Abstract
The human adaptive immune system is a very complex network of different types of cells, cytokines, and signaling molecules. This complex network makes it difficult to understand the system level regulations. To properly explain the immune system, it is necessary to explicitly investigate the presence of different feedback and feedforward loops (FFLs) and their crosstalks. Considering that these loops increase the complexity of the system, the mathematical modeling has been proved to be an important tool to explain such complex biological systems. This review focuses on these regulatory loops and discusses their importance on systems modeling of the immune system.
Drug‐induced immunogenicity results from engagement with both innate and adaptive immune system responses. Although the innate immune response is a nonspecific first line of defense, the adaptive response is more antigen‐specific and develops later. The adaptive immune system is a complex system involving different immune cells, signaling molecules and cytokines, however, the importance of different feedback and feedforward loops (FFLs) and the underlying crosstalks are often overlooked. In particular, negative feedback loops are very important in regulating the response that determines immunity vs. tolerance. Positive feedbacks bolster the influence of input signal, and FFLs may lead to a biphasic or ultrasensitive response. Considerations of these loops are also very important in terms of mathematical modeling for a mechanistic understanding of the system. Mathematical modeling has been proved to be an important tool to explain complex system behavior containing interconnected feedback‐FFLs in other cellular systems and signaling networks. This review focuses on this underinvestigated area and provides new insights into the current understanding of the cellular and molecular mechanisms associated with feedback‐feedforward regulation and how these interconnected loops give rise to complex immune system behavior.
Immune system and its complexity
The study of immune systems dates back centuries. Contemporary immunology uses advanced experimental and analytical techniques to research the mechanisms by which the complex interaction of different cell types and vast array of signaling molecules and cytokines are regulated to produce effective immune responses. When the immune system encounters a foreign molecule, it activates innate and adaptive immune pathways to take necessary measures to eliminate the foreign element.1 Once this has been achieved, the immune response needs to be attenuated or switched off to maintain homeostasis. Immune response is partly regulated through the natural death of immune cells, but one of the dominant regulatory mechanisms that is often less scrutinized is the presence of feedback and FFLs.2 Feedback and FFLs have been shown to play a crucial role in shaping response‐adaptation in several cellular systems and signaling pathways, such as MEK/ERK, Akt/mTOR, JAK/STAT, and other kinase cascades.3, 4, 5, 6, 7 They have also been shown to control the cell fate during early development.8 The importance of positive and negative feedback loops in intracellular T cell signaling is discussed previously and shows how foreign vs. self‐antigens are discriminated by activation of different intracellular feedback loops.9 However, a clear description of such loops in the adaptive immune system in combined cellular and molecular level is lacking.
Simplified models of tumor immune response considering important regulatory motifs, such as negative feedback and incoherent FFLs, show the models could successfully predict the tumor growth kinetics.10, 11, 12 In general, a biological system with a negative or positive feedback can be depicted, as in Figure 1 a, in which the input signal X activates A, A activates B, and, finally, B regulates A via a feedback. Figure 1 b shows the response kinetics for three different cases: no feedback, positive feedback, and negative feedback. The presence of a negative feedback produces a pulsatile response in A, which is not observed in the cases of no feedback and positive feedback. A negative feedback can speed up the response and can cause overshoot whereas a positive feedback can slow down the response.13, 14 A complex scenario arises when there is a presence of coupled feedback loops.5, 15 Kinetics in the presence of FFLs can also be drawn in a similar manner, Figure 1 c, in which input signal X activates A, A activates B, and an activation or inhibition control of B from X in coherent or incoherent FFL, respectively. FFLs can be generated with different combinations of activation‐inhibition pathways and are discussed in more details in refs. 16, 17, 18 Figure 1 d shows sigmoidal and biphasic input‐output relationship for coherent and incoherent FFLs, respectively.
Figure 1.
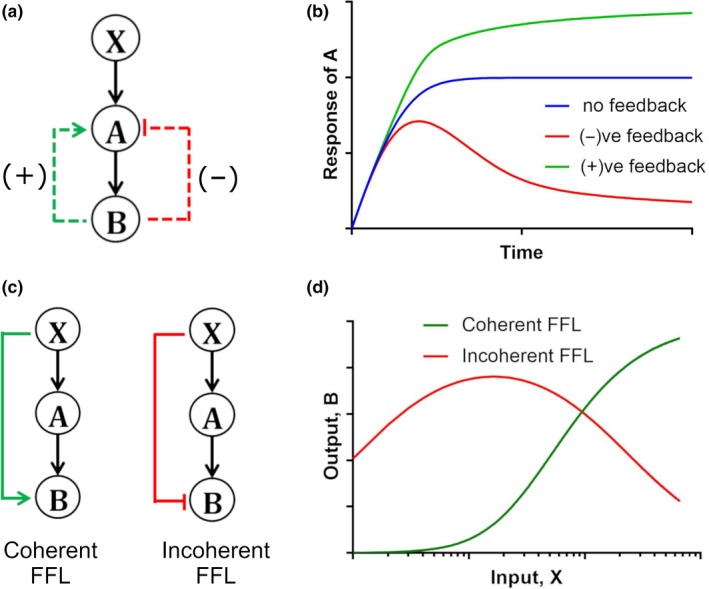
(a) Feedback loops in a biological system. The canonical pathway is X → A → B, positive feedback (green) has activating contribution from B → A and negative feedback (red) has inhibitory contribution from B → A. (b) Response kinetics of A to a step function in signal X in systems with and without feedback loops (FFLs). (c) Coherent vs. incoherent FFLs. For coherent FFLs, along with the canonical X → A → B pathway, there is a direct activating signal from X → B, whereas for incoherent FFL there is a direct inhibitory signal from X → B. (d) Sigmoidal (green) and bi‐phasic (red) input‐output response for coherent and incoherent FFL, respectively, for varying input strengths; X represents input signal, and A and B are the downstream molecules. All the kinetic profiles are plotted by solving a set of ordinary differential equations (ODEs) representing the biological systems a and c. The ODEs are solved in Matlab (MathWorks, Natick, MA). The ODE expressions, parameter values, and Matlab codes are provided in Supplementary Text S1.
