Abstract
Although defects in the RB1 tumour suppressor are one of the more common driver alterations found in triple-negative breast cancer (TNBC), therapeutic approaches that exploit this have not been identified. By integrating molecular profiling data with data from multiple genetic perturbation screens, we identified candidate synthetic lethal (SL) interactions associated with RB1 defects in TNBC. We refined this analysis by identifying the highly penetrant effects, reasoning that these would be more robust in the face of molecular heterogeneity and would represent more promising therapeutic targets. A significant proportion of the highly penetrant RB1 SL effects involved proteins closely associated with RB1 function, suggesting that this might be a defining characteristic. These included nuclear pore complex components associated with the MAD2 spindle checkpoint protein, the kinase and bromodomain containing transcription factor TAF1, and multiple components of the SCFSKP Cullin F box containing complex. Small-molecule inhibition of SCFSKP elicited an increase in p27Kip levels, providing a mechanistic rationale for RB1 SL. Transcript expression of SKP2, a SCFSKP component, was elevated in RB1-defective TNBCs, suggesting that in these tumours, SKP2 activity might buffer the effects of RB1 dysfunction.
Introduction
Patients who develop triple-negative breast cancer (TNBC), i.e., those breast cancers that lack amplification of the ERBB2 gene as well as expression of both the oestrogen and progesterone receptors, tend to have a relatively poor prognosis and represent a significant area of unmet clinical need, where novel therapeutic approaches are acutely needed (recently reviewed in ref. [1]). Although some targeted approaches have been proposed for molecularly defined subsets of TNBC patients, for most, classical chemotherapy regimens still represent the mainstay of treatment, making the requirement to identify novel targets in this disease critical. One approach to this problem has been to define the molecular composition of TNBCs and then to use this information to help identify therapeutic vulnerabilities that might operate in the disease. Already, the delineation of genomic, transcriptomic and proteomic profiles of tumours has identified a series of distinct molecular subtypes of TNBC, as well as identifying likely cancer driver gene mutations that operate in the disease [2].
One of the more frequent driver alterations in TNBCs involves deleterious mutations (e.g., truncating mutation, gene deletions, etc.) in the Retinoblastoma tumour suppressor gene (RB1 aka Rb). In non-tumour cells, Rb’s canonical role is in cell cycle progression, a function mediated in part by the repressive effect Rb has on the E2F family of transcription factors [3]. A somewhat reductionist model of Rb’s role in tumour suppression suggests that loss of Rb’s E2F repressive function allows premature transition of cells through the G1 cell cycle checkpoint; it also seems likely that loss of Rb function in breast cancer also influences additional processes that contribute to the development of the disease, including the differentiation of stem and progenitor cells and the transition of cells from an epithelial to a mesenchymal phenotype [3].
Commensurate with its key role in cell cycle control, genomic alterations in the RB1 gene are relatively common in TNBCs [4–6] as well as in a series of other malignancies [7–9]. In TNBC, loss of Rb protein expression is found in >40 % of cases [10, 11] (and reviewed in ref. [12]). Although patients with Rb-defective tumours (as defined by immunohistochemistry and/or gene expression) tend to have a relatively favourable response to classical chemotherapy [13–15] many either fail to respond to therapy or later relapse with therapy-resistant disease, suggesting that novel therapeutic approaches are required to target this patient subset.
One approach to identifying novel therapeutic targets that could be exploited in patients with specific tumour suppressor gene defects has been to identify synthetic lethal interactions associated with these genes. For example, the identification of synthetic lethal interactions between BRCA1 or BRCA2 tumour suppressor genes and inhibition of the PARP1 DNA repair protein has driven the clinical development and approval for use of PARP inhibitors for the treatment of cancer [16]. One notable feature of the BRCA/PARP1 synthetic lethal effect, which contributes to its translational value, is that it is highly penetrant [17]; i.e., in otherwise molecularly diverse pre-clinical models, and cancer patients, the presence of the predictive biomarker, in this case BRCA1 or BRCA2 mutation, more often than not predicts a profound antitumour cell response to a PARP inhibitor. This highly penetrant nature suggests that this particular synthetic lethal effect is robust in the face of the molecular heterogeneity that exists between different human cancers. Here we describe efforts to identify highly penetrant synthetic lethal effects associated with deleterious Rb alterations in TNBC; we reasoned that those with greatest penetrance will be more likely to operate in the diverse molecular contexts within the TNBC subtype and thus represent more promising therapeutic targets. Although genes such as TSC2 [18] and elements of the Dicer pathway [19] have been shown to be synthetic lethal with Rb defects, as far as we are aware, the penetrance of these effects, or whether these operate in models of TNBC, has not as yet been assessed. The availability of several, large-scale, short hairpin RNA (shRNA) and small interfering RNA (siRNA) screens [20–23], conducted in multiple tumour cell lines (TCLs), some of which are derived from TNBCs, now make a detailed identification of highly penetrant RB1-related synthetic lethal effects now possible. For this reason, we describe here a detailed molecular analysis of Rb status in TCLs derived from TNBC. We then designed a straightforward data analysis pipeline that allowed us to use this Rb annotation to interrogate both in-house and publically available large-scale, shRNA and siRNA screens to identify candidate Rb-related synthetic lethal effects. Within this data analysis pipeline, we included an estimate of penetrance. In triaging the candidate Rb-synthetic lethal effects that operated in TNBC tumour cells to identify those with greatest penetrance, we identified a series of pharmacologically tractable effects, one of which, SKP2, we validated using both genetic and pharmacological methods. We also noted that a significant proportion of the highly penetrant Rb SL effects in TNBC involved proteins closely associated with Rb function, suggesting that this might be a defining characteristic.
Results
Annotation of Rb status in TNBC TCLs
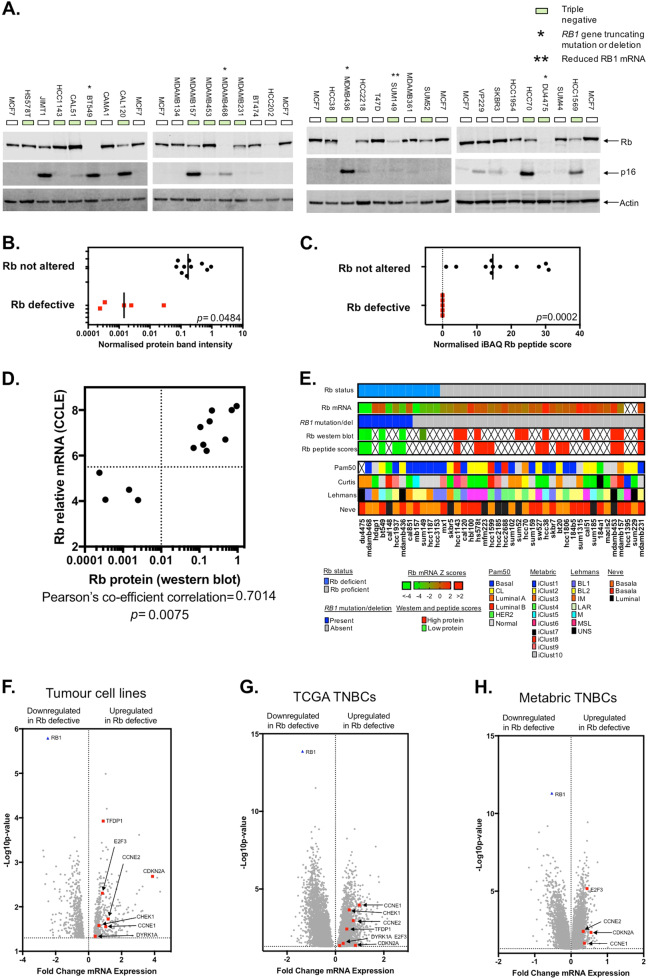
To identify highly robust synthetic lethal effects associated with Rb defects in TNBC, we classified a molecularly diverse panel of TNBC TCLs according to Rb status and then used this Rb classification to interrogate publically available genetic screen data using a data analysis pipeline (described later) that identified highly penetrant synthetic lethal effects. To do this, we first classified TNBC TCLs using a combination of genomic, transcriptomic and proteomic data to identify those with Rb genetic or epigenetic defects. We used western blotting to assess Rb protein expression in 30 breast TCLs, including 16 TNBCs (Fig. 1a, b). In this analysis, we found that TCLs with deleterious mutations in the RB1 gene (BT549, RB1 c.265_607del343; MDAMB468, RB1 c.265_2787del2523; DU4475, RB1 c.1_2787del2787; and MDAMB436, RB1 c.607_608ins107) lacked Rb protein expression, suggesting that the mutational status of RB1 correlated with protein expression (Fig. 1a, b). We also found that SUM149 cells exhibited low Rb protein expression, an observation we found to be consistent with reduced RB1 mRNA levels (Supplementary Data 1). To assess Rb protein status by orthogonal means, we compared our western blot data with publically available mass spectrometry data describing the proteomic profiles of 16 TNBC TCLs [24]. Using average intensity-based absolute protein abundance (iBAQ) data for Rb from mass spectrometry profiling [24] (Supplementary Data 1), we found that TNBC TCLs classified by western blotting as being Rb-defective exhibited no Rb peptides (MDAMB468, MDAMB436, HCC1937, DU4475, BT549) when compared to those TCLs we had classified as Rb-proficient (Fig. 1c, p = 0.0002), giving us some confidence in our classification. Examination of transcriptomic profiles of TNBC TCLs [25] revealed that TCLs with reduced levels of RB1 mRNA exhibited low Rb protein expression (Fig. 1d, p = 0.0075), suggesting that RB1 mRNA expression levels could be used as a reasonable proxy for protein expression. Taking proteomic, genomic and transcriptomic data into consideration (Fig. 1e), we then classified a total of 42 TNBC TCLs according to Rb status, identifying 12 with one or more defects in Rb (“Rb-defective”, e.g., low protein expression, truncating mutation/gene deletion and reduced mRNA levels: BT549, HCC1937, DU4475, MDAMB436, MDAMB468, CAL148, HDQP1, MB157, SUM149, HCC1187, HCC3153 and CAL851) and 30 TNBC TCLs as not exhibiting such defects (“Rb not altered” TCLs).
