptional profiles associated with AI for Patterns A–C. A key for these plots is shown to the right.
Motif Analysis Reveals Nuclear Receptors as Important Regulators of Transcription After a History of Cocaine SA
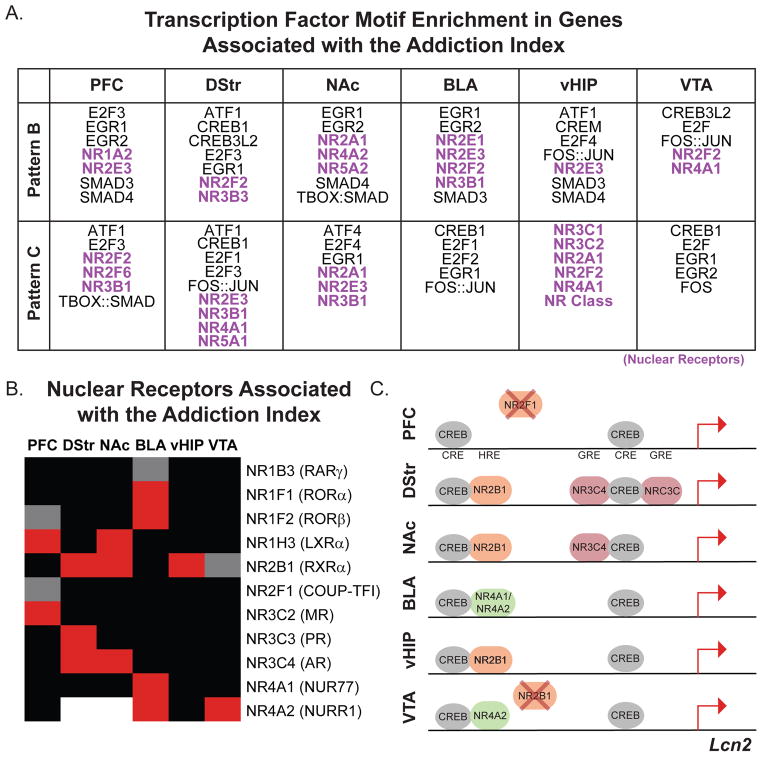
We conducted HOMER motif analysis on genes associated with AI and categorized as either Pattern B or C for each brain region (Figures 1F and 7A; Supplemental Table S6). We found enrichment of several putative transcription factor binding sites implicated previously in reward-associated behaviors (SMAD, E2F, CREB, EGR, and AP1 families) across multiple brain regions (7, 8, 28–33). Interestingly, the nuclear receptor (NR) family was predicted in every brain region. HNF4A (NR2A1) was a predicted regulator in Patterns associated with a history of cocaine SA (Figure 3J–L; Patterns B and C) and genes associated with AI (Figure 5I). NRs have recently been identified as critical for CREB-regulated learning and memory in hippocampus (34) and important for aspects of cocaine SA in NAc (35). This, in combination with the prediction of CREB as an upstream regulator across all 3 Patterns and AI, raised the hypothesis that NRs may influence CREB transcriptional regulation in a context-dependent manner throughout the brain.
Figure 7. Motif analysis reveals putative role for NRs in controlling region-specific cocaine-induced gene expression.
(A) HOMER motif analysis was conducted on genes defined as either Pattern B or C and significantly associated with the AI (lists from Figure 6 enrichment tests). Table of putative transcription factor families whose motifs were enriched in at least 4 of 6 brain regions. Members of the NR family were predicted upstream regulators in all brain regions and were Pattern-specific. (B) NR family members are positively (red) and negatively (gray) associated with the AI in a region-specific manner. Black indicates no association and white indicates no detectable expression. Only NRs with a significant association in at least one brain region are displayed. (C) Hypothetical model of transcriptional co-regulation by CREB and NRs in a gene positively associated with AI across all brain regions (VTA = trend). In silico analysis of transcription factor binding sites, identified using MatInspector, indicate motifs in close proximity to each other (less than 50 bp), and binding data from the MatInspector database indicate binding of specific NRs within the Lcn2 promoter. Based on our AI data, we extrapolated possible region-specific binding states that could be regulating the transcriptional response to drug or context re-exposure. Color indicates subfamily of NRs: orange = NR2 subfamily; pink = NR3 subfamily; green = NR4 subfamily. X = negative association with AI.
Because NR family members are associated with AI across all brain regions and show region-specific alterations in expression (Figure 7B), we considered the possibility that the region-specific association of NRs with AI, coupled with known regulation of CREB activity and binding, could influence the magnitude of expression of addiction-related genes after a history of cocaine SA. We used in silico analysis to test the hypothesis that CREB and NRs could potentially interact to influence expression in a context-specific manner. We identified proximally located CREB and NR binding sites (MatInspector, Genomatix, Germany) in a representative gene, Lcn2, that was positively associated with AI across multiple brain regions (Figure 7C;). Hypothetical transcription factor binding states in each brain region are presented based on region-specific NR expression, association with AI, and known binding data from the MatInspector database (Figure 7C & D). This illustrates the concept that different NRs could influence CREB-induced transcriptional regulation in a region-specific manner. For example, there are two regions in the promoter of Lcn2 where CREB and NR binding motifs occur within 50 bp of each other. In NAc and VTA, different NRs are expressed and/or associated with AI. Thus, two putative binding states are represented: 1) In NAc, NR2B1 binds near CREB in the more distal binding zone, while both NR3C4 and NRC3C bind near CREB in the more proximal binding zone; 2) in VTA, because NR2B1 is negatively associated with AI, it is not available to bind, while NR4A2 is positively associated and available (Figure 7C). This Figure serves to illustrate just one hypothetical mechanism by which the same upstream regulator (e.g., CREB) can have different downstream effects across brain regions and behavioral histories. Furthermore, this analysis serves as an example of how our extensive datasets can be used moving forward.
