Abstract
The aim of this project was to identify candidate novel therapeutic targets to facilitate the treatment of COPD using machine-based learning (ML) algorithms and penalized regression models. In this study, 59 healthy smokers, 53 healthy non-smokers and 21 COPD smokers (9 GOLD stage I and 12 GOLD stage II) were included (n = 133). 20,097 probes were generated from a small airway epithelium (SAE) microarray dataset obtained from these subjects previously. Subsequently, the association between gene expression levels and smoking and COPD, respectively, was assessed using: AdaBoost Classification Trees, Decision Tree, Gradient Boosting Machines, Naive Bayes, Neural Network, Random Forest, Support Vector Machine and adaptive LASSO, Elastic-Net, and Ridge logistic regression analyses. Using this methodology, we identified 44 candidate genes, 27 of these genes had been previously been reported as important factors in the pathogenesis of COPD or regulation of lung function. Here, we also identified 17 genes, which have not been previously identified to be associated with the pathogenesis of COPD or the regulation of lung function. The most significantly regulated of these genes included: PRKAR2B, GAD1, LINC00930 and SLITRK6. These novel genes may provide the basis for the future development of novel therapeutics in COPD and its associated morbidities.
Introduction
Chronic obstructive pulmonary disease (COPD) is a progressive inflammatory disease characterized by airway obstruction and is predicted to be among the first three causes of death worldwide1,2. Clinical presentations include emphysema, small airway obstructions and chronic bronchitis. COPD has been shown to develop in 30% of smokers and smoking history, combined with reduced daily physical activity, may be the main risk factor associated with the development of COPD3. Additional risk factors in COPD, in genetically susceptible individuals, include a history of maternal smoking, second hand smoke, polluted air, maternal/paternal asthma, childhood asthma or respiratory infections and malnutrition4. Although COPD archetypically manifests itself in males, recent studies have demonstrated an increased incidence and mortality rates in females. Furthermore, female patients with COPD are more often misdiagnosed and/or underdiagnosed5,6.
From a genetic perspective, COPD is a complex disease arising from mutations in multiple alleles and the lack of integration of data in this disease has been attributed to dispersed, independent genome-wide association studies (GWAS)7. DNA microarrays now permit scientists to screen thousands of genes simultaneously in order to determine which genes are active, hyperactive or silent in normal or COPD tissue. Furthermore, network-based medicine has also been recently employed to facilitate the investigation of genomics, transcriptomics, proteomics and other “–omics” in order to better understand complex diseases, such as COPD8. However, from a biological perspective, only a only a small subset of genes identified by these methodologies will be strongly indicative of the target disease9. Therefore, in this study, we employed a novel methodology, namely machine-based learning algorithms combined with penalized regression models, in order to study genomic change in COPD in a more selective manner. Furthermore, we have also had a longstanding interest in the genetics of COPD, formally as part of a European Union consortium10–13. Here, we now extend on these initial observations.
This study was designed to apply signaling-network methodology with machine-based learning methods to better understand the genetic etiology of smoking exposure and COPD in 59 healthy smokers, 53 healthy non-smokers and 21 COPD smokers (9 of GOLD stage I and 12 of GOLD stage II) were included (Total: n = 133). Furthermore, AdaBoost Classification Trees, Decision Tree, Gradient Boosting Machines, Naive Bayes, Neural Network, Random Forest, Support Vector Machine (as machine learning algorithms) and adaptive LASSO, elastic-net, and ridge logistic regression (as statistical models) were also applied.
In summary, we identified 44 candidate genes associating with smoking exposure and the incidence/progression of COPD. We also identified 17 novel genes, which were not previously associated with COPD, the regulation of lung function or smoking exposure. The most significantly regulated of these genes included: PRKAR2B, GAD1, LINC00930, and SLITRK6. These novel genes may provide the basis for the future development of novel therapeutics in COPD and warrant further investigation and validation.
Results
Differential analysis of gene expression data
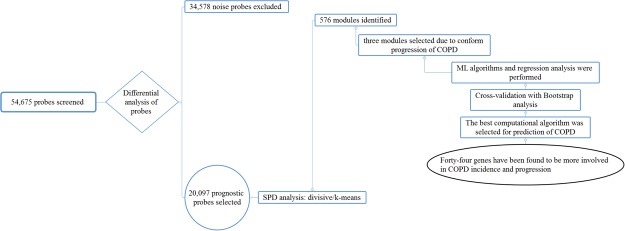
In this study, 54,675 probes were screened using the microarray dataset generated from SAE cells previously from: 59 healthy smokers, 53 healthy non-smokers and 21 COPD smokers (42.8% of GOLD stage I and 57.2% of GOLD stage II) (Table 1)14. Differential analysis was subsequently performed in order to select 20,097 probes. Subsequently, 718 probes and 544 genes (Fig. 1) were identified which were significantly changed (all p values < 0.0001) in COPD patients compared with healthy non-smokers. These genes, which include USP27X, PPP4R4, AHRR, PRKAR2B, GAD1, CYP1A1 and CYP1B1, are listed in the Supplementary File S1.
Table 1.
Basic characteristics of the study samples.
| Characteristics | COPD Smoker (N = 21) | Healthy Smoker (N = 59) | Healthy Non-smoker (N = 53) | P-value | |
|---|---|---|---|---|---|
| Age (Year)* | 50.38 ± 7.081 | 42.93 ± 7.267 | 41.0 ± 11.30 | <0.001 | |
| Smoking (pack per year)* | 36.98 ± 23.953 | 27.6 ± 16.975 | — | 0.078 | |
| FVC* | 97 ± 20 | 109 ± 13 | 107 ± 13 | 0.004 | |
| FEV1* | 74 ± 20 | 107 ± 14 | 105 ± 14 | <0.001 | |
| FEV1/FVC* | 61 ± 8 | 80 ± 5 | 81 ± 6 | <0.001 | |
| Sex+ | Male | 17 (81) | 39 (66.1) | 38 (71.7) | 0.535 |
| Female | — | — | — | Ref. | |
| Ethnic+ | Caucasian | 14 (66.6) | 14 (23.7) | 20 (37.7) | 0.038 |
| Black | — | — | — | Ref. | |
| Stage+ (Gold) of COPD | II | 12 (57.2) | — | — | NA |
| I | 9 (42.8) | — | — | Ref. | |
* indicated as mean ± standard deviation, + indicated as N (%), Ref. considered as the reference level for each categorical variable, NA: not applicable.
Figure 1.
Schematic demonstrating study plan and flowchart.
Module identification
Normalized gene expression data was used for module identification in the SPD algorithm. In total, 576 modules were identified. Three modules were biologically more related to the progression and phenotype of COPD including, 119, 242 and 324. The minimal spanning trees obtained from the SPD algorithm are shown in Fig. 2. All the genes involved in COPD progression are presented in Table 2 and then included in machine-learning and statistical modeling approaches. From these three selected modules, gene expression within two of the modules (Fig. 2a,b), associated with COPD-progression, was increased in SAE cells. In contrast, gene expression within the third module (Fig. 2c), associated with COPD-progression, was decreased in SAE cells. In Fig. 2d, classification of samples was shown based on the disease stage (dark blue = healthy non-smoker, light blue = healthy smoker, light brown = COPD stage I and dark brown = COPD stage II).
Figure 2.
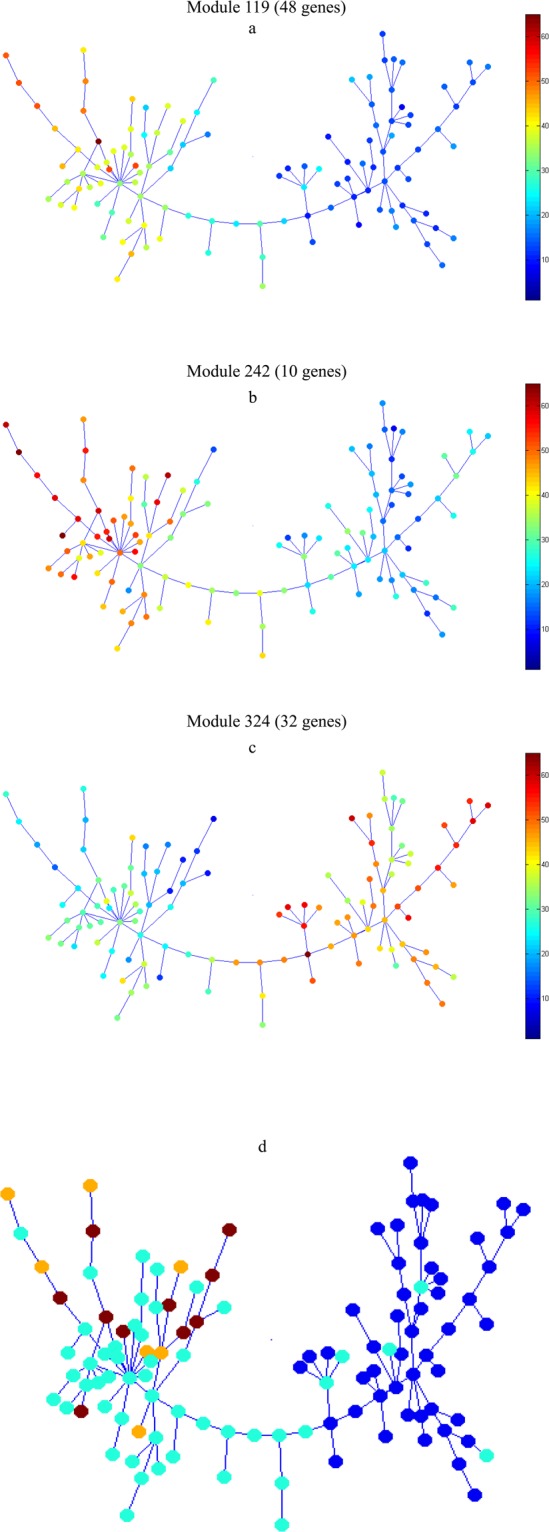
Genes involved in the progression of the COPD based on the minimal- inclusive trees were obtained from SPD algorithm (dark blue = healthy non-smoker, light blue = healthy smoker, light brown = stage I of COPD smoker and dark brown = stage II of COPD smoker).
Table 2.
List of the genes involved in the progression of COPD by sample progression discovery (SPD) algorithm.
| Related Modules with progression of COPD | Number of involved Genes | Genes Symbol |
|---|---|---|
| Module 119 | 48 | MUCL1, LOC652993, LINC00639, LINC00942, TXNRD1, CYP1B1, ME1, GAD1, CBR3, CYP1A1, NRG1, CYP4F3, AKR1B10, HTR2B, NR0B1, GRM1, ABCC3, CDRT1, AKR1C3, CBR1, TRIM9, SPP1, ADH7, FTH1P5, FTL, ADD3-AS1, AKR1C1, SLC7A11, CACNA2D3, LHX6, CABYR, HS3ST3A1, PLEKHA8P1, BACH2, SFRP2, RPSA, CLIP4, ST3GAL4-AS1, SAMD5, AHRR, ANKDD1A, LINC00589, TMCC3, RNF175, RIMKLA, LOC100652994, GPX2, LOC344887 |
| Module 242 | 10 | LINC00930, UCHL1, REEP1, EGF, CLEC11A, TMEM74B, DNHD1, C4orf48, C6orf164, JAKMIP3 |
| Module 324 | 32 | ZSCAN4, LOC338667, PRKAR2B, PLAG1, ZNF211, SCGB1A1, TLR5, KANK1, PPP4R4, THSD7A, CYB5A, GMNN, GPRC5A, PIEZO2, GFOD1, ZNF419, THSD4, CCDC37, PAPLN, GLI3, PRKAG2-AS1, PRDM11, LOC285812, SCGB3A1, USP27X, KCNA1, LOC100507560, PRDM16, SLITRK6, CYP4Z1, GPR115, RASSF10 |
Gene selection and prediction
Based on the machine-learning and statistical penalized algorithms, and after adjustment of the effect of pack per year of smoking, elastic-net logistic regression had the highest AUC (82%), sensitivity (85%), specificity (51%) and lowest misclassification error rate (25%). In reverse, decision trees method has lowest AUC (57%), sensitivity (69%), specificity (43%) and highest misclassification error rate (39%) than other algorithms. Based on the elastic-net logistic regression, the most important selected genes included, THSD4, PPP4R4, JAKMIP3, LINC00930, DNHD1, TMCC3, CCDC37, PRDM11, GLI3, ABCC3, ADH7, SAMD5, RASSF10, USP27X, GAD1, CYP1A1, NR0B1, CYP1B1, PLAG1, PIEZO2, SCGB1A1, LOC100507560.
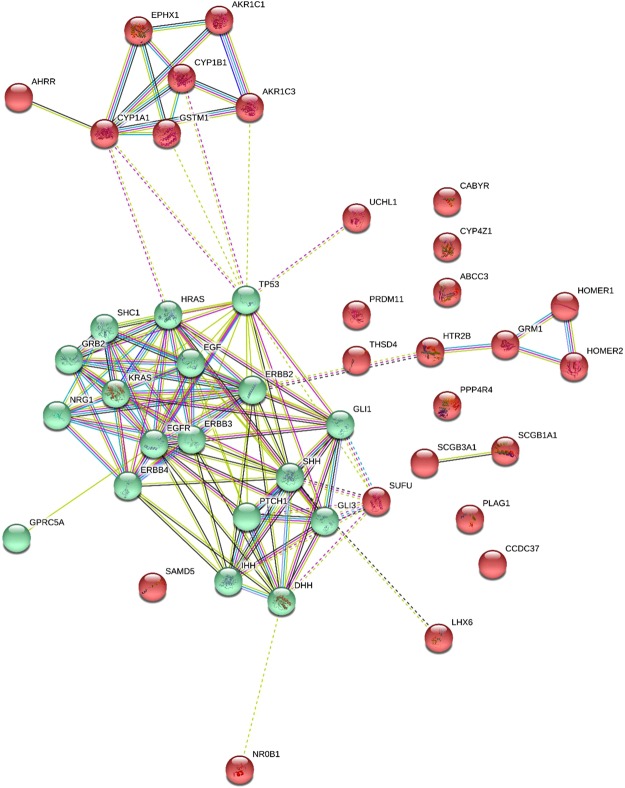
Consequently, 44 candidate genes identified here are associated with either the occurrence or progression of COPD, or lung function (Table 3). According to the results of each computational method, 44 were selected and the computational methods were hierarchically clustered, simultaneously (Figs 3 and 4). Of these 44 genes, 27 have been previously reported in the literature to be associated with COPD, lung function (FVC, FEV1 or the FEV1/FVC ratio) or other lung diseases. These 27 genes also include the genes of THSD4, PPP4R4, SCGB1A1, and NRG1, already detected in GWA studies to determine single nucleotide polymorphisms (SNPs) specifically for COPD (Table 4). Furthermore, in our study, SNPs within 4 additional genes have been detected in GWAS studies carried out previously in lung-related studies including: PRDM11 and AHRR FVC, smoking15,16, CYP1A1 childhood bronchitis17 and CYP1B1 lung cancer18. In this study, we have identified 17 genes which have not previously been detected in COPD studies, these include: LINC00942, REEP1, C6orf164, LINC00589, JAKMIP3, LINC00930, DNHD1, TMCC3, ADH7, PRKAR2B, GAD1, LOC338667, CYB5A, PIEZO2, SLITRK6, KCNA1 and LOC100507560 (Table 4). These genes may represent novel biomarkers in the diagnosis and prognosis of COPD. Figure 5 depicts the functional protein-association networks for the 44 selected genes, as shown by STRING.
Table 3.
Probes and corresponding 44 genes selected by ML algorithms and penalized regression models for association between the genes with occurrence and progression of COPD. The effect of smoking (pack per year) was adjusted in all of the methods.
| Gene Symbol | Probe ID | Number of Methods | LASSO | Adapt. LASSO | Elastic net | Ridge | SVM | GBM | NB | RF | ANN | RT | ABCT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | PPP4R4 | 233002_at | 3 | 80% | 78% | 96% | — | — | — | — | — | — | — | — |
| 2. | THSD4 | 222835_at | 2 | — | — | 90% | — | — | — | — | — | 43% | — | — |
| 3. | NRG1 | 206343_s_at | 3 | — | — | — | — | 55% | — | 55% | — | — | — | 65% |
| 4. | SCGB1A1 | 205725_at | 6 | — | — | 30% | 61% | 54% | — | 54% | — | 78% | — | 64% |
| 5. | AHRR | 229354_at | 8 | 98% | 96% | — | 77% | 76% | — | 76% | 68% | 48% | — | 76% |
| 6. | CYP1A1 | 205749_at | 11 | 90% | 82% | 20% | 65% | 73% | 11% | 72% | 74% | 43% | 77% | 73% |
| 7. | CYP1B1 | 202437_s_at | 9 | 88% | 80% | 32% | 65% | 64% | 35% | 64% | 58% | — | — | 65% |
| 8. | PRDM11 | 229687_s_at | 1 | — | — | 50% | — | — | — | — | — | — | — | — |
| 9. | CBR3 | 205379_at | 1 | — | — | — | — | — | 14% | — | — | — | — | — |
| 10. | AKR1C1 | 217626_at | 1 | — | — | — | — | — | 10% | — | — | — | — | — |
| 11. | AKR1C3 | 209160_at | 1 | — | — | — | — | — | 5% | — | — | — | — | — |
| 12. | GRM1 | 207299_s_at | 1 | — | — | — | — | — | 4% | — | — | — | — | — |
| 13. | CYP4Z1 | 237395_at | 1 | — | — | — | — | — | — | — | 67% | — | — | — |
| 14. | UCHL1 | 201387_s_at | 1 | — | — | — | — | — | — | — | 57% | — | — | — |
| 15. | CABYR | 219928_s_at | 1 | — | — | — | — | — | — | — | 54% | — | — | — |
| 16. | GPRC5A | 203108_at | 2 | 100% | 100% | — | — | — | — | — | — | — | — | — |
| 17. | CCDC37 | 243758_at | 1 | — | — | 50% | — | — | — | — | — | — | — | — |
| 18. | GLI3 | 227376_at | 3 | — | — | 38% | — | — | 12% | — | — | 43% | — | — |
| 19. | ABCC3 | 208161_s_at | 3 | — | — | 30% | — | — | — | — | 58% | 52% | — | — |
| 20. | SAMD5 | 228653_at | 3 | — | — | 24% | — | — | 41% | — | 57% | — | — | — |
| 21. | RASSF10 | 238755_at | 5 | — | — | 23% | — | 75% | — | 75% | 68% | 64% | — | — |
| 22. | USP27X | 230620_at | 11 | 99% | 94% | 31% | 100% | 100% | 100% | 100% | 100% | 49% | 100% | 100% |
| 23. | HTR2B | 206638_at | 1 | — | — | — | — | — | 5% | — | — | — | — | — |
| 24. | NR0B1 | 206645_s_at | 5 | — | — | 33% | — | 66% | — | 66% | — | 58% | — | 66% |
| 25. | PLAG1 | 205372_at | 5 | — | — | 26% | 61% | 61% | — | 61% | — | — | — | 61% |
| 26. | SCGB3A1 | 230378_at | 5 | — | — | — | 65% | 58% | — | 58% | 65% | — | — | 58% |
| 27. | LHX6 | 219884_at | 1 | — | — | — | 55% | — | — | — | — | — | — | — |
| 28. | LINC00942 | 1558308_at | 1 | — | — | — | — | — | — | — | — | 52% | — | — |
| 29. | REEP1 | 204364_s_at | 1 | — | — | — | — | — | — | — | — | 45% | — | — |
| 30. | C6orf164 | 230506_at | 1 | — | — | — | — | — | 44% | — | — | — | — | — |
| 31. | LINC00589 | 232718_at | 1 | — | — | — | — | — | 13% | — | — | — | — | — |
| 32. | JAKMIP3 | 233076_at | 4 | — | — | 100% | — | — | 64% | — | 98% | 56% | — | — |
| 33. | LINC00930 | 1556768_at | 3 | — | — | 78% | — | — | 4% | — | — | 100% | — | — |
| 34. | DNHD1 | 229631_at | 1 | — | — | 53% | — | — | — | — | — | — | — | — |
| 35. | TMCC3 | 235146_at | 7 | — | — | 52% | — | 82% | 87% | 82% | 64% | 73% | 84% | — |
| 36. | ADH7 | 210505_at | 3 | — | — | 27% | — | — | 27% | — | — | 54% | — | — |
| 37. | PRKAR2B | 203680_at | 7 | 96% | 96% | — | 76% | 74% | — | 73% | 76% | — | — | 74% |
| 38. | GAD1 | 205278_at | 9 | — | — | 23% | 74% | 67% | 48% | 67% | 73% | 46% | 84% | 67% |
| 39. | LOC338667 | 1564786_at | 3 | — | — | — | — | 65% | — | 65% | — | 43% | — | — |
| 40. | CYB5A | 217021_at | 6 | — | — | — | 65% | 63% | 3% | 63% | 87% | — | — | 64% |
| 41. | PIEZO2 | 219602_s_at | 6 | — | — | 56% | 65% | 60% | — | 60% | — | 68% | — | 60% |
| 42. | SLITRK6 | 235976_at | 4 | — | — | — | 58% | 57% | — | 57% | — | — | — | 57% |
| 43. | KCNA1 | 230849_at | 3 | — | — | — | — | 52% | — | 53% | — | — | — | 53% |
| 44. | LOC100507560 | 231379_at | 9 | — | — | 38% | 48% | 74% | 82% | 74% | 62% | 41% | 100% | 50% |
| AUC% Sensitivity (SD) Specificity (SD) Misclassification Error Rate (SD) |
79% | 74% | 82% | 76.6% | 61.6% | 76% | 77% | 80% | 70% | 57% | 74.7% | |||
| 0.83 (0.14) | 0.81 (0.16) | 0.85 (0.13) | 1 | 0.92 (0.10) | 0.98 (0.04) | 0.84 (0.12) | 0.95 (0.08) | 0.68 (0.17) | 0.69 (0.20) | 0.81 (0.14) | ||||
| 0.5 (0.30) | 0.37 (0.10) | 0.51 (0.29) | 0 | 0.15 (0.13) | 0.02 (0.07) | 0.49 (0.26) | 0.07 (0.15) | 0.66 (0.24) | 0.43 (0.24) | 0.39 (0.14) | ||||
| 0.27 (0.14) | 0.31 (0.15) | 0.25 (0.10) | 0.30 (0.03) | 0.31 (0.06) | 0.30 (0.05) | 0.26 (0.09) | 0.31 (0.09) | 0.32 (0.13) | 0.39 (0.12) | 0.31 (0.11) | ||||
Important index (value) for each gene in any method was reported. The third column indicated number of studies that it confirmed the association of each gene with progression of the COPD. Third column indicated sum of number of methods that it confirmed each gene (Range score: 0 to 11).
