Abstract
Mobile genetic elements (MGEs) that frequently transfer within and between bacterial species play a critical role in bacterial evolution, and often carry key accessory genes that associate with a bacteria’s ability to cause disease. MGEs carrying antimicrobial resistance (AMR) and/or virulence determinants are common in the opportunistic pathogen Klebsiella pneumoniae, which is a leading cause of highly drug-resistant infections in hospitals. Well-characterised virulence determinants in K. pneumoniae include the polyketide synthesis loci ybt and clb (also known as pks), encoding the iron-scavenging siderophore yersiniabactin and genotoxin colibactin, respectively. These loci are located within an MGE called ICEKp, which is the most common virulence-associated MGE of K. pneumoniae, providing a mechanism for these virulence factors to spread within the population. Here we apply population genomics to investigate the prevalence, evolution and mobility of ybt and clb in K. pneumoniae populations through comparative analysis of 2498 whole-genome sequences. The ybt locus was detected in 40 % of K. pneumoniae genomes, particularly amongst those associated with invasive infections. We identified 17 distinct ybt lineages and 3 clb lineages, each associated with one of 14 different structural variants of ICEKp. Comparison with the wider population of the family Enterobacteriaceae revealed occasional ICEKp acquisition by other members. The clb locus was present in 14 % of all K. pneumoniae and 38.4 % of ybt+ genomes. Hundreds of independent ICEKp integration events were detected affecting hundreds of phylogenetically distinct K. pneumoniae lineages, including at least 19 in the globally-disseminated carbapenem-resistant clone CG258. A novel plasmid-encoded form of ybt was also identified, representing a new mechanism for ybt dispersal in K. pneumoniae populations. These data indicate that MGEs carrying ybt and clb circulate freely in the K. pneumoniae population, including among multidrug-resistant strains, and should be considered a target for genomic surveillance along with AMR determinants.
Keywords: Klebsiella pneumoniae, yersiniabactin, colibactin, ICEKp, mobile genetic elements, virulence
Impact Statement.
Klebsiella pneumoniae infections are becoming increasingly difficult to treat with antibiotics. Some K. pneumoniae strains also carry extra genes that allow them to synthesise yersiniabactin, an iron-scavenging molecule, which enhances their ability to cause disease. These genes are located on a genetic element that can easily transfer between strains. Here, we screened 2498 K. pneumoniae genome sequences and found substantial diversity in the yersiniabactin genes and the associated genetic elements, including a novel mechanism of transfer, and detected hundreds of distinct yersiniabactin acquisition events between strains of K. pneumoniae. We show that these yersiniabactin mobile genetic elements are specifically adapted to the K. pneumoniae population but also occasionally acquired by other bacterial members of the family Enterobacteriaceae such as Escherichia coli. These insights into the movement and genetics of yersiniabactin genes allow tracking of the evolution and spread of yersiniabactin in global K. pneumoniae populations and monitoring for acquisition of yersiniabactin in antibiotic-resistant strains.
Introduction
Mobile genetic elements (MGEs) including plasmids, transposons and integrative conjugative elements (ICEs) can generate significant genotypic and phenotypic variation within bacterial populations, driving the emergence of niche- or host-adapted lineages or pathotypes [1, 2]. Despite the risk that acquisition of virulence-associated MGEs can pose to pathogen emergence, few studies have explored the diversity, distribution and dynamics of such MGEs within their host bacterial populations.
ICEKp is an integrative conjugative element (ICE) that mobilises the ybt locus, which encodes biosynthesis of the siderophore yersiniabactin and its receptor [3]. Yersiniabactin and other siderophore systems are considered to be key bacterial virulence factors as they provide mechanisms for scavenging iron (an essential nutrient) from host transport proteins, thereby enhancing the ability of bacteria to survive and replicate within the host [4–6]. Nearly all Klebsiella pneumoniae produce the siderophore enterobactin, however its scavenging mechanisms are inhibited by human lipocalin-2 (Lcn2), which has a strong binding affinity for ferric and aferric enterobactin [7] and induces an inflammatory response upon binding [8]. Yersiniabactin escapes Lcn2 binding, thus avoiding the inflammatory response and enhancing bacterial growth and dissemination to the spleen, although it does not provide sufficient iron to allow growth in human serum or urine [8–11]. Yersiniabactin can also bind other heavy metals besides iron; for example, yersiniabactin expressed by uropathogenic Escherichia coli has been shown to bind Cu2+, providing protection against copper toxicity and redox-based phagocyte defences [12]. Yersiniabactin is by far the most common K. pneumoniae high-virulence determinant, present in roughly a third of clinical isolates, and is significantly associated with strains isolated from bacteraemia and tissue-invasive infections such as liver abscess, compared with those from non-invasive infections or asymptomatic colonisation [3, 13]. In contrast, the virulence plasmid pK2044 described in strain NTUH-K2044, which encodes hypermucoidy and the acquired siderophores salmochelin (which modifies enterobactin to escapes Lcn2 binding [14]) and aerobactin (which can scavenge iron from the host blood protein transferrin [15]), is present in less than 5 % of K. pneumoniae isolates sampled from infections [13].
The ybt locus was first described in the Yersinia high pathogenicity island (HPI), variants of which have since been reported in other species of the family Enterobacteriaceae [16], including K. pneumoniae, where ybt is located within ICEKp [3, 17, 18]. ICEKp is self-transmissible, involving excision (requiring the gene xis), formation of an extrachromosomal circular intermediate (requiring the integrase gene int and 17 bp direct repeats at both outer ends), mobilization to recipient cells (requiring virB1, mobB and oriT) and integration at attO sites present in four closely-located tRNA-Asn copies in the K. pneumoniae chromosome [3, 19]. ICEKp sometimes carries additional virulence determinants, including iro (encoding salmochelin synthesis) or clb (also known as pks, encoding synthesis of the genotoxic polyketide colibactin [3, 17, 20], which can induce double-strand DNA breaks in eukaryotic cells [21]). Therefore, ICEKp represents a prominent virulence element that strongly influences the pathogenicity of strains of K. pneumoniae. Although it is significantly associated with invasive infections, the diversity of ICEKp structures and their transmission dynamics within the host bacterial population have not yet been characterized. Here we address this important gap in K. pneumoniae virulence evolutionary dynamics by using comparative genomics. We investigate ybt phylogenetics, ICEKp structure, integration sites and chromosomal genotypes, and use these signals to track the movement of ICEKp in K. pneumoniae populations, and explore the relationship of this MGE with its bacterial host.