DISCUSSION
These data provide the first unbiased assessment of gene regulation across various time-points of cocaine SA—short- and long-term WD— and two different re-exposure paradigms in six interconnected brain reward regions. While prior studies have investigated transcriptional responses to cocaine re-exposure after SA (6–12), these have not done so transcriptome-wide across a range of brain regions. Furthermore, this study is particularly powerful as we used individual variability to identify transcripts associated with aspects of cocaine SA behavior. We leveraged two statistical approaches (pattern identification and factor analysis) to characterize novel gene expression patterns throughout the reward circuitry that are sensitive to drug re-exposure after prolonged WD from cocaine SA.
Traditional methods of analyzing RNA-seq data have focused on pair-wise comparisons to identify DEGs when compared to a single control group. Our dataset contained two control groups, so pair-wise comparison using each condition’s control (S24 and SS) could not uncover all transcriptional differences. Therefore, we utilized a novel approach to identify patterns of expression that reflect differences from both baselines and identified transcripts that were uniquely altered by either context re-exposure alone or context + drug re-exposure. This revealed that many genes associated with long-term WD and re-exposure were altered in magnitude but not direction. Pattern identification therefore allowed us to detect genes that were uniquely altered by acute cocaine (Pattern A), cocaine-paired context (Pattern B), or context + cocaine re-exposure (Pattern C) independent of baseline changes. Furthermore, each gene was only characterized as one pattern per brain region, thus revealing those genes associated uniquely with context- and/or drug-induced relapse.
This pattern identification analysis revealed individual transcripts that are regulated across multiple brain regions and may serve as therapeutic targets for addiction. For example, in Pattern C, two protein kinases (Lmtk3 and Map4k2) are downregulated in DStr, NAc, BLA, and VTA. Knockout of Lmtk3 increases locomotor activity and dopamine turnover in striatum (36). Both are involved in actin cytoskeletal remodeling (37, 38) and Map4k2 has been linked to inflammatory responses (39), two key processes in synaptic plasticity (6, 40). Similarly, transcripts were identified that were associated with AI across multiple brain regions (Figure 5H). Notably, Lcn2 was positively associated with AI across all 6 brain regions (VTA = trend). LCN2 forms a complex with matrix metalloproteinase 9 (MMP9) and protects it from degradation, thus prolonging its activity (41). MMP9 activity has been shown to be critical for cue- and cocaine-induced reinstatement (42). These transcripts provide valuable information regarding biological processes important for cocaine addiction, and serve as potential brain-wide therapeutic targets.
One key finding of the pattern analysis came from upstream regulator analysis, which showed that many predicted transcriptional regulators were consistent across Patterns and brain regions (Figure 3J–L). This is significant because each gene list is unique for a Pattern within a brain region, suggesting that the targets of these predicted regulators change depending on cocaine history and re-exposure paradigm. This provides a potential mechanism for our hypothesis that a history of cocaine SA “primes” the reward circuitry at the transcriptional level to respond to context/drug re-exposure.
We identified CREB1 as a predicted upstream regulator in Patterns A, B, and C in PFC, BLA, and NAc – brain regions implicated in cue-induced reinstatement (43–45). CREB1 has long been implicated in addiction-related phenomena (24, 25, 46) and is critical for synaptic plasticity and reward learning. Prediction of CREB1 as a regulator of expression in all brain regions upon initial exposure to cocaine validates our pattern identification methodology.
Individual differences in SA behavioral responses correlate with gene expression changes following WD. To date, those correlations have been restricted to drug-taking animals without including saline controls, and none have been performed transcriptome-wide (47, 48). Two limitations of previous analyses are: 1) false positives/negatives due to constraints in statistical analysis of small sample sizes typical of RNA-seq experiments, and 2) the inability to use all available SA behavioral data in correlation analysis (e.g., saline animals cannot be correlated with intake). To understand how individual differences in cocaine SA behavior might influence the transcriptional landscape after long-term WD and re-exposure, we used factor analysis to generate a composite AI that incorporates variability in SA behaviors associated with addiction-like outcomes and discriminates between saline and cocaine animals (Figure 4). This allowed us to use the saline controls in our linear model to account for baseline differences in behavior and substantially increased our sample size, reducing the likelihood of false discovery.
The greater transcriptional response in Patterns B and C drive association with the AI in a region-specific manner (Figure 5B–G). This is further reflected in the RRHO analyses (Figure 6). Thus, context is exceptionally important for the transcriptional component of relapse, and the response appears to be region-specific. RRHOs highlight which Pattern of gene expression contributes to AI in each brain region, thus showing which Pattern most reflects addiction-related behaviors. Together, our data suggest that transcriptional reprogramming occurs during long-term WD and is associated with the degree of the addictive phenotype.