By comparison, the immune system involves complex interactions between immune cells and cytokines, and includes several different feedback and FFLs. The immunogenic vs. tolerogenic response of the immune system may depend on the complex interplay of these activating and inhibitory loops. This review focuses on the intercellular interactions and the associated positive and negative feedback and FFLs in the adaptive immune system. An overview of these connections, derived by considering evidence from both human and mouse immunology, is depicted in Figure 2 and described in detail in the following sections. Identification of these regulatory loops will help better understand the immune system and will also facilitate improved mathematical modeling of the immune system to biotherapeutics.
Figure 2.
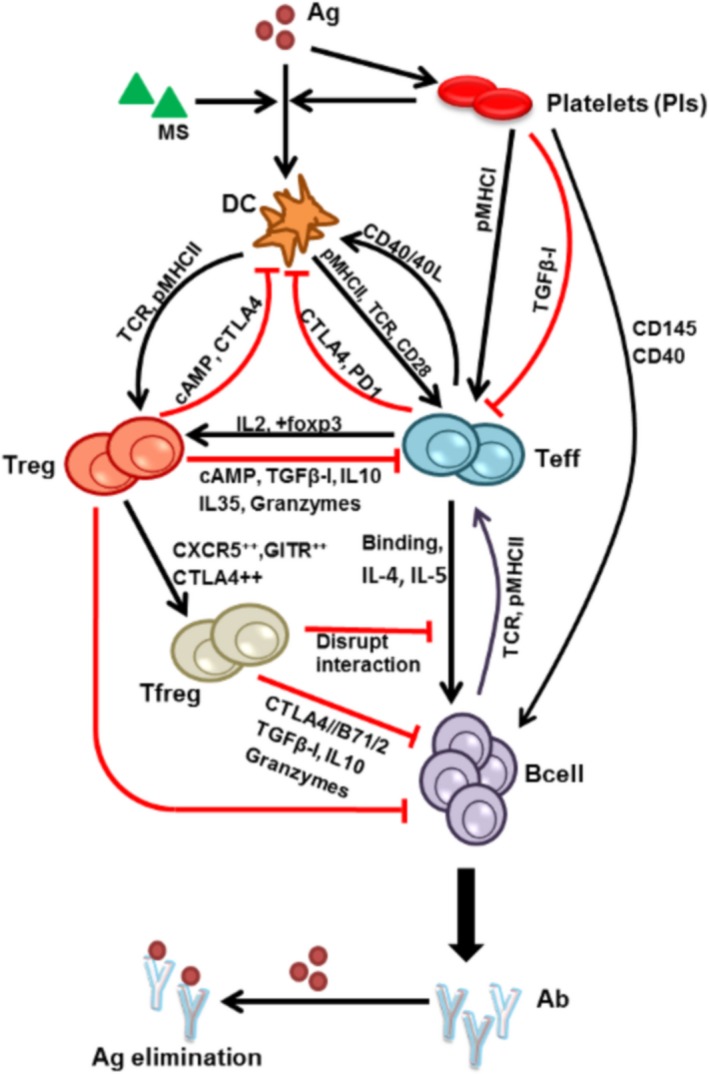
Adaptive immune responses mediated by foreign antigens through activation of immune cells and secretion of cytokines. Black lines represent activation or positive regulations, and the red lines represent inhibition or negative regulations. Ab, antibody; Ag, antigen; cAMP, cyclic adenosine monophosphate; CTLA, cytotoxic T‐lymphocyte antigen; DC, dendritic cells; IL, interleukin; MS, maturation signal; pMHC‐II, peptide‐major histocompatibility complex II; TGFβ, transforming growth factor‐beta.
Dendritic cell activation
The adaptive immune system is activated when exposed to foreign elements, such as pathogens, vaccines, biological drugs (antibodies, Fc fusion proteins), or peptide sequences. In peripheral tissue, toll‐like receptors (TLRs), Fc receptors on dendritic cells (DCs) recognize these foreign elements or antigens (Ags) and internalize them via endocytosis.19, 20 The internalized Ags are then processed by the cell's internal machinery (protease enzymes, such as cathepsins) to generate small peptides. These peptides then bind to the major histocompatibility complex (MHC)‐II molecules and are presented on the surface of DCs. There is also evidence showing MHC‐II binding and processing of Ag may occur simultaneously.21 The expression of MHC‐II and other costimulatory molecules increases with Ag exposure to DCs.21 TLR ligands, such as lipopolysaccharides, act as maturation signals for the DCs.22 The detail on the mechanism of recognizing and presenting self vs. non‐self Ags is beyond the scope of this review and the reader is referred to refs. 23, 24, 25.
T cell activation
Dendritic cells present the peptide‐MHC‐II complex on their surface to the T cells. The complex binds to and activates the T cell receptor (TCR). Although TCR activation is not sufficient to fully activate T cells, it provides specificity to the immune response.26, 27 Full activation of T cells require a secondary costimulatory signal from DCs. DCs express several costimulatory signaling molecules on their surface, such as CD80/86 (B7‐1/2), CD40, ICOSL, and OX40L that bind to costimulatory receptors, such as CD28, CD40L, ICOS, OX40, and cytotoxic T‐lymphocyte antigen‐4 (CTLA‐4) on the surface of T cells.28 This binding results in full activation of T cells (effector T cells (Teff)), which in turn leads to the production of interleukin (IL)‐2 and other cytokines, whereas the absence of a secondary signal leads to T cell anergy.29, 30 The kinetics of the signal mediated by the costimulatory molecules depends on their expression dynamics—some molecules, such as CD80/86 and CD28, are expressed at the early stage of activation, whereas others, such as CTLA‐4, are expressed at the late stage. Among the various costimulatory molecules, CD28 is the most prominent and most studied. Contrasting evidence supports constitutive or induced expression of CD28 on the T cell surfaces.31 CD28 binds to the costimulatory molecules CD80/86 on DCs thereby aiding in TCR‐mediated T cell activation.32 Mice deficient in CD28 or CD80/86 show reduced T cell activation and proliferation.33, 34
Positive feedback from Teff to DCs
There is a second wave of costimulatory signal that further enhances T cell activation and is crucial for production of long‐term memory cells. This second wave is a positive feedback signal from activated T cells to DCs.22, 35 For example, initial T cell activation promotes enhanced expression of CD40L, which binds to CD40 on activated DCs and thereby results in further increase in expression of CD80/86, OX40L, and ICOSL.36 The increased expression of CD80/86, OX40L, and ICOSL further contributes to the activation of T cells. Through this positive feedback, T cells enhance activation of DCs and, thus, increase their own proliferation (Figure 2). The simultaneous play of TCR activation, costimulation, and cytokine inputs to amplify T cell proliferation and differentiation is discussed in ref. 37.