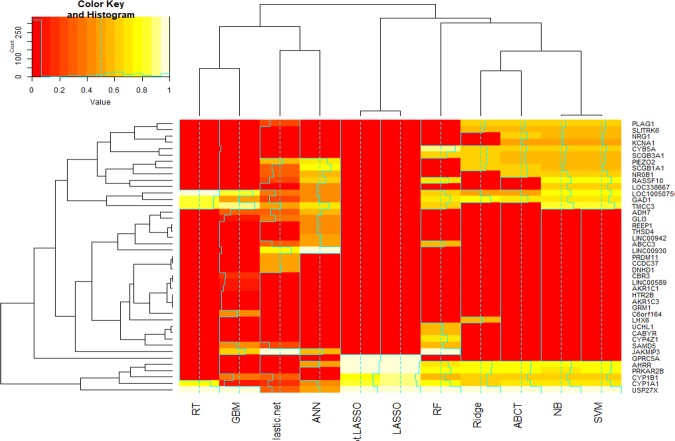
Figure 3.
Interactive cluster heatmap displaying importance index of the forty-four candidate genes (as columns) in each of the machine learning and statistical methods (as rows), rows and columns of the heatmap have been reordered according to a hierarchical clustering, represented by the dendrogram, colors represent importance index of the genes (red to yellow: lower to higher of importance value).
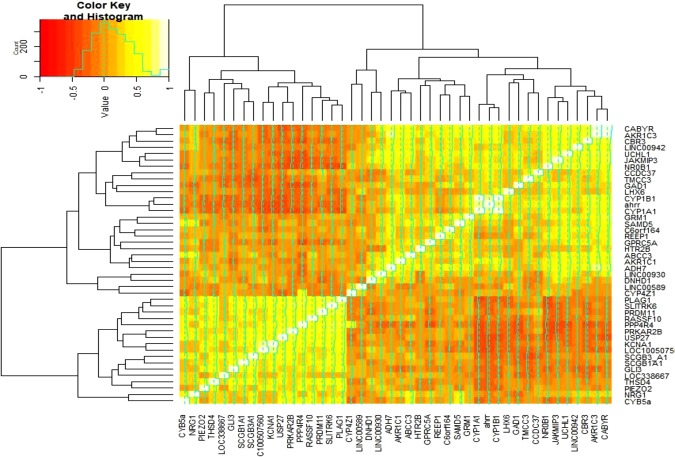
Figure 4.
Spearman’s rank correlation, co-expression, matrix between the selected genes: heatmap for hierarchical clustering the forty-four candidate genes based on their pattern of gene expression.
Table 4.
Confirmation of the association of selected genes with COPD/or lung function by literature reviewing in PubMed databank with ((“COPD” OR “Lung Function”) AND “name of each selected gene”).
| Gene Symbol | Probe ID | Number of studies | References (PMIDs) | |
|---|---|---|---|---|
| 1. | PPP4R4 | 233002_at | 1 | 28170284 |
| 2. | THSD4 | 222835_at | 6 | 27564456, 24286382, 23932459, 22461431, 21965014, 20010834 |
| 3. | NRG1 | 206343_s_at | 15 | 28950338, 28901268, 28604730, 28396363, 28391773, 27626312, 26837769, 26200269, 25870798, 25531467, 25501131, 25384085, 24469108, 23390248, 22665269 |
| 4. | SCGB1A1 | 205725_at | 4 | 27081700, 26937342, 26159408, 23144326 |
| 5. | AHRR | 229354_at | 9 | 28854564, 29262847, 28100713, 28056099, 27924164, 27632354, 26667048, 22232023, 18172554 |
| 6. | CYP1A1 | 205749_at | 108 | 29212267, 29076184, 28827732, 28283091, and etc. |
| 7. | CYP1B1 | 202437_s_at | 38 | 29110844, 28858732, and etc. |
| 8. | PRDM11 | 229687_s_at | 1 | 28938616 |
| 9. | CBR3 | 205379_at | 1 | 26916823 |
| 10. | AKR1C1 | 217626_at | 8 | 29344298, 28210161, 26338969, 24976539, 23534707, 23474755, 17266043, 16915569 |
| 11. | AKR1C3 | 209160_at | 7 | 23534707, 28704416, 27629782, 25603868, 23665002, 23519145, 15284179 |
| 12. | HTR2B | 206638_at | 1 | 27301951 |
| 13. | GRM1 | 207299_s_at | 1 | 23303475 |
| 14. | CYP4Z1 | 237395_at | 1 | 19473719 |
| 15. | UCHL1 | 201387_s_at | 5 | 28688920, 25615526, 23534707, 21143527, 17108109 |
| 16. | CABYR | 219928_s_at | 5 | 26938915, 26843620, 24362251, 17317841, 21274509 |
| 17. | GPRC5A | 203108_at | 10 | 29382653, 28849235, 28088789, 26447616, 25621293, 25311788, 23154545, 22239913, 20686609, 20563252 |
| 18. | CCDC37 | 243758_at | 2 | 26200272, 22011669 |
| 19. | GLI3 | 227376_at | 3 | 27146893, 23736020, 23667589 |
| 20. | ABCC3 | 208161_s_at | 4 | 24176985, 23369236, 22699933, 19107936 |
| 21. | SAMD5 | 228653_at | 1 | 25411851 |
| 22. | RASSF10 | 238755_at | 1 | 24433832 |
| 23. | USP27X | 230620_at | 1 | 27013495 |
| 24. | NR0B1 | 206645_s_at | 1 | 28965760 |
| 25. | PLAG1 | 205372_at | 2 | 29305497, 29249655 |
| 26. | SCGB3A1 | 230378_at | 5 | 26937342, 21636547, 20849603, 20660313, 19334046 |
| 27. | LHX6 | 219884_at | 4 | 28900494, 28396596, 27610375, 24157876 |
| 28. | LINC00942 | 1558308_at | 0 | — |
| 29. | REEP1 | 204364_s_at | 0 | — |
| 30. | C6orf164 | 230506_at | 0 | — |
| 31. | LINC00589 | 232718_at | 0 | — |
| 32. | JAKMIP3 | 233076_at | 0 | — |
| 33. | LINC00930 | 1556768_at | 0 | — |
| 34. | DNHD1 | 229631_at | 0 | — |
| 35. | TMCC3 | 235146_at | 0 | — |
| 36. | ADH7 | 210505_at | 0 | — |
| 37. | PRKAR2B | 203680_at | 0 | — |
| 38. | GAD1 | 205278_at | 0 | — |
| 39. | LOC338667 | 1564786_at | 0 | — |
| 40. | CYB5A | 217021_at | 0 | — |
| 41. | PIEZO2 | 219602_s_at | 0 | — |
| 42. | SLITRK6 | 235976_at | 0 | — |
| 43. | KCNA1 | 230849_at | 0 | — |
| 44. | LOC100507560 | 231379_at | 0 | — |
Figure 5.
STRING protein–protein interaction networks for the forty-four candidate genes.
Investigation of the differential expression of genes in healthy non-smokers (HNS; control subjects), healthy smokers, COPD patients, and COPD Stage I and II patients
In this study, we also investigated the differential expression of our 44 candidate genes in healthy non-smokers (HNS; control subjects; n = 53), healthy smokers (HS; n = 59), COPD patients (n = 21), and COPD stage I (COPD I; n = 9) and II (COPD II; n = 12) patients, respectively. We investigated the differential gene expression between HNS and HS and found significant differences in expression in 39/44 (88.6%) of all genes. In addition, 16/17 (94.1%) of the genes, not previously detected associating with COPD or lung function, were differentially expressed (Table 5; column HS v HNS). We then investigated the differential expression of these 44 genes in HS and COPD patients. Here, 24/44 (54.5%) of all genes studies were significantly regulated. Furthermore, 10/17 previously undetected genes in COPD/lung function were differentially regulated (Table 5; column COPD v HS). Finally, we investigated the regulation of these 44 genes in COPD Stage I and II patients compared with HS (Table 5; columns stage I v HS and stage II v HS). Here, we observed that 5/44 (11.4%; COPD stage I) and 16/44 (36.3%; COPD stage II) were differentially regulated. Among the previously undetected genes in COPD/lung function, 10/17 (58.8%) and 6/17 (35.3%) were significantly different in COPD stage I and II, respectively, compared with HS. A number of genes were significantly different in all four analyses (HS v HNS; HS v COPD; HS v COPD I; HS v COPD II), including: USP27X, AHRR, CYP1A1 and CYP1B1. Interestingly, of these genes, not previously identified to associate with COPD/lung function, PRKAR2B and GAD1 were significantly different in all four analyses. Therefore, this study reveals for the first time the potential role of PRKAR2B and GAD1 in COPD and smoking-related dysfunction in lung.
Table 5.
Relative expression of 44 candidate genes in healthy controls (smokers and non-smokers) and COPD smoker patients (stage I and stage II).
| Gene Symbol | Healthy Non-Smoker (N = 53) | Healthy Smoker (N = 59) | COPD smoker (N = 21) | COPD stage I smoker (N = 9) | COPD stage II smoker (N = 12) | Fold Regulation, adjusted p-value (HS vs. HNS) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | PPP4R4 | 6.56 ± 0.75 | 6.01 ± 0.88 | 5.56 ± 0.69 | 5.88 ± 0.44 | 5.32 ± 0.77 | −1.091, 0.004 | −1.081, 0.026 | −1.022, 0.99 | −1.130, 0.036 |
| 2. | THSD4 | 7.59 ± 0.50 | 7.34 ± 0.54 | 7.11 ± 0.51 | 7.47 ± 0.31 | 6.84 ± 0.46 | −1.034, 0.052 | −1.032, 0.113 | 1.018, 0.99 | −1.073, 0.013 |
| 3. | NRG1 | 3.79 ± 0.25 | 4.05 ± 0.47 | 3.80 ± 0.38 | 3.94 ± 0.48 | 3.69 ± 0.25 | 1.069, 0.002 | −1.066, 0.005 | −1.028, 0.99 | −1.097, 0.021 |
| 4. | SCGB1A1 | 14.48 ± 0.15 | 14.36 ± 0.20 | 14.15 ± 0.32 | 14.18 ± 0.24 | 14.11 ± 0.37 | −1.008, 0.015 | −1.015, 0.002 | −1.013, 0.103 | −1.018, 0.002 |
| 5. | AHRR | 3.86 ± 0.16 | 4.58 ± 0.65 | 5.23 ± 0.76 | 5.42 ± 0.81 | 5.08 ± 0.71 | 1.186, 0.001 | 1.142, 0.001 | 1.183, 0.001 | 1.109, 0.025 |
| 6. | CYP1A1 | 4.61 ± 0.24 | 6.12 ± 1.84 | 7.72 ± 1.80 | 8.04 ± 1.52 | 7.48 ± 2.02 | 1.327, 0.001 | 1.261, 0.001 | 1.314, 0.003 | 1.222, 0.018 |
| 7. | CYP1B1 | 3.64 ± 0.51 | 7.74 ± 2.04 | 9.50 ± 1.16 | 9.92 ± 0.59 | 9.19 ± 1.40 | 2.126, 0.001 | 1.227, 0.002 | 1.282, 0.003 | 1.187, 0.045 |
| 8. | PRDM11 | 5.67 ± 0.36 | 5.35 ± 0.32 | 5.28 ± 0.29 | 5.34 ± 0.22 | 5.23 ± 0.33 | −1.059, 0.001 | −1.013, 0.363 | −1.002, 0.99 | −1.023, 0.99 |
| 9. | CBR3 | 7.23 ± 0.43 | 8.15 ± 0.68 | 8.35 ± 0.64 | 8.47 ± 0.50 | 8.26 ± 0.73 | 1.127, 0.001 | 1.025, 0.287 | 1.039, 0.819 | 1.013, 0.99 |
| 10. | AKR1C1 | 6.82 ± 0.68 | 8.56 ± 1.11 | 8.32 ± 1.20 | 8.44 ± 1.23 | 8.22 ± 1.21 | 1.255, 0.001 | −1.029, 0.542 | −1.014, 0.99 | −1.041, 0.99 |
| 11. | AKR1C3 | 10.30 ± 0.43 | 11.85 ± 0.72 | 11.96 ± 0.66 | 12.20 ± 0.47 | 11.78 ± 0.74 | 1.150, 0.001 | 1.009, 0.748 | 1.029, 0.783 | −1.006, 0.99 |
| 12. | HTR2B | 3.92 ± 0.21 | 4.21 ± 0.30 | 4.25 ± 0.31 | 4.19 ± 0.40 | 4.29 ± 0.23 | 1.074, 0.001 | 1.010, 0.618 | −1.005, 0.99 | 1.019, 0.99 |
| 13. | GRM1 | 3.74 ± 0.11 | 4.04 ± 0.34 | 4.16 ± 0.55 | 4.21 ± 0.36 | 4.13 ± 0.66 | 1.080, 0.001 | 1.030, 0.571 | 1.042, 0.99 | 1.022, 0.99 |
| 14. | CYP4Z1 | 6.83 ± 0.57 | 6.24 ± 0.57 | 5.92 ± 0.38 | 5.89 ± 0.45 | 5.94 ± 0.33 | −1.094, 0.001 | −1.054, 0.02 | −1.059, 0.445 | −1.050, 0.404 |
| 15. | UCHL1 | 5.30 ± 0.56 | 8.79 ± 1.50 | 9.27 ± 1.81 | 9.24 ± 1.69 | 9.28 ± 1.97 | 1.658, 0.001 | 1.055, 0.183 | 1.051, 0.99 | 1.055, 0.97 |
| 16. | CABYR | 4.86 ± 0.28 | 6.92 ± 1.23 | 7.46 ± 1.37 | 7.98 ± 1.13 | 7.09 ± 1.46 | 1.424, 0.001 | 1.078, 0.162 | 1.153, 0.003 | 1.024, 0.99 |
| 17. | GPRC5A | 7.58 ± 0.63 | 7.50 ± 0.43 | 8.01 ± 0.61 | 7.89 ± 0.71 | 8.10 ± 0.54 | −1.010, 0.99 | 1.068, 0.001 | 1.052, 0.262 | 1.080, 0.005 |
| 18. | CCDC37 | 9.44 ± 0.54 | 9.35 ± 0.53 | 9.26 ± 0.63 | 9.21 ± 0.80 | 9.29 ± 0.50 | −1.009, 0.99 | −1.010, 0.381 | −1.015, 0.99 | −1.006, 0.99 |
| 19. | GLI3 | 7.59 ± 0.38 | 6.74 ± 0.57 | 6.62 ± 0.38 | 6.69 ± 0.43 | 6.56 ± 0.34 | −1.126, 0.001 | −1.018, 0.292 | −1.007, 0.99 | −1.027, 0.99 |
| 20. | ABCC3 | 6.95 ± 0.44 | 7.88 ± 0.61 | 7.62 ± 0.78 | 7.68 ± 0.76 | 7.57 ± 0.82 | 1.134, 0.001 | −1.034, 0.226 | −1.026, 0.99 | −1.041, 0.733 |
| 21. | SAMD5 | 3.74 ± 0.14 | 3.98 ± 0.28 | 3.93 ± 0.49 | 4.0 ± 0.28 | 3.87 ± 0.62 | 1.064, 0.001 | −1.013, 0.196 | 1.005, 0.99 | −1.028, 0.99 |
| 22. | RASSF10 | 7.67 ± 0.49 | 7.06 ± 0.59 | 6.62 ± 0.55 | 6.80 ± 0.31 | 6.47 ± 0.66 | −1.086, 0.001 | −1.066, 0.001 | −1.038, 0.99 | −1.091, 0.006 |
| 23. | USP27X | 7.43 ± 0.28 | 7.13 ± 0.40 | 6.65 ± 0.40 | 6.70 ± 0.27 | 6.60 ± 0.48 | −1.042, 0.001 | −1.072, 0.001 | −1.064, 0.007 | −1.080, 0.001 |
| 24. | NR0B1 | 3.93 ± 0.24 | 4.39 ± 0.77 | 4.76 ± 0.77 | 4.87 ± 0.89 | 4.67 ± 0.69 | 1.117, 0.001 | 1.084, 0.025 | 1.109, 0.152 | 1.064, 0.960 |
| 25. | PLAG1 | 5.79 ± 0.56 | 5.25 ± 0.55 | 4.81 ± 0.54 | 4.95 ± 0.54 | 4.70 ± 0.54 | −1.103, 0.001 | −1.091, 0.002 | −1.060, 0.743 | −1.117, 0.025 |
| 26. | SCGB3A1 | 14.21 ± 0.41 | 13.92 ± 0.60 | 13.16 ± 0.83 | 13.53 ± 0.91 | 12.87 ± 0.65 | −1.021, 0.061 | −1.058, 0.001 | −1.029, 0.412 | −1.081, 0.001 |