Results
Diversity of the ybt locus in K. pneumoniae
First we screened for ybt genes in 2498 genomes of members of the K. pneumoniae complex (data sources are listed in Table S1, available in the online version of this article), and found ybt in 39.5 % of 2289 K. pneumoniae, but only 2 out of 146 Klebsiella variicola and none out of 63 Klebsiella quasipneumoniae. Prevalence was 40.0 % in the carbapenemase-associated K. pneumoniae clonal group (CG) 258, 87.8 % in the hypervirulent K. pneumoniae CG23, and 32.2 % in the wider K. pneumoniae population. Consistent with previous reports [3, 13], amongst human isolates with reliable clinical source information (Table S1), the presence of ybt was significantly associated with infection isolates [odds ratio (OR)=3.6, P<1×10−7], particularly those from invasive infections (OR=28.6 for liver abscess, OR=4.1 for blood isolates; see Table S2).
Each of the 11 ybt locus genes displayed substantial diversity within the K. pneumoniae population (see Supplementary Text; Fig. S1, Table S3). The majority of ybt loci grouped into 17 ybt phylogenetic lineages, with a mean nucleotide divergence of 0.034 % and mean 8 out of 11 shared loci within lineages, compared with a mean 0.478 % nucleotide divergence and mean 0 out of 11 shared loci between lineages. A total of 11 recombination events were identified in the ybt locus (Fig. S2). Nine appeared to involve import of divergent alleles from outside the K. pneumoniae population (1.5–8.37 % nucleotide divergence from ybt alleles detected in other K. pneumoniae) and two involved exchange of irp2 alleles within K. pneumoniae (0.40–0.41 % nucleotide divergence; see Fig. S2). We further explored the genetic diversity of ybt using phylogenetic and multi-locus sequence typing (MLST) analyses (see Methods). Ybt locus sequence types (YbSTs), defined by unique combinations of ybt gene alleles, were assigned to 842 ybt+ isolates (Table S4). A total of 329 distinct YbSTs (Table S5) were identified, which clustered into 17 phylogenetic lineages (referred to hereafter as ybt 1, ybt 2, etc.; see Figs 1, S2 and S3); the phylogenetic tree of translated amino acid sequences was concordant with these lineages (see Fig. S4, Supplementary Text).
Fig. 1.
Recombination-filtered phylogenetic analysis of 329 yersiniabactin sequence types identified across 842 genomes. Each leaf represents a single yersiniabactin sequence type (YbST) and these YbST sequences cluster into 17 lineages, as labelled. Tracks (from inner to outer): (1) lineage (key as labelled above the tree nodes; white, unassigned), (2) ICEKp structure (white, undetermined), (3) presence or absence of KpZM module and (4) tRNA-Asn insertion site (white, undetermined). Recombination events (see Fig. S2) are depicted with a red asterisk next to the relevant branch or a single YbST.
Analysis of protein coding sequences yielded dN/dS values below 0.6 for all genes (Table S3), consistent with moderate purifying selection. Nonsense or frameshift mutations were identified in ybt genes in 11 % of isolates carrying the ybt locus; these mostly affected irp2 (6.5 %) or irp1 (4 %) (see Table S3), which encode key structural components in the yersiniabactin biosynthesis (Fig. 2a) [22, 23]. Inactivation of either of these genes prevents synthesis of yersiniabactin in Yersinia enterocolitica [24], and is predicted to have the same effect in K. pneumoniae. Most of these mutations (85 %) were only observed in a single isolate, indicating that they are not conserved and potentially arose during storage or culture. Consistent with this hypothesis, the presence of inactivating mutations in ybt genes was significantly associated with historical isolates (70 % amongst ybt+ isolates stored since at least 1960, 8 % amongst ybt+ isolates stored since 2000; OR 27 [95 % CI 11–74], P<10−13 using Fisher’s exact test). The notable exceptions were ST67 K. pneumoniae subspecies rhinoscleromatis genomes (conserved frameshift in irp2) and ST3 (conserved nonsense mutation in irp1), indicating negative selection in these lineages.
Fig. 2.
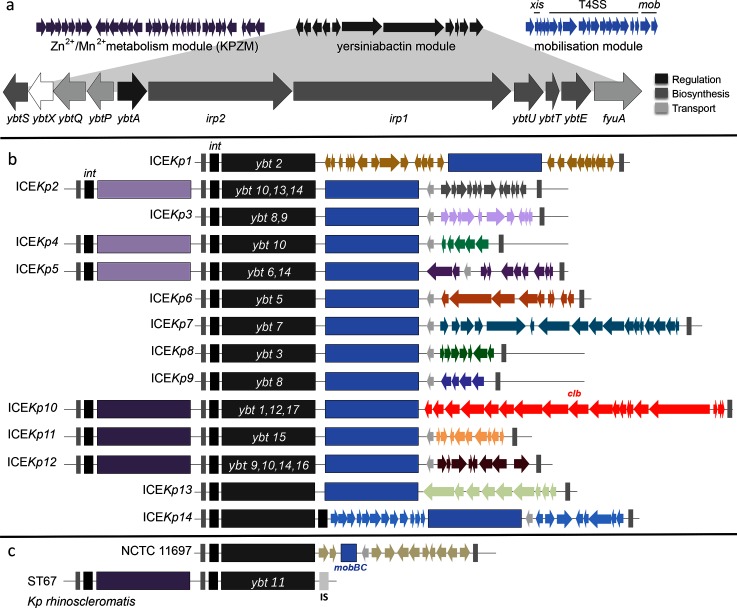
ICEKp structures. (a) Gene structure for core modules, which are shown in b and c as coloured blocks: yersiniabactin synthesis locus ybt (dark grey, labelled with the most commonly associated ybt lineage if one exists), mobilisation module (blue) and Zn2+/Mn2+ module (purple, usually present, light purple, rarely present). In a and b, the variable gene content unique to each ICEKp structure, which is typically separated from the mobilisation module by an antirestriction protein (light grey arrow), is shown in a unique colour as per Fig. 1. Grey rectangles represent direct repeats; black rectangles, P4-like integrase genes. Genes that make up the ybt locus are also shown and shaded according to their overall role in yersiniabactin synthesis (further details provided in Table S3). (b) Genetic structures of apparently intact ICEKp variants (see Table S6 and sequences deposited in GenBank for details of specific genes). (c) Disrupted ICEKp loci.