Fig. 1.
Rb status in TNBC tumour cell lines. a Western blot illustrating Rb and p16 protein expression in 30 breast tumour cell lines (TCLs). BT549, MDAMB436, MDAMB468 and DU4475 each possess loss-of-function mutations in the RB1 gene. SUM149 cells express reduced RB1 mRNA. b Scatter plot illustrating quantification of Rb protein levels delineated from a. Protein expression in Rb-defective vs. not altered, p = 0.0484, Student’s t test. c Scatter plot illustrating Rb protein expression in defective and proficient TNBC TCLs estimated by mass spectrometry data from ref. [24]. TNBC TCLs were classified into “Rb-defective” and “not altered” groups according to western blot data from a; using this classification, normalised iBAQ Rb peptide scores were compared and are shown. p = 0.0002, Fishers exact test. d Scatter plot illustrating the correlation between Rb protein and mRNA transcript levels in TNBC TCLs. Rb protein levels from a and b are compared to Rb mRNA transcript levels obtained from the CCLE database [25]. Correlation r = 0.7, p = 0.0075, Pearson’s correlation. e Oncoprint illustrating Rb and breast cancer subtype status amongst 42 TNBC TCLs. f Volcano plot illustrating mRNAs that are differentially expressed (limma analysis p < 0.05) in Rb-defective (vs. Rb not altered) TNBC TCLs. Genes functionally related to Rb are highlighted, as is Rb itself. g Volcano plot of mRNAs that are differentially expressed (limma analysis p < 0.05) in 48 Rb-defective (vs. Rb not altered) triple-negative breast tumours from the TCGA study [27]. Genes functionally related to Rb are highlighted, as is Rb itself. h.Volcano plot of mRNAs that are differentially expressed (limma analysis p < 0.05) in 55 Rb-defective (vs. Rb not altered) triple-negative breast tumours from the Metabric study [28]. Genes functionally related to Rb are highlighted, as is Rb itself
To further assess the validity of our Rb classification, we assessed the transcriptomic profiles of Rb-defective TNBC TCLs to assess whether these reflected loss of Rb function (Fig. 1f). Using the Rb-defective and Rb not altered classification described above for 42 TNBC TCLs, we identified 839 differentially expressed genes (452 with reduced expression in Rb-defective TCLs, 387 with elevated expression (p < 0.05, limma analysis, Supplementary Data 2)). As expected, we found RB1 itself to be the fifth most downregulated gene in the Rb-defective TCLs compared to Rb not altered TCLs (log fold change of −2.4, p value = 1.6 × 10−6 (limma analysis); Supplementary Data 2 and Fig. 1f). We also found that Rb-defective TCLs exhibited elevated expression of cyclins associated with G1 checkpoint control and S phase progression (cyclin E1 (p = 0.03), E2 (p = 0.02) and CDKN2A (p16; p = 0.002)), as well as elevated expression of the Rb-regulated E2F3 transcription factor and its binding partner TFDP1 (Fig. 1f (p = 0.005 and 0.0001, respectively)). Using the ENRICHR annotation tool [26] to identify pathways rather than individual genes that were differentially expressed in the Rb-defective group, we found that pathways related to Rb and G1 to S cell cycle control to be among the most significantly dysregulated in Rb-defective TNBC TCLs, including “Retinoblastoma (RB) in Cancer WP2446” p = 1.7 × 10−11 and “G1 to S cell cycle control WP45” p = 1.4 × 10−9, both of which included genes such as MCM4, 6 and 7, TFDP1, CCNE1, CCNE2, CHEK1, E2F3 and RBP1. Using the same annotation tool we also searched for the key transcription factors that regulated genes that were differentially expressed in Rb-defective TNBC TCLs. We found that a significant proportion of the differentially expressed genes in Rb-defective TNBC TCLs were targets of Rb-related E2F-family transcription factors, including E2F7, E2F4 and E2F1 (e.g., p = 0.001, 0.005 and 9.3 × 10−7 for E2F7, E2F4 and E2F1, respectively).
To compare the observations made in TCLs with TNBC tumours, we used the same approach of exploiting RB1 gene mutation/copy number status and RB1 mRNA expression profiles to classify 140 The Cancer Genome Atlas (TCGA) triple-negative breast tumours [27] according to Rb status; this approach identified 48 Rb-defective TNBC tumours and 92 where Rb was not altered. Assessing the transcriptomic profiles of these TNBCs we again found that genes associated with Rb and Rb-related G1 to S cell cycle control were frequently dysregulated in Rb-defective TNBCs, including CDKN2A, TFDP1, CCNE1, CCNE2, E2F3, CHEK1 and DYRK1A, a recently identified RB1 synthetic lethal gene [21] (Fig. 1g, Supplementary Data 3), consistent with the observations made in TNBC TCLs. We also applied the same approach to classify 182 Metabric TNBC tumours [28] according to Rb status; this approach identified 55 Rb-defective and 132 Rb not altered TNBC samples. Assessing the transcriptomic profiles of these TNBCs we again found that genes associated with Rb and Rb-related G1 to S cell cycle control were frequently dysregulated in Rb-defective TNBCs (Fig. 1h, Supplementary Data 4). These global transcriptional patterns in Rb-defective TNBC TCLs and human tumours suggested that our Rb classification approach was somewhat valid.
A pipeline for the identification of highly penetrant Rb synthetic lethal effects in TNBC
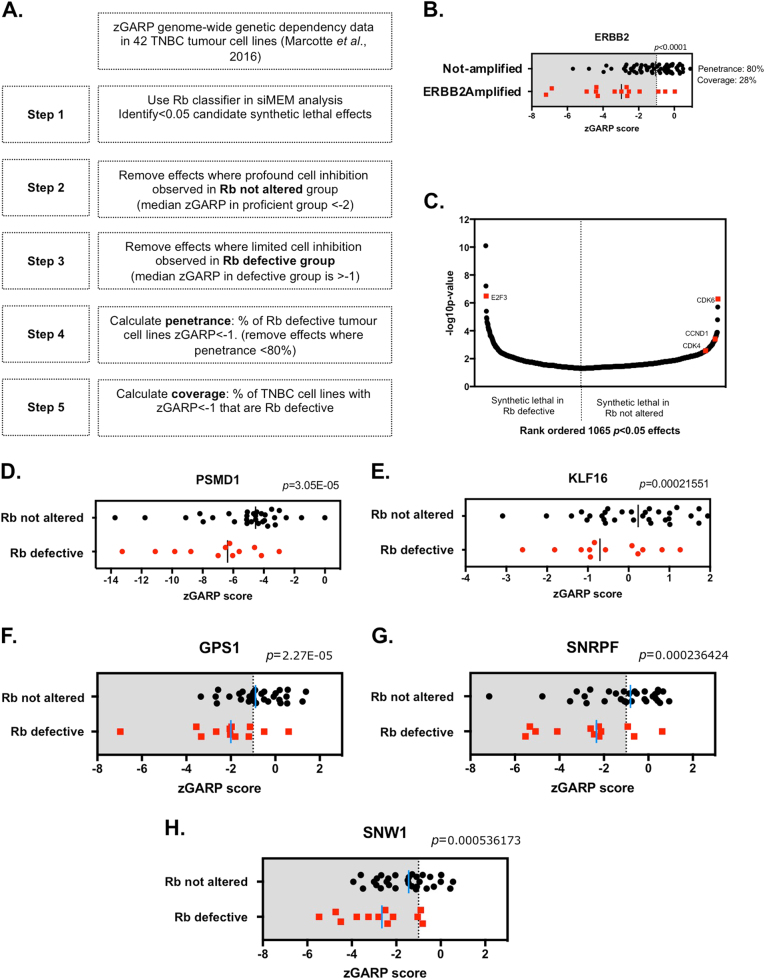
Using the Rb classification of TNBC TCLs described above, we re-analysed publically available genetic screen data (e.g., genome-wide shRNA screen data [22, 23], gene subset shRNA screen data [29] or gene subset siRNA data [20, 21]) from TNBC TCLs, using a series of iterative data-processing steps designed to identify highly penetrant synthetic lethal effects (Fig. 2a). In summary, these steps involved: (i) step one—identification of candidate synthetic lethal effects: using shRNA screen data [22] for 12 Rb-defective TNBC cell lines and 30 Rb not altered cell lines we used the si/shRNA mixed effect model (siMEM) algorithm [22] to identify those genes whose inhibition appeared to target the Rb-defective models to a greater extent than Rb-proficient TNBC TCLs (p < 0.05). (ii) Steps two and three—apply Z score thresholds to identify profound cell inhibitory effects: although step one allowed us to identify genes whose inhibition selectively targeted Rb-defective TCLs to a greater extent than Rb not altered TCLs, we reasoned that the scale of cell inhibition in these two TCL cohorts might also be critical in identifying the most suitable therapeutic targets. For example, for the purposes of identifying novel therapeutic targets we were less interested in genes whose inhibition profoundly inhibited both Rb-defective and Rb not altered groups, even if the Rb-defective models exhibited significantly greater sensitivity; we assumed that inhibition of these targets would likely cause significant normal cell toxicity. Similarly, we also assumed that candidate synthetic lethal effects that did not elicit profound cell inhibitory effects in Rb-defective TCLs would be less likely to elicit profound anticancer therapeutic effects when exploited clinically. For these reasons, we triaged the list of candidate synthetic lethal effects identified in step one to remove from further analysis: (a) those genes that appeared to be synthetic lethal with Rb but whose targeting elicited profound cell inhibition in Rb not altered TCLs; and (b) those genes that appeared to be synthetic lethal with Rb but whose targeting did not elicit profound cell inhibitory effects in Rb-defective TCLs. To do this we used shRNA/siRNA Z score thresholds to estimate the effect of each RNAi reagent. (iii) Steps four and five—identify effects with greatest penetrance: we reasoned that the most clinically effective synthetic lethal targets are likely to be those that have complete or highly penetrant effects, i.e., the presence of the predictive biomarker (in this case an Rb defect) is more often than not associated with profound sensitivity to target inhibition. For this reason, we finally triaged synthetic lethal effects by calculating synthetic lethal penetrance (SLP) scores; in this case, SLP for each synthetic lethal effect was calculated as the percentage of Rb-defective TCLs that exhibited a Z score of <−1 when targeted with shRNA. We also calculated synthetic lethal coverage (SLC) scores, i.e., the percentage of TNBC TCLs that were sensitive to shRNA that were Rb-defective, to estimate the specificity of synthetic lethal effects for Rb defects.