Feedback inhibition through co‐inhibition
After T cells have effectively controlled or eliminated foreign Ag, the immune response is terminated via T cell contraction.38 The contraction is mediated by cell apoptosis, feedback inhibition by regulatory T cells (Tregs), or feedback inhibition by co‐inhibitory molecules. Like costimulatory molecules, co‐inhibitory molecules are expressed on the surface of T cells and vary in their expression kinetics.39 The repertoire of costimulatory and co‐inhibitory receptors on T cells is highly diverse. The fate of T cells is determined by the strength and kinetics of different costimulatory and co‐inhibitory molecules. Most studied co‐inhibitory molecules include CTLA‐4, programmed cell death protein 1 (PD‐1), and B‐ and T‐lymphocyte attenuator (BTLA).40, 41
CTLA4 is a CD28 homologue that binds to CD80/86 on DCs with higher affinity than CD28 and thereby causes dissociation and endocytosis, which inhibits TCR signaling through CD28.42 Mice deficient in CTLA4 showed higher T cell activation and proliferation.43
Another important co‐inhibitory molecule is PD‐1, which also inhibits CD28 mediated costimulation by binding to ligands PDL1 and PDL2 on DCs.40 Thus, the CTLA4 and PD‐1 mediated negative feedback loops and the costimulatory positive feedback loops regulate the balance between costimulation and co‐inhibition needed for activation or inhibition of T cells. CTLA4 and PD‐1 are crucial for the maintenance of central and peripheral tolerance and development of T cell contraction through co‐inhibition. Tolerance is the state of unresponsiveness of the immune system to immune stimulating molecules. Thereby, positive and negative feedback loops mediated by costimulatory and co‐inhibitory molecules are of particular interest in drug development for treatment of immunodeficiency or autoimmune disease therapies.41
Negative regulations through Tregs
TCR activation and CD80/86‐CD28 interaction is important in the development and survival of another class of T cells, called Tregs.44, 45 Tregs can also be produced in an IL‐2 dependent fashion from activated Teff cells with increased expression of CD25 and Foxp3.46, 47 The primary function of Tregs is to inhibit activated T cells and DCs, thereby giving rise to two cellular‐level negative feedback loops.26, 48, 49 In accordance, dysregulation of Tregs function via mutation or deletion of Foxp3 leads to autoimmunity in humans and mice.50, 51 Thus, owing to feedback control, Tregs should be considered as part of the system rather than a genetically distinct programmed lineage.49 Several mechanisms for Treg‐mediated inhibition have been proposed.48, 52 These can be divided into three main classes: (1) cell‐cell contact, (2) inhibitory cytokine secretion, and (3) competition. Tregs can inhibit DCs through mechanisms 1 and 2, and Teff cells through mechanisms 1, 2, and 3. In the cell‐cell contact model, Tregs directly bind to the DCs or effector T cells through receptor‐ligand interactions, such as peptide‐major histocompatibility complex (pMHC)II‐TCR and CTLA4/B7‐1, and mediate inhibition, cytolysis, or apoptosis of the cells by delivering suppressive factors, such as cyclic adenosine monophosphate and transforming growth factor‐beta (TGFβ)1 via gap junctions48, 53, 54 or by membrane‐bound TGFβ. Next, in the inhibitory cytokine secretion model, Tregs upon activation can directly secrete IL‐10, TGFβ, and IL‐35 that mediate suppression of DCs and Teff cells.55, 56, 57 Last, in the competition model, Tregs either compete for cytokines (such as IL‐2), which leads to cytokine‐deprived cell apoptosis58, 59, 60 or may bind to DCs through CTLA4/B7‐1 interaction thereby decreasing the amount of costimulatory molecule (B7‐1/2) available to bind CD28 on naïve T cells, thus inhibiting T cell activation. It is not known whether all these mechanisms are needed simultaneously to exert Treg‐mediated inhibition. In a particular immune setting, a single mechanism might be sufficient or all mechanisms may be necessary in an additive manner.61
Tregs also inhibit B cell antibody production by secreting inhibitory factors IL‐10, TGFβ, and granzymes, or by binding through CTLA4/B7‐1 interaction.53 It has been shown that increased presence of Tregs decreases the production of antibodies, whereas their absence increases production.62, 63, 64 Recently, a new subset of Tregs called T‐follicular regulatory cells (Tfregs) has also been shown to inhibit B cell activity by directly entering the germinal center and secreting inhibitory cytokines. Natural Tregs differentiate into Tfregs with expressing high levels of CLTA4, GITR, and CXCR5.65, 66 Tfregs may also inhibit B cell activity by disrupting the mechanical interaction between T cells and B cells.66
Feedback and feedforward regulation by platelets
Platelets are best known for their role in hemostasis,67 but their function in the immune response is an emerging research area. After antigen exposure, platelets get activated as part of the innate immune response and their activation is mediated by surface TLRs and danger signal lipopolysaccharides.68, 69, 70 As part of the innate immune response, platelets can directly endocytose or internalize pathogens. Activated platelets modulate adaptive immune response via numerous direct or indirect mechanisms. They act as a catalyst of adaptive immune response through a positive feedback that promotes antigen aggregation through activated integrin‐fibrinogen interactions on the platelet's surface, increases antigen presentation,71, 72 and enhances DC activation.73, 74, 75, 76 The role of antigen aggregates in increasing immune response has been shown earlier.77
Platelets are often called antigen presenting cells (APCs) as they activate T cells through pMHCI.78 The pMHCI is constitutively expressed by platelets and their expression increases significantly during inflammation.79
Platelets also negatively regulate T cells and suppress their activation through production and activation of immunosuppressive factors, such as TGFβ and lactic acid. Platelets activate TGFβ by binding the TGFβ‐docking receptor, GARP.80 Platelets activate B cells through CD145‐CD40 interactions and thereby increase production of antibodies.81, 82, 83 Thus, platelets have a complex mechanism of regulating immunity vs. tolerance. Figure 3 shows a simplified depiction of the effect of platelets on innate and adaptive immune systems.