Diversity of ICEKp structures and integration sites in K. pneumoniae
With the exception of ybt 4 (which we found to be plasmid-borne and representing a novel mechanism of ybt transfer in K. pneumoniae; see Supplementary Text), all ybt loci detected in the K. pneumoniae genomes were located within an ICEKp structure integrated into one of four copies of tRNA-Asn located in a chromosomal region that spans 16.4 kbp in size in strains lacking MGE insertions at these sites (Fig. S5). Examples of ICEKp integration were observed at all four tRNA-Asn sites (Fig. 1), but the frequency of integration differed substantially by site: 35.7, 44.7 and 19.5 % for sites 1, 3 and 4 respectively, and just one integration at site 2. Multiple ICEKp integration sites were observed for most ybt lineages (Fig. 1); thus, there is no evidence that ICEKp variants target specific tRNA-Asn copies.
The boundaries of each ICEKp variant were identified by the 17 bp direct repeats formed upon integration [3], and their structures were compared. This confirmed that ICEKp structures identified in K. pneumoniae share several features: (i) a P4-like integrase gene, int, at the left end; (ii) the 29 kbp ybt locus; (iii) a 14 kbp sequence encoding the xis excisionase, virB-type 4 secretion system (T4SS), oriT transfer origin and mobBC proteins (responsible for mobilisation) [3]. In addition, we found that each ICEKp carried a distinct cluster of cargo genes at its right end, which we used to classify the ICEs into 14 distinct structures which we labelled as ICEKp2, ICEKp3, etc., preserving the original nomenclature of ICEKp1 [3, 25] (see Fig. 2b; Table S6). We detected occasional additional gene content variation between ICEKp sequences, arising from transposases and other insertion or deletion events. Most of the 14 ICEKp structures were uniquely associated with a monophyletic group in the ybt nucleotide-based phylogeny (i.e. a single ybt lineage; see Figs 1 and 2; Table S6); the exception was the ICEKp10 structure, which carries the clb locus in the cargo region and was associated with ybt lineages 1, 12 and 17 (details below).
All ICEKp carried the integrase gene int; however we identified two variant forms of ICEKp lacking the mobilisation genes. First, K. pneumoniae subspecies rhinoscleromatis (ST67) genomes carried ybt 11 but lacked the entire mobilisation module (Fig. 2c); as noted above, they also carried nonsense mutations in irp2. Second, strain NCTC 11697 carried a highly divergent ybt locus (>2 % nucleotide divergence from all other ybt sequences, marked with ** in Fig. 1) and lacked the virB-T4SS and xis genes (Fig. 2c). An approximately 34 kbp Zn2+ and Mn2+ metabolism module (KpZM) was identified upstream of six different ICEKp structures (Fig. 2). This module includes an integrase at the left end that shares 97.5 % amino acid identity with that of ICEKp, and the same 17 bp direct repeat was found upstream of both integrases and downstream of ICEKp. It is therefore likely that the entire sequence between the outermost direct repeats (grey bars in Fig. 2b), including the KpZM module, ybt locus, mobility and cargo regions, can be mobilised together as a single MGE (see Supplementary Text).
ICEKp1 was the first yersiniabactin ICE reported in K. pneumoniae [3, 25] and carries a 18 kbp insertion between the ybt and mobilisation genes, which our comparative analyses showed to be quite atypical (see Fig. 2b). As previously reported, the inserted sequence encodes iro and rmpA (which upregulates capsule production and is associated with hypermucoid phenotype) and is homologous to a region on the virulence plasmid pLVPK [3]. The only other ICEKp structure in which we identified known K. pneumoniae virulence determinants was ICEKp10, whose cargo region harbours the approximately 51 kbp colibactin (clb) locus. The ICEKp10 structure corresponds to the genomic island described in ST23 strain 1084 as GM1–GM3 of genomic island KPHPI208, and in ST66 strain Kp52.145 as an ICE-Kp1-like region [17, 18]. We detected ICEKp10 in 40 % of ST258 (31 % of CG258), 77 % of ST23 (61 % of CG23) and 4.0 % of other K. pneumoniae genomes including 25 other STs (total 13.85 and 38.43 % of all, and ybt+, K. pneumoniae genomes respectively). Notably, all but three of the ICEKp10 strains carried the KpZM module at the left end, indicating that the clb locus (Table S7) is usually mobilized within a larger structure including KpZM. MLST analysis of clb genes identified 65 CbSTs (Table S8), similar to the number of YbSTs detected in ICEKp10 (n=86). Phylogenetic analysis of the clb locus (excluding clbJ and clbK (due to a 4153 bp deletion that commonly spans the two genes; see Fig. S6, Supplementary Text), revealed three clb lineages that were each associated with a phylogenetically distinct ybt lineage: clb 1 (ybt 12), clb 2A (ybt 1) and clb 2B (ybt 17), consistent with multiple acquisitions of clb into the ICE (see Figs 1 and 3 and Supplementary Text).
Fig. 3.
Colibactin diversity. (a) ClbJ/K deletion. A 5.6 kbp in-frame deletion between the clbJ (6501 kbp) and clbK (6465 kbp) genes of the colibactin locus observed in a large number of ICEKp10 structures. (b) Recombination-free phylogeny of genes in the colibactin locus (excluding clbJ and clbK). Each leaf represents a unique colibactin locus found across 314 isolates. Tracks (from inner to outer): (1) clb lineage, (2) ybt lineage (grey, not part of any main ybt lineage; white, ybt-negative), (3) presence of clbJ/K deletion (white, no deletion) and (4) occurrence of intragenic transposase insertions within clb locus (white, no insertion).
Transmission of ICEKp in the K. pneumoniae host population
We identified n=206 unique combinations of ICEKp structure, chromosomal ST and integration site. These probably represent distinct ybt acquisition events, however it is possible that the ICEKp could migrate to another insertion site following initial integration; hence a more conservative estimate for distinct ybt acquisition events is the number of unique combinations of ICEKp structure and chromosomal ST, n=189 (ST phylogenetic relationships are given on Fig. S7). The most widely distributed variant was ICEKp4 (found in 37 chromosomal STs) followed by ICEKp10, ICEKp5 and ICEKp3 (n=24, 23 and 23 chromosomal STs, respectively). Conversely, ICEKp7, ICEKp8 and ICEKp13 were found in one K. pneumoniae host strain each (ST111, site 1, n=2; ST37, site 4, n=7; ST1393, site 1, n=1; respectively); ICEKp14 was found only in K. variicola (ST1986, site 4, n=1).