Fig. 2.
Identifying highly penetrant Rb synthetic lethal effects that operate in TNBC. a Schematic illustrating the data analysis workflow used. In the first instance, 16 056 gene zGARP scores from shRNA screens in 42 TNBC cell lines described in the Colt2 data set were analysed; parallel analyses were carried out using data from the DRIVE and Achilles data sets (see main text). b Scatter plot illustrating ERBB2 zGARP scores in 77 breast tumour cell lines partitioned according to effects in ERBB2-amplified and -non-amplified TCLs. ERBB2 shRNA elicits a p < 0.0001 oncogene addiction effect (siMEM) with 80% penetrance in ERBB2-amplified tumour cell lines (red). Coverage is also shown. c Scatter plot illustrating 1065 p < 0.05 significant siMEM Rb synthetic lethal effects identified from the siMEM analysis of TNBC TCLs in the Colt2 study (step one in a). p < 0.05 effects are ranked ordered by siMEM p value. E2F3 (synthetic lethal in Rb-defective), CDK6, CDK4 and the CDK4,6 cyclin partner, Cyclin D1 (CCND1) (dependencies in Rb not altered) are highlighted. d, e. Scatter plots illustrating Z scores in 42 TNBC TCLs for two siMEM p < 0.05 candidate Rb synthetic lethal effects, PSMD1 (D) and KLF16 (E), removed from further analysis by the use of Z score filters (steps two and three in a). zGARP scores for PSMD1 (preferentially target Rb-defective, siMEM p value 3 × 10−5) indicate all but two Rb not altered tumour cell lines exhibit Z score of <−2 (median Z in not altered group < −4). zGARP scores for KLF16 (preferentially target Rb-defective, siMEM p value 0.0002) indicate that the majority of Rb-defective TCLs exhibit Z score of >−1 (median Z in defective group = −0.8), despite median Z scores being significantly different in Rb not altered vs. deficient TCLs. f–h Scatter plots illustrating Z scores in 42 TNBC TCLs for three siMEM p < 0.05 candidate Rb synthetic lethal effects, GPS1, SNRPF and SNW1, where median Z in defective group < −1 and median Z in proficient group > −2 (steps two and three in a)
To illustrate the concepts of penetrance and coverage as applied to functional genomic screens in TCLs, we examined a well-characterised and therapeutically actionable oncogene addiction effect that operates in breast cancer, namely that associated with amplification of the epidermal growth factor receptor oncogene, ERBB2. Amplification and overexpression of ERBB2 is used clinically to stratify breast cancer patients for treatment with ERBB2-targeting agents such as the monoclonal antibody trastuzumab [30]. In examining the Colt2/Marcotte et al. shRNA data set (78 432 shRNAs targeting 16 056 genes in 77 breast TCLs representing TNBC and non-TNBC subtypes [22]), we found that as shown before [22], that shRNA targeting of ERBB2 selectively targeted ERBB2-amplified breast TCLs (siMEM, p < 0.0001), elicited minimal inhibition in ERBB2 non-amplified TCLs (median Z −1), profound inhibition in ERBB2-amplified TCLs (median Z -3) and had a SLP (penetrance) score of 80% and a SLC (coverage) score of 28% (Fig. 2b); the high penetrance score in this case, taken together with ERBB2 fulfilling the other selection criteria, reconfirms ERBB2 as a suitable therapeutic target in appropriately stratified breast cancer patients.
Identification of Rb synthetic lethal effects from shRNA screens
To identify Rb synthetic lethal effects, we first used genome-wide shRNA data from 42 TNBC TCLs described in the Colt2/Marcotte et al. study [22]. In this data set, the combined effects of multiple shRNAs targeting a single gene are described as Z normalised Gene Activity Ranking Profile (zGARP) scores [31]. We re-analysed this shRNA screen data using the siMEM mixed effect linear model method (see step one, above), which identifies synthetic lethal effects by estimating the depletion or “dropout” rate of individual shRNAs in cohorts of cell lines classified according to a molecular feature [22], in this case Rb deficiency. Using the siMEM approach with our Rb classification of 42 TNBC TCLs, we identified 1065 Rb-specific dependencies (p < 0.05, siMEM, Fig. 2c): 437 genes, where shRNA preferentially inhibited Rb-defective TNBC (i.e. Rb synthetic lethal effects) and 628 genes, where shRNAs preferentially targeted Rb-proficient TNBC TCLs (Supplementary Data 5). Amongst these, we noted that shRNAs targeting CDK4, CDK6 or the CDK4,6 cyclin partner gene Cyclin D1 (CCND1) preferentially inhibited Rb-proficient TNBC TCLs (Fig. 2c), consistent with the hypothesis that inhibition of CDK4,6 activity restores cell cycle control in Rb-proficient TNBC tumour cells and elicits cell inhibition [32]. In terms of identifying Rb dependencies, we noted that the siMEM analysis of the Colt2 data set identified shRNA targeting the E2F family transcription factor, E2F3, as being one of the most significant Rb synthetic lethal effects (Fig. 2c). These Rb-related observations thus gave us some confidence in the approach. We also carried out similar analyses in siRNA/shRNA data sets that included TNBC TCLs, from other sources: the DRIVE data set [29]; the Achilles data set [23]; and the ICR-Intercell data set [20, 21], and provide the lists of Rb dependencies identified in these data sets in Supplementary Data 6, 7, 8, respectively.
Application of Z score thresholds identifies profound cell inhibitory effects
As described above, we were interested in identifying Rb synthetic lethal effects that had profound effects in Rb-defective TNBCs but had minimal effects in cells without Rb defects. Although approaches such as siMEM are useful in identifying putative vulnerabilities, they often do not, when used in isolation, identify synthetic lethal effects with these favoured characteristics. For example, Fig. 2d, e illustrates a pair of p < 0.05 Rb dependencies identified by siMEM analysis (step one), which had either profound cell inhibitory effects in both Rb not altered as well as Rb-defective TNBC TCLs, e.g., PSMD1 (Fig. 2d, siMEM p = 3 × 10−5, median Z in Rb-defective of −6, median Z in Rb not altered of −4) or relative paucity of profound cell inhibition effects in the Rb-defective TCL cohort, e.g., KLF16 (Fig. 2e, siMEM p = 0.0002, median Z in Rb-defective of −0.8). To eliminate such effects from further study, we applied a pragmatic approach that removed from further assessment p < 0.05 synthetic lethal effects where the median zGARP score in the Rb-defective TCLs was >−1 (i.e., effects, where profound cell inhibition in Rb TCLs not observed) and those effects where median zGARP score in Rb-proficient effects was <−2 (i.e., dependencies that still elicited profound cell inhibition in Rb-proficient TCLs); three examples that fulfilled these criteria, GPS1, SNRPF and SNW1, are shown in Fig. 2f, g, h. This triage step identified 122 Rb synthetic lethal effects in the Colt2 data set that fulfilled these criteria (Supplementary Data 9). Similarly, triaged dependencies were identified in the DRIVE [29], Achilles [23] and ICR-Intercell data sets [20, 21] (Supplementary Data 10, 11 and 12, respectively).
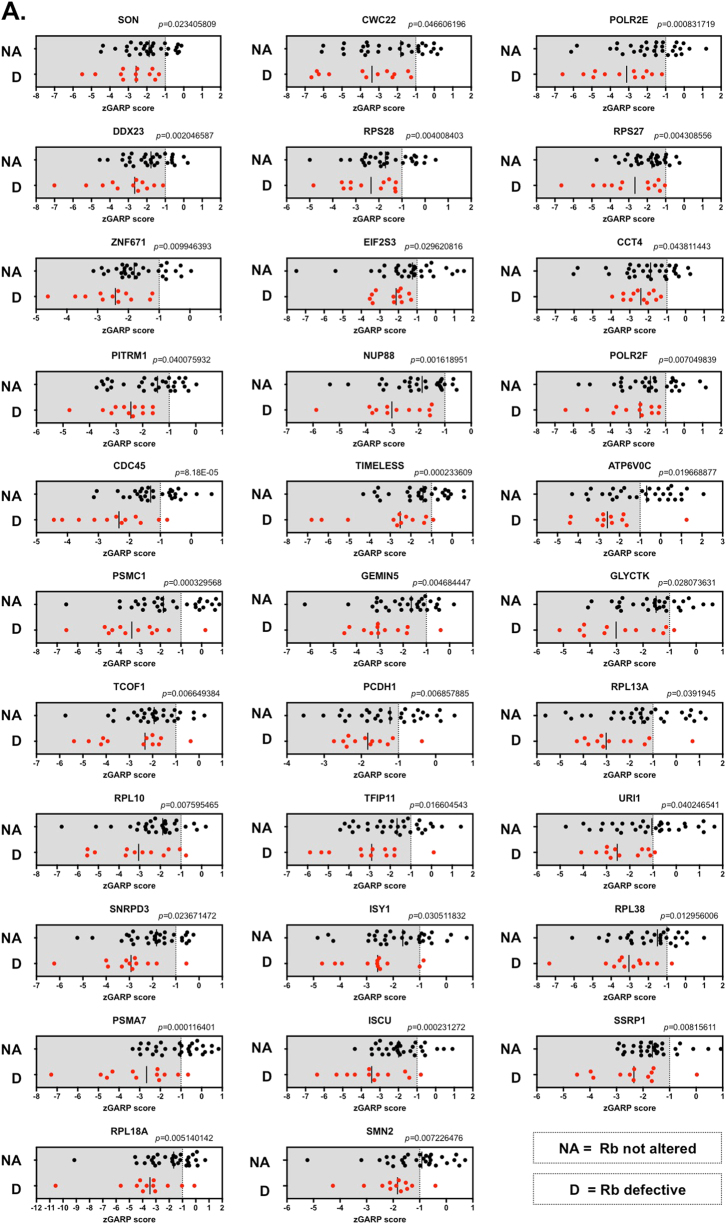
Highly penetrant Rb synthetic lethal effects in TNBC include TAF1, TAF1 target genes, nuclear pore complex proteins and the SCFSKP-COP9 signalosome complexes
We next calculated SLP and SLC scores for each Rb synthetic lethal effect triaged in steps two and three. In our analysis of the Colt2 data set, we calculated SLP and SLC scores for 122 Rb synthetic lethal effects, identifying 54 effects that had >80% penetrance (Fig. 3a (SLP > 90% shown), Supplementary Data 9) and also identified highly penetrant effects from our analysis of the DRIVE and Achilles and ICR-Intercell data sets (Supplementary Data 10, 11 and 12, respectively). Amongst the highly penetrant Rb SL candidates (>80% penetrant), a significant number is known to be involved in RNA splicing (Supplementary Figure 1, p = 5.14 × 10−12, GO Biological Processes 2017, ‘mRNA splicing, via spliceosome’, Enrichr [26], Supplementary Data 13), including: ISY1 (SLP = 92%); SON (SLP = 100%); CWC22 (SLP = 100%); DDX23 (SLP = 100%); POLR2E (SLP = 100%); GEMIN5 (SLP = 92%); POLR2F (SLP = 100%); TFIP11 (SLP = 92%); SRRM2 (SLP = 83%); LSM2 (SLP = 83%); SNW1 (SLP = 83%); SART3 (SLP = 83%); FIP1L1 (SLP = 83%); and SNRPD3 (SLP = 92). Interestingly, a RNAi screen in Caenorhabditis elegans also identified a synthetic lethal interaction between Rb loss and components of the splicing machinery [33]. Similarly, a number of proteins involved in the regulation of translation were also identified as candidate Rb SL targets (p = 1.7 × 10−7, GO Biological Processes 2017, ‘regulation of translation by machinery localisation’, Enrichr [26], Supplementary Figure 1, Supplementary Data 13). These included: RPS24 (SLP = 83%); RPS27 (SLP = 100%); RPS28 (SLP = 100%); RPL18A (SLP = 92%); RPL13A (SLP = 92%); RPL10 (SLP = 92%); TCOF1 (SLP = 92%); GEMIN5 (SLP = 92%); and RPL38 (SLP = 92%).