Figure 3.
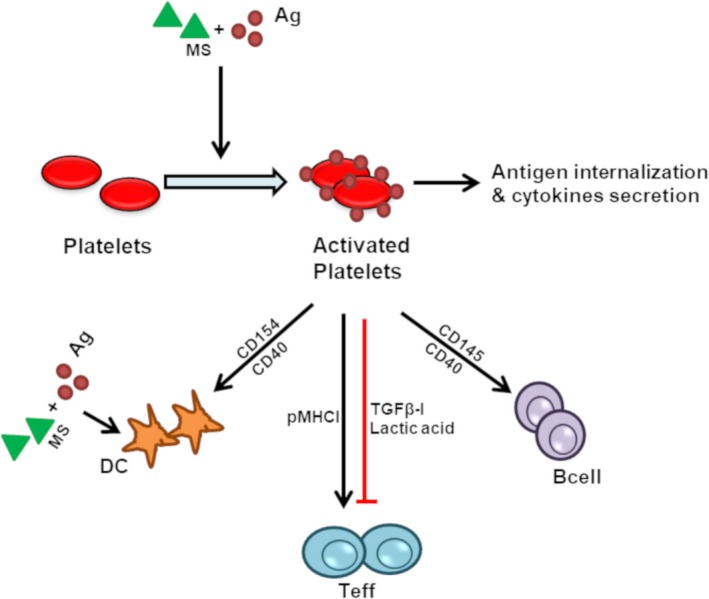
Role of platelets in regulating innate and adaptive immune responses. Antigens (Ag) and maturation signal (MS) mediated activation of platelets leads to aggregation, endocytosis/internalization of the antigen, secretion of the cytokines (sCD40L, interleukin (IL)‐1β) and subsequent activation of the immune cells (dendritic cells (DC), T cells, and B cells). Teff, effector T cells; TGFβ, transforming growth factor‐beta.
B cell activation
B cells can get activated through a T‐cell dependent or independent manner. In T‐cell independent B cell activation mechanism, the antigens (known as thymus‐independent antigens) with their repetitive structure make cross‐linking of the B‐cell receptors and elicit B cell activation. The repetitive structure of the antigen may arise due to aggregation.84 In T‐cell dependent activation mechanism, activated T cells may directly bind to B cells forming immunological synapses or secrete stimulatory cytokines (IL‐4, IL‐5, etc.) to activate B cells. In many immune settings, B cells function as APCs for T cells,85, 86, 87 thus creating a positive feedback loop. As an APC, B cells bind antigens to its surface expressed immunoglobulin (B‐cell receptors), internalizes antigen, processes antigen by internal machinery, binds antigenic peptides to MHC‐II molecules, and present them on the cell surface like DCs.85, 86, 87 In the T‐cell dependent mechanism, binding of antigen is not sufficient to fully activate B cells and it requires a second signal through T‐cell dependent pathways.88, 89
Antidrug antibody production and increased antigen clearance
Activated B cells undergo clonal expansion and they proliferate and differentiate into short and long‐lived plasma cells, which are responsible for producing antibodies against the antigens the B cells experienced. Thus, all biological drugs or therapeutic proteins have the ability to induce production of antidrug antibodies (ADAs) to some extent. ADA production affects clinical outcomes by impacting drug pharmacokinetics, and consequently its pharmacodynamics and efficacy profiles. The binding of ADAs to drugs depends on their affinities, dose, and duration of exposure.90, 91, 92
A summary of all the feedback and FFLs described in this section and in Figure 2 is given in Table 1.
Table 1.
Presence of feedback and feedforward loops in the adaptive immune systema
| Feedback loops | Feedforward loops |
|---|---|
| DC→Teff→DC (+) (−) | Ag → DC → Teff & Ag → Pl → Teff (C) (I) |
| DC → Treg → DC (−) | DC → Treg & DC → Teff → Treg (C) |
| Teff → Treg → Teff (−) | DC → Teff & DC → Treg → Teff (I) |
| Teff → Bcell → Teff (+) | Pl → Teff → Bcell & Pl → Bcell (C) (I) |
| Ag → Pl → Ag‐DC (+) | Treg → Teff → Bcell & Treg → Bcell (C) |
| Bcell → Teff → Bcell | Teff → Treg → Bcell & Teff → Bcell (I) |
| Ag → DC → TC → Bcell → ADA → Ag (−) | DC → Teff → Bcell & DC → Treg → Bcell (I) |
Ag, antigen; ADA, antidrug‐antibody; DC, dendritic cell; Pl, platelet; Teff, effector T cell; Treg, regulatory T cell. a(+) and (−) are positive and negative feedbacks, respectively. (C) represents coherent and (I) represents incoherent feedforward loops. For feedforward loops, two pathways are indicated for each loop.
Discussion
Viewing the immune system through the lens of interconnected feedback‐FFLs will open new perspective on the system. Although many studies have been conducted to understand the underlying biology of immune responses, the new perspective proposed here will enable deeper understanding of the dynamics of immune responses. This study will help investigate the immune system kinetics and understand system level regulation and also help build a consistent mathematical model of the immune system network.