| 27. | LHX6 | 5.36 ± 0.34 | 5.80 ± 0.44 | 6.12 ± 0.51 | 6.11 ± 0.51 | 6.12 ± 0.53 | 1.082, 0.001 | 1.055, 0.029 | 1.053, 0.246 | 1.055, 0.147 |
| 28. | LINC00942 | 3.58 ± 0.17 | 3.87 ± 0.71 | 4.11 ± 0.99 | 4.24 ± 1.23 | 4.01 ± 0.79 | 1.081, 0.035 | 1.062, 0.258 | 1.096, 0.251 | 1.036, 0.99 |
| 29. | REEP1 | 9.34 ± 0.63 | 9.82 ± 0.56 | 9.61 ± 0.77 | 9.38 ± 0.93 | 9.78 ± 0.62 | 1.051, 0.001 | −1.022, 0.329 | −1.047, 0.408 | −1.004, 0.99 |
| 30. | C6orf164 | 5.49 ± 0.50 | 6.09 ± 0.55 | 6.21 ± 0.61 | 6.17 ± 0.43 | 6.23 ± 0.73 | 1.109, 0.001 | 1.020, 0.315 | 1.013, 0.99 | 1.023, 0.99 |
| 31. | LINC00589 | 5.15 ± 0.26 | 5.38 ± 0.31 | 5.40 ± 0.31 | 5.33 ± 0.17 | 5.44 ± 0.38 | 1.044, 0.001 | 1.004, 0.706 | −1.009, 0.99 | 1.011, 0.99 |
| 32. | JAKMIP3 | 4.51 ± 0.21 | 5.27 ± 0.61 | 5.68 ± 0.85 | 5.71 ± 0.98 | 5.65 ± 0.79 | 1.168, 0.001 | 1.078, 0.048 | 1.083, 0.099 | 1.072, 0.160 |
| 33. | LINC00930 | 6.01 ± 0.43 | 6.91 ± 0.57 | 6.54 ± 0.67 | 6.54 ± 0.82 | 6.53 ± 0.58 | 1.150, 0.001 | −1.057, 0.059 | −1.056, 0.503 | −1.058, 0.186 |
| 34. | DNHD1 | 6.64 ± 0.48 | 7.14 ± 0.53 | 7.14 ± 0.74 | 7.14 ± 0.92 | 7.13 ± 0.61 | 1.075, 0.001 | 1.000, 0.912 | 1, 0.99 | −1.001, 0.99 |
| 35. | TMCC3 | 3.82 ± 0.38 | 4.09 ± 0.48 | 4.45 ± 0.43 | 4.49 ± 0.47 | 4.42 ± 0.42 | 1.071, 0.007 | 1.088, 0.001 | 1.098, 0.088 | 1.081, 0.149 |
| 36. | ADH7 | 8.01 ± 0.93 | 10.81 ± 0.69 | 10.70 ± 0.62 | 10.81 ± 0.56 | 10.61 ± 0.68 | 1.350, 0.001 | −1.010, 0.287 | 1, 0.99 | −1.019, 0.99 |
| 37. | PRKAR2B | 7.35 ± 0.58 | 6.45 ± 0.72 | 5.86 ± 0.59 | 5.89 ± 0.66 | 5.83 ± 0.56 | −1.139, 0.001 | −1.101, 0.001 | −1.095, 0.096 | −1.106, 0.015 |
| 38. | GAD1 | 4.91 ± 0.61 | 6.31 ± 1.0 | 7.25 ± 0.86 | 7.07 ± 1.10 | 7.39 ± 0.66 | 1.285, 0.001 | 1.149, 0.001 | 1.120, 0.076 | 1.171, 0.001 |
| 39. | LOC338667 | 5.39 ± 0.32 | 5.19 ± 0.23 | 5.04 ± 0.25 | 5.03 ± 0.17 | 5.04 ± 0.30 | −1.038, 0.002 | −1.030, 0.014 | −1.032, 0.647 | −1.030, 0.675 |
| 40. | CYB5A | 4.72 ± 0.19 | 4.71 ± 0.18 | 4.53 ± 0.24 | 4.62 ± 0.23 | 4.45 ± 0.23 | −1.003, 0.99 | −1.040, 0.002 | −1.019, 0.99 | −1.058, 0.010 |
| 41. | PIEZO2 | 7.24 ± 0.85 | 6.48 ± 0.82 | 5.89 ± 0.63 | 5.77 ± 0.72 | 5.96 ± 0.58 | −1.117, 0.001 | −1.100, 0.004 | −1.123, 0.090 | −1.087, 0.225 |
| 42. | SLITRK6 | 7.97 ± 0.61 | 6.86 ± 0.84 | 6.24 ± 0.78 | 6.31 ± 1.04 | 6.18 ± 0.55 | −1.162, 0.001 | −1.099, 0.004 | −1.087, 0.322 | −1.110, 0.041 |
| 43. | KCNA1 | 6.87 ± 1.03 | 5.79 ± 0.94 | 5.19 ± 0.77 | 5.49 ± 0.89 | 4.97 ± 0.51 | −1.186, 0.001 | −1.116, 0.011 | −1.054, 0.99 | −1.165, 0.043 |
| 44. | LOC100507560 | 5.85 ± 0.69 | 5.42 ± 0.61 | 4.92 ± 0.48 | 4.92 ± 0.55 | 4.91 ± 0.44 | −1.079, 0.002 | −1.102, 0.001 | −1.102, 0.155 | −1.104, 0.080 |
The Adj. P is based on the marginally adjusted 𝑝 values by the Benjamini-Hochberg-FDR correction at α = 0.05; Median ± Interquartile range.
Investigation of the gender effect on differential gene expression in HNS, HS and COPD (Stage I and II) patients
Here we examined the effects of gender on the expression of our 44 candidate genes. We demonstrated that the expression of 40/44 (90.9%) of these genes is significantly different in HS men compared to HNS men (Table 6; HS v HNS). In addition, 15/17 (88.2%) of the novel genes previously undetected in COPD/lung function had significantly different expression levels (Table 6; HS v HNS) in men. Investigation of the expression levels of the 44 candidate genes in men with COPD versus HS revealed that 21/44 (47.7%) of genes were significantly different (Table 6; COPD v HS) and 10/17 (58.8%) of previously undetected genes in COPD/lung function were also significantly different. When HS were compared to COPD Stage I and II patients, respectively, 4/44 (Stage I; 9.0%) and 7/44 (Stage II; 15.9%) of the total candidate genes were significantly different in male HS compared to HNS. Of the 17 novel genes detected in this study, 1/17 (Stage I; 5.9%) and 3/17 (Stage II; 17.6%) were significantly different in males compared to HS (Table 6; Stage I or Stage II v HS). A number of the 44 candidate genes were significantly different in males across all four analyses, these included USP27X, AHRR, and the novel gene, JAKMIP3.
Table 6.
Comparison of relative expression of 44 candidate genes between healthy controls (smokers and non-smokers) and COPD smoker patients (stage I and stage II) in men and women groups separately.
| Sex group | Gene Symbol | Healthy Non-Smoker (N = 37) | Healthy Smoker (N = 38) | COPD smoker (N = 17) | COPD stage I smoker (N = 9) | COPD stage II smoker (N = 8) | Fold Regulation, adjusted p-value (HS vs. HNS) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Men | 1. | PPP4R4 | 6.45 ± 0.77 | 6.10 ± 0.84 | 5.66 ± 0.65 | 5.88 ± 0.44 | 5.41 ± 0.79 | −1.057, 0.395 | −1.078, 0.061 | −1.038, 0.99 | −1.128, 0.14 |
| 2. | THSD4 | 7.61 ± 0.49 | 7.28 ± 0.49 | 7.22 ± 0.44 | 7.47 ± 0.31 | 6.95 ± 0.41 | −1.044, 0.022 | −1.008, 0.61 | 1.026, 0.99 | −1.048, 0.40 | |
| 3. | NRG1 | 3.76 ± 0.22 | 4.04 ± 0.51 | 3.82 ± 0.39 | 3.94 ± 0.48 | 3.69 ± 0.23 | 1.073, 0.010 | −1.058, 0.032 | −1.024, 0.99 | −1.095, 0.10 | |
| 4. | SCGB1A1 | 14.49 ± 0.13 | 14.35 ± 0.21 | 14.17 ± 0.32 | 14.19 ± 0.25 | 14.15 ± 0.41 | −1.010, 0.030 | −1.013, 0.035 | −1.011, 0.260 | −1.014, 0.138 | |
| 5. | AHRR | 3.87 ± 0.17 | 4.67 ± 0.67 | 5.33 ±± 0.79 | 5.43 ± 0.81 | 5.23 ± 0.81 | 1.206, 0.001 | 1.141, 0.003 | 1.162, 0.002 | 1.120, 0.064 | |
| 6. | CYP1A1 | 4.63 ± 0.24 | 6.31 ± 1.84 | 7.73 ± 1.61 | 8.04 ± 1.52 | 7.40 ± 1.75 | 1.364, 0.001 | 1.225, 0.005 | 1.274, 0.011 | 1.172, 0.318 | |
| 7. | CYP1B1 | 3.70 ± 0.56 | 8.04 ± 2.05 | 9.68 ± 0.88 | 9.92 ± 0.60 | 9.42 ± 1.11 | 2.174, 0.001 | 1.204, 0.013 | 1.234, 0.019 | 1.172, 0.246 | |
| 8. | PRDM11 | 5.67 ± 0.35 | 5.33 ± 0.34 | 5.31 ± 0.30 | 5.35 ± 0.22 | 5.27 ± 0.39 | −1.062, 0.001 | −1.004, 0.827 | 1.003, 0.99 | −1.012, 0.99 | |
| 9. | CBR3 | 7.27 ± 0.44 | 8.27 ± 0.65 | 8.44 ± 0.64 | 8.48 ± 0.50 | 8.40 ± 0.80 | 1.137, 0.001 | 1.021, 0.392 | 1.026, 0.99 | 1.017, 0.99 | |
| 10. | AKR1C1 | 6.74 ± 0.62 | 8.61 ± 1.10 | 8.55 ± 1.09 | 8.44 ±± 1.24 | 8.67 ± 0.97 | 1.277, 0.001 | −1.007, 0.884 | −1.019, 0.99 | 1.007, 0.99 | |
| 11. | AKR1C3 | 10.28 ± 0.41 | 11.9 ± 50.76 | 12.06 ± 0.60 | 12.20 ± 0.47 | 11.91 ± 0.73 | 1.162, 0.001 | 1.013, 0.956 | 1.021, 0.99 | −1.004, 0.99 | |
| 12. | HTR2B | 3.86 ± 0.17 | 4.26 ± 0.30 | 4.24 ± 0.32 | 4.19 ± 0.40 | 4.30 ± 0.21 | 1.104, 0.001 | −1.005, 0.884 | −1.016, 0.99 | 1.009, 0.99 | |
| 13. | GRM1 | 3.74 ± 0.12 | 4.06 ± 0.35 | 4.19 ± 0.59 | 4.20 ± 0.37 | 4.17 ± 0.81 | 1.083, 0.002 | 1.032, 0.478 | 1.036, 0.99 | 1.028, 0.99 | |
| 14. | CYP4Z1 | 6.81 ± 0.59 | 6.18 ± 0.54 | 5.94 ± 0.35 | 5.90 ± 0.45 | 5.99 ± 0.21 | −1.102, 0.002 | −1.040, 0.101 | −1.048, 0.88 | −1.032, 0.99 | |
| 15. | UCHL1 | 5.30 ± 0.62 | 8.67 ± 1.37 | 9.46 ± 1.63 | 9.25 ± 1.69 | 9.72 ± 1.63 | 1.635, 0.001 | 1.091, 0.061 | 1.066, 0.99 | 1.121, 0.13 | |
| 16. | CABYR | 4.86 ± 0.28 | 7.14 ± 1.18 | 7.60 ± 1.39 | 7.98 ± 1.13 | 7.20 ± 1.63 | 1.470, 0.001 | 1.064, 0.308 | 1.118, 0.15 | 1.008, 0.99 | |
| 17. | GPRC5A | 7.64 ± 0.61 | 7.57 ± 0.43 | 8.02 ± 0.62 | 7.89 ± 0.71 | 8.17 ± 0.52 | −0.990, 0.99 | 1.059, 0.006 | 1.043, 0.57 | 1.080, 0.032 | |
| 18. | CCDC37 | 9.48 ± 0.55 | 9.29 ± 0.57 | 9.25 ± 0.65 | 9.22 ± 0.80 | 9.28 ± 0.47 | −1.021, 0.969 | −1.004, 0.61 | −1.008, 0.99 | −1.001, 0.99 | |
| 19. | GLI3 | 7.55 ± 0.39 | 6.62 ± 0.55 | 6.60 ± 0.41 | 6.70 ± 0.43 | 6.50 ± 0.39 | −1.141, 0.001 | −1.003, 0.927 | 1.013, 0.99 | −1.018, 0.99 | |
| 20. | ABCC3 | 6.97 ± 0.46 | 7.95 ± 0.54 | 7.72 ± 0.77 | 7.68 ± 0.76 | 7.77 ± 0.83 | 1.140, 0.001 | −1.030, 0.412 | −1.034, 0.99 | −1.023, 0.99 | |
| 21. | SAMD5 | 3.74 ± 0.14 | 4.02 ± 0.30 | 3.94 ± 0.54 | 4.00 ± 0.28 | 3.87 ± 0.76 | 1.074, 0.002 | −1.020, 0.133 | −1.003, 0.99 | −1.037, 0.99 | |
| 22. | RASSF10 | 7.70 ± 0.44 | 7.12 ± 0.57 | 6.67 ± 0.52 | 6.81 ± 0.31 | 6.51 ± 0.67 | −1.082, 0.001 | −1.067, 0.004 | −1.045, 0.552 | −1.093, 0.02 | |
| 23. | USP27X | 7.43 ± 0.29 | 7.12 ± 0.39 | 6.68 ± 0.28 | 6.71 ± 0.27 | 6.65 ± 0.30 | −1.043, 0.001 | −1.066, 0.001 | −1.061, 0.007 | −1.071, 0.002 | |
| 24. | NR0B1 | 3.94 ± 0.24 | 4.39 ± 0.78 | 4.84 ± 0.76 | 4.88 ± 0.89 | 4.80 ± 0.64 | 1.114, 0.005 | 1.103, 0.011 | 1.112, 0.181 | 1.094, 0.627 | |
| 25. | PLAG1 | 5.73 ± 0.61 | 5.22 ± 0.56 | 4.83 ± 0.57 | 4.95 ± 0.55 | 4.69 ± 0.60 | −1.098, 0.002 | −1.081, 0.018 | −1.055, 0.99 | −1.113, 0.133 | |
| 26. | SCGB3A1 | 14.25 ± 0.34 | 13.86 ± 0.64 | 13.31 ± 0.84 | 13.54 ± 0.91 | 13.07 ± 0.72 | −1.028, 0.036 | −1.041, 0.025 | −1.024, 0.93 | −1.060, 0.004 | |
| 27. | LHX6 | 5.36 ± 0.33 | 5.85 ± 0.38 | 6.10 ± 0.49 | 6.12 ± 0.52 | 6.07 ± 0.48 | 1.091, 0.001 | 1.043, 0.071 | 1.046, 0.336 | 1.038, 0.754 | |
| 28. | LINC00942 | 3.58 ± 0.19 | 3.88 ± 0.80 | 4.21 ± 1.05 | 4.25 ± 1.24 | 4.17 ± 0.87 | 1.084, 0.172 | 1.085, 0.155 | 1.094,0.551 | 1.073, 0.99 | |
| 29. | REEP1 | 9.29 ± 0.66 | 9.81 ± 0.56 | 9.62 ± 0.78 | 9.39 ± 0.93 | 9.88 ±0.54 | 1.056, 0.005 | −1.020, 0.434 | −1.045, 0.581 | 1.007, 0.99 | |
| 30. | C6orf164 | 5.45 ± 0.42 | 6.14 ± 0.55 | 6.23 ± 0.52 | 6.17 ± 0.43 | 6.29 ± 0.62 | 1.126, 0.001 | 1.015, 0.489 | 1.005, 0.99 | 1.024, 0.99 | |
| 31. | LINC00589 | 5.09 ± 0.22 | 5.36 ± 0.34 | 5.34 ± 0.28 | 5.34 ± 0.17 | 5.35 ± 0.39 | 1.053, 0.001 | −1.004, 0.899 | −1.004, 0.99 | −1.002, 0.99 | |
| 32. | JAKMIP3 | 4.50 ± 0.20 | 5.28 ± 0.56 | 5.75 ± 0.85 | 5.71 ± 0.98 | 5.79 ± 0.74 | 1.173, 0.001 | 1.089, 0.049 | 1.082, 0.09 | 1.096, 0.07 | |
| 33. | LINC00930 | 6.01 ± 0.42 | 6.76 ± 0.53 | 6.56 ± 0.64 | 6.55 ± 0.82 | 6.58 ± 0.41 | 1.126, 0.001 | −1.030, 0.334 | −1.033, 0.99 | −1.028, 0.99 | |
| 34. | DNHD1 | 6.66 ± 0.50 | 7.07 ± 0.55 | 7.16 ± 0.80 | 7.15 ± 0.93 | 7.18 ± 0.69 | 1.061, 0.020 | 1.013, 0.662 | 1.011, 0.99 | 1.015, 0.99 | |
| 35. | TMCC3 | 3.83 ± 0.43 | 4.15 ± 0.52 | 4.55 ± 0.42 | 4.49 ± 0.47 | 4.61 ± 0.38 | 1.083, 0.021 | 1.096, 0.002 | 1.084, 0.323 | 1.112, 0.095 | |
| 36. | ADH7 | 7.94 ± 0.88 | 10.9 ± 10.74 | 10.78 ± 0.54 | 10.82 ± 0.56 | 10.73 ± 0.56 | 1.375, 0.001 | −1.011, 0.166 | −1.009, 0.99 | −1.017, 0.99 | |