Fig. 3.
Highly penetrant Rb synthetic lethal effects. Scatter plots illustrating Z scores in 42 TNBC TCLs for 30 candidate Rb synthetic lethal effects, which pass Z score threshold assessment and demonstrate a penetrance score of >90%, as summarised in steps 1–5 of Fig. 2a
We also identified two nuclear pore complex (NPC) components [34, 35] NUP88 (SLP = 100%) and NUP214 (SLP = 83%) as highly penetrant Rb synthetic lethal partners (Supplementary Figure 1, Supplementary Data 13). NPCs are responsible for trafficking proteins between the nucleus and the cytoplasm. In particular, NPCs control spindle assembly, faithful chromosome segregation and mitotic progression by controlling the temporal and spatial localisation of proteins [36–38], including the E2F transcriptional target and spindle assembly checkpoint protein, MAD2, whose elevated expression is required for chromosomal instability in Rb-defective cells [39, 40]. It seems possible that the highly penetrant synthetic lethalities between Rb and NUP88 or NUP214 could be related to the temporal and spatial localisation of MAD2, perhaps by causing a level of excessive genomic instability that is incompatible with cell viability.
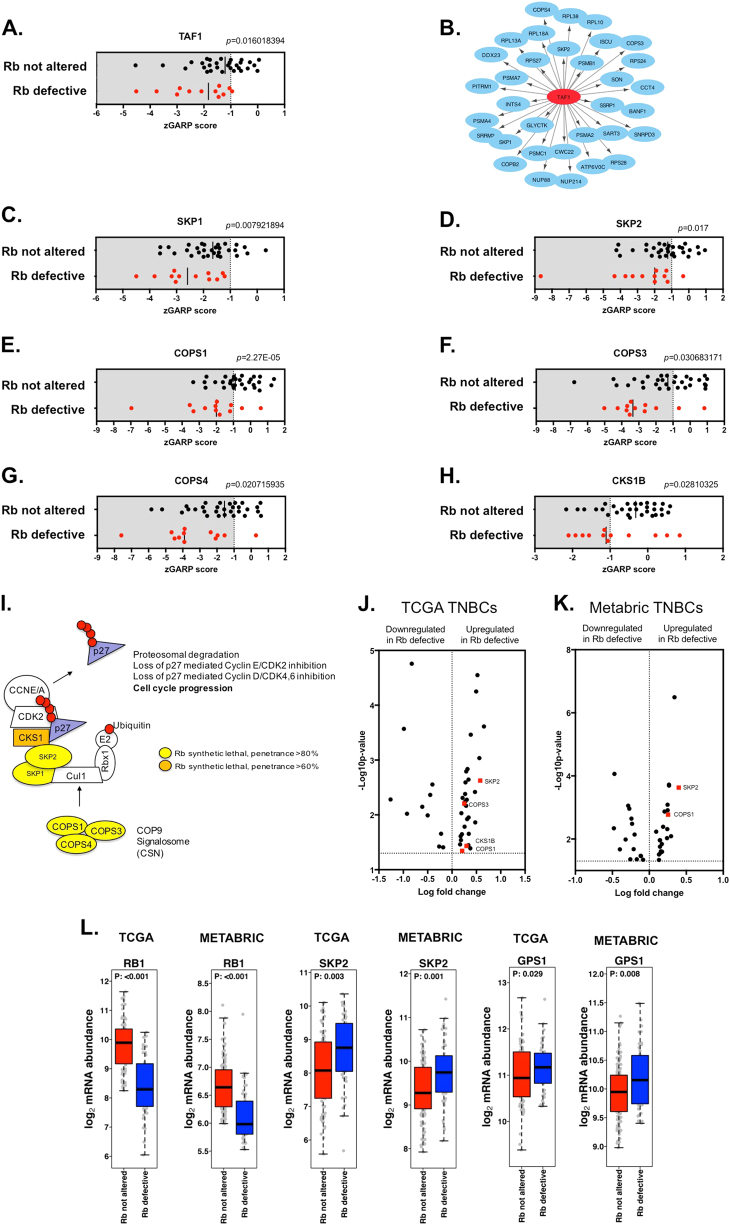
When taking all of the highly penetrant Rb synthetic lethal effects identified in the analysis of the Colt2 data set into account, a significant number of these included genes/proteins known to be associated with Rb function, including the Rb-interacting protein TAF1 (siMEM p = 0.016, SLP 92% and SLC 38%, Fig. 4a) [41]. TAF1 encodes the major 250 kDa subunit of the transcription initiation factor, TFIID [42], which binds core promoter regions, including promoter start sites in genes implicated in cell cycle control [43]. We found that over half (n = 33) of the highly penetrant Rb synthetic lethal genes encompassed putative TAF1-binding sites, as defined by TAF1 Chip-Seq data [26, 44] a significant enrichment over chance alone (p = 2.72 × 10−13, Fig. 4b). One mechanistic explanation for these observations might be that many of the highly penetrant Rb synthetic lethal interactions that operate in TNBC are caused by aberrant TAF1 activity, a downstream effect of Rb dysfunction. TAF1 encompasses two protein domains, a kinase and bromodomain, which in principle are pharmacologically tractable. When taken together with the highly penetrant Rb synthetic lethal interaction, this might make TAF1 an attractive target for cancer drug discovery [45–47].
Fig. 4.
TAF1 and SKP2 as central nodes in highly penetrant Rb synthetic lethal networks. a Scatter plot illustrating Z scores in 42 TNBC TCLs for TAF1 from the data analysis illustrated in Fig. 2a. b Cytoscape network plot illustrating 33 highly penetrant (>80% penetrance) Rb synthetic lethal effects identified as TAF1 transcription factor target genes, as annotated by ENCODE data [26, 44]. c–h Scatter plots illustrating Z scores in 42 TNBC TCLs for SKP1, SKP2, COPS1,3,4 and CKS1B from the data analysis in Fig. 2a. i Pathway diagram highlighting the role of multiple highly penetrant Rb synthetic lethal effects in the control of p27 activity. j Volcano plot illustrating mRNAs from highly penetrant Rb SL genes that are differentially expressed (limma analysis p < 0.05) in 48 Rb-defective vs. 92 Rb not altered triple-negative breast tumours from the TCGA study [27]. Four highly SCFSKP/COP9 complex genes, highlighted in red, demonstrate significantly higher mRNA expression levels in the Rb-defective cell lines. k As per j, using data from the Metabric study [28]. l Box plots illustrating elevated SKP2 or GPS1 (COPS1) mRNA expression in Rb-defective TNBC from both the TCGA [27] and Metabric studies [28]. p-values shown calculated with Wilcox rank sum test
SKP1 and SKP2 were also identified as highly penetrant Rb synthetic lethal effects (Fig. 4c, d, SLP 100% and 92% for SKP1 and SKP2, respectively). Both SKP1 and SKP2 are part of an E3 ubiquitin ligase complex, Skp, Cullin F-box containing complex (SCFSKP2), whose activity is greatest during late G1/early S phase of the cell cycle. Using ubiquitin ligation, the SCFSKP2 complex targets proteins for proteasomal degradation, including the cyclin-dependent kinase inhibitors, p21 and p27 [48]. Three other highly penetrant Rb synthetic lethal effects identified were also associated with the SCF complex (Fig. 4e–g); COPS1 (aka GPS1, SLP 83%, Fig. 4e), COPS3 (SLP 83%, Fig. 4f) and COPS4 (SLP 92%, Fig. 4g) encode components of the constitutive photomorphogenesis 9 (COP9) signalosome complex (CSN [49, 50]). CSN regulates the ubiquitin ligase activity of SCF complexes via the deneddylation of the ring finger subunits (e.g., Rbx1) within SCF [51] (Fig. 4i). We also noted that CKS1B (CDC28 protein kinase regulatory subunit 1B) also represented a penetrant Rb synthetic lethal partner (SLP 58%, Fig. 4h); together with its co-factor SKP2, CKS1B provides the substrate-specific targeting of p27 by SCFSKP2 [52]. When we compared transcriptomic data from RB1-defective TNBCs to those from TNBC with no apparent RB1 defect (Supplementary Data 3 and 4), we noted that RB1-defective TNBCs expressed significantly elevated levels of SKP2 and COPS1 (GPS1) mRNA (Fig. 4j, k, l, p < 0.05 for both TCGA [27] and Metabric [28] data, Wilcox rank sum test), suggesting that in these particular tumours, elevated SKP2 activity might buffer the effects of RB1 dysfunction.