Previously, mathematical modeling has been successfully used to explain intracellular interactions, cell‐cell interactions, protein signaling networks, and physiological interactions.4, 7, 93, 94 Modeling helps to discover complex phenomena in a biological system, which is very difficult to identify otherwise; such as experimentally. One potential application of the mathematical modeling of the immune system will be on modeling immunogenicity of therapeutic proteins. It is very challenging to predict immunogenicity of protein products or drugs, and recently the immunogenicity has brought much attention of the pharmaceutical research communities. Development of the preclinical predictive immunogenicity models will allow us to screen for the potential immunogenic drugs. Immunogenicity models will help design clinical trials prior to administration of the drugs to humans. It will also help designing the de‐immunization strategy to reduce the overall immunogenicity. Recently, several mathematical models of the immune system have been developed to predict immunogenicity of drug therapeutics.93, 95, 96, 97 The investigation of feedback‐FFLs in the immune system is lacking, with some notable exceptions.98, 99, 100 The simplistic representation of the adaptive immune system in Figure 4 resembles the various biological systems shown in Figure 1. The left panels show different feedback and FFLs in the simplified immune system, whereas the right panels show the illustration of the kinetics of Ag, DCs, and ADA responses. Simplified mathematical models are used to plot the kinetic profiles (Supplementary Text S1). Figure 4 a demonstrates that the presence of a negative feedback from ADA to Ag can explain the increase in the ADA‐mediated antigen clearance. Figure 4 b,c, respectively, show the possible effect of positive and negative feedback loops between DCs and T cells, and how the activated DC concentration is affected at later timepoints. The effect of FFLs can also be observed in a similar fashion (Figure 4 d). The presence of an incoherent FFL may introduce transient kinetics in ADA production. The effect of combined positive‐negative feedback‐FFLs will be much more complex, as shown in Figure 2 and Table 1. With different signal strengths of the feedback and FFLs, it will be possible to investigate and explain a particular immune setting.
Figure 4.
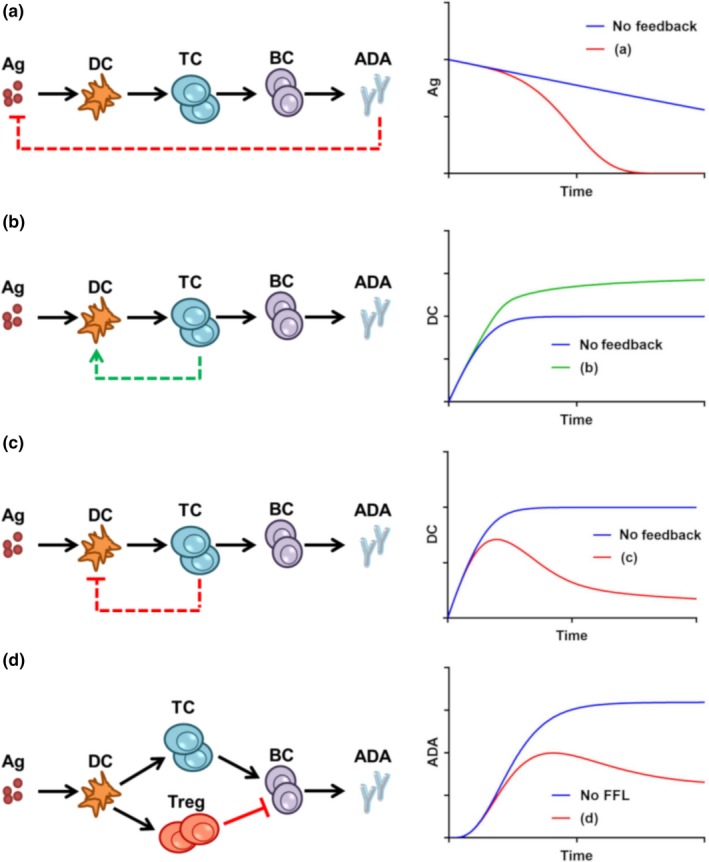
Feedback and feedforward loops (FFLs) in the adaptive immune system and their potential impact on response kinetics of corresponding species (a) negative feedback from antidrug‐antibody (ADA) to antigen (Ag); (b) positive feedback from T cells to dendritic cells (DCs); (c) negative feedback from T cells to DCs; and (d) an incoherent FFL affecting B‐cell number and ADA production. Blue = no feedback or no FFL; green = positive feedback; and red = negative feedback or incoherent FFL. All the kinetic profiles are plotted by solving a set of ordinary differential equations (ODEs) representing the different biological systems. ODEs are solved in Matlab. The ODE expressions, parameter values, and Matlab codes are provided in Supplementary Text S1. BC, B cells; TC, T cells.
In conclusion, in this review, we revisited the immune system emphasizing the different regulatory loops associated with it and simulated different hypothetical scenario. This review sheds light on the importance of investigating the role of these interconnecting loops in the immune system. Considering these regulatory loops in mathematical models has been shown to successfully explain the complex system behavior in other research areas and we hope consideration of these loops in immune system modeling will greatly enhance our understanding of the immune system and drug‐induced immunogenicity.
Funding
This work was supported by the Pfizer WRD program.
Conflict of Interest
The authors declared no competing interests for this work.
Supporting information
Supplementary Text S1. Importance of feedback and feedforward loops to adaptive two‐immune response modeling.
Acknowledgments
The authors thank Andrzej Kierzek and Piet van der Graaf of Certara for their valuable feedback and suggestions.