| 37. | PRKAR2B | 7.26 ± 0.63 | 6.43 ± 0.71 | 5.84 ± 0.57 | 5.89 ± 0.66 | 5.79 ± 0.50 | −1.128, 0.001 | −1.101, 0.004 | −1.092, 0.161 | −1.112, 0.066 | |
| 38. | GAD1 | 4.89 ± 0.60 | 6.33 ± 1.09 | 7.23 ± 0.89 | 7.07 ± 1.10 | 7.41 ± 0.61 | 1.295, 0.001 | 1.142, 0.009 | 1.117, 0.162 | 1.171, 0.024 | |
| 39. | LOC338667 | 5.37 ± 0.33 | 5.17 ± 0.23 | 4.98 ± 0.21 | 5.03 ± 0.18 | 4.92 ± 0.24 | −1.040, 0.006 | −1.038, 0.012 | −1.027, 0.99 | −1.050, 0.134 | |
| 40. | CYB5A | 4.7 ± 0.20 | 4.72 ± 0.18 | 4.58 ± 0.20 | 4.62 ± 0.23 | 4.53 ± 0.17 | 1.0, 0.99 | −1.031, 0.014 | −1.021, 0.99 | −1.041, 0.089 | |
| 41. | PIEZO2 | 7.13 ± 0.86 | 6.34 ± 0.73 | 5.83 ± 0.63 | 5.78 ± 0.72 | 5.89 ± 0.55 | −1.125, 0.001 | −1.087, 0.02 | −1.097, 0.326 | −1.077, 0.735 | |
| 42. | SLITRK6 | 7.95 ± 0.65 | 6.89 ± 0.78 | 6.34 ± 0.83 | 6.32 ± 1.04 | 6.36 ± 0.57 | −1.155, 0.001 | −1.087, 0.028 | −1.090, 0.317 | −1.082, 0.379 | |
| 43. | KCNA1 | 6.80 ± 1.00 | 5.84 ± 0.90 | 5.34 ± 0.73 | 5.50 ± 0.90 | 5.16 ± 0.45 | −1.166, 0.001 | −1.094, 0.041 | −1.062, 0.99 | −1.131, 0.271 | |
| 44. | LOC100507560 | 5.85 ± 0.69 | 5.39 ± 0.60 | 4.95 ± 0.52 | 4.92 ± 0.56 | 4.98 ± 0.50 | −1.084, 0.012 | −1.089, 0.004 | −1.096, 0.263 | −1.083, 0.507 | |
| Gene Symbol | Healthy Non-Smoker (N = 16) | Healthy Smoker (N = 21) | COPD smoker (N = 4) | COPD stage I smoker (N = 0) | COPD stage II smoker (N = 4) | Fold Regulation, adjusted p-value (HS vs. HNS) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | ||
| Women | 1. | PPP4R4 | 6.87 ±± 0.63 | 5.84 ± 0.96 | 5.15 ± 0.81 | — | 5.15 ± 0.81 | −1.175, 0.005 | −1.135, 0.034 | — | −1.135, 0.034 |
| 2. | THSD4 | 7.58 ± 0.54 | 7.44 ± 0.63 | 6.63 ± 0.56 | — | 6.63 ± 0.56 | −1.018, 0.99 | −1.123, 0.047 | — | −1.123, 0.047 | |
| 3. | NRG1 | 3.87 ± 0.30 | 4.07 ± 0.39 | 3.71 ± 0.34 | — | 3.71 ± 0.34 | 1.051, 0.291 | −1.099, 0.184 | — | −1.099, 0.184 | |
| 4. | SCGB1A1 | 14.48 ± 0.20 | 14.40 ± 0.16 | 14.04 ± 0.31 | — | 14.04 ± 0.31 | −1.006, 0.682 | −1.025, 0.006 | — | −1.025, 0.006 | |
| 5. | AHRR | 3.84 ± 0.16 | 4.43 ± 0.61 | 4.81 ± 0.38 | — | 4.81 ± 0.38 | 1.154, 0.002 | 1.085, 0.542 | — | 1.085, 0.542 | |
| 6. | CYP1A1 | 4.59 ± 0.26 | 5.79 ± 1.85 | 7.66 ± 2.80 | — | 7.66 ± 2.80 | 1.260, 0.037 | 1.324, 0.081 | — | 1.324, 0.081 | |
| 7. | CYP1B1 | 3.52 ± 0.35 | 7.21 ± 1.96 | 8.77 ± 2.04 | — | 8.77 ± 2.04 | 2.050, 0.001 | 1.216, 0.27 | — | 1.216, 0.27 | |
| 8. | PRDM11 | 5.70 ± 0.39 | 5.39 ± 0.31 | 5.15 ± 0.18 | — | 5.15 ± 0.18 | −1.057, 0.029 | −1.047, 0.56 | — | −1.047, 0.56 | |
| 9. | CBR3 | 7.14 ± 0.39 | 7.93 ± 0.70 | 7.98 ± 0.53 | — | 7.98 ± 0.53 | 1.111, 0.002 | 1.006, 0.99 | — | 1.006, 0.99 | |
| Women | 10. | AKR1C1 | 7.00 ± 0.83 | 8.48 ± 1.17 | 7.40 ± 1.35 | — | 7.40 ± 1.35 | 1.211, 0.002 | −1.146, 0.224 | — | −1.146, 0.224 |
| 11. | AKR1C3 | 10.36 ± 0.52 | 11.68 ± 0.61 | 11.55 ± 0.83 | — | 11.55 ± 0.83 | 1.127, 0.002 | −1.011, 0.99 | — | −1.011, 0.99 | |
| 12. | HTR2B | 4.08 ± 0.22 | 4.13 ± 0.31 | 4.29 ± 0.31 | — | 4.29 ± 0.31 | 1.012, 0.99 | 1.037, 0.97 | — | 1.037, 0.97 | |
| 13. | GRM1 | 3.75 ± 0.11 | 4.04 ± 0.34 | 4.05 ± 0.32 | — | 4.05 ± 0.32 | 1.076, 0.010 | 1.004, 0.99 | — | 1.004, 0.99 | |
| 14. | CYP4Z1 | 6.91 ± 0.54 | 6.36 ± 0.63 | 5.84 ± 0.52 | — | 5.84 ± 0.52 | −1.085, 0.037 | −1.090, 0.321 | — | −1.090, 0.321 | |
| 15. | UCHL1 | 5.32 ± 0.41 | 9.03 ± 1.72 | 8.49 ± 2.66 | — | 8.49 ± 2.66 | 1.698, 0.001 | −1.063, 0.99 | — | −1.063, 0.99 | |
| 16. | CABYR | 4.87 ± 0.32 | 6.55 ± 1.26 | 6.88 ± 1.26 | — | 6.88 ± 1.26 | 1.343, 0.001 | 1.051, 0.99 | — | 1.051, 0.99 | |
| 17. | GPRC5A | 7.42 ± 0.69 | 7.39 ± 0.43 | 7.96 ± 0.64 | — | 7.96 ± 0.64 | −1.005, 0.99 | 1.077, 0.216 | — | 1.077, 0.216 | |
| 18. | CCDC37 | 9.36 ± 0.54 | 9.48 ± 0.44 | 9.34 ± 0.62 | — | 9.34 ± 0.62 | 1.013, 0.99 | −1.015, 0.99 | — | −1.015, 0.99 | |
| 19. | GLI3 | 7.70 ± 0.36 | 6.99 ± 0.55 | 6.71 ± 0.22 | — | 6.71 ± 0.22 | −1.101, 0.001 | −1.042, 0.723 | — | −1.042, 0.723 | |
| 20. | ABCC3 | 6.91 ± 0.43 | 7.76 ± 0.72 | 7.20 ± 0.78 | — | 7.20 ± 0.78 | 1.122, 0.002 | −1.077, 0.345 | — | −1.077, 0.345 | |
| 21. | SAMD5 | 3.78 ± 0.17 | 3.91 ± 0.25 | 3.87 ± 0.21 | — | 3.87 ± 0.21 | 1.037, 0.20 | −1.010, 0.99 | — | −1.010, 0.99 | |
| 22. | RASSF10 | 7.64 ± 0.62 | 6.96 ± 0.62 | 6.41 ± 0.74 | — | 6.41 ± 0.74 | −1.097, 0.011 | −1.086, 0.374 | — | −1.086, 0.374 | |
| 23. | USP27X | 7.47 ± 0.30 | 7.16 ± 0.44 | 6.51 ± 0.81 | — | 6.51 ± 0.81 | −1.043, 0.158 | −1.10, 0.04 | — | −1.10, 0.04 | |
| 24. | NR0B1 | 3.94 ± 0.24 | 4.40 ± 0.77 | 4.44 ± 0.84 | — | 4.44 ± 0.84 | 1.119, 0.065 | 1.009, 0.99 | — | 1.009, 0.99 | |
| 25. | PLAG1 | 5.95 ± 0.40 | 5.32 ± 0.55 | 4.74 ± 0.49 | — | 4.74 ± 0.49 | −1.120, 0.002 | −1.121, 0.115 | — | −1.121, 0.115 | |
| 26. | SCGB3A1 | 14.13 ± 0.55 | 14.05 ± 0.51 | 12.51 ± 0.26 | — | 12.51 ± 0.26 | −1.006, 0.99 | −1.123, 0.001 | — | −1.123, 0.001 | |
| 27. | LHX6 | 5.37 ± 0.40 | 5.73 ± 0.55 | 6.22 ± 0.68 | — | 6.22 ± 0.68 | 1.067, 0.116 | 1.086, 0.253 | — | 1.086, 0.253 | |
| 28. | LINC00942 | 3.59 ± 0.14 | 3.85 ± 0.51 | 3.73 ± 0.63 | — | 3.73 ± 0.63 | 1.074, 0.154 | −1.034, 0.99 | — | −1.034, 0.99 | |
| 29. | REEP1 | 9.47 ± 0.58 | 9.85 ± 0.57 | 9.59 ± 0.81 | — | 9.59 ± 0.81 | 1.040, 0.217 | −1.028, 0.99 | — | −1.028, 0.99 | |
| 30. | C6orf164 | 5.58 ± 0.67 | 6.01 ± 0.57 | 6.14 ± 1.02 | — | 6.14 ± 1.02 | 1.077, 0.224 | 1.021, 0.99 | — | 1.021, 0.99 | |
| 31. | LINC00589 | 5.30 ± 0.29 | 5.43 ± 0.25 | 5.64 ± 0.34 | — | 5.64 ± 0.34 | 1.025, 0.524 | 1.037, 0.549 | — | 1.037, 0.549 | |
| 32. | JAKMIP3 | 4.54 ± 0.23 | 5.28 ± 0.72 | 5.39 ± 0.94 | — | 5.39 ± 0.94 | 1.164, 0.002 | 1.020, 0.99 | — | 1.020, 0.99 | |
| 33. | LINC00930 | 6.04 ± 0.47 | 7.20 ± 0.56 | 6.45 ± 0.92 | — | 6.45 ± 0.92 | 1.191, 0.001 | −1.116, 0.07 | — | −1.116, 0.07 | |
| 34. | DNHD1 | 6.61 ± 0.46 | 7.28 ± 0.47 | 7.05 ± 0.47 | — | 7.05 ± 0.47 | 1.102, 0.001 | −1.033, 0.99 | — | −1.033, 0.99 | |
| 35. | TMCC3 | 3.82 ± 0.27 | 4.01 ± 0.40 | 4.07 ± 0.23 | — | 4.07 ± 0.23 | 1.051, 0.283 | 1.016, 0.99 | — | 1.016, 0.99 | |
| 36. | ADH7 | 8.22 ± 1.05 | 10.63 ± 0.57 | 10.37 ± 0.92 | — | 10.37 ± 0.92 | 1.294, 0.001 | −1.025, 0.99 | — | −1.025, 0.99 | |
| 37. | PRKAR2B | 7.59 ± 0.37 | 6.49 ± 0.75 | 5.92 ± 0.76 | — | 5.92 ± 0.76 | −1.169, 0.001 | —1.096, 0.325 | — | −1.096, 0.325 | |
| 38. | GAD1 | 4.98 ± 0.66 | 6.27 ± 0.86 | 7.35 ± 0.87 | — | 7.35 ± 0.87 | 1.260, 0.001 | 1.171, 0.058 | — | 1.171, 0.058 | |
| 39. | LOC338667 | 5.45 ± 0.33 | 5.24 ± 0.25 | 5.31 ± 0.26 | — | 5.31 ± 0.26 | −1.040, 0.11 | 1.013, 0.99 | — | 1.013, 0.99 | |
| 40. | CYB5A | 4.74 ± 0.19 | 4.69 ± 0.20 | 4.31 ± 0.30 | — | 4.31 ± 0.30 | −1.010, 0.99 | −1.089, 0.005 | — | −1.089, 0.005 | |
| 41. | PIEZO2 | 7.53 ± 0.82 | 6.78 ±± 0.92 | 6.14 ± 0.69 | — | 6.14 ± 0.69 | −1.110, 0.054 | −1.105, 0.488 | — | −1.105, 0.488 | |
| 42. | SLITRK6 | 8.02 ± 0.51 | 6.82 ± 0.99 | 5.85 ± 0.34 | — | 5.85 ± 0.34 | −1.176, 0.001 | −1.166, 0.069 | — | −1.166, 0.069 | |
| 43. | KCNA1 | 7.04 ± 1.15 | 5.72 ± 1.02 | 4.63 ± 0.77 | — | 4.63 ± 0.77 | −1.231, 0.002 | −1.234, 0.184 | — | −1.234, 0.184 | |
| 44. | LOC100507560 | 5.88 ± 0.73 | 5.48 ± 0.63 | 4.81 ± 0.31 | — | 4.81 ± 0.31 | −1.073, 0.227 | −1.141, 0.171 | — | −1.141, 0.171 |
The Adj. P is based on the marginally adjusted 𝑝 values by the Benjamini-Hochberg-FDR correction at α = 0.05; Median ± Interquartile range.
We then investigated the expression of our 44 candidate genes, including our 17 novel genes, in HNS, HS, COPD, and COPD Stage I and II. Here, we determined that 52.3% of the 44 candidate genes were significantly differentially expressed in HS compared to HNS females (Table 6; HS v HNS). In addition, 47.1% of the 17 novel genes were significantly different in HS females compared to HNS females. A comparison of female COPD patients to HS females revealed that expression of 7/44 (15.9%) of the 44 candidate genes and 2/17 (11.8%) of the 17 novel genes were significantly different in COPD patients compared to HS (Table 6; COPD v HS). Furthermore, we also observed of significant difference in COPD Stage II compared with HS in 6/44 (13.6%) of the 44 candidate genes and 2/17 (11.8%) of the novel genes detected in this study in females (Table 6; Stage II v HS). A number of the 44 candidate genes were significantly different in females across all four analyses, these included CYP1A1 and the novel genes, LINC00930, GAD1 and SLITRK6.
Investigation of the age effect on differential gene expression in HNS, HS and COPD (Stage I and II) patients
In this study, we also investigated the effect of age (i.e. subject or patient age: < or ≥50 years) on gene expression in HNS, HS, COPD patients, and stage I and II patients. We observed a significant change in the gene expression of 40/44 (90.9%) total candidate genes and 15/17 (88.2%) novel genes in subjects ≤ 50 years (Table 7; <50 years; HS v HNS). Comparison of HS to COPD patients revealed that expression of 20/44 (45.5%) of our candidate genes were significantly different in COPD patients ≤ 50 years. In addition, expression of 8/17 (47.1%) of our novel genes were significantly different in COPD patients compared to HS ≤ 50 years (Table 7; <50 years; COPD v HS). We also investigated differential gene expression in Stage I and II COPD patients ≤ 50 years and determined that 6/44 (13.6%; Stage I) and 9/44 (20.5%; Stage II) candidate genes, respectively, were significantly different in patients ≤ 50 years. In our cohort of 17 novel genes, we determined that 2/17 (11.8%; Stage I) and 3/17 (17.6%; Stage II) among our total 44 candidate genes, respectively, were significantly different in patients ≤ 50 years (Table 7; <50 years; Stage I and Stage II v HS). Furthermore, a certain number of these candidate genes were significantly different in subjects ≤ 50 years, across all four analysis groups, which included USP27X, CYP1A1 and the novel genes of JAKMIP3 and GAD1.
Table 7.
Comparison of relative expression of 44 candidate genes between healthy controls (smokers and non-smokers) and COPD smoker patients (stage I and stage II) in age groups separately.