The high degree of overlap of transcripts associated with AI across brain regions (Figure 5I) suggests once again that there is a suite of transcripts throughout the reward circuitry being targeted by similar upstream regulators. As in the Patterns, CREB1 was a predicted upstream regulator in PFC, NAc, BLA, and vHIP of genes associated with AI (Figure 5J). HNF4A was also a predicted upstream regulator of genes associated with AI and was one of two upstream regulators (TCF7L2) predicted for both Patterns B and C. HNF4A is implicated in epigenetic mechanisms (49–52) and dendritic spine morphology (51). While expression of Hnf4a was not detected in our sequencing data, other NRs were. Additionally, many NRs share a consensus sequence and compete for DNA binding (53).
Based on this knowledge, we used HOMER de novo motif analysis to identify putative transcription factor binding sites across genes in Patterns B or C that were also associated with AI. Strikingly, NRs were present in every brain region in a similar Pattern-specific manner as seen by RRHO. Furthermore, CREB1 and other CREB family members were predicted in all brain regions. Thus, we posit that NRs might influence transcriptional regulation by CREB proteins in response to drug/context re-exposure in a region-specific manner.
Using novel analytic approaches followed by upstream-regulator, motif and other in silico analyses, we present here candidate genes and transcriptional regulators that might serve as therapeutics for addiction-related disorders. While CREB and NRs are highlighted for follow-up, this serves as just one example for how this vast dataset can be mined in future studies. To conclude, our datasets provide a highly unique resource of transcriptional regulation throughout the brain’s reward circuitry and across cocaine SA, WD, and re-exposure. The transcriptional reprogramming that occurs offers valuable information regarding gene expression correlating with high performance on a highly ethologically relevant model of addiction. Thus, this work provides an increasingly complete understanding of the molecular basis of cocaine addiction and allows us to work toward individualized therapeutics.
Supplementary Material
Figure S1: qPCR validation of Patterns in three brain regions reveals that fold changes of at least 15% are replicable. (A) List of 8 genes categorized as Patterns A, B, or C were validated using qPCR on technical replicates of the samples used in the RNA-seq experiment. Only those genes with a fold change of at least 15% were validated. (B–D) Expression of representative transcripts measured by RNA-seq and qPCR. Changes in expression of at least 15% in the RNA-seq data were validated by qPCR. This is exemplified by those changes in Zfp763 (categorized as Pattern A but with <15% change in expression); Sox18 and Creb1 (Categorized as Pattern A or C, respectively with >15% change in expression). Gray shaded area on graphs indicates 15% change from S24. * = p<0.05
Figure S2: Similar pathways are associated with Patterns of gene expression across brain regions. (A) Pattern A was associated with protein kinase A signaling, while (B) Pattern B was dominated by NFκB and PPAR, a nuclear receptor, signaling. (C) Pathways associated with Pattern C included synaptic long-term depression and NFκB signaling. Pathways associated with both Patterns B and C are highlighted in purple. Only those pathways that met the following criteria were included: at least 1 brain region with an activation z-score>2 and p-value<0.01. Activation z-Scores: positive (yellow) = overrepresentation of targets activated in pathway; negative (blue) = overrepresentation of targets repressed in pathway; no direction (black) = no significant enrichment of activated or repressed targets; white = not a predicted pathway.
Figure S3: Factor loading for behavioral endpoints used in factor analysis. (A) Behavioral data represented in the factor analysis. All lever pressing data (food training, FR1, FR2; active vs. inactive) were included as variables in the factor analysis. Here we present a subset of the data aligned to the first day of each phase of self-administration. Because all animals had differing numbers of days in each phase, only those days in which the majority (>70%) of the animals in the study are presented. An image of the complete data set is presented in Figure 1D. (B) Factor analysis was used to reduce multidimensional behavioral endpoints to factors. The association of each factor with each behavioral endpoint included in the analysis is displayed. Factors were positively (yellow), negatively (blue), or not associated (black) with each endpoint. These particular associations allowed for the interpretation of the how each factor related to various SA behaviors.
Figure S4: Factor 2 discriminates between baseline differences in saline animals. (A) Factor 2, in the factor analysis, was positively associated with both active and inactive lever pressing and negatively associated with intake. (B) Individual data for total number of lever presses in saline (left) and cocaine (right) for the entire SA experiment, including food training. (C) Individual factor values for Factor 2 for saline (left) or cocaine (right) animals. Animals with the greatest number of lever presses, but no intake, had highest factor value (
 in saline group). Animals with increased lever pressing coupled with high intake (
in saline group). Animals with increased lever pressing coupled with high intake (
 in cocaine group) had lower factor values. Finally, those animals with few lever presses and no intake (
in cocaine group) had lower factor values. Finally, those animals with few lever presses and no intake (
 in saline group) had the lowest factor values. (D) Linear modeling was used to identify genes associated with Factor 2 within each brain region. Only genes with a |slope|>0.2 and a nominal p-value of <0.05 were investigated. (D) Genes were ranked by -log p-value signed by the slope of the association with Factor 2. Negative associations with Factor 2 are presented in gray and genes positively associated with Factor 2 are presented in red. (D) Heatmaps presented are transformed to indicate change in expression from SS controls. Blue = fold change in the negative direction from SS vs S24 and yellow = fold change in the positive direction from SS vs S24. These data indicate that changes in expression in transcripts associated with Factor 2 are most robust in the SS vs S24. This highlights the power of factor analysis to extract important information related to baseline behaviors and indicates that those differences are reflected in our transcriptomic data as well. (E) Overlap of genes positively (left) or negatively (right) associated with Factor 2 across brain regions, color-coded for significance. Total number of genes in each brain region listed in parentheses and total number of genes overlapping between regions indicated in corresponding boxes. There is a high degree of overlap of transcripts associated with Factor 2 in all brain regions.