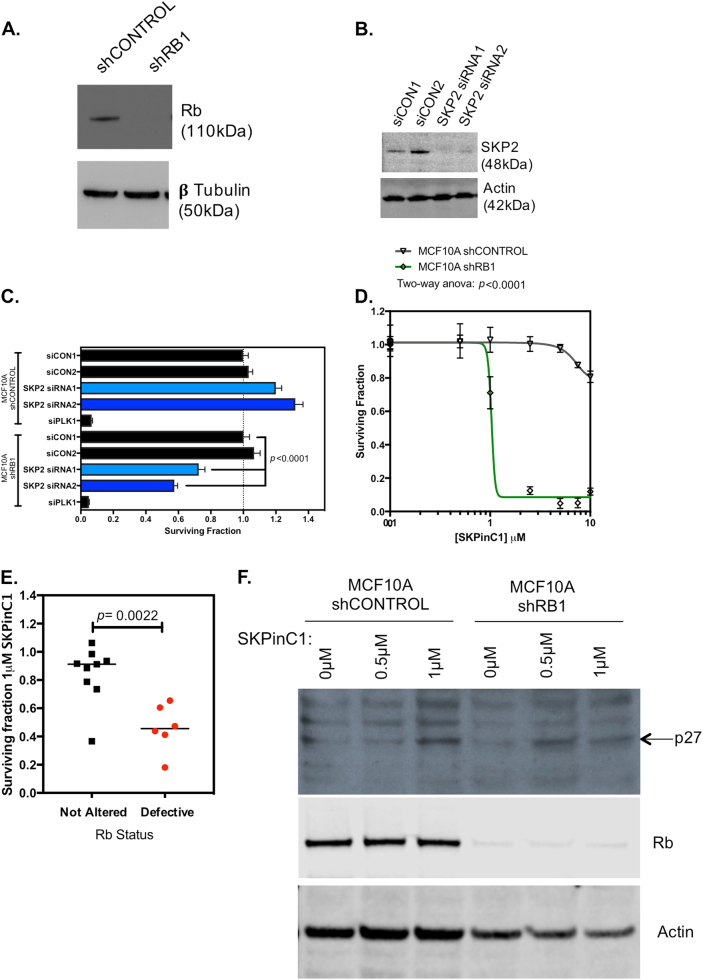
SKP2 directly interacts with Rb [53] and has previously been shown to be required for the hyper-proliferative phenotype of Rb-depleted human retinoblastoma cells, via its regulatory control over p27 levels [48]. In cells with normal G1/S cell cycle control, Rb binds SKP2, impairing its activity within the SCFSKP2 complex; loss of Rb however results in elevated SKP2 activity, a resultant reduction in p27 and p21 protein levels, loss of Cyclin E-CDK2 and Cyclin D-CDK4,6 inhibition and thus progression of cells through the G1 restriction point and into S phase [54]. To directly test whether Rb loss in a breast epithelial cell could cause synthetic lethality with SKP2 inhibition, we silenced SKP2 using two different siRNAs in non-tumour breast epithelial MCF10A cells expressing either a shRNA silencing RB1 or a non-silencing control shRNA (Fig. 5a). Silencing of SKP2 (Fig. 5b) elicited synthetic lethality in Rb-defective cells but not in Rb-proficient cells (Fig. 5c, Student’s t test p < 0.0001, Supplementary Figure 2). We also found that the toolbox SKP2 inhibitor SKPinC1, which impairs the binding of phosphorylated p27 to the substrate recognition pocket formed by SKP2 and CKS1B [55], had a profound synthetic lethal effect on Rb-defective MCF10A cells, but minimal effects in Rb wild-type cells (p < 0.001 two-way analysis of variance (ANOVA), Fig. 5d) and induced apoptosis in Rb-defective cells (Supplementary Figure 3). We also assessed SKPinC1 sensitivity in 13 TNBC TCLs, and found that Rb-defective TCLs were more sensitive than TCLs with no apparent Rb defect (Fig. 5e and p < 0.0022, Student’s t test). In addition, we confirmed that exposure of MCF10A cells with SKPinC1 inhibitor led to a stabilisation of p27 protein levels (Fig. 5f). Targeting of p27 by siRNA also partially reversed the inhibitory effect of SKPinC1 (Supplementary Figure 4), suggesting that the effect of SKPinC1 was p27-dependent. Taken together, these observations suggested that SKP2 small-molecule inhibition, could in principle, elicits the highly penetrant Rb synthetic lethal effect seen in TNBC tumour cells with RNA interference reagents.
Fig. 5.
Small-molecule inhibition of SKP2 in Rb-defective breast cell lines is synthetic lethal. a Western blot illustrating loss of Rb expression in isogenic MCF10A non-tumour breast epithelial cells with constitutive expression of either a control non-targeting shRNA (shCONTROL) or an Rb-targeting shRNA (shRB1). b Western blot illustrating loss of SKP2 protein expression in MCF10A cells 48 h after transfection with SKP2 or control, non-targeting, siRNAs (siCON1 or siCON2). c Bar chart illustrating cell inhibitory effects in isogenic MCF10A cells with/without stable expression of Rb shRNA transfected with SKP2 siRNA. Cells were reverse transfected with siRNAs as shown and cultured for 5 continuous days, at which point cell viability was assessed by use of CellTitre-Glo reagent. SKP2 siRNA caused significant cell inhibition (p < 0.001, Student’s t test) in cells with stable Rb silencing, but not in cells with wild-type Rb expression. d Dose response survival curves illustrating the cell inhibitory effects of the SKP2 small-molecule inhibitor, SKPinC1, in isogenic MCF10A cells with/without stable expression of Rb shRNA. Cells were exposed to SKPinC1 for 5 continuous days at which point cell viability was assessed by use of CellTitre-Glo reagent. Rb-defective cells demonstrated a profound sensitivity, compared to Rb wild-type cells (p < 0.0001, two-way ANOVA, SF50 = 1 and >10 μM in Rb-defective and wild-type cells, respectively). e Tumour cell inhibitory effect of SKPinC1 in 13 TNBC TCLs classified according to Rb status. Cells were exposed to 1 μM SKPinC1 as in h. Surviving fractions are shown (p = 0.0022, Student’s t test). f Western blot illustrating p27 protein levels in Rb-defective cells exposed to 0.5 and 1 μM SKPinC1 for 24 h
We also assessed whether other highly penetrant synthetic lethal effects operated in the RB1 isogenic MCF10A system. Using an arrayed siRNA screen we tested all 54 genes that we identified as highly penetrant (>80% penetrance) RB-synthetic lethal effects in TNBC cell lines in the Colt2 study along with an additional four controls (e.g., E2F3) for a total of 58 genes. We found that over half of the identified dependencies (55%) were observed in the isogenic system (Supplementary Figure 5), including profound synthetic lethal effects associated with TIMELESS, PCDH1, PITRM1, E2F3, SMN2 and TCOF1. This suggests that these effects can be specifically associated with RB loss, and that they are not an artefact of either the shRNA library used in Colt2 or the pooled screening approach. It seemed possible that differences in the proliferative rate of RB1-defective vs. wild-type MCF10A cells could account for the synthetic lethal effects observed. However, we found that the proliferative rate of RB1-defective and wild-type MCF10A cells was similar (Supplementary Figure 6), suggesting this might not have a significant bearing on the synthetic lethal effects identified. Moreover, analysis of previously published doubling times for 17 TNBC cell lines identified no significant difference between RB1-defective and RB1-proficient models (Mann-Whitney U-test p = 0.4) [56].
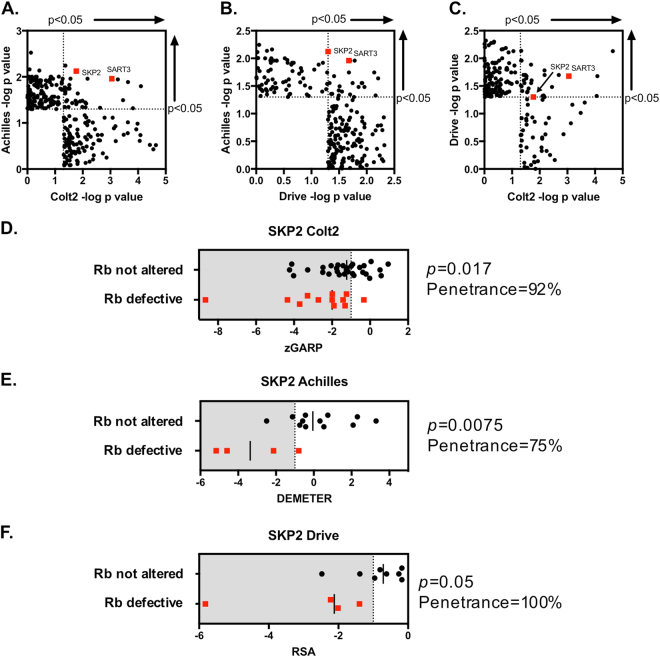
We also assessed which of the highly penetrant Rb synthetic lethal effects identified in our analysis of the Colt2 data were also identified as highly penetrant effects in the TNBC TCLs in two other shRNA screens: Achilles [23] and DRIVE [29]. Comparing the p < 0.05 Rb penetrant synthetic lethal effects between the three data sets, we noted that SKP2 was one of two synthetic lethal effects identified in all three screens, the other effect being associated with SART3, the RNA splicing factor (Fig. 6a-c, Supplementary Data 9 and 10, 11). In each screen, SKP2/Rb synthetic lethality was highly penetrant with SLP scores of 92% (Colt2), 75% (Achilles) and 100% (DRIVE data set, Fig. 6d–f). Whilst the TNBC TCLs described in these three data sets are partially overlapping, the shRNA libraries used were independently designed and synthesised, and the screens independently carried out. As such, identifying SKP2 as a highly penetrant Rb synthetic lethal effect in all three data sets suggested that this effect was somewhat independent of the shRNA reagents used.