| Age group | Gene Symbol | Healthy Non-Smoker (N = 45) | Healthy Smoker (N = 50) | COPD smoker (N = 10) | COPD stage I smoker (N = 5) | COPD stage II smoker (N = 5) | Fold Regulation, adjusted p-value (HS vs. HNS) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| <50 years old | 1. | PPP4R4 | 6.56 ± 0.77 | 6.01 ± 0.86 | 5.23 ± 0.60 | 5.64 ± 0.43 | 4.84 ± 0.48 | −1.091,0.009 | −1.149, 0.005 | −1.065, 0.99 | −1.242, 0.009 |
| 2. | THSD4 | 7.65 ± 0.47 | 7.36 ± 0.56 | 7.24 ± 0.47 | 7.47 ± 0.33 | 7.02 ± 0.51 | −1.039, 0.049 | −1.017, 0.579 | 1.015, 0.99 | −1.049, 0.91 | |
| 3. | NRG1 | 3.81 ± 0.26 | 4.05 ± 0.45 | 3.89 ± 0.50 | 4.06 ± 0.62 | 3.72 ± 0.29 | 1.064, 0.008 | −1.041, 0.131 | 1.002, 0.99 | −1.090, 0.336 | |
| 4. | SCGB1A1 | 14.50 ± 0.15 | 14.36 ± 0.19 | 14.01 ± 0.41 | 14.15 ± 0.33 | 13.88 ± 0.47 | −1.010, 0.005 | −1.025, 0.009 | −1.015, 0.149 | −1.035, 0.001 | |
| 5. | AHRR | 3.87 ± 0.15 | 4.53 ± 0.63 | 5.27 ± 0.73 | 5.66 ± 0.71 | 4.91 ± 0.59 | 1.168, 0.001 | 1.163, 0.004 | 1.250, 0.001 | 1.085, 0.618 | |
| 6. | CYP1A1 | 4.61 ± 0.24 | 5.95 ± 1.79 | 7.91 ± 2.16 | 8.22 ± 1.81 | 7.61 ± 2.67 | 1.290, 0.001 | 1.329, 0.008 | 1.381, 0.007 | 1.278, 0.046 | |
| 7. | CYP1B1 | 3.64 ± 0.53 | 7.54 ± 2.04 | 9.75 ± 1.17 | 10.23 ± 0.54 | 9.30 ± 1.53 | 2.072, 0.001 | 1.293, 0.003 | 1.357, 0.006 | 1.234, 0.17 | |
| 8. | PRDM11 | 5.72 ± 0.36 | 5.36 ± 0.34 | 5.20 ± 0.31 | 5.30 ± 0.18 | 5.11 ± 0.40 | −1.067, 0.001 | −1.031, 0.193 | −1.011, 0.99 | −1.048, 0.82 | |
| 9. | CBR3 | 7.26 ± 0.43 | 8.10 ± 0.68 | 8.58 ± 0.59 | 8.74 ± 0.41 | 8.44 ± 0.75 | 1.116, 0.001 | 1.059, 0.039 | 1.079, 0.155 | 1.042, 0.99 | |
| 10. | AKR1C1 | 6.92 ± 0.67 | 8.54 ± 1.14 | 8.71 ± 1.18 | 8.58 ± 1.42 | 8.85 ± 1.04 | 1.235, 0.001 | 1.020, 0.491 | 1.004, 0.99 | 1.036, 0.99 | |
| 11. | AKR1C3 | 10.31 ± 0.46 | 11.82 ± 0.73 | 12.15 ± 0.67 | 12.42 ± 0.42 | 11.88 ± 0.82 | 1.146, 0.001 | 1.028, 0.275 | 1.051, 0.266 | 1.006, 0.99 | |
| 12. | HTR2B | 3.94 ± 0.21 | 4.22 ± 0.32 | 4.29 ± 0.36 | 4.31 ± 0.51 | 4.26 ± 0.18 | 1.072, 0.001 | 1.017, 0.421 | 1.021, 0.99 | 1.010, 0.99 | |
| 13. | GRM1 | 3.74 ± 0.11 | 4.01 ± 0.30 | 4.37 ± 0.73 | 4.38 ± 0.40 | 4.36 ± 1.01 | 1.072, 0.001 | 1.090, 0.112 | 1.091, 0.068 | 1.087, 0.028 | |
| 14. | CYP4Z1 | 6.88 ± 0.58 | 6.27 ± 0.59 | 5.90 ± 0.35 | 5.89 ± 0.45 | 5.91 ± 0.27 | −1.097, 0.001 | −1.063, 0.058 | −1.065, 0.834 | −1.062, 0.903 | |
| 15. | UCHL1 | 5.33 ± 0.59 | 8.71 ± 1.50 | 10.11 ± 1.79 | 10.35 ± 1.36 | 9.88 ± 2.31 | 1.636, 0.001 | 1.161, 0.008 | 1.188, 0.049 | 1.134, 0.181 | |
| 16. | CABYR | 4.86 ± 0.29 | 6.85 ± 1.20 | 7.84 ± 1.64 | 8.52 ± 1.12 | 7.22 ± 1.99 | 1.409, 0.001 | 1.145, 0.037 | 1.244, 0.003 | 1.054, 0.99 | |
| 17. | GPRC5A | 7.58 ± 0.66 | 7.50 ± 0.45 | 8.16 ± 0.61 | 7.98 ± 0.68 | 8.35 ± 0.55 | −1.011, 0.99 | 1.088, 0.003 | 1.065, 0.392 | 1.114, 0.010 | |
| 18. | CCDC37 | 9.47 ± 0.55 | 9.34 ± 0.55 | 9.54 ± 0.66 | 9.40 ± 0.80 | 9.68 ± 0.55 | −1.013, 0.99 | 1.021, 0.433 | 1.006, 0.99 | 1.036, 0.99 | |
| 19. | GLI3 | 7.62 ± 0.39 | 6.72 ± 0.60 | 6.51 ± 0.48 | 6.61 ± 0.49 | 6.41 ± 0.49 | −1.133, 0.001 | −1.032, 0.221 | −1.017, 0.99 | −1.050, 0.99 | |
| 20. | ABCC3 | 6.96 ± 0.43 | 7.91 ± 0.62 | 7.98 ± 0.69 | 8.14 ± 0.66 | 7.82 ± 0.76 | 1.136, 0.001 | 1.009, 0.716 | 1.029, 0.99 | −1.011, 0.99 | |
| 21. | SAMD5 | 3.74 ± 0.15 | 3.97 ± 0.28 | 4.13 ± 0.62 | 4.13 ± 0.24 | 4.13 ± 0.89 | 1.060, 0.001 | 1.040, 0.455 | 1.041, 0.99 | 1.041, 0.614 | |
| 22. | RASSF10 | 7.71 ± 0.50 | 7.08 ± 0.57 | 6.41 ± 0.60 | 6.68 ± 0.36 | 6.15 ± 0.72 | −1.089, 0.001 | −1.105, 0.002 | −1.060, 0.62 | −1.152, 0.002 | |
| 23. | USP27X | 7.46 ± 0.28 | 7.16 ± 0.40 | 6.52 ± 0.50 | 6.74 ± 0.34 | 6.31 ± 0.58 | −1.041, 0.002 | −1.098, 0.001 | −1.063, 0.075 | −1.136, 0.001 | |
| 24. | NR0B1 | 3.95 ± 0.25 | 4.36 ± 0.78 | 5.15 ± 0.84 | 5.35 ± 0.90 | 4.97 ± 0.81 | 1.103, 0.003 | 1.181, 0.005 | 1.226, 0.005 | 1.139, 0.23 | |
| 25. | PLAG1 | 5.87 ± 0.55 | 5.26 ± 0.57 | 4.74 ± 0.57 | 4.93 ± 0.64 | 4.55 ± 0.49 | −1.116, 0.001 | −1.110, 0.009 | −1.067, 0.99 | −0.866, 0.045 | |
| 26. | SCGB3A1 | 14.25 ± 0.40 | 13.93 ± 0.59 | 13.48 ± 0.78 | 13.89 ± 0.51 | 13.09 ± 0.85 | −1.023, 0.025 | −1.033, 0.092 | −1.002, 0.99 | −1.064, 0.006 | |
| 27. | LHX6 | 5.33 ± 0.35 | 5.80 ± 0.44 | 6.24 ± 0.57 | 6.28 ± 0.61 | 6.20 ± 0.59 | 1.087, 0.001 | 1.076, 0.016 | 1.084, 0.084 | 1.069, 0.25 | |
| 28. | LINC00942 | 3.57 ± 0.17 | 3.85 ± 0.69 | 4.45 ± 1.28 | 4.67 ± 1.53 | 4.24 ± 1.09 | 1.079, 0.073 | 1.156, 0.18 | 1.212, 0.008 | 1.101, 0.828 | |
| 29. | REEP1 | 9.40 ± 0.60 | 9.79 ± 0.55 | 9.81 ± 0.74 | 9.91 ± 0.91 | 9.71 ± 0.63 | 1.041, 0.013 | 1.002, 0.716 | 1.013, 0.99 | −1.009, 0.99 | |
| 30. | C6orf164 | 5.53 ± 0.51 | 6.06 ± 0.56 | 6.08 ± 0.52 | 6.17 ± 0.53 | 5.99 ± 0.56 | 1.097, 0.001 | 1.003, 0.66 | 1.017, 0.99 | −1.012, 0.99 | |
| 31. | LINC00589 | 5.15 ± 0.27 | 5.38 ± 0.32 | 5.32 ± 0.29 | 5.33 ± 0.16 | 5.30 ± 0.41 | 1.045, 0.001 | −1.011, 0.848 | −1.010, 0.99 | −1.015, 0.99 | |
| 32. | JAKMIP3 | 4.50 ± 0.22 | 5.25 ± 0.61 | 6.18 ± 0.81 | 6.20 ± 0.92 | 6.16 ± 0.80 | 1.166, 0.001 | 1.177, 0.002 | 1.180, 0.001 | 1.174, 0.001 | |
| 33. | LINC00930 | 6.02 ± 0.36 | 6.90 ± 0.59 | 6.77 ± 0.71 | 6.87 ± 0.79 | 6.66 ± 0.69 | 1.146, 0.001 | −1.019, 0.716 | −1.004, 0.99 | −1.035, 0.99 | |
| 34. | DNHD1 | 6.68 ± 0.50 | 7.15 ± 0.54 | 7.37 ± 0.83 | 7.47 ± 0.98 | 7.27 ± 0.75 | 1.070, 0.001 | 1.031, 0.358 | 1.044, 0.99 | 1.017, 0.99 | |
| 35. | TMCC3 | 3.80 ± 0.28 | 4.11 ± 0.48 | 4.48 ± 0.53 | 4.56 ± 0.62 | 4.40 ± 0.48 | 1.082, 0.001 | 1.090, 0.019 | 1.109, 0.12 | 1.071, 0.85 | |
| 36. | ADH7 | 8.08 ± 0.94 | 10.78 ± 0.72 | 10.87 ± 0.60 | 11.09 ± 0.59 | 10.66 ± 0.58 | 1.335, 0.001 | 1.008, 0.804 | 1.029, 0.99 | −1.012, 0.99 | |
| 37. | PRKAR2B | 7.35 ± 0.61 | 6.46 ± 0.74 | 5.74 ± 0.51 | 5.83 ± 0.46 | 5.64 ± 0.59 | −1.137, 0.001 | −1.125, 0.004 | −1.107, 0.239 | −1.145, 0.054 | |
| 38. | GAD1 | 4.87 ± 0.56 | 6.25 ± 0.99 | 7.44 ± 0.87 | 7.26 ± 1.01 | 7.62 ± 0.78 | 1.283, 0.001 | 1.190, 0.002 | 1.162, 0.068 | 1.219, 0.005 | |
| 39. | LOC338667 | 5.41 ± 0.34 | 5.20 ± 0.24 | 5.03 ± 0.22 | 5.04 ± 0.20 | 5.03 ± 0.26 | −1.040, 0.002 | −1.034, 0.071 | −1.031, 0.99 | −1.034, 0.99 | |
| 40. | CYB5A | 4.73 ± 0.20 | 4.73 ± 0.18 | 4.61 ± 0.23 | 4.70 ± 0.19 | 4.53 ± 0.26 | −1.000, 0.99 | −1.026, 0.235 | −1.006, 0.99 | −1.043, 0.99 | |
| 41. | PIEZO2 | 7.34 ± 0.84 | 6.49 ± 0.87 | 5.71 ± 0.64 | 5.71 ± 0.79 | 5.71 ± 0.54 | −1.132, 0.001 | −1.137, 0.008 | −1.136, 0.291 | −1.135, 0.255 | |
| 42. | SLITRK6 | 8.05 ± 0.59 | 6.89 ± 0.84 | 6.01 ± 0.99 | 6.10 ± 1.31 | 5.93 ± 0.67 | −1.169, 0.001 | −1.146, 0.015 | −1.129, 0.249 | −1.161, 0.043 | |
| 43. | KCNA1 | 6.95 ± 1.04 | 5.79 ± 0.96 | 5.19 ± 0.72 | 5.27 ± 0.96 | 5.11 ± 0.47 | −1.200, 0.001 | −1.116, 0.031 | −1.098, 0.99 | −1.133, 0.658 | |
| 44. | LOC100507560 | 5.90 ± 0.71 | 5.45 ± 0.62 | 5.03 ± 0.65 | 5.01 ± 0.75 | 5.05 ± 0.63 | −1.083, 0.006 | −1.083, 0.041 | −1.087, 0.99 | −1.079, 0.99 | |
| Age group | Gene Symbol | Healthy Non-Smoker (N = 8) | Healthy Smoker (N = 9) | COPD smoker (N = 11) | COPD stage I smoker (N = 4) | COPD stage II smoker (N = 7) | Fold Regulation, adjusted p-value (HS vs. HNS) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | |
| ≥50 years old | 1. | PPP4R4 | 6.64 ± 0.64 | 6.05 ± 1.16 | 5.87 ± 0.63 | 6.20 ± 0.23 | 5.70 ± 0.75 | −1.098, 0.99 | −1.031, 0.191 | 1.024, 0.99 | −1.062, 0.99 |
| 2. | THSD4 | 7.29 ± 0.62 | 7.15 ± 0.33 | 6.99 ± 0.53 | 7.48 ± 0.33 | 6.72 ± 0.42 | −1.019, 0.99 | −1.023, 0.366 | 1.046, 0.99 | −1.065, 0.663 | |
| 3. | NRG1 | 3.71 ± 0.18 | 4.02 ± 0.63 | 3.72 ± 0.22 | 3.80 ± 0.15 | 3.68 ± 0.25 | 1.084, 0.606 | −1.081, 0.315 | −1.059, 0.99 | −1.095, 0.447 | |
| 4. | SCGB1A1 | 14.38 ± 0.13 | 14.38 ± 0.30 | 14.27 ± 0.13 | 14.23 ± 0.10 | 14.28 ± 0.16 | 1.000, 0.99 | −1.008, 0.132 | −1.010, 0.99 | −1.007, 0.99 | |
| 5. | AHRR | 3.79 ± 0.23 | 5.13 ± 0.74 | 5.19 ± 0.82 | 5.15 ± 0.96 | 5.21 ± 0.81 | 1.353, 0.012 | 1.012, 0.92 | 1.004, 0.99 | 1.016, 0.99 | |
| 6. | CYP1A1 | 4.64 ± 0.23 | 7.88 ± 1.63 | 7.55 ± 1.46 | 7.83 ± 1.26 | 7.40 ± 1.64 | 1.698, 0.001 | −1.044, 0.763 | −1.007, 0.99 | −1.065, 0.99 | |
| 7. | CYP1B1 | 3.68 ± 0.43 | 9.76 ± 1.12 | 9.28 ± 1.15 | 9.55 ± 0.47 | 9.12 ± 1.43 | 2.650, 0.001 | −1.052, 0.546 | −1.022, 0.99 | −1.069, 0.99 | |
| 8. | PRDM11 | 5.42 ± 0.18 | 5.32 ± 0.12 | 5.35 ± 0.26 | 5.41 ± 0.27 | 5.32 ± 0.27 | −1.019, 0.99 | 1.006, 0.841 | 1.018, 0.99 | −1.000, 0.99 | |
| 9. | CBR3 | 7.06 ± 0.44 | 8.60 ± 0.54 | 8.15 ± 0.63 | 8.17 ± 0.47 | 8.14 ± 0.74 | 1.218, 0.001 | −1.055, 0.159 | −1.052, 0.99 | −1.056, 0.99 | |
| ≥50 years old | 10. | AKR1C1 | 6.20 ± 0.40 | 8.75 ± 0.90 | 7.97 ± 1.15 | 8.28 ± 1.14 | 7.80 ± 1.21 | 1.412, 0.001 | −1.098, 0.159 | −1.057, 0.99 | −1.121, 0.596 |
| 11. | AKR1C3 | 10.24 ± 0.28 | 12.18 ± 0.59 | 11.79 ± 0.63 | 11.92 ± 0.42 | 11.72 ± 0.75 | 1.190, 0.001 | −1.033, 0.269 | −1.022, 0.99 | −1.040, 0.911 | |
| 12. | HTR2B | 3.85 ± 0.21 | 4.17 ± 0.22 | 4.22 ± 0.28 | 4.05 ± 0.18 | 4.32 ± 0.28 | 1.083, 0.142 | 1.012, 0.99 | −1.028, 0.99 | 1.036, 0.99 | |
| 13. | GRM1 | 3.75 ± 0.15 | 4.37 ± 0.54 | 3.98 ± 0.17 | 3.99 ± 0.17 | 3.97 ± 0.19 | 1.164, 0.008 | −1.098, 0.056 | −1.095, 0.328 | −1.099, 0.141 | |
| 14. | CYP4Z1 | 6.58 ± 0.45 | 6.02 ± 0.45 | 5.95 ± 0.42 | 5.92 ± 0.53 | 5.96 ± 0.38 | −1.094, 0.20 | −1.012, 0.688 | −1.017, 0.99 | −1.009, 0.99 | |
| 15. | UCHL1 | 5.18 ± 0.35 | 9.51 ± 1.42 | 8.56 ± 1.54 | 8.03 ± 1.04 | 8.89 ± 1.74 | 1.836, 0.001 | −1.111, 0.269 | −1.185, 0.467 | −1.070, 0.99 | |
| 16. | CABYR | 4.89 ± 0.32 | 7.69 ± 1.35 | 7.13 ± 0.99 | 7.36 ± 0.83 | 7.00 ± 1.11 | 1.575, 0.001 | −1.079, 0.228 | −1.045, 0.99 | −1.099, 0.99 | |
| 17. | GPRC5A | 7.60 ± 0.45 | 7.60 ± 0.20 | 7.88 ± 0.61 | 7.79 ± 0.84 | 7.93 ± 0.50 | 1.000, 0.99 | 1.037, 0.108 | 1.024, 0.99 | 1.042, 0.99 | |
| 18. | CCDC37 | 9.33 ± 0.52 | 9.48 ± 0.36 | 9.02 ± 0.50 | 9.00 ± 0.86 | 9.04 ± 0.22 | 1.016, 0.99 | −1.051, 0.044 | −1.054, 0.982 | −1.049, 0.701 | |
| 19. | GLI3 | 7.39 ± 0.36 | 6.91 ± 0.25 | 6.73 ± 0.25 | 6.81 ± 0.39 | 6.68 ± 0.15 | −1.069, 0.044 | −1.027, 0.269 | −1.015, 0.99 | −1.034, 0.99 | |
| 20. | ABCC3 | 6.92 ± 0.59 | 7.66 ± 0.52 | 7.31 ± 0.75 | 7.16 ± 0.48 | 7.40 ± 0.89 | 1.106, 0.379 | −1.048, 0.315 | −1.070, 0.99 | −1.034, 0.99 | |
| 21. | SAMD5 | 3.77 ± 0.15 | 4.08 ± 0.33 | 3.75 ± 0.24 | 3.85 ± 0.28 | 3.70 ± 0.22 | 1.082, 0.183 | −1.088, 0.044 | −1.059, 0.953 | −1.104, 0.06 | |
| 22. | RASSF10 | 7.48 ± 0.40 | 6.90 ± 0.79 | 6.81 ± 0.46 | 6.97 ± 0.17 | 6.72 ± 0.56 | −1.083, 0.534 | −1.013, 0.366 | 1.010, 0.99 | −1.027, 0.99 | |
| 23. | USP27X | 7.32 ± 0.36 | 6.88 ± 0.39 | 6.77 ± 0.27 | 6.67 ± 0.19 | 6.82 ± 0.31 | −1.064, 0.168 | −1.016, 0.482 | −1.031, 0.99 | −1.008, 0.99 | |
| 24. | NR0B1 | 3.84 ± 0.18 | 4.68 ± 0.72 | 4.43 ± 0.52 | 4.35 ± 0.47 | 4.48 ± 0.57 | 1.217, 0.041 | −1.056, 0.421 | −1.076, 0.99 | −1.044, 0.99 | |
| 25. | PLAG1 | 5.35 ± 0.50 | 5.23 ± 0.41 | 4.88 ± 0.53 | 4.98 ± 0.50 | 4.82 ± 0.58 | −1.023, 0.99 | −1.072, 0.159 | −1.050, 0.99 | −1.085, 0.99 | |
| 26. | SCGB3A1 | 13.96 ± 0.39 | 13.88 ± 0.75 | 12.87 ± 0.79 | 13.10 ± 1.20 | 12.73 ± 0.49 | −1.006, 0.99 | −1.078, 0.044 | −1.059, 0.642 | −1.090, 0.042 | |
| 27. | LHX6 | 5.55 ± 0.26 | 5.89 ± 0.46 | 6.01 ± 0.46 | 5.91 ± 0.33 | 6.07 ± 0.53 | 1.062, 0.863 | 1.020, 0.269 | 1.003, 0.99 | 1.030, 0.99 | |
| 28. | LINC00942 | 3.64 ± 0.20 | 4.02 ± 0.93 | 3.83 ± 0.44 | 3.77 ± 0.27 | 3.86 ± 0.53 | 1.105, 0.987 | −1.050, 0.841 | −1.066, 0.99 | −1.042, 0.99 | |
| 29. | REEP1 | 8.96 ± 0.75 | 10.11 ± 0.61 | 9.43 ± 0.78 | 8.77 ± 0.41 | 9.84 ± 0.66 | 1.129, 0.028 | −1.072, 0.088 | −1.153, 0.024 | −1.028, 0.99 | |
| 30. | C6orf164 | 5.27 ± 0.44 | 6.38 ± 0.51 | 6.33 ± 0.68 | 6.18 ± 0.35 | 6.42 ± 0.82 | 1.212, 0.016 | −1.008, 0.841 | −1.033, 0.99 | 1.006, 0.99 | |
| 31. | LINC00589 | 5.14 ± 0.20 | 5.42 ± 0.33 | 5.47 ± 0.32 | 5.35 ± 0.20 | 5.55 ± 0.36 | 1.053, 0.609 | 1.009, 0.763 | −1.012, 0.99 | 1.024, 0.99 | |
| 32. | JAKMIP3 | 4.55 ± 0.17 | 5.52 ± 0.60 | 5.26 ± 0.63 | 5.16 ± 0.78 | 5.31 ± 0.58 | 1.214, 0.022 | −1.049, 0.482 | −1.069, 0.99 | −1.039, 0.99 | |
| 33. | LINC00930 | 5.98 ± 0.79 | 7.01 ± 0.41 | 6.34 ± 0.59 | 6.16 ± 0.76 | 6.44 ± 0.52 | 1.172, 0.061 | −1.106, 0.044 | −1.137, 0.348 | −1.088, 0.767 | |
| 34. | DNHD1 | 6.42 ± 0.27 | 7.07 ± 0.51 | 6.94 ± 0.61 | 6.77 ± 0.80 | 7.04 ± 0.53 | 1.100, 0.202 | −1.019, 0.92 | −1.045, 0.99 | −1.004, 0.99 | |
| 35. | TMCC3 | 4.03 ± 0.80 | 4.02 ± 0.55 | 4.43 ± 0.35 | 4.42 ± 0.23 | 4.44 ± 0.42 | −1.002, 0.99 | 1.102, 0.035 | 1.098, 0.99 | 1.105, 0.99 | |
| 36. | ADH7 | 7.63 ± 0.89 | 11.08 ± 0.44 | 10.54 ± 0.63 | 10.48 ± 0.29 | 10.58 ± 0.78 | 1.453, 0.001 | −1.051, 0.044 | −1.058, 0.99 | −1.048, 0.99 | |
| 37. | PRKAR2B | 7.38 ± 0.42 | 6.38 ± 0.55 | 5.97 ± 0.67 | 5.96 ± 0.93 | 5.97 ± 0.55 | −1.158, 0.04 | −1.069, 0.228 | −1.070, 0.99 | −1.068, 0.99 | |
| 38. | GAD1 | 5.22 ± 0.87 | 6.89 ± 1.08 | 7.09 ± 0.86 | 6.85 ± 1.32 | 7.23 ± 0.57 | 1.321, 0.025 | 1.029, 0.688 | −1.007, 0.99 | 1.049, 0.99 | |
| 39. | LOC338667 | 5.32 ± 0.20 | 5.14 ± 0.22 | 5.05 ± 0.29 | 5.02 ± 0.17 | 5.06 ± 0.35 | −1.035, 0.99 | −1.018, 0.315 | −1.024, 0.99 | −1.015, 0.99 | |
| 40. | CYB5A | 4.70 ± 0.17 | 4.58 ± 0.18 | 4.45 ± 0.23 | 4.53 ± 0.27 | 4.41 ± 0.21 | −1.026, 0.99 | −1.029, 0.228 | −1.012, 0.99 | −1.040, 0.81 | |
| 41. | PIEZO2 | 6.64 ± 0.77 | 6.52 ± 0.33 | 6.05 ± 0.61 | 5.86 ± 0.74 | 6.16 ± 0.57 | −1.019, 0.99 | −1.078, 0.159 | −1.111, 0.81 | −1.058, 0.99 | |
| 42. | SLITRK6 | 7.50 ± 0.62 | 6.67 ± 0.96 | 6.46 ± 0.51 | 6.60 ± 0.69 | 6.38 ± 0.42 | −1.125, 0.298 | −1.033, 0.482 | −1.010, 0.99 | −1.046, 0.99 | |
| 43. | KCNA1 | 6.40 ± 1.00 | 5.83 ± 0.82 | 5.20 ± 0.85 | 5.79 ± 0.86 | 4.89 ± 0.69 | −1.098, 0.99 | −1.121, 0.159 | −1.007, 0.99 | −1.193, 0.356 | |
| 44. | LOC100507560 | 5.59 ± 0.52 | 5.20 ± 0.52 | 4.82 ± 0.22 | 4.81 ± 0.18 | 4.83 ± 0.25 | −1.075, 0.638 | −1.079, 0.088 | −1.081, 0.85 | −1.077, 0.655 |
The Adj. P is based on the marginally adjusted 𝑝 values by the Benjamini-Hochberg-FDR correction at α = 0.05; Median ± Interquartile range.