A total of 26 K. pneumoniae chromosomal STs showed evidence of multiple insertion sites and/or ICEKp structures, indicative of multiple integrations of ICEKp within the evolutionary history of these clones (Figs 4, S7 and Table S4). Most unique acquisition events defined by unique combinations of ICEKp structure and chromosomal STs (65 %) were identified in a single genome sequence. The frequency of ybt carriage and unique ybt acquisitions per ST was correlated with the number of genomes observed per ST (R2=0.71, P<1×10−8 for log-linear relationship; see Fig. 5(c)), indicating that the discovery of novel integrations within lineages is largely a function of sampling. This implies that ICEKp may frequently be gained and lost from all lineages and that deeper sampling would continue to uncover further acquisitions and losses. Notably, of the 35 clonal groups that were represented by at least 10 genomes, 30 (86 %) included at least one ICEKp acquisition (Figs 4 and S7). The five other common clonal groups each consisted mostly of isolates from a localised hospital cluster (ST323, Melbourne; ST490, Oxford; ST512, Italy; ST681, Melbourne; ST874, Cambridge); and we predict that more diverse sampling of these clonal groups would detect ICEKp acquisition events.
Fig. 4.
Frequency and diversity of ybt sequences and acquisition events in common K. pneumoniae clonal groups. This analysis includes all clonal groups for which there were at least ten genomes available for analysis; each represents a deep branching lineage of the K. pneumoniae core genome phylogeny as shown in Fig. S7. (a) Frequency of ybt presence within each clonal group, coloured by ybt lineage according to the scheme in Figs 1 and 2 and b (white, proportion of genomes without ybt). (b) Number of ybt-positive K. pneumoniae genomes designated to a particular ybt lineage, with the colours corresponding to a particular ICEKp variant as indicated. Lineages associated with ICEKp10 are marked with asterisks (i.e. ybt1, 12 and 17). The sizes correspond to sample size of genomes. (c) Number of genomes (black bars, bottom axis) and number of independent ybt acquisition events (red, top axis) per clonal group. Independent ybt acquisition events were defined as unique combinations of ICEKp structure and insertion site in each ST.
Fig. 5.
Diversity of ybt loci acquired by CG258. The tree shown is an outgroup-rooted maximum likelihood phylogeny inferred from recombination-filtered core gene SNPs for representative ybt-positive CG258 isolates. Clades are labelled by chromosomal MLST and the two ST258 clades identified in Deleo et al. [51]; tracks indicate (1) ybt lineage, (2) ICEKp structure, (3) ICEKp insertion site, (4) clb presence/absence and (5) clb locus disruptions, coloured according to the inset legend. The K-locus, as determined by Kaptive, and wzi allele number are also listed.
Of the unique ICEKp acquisition events that were detected in more than one genome, 68 % (n=50 out of 73) showed diversity in the YbST. This diversity was attributable to minor allelic changes (SNPs in a median of 2.5 loci), consistent with clonal expansion of ICEKp-positive K. pneumoniae strains and diversification of the ybt locus in situ. The greatest amount of YbST diversity within such groups was observed in hypervirulent clones ST23 (18 YbSTs of ICEKp10/ybt 1 in site 1), ST86 (12 YbSTs of ICEKp3 in site 3) and ST67 K. pneumoniae subspecies rhinoscleromatis (five YbSTs in site 1); followed by hospital-outbreak-associated MDR clones ST15 (six YbSTs of ICEKp4 in site 1 and five in site 3), ST45 (five YbSTs of ICEKp4), ST101 (five YbSTs of ICEKp3 in site 3) and ST258 (detailed below). This level of diversity is indicative of long-term maintenance of the ICEKp in these lineages, allowing time for the ybt genes to accumulate mutations.
Given the clinical significance of the carbapenemase-associated CG258 [26], we explored ICEKp acquisition in these genomes in greater detail. Ybt was detected in 269 CG258 isolates (40 %) from 17 countries; 218 isolates also carried clb (nearly all from the USA; see Table S4). A set of 58 YbSTs were identified amongst CG258 isolates and clustered into seven ybt lineages associated with six ICEKp structures. Comparison of ybt lineage, ICEKp structure and insertion site with a recombination-filtered core genome phylogeny for representative CG258 strains with yersiniabactin indicated dozens of independent acquisitions of ICEKp sequence variants in this clonal complex (Fig. 5). Near-identical clb 2B (ICEKp10/ybt 17) sequences were identified in 212 ST258 strains (40 %), mostly at tRNA-Asn site 3, isolated in the USA during 2003–2014. Most of these isolates carried the clbJ/clbK deletion (n=175, 83 %), and also transposase insertions within other clb genes (n=173, see Table S4) that may prevent colibactin production (Fig. 5). A total of 27 ybt+clb+ ST258 isolates (5 %) had an apparently intact clb locus; two were isolated in Colombia in 2009 and the rest from the USA during 2004–2010 (Table S4), including the previously reported KPNIH33 [27]. The results demonstrate the very high genetic and functional dynamics of ICEKp within a very recently emerged K. pneumoniae epidemic lineage, estimated to have emerged in the mid-1990s [28].