References
- 1. Abbas, A.K. , Lichtman, A.H. & Pillai, S. Basic Immunology: Functions and Disorders of the Immune System. 4th edn (Elsevier/Saunders, Philadelphia, PA, 2014). [Google Scholar]
- 2. Carrera Silva, E.A. et al T cell‐derived protein S engages TAM receptor signaling in dendritic cells to control the magnitude of the immune response. Immunity 39, 160–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Behar, M. , Hao, N. , Dohlman, H.G. & Elston, T.C. Mathematical and computational analysis of adaptation via feedback inhibition in signal transduction pathways. Biophys. J. 93, 806–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cirit, M. , Wang, C.C. & Haugh, J.M. Systematic quantification of negative feedback mechanisms in the extracellular signal‐regulated kinase (ERK) signaling network. J. Biol. Chem. 285, 36736–36744 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim, J.R. , Yoon, Y. & Cho, K.H. Coupled feedback loops form dynamic motifs of cellular networks. Biophys. J. 94, 359–365 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandman, O. & Meyer, T. Feedback loops shape cellular signals in space and time. Science 322, 390–395 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahman, A. & Haugh, J.M. Kinetic modeling and analysis of the Akt/mechanistic target of rapamycin complex 1 (mTORC1) signaling axis reveals cooperative. Feedforward regulation. J. Biol. Chem. 292, 2866–2872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Freeman, M. Feedback control of intercellular signalling in development. Nature 408, 313–319 (2000). [DOI] [PubMed] [Google Scholar]
- 9. Mueller, D.L. Tuning the immune system: competing positive and negative feedback loops. Nat. Immunol. 4, 210–211 (2003). [DOI] [PubMed] [Google Scholar]
- 10. Sontag, E.D. A dynamic model of immune responses to antigen presentation predicts different regions of tumor or pathogen elimination. Cell System 4, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parra‐Guillen, Z.P. , Berraondo, P. , Grenier, E. , Ribba, B. & Troconiz, I.F. Mathematical model approach to describe tumour response in mice after vaccine administration and its applicability to immune‐stimulatory cytokine‐based strategies. AAPS J. 15, 797–807 (2013a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parra‐Guillen, Z.P. , Berraondo, P. , Ribba, B. & Troconiz, I.F. Modeling tumor response after combined administration of different immune‐stimulatory agents. J. Pharmacol. Exp. Ther. 346, 432–442 (2013b). [DOI] [PubMed] [Google Scholar]
- 13. Rosenfeld, N. , Elowitz, M.B. & Alon, U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323, 785–793 (2002). [DOI] [PubMed] [Google Scholar]
- 14. Ray, J.C. & Igoshin, O.A. Adaptable functionality of transcriptional feedback in bacterial two‐component systems. PLoS Comput. Biol. 6, e1000676 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tiwari, A. & Igoshin, O.A. Coupling between feedback loops in autoregulatory networks affects bistability range, open‐loop gain and switching times. Phys. Biol. 9, 055003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mangan, S. & Alon, U. Structure and function of the feed‐forward loop network motif. Proc. Natl. Acad. Sci. USA 100, 11980–11985 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alon, U. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461 (2007). [DOI] [PubMed] [Google Scholar]
- 18. Rahman, A. & Haugh, J.M. Deactivation of a negative regulator: a distinct signal transduction mechanism, pronounced in Akt signaling. Biophys. J. 107, L29–L32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Platt, C.D. et al Mature dendritic cells use endocytic receptors to capture and present antigens. Proc. Natl. Acad. Sci. USA 107, 4287–4292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amigorena, S. & Bonnerot, C. Fc receptor signaling and trafficking: a connection for antigen processing. Immunol. Rev. 172, 279–284 (1999). [DOI] [PubMed] [Google Scholar]
- 21. Sadegh‐Nasseri, S. & Kim, A. Exogenous antigens bind MHC class II first, and are processed by cathepsins later. Mol. Immunol. 68, 81–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdi, K. , Singh, N.J. & Matzinger, P. Lipopolysaccharide‐activated dendritic cells: “exhausted” or alert and waiting? J. Immunol. 188, 5981–5989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang, H. & Chess, L. How the immune system achieves self‐nonself discrimination during adaptive immunity. Adv. Immunol. 102, 95–133 (2009). [DOI] [PubMed] [Google Scholar]
- 24. Hopp, A.K. , Rupp, A. & Lukacs‐Kornek, V. Self‐antigen presentation by dendritic cells in autoimmunity. Front. Immunol. 5, 55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. ten Broeke, T. , Wubbolts, R. & Stoorvogel, W. MHC class II antigen presentation by dendritic cells regulated through endosomal sorting. Cold Spring Harb. Perspect. Biol. 5, a016873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharpe, A.H. & Abbas, A.K. T‐cell costimulation–biology, therapeutic potential, and challenges. N. Engl. J. Med. 355, 973–975 (2006). [DOI] [PubMed] [Google Scholar]
- 27. Labrecque, N. et al How much TCR does a T cell need? Immunity 15, 71–82 (2001). [DOI] [PubMed] [Google Scholar]
- 28. Sharpe, A.H. Mechanisms of costimulation. Immunol. Rev. 229, 5–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen, L. & Flies, D.B. Molecular mechanisms of T cell co‐stimulation and co‐inhibition. Nat. Rev. Immunol. 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janardhan, S.V. , Praveen, K. , Marks, R. & Gajewski, T.F. Evidence implicating the Ras pathway in multiple CD28 costimulatory functions in CD4+ T cells. PLoS One 6, e24931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Turka, L.A. , Ledbetter, J.A. , Lee, K. , June, C.H. & Thompson, C.B. CD28 is an inducible T cell surface antigen that transduces a proliferative signal in CD3+ mature thymocytes. J. Immunol. 144, 1646–1653 (1990). [PubMed] [Google Scholar]
- 32. Greene, J.L. et al Covalent dimerization of CD28/CTLA‐4 and oligomerization of CD80/CD86 regulate T cell costimulatory interactions. J. Biol. Chem. 271, 26762–26771 (1996). [DOI] [PubMed] [Google Scholar]
- 33. Sharpe, A.H. & Freeman, G.J. The B7‐CD28 superfamily. Nat. Rev. Immunol. 2, 116–126 (2002). [DOI] [PubMed] [Google Scholar]
- 34. Lucas, P.J. , Negishi, I. , Nakayama, K. , Fields, L.E. & Loh, D.Y. Naive CD28‐deficient T cells can initiate but not sustain an in vitro antigen‐specific immune response. J. Immunol. 154, 5757–5768 (1995). [PubMed] [Google Scholar]