We then investigated the differential regulation of these 44 candidate genes in HNS, HS, COPD patients, and Stage I and II patients over 50 years (Table 7; ≥50 years). In this age group, gene expression was not significantly different. This was surprising, as the symptoms of COPD worsen with age and one would expect associated gene regulation to become more dysregulated. Specifically, a comparison of HS to HNS in subjects over 50 years revealed that expression of 16/44 (36.4%) of the candidate genes and 5/17 (29.4%) 17 novel genes were significantly different (Table 7; ≥50 years; HS v HNS). Investigation of gene expression in COPD versus HS in subjects over 50 years revealed that expression of 3/44 (6.8%) of the 44 candidate genes and 1/17 (5.9%) of the novel genes were significantly different (Table 7; ≥ 50 years; COPD v HS). Subsequently, we investigated the differential gene expression in Stage I and II COPD patients over 50 years and determined that 1/44% (2.3%; Stage I) and 0/44 (0%; Stage II), respectively, were significantly different in patients ≤50 years. In our cohort of 17 novel genes, we determined that expression of only 1/17% (5.9%; Stage I) and 0/17 (0%; Stage II) genes, respectively, were significantly different in patients ≤50 years (Table 7; ≥50 years; Stage I and Stage II v HS). Furthermore, in COPD patients over 50 years, no genes were significantly different across all four analysis groups (i.e. HS v HNS; COPD v HS; Stage I v HS and Stage II v HS).
Investigation of the effect of cigarette pack number per year on differential gene expression in HS and COPD (Stage I and II) patients
Here, we also investigated the effect of cigarette pack number per year (i.e. < or ≥50 cigarette packs/year) on the gene expression in HS, COPD patients, and Stage I and II patients. We analyzed the differential regulation of the 44 candidate genes in HS, COPD patients, and Stage I and II patients who consumed less than 50 packs/year (Table 8; <50 packs/year). In this age group, gene expression was not significantly different. Investigation of gene expression in COPD versus HS in subjects who consumed less than 50 packs/year revealed that expression of 4/44 (9.1%) candidate genes and 2/17 (11.8%) novel genes were significantly different (Table 8; <50 packs/year; COPD v HS). Subsequently, we studied the differential gene expression in Stage II COPD patients who consumed ≥50 packs/year and determined that 4/44 (9.1%) candidate genes, were significantly different in patients ≤50 years. In our cohort of 17 novel genes, we determined that the expression of only 1/17 (5.9%) of Stage II genes was significantly different in patients who consumed ≥50 packs/year compared to HS (Table 8; <50 packs/year; Stage I and Stage II v HS). Furthermore, a certain number of candidate genes were significantly different in subjects who consumed ≥50 packs/year, across in both analysis groups, which included SAMD5, PLAG1 and the novel gene, SLITRK6.
Table 8.
Comparison of relative expression of 44 candidate genes between healthy controls (smokers and non-smokers) and COPD smoker patients (stage I and stage II) in number of pack of cigarette per year, separately.
| Smoking group | Gene Symbol | Healthy Smoker (N = 37) | COPD smoker (N = 6) | COPD stage I smoker (N = 0) | COPD stage II smoker (N = 6) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | |
|---|---|---|---|---|---|---|---|---|---|
| <50 packs per year | 1. | PPP4R4 | 6.69 ± 0.53 | 5.81 ± 0.10 | — | 5.81 ± 0.10 | −1.153, 0.074 | — | −1.153, 0.074 |
| 2. | THSD4 | 7.28 ± 0.38 | 6.99 ± 0.26 | — | 6.99 ± 0.26 | −1.042, 0.369 | — | −1.042, 0.369 | |
| 3. | NRG1 | 3.97 ± 0.52 | 3.66 ± 0.01 | — | 3.66 ± 0.01 | −1.086, 0.428 | — | −1.086, 0.428 | |
| 4. | SCGB1A1 | 14.31 ± 0.36 | 14.40 ± 0.23 | — | 14.40 ± 0.23 | 1.007, 0.762 | — | 1.007, 0.762 | |
| 5. | AHRR | 5.01 ± 0.44 | 5.74 ± 1.43 | — | 5.74 ± 1.43 | 1.145, 0.572 | — | 1.145, 0.572 | |
| 6. | CYP1A1 | 5.93 ± 1.39 | 7.84 ± 1.61 | — | 7.84 ± 1.61 | 1.321, 0.18 | — | 1.321, 0.18 | |
| 7. | CYP1B1 | 8.60 ± 1.25 | 10.10 ± 0.43 | — | 10.10 ± 0.43 | 1.175, 0.189 | — | 1.175, 0.189 | |
| 8. | PRDM11 | 5.41 ± 0.21 | 5.41 ± 0.29 | — | 5.41 ± 0.29 | 1.001, 0.971 | — | 1.001, 0.971 | |
| 9. | CBR3 | 8.18 ± 0.48 | 8.89 ± 0.59 | — | 8.89 ± 0.59 | 1.086, 0.159 | — | 1.086, 0.159 | |
| 10. | AKR1C1 | 9.19 ± 0.39 | 8.91 ± 0.86 | — | 8.91 ± 0.86 | −1.031, 0.737 | — | −1.031, 0.737 | |
| 11. | AKR1C3 | 12.26 ± 0.38 | 12.45 ± 0.12 | — | 12.45 ± 0.12 | 1.015, 0.555 | — | 1.015, 0.555 | |
| 12. | HTR2B | 4.04 ± 0.27 | 4.33 ± 0.36 | — | 4.33 ± 0.36 | 1.072, 0.282 | — | 1.072, 0.282 | |
| 13. | GRM1 | 4.30 ± 0.62 | 4.14 ± 0.25 | — | 4.14 ± 0.25 | −1.038, 0.708 | — | −1.038, 0.708 | |
| 14. | CYP4Z1 | 6.13 ± 0.34 | 6.10 ± 0.05 | — | 6.10 ± 0.05 | −1.006, 0.876 | — | −1.006, 0.876 | |
| 15. | UCHL1 | 10.40 ± 0.84 | 10.34 ± 0.52 | — | 10.34 ± 0.52 | −1.006, 0.908 | — | −1.006, 0.908 | |
| 16. | CABYR | 8.02 ± 0.55 | 8.08 ± 1.23 | — | 8.08 ± 1.23 | 1.007, 0.939 | — | 1.007, 0.939 | |
| 17. | GPRC5A | 7.21 ± 0.38 | 7.80 ± 0.07 | — | 7.80 ± 0.07 | 1.082, 0.094 | — | 1.082, 0.094 | |
| 18. | CCDC37 | 9.24 ± 0.23 | 8.88 ± 0.01 | — | 8.88 ± 0.01 | −1.040, 0.086 | — | −1.040, 0.086 | |
| 19. | GLI3 | 6.57 ± 0.44 | 6.72 ± 0.03 | — | 6.72 ± 0.03 | 1.023, 0.685 | — | 1.023, 0.685 | |
| 20. | ABCC3 | 8.13 ± 0.40 | 7.80 ± 0.39 | — | 7.80 ± 0.39 | −1.042, 0.371 | — | −1.042, 0.371 | |
| 21. | SAMD5 | 4.31 ± 0.34 | 3.44 ± 0.08 | — | 3.44 ± 0.08 | −1.252, 0.003 | — | −1.252, 0.003 | |
| 22. | RASSF10 | 7.33 ± 0.37 | 6.52 ± 1.20 | — | 6.52 ± 1.20 | −1.125, 0.529 | — | −1.125, 0.529 | |
| 23. | USP27X | 7.12 ± 0.44 | 6.61 ± 0.08 | — | 6.61 ± 0.08 | −1.077, 0.18 | — | −1.077, 0.18 | |
| 24. | NR0B1 | 4.41 ± 0.43 | 5.12 ± 0.70 | — | 5.12 ± 0.70 | 1.160, 0.145 | — | 1.160, 0.145 | |
| 25. | PLAG1 | 5.37 ± 0.15 | 4.56 ± 0.34 | — | 4.56 ± 0.34 | −1.178, 0.005 | — | −1.178, 0.005 | |
| 26. | SCGB3A1 | 13.83 ± 0.81 | 13.18 ± 0.05 | — | 13.18 ± 0.05 | −1.049, 0.319 | — | −1.049, 0.319 | |
| 27. | LHX6 | 5.86 ± 0.34 | 6.27 ± 0.70 | — | 6.27 ± 0.70 | 1.070, 0.549 | — | 1.070, 0.549 | |
| 28. | LINC00942 | 4.20 ± 1.01 | 3.86 ± 0.22 | — | 3.86 ± 0.22 | −1.088, 0.417 | — | −1.088, 0.417 | |
| 29. | REEP1 | 10.30 ± 0.30 | 10.13 ± 0.37 | — | 10.13 ± 0.37 | −1.017, 0.629 | — | −1.017, 0.629 | |
| 30. | C6orf164 | 6.39 ± 0.44 | 6.60 ± 0.37 | — | 6.60 ± 0.37 | 1.033, 0.587 | — | 1.033, 0.587 | |
| 31. | LINC00589 | 5.34 ± 0.11 | 5.55 ± 0.21 | — | 5.55 ± 0.21 | 1.040, 0.115 | — | 1.040, 0.115 | |
| 32. | JAKMIP3 | 5.44 ± 0.26 | 5.73 ± 0.11 | — | 5.73 ± 0.11 | 1.052, 0.222 | — | 1.052, 0.222 | |
| 33. | LINC00930 | 7.36 ± 0.41 | 6.71 ± 0.05 | — | 6.71 ± 0.05 | −1.097, 0.022 | — | −1.097, 0.022 | |
| 34. | DNHD1 | 7.08 ± 0.45 | 6.97 ± 0.47 | — | 6.97 ± 0.47 | −1.016, 0.774 | — | −1.016, 0.774 | |
| 35. | TMCC3 | 4.39 ± 0.64 | 4.74 ± 0.44 | — | 4.74 ± 0.44 | 1.081, 0.548 | — | 1.081, 0.548 | |
| 36. | ADH7 | 11.06 ± 0.20 | 11.17 ± 0.31 | — | 11.17 ± 0.31 | 1.010, 0.582 | — | 1.010, 0.582 | |
| 37. | PRKAR2B | 6.48 ± 0.51 | 5.64 ± 0.57 | — | 5.64 ± 0.57 | −1.149, 0.11 | — | −1.149, 0.11 | |
| 38. | GAD1 | 7.65 ± 0.56 | 7.33 ± 0.04 | — | 7.33 ± 0.04 | −1.044, 0.46 | — | −1.044, 0.46 | |
| 39. | LOC338667 | 5.06 ± 0.25 | 4.83 ± 0.07 | — | 4.83 ± 0.07 | −1.047, 0.276 | — | −1.047, 0.276 | |
| 40. | CYB5A | 4.67 ± 0.22 | 4.38 ± 0.02 | — | 4.38 ± 0.02 | −1.066, 0.139 | — | −1.066, 0.139 | |
| 41. | PIEZO2 | 6.46 ± 0.34 | 6.44 ± 0.32 | — | 6.44 ± 0.32 | −1.003, 0.931 | — | −1.003, 0.931 | |
| 42. | SLITRK6 | 7.17 ± 0.10 | 6.61 ± 0.05 | — | 6.61 ± 0.05 | −1.085, 0.001 | — | −1.085, 0.001 | |
| 43. | KCNA1 | 5.76 ± 0.52 | 5.45 ± 0.56 | — | 5.45 ± 0.56 | −1.056, 0.517 | — | −1.056, 0.517 | |
| 44. | LOC100507560 | 5.09 ± 0.46 | 4.90 ± 0.30 | — | 4.90 ± 0.30 | −1.038, 0.607 | — | −1.038, 0.607 | |
| Smoking group | Gene Symbol | Healthy Smoker (N = 22) | COPD smoker (N = 15) | COPD stage I smoker (N = 9) | COPD stage II smoker (N = 6) | Fold Regulation, adjusted p-value (COPD vs. HS) | Fold Regulation, adjusted p-value (stage I vs. HS) | Fold Regulation, adjusted p-value (stage II vs. HS) | |
| ≥50 packs per year | 1. | PPP4R4 | 5.95 ± 0.89 | 5.49 ± 0.71 | 5.82 ± 0.43 | 5.23 ± 0.82 | −1.084, 0.047 | −1.023, 0.99 | −1.138, 0.054 |
| 2. | THSD4 | 7.34 ± 0.56 | 7.11 ± 0.54 | 7.51 ± 0.32 | 6.81 ± 0.50 | −1.032, 0.178 | 1.022, 0.99 | −1.078, 0.02 | |
| 3. | NRG1 | 4.06 ± 0.47 | 3.82 ± 0.41 | 3.97 ± 0.50 | 3.70 ± 0.28 | −1.063, 0.014 | −1.021, 0.99 | −1.096, 0.03 | |
| 4. | SCGB1A1 | 14.37 ± 0.18 | 14.12 ± 0.33 | 14.20 ± 0.26 | 14.06 ± 0.37 | −1.018, 0.001 | −1.012, 0.135 | −1.022, 0.001 | |
| 5. | AHRR | 4.55 ± 0.67 | 5.18 ± 0.72 | 5.47 ± 0.86 | 4.96 ± 0.52 | 1.138, 0.001 | 1.203, 0.001 | 1.092, 0.169 | |
| 6. | CYP1A1 | 6.15 ± 1.89 | 7.78 ± 1.88 | 8.25 ± 1.49 | 7.42 ± 2.18 | 1.265, 0.003 | 1.342, 0.002 | 1.207, 0.059 | |
| 7. | CYP1B1 | 7.67 ± 2.10 | 9.43 ± 1.23 | 9.95 ± 0.63 | 9.03 ± 1.49 | 1.229, 0.006 | 1.298, 0.004 | 1.177, 0.15 | |
| 8. | PRDM11 | 5.35 ± 0.34 | 5.25 ± 0.29 | 5.32 ± 0.22 | 5.19 ± 0.34 | −1.019, 0.239 | −1.005, 0.99 | −1.030, 0.99 | |
| 9. | CBR3 | 8.15 ± 0.70 | 8.33 ± 0.63 | 8.57 ± 0.46 | 8.14 ± 0.71 | 1.022, 0.383 | 1.052, 0.428 | −1.001, 0.99 | |
| ≥50 packs per year | 10. | AKR1C1 | 8.51 ± 1.15 | 8.21 ± 1.25 | 8.36 ± 1.30 | 8.09 ± 1.27 | −1.037, 0.475 | −1.018, 0.99 | −1.051, 0.99 |
| 11. | AKR1C3 | 11.8 ± 20.73 | 11.88 ± 0.68 | 12.16 ± 0.49 | 11.66 ± 0.76 | 1.007, 0.968 | 1.029, 0.92 | −1.014, 0.99 | |
| 12. | HTR2B | 4.23 ± 0.31 | 4.24 ± 0.33 | 4.18 ± 0.43 | 4.29 ± 0.23 | 1.002, 0.884 | −1.012, 0.99 | 1.013, 0.99 | |
| 13. | GRM1 | 4.03 ± 0.30 | 4.16 ± 0.59 | 4.20 ± 0.39 | 4.13 ± 0.74 | 1.032, 0.771 | 1.042, 0.811 | 1.026, 0.99 | |
| 14. | CYP4Z1 | 6.25 ± 0.60 | 5.92 ± 0.40 | 5.93 ± 0.48 | 5.91 ± 0.35 | −1.056, 0.034 | −1.055, 0.706 | −1.059, 0.379 | |
| 15. | UCHL1 | 8.65 ± 1.48 | 9.28 ± 1.87 | 9.52 ± 1.61 | 9.09 ± 2.13 | 1.073, 0.11 | 1.100, 0.463 | 1.051, 0.9 | |
| 16. | CABYR | 6.83 ± 1.24 | 7.40 ± 1.44 | 8.06 ± 1.18 | 6.91 ± 1.50 | 1.083, 0.157 | 1.180, 0.01 | 1.011, 0.99 | |
| 17. | GPRC5A | 7.54 ± 0.43 | 8.05 ± 0.65 | 7.92 ± 0.75 | 8.16 ± 0.58 | 1.068, 0.003 | 1.051, 0.353 | 1.083, 0.008 | |
| 18. | CCDC37 | 9.37 ± 0.55 | 9.39 ± 0.56 | 9.40 ± 0.66 | 9.38 ± 0.51 | 1.002, 0.802 | 1.003, 0.99 | 1.002, 0.99 | |
| 19. | GLI3 | 6.76 ± 0.59 | 6.62 ± 0.41 | 6.73 ± 0.45 | 6.54 ± 0.37 | −1.021, 0.25 | −1.004, 0.99 | −1.034, 0.946 | |
| 20. | ABCC3 | 7.86 ± 0.62 | 7.65 ± 0.82 | 7.79 ± 0.74 | 7.53 ± 0.90 | −1.027, 0.428 | −1.008, 0.99 | −1.044 0.82 | |
| 21. | SAMD5 | 3.95 ± 0.26 | 3.97 ± 0.50 | 3.98 ± 0.29 | 3.96 ± 0.64 | 1.005, 0.53 | 1.007, 0.99 | 1.003, 0.99 | |
| 22. | RASSF10 | 7.04 ± 0.60 | 6.61 ± 0.52 | 6.79 ± 0.33 | 6.47 ± 0.62 | −1.065, 0.002 | −1.036, 0.99 | −1.088, 0.018 | |
| 23. | USP27X | 7.13 ± 0.40 | 6.65 ± 0.44 | 6.72 ± 0.29 | 6.60 ± 0.54 | −1.072, 0.001 | −1.062, 0.017 | −1.081, 0.001 | |
| 24. | NR0B1 | 4.39 ± 0.80 | 4.77 ± 0.79 | 5.00 ± 0.87 | 4.59 ± 0.70 | 1.087, 0.032 | 1.139, 0.058 | 1.047, 0.99 | |
| 25. | PLAG1 | 5.24 ± 0.58 | 4.84 ± 0.57 | 4.98 ± 0.58 | 4.74 ± 0.58 | −1.083, 0.012 | −1.054, 0.99 | −1.106, 0.07 | |
| 26. | SCGB3A1 | 13.93 ± 0.59 | 13.25 ± 0.79 | 13.81 ± 0.51 | 12.82 ± 0.71 | −1.051, 0.002 | −1.008, 0.99 | −1.087, 0.001 | |
| 27. | LHX6 | 5.80 ± 0.45 | 6.09 ± 0.52 | 6.08 ± 0.54 | 6.10 ± 0.53 | 1.050, 0.088 | 1.048, 0.498 | 1.050, 0.271 | |
| 28. | LINC00942 | 3.84 ± 0.68 | 4.16 ± 1.06 | 4.31 ± 1.30 | 4.05 ± 0.87 | 1.083, 0.29 | 1.121, 0.093 | 1.053, 0.99 | |
| 29. | REEP1 | 9.78 ± 0.56 | 9.64 ± 0.73 | 9.55 ± 0.86 | 9.71 ± 0.65 | −1.015, 0.484 | −1.024, 0.99 | −1.007, 0.529 | |
| 30. | C6orf164 | 6.07 ± 0.56 | 6.19 ± 0.64 | 6.21 ± 0.45 | 6.17 ± 0.78 | 1.020, 0.376 | 1.023, 0.99 | 1.016, 0.99 | |
| 31. | LINC00589 | 5.39 ± 0.33 | 5.39 ± 0.33 | 5.35 ± 0.18 | 5.42 ± 0.41 | 1.000, 0.843 | −1.008, 0.99 | 1.006, 0.99 | |
| 32. | JAKMIP3 | 5.26 ± 0.63 | 5.74 ± 0.88 | 5.86 ± 0.94 | 5.64 ± 0.87 | 1.091, 0.036 | 1.113, 0.019 | 1.071, 0.233 | |
| 33. | LINC00930 | 6.87 ± 0.57 | 6.60 ± 0.65 | 6.73 ± 0.70 | 6.50 ± 0.63 | −1.041, 0.204 | −1.021, 0.99 | −1.057, 0.29 | |
| 34. | DNHD1 | 7.15 ± 0.54 | 7.25 ± 0.70 | 7.36 ± 0.78 | 7.17 ± 0.65 | 1.014, 0.721 | 1.029, 0.99 | 1.003, 0.99 | |
| 35. | TMCC3 | 4.07 ± 0.47 | 4.43 ± 0.45 | 4.51 ± 0.50 | 4.36 ± 0.41 | 1.088, 0.001 | 1.107, 0.055 | 1.072, 0.348 | |
| 36. | ADH7 | 10.79 ± 0.72 | 10.65 ± 0.65 | 10.84 ± 0.59 | 10.50 ± 0.69 | −1.013, 0.245 | 1.005, 0.99 | −1.027, 0.99 | |
| 37. | PRKAR2B | 6.45 ± 0.74 | 5.85 ± 0.61 | 5.83 ± 0.68 | 5.87 ± 0.59 | −1.103, 0.003 | −1.106, 0.082 | −1.099, 0.062 | |
| 38. | GAD1 | 6.20 ± 0.96 | 7.39 ± 0.75 | 7.38 ± 0.82 | 7.40 ± 0.74 | 1.192, 0.001 | 1.190, 0.001 | 1.195, 0.002 | |
| 39. | LOC338667 | 5.20 ± 0.23 | 5.07 ± 0.26 | 5.05 ± 0.18 | 5.09 ± 0.31 | −1.026, 0.056 | −1.031, 0.865 | −1.022, 0.99 | |
| 40. | CYB5A | 4.72 ± 0.19 | 4.54 ± 0.25 | 4.63 ± 0.25 | 4.47 ± 0.25 | −1.040, 0.012 | −1.018, 0.99 | −1.054, 0.004 | |
| 41. | PIEZO2 | 6.49 ± 0.86 | 5.87 ± 0.63 | 5.86 ± 0.72 | 5.88 ± 0.59 | −1.106, 0.007 | −1.107, 0.268 | −1.104, 0.168 | |
| 42. | SLITRK6 | 6.83 ± 0.88 | 6.18 ± 0.83 | 6.28 ± 1.11 | 6.11 ± 0.58 | −1.105, 0.011 | −1.088, 0.443 | −1.119, 0.03 | |
| 43. | KCNA1 | 5.80 ± 0.97 | 5.10 ± 0.76 | 5.39 ± 0.89 | 4.89 ± 0.58 | −1.137, 0.007 | −1.076, 0.99 | −1.186, 0.032 | |
| 44. | LOC100507560 | 5.46 ± 0.61 | 4.93 ± 0.51 | 4.94 ± 0.59 | 4.92 ± 0.48 | −1.108, 0.001 | −1.104, 209 | −1.108, 0.087 |
The Adj. P is based on the marginally adjusted 𝑝 values by the Benjamini-Hochberg-FDR correction at α = 0.05; Median ± Interquartile range.