Relationship to ybt in other species
Finally, we sought to understand the relationship between ICEKp, which we have shown to circulate within K. pneumoniae, and the ybt loci found in other bacterial species. blast searching NCBI GenBank identified the ybt locus in n=242 genomes from 11 species outside the K. pneumoniae complex (Table S9); all belonged to the family Enterobacteriaceae. The phylogenetic and structural relationships of these loci with those found in K. pneumoniae are shown in Fig. 6, which indicates that ybt sequences mobilised by ICEKp form a subclade that is strongly associated with the species K. pneumoniae. The vast majority of ICEKp sequences were found in K. pneumoniae (97 %); the exceptions were 15 E. coli, 6 Klebsiella aerogenes, 4 Citrobacter koseri, 2 K. variicola and 1 Enterobacter hormaechei [29]; see Fig. 6, Table S9). These include one novel ICEKp variant in E. coli strain C8 (accession CP010125.1), however this was also detected in a recently sequenced K. pneumoniae draft genome (accession GCF_002248635.1). ICEKp accounted for just 11 % of ybt sequences detected outside K. pneumoniae, and in the remaining 89 % it was not associated with ICEKp or any other identifiable conjugative machinery. Notably, ybt sequences from the Yersinia HPI of the Yersinia pseudotuberculosis/pestis complex clustered within the ICEKp clade of ybt sequences, and not with the ybt sequences from Yersinia enterocolitica (Fig. 6). This indicates that the HPI found in the Y. pseudotuberculosis complex is derived from ICEKp. Furthermore, the HPI shares the int (Fig. S8) and xis of ICEKp, facilitating integration and excision of the HPI, but appears to have lost the conjugative machinery that enables its spread between cells from distinct lineages of yersiniae, consistent with the results of previous investigations that showed that the HPI is unable to self-transmit [30, 31]. Additionally, the int in the HPI of Y. enterocolitica and Y. pseudotuberculosis are unlikely to encode a functional integrase due to frameshift and nonsense mutations.
Fig. 6.
Phylogenetic and structural comparison of ybt sequences and corresponding mobile genetic elements in K. pneumoniae and other members of the Enterobacteriaceae. (a) Tree shows the midpoint-rooted maximum likelihood phylogeny for DNA sequences of complete ybt loci representative of all those found by blast search of GenBank, and representatives of the ybt lineages identified in this work amongst the K. pneumoniae population (shown in Fig. 1). Columns on the right are binary indicators of the genetic context of the ybt locus in each case: (i) located on the chromosome, (ii) located at a tRNA-Asn site, (iii) presence of an integrase (grey indicates non-functional integrase caused by a frameshift or nonsense mutation), excisionase and/or virB-T4SS+mobBC portion of the mobility module. For ybt sequences in the clade associated with ICEKp, subclades corresponding to the ybt lineages (defined in Fig. 1) are shaded and additional indicator columns are included to show the corresponding ICEKp structure where present (defined in Fig. 2) coloured according to the inset legend (* indicates ICEKp structure not resolvable from available draft genome sequences, ** indicates novel ICE structure). The number of genomes of each species in which the defined ybt lineages was detected is printed on the right. (b) Taxonomic relationships between Enterobacteriaceae species in which ybt was detected, in the form of a neighbour-joining tree of concatenated gyrA and rpoB sequences extracted from ybt-positive genomes.
The most genetically distant ybt loci were found in the chromosomes of Klebsiella oxytoca and the related species Klebsiella michiganensis and Klebsiella grimontii (Fig. 6), with no identifiable mobility-associated genes in their proximity; we propose the hypothesis that the locus may have originated in the ancestor of these species before becoming mobilised. A related form, situated next to a tRNA-Asn but with no identifiable integrase gene, was also present in Klebsiella ornithinolytica (also known as Raoultella ornithinolytica) (Fig. 6). The ybt sequences in members of the genus Salmonella formed two related clades, one chromosomal and one plasmid-borne, but lacked proximal mobility-associated genes (Fig. 6); notably the plasmid-borne form of ybt in K. pneumoniae appears to be derived from that of ICEKp and not the plasmid-borne form from members of the genus Salmonella (Fig. 6). Finally, this analysis sheds some light on the origins of the atypical form of ybt in K. pneumoniae NCTC 11697, which includes the integrase but lacks the T4SS mobility region (Fig. 2), and whose position in the ybt tree indicates that it may be related to a progenitor element from which ICEKp evolved via acquisition of the excisionase and conjugative machinery.
Discussion
This study provides key insights into the evolution, dynamics and structure of ICEKp, the most frequent MGE associated with hypervirulent/invasive K. pneumoniae infections. The population structure of ICEKp comprises numerous sublineages that are each associated with unique complements of cargo genes in addition to the yersiniabactin synthesis locus ybt (Figs 1 and 2). With the exception of inactivating mutations that appear to arise in culture and occasionally in hospital-associated lineages (discussed below), the ybt and clb loci appear to be under strong purifying selection in the K. pneumoniae population (low dN/dS, see Tables S3 and S7) and all variants are predicted to synthesise the same yersiniabactin and colibactin polyketide molecules.
The data demonstrate that ICEKp circulates dynamically within the K. pneumoniae population. The sheer number of distinct ICEKp acquisitions detected in K. pneumoniae (n≥189), and the scale of distinct acquisition events within individual K. pneumoniae clonal groups (Figs 4 and 5), indicates this MGE is highly transmissible within the host bacterial population. Genetic separation of the ICEKp form of ybt compared with those in other members of the family Enterobacteriaceae (Fig. 6), and lack of ICEKp outside K. pneumoniae, indicates that ICEKp may be specifically adapted to circulate in K. pneumoniae. We propose that this MGE has been a feature of the K. pneumoniae population that predates K. pneumoniae sublineage diversification, because (i) the ybt genes of ICEKp displayed a similar degree of nucleotide diversity as K. pneumoniae core chromosomal genes (mean 0.5 %) [13] and (ii) ICEKp was notably rare or absent from K. pneumoniae’s closest relatives, K. variicola and K. quasipneumoniae.
The data indicate that the population prevalence of ICEKp within K. pneumoniae (around one third of the population) is sustained through highly dynamic horizontal gene transfer events rather than stable maintenance within K. pneumoniae lineages by vertical inheritance. The intermediate frequency of ICEKp in K. pneumoniae is indicative of the existence of some form of balancing selection for the encoded traits. This typically occurs when a trait is most beneficial only when it is not shared by the entire population (e.g. antigenic variation resulting in variable susceptibility to predators or host immunity [32]), or the trait has selective advantages only under certain conditions but high costs in others. Acquisition of ybt has benefits in certain iron-depleted conditions, which are presumably encountered in a wide range of environmental and host-associated niches [6]; siderophores including yersiniabactin also confer significant growth advantage in heavy metal contaminated soil irrespective of iron content [33]. Hence the dynamics of ICEKp/ybt may reflect the diverse lifestyles of K. pneumoniae, which can vary between hosts and in the environment. However loss of ybt also appears to be common, probably occurring due to the high-energy costs from synthesising the polyketide hybrid molecule. Inactivation of irp1 or irp2 in historical isolates (70 %) supports strong negative selection against yersiniabactin production in rich media. Notably, K. pneumoniae almost universally can synthesise enterobactin, hence the benefit of ybt depends on not only the availability of iron but also the form of iron, and other factors such as ability to compete with host iron-binding proteins and evasion of the mammalian immune system’s targeting of enterobactin via Lcn2 [6, 8, 9]. ICEKp cargo genes probably contribute additional costs and benefits to host cells, modifying the fitness equation for their bacterial host; further work will be needed to explore the differential effects and functional relevance of these genes. It is notable that clb-carrying ICEKp10 was widespread (detected across 32 K. pneumoniae lineages) but the clb genes were frequently disrupted, indicating subjection to balancing selection. These disruptions were particularly high amongst the hospital-associated MDR clone ST258, and may indicate selection against costly colibactin production in hospital-adapted strains that already benefit from positive selection under antimicrobial exposure.