in saline group) had the lowest factor values. (D) Linear modeling was used to identify genes associated with Factor 2 within each brain region. Only genes with a |slope|>0.2 and a nominal p-value of <0.05 were investigated. (D) Genes were ranked by -log p-value signed by the slope of the association with Factor 2. Negative associations with Factor 2 are presented in gray and genes positively associated with Factor 2 are presented in red. (D) Heatmaps presented are transformed to indicate change in expression from SS controls. Blue = fold change in the negative direction from SS vs S24 and yellow = fold change in the positive direction from SS vs S24. These data indicate that changes in expression in transcripts associated with Factor 2 are most robust in the SS vs S24. This highlights the power of factor analysis to extract important information related to baseline behaviors and indicates that those differences are reflected in our transcriptomic data as well. (E) Overlap of genes positively (left) or negatively (right) associated with Factor 2 across brain regions, color-coded for significance. Total number of genes in each brain region listed in parentheses and total number of genes overlapping between regions indicated in corresponding boxes. There is a high degree of overlap of transcripts associated with Factor 2 in all brain regions.
Figure S5: Raw heatmap of addiction index associated genes. (A–F) Raw expression of genes associated with AI in all brain regions for all groups when compared to the same baseline (S24). Log fold-change in expression of genes associated with AI and ranked by the sign of the association and -log(p-value) (gray = negative associations; red = positive associations). In all groups but C24, genes that were negatively associated with AI (gray bar) were downregulated and genes positively associated with AI (red bar) were upregulated. In all brain regions, the strongest response was in comparisons representing either Pattern B or C, suggesting that that transcriptional response to re-exposure to context/cocaine is influenced by addiction-related behaviors during cocaine SA.
Figure S6: Pathways associated with the addiction index (AI). Ingenuity Pathway Analysis revealed genes associated with the AI, which were enriched for cAMP-mediated signaling, PPARα/RXRα activation, and PI3K/AKT signaling among others. Activation z-Scores: positive (yellow) = overrepresentation of targets activated by regulator; negative (blue) = overrepresentation of targets repressed by regulator; no direction (black) = no significant enrichment of activated or repressed targets; white = not a predicted upstream regulator. Behavioral data analyzed using Kruskal-Wallis followed by Mann-Whitney Nonparametric Test; *p<0.05; **p<0.001; data presented as mean ± SEM.
Figure S7: Overlap of transcriptional profiles related to the AI and saline controls. (A–F) RRHO plots reveal little overlap of genes positively or negatively associated with AI and up- or downregulated in saline control animals (SS vs S24). As predicted, little to no overlap of expression profiles was observed in NAc, vHIP and VTA. Overlap of expression was weak in PFC, DStr and BLA and similar to that observed in the comparisons with Pattern A. A key for these plots is provided.
Figure S8: Full list of NR family members associated with the AI. Heatmap of association of all known nuclear receptors with the AI. Members of NR1 – 4 subfamilies are expressed throughout the reward circuitry. Strongest associations are found within NR2B and NR4A subfamilies. Yellow = positive association; Blue = negative association; Black = no significant association; white = expression not detected in our dataset.
A complete list of DEGs (nominal p-value < 0.05; fold-change ± 15%) in relation to saline controls (either S24 or SS) presented in Figure 1B. Each comparison is presented on a separate tab.
A complete list of genes categorized as Pattern A, B, or C in each brain region. Log fold-change for all conditions when compared to the same baseline (S24) are included. Genes were categorized by their expression patterns and a fold-change cut off of ± 15% was applied to each list to identify genes uniquely altered under each re-exposure condition. Each Pattern is presented on a separate tab.
A complete list of genes categorized as Patterns A, B, or C that overlap across multiple brain regions. Fisher’s exact tests revealed significant enrichment across lists. Comparisons reaching significance after multiple comparison correction (FDR) are bolded. Each pattern and direction of regulation is presented on a separate tab of the table.
A complete list of genes associated with AI that overlap across multiple brain region. Fisher’s exact tests revealed significant enrichment across lists. Comparisons reaching significance after multiple comparison correction (FDR) are bolded. Positive and negative associations are presented on separate tabs.
A full list of predicted regulators of genes associated with AI and their activation z-scores. Activation z-Scores: positive = overrepresentation of targets activated by regulator; negative = overrepresentation of targets repressed by regulator; no direction = no significant enrichment of activated or repressed targets; white = not a predicted upstream regulator.