In COPD patients who consumed ≥50 packs/year, we observed a significant change in gene expression in 22/44 (50%) candidate genes and 9/17 (52.9%) novel genes compared with HS (Table 8; ≥50 packs/year; COPD v HS). We also investigated differential gene expression in Stage I and II COPD patients who consumed ≥50 packs/year and determined that 7/44 (15.9%; Stage I) and 10/44 (22.7%; Stage II) candidate genes were significantly different compared to gene expression in HSs. In our cohort of 17 novel genes, we determined that gene expression in 2/17% (11.8%; Stage I) and 4/17% (23.5%; Stage II) was significantly different in COPD patients who consumed ≥50 packs/year (Table 8; ≥50 packs/year; Stage I and Stage II v HS). In addition, a certain number of the 44 candidate genes were significantly different across all four-analysis groups, which included USP27X, CYP1A1 and the novel genes, PRKAR2B and GAD1.
Discussion
Chronic obstructive pulmonary disease (COPD) is a progressive inflammatory disease characterized by airway obstruction and is predicted to be among the first three causes of death worldwide1,2. A significant degree of clinical heterogeneity has been observed in COPD patients. In functional terms, all COPD patients experience a loss in lung function, as measured using FEV1 and FVC. However, these clinical parameters are not optimal and FEV1 has been shown to correlate weakly with clinical outcome and health status19,20. Currently, there is an unmet clinical need to identify novel biomarkers that will facilitate improved diagnosis and prognosis in COPD.
To date, COPD has been shown to develop in 30% of smokers, with smoking being one of the main risk factors associated with the development of COPD3. The aim of this project was to identify candidate novel biomarkers, which may provide future novel therapeutic targets, in order to facilitate the treatment of COPD using machine-based learning algorithms and penalized regression models. In this study, 59 healthy smokers, 53 healthy non-smokers and 21 COPD smokers (9 GOLD stage I and 12 GOLD stage II) were included (n = 133). 20,097 probes were generated from SAE microarray data obtained from these subjects previously14. Consequently, 44 candidate genes were identified to be associated with the occurrence or progression of COPD, or lung function. Of these 44 genes, 27 have been previously reported in the literature to be associated with COPD or lung function (FVC, FEV1 or the FEV1/FVC ratio). In this study, we also identified 17 genes not previously detected in COPD studies that may represent novel biomarkers in the diagnosis and prognosis of COPD. In our analyses among healthy non-smokers and healthy smokers and COPD patients (GOLD stage I and II), the most significantly regulated novel genes were: PRKAR2B, GAD1, LINC00930 and SLITRK6.
PRKAR2B is a protein kinase type II-beta regulatory subunit dependent on cAMP and encoded by the PRKAR2B gene in human21. In our overall analyses, expression of PRKAR2B was significantly downregulated in healthy smokers and in COPD patients (and in COPD stage II) compared to healthy non-smokers. Furthermore, in males, PRKAR2B expression was also significantly downregulated in healthy smokers and in COPD patients compared to healthy non-smokers. In females, these differences were less pronounced. In subjects less than 50 years, PRKAR2B expression was significantly downregulated in healthy smokers and in COPD patients compared to healthy non-smokers. In patients over 50 years, these differences were less pronounced. With regards to smoking exposure, COPD patients who smoked more than 50 packs per year had significantly lower PRKAR2B gene expression than healthy non-smokers. This decrease was not evident in COPD patients who smoked less than 50 cigarette packs per year. Thus, we hypothesise that PRKAR2B may represent a previously unknown factor both in pathogenesis of COPD and smoking exposure. PRKAR2B is an important protein kinase in cAMP signaling, and other researchers have demonstrated that cAMP is a protective factor in the lung and COPD. Furthermore, cAMP has been shown to attenuate pro-inflammatory responses whilst concomitantly increasing anti-inflammatory responses in a number of innate immune cells22. The reduced PRKAR2B gene expression observed in this study may reveal PRKAR2B as a novel target in the treatment of COPD.
In contrast, in this study we also observed a significant upregulation of the novel gene, GAD1, in healthy smokers and COPD patients (and in stage II) compared to healthy non-smokers. In male only subjects, this pattern was replicated. The increase in expression of GAD1 in healthy smokers and COPD patients compared to healthy non-smokers was marginally less significant than in male subjects, as expected. In subjects younger than 50 years, there was a more significant increase in GAD1 expression in healthy smokers and COPD patients compared to subjects over 50 years and also the non-smokers. Smoking exposure only significantly increased GAD1 levels in healthy smokers and COPD patients who smoked more than 50 packs per year. There were no significant changes in GAD1 expression in subjects who smoked less than 50 packs per year. Other studies have shown that levels of γ-aminobutyric acid (GABA) and glutamic acid decarboxylase 1 (GAD1), the enzyme that synthesizes GABA, are significantly increased in neoplastic tissues23. Furthermore, other researchers have shown that the GAD1 promoter is hypermethylated in a number of cancer cells. This effect was shown to lead to the production of high levels of GAD1, as opposed to gene silencing which one would expect. The GAD1 promoter contains a number of CpG island motifs which facilitate this hypermethylation. In this study, we hypothesise that the increased levels of GAD1 detected following smoking exposure and COPD could mean that GAD1 is an important target in the treatment of this smoking-related disease. Previous studies have demonstrated that patients with COPD are at an increased risk for both the development of primary lung cancer, as well as poor outcome after lung cancer diagnosis and treatment24. Targeting the knockdown of GAD1 in COPD may attenuate the increased risk of lung cancer in COPD patients.
LINC00930 was an additional novel gene detected in this study using machine-based learning. This is a “long intergenic non-protein coding (linc) RNA 930” (https://www.genecards.org/cgi-bin/carddisp.pl?gene=LINC00930). Interestingly, some other novel genes including LINC00942 and LINC00589 were also detected in this study. However, they were not as significantly regulated by smoking exposure or in COPD patients. In general, lincRNAs are found in between coding genes rather than antisense to them or within introns. Although the specific function of lincRNAs is not well known, they are thought to contribute to RNA stability in cells and hence gene expression. In our study, LINC009030 expression was significantly increased in healthy smokers overall, and in males and females, compared to healthy non-smokers. LINC009030 expression was significantly increased in smokers less than 50 years when only compared to non-smokers. In the over-50 age group, COPD patients had significantly less LINC009030 expression compared to healthy non-smokers. Regarding the smoking exposure, COPD patients and Stage II COPD patients who smoked less than 50 packs per year had significantly less LINC009030 expression compared to healthy non-smokers. In this study, LINC009030 was the only novel gene whose expression was significantly upregulated in smokers while significantly downregulated in COPD patients. Additional experimentation is required for elucidating the mechanism underlying this result in order to evaluate the therapeutic potential of targeting LINC009030 in smoking-related morbidities and in COPD patients.
SLITRK6 was the last novel gene, which we determined to be significantly regulated in smoking and in COPD in this study. SLITRK6 is a member of the SLITRK family of neuronal transmembrane proteins that was discovered as a bladder tumor antigen using suppressive subtractive hybridization25. Using immunohistochemistry, SLITRK6 has been shown to be extensively expressed in multiple epithelial tumors, including lung, bladder and breast cancer, as well as in glioblastoma25. In our study, we demonstrated that SLITRK6 was significantly downregulated in smokers and COPD patients (including stage II) compared to healthy non-smokers. Male smokers and COPD patients also had significantly lower SLITRK6 expression compared to non-smokers. In females, SLITRK6 expression was significantly less in smokers, but not COPD patients, compared to non-smokers. Smokers and COPD patients (including stage II) aged less than 50 years had significantly lower SLITRK6 expression compared to non-smokers. These effects were not evident in subjects over 50 years. Concerning the smoking exposure, COPD patients (including stage II) who smoked more than 50 packs per year had significantly lower SLITRK6 expression compared to non-smokers. This effect was also evident in COPD patients (including stage II) who smoked less than 50 packs per year. The highlight of this study was that the expression of the oncogenesis-promoting enzyme, GAD1, is significantly increased in response to smoking exposure and in COPD. Although SLITRK6 is considered a tumourogenesis promoting factor, it was significantly decreased in this study in responses to smoking exposure and in COPD. Here, we hypothesise that GAD1 may represent a better target in order to attenuate the incidence of cancer following COPD. However, further investigations are needed to explain the exact function of SLITRK6 in smoking-associated morbidities and in COPD.
More recently, machine-based learning algorithms have gained increasing attention in bioinformatics and biology research26,27. In contrast, regularization-based regression models (e.g. LASSO logistic regression) have already been used widely in microarray analysis28. Microarray analysis has a number of limitations including overfitting and multi-collinearity. In order to address these issues, regularization of parameters is required29. In this study, the area under the receiver operator characteristic curve (AUC), the sensitivity and specificity, and the misclassification error rate were quantitated for machine-based learning algorithm and penalty-based statistical method used. In this study based on repeated 5-CV, the elastic-net, random forest, and LASSO regularized logistic regression models were found to perform better than the naive Bayes, ridge, gradient boosting machines, adaptive boosting classification trees, extension of LASSO, artificial neural network, support vector machines, and decision tree models, respectively. Elastic-net regularization produced a sparse model with good prediction accuracy and good grouping-capability. This result is in keeping with those from previous studies, which demonstrated that elastic-net frequently performs better than ridge and LASSO for model selection consistency and prediction accuracy in microarray datasets28,30,31. Therefore, the results of this study are in agreement with those from previous ones.
In summary, we employed machine-based learning algorithms and penalized regression models in order to identify 44 candidate genes, whose expression was significantly regulated by smoking exposure and/or COPD. We also identified 17 novel genes which were not previously determined to be associated with smoking exposure or COPD. We determined that four of these novel genes, namely PRKARB2, GAD1, LINC00930 and SLITKR6, were the most significantly regulated by smoking exposure or in COPD. We also determined that elastic-net logistic regression in our dataset had a higher accuracy rate compared to the other algorithms. Therefore, in microarray data, elastic-net logistic regression may provide a useful methodology for future studies in the discovery of novel diagnostic- and prognostic-biomarkers, and novel therapeutic targets in the treatment of COPD and other smoking-related diseases.
The strengths of this study include the use of modern and accepted computational methods, the address of potential sources of bias, validation of all of the results by literature review, the use of appropriate cross-validation method (repeated 5-CV), enrichment analysis and the use of STRING networks. The main limitations of the study are the small sample sizes and that this is a case-control study only. Specifically, this study does not establish the respective associations between the 44 candidate genes and any COPD outcomes (e.g. lung function changes, exacerbations or mortality). Therefore, future studies by us will investigate and validate the candidacy of our 44 novel genes as novel therapeutic targets in COPD using larger patient cohorts. Furthermore, these studies, will investigate the association between these 44 novel genes and the change in lung function over time (FEV1 or FEV1/FVC ratio), incidence of exacerbations and mortality in COPD patients.
Methods
Study subjects and dataset
In this study, 59 healthy smokers, 53 healthy non-smokers and 21 COPD smokers (9 of stage I, GOLD I and 12 of stage II, GOLD II) were included (Total: n = 133). Subjects were predominantly male (n = 95; 71.4%) and Caucasian (n = 48; 36.1%; Table 1). Pulmonary function tests from COPD patients revealed that forced vital capacity (FVC), forced expiratory volume 1 (FEV1) and the FEV1/FVC ratio were significantly lower in these patients compared with healthy smokers and healthy non-smokers (Table 1). From these subjects, the raw data of gene expression architecture in the small airway epithelium (SAE) cells of COPD was used14. 20,097 probes from 133 subjects were generated. Genome-wide gene expression analysis (GWAS) was performed using HG-U133 Plus 2.0 array (Affymetrix, Santa Clara, CA)14. Overall microarray quality was verified by the criteria: (1) 3′/5′ ratio for GAPDH ≤3; and (2) scaling factor ≤10.0. The captured image data from the HG-U133 Plus 2.0 arrays was processed using MAS5 algorithm. The data was normalized using GeneSpring version 7.3.1 (Agilent technologies, Palo Alto, CA). See Supplemental Methods for further details. The raw data is available at the Gene Expression Omnibus (GEO) site (http://www.ncbi.nlm.nih.gov/geo/), accession number for this dataset is GSE20257.