Concerningly, our data highlights the possibility that the rate of ICEKp transmission in the population may be sufficiently high that ybt is readily available to most K. pneumoniae lineages. Hence new clinically important high-pathogenicity lineages could theoretically arise at any time following introduction of ybt to a strain background that already has features favourable for transmission or pathogenicity in humans, including antimicrobial resistance (AMR). Indeed we found ICEKp to be frequent amongst many of the recognised MDR K. pneumoniae clones, such as CG258, indicating that the convergence of AMR and yersiniabactin production is happening frequently in K. pneumoniae, potentiating the emergence of lineages that pose substantially greater risk to human health than the broader K. pneumoniae population, which typically behaves as an opportunistic, mostly susceptible, pathogen.
FIBK plasmid-borne ybt constitutes an entirely novel mechanism for ybt mobilisation in K. pneumoniae. The FIBK plasmid replicon is very common and highly stable in K. pneumoniae but not E. coli [34, 35], indicating that these plasmids are adapted to K. pneumoniae hosts and have the potential to readily transmit ybt within the population. Worryingly, many FIBK plasmids have already acquired AMR transposons [36], indicating that there may be few barriers to convergence of AMR and virulence genes in a single FIBK plasmid replicon. Given its potential transmissibility and stability in K. pneumoniae hosts, this forms another substantial public health threat and warrants careful monitoring.
The extensive diversity uncovered amongst ybt and clb sequences and ICEKp structures in this study provides several epidemiological markers with which to track their movements in the K. pneumoniae population through analysis of whole-genome sequence data, which is increasingly being generated for infection control and AMR surveillance purposes [37, 38]. The work presented here provides a clear framework for straightforward detection, typing and interpretation of ybt and clb sequences via the YbST and CbST schemes (Figs 1 and 3), which are publicly available in the BIGSdb K. pneumoniae database and can be easily interrogated using the BIGSdb web application or using common tools such as blast (https://github.com/katholt/Kleborate) or SRST2 [39]. In doing so, detection of these key virulence loci provides much-needed insights into the emergence and spread of pathogenic K. pneumoniae lineages, which will be particularly important for tracking the convergence of virulence and AMR in this troublesome pathogen.
Of broader relevance, the data show that the deepest diversity of ybt sequences is present in the members of the genus Klebsiella and that MGE-borne ybt emerged within K. pneumoniae before spreading to other Enterobacteriaceae (Fig. 6). In particular the HPI of Y. pestis and Y. pseudotuberculosis, where yersiniabactin was first identified and from which it draws its name, is derived from the ICEKp of K. pneumoniae; hence the name klebsibactin may have been more appropriate. This adds to the growing body of evidence that members of the genus Klebsiella act as a reservoir of AMR and pathogenicity genes for other members of the family Enterobacteriaceae; KPC and NDM-1 being recent examples of AMR genes first identified in members of the genus Klebsiella that have rapidly become widespread [40, 41]. We suggest that this unique role of Klebsiella is linked to its more generalist lifestyle, which offers more opportunities to sample accessory genes from a wide array of gene pools. In support of this, members of the genus Klebsiella exhibit extreme differences in gene content within and between species, their accessory genes display a wide range of DNA G+C contents and taxonomic sources [13], and strains from environmental niches, such as K. oxytoca, can have very large genomes that exceed 6 Mbp. The present work further emphasises the clinical importance of the unique position that members of the genus Klebsiella occupy in the broader microbial sphere as a source of important pathogenicity as well as AMR genes for other members of the family Enterobacteriaceae, and should be a motivating factor for further exploration of the ecological and evolutionary mechanisms behind this phenomenon.
Methods
Bacterial genome sequences
We analysed a total of 2498 K. pneumoniae genomes (2284 K. pneumoniae sensu stricto, 63 K. quasipneumoniae, 146 K. variicola, 5 undefined or hybrid [13]) obtained from various sources representing a diverse geographical and clonal distribution (Table S1; see Table S4 for full list of isolates and their properties). Just under a third of these genomes had been collected and sequenced in-house during four previous studies of human hospital isolates [13, 42–44]. These isolates from genotypically and geographically diverse backgrounds, which had clinical source information and were not associated with outbreaks, were used to estimate the distribution of the yersiniabactin locus amongst human isolates associated with the different types of infections listed in Table S2.
Where available, Illumina short reads were analysed directly and assembled using SPAdes v3.6.1, storing the assembly graphs for further analysis of genetic context. Where reads were unavailable (n=921), publicly available pre-assembled contigs were used. These had been generated using various strategies and assembly graphs were not available for inspection.
One isolate from our collection (strain INF167, isolated from a patient at the Alfred Hospital, Melbourne, Australia in 2013) was subjected to further sequencing using a MinION Mk1B and R9 Mk1 flow cell (Oxford Nanopore Technologies). A 2D MinION library was generated from 1.5 µg purified genomic DNA using the Nanopore Sequencing Kit (SQK-NSK007). DNA was repaired (NEBNext FFPE RepairMix), prepared for ligation (NEBNextUltra II End-Repair/dA-tailing Module) and ligated with adapters (NEB Blunt/TA Ligase Master Mix). We sequenced the library for 48 h, obtaining 3862 reads (mean length 3049 bp, maximum 44 026 bp) that were used to scaffold the SPAdes assembly graph using a novel hybrid assembly algorithm (http://github.com/rrwick/Unicycler). The resulting assembly included one circular plasmid, which was annotated using Prokka [45] and submitted to GenBank under the accession number KY454639.