A complete list of genes categorized as Pattern A, B, or C that overlap with those associated with AI within each brain region. Fisher’s exact tests revealed significant enrichment across lists. Comparisons reaching significance after multiple comparison correction (FDR) are bolded. Comparison of Pattern/AI for each brain region are presented on a separate tab.
Fisher’s exact tests revealed significant enrichment of cell-type specific genes in those lists of genes categorized as Pattern A, B, or C or genes associated with AI within each brain region. Only comparisons reaching significance after multiple comparison correction (FDR) are presented.
A complete list of the associations and p-values for each gene and Factor across all brain regions is presented. Each brain region is provided on a separate tab.
Acknowledgments
This work was funded by grants from the National Institute on Drug Abuse (NIDA) P01DA008227 (EJN), R01DA007359 (EJN), K99DA04211 (ESC), K01MH103473 (MLS), K01DA038654 (RWL). We would like to thank Drs. David Self and Erin Larson for their thoughtful comments regarding the design and execution of mouse self-administration experiments.
Footnotes
FINANCIAL DISCLOSURES
The authors reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 2.Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 2016;3:760–773. doi: 10.1016/S2215-0366(16)00104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koob GF, Caine SB, Parsons L, Markou A, Weiss F. Opponent process model and psychostimulant addiction. Pharmacol Biochem Behav. 1997;57:513–521. doi: 10.1016/s0091-3057(96)00438-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang JQ, McGinty JF. Glutamate-dopamine interactions mediate the effects of psychostimulant drugs. Addict Biol. 1999;4:141–150. doi: 10.1080/13556219971641. [DOI] [PubMed] [Google Scholar]
- 5.White FJ, Kalivas PW. Neuroadaptations involved in amphetamine and cocaine addiction. Drug Alcohol Depend. 1998;51:141–153. doi: 10.1016/s0376-8716(98)00072-6. [DOI] [PubMed] [Google Scholar]
- 6.Cahill ME, Bagot RC, Gancarz AM, Walker DM, Sun H, Wang ZJ, et al. Bidirectional Synaptic Structural Plasticity after Chronic Cocaine Administration Occurs through Rap1 Small GTPase Signaling. Neuron. 2016;89:566–582. doi: 10.1016/j.neuron.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra R, Francis TC, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM, et al. Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J Neurosci. 2015;35:7927–7937. doi: 10.1523/JNEUROSCI.0548-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gancarz-Kausch AM, Schroeder GL, Panganiban C, Adank D, Humby MS, Kausch MA, et al. Transforming growth factor beta receptor 1 is increased following abstinence from cocaine self-administration, but not cocaine sensitization. PLoS One. 2013;8:e83834. doi: 10.1371/journal.pone.0083834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, Self DW. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci. 2007;10:1029–1037. doi: 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- 10.Freeman WM, Patel KM, Brucklacher RM, Lull ME, Erwin M, Morgan D, et al. Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharmacology. 2008;33:1807–1817. doi: 10.1038/sj.npp.1301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu L, Grimm JW, Shaham Y, Hope BT. Molecular neuroadaptations in the accumbens and ventral tegmental area during the first 90 days of forced abstinence from cocaine self-administration in rats. J Neurochem. 2003;85:1604–1613. doi: 10.1046/j.1471-4159.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- 12.Larson EB, Akkentli F, Edwards S, Graham DL, Simmons DL, Alibhai IN, et al. Striatal regulation of DeltaFosB, FosB, and cFos during cocaine self-administration and withdrawal. J Neurochem. 2010;115:112–122. doi: 10.1111/j.1471-4159.2010.06907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadakierska-Chudy A, Frankowska M, Jastrzebska J, Wydra K, Miszkiel J, Sanak M, et al. Cocaine Administration and Its Withdrawal Enhance the Expression of Genes Encoding Histone-Modifying Enzymes and Histone Acetylation in the Rat Prefrontal Cortex. Neurotox Res. 2017;32:141–150. doi: 10.1007/s12640-017-9728-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallender EJ, Goswami DB, Shinday NM, Westmoreland SV, Yao WD, Rowlett JK. Transcriptomic profiling of the ventral tegmental area and nucleus accumbens in rhesus macaques following long-term cocaine self-administration. Drug Alcohol Depend. 2017;175:9–23. doi: 10.1016/j.drugalcdep.2017.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman WM, Lull ME, Patel KM, Brucklacher RM, Morgan D, Roberts DC, et al. Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neurosci. 2010;11:29. doi: 10.1186/1471-2202-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagot RC, Cates HM, Purushothaman I, Lorsch ZS, Walker DM, Wang J, et al. Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron. 2016;90:969–983. doi: 10.1016/j.neuron.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagot RC, Cates HM, Purushothaman I, Vialou V, Heller EA, Yieh L, et al. Ketamine and Imipramine Reverse Transcriptional Signatures of Susceptibility and Induce Resilience-Specific Gene Expression Profiles. Biol Psychiatry. 2017;81:285–295. doi: 10.1016/j.biopsych.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labonte B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pena CJ, Kronman HG, Walker DM, Cates HM, Bagot RC, Purushothaman I, et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356:1185–1188. doi: 10.1126/science.aan4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seney ML, Huo Z, Cahill K, French L, Puralewski R, Zhang J, et al. Opposite molecular signatures of depression in men and women. Biological Psychiatry. doi: 10.1016/j.biopsych.