- 35. Acuto, O. & Michel, F. CD28‐mediated co‐stimulation: a quantitative support for TCR signalling. Nat. Rev. Immunol. 3, 939–951 (2003). [DOI] [PubMed] [Google Scholar]
- 36. Walker, L.S. , Gulbranson‐Judge, A. , Flynn, S. , Brocker, T. & Lane, P.J. Co‐stimulation and selection for T‐cell help for germinal centres: the role of CD28 and OX40. Immunol. Today 21, 333–337 (2000). [DOI] [PubMed] [Google Scholar]
- 37. Marchingo, J.M. et al T cell signaling. Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science 346, 1123–1127 (2014). [DOI] [PubMed] [Google Scholar]
- 38. Vigano, S. , Perreau, M. , Pantaleo, G. & Harari, A. Positive and negative regulation of cellular immune responses in physiologic conditions and diseases. Clin. Dev. Immunol. 2012, 485781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu, Y. , Yao, S. & Chen, L. Cell surface signaling molecules in the control of immune responses: a tide model. Immunity 34, 466–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hubo, M. et al Costimulatory molecules on immunogenic versus tolerogenic human dendritic cells. Front. Immunol. 4, 82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Podojil, J.R. & Miller, S.D. Molecular mechanisms of T‐cell receptor and costimulatory molecule ligation/blockade in autoimmune disease therapy. Immunol. Rev. 229, 337–355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van der Merwe, P.A. , Bodian, D.L. , Daenke, S. , Linsley, P. & Davis, S.J. CD80 (B7‐1) binds both CD28 and CTLA‐4 with a low affinity and very fast kinetics. J. Exp. Med. 185, 393–403 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tivol, E.A. et al Loss of CTLA‐4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA‐4. Immunity 3, 541–547 (1995). [DOI] [PubMed] [Google Scholar]
- 44. Zou, T. , Caton, A.J. , Koretzky, G.A. & Kambayashi, T. Dendritic cells induce regulatory T cell proliferation through antigen‐dependent and ‐independent interactions. J. Immunol. 185, 2790–2799 (2010). [DOI] [PubMed] [Google Scholar]
- 45. Campbell, D.J. & Koch, M.A. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol. 11, 119–130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li, X. & Zheng, Y. Regulatory T cell identity: formation and maintenance. Trends Immunol. 36, 344–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sakaguchi, S. , Miyara, M. , Costantino, C.M. & Hafler, D.A. FOXP3+ regulatory T cells in the human immune system. Nat. Rev. Immunol. 10, 490–500 (2010). [DOI] [PubMed] [Google Scholar]
- 48. Sojka, D.K. , Huang, Y.H. & Fowell, D.J. Mechanisms of regulatory T‐cell suppression ‐ a diverse arsenal for a moving target. Immunology 124, 13–22 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ono, M. & Tanaka, R.J. Controversies concerning thymus‐derived regulatory T cells: fundamental issues and a new perspective. Immunol. Cell Biol. 94, 3–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bennett, C.L. et al The immune dysregulation, polyendocrinopathy, enteropathy, X‐linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27, 20–21 (2001). [DOI] [PubMed] [Google Scholar]
- 51. Kim, J.M. , Rasmussen, J.P. & Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007a). [DOI] [PubMed] [Google Scholar]
- 52. Vignali, D.A. , Collison, L.W. & Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakamura, K. , Kitani, A. & Strober, W. Cell contact‐dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface‐bound transforming growth factor beta. J. Exp. Med. 194, 629–644 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rueda, C.M. , Jackson, C.M. & Chougnet, C.A. Regulatory T‐cell‐mediated suppression of conventional T‐cells and dendritic cells by different cAMP intracellular pathways. Front. Immunol. 7, 216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Powrie, F. , Carlino, J. , Leach, M.W. , Mauze, S. & Coffman, R.L. A critical role for transforming growth factor‐beta but not interleukin 4 in the suppression of T helper type 1‐mediated colitis by CD45RB(low) CD4+ T cells. J. Exp. Med. 183, 2669–2674 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Asseman, C. , Read, S. & Powrie, F. Colitogenic Th1 cells are present in the antigen‐experienced T cell pool in normal mice: control by CD4+ regulatory T cells and IL‐10. J. Immunol. 171, 971–978 (2003). [DOI] [PubMed] [Google Scholar]
- 57. Arce‐Sillas, A. et al Regulatory T cells: molecular actions on effector cells in immune regulation. J. Immunol. Res. 2016, 1720827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. de la Rosa, M. , Rutz, S. , Dorninger, H. & Scheffold, A. Interleukin‐2 is essential for CD4+ CD25+ regulatory T cell function. Eur. J. Immunol. 34, 2480–2488 (2004). [DOI] [PubMed] [Google Scholar]
- 59. Barthlott, T. et al CD25+ CD4+ T cells compete with naive CD4+ T cells for IL‐2 and exploit it for the induction of IL‐10 production. Int. Immunol. 17, 279–288 (2005). [DOI] [PubMed] [Google Scholar]
- 60. Pandiyan, P. , Zheng, L. , Ishihara, S. , Reed, J. & Lenardo, M.J. CD4+ CD25+ Foxp3+ regulatory T cells induce cytokine deprivation‐mediated apoptosis of effector CD4+ T cells. Nat. Immunol. 8, 1353–1362 (2007). [DOI] [PubMed] [Google Scholar]
- 61. Vignali, D. How many mechanisms do regulatory T cells need? Eur. J. Immunol. 38, 908–911 (2008). [DOI] [PubMed] [Google Scholar]
- 62. Lim, H.W. , Hillsamer, P. , Banham, A.H. & Kim, C.H. Cutting edge: direct suppression of B cells by CD4+ CD25+ regulatory T cells. J. Immunol. 175, 4180–4183 (2005). [DOI] [PubMed] [Google Scholar]
- 63. Zhao, D.M. , Thornton, A.M. , DiPaolo, R.J. & Shevach, E.M. Activated CD4+ CD25+ T cells selectively kill B lymphocytes. Blood 107, 3925–3932 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Curotto de Lafaille, M.A. et al Adaptive Foxp3+ regulatory T cell‐dependent and ‐independent control of allergic inflammation. Immunity 29, 114–126 (2008). [DOI] [PubMed] [Google Scholar]