Multi-locus sequence typing (MLST) analysis
Genomes were assigned chromosomal K. pneumoniae sequence types by comparison to the seven-locus K. pneumoniae MLST scheme [46] in the K. pneumoniae BIGSdb database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) [47] using SRST2 to analyse reads [39] and BLAST+ to analyse assemblies.
In order to produce novel MLST schemes [48] for the yersiniabactin and colibactin loci, sequences of the alleles for genes belonging to the yersiniabactin (ybtS, ybtX, ybtQ, ybtP, ybtA, irp2, irp1, ybtU, ybtT, ybtE, fyuA) and colibactin (clbABCDEFGHIJKLMNOPQR) synthesis loci were extracted from the K. pneumoniae genome, by comparison to known alleles in the K. pneumoniae BIGSdb database. To maximise resolution for the novel virulence locus MLST schemes, we included in the definition of sequence types alleles for all 11 genes of the ybt locus and 16 out of 18 genes of the clb locus (clbJ and clbK were excluded as they are subject to a common deletion as described in the Results). Each observed combination of alleles was assigned a unique yersiniabactin sequence type (YbST, listed in Table S5) or colibactin sequence type (CbST, listed in Table S8). The schemes and allele sequences are available from the BIGSdb-K. pneumoniae website and in the Kleborate repository (https://github.com/katholt/Kleborate), which includes a command-line tool for genotyping new genomes. All genomes with detectable ybt or clb sequences were included in the definition of YbSTs or CbSTs, with the exception of 61 genomes for which data quality was too low for accurate calling of all alleles (criteria: read depth <20×;<90 % agreement of alleles at the read level; and/or incomplete assembly of the ybt or clb region, which usually was associated with low read depth and generally poor assembly quality with N50 <100 000 bp).
Phylogenetic analyses
For each YbST, alignments of the concatenated corresponding allele sequences were produced using Muscle v3.8.31. Recombination events were identified using Gubbins v2.0.0 [49], which screens for regions with a high density of single nuclear polymorphisms (SNPs) that are likely to represent an imported sequence variant. The initial alignment was 28 214 bp long with 2234 variant sites, of which 232 were identified as recombinant and masked from the alignment (regions shown in Fig. S2, visualised using Phandango (https://github.com/jameshadfield/phandango/). Maximum likelihood (ML) trees were inferred from the recombination-masked alignment by running RAxML v7.7.2 [50] five times with the generalised time-reversible (GTR) model and a Gamma distribution, selecting the final tree with the highest likelihood. Lineages were defined as monophyletic groups of YbSTs whose members shared features within the group (at least six shared YbST loci, same ICEKp structures) but were distinguished from other groups (zero or one shared YbST loci, different ICEKp structures). The same approach was used to generate a colibactin ML tree. ML phylogenies for the ybt loci and concatenated rpoB and gyrA sequences from representative K. pneumoniae and other Enterobacteriaceae bacteria were also generated by running RAxML v7.7.2 [50]. Nodes with lower than a bootstrap value of 75 in the Enterobacteriaceae ybt phylogeny were collapsed to polytomies with TreeCollapserCL 4 v3.2 (http://emmahodcroft.com/TreeCollapseCL.html).
Core genome SNP trees for K. pneumoniae (using one representative genome per each unique ST) and for CG258 (which includes ST258, ST11, ST340 and ST512; using a selection of representative CG258 isolates, these were subsampled from a maximum-likelihood phylogeny encompassing all CG258 isolates to remove near-identical isolates with the same year and country of isolation that were probably sequenced from an outbreak) were inferred using the mapping pipeline RedDog v1b5 (https://github.com/katholt/reddog) to (i) map short reads against K. pneumoniae ST23 strain NTUH-K2044 [25] and ST258 strain NJST258-1 [51], respectively, using Bowtie 2 v2.2.3, and (ii) identify core gene SNPs using SAMtools v1.1. The resulting SNP alignments were subjected to analysis with Gubbins v2.0.0 (to filter recombinant sites), and RAxML v7.7.2 to infer clonal phylogenies.
Translation of the nucleotide sequence into amino acid sequence, inspection of non-synonymous, frameshift and nonsense mutations, and calculations for dN/dS ratios and significance testing for conservation or positive selection were conducted using mega 6.06 [52].
Chromosomal insertion sites and ICE structures
For each ybt-positive (ybt+) genome, the annotated assembly was manually inspected to determine which of the four tRNA-Asn sites was occupied by ICEKp. This was done with reference to the MGH78578 genome, which lacks any genomic islands at tRNA-Asn sites. The Artemis genome viewer was used to inspect the annotation of the region; BLAST+ was used for genome comparison; and when the region failed to assemble into a single contig, Bandage [53] was used to inspect the locus in the assembly graph where available. Once the insertion site was determined, the structure of the ICEKp was inferred by extracting the sequence between the flanking direct 17 bp repeats ‘CCAGTCAGAGGAGCCAA’, either directly from the contigs using Artemis or from the assembly graph using Bandage. Representative sequences for each ICEKp structure (unless derived from previously assembled genomes) were annotated and deposited in GenBank (accession numbers KY454627–KY454638).
Data availability
Annotated ICE and plasmid sequences generated in this study are available in NCBI Genbank under the accession numbers specified in the text. Yersiniabactin and colibactin MLST schemes are available in the K. pneumoniae BIGSdb database at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. All whole-genome sequences analysed in this study are freely available in NCBI, accession numbers are given in Table S4.