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damez-Werno D, LaPlant Q, Sun H, Scobie KN, Dietz DM, Walker IM, et al. Drug experience epigenetically primes Fosb gene inducibility in rat nucleus accumbens. J Neurosci. 2012;32:10267–10272. doi: 10.1523/JNEUROSCI.1290-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nestler EJ. Reflections on: “A general role for adaptations in G-Proteins and the cyclic AMP system in mediating the chronic actions of morphine and cocaine on neuronal function”. Brain Res. 2016;1645:71–74. doi: 10.1016/j.brainres.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plaisier SBTR, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein JL, de la Torre-Ubieta L, Tian Y, Parikshak NN, Hernandez IA, Marchetto MC, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cates HM, Heller EA, Lardner CK, Purushothaman I, Pena CJ, Walker DM, et al. Transcription Factor E2F3a in Nucleus Accumbens Affects Cocaine Action via Transcription and Alternative Splicing. Biol Psychiatry. 2017 doi: 10.1016/j.biopsych.2017.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cates HM, Thibault M, Pfau M, Heller E, Eagle A, Gajewski P, et al. Threonine 149 phosphorylation enhances DeltaFosB transcriptional activity to control psychomotor responses to cocaine. J Neurosci. 2014;34:11461–11469. doi: 10.1523/JNEUROSCI.1611-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 32.Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vialou V, Maze I, Renthal W, LaPlant QC, Watts EL, Mouzon E, et al. Serum response factor promotes resilience to chronic social stress through the induction of DeltaFosB. J Neurosci. 2010;30:14585–14592. doi: 10.1523/JNEUROSCI.2496-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bridi MS, Hawk JD, Chatterjee S, Safe S, Abel T. Pharmacological Activators of the NR4A Nuclear Receptors Enhance LTP in a CREB/CBP-Dependent Manner. Neuropsychopharmacology. 2017;42:1243–1253. doi: 10.1038/npp.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Kong F, Crofton EJ, Dragosljvich SN, Sinha M, Li D, et al. Transcriptomics of Environmental Enrichment Reveals a Role for Retinoic Acid Signaling in Addiction. Front Mol Neurosci. 2016;9:119. doi: 10.3389/fnmol.2016.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue T, Hoshina N, Nakazawa T, Kiyama Y, Kobayashi S, Abe T, et al. LMTK3 deficiency causes pronounced locomotor hyperactivity and impairs endocytic trafficking. J Neurosci. 2014;34:5927–5937. doi: 10.1523/JNEUROSCI.1621-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyriakis JM. Signaling by the germinal center kinase family of protein kinases. J Biol Chem. 1999;274:5259–5262. doi: 10.1074/jbc.274.9.5259. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Zhang H, Lit LC, Grothey A, Athanasiadou M, Kiritsi M, et al. The kinase LMTK3 promotes invasion in breast cancer through GRB2-mediated induction of integrin beta(1) Sci Signal. 2014;7:ra58. doi: 10.1126/scisignal.2005170. [DOI] [PubMed] [Google Scholar]
- 39.Chuang HC, Wang X, Tan TH. MAP4K Family Kinases in Immunity and Inflammation. Adv Immunol. 2016;129:277–314. doi: 10.1016/bs.ai.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Yirmiya R, Goshen I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain Behav Immun. 2011;25:181–213. doi: 10.1016/j.bbi.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Yan L, Borregaard N, Kjeldsen L, Moses MA. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J Biol Chem. 2001;276:37258–37265. doi: 10.1074/jbc.M106089200. [DOI] [PubMed] [Google Scholar]
- 42.Smith AC, Kupchik YM, Scofield MD, Gipson CD, Wiggins A, Thomas CA, et al. Synaptic plasticity mediating cocaine relapse requires matrix metalloproteinases. Nat Neurosci. 2014;17:1655–1657. doi: 10.1038/nn.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalivas PW. The glutamate homeostasis hypothesis of addiction. Nat Rev Neurosci. 2009;10:561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- 44.Pickens CL, Airavaara M, Theberge F, Fanous S, Hope BT, Shaham Y. Neurobiology of the incubation of drug craving. Trends Neurosci. 2011;34:411–420. doi: 10.1016/j.tins.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.See RE. Neural substrates of cocaine-cue associations that trigger relapse. Eur J Pharmacol. 2005;526:140–146. doi: 10.1016/j.ejphar.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 46.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Tacelosky DM, Alexander DN, Morse M, Hajnal A, Berg A, Levenson R, et al. Low expression of D2R and Wntless correlates with high motivation for heroin. Behav Neurosci. 2015;129:744–755. doi: 10.1037/bne0000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Leri F, Cummins E, Kreek MJ. Individual differences in gene expression of vasopressin, D2 receptor, POMC and orexin: vulnerability to relapse to heroin-seeking in rats. Physiol Behav. 2015;139:127–135. doi: 10.1016/j.physbeh.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cicchini C, de Nonno V, Battistelli C, Cozzolino AM, De Santis Puzzonia M, Ciafre SA, et al. Epigenetic control of EMT/MET dynamics: HNF4alpha impacts DNMT3s through miRs-29. Biochim Biophys Acta. 2015;1849:919–929. doi: 10.1016/j.bbagrm.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Santiago R, Carballo-Carbajal I, Castellano G, Torrent R, Richaud Y, Sanchez-Danes A, et al. Aberrant epigenome in iPSC-derived dopaminergic neurons from Parkinson’s disease patients. EMBO Mol Med. 2015;7:1529–1546. doi: 10.15252/emmm.201505439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D, Tang H, Li XY, Deng MF, Wei N, Wang X, et al. Targeting the HDAC2/HNF-4A/miR-101b/AMPK Pathway Rescues Tauopathy and Dendritic Abnormalities in Alzheimer’s Disease. Mol Ther. 2017;25:752–764. doi: 10.1016/j.ymthe.