Gene expression analysis
Raw data (.CEL format) files were qualified, normalized, statistical comparison, removing batch effects and other unwanted variation were performed by “Affy,” “Limma” and “SVA” R packages, respectively. The cutoff of false discovery rate and fold-change for differentially expressed genes was considered at level of 0.10, and more than 2, respectively.
Module identification
Sample progression discovery (SPD) as a novel unsupervised computational approach to identify patterns of biological progression underlying microarray gene expression data. SPD assumes that individual samples of a microarray dataset are related by an unknown biological process, and that each sample represents one unknown point along the progression of that process. SPD aims to organize the samples in a manner that reveals the underlying progression and to simultaneously identify subsets of genes that are responsible for that progression. This method does not depend on prior knowledge and only uses gene expression information32. In this method, divisive/consensus k-means as a clustering gene algorithm was used (200 iterations in each consensus k-means partitioning, and 0.7 threshold for module coherence). Also, least number of genes in each modules was 10. SPD analysis was done by MATLAB 7 software.
Machine learning (ML) algorithms
Various ML algorithms including AdaBoost classification trees, decision tree, Gradient Boosting machines, Naive Bayes, neural network, random forest, support vector machine were performed in order to find genes associated with occurrence and progression of COPD, and the best ML method which has best accuracy and performance to predict COPD. All ML methods were applied using “adabag”, “CART”, “gbm”, “naivebayes”, “neuralnet”, “‘randomForest”, and “e1071” R packages.
Adaptive LASSO, elastic-net, and ridge logistic regression
The ridge regression uses an L2 penalty to regularize parameters, all of the estimated coefficients are nonzero, and hence no gene selection is performed. But, LASSO regression use the L1 penalty instead, and hence provide automatic gene selection. In other hand, ridge penalty tends to shrink the coefficients of correlated variables toward each other, good for multi-collinearity, grouped selection. But, the lasso penalty is somewhat indifferent to the choice among a set of strong but correlated variables. Therefore, LASSO is good for simultaneous estimation and eliminating trivial genes but not good for grouped selection. Elastic-net is introduced as a compromise between these two techniques, and has a penalty which is a mix of L1 and L2 penalty, combine strength between ridge and lasso33. In adaptive LASSO regression where adaptive weights, inverse absolute value of LASSO coefficient was used for each variable as its weight in adaptive LASSO, are used for penalizing different coefficients in the L1 penalty. Similar to the lasso, the adaptive lasso is shown to be near-minimax optimal. Unlike to the LASSO, the adaptive LASSO is consistent for gene selection34. The mentioned penalized logistics regression methods were done by “glmnet” R package (https://cran.r-project.org/package=glmnet). In Table 9, the summary of each machine-learning and penalized statistical methods with some of advantages and limitations were mentioned.
Table 9.
Gene selection methods: Definitions, acronyms and main advantages and limitations.
| Gene Selection Method Name | Gene Selection Method Acronym | Main Advantages | Main Limitations |
|---|---|---|---|
| Least Absolute Shrinkage and Selection Operator | LASSO | (1) Smaller mean squared error (MSE) than conventional methods; (2) It is good for simultaneous estimation and eliminating trivial genes; (3) Coefficients being easy to implement is another of the merits. |
(1) It is not good for grouped selection; (2) For highly correlated variables, conventional methods have predictive performance empirically observed to be better than LASSO; (3) This method has shown to not always provide consistent variable selection; (4) Its estimators are biased always; (5) Its efficiency depends greatly on the number of dimension of genes. |
| Adaptive Least Absolute Shrinkage and Selection Operator | Adapt. LASSO | (1) This method has all of advantages of the LASSO. (2) This method uses adaptive weights to penalize coefficients differently; (3) Adaptive LASSO provides a more consistent solution than LASSO. |
(1) It is not good for grouped selection; (2) For highly correlated variables, conventional methods have predictive performance empirically observed to be better than adapt. LASSO; (3) Its estimators are biased always; (4) Its efficiency depends greatly on the number of dimension of genes. |
| Elastic net regularization | Elastic net | (1) This method selects groups of correlated variables together, shares nice properties of both the LASSO and ridge; (2) It can be considered for situations with p > n, it allows the number of selected features to exceed the sample size; (3) This method has predictive performance better than LASSO and ridge. |
(1) It can only apply to two-class feature selection problems, it cannot resolve multi-class feature selection problems directly; (2) Its estimators aren’t robust against outliers |
| Ridge Logistic Regression | Ridge | (1) It handles the multi-collinearity problem (2) Ridge regression can reduce the variance (with an increasing bias); (3) Can improve predictive performance than ordinary least square approach. |
(1) It is not able to shrink coefficients to exactly zero; (2) It cannot perform variable selection; it includes all of predictors (e.g. genes) in the final model; (3) It cannot handles the overfitting problem. |
| Support Vector Machines | SVM | (1) It has a regularization parameter for avoiding overfitting; (2) It uses the kernel trick; (3) It is defined by a convex optimization problem (no local optimization); (4) It is a powerful classifier that works well on a wide range of classification problems, in other words, it is very good when we have no idea on the data; (5) It can apply for high dimensional and not linearly separable situations. |
(1) Choosing a good kernel function is not easy; (2) It has several key parameters that need to be set correctly to achieve the best classification results for any given problem; (3) Long training time for large datasets and large amount of training data; it was computationally intensive, especially the grid search for tuning its parameters; (4) Difficult to understand and interpret the final model, variable weights and individual impact. |
| Gradient Boosting Machines (stochastic) | GBM | (1) It can apply for high dimensional situations; (2) It works well in the situation with a lot of main and interaction parameters; (3) It can automatically select variables; (4) It is robust to outliers and missing data; (5) It can handle the numerous correlated and irrelevant variables problems; (6) It is an ensemble learning. |
(1) Long training time for large datasets; (2) Difficult to understand and interpret the model; (3) Prone to overfitting. |
| Naive Bayes | NB | (1) It is easy to implement as a single learning; (2) If the its conditional independence assumption actually holds, a Naive Bayes classifier will converge quicker than discriminative models (e.g. logistic regression); (3) It needs less training data than other algorithms; |
(1) Class conditional independence assumption for all of variables (e.g. genes); (2) It is defined by a local optimization problem. |
| Random Forest | RF | (1) It can apply for high dimensional situations; (2) It is robust to outliers and missing data; (3) It has less variance than a single decision tree; (4) Training each tree perform independently. |
(1) It is complex; (2) It requires more computational resources and are also less intuitive; (3) Its prediction process using random forests is time-consuming than decision trees; (4) It assumes that model errors are uncorrelated and uniform. |
| Artificial Neural Network | ANN | (1) It is easy to implement; (2) It can approximate any function between the independent and dependent variables; (3) It handles all possible interactions between the dependent variables; (4) It does not require any assumptions, in other words, it is very good when we have no idea on the data. |
(1) It solved for local optimization; (2) Parameters are hard to interpret; (3) Long training time for large neural networks. |
| Decision Trees | RT | (1) Easy to interpret and explain as a single learning; (2) It is very fast; (3) Its estimators are robust against outliers; (4) Can be combined with other decision techniques; (5) It handles missing values and filling them in with the most probable value. |
(1) Prone to overfitting; (2) Instability; (3) This method has predictive performance worse than random forest; (4) It solved for local optimization. |
| AdaBoost Classification Trees (Adaptive Boosting) | ABCT | (1) It can be less susceptible to the overfitting problem than most learning algorithms; (2) It combines a set of weak learners in order to form a strong classifier and selection of weak classifier is easy; (3) It is a machine learning meta-algorithm. |
(1) It can be sensitive to noisy data and outliers; (2) Requirement of a large amount of training data and long training time. |
Gene set enrichment analysis
Gene set enrichment analysis is a method to identify classes of genes that are over-represented in a large set of genes and may have an association with disease phenotypes (e.g. occurrence of COPD). The Comprehensive gene set enrichment analysis web server 2016 update called “Enrichr” was applied35.
Cross-validation, stability and accuracy
K-fold cross-validation scheme (k-cv) is a very commonly employed technique used to evaluate classifier performance. K-CV estimation of the error is the average value of the errors committed in each fold. Thus, the K-CV error estimator depends on two factors: the training set and the partition into folds. Sensitivity analysis was performed to changes in the training set and sensitivity to changes in the folds36. The bootstrap (or subsampling) is another way to bring down the high variability of cross-validation, to aims stability selection37. Repeated cross-validation is a good strategy for (a) optimizing the complexity of regression models and (b) for a realistic estimation of prediction errors when the model is applied to new cases38,39. In the present study, the algorithms split the data set by using repeated random 100 times sub-sampling in 5-fold cross-validation, permuting the sample labels every time. Cross-validated performance was summarized by observed sensitivity and specificity, and misclassification error rate. Furthermore, the area under the Receiver Operator Characteristic (ROC) curve (AUC), was used to calculate of classifiers performance40,41. Also, in order to assessing literature validation for any results, literature mining was used in the PubMed databank. Interactive cluster heatmaps was applied by “heatmaply” R package (https://cran.r-project.org/web/packages/heatmaply)42. A heatmap is a popular graphical method for visualizing high-dimensional data. A static heatmap, as an interactive heatmaps, was used to represent biological data, in which colors are used to represent the values (importance index) in a matrix where columns and rows are the machine learning and statistical methods (instances) and genes selected (attributes), respectively. Rows and columns are sorted using a hierarchical clustering technique43. The study’s follow chart is shown in Fig. 1.
Electronic supplementary material
Acknowledgements
This work was supported by research grants from Tarbiat Modares University (TMU; Grant No. 1287063) and the National Institute for Medical Research Development (NIMAD; Grant No. 958812) awarded to A.K., S.M. and S.A.
Author Contributions
S.M., A.K. and S.A. performed experiments and analysis. A.K. and S.A. contributed to experimental design. S.M. and S.A. contributed to statistical analysis. M.D. and M.E.A. analysed data and wrote the paper. S.C.D. and A.K. contributed to manuscript drafting.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33986-8.
References
- 1.Zhao J, et al. Smoking status and gene susceptibility play important roles in the development of chronic obstructive pulmonary disease and lung function decline: A population-based prospective study. Medicine. 2017;96:e7283. doi: 10.1097/md.0000000000007283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. The lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remoortel HV, et al. Risk Factors and Comorbidities in the Preclinical Stages of Chronic Obstructive Pulmonary Disease. American Journal of Respiratory and Critical Care Medicine. 2014;189:30–38. doi: 10.1164/rccm.201307-1240OC. [DOI] [PubMed] [Google Scholar]
- 4.Postma DS, Bush A, van den Berge M. Risk factors and early origins of chronic obstructive pulmonary disease. Lancet (London, England) 2015;385:899–909. doi: 10.1016/s0140-6736(14)60446-3. [DOI] [PubMed] [Google Scholar]
- 5.Raghavan D, Varkey A, Bartter T. Chronic obstructive pulmonary disease: the impact of gender. Current opinion in pulmonary medicine. 2017;23:117–123. doi: 10.1097/mcp.0000000000000353. [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg SR, Kalhan R, Mannino DM. Epidemiology of Chronic Obstructive Pulmonary Disease: Prevalence, Morbidity, Mortality, and Risk Factors. Seminars in respiratory and critical care medicine. 2015;36:457–469. doi: 10.1055/s-0035-1555607. [DOI] [PubMed] [Google Scholar]
- 7.Rajput C. Chronic Obstructive Pulmonary Disease Meta Genome-Wide Association Studies. New Insights into the Genetics of Chronic Obstructive Pulmonary Disease. American journal of respiratory cell and molecular biology. 2017;57:1–2. doi: 10.1165/rcmb.2017-0070ED. [DOI] [PubMed] [Google Scholar]
- 8.Silverman EK, Loscalzo J. Network medicine approaches to the genetics of complex diseases. Discovery medicine. 2012;14:143–152. [PMC free article] [PubMed] [Google Scholar]
- 9.Hardin M, Silverman EK. Chronic Obstructive Pulmonary DiseaseGenetics: A Review of the Past and a Look Into the Future. Chronic obstructive pulmonary diseases (Miami, Fla.) 2014;1:33–46. doi: 10.15326/jcopdf.1.1.2014.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappell S, et al. Variation in the tumour necrosis factor gene is not associated with susceptibility to COPD. European Respiratory Journal. 2007;30:810–812. doi: 10.1183/09031936.00057107. [DOI] [PubMed] [Google Scholar]
- 11.Haq I, et al. Association of MMP - 12 polymorphisms with severe and very severe COPD: A case control study of MMPs - 1, 9 and 12in a European population. BMC Medical Genetics. 2010;11:7. doi: 10.1186/1471-2350-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappell S, et al. Genetic variants of microsomal epoxide hydrolase and glutamate-cysteine ligase in COPD. European Respiratory Journal. 2008;32:931–937. doi: 10.1183/09031936.00065308. [DOI] [PubMed] [Google Scholar]
- 13.Chappell SL, et al. The role of IREB2 and transforming growth factor beta-1 genetic variants in COPD: a replication case-control study. BMC Medical Genetics. 2011;12:24. doi: 10.1186/1471-2350-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaykhiev R, et al. Cigarette smoking reprograms apical junctional complex molecular architecture in the human airway epithelium in vivo. Cellular and Molecular Life Sciences. 2011;68:877–892. doi: 10.1007/s00018-010-0500-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loth DW, et al. Genome-wide association analysis identifies six new loci associated with forced vital capacity. Nature genetics. 2014;46:669–677. doi: 10.1038/ng.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeilinger S, et al. Tobacco Smoking Leads to Extensive Genome-Wide Changes in DNA Methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh R, et al. Air pollutants, genes and early childhood acute bronchitis. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2013;749:80–86. doi: 10.1016/j.mrfmmm.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, et al. Genetic polymorphisms and lung cancer risk: Evidence from meta-analyses and genome-wide association studies. Lung Cancer. 2017;113:18–29. doi: 10.1016/j.lungcan.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Doherty DE. A Review of the Role of FEV1 in the COPD Paradigm. COPD: Journal of Chronic Obstructive Pulmonary Disease. 2008;5:310–318. doi: 10.1080/15412550802363386. [DOI] [PubMed] [Google Scholar]
- 20.Jones PW. Health Status and the Spiral of Decline. COPD: Journal of Chronic Obstructive Pulmonary Disease. 2009;6:59–63. doi: 10.1080/15412550802587943. [DOI] [PubMed] [Google Scholar]
- 21.Solberg R, et al. Mapping of the regulatory subunits RIβ and RIIβ of cAMP-Dependent protein kinase genes on human chromosome 7. Genomics. 1992;14:63–69. doi: 10.1016/S0888-7543(05)80284-8. [DOI] [PubMed] [Google Scholar]
- 22.Oldenburger A, Maarsingh H, Schmidt M. Multiple Facets of cAMP Signalling and Physiological Impact: cAMP Compartmentalization in the Lung. Pharmaceuticals. 2012;5:1291–1331. doi: 10.3390/ph5121291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan H, Tang G, Wang H, Hao L, He T, Sun X, Ting A H, Deng A, Sun S. DNA methylation reactivates GAD1 expression in cancer by preventing CTCF-mediated polycomb repressive complex 2 recruitment. Oncogene. 2015;35(30):3995–4008. doi: 10.1038/onc.2015.423. [DOI] [PubMed] [Google Scholar]
- 24.Raviv S, Hawkins KA, Malcolm M, DeCamp J, Kalhan R. Lung Cancer in Chronic Obstructive Pulmonary Disease. American Journal of Respiratory and Critical Care Medicine. 2011;183:1138–1146. doi: 10.1164/rccm.201008-1274CI. [DOI] [PubMed] [Google Scholar]
- 25.Morrison K, et al. Development of ASG-15ME, a Novel Antibody–Drug Conjugate Targeting SLITRK6, a New Urothelial Cancer Biomarker. Molecular Cancer Therapeutics. 2016;15:1301–1310. doi: 10.1158/1535-7163.mct-15-0570. [DOI] [PubMed] [Google Scholar]
- 26.Peng Y. A novel ensemble machine learning for robust microarray data classification. Computers in Biology and Medicine. 2006;36:553–573. doi: 10.1016/j.compbiomed.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Cho, S.-B. & Won, H.-H. In Proceedings of the First Asia-Pacific bioinformatics conference on Bioinformatics2003-Volume 19. 189–198 (Australian ComputerSociety, Inc.).
- 28.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 29.Pan W, Xie B, Shen X. Incorporating predictor network in penalized regression with application to microarray data. Biometrics. 2010;66:474–484. doi: 10.1111/j.1541-0420.2009.01296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou H, Hastie T. Regression shrinkage and selection via the elastic net, with applications to microarrays. Journal of the Royal Statistical Society: Series B. 2003;301-320:v67. [Google Scholar]
- 31.Ogutu, J. O., Schulz-Streeck, T. & Piepho, H.-P. In BMC proceedings. S10 (BioMed Central). [DOI] [PMC free article] [PubMed]
- 32.Qiu P, Gentles AJ, Plevritis SK. Discovering biological progression underlying microarray samples. PLoS computational biology. 2011;7:e1001123. doi: 10.1371/journal.pcbi.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman, J., Hastie, T. & Tibshirani, R. The elements of statistical learning. Vol. 1 (Springer series in statistics New York, 2001).
- 34.Zou H. The adaptive lasso and its oracle properties. Journal of the American statistical association. 2006;101:1418–1429. doi: 10.1198/016214506000000735. [DOI] [Google Scholar]
- 35.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez JD, Perez A, Lozano JA. Sensitivity analysis of k-fold cross validation in prediction error estimation. IEEE transactions on pattern analysis and machine intelligence. 2010;32:569–575. doi: 10.1109/TPAMI.2009.187. [DOI] [PubMed] [Google Scholar]
- 37.Kim J-H. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Computational statistics & data analysis. 2009;53:3735–3745. doi: 10.1016/j.csda.2009.04.009. [DOI] [Google Scholar]
- 38.Filzmoser P, Liebmann B, Varmuza K. Repeated double cross validation. Journal of Chemometrics. 2009;23:160–171. doi: 10.1002/cem.1225. [DOI] [Google Scholar]
- 39.Braga-Neto UM, Dougherty ER. Is cross-validation valid for small-sample microarray classification? Bioinformatics. 2004;20:374–380. doi: 10.1093/bioinformatics/btg419. [DOI] [PubMed] [Google Scholar]
- 40.Chang JC, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. The Lancet. 2003;362:362–369. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 41.Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. Journal of biomedical informatics. 2002;35:352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 42.Galili, T. Heatmaply: interactive heat maps (with R). Month545 (2016).
- 43.Bacardit J, Llorà X. Large‐scale data mining using genetics‐based machine learning. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. 2013;3:37–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.