Fig. 6.
SKP2 identified as a highly penetrant Rb synthetic lethal effect in multiple, independently derived, data sets. a–c Scatter plots comparing p-values (−log10) for Rb synthetic lethal effects identified in Colt2 [22], Achilles [23] and DRIVE [29] data sets. p < 0.05 effects in any single screen are shown. p < 0.05 effects in two screens are shown in the top right hand quadrant of each plot. SKP2 and SART3, which were identified in all three screens as synthetically lethal with Rb defects, are highlighted in red. p < 0.05 effects in Colt2 data were identified by siMEM, followed by Z score and penetrance filtering (Fig. 2a); p < 0.05 effects in DRIVE and Achilles data were identified by median permutation t test, followed by Z score and penetrance filtering (Fig. 2a). d Scatter plots illustrating Z scores in 42 TNBC TCLs for SKP2 from the Colt2 data analysis. e Scatter plots illustrating Z scores in 16 TNBC TCLs for SKP2 from the Achilles data analysis. f Scatter plots illustrating Z scores in 12 TNBC TCLs for SKP2 from the Drive data analysis
Rb/SKP2 synthetic lethality operates in tumour cells from lung and other cancer histologies
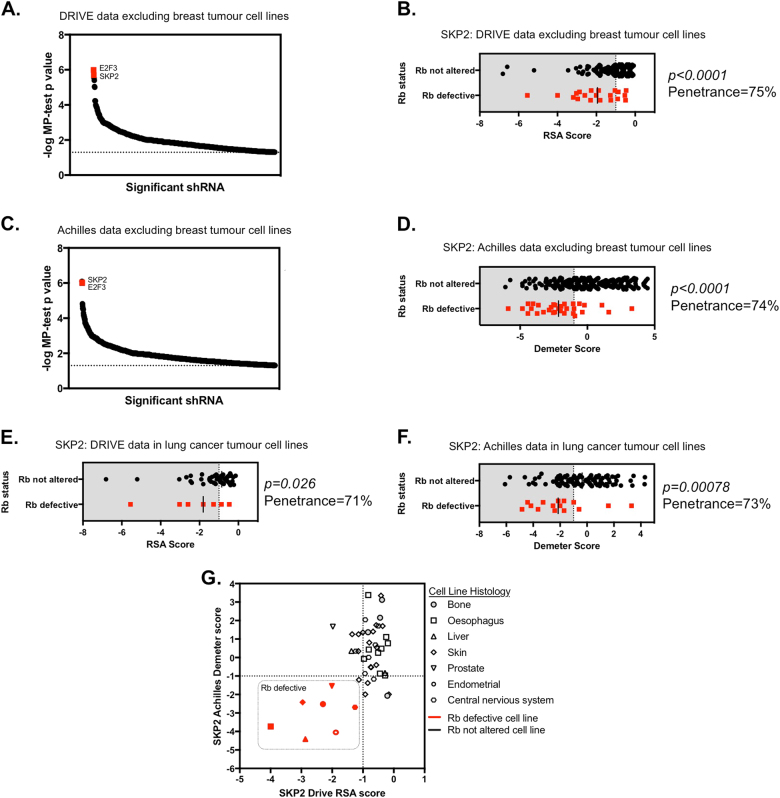
Rb defects are relatively prevalent in TNBC, but are not unique to this cancer subtype and are also evident in in many tumour types, including, for example, small-cell lung cancers, bladder carcinomas, osteosarcomas, glioblastomas, endometrial carcinomas and retinoblastomas [7, 8, 57–59]. To estimate whether the highly penetrant Rb/SKP2 synthetic lethality in TNBC extended to other cancer histologies, we re-analysed the results of two recent large-scale shRNA screens that encompass TCLs derived from non-breast cancer histologies. Project DRIVE includes 373 non-breast TCLs with available Rb mutation and copy number status that were derived from 18 different cancer histologies [29]. In an analysis of the DRIVE data set that excluded the breast TCLs, we found a significant association between RB1 mutation/deletion and sensitivity to SKP2 shRNA (Fig. 7a, b; median permutation (MP)-test p < 0.0001, SLP = 75%). Of over 6000 genes tested, only E2F3 (MP test p < 0.0001) showed a stronger association with Rb status (Fig. 7a). Similarly, project Achilles includes 467 non-breast TCLs with available Rb mutation and copy number status, derived from 20 different cancer histologies [23]. In an analysis of the Achilles data set that excluded the breast TCLs, we also found a significant association between RB1 mutation/deletion and sensitivity to SKP2 shRNA (Fig. 7c, d; MP test p < 0.0001, SLP = 74%). Of over 17 000 genes, SKP2 synthetic lethality was the second most significant effect associated with Rb loss, after CDK2 (MP test p < 0.0001) with E2F3 being the third most significant effect (Fig. 7c). The elevated penetrance of the Rb/SKP2 synthetic lethal effect in tumour cell models other than those derived from breast cancer suggested that this genetic dependency might operate in multiple histologies. When we considered the specific cancer histology of TCLs, rather than analysing these en mass, we found in both the Achilles and DRIVE data sets the Rb/SKP2 synthetic lethality was detectable in TCLs derived from lung cancers (p = 0.026/SLP = 71% and p = 0.0008/SLP = 73% for DRIVE and Achilles, respectively; Fig. 7e, f). However, we do note that the relatively small number of Rb-defective TCLs from non-lung cancer histologies in these data sets might impair the ability to detect the Rb/SKP2 synthetic lethality. For example, in both DRIVE and Achilles data sets, we noted that Rb-defective TCLs derived from prostate cancer, osteosarcoma, liver cancer and oesophageal cancer, exhibited sensitivity to SKP2 shRNA (Fig. 7g).
Fig. 7.
SKP2 identified as a highly penetrant Rb synthetic lethal effect in other histologies in two independently derived data sets. a Scatter plot of 775 p < 0.05 significant Rb synthetic lethal effects identified from the MP test analysis of 373 non-breast cancer TCLs in the Drive study (step one in Fig. 1a). All 775 p < 0.05 effects are ranked ordered by MP test p value. SKP2 and E2F3 are highlighted. b Scatter plot illustrating RSA scores in 373 non-breast TCLs with Rb annotation for SKP2 sensitivity from the Drive data analysis. c Scatter plot of 1467 p < 0.05 significant Rb synthetic lethal effects identified from the MP test analysis of 467 non-breast TCLs in the Achilles study (step one in Fig. 1a). All 1467 p < 0.05 effects are ranked ordered by MP test p value. SKP2 and E2F3 are highlighted. d Scatter plots illustrating Demeter scores in 1467 non-breast TCLs with Rb annotation for SKP2 sensitivity from the Achilles data analysis. e, f Scatter plots illustrating RSA and Demeter scores in 63 and 115 lung TCLs with Rb annotation for SKP2 sensitivity from the Drive and Achilles studies, respectively. g Scatter plot of intersect of cell line between the two data sets showing SKP2 sensitivity in Drive RSA scores (x axis) and Achilles Demeter scores (y axis) for selected histologies with only a single Rb-defective line. This graph illustrates a trend between Rb defects and sensitivity to SKP2 shRNA across seven different histotypes
Discussion
There is now a relatively long-standing history of using genetic concepts such as synthetic lethality to identify novel therapeutic targets for the treatment of cancer [60]. In part these efforts have been successful, with synthetic lethal treatments [16] and drugs that exploit oncogene addiction effects now being approved for the treatment of the clinical disease [61]. However, despite these advances, one challenge to this approach has been in identifying highly penetrant synthetic lethal effects that associate with the presence of a molecular biomarker. Here we describe efforts to identify highly penetrant synthetic lethal effects associated with loss of the tumour suppressor Rb in TNBC. Following the classification of 42 TNBC TCLs according to Rb status, we interrogated genome-scale and focussed gene set shRNA screening data sets, identified candidate synthetic lethal effects and then used stringent filters to triage from the list of candidate synthetic lethal effects those most likely to represent highly penetrant effects. This approach allowed us to identify a number of highly penetrant synthetic lethal effects, many of which are associated with known functions of Rb and/or associated with Rb-interacting proteins. These included TAF1 and TAF1 target genes as well as members of the SCFSKP2 complex.
We note that there are considerable caveats associated with the approach we have taken to identifying highly penetrant Rb-related synthetic lethal effects in TNBC and the interpretation of the data; understanding these caveats is critical to the use of the information we provide in this work. Primary amongst these is that we have exclusively used data from in vitro genetic screens to identify synthetic lethal effects and assess their penetrance. It seems likely that some of the effects identified in our analysis only operate in the context of two-dimensional in vitro culture and are abrogated, and therefore appear less penetrant, in three-dimensional and/or in vivo settings. Such a possibility thus provides the rationale for also assessing synthetic lethal effects, and assessing their penetrance, in in vivo models of cancer. Second, there is little way of effectively estimating the true false negative rate of our approach; it is entirely possible that we have not identified real, highly penetrant, Rb-related synthetic lethal effects either because the RNA interference reagents used in genetic screens ineffectively inactive the genes they are designed to target or because some other form of gene/protein inactivation (e.g., catalytic inhibition of the target protein rather than gene silencing) is required to elicit a synthetic lethal effect. Nevertheless, the identification in multiple, independently conducted, genetic screens of the highly penetrant Rb/SKP2 synthetic lethal effect, and its recapitulation with a small-molecule inhibitor, suggests that this highly penetrant synthetic lethality effect is unlikely to be a false positive.