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanishi K, Doe N, Sumida M, Watanabe Y, Yoshida M, Yamamoto H, et al. Hepatocyte nuclear factor 4 alpha is a key factor related to depression and physiological homeostasis in the mouse brain. PLoS One. 2015;10:e0119021. doi: 10.1371/journal.pone.0119021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olivares AM, Moreno-Ramos OA, Haider NB. Role of Nuclear Receptors in Central Nervous System Development and Associated Diseases. J Exp Neurosci. 2015;9:93–121. doi: 10.4137/JEN.S25480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: qPCR validation of Patterns in three brain regions reveals that fold changes of at least 15% are replicable. (A) List of 8 genes categorized as Patterns A, B, or C were validated using qPCR on technical replicates of the samples used in the RNA-seq experiment. Only those genes with a fold change of at least 15% were validated. (B–D) Expression of representative transcripts measured by RNA-seq and qPCR. Changes in expression of at least 15% in the RNA-seq data were validated by qPCR. This is exemplified by those changes in Zfp763 (categorized as Pattern A but with <15% change in expression); Sox18 and Creb1 (Categorized as Pattern A or C, respectively with >15% change in expression). Gray shaded area on graphs indicates 15% change from S24. * = p<0.05
Figure S2: Similar pathways are associated with Patterns of gene expression across brain regions. (A) Pattern A was associated with protein kinase A signaling, while (B) Pattern B was dominated by NFκB and PPAR, a nuclear receptor, signaling. (C) Pathways associated with Pattern C included synaptic long-term depression and NFκB signaling. Pathways associated with both Patterns B and C are highlighted in purple. Only those pathways that met the following criteria were included: at least 1 brain region with an activation z-score>2 and p-value<0.01. Activation z-Scores: positive (yellow) = overrepresentation of targets activated in pathway; negative (blue) = overrepresentation of targets repressed in pathway; no direction (black) = no significant enrichment of activated or repressed targets; white = not a predicted pathway.
Figure S3: Factor loading for behavioral endpoints used in factor analysis. (A) Behavioral data represented in the factor analysis. All lever pressing data (food training, FR1, FR2; active vs. inactive) were included as variables in the factor analysis. Here we present a subset of the data aligned to the first day of each phase of self-administration. Because all animals had differing numbers of days in each phase, only those days in which the majority (>70%) of the animals in the study are presented. An image of the complete data set is presented in Figure 1D. (B) Factor analysis was used to reduce multidimensional behavioral endpoints to factors. The association of each factor with each behavioral endpoint included in the analysis is displayed. Factors were positively (yellow), negatively (blue), or not associated (black) with each endpoint. These particular associations allowed for the interpretation of the how each factor related to various SA behaviors.
Figure S4: Factor 2 discriminates between baseline differences in saline animals. (A) Factor 2, in the factor analysis, was positively associated with both active and inactive lever pressing and negatively associated with intake. (B) Individual data for total number of lever presses in saline (left) and cocaine (right) for the entire SA experiment, including food training. (C) Individual factor values for Factor 2 for saline (left) or cocaine (right) animals. Animals with the greatest number of lever presses, but no intake, had highest factor value (
 in saline group). Animals with increased lever pressing coupled with high intake (
in saline group). Animals with increased lever pressing coupled with high intake (
 in cocaine group) had lower factor values. Finally, those animals with few lever presses and no intake (
in cocaine group) had lower factor values. Finally, those animals with few lever presses and no intake (
 in saline group) had the lowest factor values. (D) Linear modeling was used to identify genes associated with Factor 2 within each brain region. Only genes with a |slope|>0.2 and a nominal p-value of <0.05 were investigated. (D) Genes were ranked by -log p-value signed by the slope of the association with Factor 2. Negative associations with Factor 2 are presented in gray and genes positively associated with Factor 2 are presented in red. (D) Heatmaps presented are transformed to indicate change in expression from SS controls. Blue = fold change in the negative direction from SS vs S24 and yellow = fold change in the positive direction from SS vs S24. These data indicate that changes in expression in transcripts associated with Factor 2 are most robust in the SS vs S24. This highlights the power of factor analysis to extract important information related to baseline behaviors and indicates that those differences are reflected in our transcriptomic data as well. (E) Overlap of genes positively (left) or negatively (right) associated with Factor 2 across brain regions, color-coded for significance. Total number of genes in each brain region listed in parentheses and total number of genes overlapping between regions indicated in corresponding boxes. There is a high degree of overlap of transcripts associated with Factor 2 in all brain regions.