- 65. Linterman, M.A. et al Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 17, 975–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sage, P.T. & Sharpe, A.H. T follicular regulatory cells in the regulation of B cell responses. Trends Immunol. 36, 410–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Menter, D.G. et al Platelet “first responders” in wound response, cancer, and metastasis. Cancer Metastasis Rev. 36, 199–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garraud, O. & Cognasse, F. Are platelets cells? And if yes, are they immune cells? Front. Immunol. 6, 70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rondina, M.T. & Garraud, O. Emerging evidence for platelets as immune and inflammatory effector cells. Front. Immunol. 5, 653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cognasse, F. et al The inflammatory role of platelets via their TLRs and Siglec receptors. Front. Immunol. 6, 83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Speth, C. , Loffler, J. , Krappmann, S. , Lass‐Florl, C. & Rambach, G. Platelets as immune cells in infectious diseases. Future Microbiol. 8, 1431–1451 (2013). [DOI] [PubMed] [Google Scholar]
- 72. Fitzgerald, J.R. , Foster, T.J. & Cox, D. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4, 445–457 (2006). [DOI] [PubMed] [Google Scholar]
- 73. Czapiga, M. , Kirk, A.D. & Lekstrom‐Himes, J. Platelets deliver costimulatory signals to antigen‐presenting cells: a potential bridge between injury and immune activation. Exp. Hematol. 32, 135–139 (2004). [DOI] [PubMed] [Google Scholar]
- 74. Hagihara, M. et al Platelets, after exposure to a high shear stress, induce IL‐10‐producing, mature dendritic cells in vitro. J. Immunol. 172, 5297–5303 (2004). [DOI] [PubMed] [Google Scholar]
- 75. Elzey, B.D. et al Platelet‐mediated modulation of adaptive immunity. A communication link between innate and adaptive immune compartments. Immunity 19, 9–19 (2003). [DOI] [PubMed] [Google Scholar]
- 76. Ratanji, K.D. , Derrick, J.P. , Dearman, R.J. & Kimber, I. Immunogenicity of therapeutic proteins: influence of aggregation. J. Immunotoxicol. 11, 99–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yin, L. , Chen, X. , Tiwari, A. , Vicini, P. & Hickling, T.P. The role of aggregates of therapeutic protein products in immunogenicity: an evaluation by mathematical modeling. J. Immunol. Res. 2015, 401956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chapman, L.M. et al Platelets present antigen in the context of MHC class I. J. Immunol. 189, 916–923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sowa, J.M. , Crist, S.A. , Ratliff, T.L. & Elzey, B.D. Platelet influence on T‐ and B‐cell responses. Arch. Immunol. Ther. Exp. (Warsz.) 57, 235–241 (2009). [DOI] [PubMed] [Google Scholar]
- 80. Rachidi, S.M.A. et al Platelets subvert T cell immunity against cancer via GARP‐TGFβ axis. Sci. Immunol. 2, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Elzey, B.D. et al Cooperation between platelet‐derived CD154 and CD4+ T cells for enhanced germinal center formation. J. Leukoc. Biol. 78, 80–84 (2005). [DOI] [PubMed] [Google Scholar]
- 82. Cognasse, F. et al Human platelets can activate peripheral blood B cells and increase production of immunoglobulins. Exp. Hematol. 35, 1376–1387 (2007). [DOI] [PubMed] [Google Scholar]
- 83. Sprague, D.L. et al Platelet‐mediated modulation of adaptive immunity: unique delivery of CD154 signal by platelet‐derived membrane vesicles. Blood 111, 5028–5036 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen, X. et al A mathematical model of the effect of immunogenicity on therapeutic protein pharmacokinetics. AAPS J. 15, 1141–1154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yuseff, M.I. , Pierobon, P. , Reversat, A. & Lennon‐Dumenil, A.M. How B cells capture, process and present antigens: a crucial role for cell polarity. Nat. Rev. Immunol. 13, 475–486 (2013). [DOI] [PubMed] [Google Scholar]
- 86. Rodriguez‐Pinto, D. & Moreno, J. B cells can prime naive CD4+ T cells in vivo in the absence of other professional antigen‐presenting cells in a CD154‐CD40‐dependent manner. Eur. J. Immunol. 35, 1097–1105 (2005). [DOI] [PubMed] [Google Scholar]
- 87. Janeway, C.A. , Travers, P. , Walport, M. & Shlomchik, M. Immunobiology: The Immune System in Health and Disease, 5th edn (Garland Publishing, New York, 2001). [Google Scholar]
- 88. Kurosaki, T. , Kometani, K. & Ise, W. Memory B cells. Nat. Rev. Immunol. 15, 149–159 (2015). [DOI] [PubMed] [Google Scholar]
- 89. Parker, D.C. T cell‐dependent B cell activation. Annu. Rev. Immunol. 11, 331–360 (1993). [DOI] [PubMed] [Google Scholar]
- 90. Tatarewicz, S.M. et al Strategic characterization of anti‐drug antibody responses for the assessment of clinical relevance and impact. Bioanalysis 6, 1509–1523 (2014). [DOI] [PubMed] [Google Scholar]
- 91. Thway, T.M. et al Impact of anti‐drug antibodies in preclinical pharmacokinetic assessment. AAPS J. 15, 856–863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Edlund, H. et al Magnitude of increased infliximab clearance imposed by anti‐infliximab antibodies in Crohn's disease is determined by their concentration. AAPS J. 19, 223–233 (2017). [DOI] [PubMed] [Google Scholar]
- 93. Chen, X. , Hickling, T.P. & Vicini, P. A mechanistic, multiscale mathematical model of immunogenicity for therapeutic proteins: part 1‐theoretical model. CPT Pharmacometrics Syst. Pharmacol. 3, e133 (2014a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ahmed, S. et al Data‐driven modeling reconciles kinetics of ERK phosphorylation, localization, and activity states. Mol. Syst. Biol. 10, 718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen, X. , Hickling, T.P. & Vicini, P. A mechanistic, multiscale mathematical model of immunogenicity for therapeutic proteins: part 2‐model applications. CPT Pharmacometrics Syst. Pharmacol. 3, e134 (2014b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Palsson, S. et al The development of a fully‐integrated immune response model (FIRM) simulator of the immune response through integration of multiple subset models. BMC Syst. Biol. 7, 95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bianca, C. , Chiacchio, F. , Pappalardo, F. & Pennisi, M. Mathematical modeling of the immune system recognition to mammary carcinoma antigen. BMC Bioinform. 13(suppl. 17), S21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kim, P.S. , Lee, P.P. & Levy, D. Modeling regulation mechanisms in the immune system. J. Theor. Biol. 246, 33–69 (2007). [DOI] [PubMed] [Google Scholar]
- 99. Khailaie, S. et al A mathematical model of immune activation with a unified self‐nonself concept. Front. Immunol. 4, 474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Su, B. , Zhou, W. , Dorman, K.K. & Jones, D.E. Mathematical modelling of immune response in tissues. Comput. Math. Methods Med. 10, 9–38 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text S1. Importance of feedback and feedforward loops to adaptive two‐immune response modeling.