As well as targeting Rb-defective TNBC, the potential for using SKP2 inhibition in other cancer histologies associated with Rb defects is considerable. For example, the hyper-proliferative phenotype of Rb-depleted human retinoblastoma cells and mouse melanotrophs is dependent upon the SKP2 [48, 62] and the SKP2/CKS1 pocket inhibitor, SKPinC1, inhibits Rb/p53-defective mouse prostate tumour cell organoids [63]. Finally, large-scale shRNA screens in TCLs from a variety of cancer histologies (and our analysis described in Fig. 6) suggest that the dependency of Rb-defective TCLs upon SKP2 might extend beyond models of TNBC. Whether the high penetrance of the Rb/SKP2 synthetic lethality seen in models of TNBC extends to each of these histologies remains to be seen, but our initial analysis in Fig. 6 suggests that highly penetrant effects might also be apparent in lung cancer.
It might be interesting to speculate what characteristics might differentiate a highly penetrant synthetic lethal effect that operates in cancer from less-penetrant effects; being able to understand the factors that distinguish one from the other might eventually allow highly penetrant synthetic lethal effects in cancer to be predicted from first principles, thus reducing the reliance upon large-scale genetic screens in multiple TCLs to empirically establish penetrance. From our relatively unbiased analysis of genome-scale shRNA screens, it is perhaps striking that many of the highly penetrant Rb synthetic lethal effects we identified (e.g., SKP1, SKP2, CKS1B, COPS1, COPS2 and COPS3) have two characteristics: (i) they are closely, rather than distally, involved in controlling an essential process in highly proliferating tumour cells, namely G1 cell cycle progression by Cyclin/CDK activity; and (ii) this process is also closely, rather than distally, controlled by the synthetic lethal partner, Rb. It seems reasonable to think that synthetic lethal interactions that control essential processes in cancer cells via small-world networks (i.e., those than contain a relatively small number of nodes between synthetic lethal partners and proteins involved in essential processes) might be less likely to be reversed, and therefore more likely highly penetrant, than synthetic lethal effects that control essential processes via distal molecular mechanisms that involve larger molecular networks with multiple intervening nodes. Whether this turns out to be a general principle or not remains to be seen but already others have started to establish that many synthetic lethal effects associated with tumour suppressor genes other than Rb can be classified into a defined, and relatively small number of classes, including those between paralogs and those between genes in the same molecular pathway [23]. This suggests that some of the principles that govern how synthetic lethal effects operate in tumour cells can indeed be established.
Materials and methods
Cell lines, compounds and siRNA
All cell lines were obtained from the American Type Culture Collection and maintained according to the supplier’s instructions. Cell lines were routinely STR typed and mycoplasma tested. Cell lines were grown and transfected with individual and SMARTpool siGENOME siRNA (Dharmacon) and transfected using RNAiMAX (Invitrogen) reagent as described in ref. [21]. Transfection efficiency was monitored using positive control (siPLK1) and two different negative control siRNAs (siCON1 and siCON2; Dharmacon, catalogue numbers D-001210-01-20 and D-001206-14-20). SKPinC1 inhibitor was purchased from Tocris (no. 4817) and was solubilised as a 10 mM stock solution in dimethyl sulfoxide (DMSO).
Western blot analysis
Whole-cell protein lysates were extracted from cells by lysis with NP250 buffer (20 mM Tris (pH 7.6), 1 mM EDTA, 0.5% NP40 and 235 mM NaCl). In gene silencing experiments, cells were cultured for 48 h after siRNA transfection, at which point lysates were generated. Following a 20 min incubation at 4 °C, lysates were centrifuged and supernatents collected. Electrophoresis was performed using Novex precast Bis-Tris gels (Invitrogen) and gels were blotted onto nitrocellulose filters as described previously [64]. Blots were immunoblotted in 5% (w/v) milk at 4 °C overnight using the following primary antibodies: anti-Rb1 (1/1000 (v/v) dilution in 5% (v/v) milk, New England Biolab, 9309); anti-p16 (1/1000, abcam); anti-SKP2 (1/1000, New England Biolab, 4358); anti-p27 (1/1000, New Engand Biolab, 2552); anti-tubulin (1/1000, abcam); and anti-actin (1/1000, Santa Cruz, sc-1616). After washing, blots were incubated 1 h at room temperature with secondary antibodies (Li-COR) diluted 1/10 000 (v/v) in 5% (w/v) milk. Protein bands were visualised and quantified using the Odyssey FC imaging system and ImageStudio software (Li-COR).
Tumour and cell line expression analysis
Limma [65] was performed to identify differentially expressed genes RB1-defective vs. RB1 not altered TCLs, using data from refs. [22, 25]. A design matrix with the cell line RB1 classifications was created and a linear model was fitted to expression values of each gene. Next, an empirical Bayes method was used to obtain more precise gene-wise variability estimates between the two groups. Differential expression between groups was represented as log fold change scores with associated p-values and adjusted p-values. Publically available cell line mRNA expression data sets used in this study include Marcotte [22] and CCLE [25], as indicated.
For the analysis of TNBC tumours, mRNA expression, DNA copy number and somatic mutation profiles for TCGA tumours were downloaded from GDAC (https://gdac.broadinstitute.org/), release 2016_01_28. GISTIC v2 level 4 data were used for copy number analysis and log2 ratios were converted to genomic gains/amplifications, neutral and loss/deletion states using threshold of ±log2(3/2). Raw Metabric data files were downloaded from the European Genome-phenome Archive (EGA; study ID EGAS00000000083). The Metabric breast cancer data set was pre-processed, summarised and quantile-normalised from the raw expression files generated by Illumina BeadStudio (R packages: beadarray v2.4.2 and illuminaHuman v3.db_1.12.2). Probe to gene-level mapping was performed by keeping the most variable (standard deviation) probe. Metabric copy number calls were used as published in the original study [66].
For TCGA breast cancer cohort, previously published [67] TNBC6 and TNBC4 calls were used resulting in 140 patients with matched mRNA, copy number and mutation profiles. For the Metabric cohort, TNBC6 calls were successfully derived from TNBC subtyping portal [68] (http://cbc.mc.vanderbilt.edu/tnbc) for 187 patients with matched mRNA and copy number profiles. In the TCGA cohort, TNBC samples with RB1 mRNA Z score < −1 were regarded as Rb-low, RB1 copy number log2 ratio < −0.585 were regarded as Rb loss and samples with RB1 truncating mutations were considered as Rb inactivated. This resulted in 48 Rb-defective and 92 Rb not altered TNBC samples. In the Metabric cohort, samples with RB1 mRNA Z score < −1 were regarded as Rb-low, whilst samples with RB1 loss/deletions were regarded as Rb loss. This resulted in 55 Rb-defective and 132 Rb not altered TNBC samples.
Differential gene expression analysis on TCGA TNBC samples was performed using limma voom [65].
Association testing
The siMEM was used to measure the essentiality of genes in the Colt2 genome-wide shRNA dropout screen [22]. siMEM uses hierarchical linear regression and considers level of each shRNA to be a regression function of its initial abundance over time, baseline trend in abundance over time and difference in abundance trend between samples sharing a common feature. siMEM results are represented as the difference between the dropout rate of hairpins in the two cell line groups being compared.
For analysis of the DRIVE and Achilles shRNA screen data sets, where only a single screen time point was collected, a one-sided MP test was used to identify associations between RB1 status of cell lines and their sensitivities to shRNA targeting of genes. For each gene, the observed difference between the median of scores of RB1-defective and RB1 not altered groups of TCLs was compared. A total of one million random samples with the same sample size as in the two groups were created. The differences in the medians of the groups were calculated for each random sample and the statistical significance of the difference was determined.
Cell viability assays
In all, 500 cells per well were seeded into 384-well plates. After 24 h cells were exposed to increasing concentrations of SKPinC1 inhibitor diluted in DMSO using an Echo 550 liquid handler (Labcyte). Cells were incubated with the inhibitor for 5 days after which cell viability was estimated with CellTitre-Glo reagent (Promega). For gene silencing experiments cells were incubated for 5 days following siRNA transfection prior to viability assessment. Luminescence readings from drug-exposed cells were normalised to those from DMSO-exposed cells to calculate surviving fractions (SFs). SFs were used to derive four-parameter logistic regression dose/response curves using Graphpad Prism software, as per the manufacturer’s instructions. Apoptosis was measure using Caspase-Glo 3/7 reagent (Promega).
Statistical analysis
Unless otherwise stated all data are represented here as mean ± standard error using Graphpad Prism Software (La Jolla, CA). All conditions were performed in triplicate in at least triplicate experiments. Statistical significance was measured using either Student’s t test, Pearson coefficient correlation, Mann-Whitney U-test or two-way ANOVA. p < 0.05 was considered significant.
Electronic supplementary material
Acknowledgements
This work was funded by Breast Cancer Now and Cancer Research UK Programme Grants to CJL. CJR was a Sir Henry Wellcome Fellow jointly funded by Science Foundation Ireland, the Health Research Board and the Wellcome Trust (grant number 103049/Z/13/Z) under the SFI-HRB-Wellcome Trust Biomedical Research Partnership. We acknowledge NHS funding to the NIHR Biomedical Research Centre at the Royal Marsden Hospital. This study makes use of data generated by the Molecular Taxonomy of Breast Cancer International Consortium, which was funded by Cancer Research UK and the British Columbia Cancer Agency Branch. The results published here are based, in part, upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Colm J. Ryan, Email: Colm.RyanRyan@ucd.ie
Christopher J. Lord, Email: Chris.Lord@icr.ac.uk
Electronic supplementary material
The online version of this article (10.1038/s41388-018-0368-z) contains supplementary material, which is available to authorized users.