in saline group) had the lowest factor values. (D) Linear modeling was used to identify genes associated with Factor 2 within each brain region. Only genes with a |slope|>0.2 and a nominal p-value of <0.05 were investigated. (D) Genes were ranked by -log p-value signed by the slope of the association with Factor 2. Negative associations with Factor 2 are presented in gray and genes positively associated with Factor 2 are presented in red. (D) Heatmaps presented are transformed to indicate change in expression from SS controls. Blue = fold change in the negative direction from SS vs S24 and yellow = fold change in the positive direction from SS vs S24. These data indicate that changes in expression in transcripts associated with Factor 2 are most robust in the SS vs S24. This highlights the power of factor analysis to extract important information related to baseline behaviors and indicates that those differences are reflected in our transcriptomic data as well. (E) Overlap of genes positively (left) or negatively (right) associated with Factor 2 across brain regions, color-coded for significance. Total number of genes in each brain region listed in parentheses and total number of genes overlapping between regions indicated in corresponding boxes. There is a high degree of overlap of transcripts associated with Factor 2 in all brain regions.
Figure S5: Raw heatmap of addiction index associated genes. (A–F) Raw expression of genes associated with AI in all brain regions for all groups when compared to the same baseline (S24). Log fold-change in expression of genes associated with AI and ranked by the sign of the association and -log(p-value) (gray = negative associations; red = positive associations). In all groups but C24, genes that were negatively associated with AI (gray bar) were downregulated and genes positively associated with AI (red bar) were upregulated. In all brain regions, the strongest response was in comparisons representing either Pattern B or C, suggesting that that transcriptional response to re-exposure to context/cocaine is influenced by addiction-related behaviors during cocaine SA.
Figure S6: Pathways associated with the addiction index (AI). Ingenuity Pathway Analysis revealed genes associated with the AI, which were enriched for cAMP-mediated signaling, PPARα/RXRα activation, and PI3K/AKT signaling among others. Activation z-Scores: positive (yellow) = overrepresentation of targets activated by regulator; negative (blue) = overrepresentation of targets repressed by regulator; no direction (black) = no significant enrichment of activated or repressed targets; white = not a predicted upstream regulator. Behavioral data analyzed using Kruskal-Wallis followed by Mann-Whitney Nonparametric Test; *p<0.05; **p<0.001; data presented as mean ± SEM.
Figure S7: Overlap of transcriptional profiles related to the AI and saline controls. (A–F) RRHO plots reveal little overlap of genes positively or negatively associated with AI and up- or downregulated in saline control animals (SS vs S24). As predicted, little to no overlap of expression profiles was observed in NAc, vHIP and VTA. Overlap of expression was weak in PFC, DStr and BLA and similar to that observed in the comparisons with Pattern A. A key for these plots is provided.
Figure S8: Full list of NR family members associated with the AI. Heatmap of association of all known nuclear receptors with the AI. Members of NR1 – 4 subfamilies are expressed throughout the reward circuitry. Strongest associations are found within NR2B and NR4A subfamilies. Yellow = positive association; Blue = negative association; Black = no significant association; white = expression not detected in our dataset.
A complete list of DEGs (nominal p-value < 0.05; fold-change ± 15%) in relation to saline controls (either S24 or SS) presented in Figure 1B. Each comparison is presented on a separate tab.
A complete list of genes categorized as Pattern A, B, or C in each brain region. Log fold-change for all conditions when compared to the same baseline (S24) are included. Genes were categorized by their expression patterns and a fold-change cut off of ± 15% was applied to each list to identify genes uniquely altered under each re-exposure condition. Each Pattern is presented on a separate tab.
A complete list of genes categorized as Patterns A, B, or C that overlap across multiple brain regions. Fisher’s exact tests revealed significant enrichment across lists. Comparisons reaching significance after multiple comparison correction (FDR) are bolded. Each pattern and direction of regulation is presented on a separate tab of the table.
A complete list of genes associated with AI that overlap across multiple brain region. Fisher’s exact tests revealed significant enrichment across lists. Comparisons reaching significance after multiple comparison correction (FDR) are bolded. Positive and negative associations are presented on separate tabs.
A full list of predicted regulators of genes associated with AI and their activation z-scores. Activation z-Scores: positive = overrepresentation of targets activated by regulator; negative = overrepresentation of targets repressed by regulator; no direction = no significant enrichment of activated or repressed targets; white = not a predicted upstream regulator.
A complete list of genes categorized as Pattern A, B, or C that overlap with those associated with AI within each brain region. Fisher’s exact tests revealed significant enrichment across lists. Comparisons reaching significance after multiple comparison correction (FDR) are bolded. Comparison of Pattern/AI for each brain region are presented on a separate tab.
Fisher’s exact tests revealed significant enrichment of cell-type specific genes in those lists of genes categorized as Pattern A, B, or C or genes associated with AI within each brain region. Only comparisons reaching significance after multiple comparison correction (FDR) are presented.
A complete list of the associations and p-values for each gene and Factor across all brain regions is presented. Each brain region is provided on a separate tab.