Supplementary Data
Funding information
This work was funded by the National Health and Medical Research Council (NHMRC) of Australia (project #1043822 and Fellowship #1061409), a Senior Medical Research Fellowship from the Viertel Foundation of Australia, and the Bill and Melinda Gates Foundation of Seattle, USA.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; CbST, colibactin sequence type; HPI, high pathogenicity island; ICE, integrative conjugative element; Lcn2, lipocalin-2; MGE, mobile genetic element; MLST, multi-locus sequence typing; OR, odds ratio; ST, sequence type; YbST, yersiniabactin sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Frost LS, Leplae R, Summers AO, Toussaint A, Edmonton A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 2.McNally A, Thomson NR, Reuter S, Wren BW. 'Add, stir and reduce': Yersinia spp. as model bacteria for pathogen evolution. Nat Rev Microbiol. 2016;14:177–190. doi: 10.1038/nrmicro.2015.29. [DOI] [PubMed] [Google Scholar]
- 3.Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol. 2008;190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wooldridge KG, Williams PH. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325-48. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 5.Koczura R, Kaznowski A. Occurrence of the Yersinia high-pathogenicity island and iron uptake systems in clinical isolates of Klebsiella pneumoniae. Microb Pathog. 2003;35:197–202. doi: 10.1016/S0882-4010(03)00125-6. [DOI] [PubMed] [Google Scholar]
- 6.Holden VI, Bachman MA. Diverging roles of bacterial siderophores during infection. Metallomics. 2015;7:986–995. doi: 10.1039/C4MT00333K. [DOI] [PubMed] [Google Scholar]
- 7.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/S1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 8.Bachman MA, Miller VL, Weiser JN. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009;5:e1000622. doi: 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachman MA, Oyler JE, Burns SH, Caza M, Lépine F, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holden VI, Breen P, Houle S, Dozois CM, Bachman MA. Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during Pneumonia. MBio. 2016;7:e01397-16-10. doi: 10.1128/mBio.01397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawlor MS, O'Connor C, Miller VL. Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun. 2007;75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh EI, Henderson JP. Microbial copper-binding siderophores at the host–pathogen interface. J Biol Chem. 2015;290:18967–18974. doi: 10.1074/jbc.R115.644328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112:E3574. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abergel RJ, Moore EG, Strong RK, Raymond KN. Microbial evasion of the immune system: structural modifications of enterobactin impair siderocalin recognition. J Am Chem Soc. 2006;128:10998–10999. doi: 10.1021/ja062476+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konopka K, Bindereif A, Neilands JB. Aerobactin-mediated utilization of transferrin iron. Biochemistry. 1982;21:6503–6508. doi: 10.1021/bi00268a028. [DOI] [PubMed] [Google Scholar]
- 16.Bach S, de Almeida A, Carniel E. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol Lett. 2000;183:289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x. [DOI] [PubMed] [Google Scholar]
- 17.Lai YC, Lin AC, Chiang MK, Dai YH, Hsu CC, et al. Genotoxic Klebsiella pneumoniae in Taiwan. PLoS One. 2014;9:e96292. doi: 10.1371/journal.pone.0096292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. doi: 10.1186/1741-7007-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcoleta AE, Berríos-Pastén C, Nuñez G, Monasterio O, Lagos R. Klebsiella pneumoniae asparagine tDNAs are integration hotspots for different genomic islands encoding microcin E492 production determinants and other putative virulence factors present in hypervirulent strains. Front Microbiol. 2016;7:1–17. doi: 10.3389/fmicb.2016.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putze J, Hennequin C, Nougayrède JP, Zhang W, Homburg S, Karch H, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carniel E, Mazigh D, Mollaret HH. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Almeida AM, Guiyoule A, Guilvout I, Iteman I, Baranton G, et al. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb Pathog. 1993;14:9–21. doi: 10.1006/mpat.1993.1002. [DOI] [PubMed] [Google Scholar]
- 24.Pelludat C, Rakin A, Jacobi CA, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conlan S, Deming C, Tsai YC, Lau AF, Dekker JP, et al. Complete genome sequence of a Klebsiella pneumoniae isolate with chromosomally encoded carbapenem resistance and colibactin synthesis loci. Genome Announc. 2014;2:e01332-14. doi: 10.1128/genomeA.01332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One. 2015;10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paauw A, Leverstein-van Hall MA, Verhoef J, Fluit AC. Evolution in quantum leaps: multiple combinatorial transfers of HPI and other genetic modules in Enterobacteriaceae. PLoS One. 2010;5:e8662. doi: 10.1371/journal.pone.0008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ch'ng SL, Octavia S, Xia Q, Duong A, Tanaka MM, et al. Population structure and evolution of pathogenicity of Yersinia pseudotuberculosis. Appl Environ Microbiol. 2011;77:768–775. doi: 10.1128/AEM.01993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert S, Rakin A, Heesemann J. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int J Med Microbiol. 2004;294:83–94. doi: 10.1016/j.ijmm.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Croucher NJ, Campo JJ, Le TQ, Liang X, Bentley SD, et al. Diverse evolutionary patterns of pneumococcal antigens identified by pangenome-wide immunological screening. Proc Natl Acad Sci USA. 2017;114:E357. doi: 10.1073/pnas.1613937114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesse E, O'Brien S, Tromas N, Bayer F, Luján AM, et al. Ecological selection of siderophore-producing microbial taxa in response to heavy metal contamination. Ecol Lett. 2018;21 doi: 10.1111/ele.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Löhr IH, Hülter N, Bernhoff E, Johnsen PJ, Sundsfjord A, et al. Persistence of a pKPN3-like CTX-M-15-encoding IncFIIK plasmid in a Klebsiella pneumoniae ST17 host during two years of intestinal colonization. PLoS One. 2015;10:e0116516. doi: 10.1371/journal.pone.0116516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandegren L, Linkevicius M, Lytsy B, Melhus Å, Andersson DI. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J Antimicrob Chemother. 2012;67:74–83. doi: 10.1093/jac/dkr405. [DOI] [PubMed] [Google Scholar]
- 36.Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 37.Kwong JC, McCallum N, Sintchenko V, Howden BP. Whole genome sequencing in clinical and public health microbiology. Pathology. 2015;47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robilotti E, Kamboj M. Integration of whole-genome sequencing into infection control practices: the potential and the hurdles. J Clin Microbiol. 2015;53:1054–1055. doi: 10.1128/JCM.00349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, et al. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. 2011;104:40–45. doi: 10.1097/SMJ.0b013e3181fd7d5a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. Characterization of a new metallo-β-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee IR, Molton JS, Wyres KL, Gorrie C, Wong J, et al. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep. 2016;6:29316. doi: 10.1038/srep29316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2 doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorrie CL, Mirceta M, Wick RR, Edwards DJ, Thomson NR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65:208–215. doi: 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 46.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 49.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 51.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci USA. 2014;111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Annotated ICE and plasmid sequences generated in this study are available in NCBI Genbank under the accession numbers specified in the text. Yersiniabactin and colibactin MLST schemes are available in the K. pneumoniae BIGSdb database at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. All whole-genome sequences analysed in this study are freely available in NCBI, accession numbers are given in Table S4.