References
- 1.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russnes HG, Lingjaerde OC, Borresen-Dale AL, Caldas C. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. Am J Pathol. 2017;187:2152–62. doi: 10.1016/j.ajpath.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Z, Jones R, Liu JC, Deng T, Robinson T, Chung PE, et al. RB1 and p53 at the crossroad of EMT and triple-negative breast cancer. Cell Cycle. 2011;10:1563–70. doi: 10.4161/cc.10.10.15703. [DOI] [PubMed] [Google Scholar]
- 4.Jones RA, Robinson TJ, Liu JC, Shrestha M, Voisin V, Ju Y, et al. RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J Clin Invest. 2016;126:3739–57. doi: 10.1172/JCI81568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knudsen ES, McClendon AK, Franco J, Ertel A, Fortina P, Witkiewicz AK. RB loss contributes to aggressive tumor phenotypes in MYC-driven triple negative breast cancer. Cell Cycle. 2015;14:109–22. doi: 10.4161/15384101.2014.967118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson TJ, Liu JC, Vizeacoumar F, Sun T, Maclean N, Egan SE, et al. RB1 status in triple negative breast cancer cells dictates response to radiation treatment and selective therapeutic drugs. PLoS ONE. 2013;8:e78641. doi: 10.1371/journal.pone.0078641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corney DC, Flesken-Nikitin A, Choi J, Nikitin AY. Role of p53 and Rb in ovarian cancer. Adv Exp Med Biol. 2008;622:99–117. doi: 10.1007/978-0-387-68969-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higashiyama M, Doi O, Kodama K, Yokouchi H, Tateishi R. Retinoblastoma protein expression in lung cancer: an immunohistochemical analysis. Oncology. 1994;51:544–51. doi: 10.1159/000227401. [DOI] [PubMed] [Google Scholar]
- 9.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8:303–16. doi: 10.1016/S1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 10.Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, et al. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics. 2011;6:638–49. doi: 10.4161/epi.6.5.15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trere D, Brighenti E, Donati G, Ceccarelli C, Santini D, Taffurelli M, et al. High prevalence of retinoblastoma protein loss in triple-negative breast cancers and its association with a good prognosis in patients treated with adjuvant chemotherapy. Ann Oncol. 2009;20:1818–23. doi: 10.1093/annonc/mdp209. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J, Thijssen B, McDermott U, Garnett M, Wessels LF, Bernards R. Targeting the RB-E2F pathway in breast cancer. Oncogene. 2016;35:4829–35. doi: 10.1038/onc.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle. 2010;9:4153–63. doi: 10.4161/cc.9.20.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witkiewicz AK, Ertel A, McFalls J, Valsecchi ME, Schwartz G, Knudsen ES. RB-pathway disruption is associated with improved response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2012;18:5110–22. doi: 10.1158/1078-0432.CCR-12-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–8. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Searle JS, Li B, Du W. Targeting Rb mutant cancers by inactivating TSC2. Oncotarget. 2010;1:228–32. doi: 10.18632/oncotarget.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nittner D, Lambertz I, Clermont F, Mestdagh P, Kohler C, Nielsen SJ, et al. Synthetic lethality between Rb, p53 and Dicer or miR-17-92 in retinal progenitors suppresses retinoblastoma formation. Nat Cell Biol. 2012;14:958–65. doi: 10.1038/ncb2556. [DOI] [PubMed] [Google Scholar]
- 20.Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I, et al. Functional viability profiles of breast cancer. Cancer Discov. 2011;1:260–73. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell J, Ryan CJ, Brough R, Bajrami I, Pemberton HN, Chong IY, et al. Large-scale profiling of kinase dependencies in cancer cell lines. Cell Rep. 2016;14:2490–501. doi: 10.1016/j.celrep.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–76.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence RT, Perez EM, Hernandez D, Miller CP, Haas KM, Irie HY, et al. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11:630–44. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald ER, 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–92.e10. doi: 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Takada M, Ishiguro H, Nagai S, Ohtani S, Kawabata H, Yanagita Y, et al. Survival of HER2-positive primary breast cancer patients treated by neoadjuvant chemotherapy plus trastuzumab: a multicenter retrospective observational study (JBCRG-C03 study) Breast Cancer Res Treat. 2014;145:143–53. doi: 10.1007/s10549-014-2907-9. [DOI] [PubMed] [Google Scholar]
- 31.Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–89. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asghar US, Barr AR, Cutts R, Beaney M, Babina I, Sampath D, et al. Single-cell dynamics determines response to CDK4/6 inhibition in triple-negative breast cancer. Clin Cancer Res. 2017;23:5561–72. doi: 10.1158/1078-0432.CCR-17-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceron J, Rual JF, Chandra A, Dupuy D, Vidal M, van den Heuvel S. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol. 2007;7:30. doi: 10.1186/1471-213X-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernad R, Engelsma D, Sanderson H, Pickersgill H, Fornerod M. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J Biol Chem. 2006;281:19378–86. doi: 10.1074/jbc.M512585200. [DOI] [PubMed] [Google Scholar]
- 35.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–16. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–34. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, et al. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–18. doi: 10.1091/mbc.e05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z, Jin Z, Zhang X, Shen N, Wang J, Zhao Y, et al. Nup62, associated with spindle microtubule rather than spindle matrix, is involved in chromosome alignment and spindle assembly during mitosis. Cell Biol Int. 2016;40:968–75. doi: 10.1002/cbin.10633. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–31. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–14. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao Z, Ruppert S, Robbins PD. The retinoblastoma-susceptibility gene product binds directly to the human TATA-binding protein-associated factor TAFII250. Proc Natl Acad Sci USA. 1995;92:3115–9. doi: 10.1073/pnas.92.8.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao Z, Siegert JL, Ruppert S, Robbins PD. Rb interacts with TAF(II)250/TFIID through multiple domains. Oncogene. 1997;15:385–92. doi: 10.1038/sj.onc.1201204. [DOI] [PubMed] [Google Scholar]
- 43.Lin CY, Tuan J, Scalia P, Bui T, Comai L. The cell cycle regulatory factor TAF1 stimulates ribosomal DNA transcription by binding to the activator UBF. Curr Biol. 2002;12:2142–6. doi: 10.1016/S0960-9822(02)01389-1. [DOI] [PubMed] [Google Scholar]
- 44.Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306:636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 45.Bouche L, Christ CD, Siegel S, Fernandez-Montalvan AE, Holton SJ, Fedorov O, et al. Benzoisoquinolinediones as potent and selective inhibitors of BRPF2 and TAF1/TAF1L bromodomains. J Med Chem. 2017;60:4002–22. doi: 10.1021/acs.jmedchem.7b00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford TD, Tsui V, Flynn EM, Wang S, Taylor AM, Cote A, et al. Diving into the water: inducible binding conformations for BRD4, TAF1(2), BRD9, and CECR2 bromodomains. J Med Chem. 2016;59:5391–402. doi: 10.1021/acs.jmedchem.6b00264. [DOI] [PubMed] [Google Scholar]
- 47.Sdelci S, Lardeau CH, Tallant C, Klepsch F, Klaiber B, Bennett J, et al. Mapping the chemical chromatin reactivation landscape identifies BRD4-TAF1 cross-talk. Nat Chem Biol. 2016;12:504–10. doi: 10.1038/nchembio.2080. [DOI] [PubMed] [Google Scholar]
- 48.Xu XL, Singh HP, Wang L, Qi DL, Poulos BK, Abramson DH, et al. Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature. 2014;514:385–8. doi: 10.1038/nature13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li P, Xie L, Gu Y, Li J, Xie J. Roles of multifunctional COP9 signalosome complex in cell fate and implications for drug discovery. J Cell Physiol. 2017;232:1246–53. doi: 10.1002/jcp.25696. [DOI] [PubMed] [Google Scholar]
- 50.Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–5. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 51.Chung D, Dellaire G. The role of the COP9 signalosome and neddylation in DNA damage signaling and repair. Biomolecules. 2015;5:2388–416. doi: 10.3390/biom5042388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed SI. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat Rev Mol Cell Biol. 2003;4:855–64. doi: 10.1038/nrm1246. [DOI] [PubMed] [Google Scholar]
- 53.Ji P, Jiang H, Rekhtman K, Bloom J, Ichetovkin M, Pagano M, et al. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 54.Lu Z, Bauzon F, Fu H, Cui J, Zhao H, Nakayama K, et al. Skp2 suppresses apoptosis in Rb1-deficient tumours by limiting E2F1 activity. Nat Commun. 2014;5:3463. doi: 10.1038/ncomms4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu L, Grigoryan AV, Li Y, Hao B, Pagano M, Cardozo TJ. Specific small molecule inhibitors of Skp2-mediated p27 degradation. Chem Biol. 2012;19:1515–24. doi: 10.1016/j.chembiol.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haverty PM, Lin E, Tan J, Yu Y, Lam B, Lianoglou S, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–7. doi: 10.1038/nature17987. [DOI] [PubMed] [Google Scholar]
- 57.Choi W, Ochoa A, McConkey DJ, Aine M, Hoglund M, Kim WY, et al. Genetic alterations in the molecular subtypes of bladder cancer: illustration in the Cancer Genome Atlas dataset. Eur Urol. 2017;72:354–65. doi: 10.1016/j.eururo.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldhoff P, Clarke J, Smirnov I, Berger MS, Prados MD, James CD, et al. Clinical stratification of glioblastoma based on alterations in retinoblastoma tumor suppressor protein (RB1) and association with the proneural subtype. J Neuropathol Exp Neurol. 2012;71:83–9. doi: 10.1097/NEN.0b013e31823fe8f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semczuk A, Schneider-Stock R, Miturski R, Skomra D, Tomaszewski J, Roessner A, et al. RB protein expression in human endometrial carcinomas--an immunohistochemical study. Pathol Res Pract. 2000;196:41–46. doi: 10.1016/S0344-0338(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 60.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–8. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 61.Lord CJ, Ashworth A. Biology-driven cancer drug development: back to the future. BMC Biol. 2010;8:38. doi: 10.1186/1741-7007-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/- mice. Nat Genet. 2010;42:83–8. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao H, Lu Z, Bauzon F, Fu H, Cui J, Locker J, et al. p27T187A knockin identifies Skp2/Cks1 pocket inhibitors for advanced prostate cancer. Oncogene. 2017;36:60–70. doi: 10.1038/onc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 65.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS ONE. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: a subtyping tool for triple-negative breast cancer. Cancer Inform. 2012;11:147–56. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.