Abstract
Accumulated research has revealed that the abnormal expression of microRNAs play a crucial role in tumorigenesis, potentially serving as therapeutic biomarkers in multiple tumors including cervical cancer. However, the expression level, biological role and the underlying mechanism of miRNA-143 in cervical cancer remain unclear. In the current study, we analyzed the miRNA-143 and golgi membrane protein 1 (GOLM1) expression in cervical cancer tissues and cells to explore their effects on cervical cancer occurrence and metastasis. Reverse transcription-quantitative PCR (RT-qPCR) was used to detect the miRNA-143 expression in cervical cancer tissues and cells. Following transfection, cell Transwell assays, western blot analysis and luciferase reporter assays were carried out in human cervical cancer cells. Results demonstrated that the miRNA-143 expression was dramatically decreased in both cervical cancer tissue samples and cells in contrast with the control group. We also found that the miRNA-143 expression negatively correlated with the GOLM1 expression in cervical cancer tissues and miRNA-143 inhibited cell invasion and migration via targeting GOLM1 in cervical cancer.
Keywords: microRNA-143, golgi membrane protein 1, cervical cancer, migration, invasion
Introduction
Cervical cancer, one leading cause of tumor-related deaths, is a common gynecological tumor globally (1). Due to delayed initial screening, the lack of early diagnosis and the unavailable widespread screening by cervical cytology (2), cervical cancer mainly occurs in developing countries and causes approximately 265,000 deaths every year worldwide (3). In addition, approximately 85% of new cervical cancer cases occur in lower socioeconomic countries (4). Emerging studies indicate that risk factors, such as infection of high-risk types of human papillomavirus (HPV), promiscuity and early sexual intercourse, have essential effects on cervical cancer progression (5). The normal cervical epithelium in cervical cancer can be transformed into a precancerous cervical intraepithelial neoplasia, and then develop into invasive cervical cancer (6). Despite the effort which has been made in the treatment, cervical cancer 5-year overall survival rate still remains unsatisfactory (7). Despite the recent published research, cervical cancer etiology remains unclear. Hence, in the current study, we strived to investigate the molecular mechanisms implicated in the cervical cancer occurrence and development and to study more effective therapeutic targets for cervical cancer treatment.
Recently, increased attention has been paid to the role of microRNAs in various human cancers. miRNAs, a sizable class of highly conserved, non-protein-coding and short RNAs of ~21–25 nucleotides (8), regulate gene expression by binding to their 3′UTRs (9). miRNAs can also act as regulators of various pathological and physiological processes, such as cell differentiation, apoptosis, proliferation, invasion and migration. An increasing number of studies indicate that miRNAs are abnormally expressed in multiple tumors, inhibiting or promoting the progression of cancers (10). miRNA-143 has been featured as cancer suppressor in various cancers (11,12). However, its possible functions and underlying mechanisms in cervical cancer have remained unknown. This study aimed to elucidate the expression and effects of miR-143 in cervical cancer, as well as investigate its underlying mechanisms.
Golgi membrane protein 1 (GOLM1), one of the transmembrane proteins of Golgi cisternae, is induced by viral infection (13) and commonly expressed in epithelial cells of normal tissues (14). TargetScan database analysis showed that GOLM1 was one potential direct target for miRNA-143. Few studies have found that GOLM1 may act as a biomarker for the hepatocellular carcinoma (HCC) early diagnosis (15). However, the mechanism and function of GOLM1 have not been investigated in cervical cancer. The current study aimed to investigate the functional significance of miRNA-143 and to identify the relationship between miR-143 and the molecular targets regulated by miRNA-143 in cervical cancer.
Materials and methods
Clinical specimens
Cervical cancer tissue samples and para-carcinoma tissues were collected from 35 patients from the Gansu Provincial Hospital (Lanzhu, China) between 2015 and 2017 (the clinical characteristics are shown in Table I). Each patient provided informed consent and the study was approved by the Ethics Committee of The Gansu Provincial Hospital. All tissue samples were snap-frozen in liquid nitrogen and saved at −80°C.
Table I.
Correlation of miRNA-143 expression with the clinicopathological characteristics of the cervical cancer patients.
| miRNA-143 expression | ||||
|---|---|---|---|---|
| Clinicopathological features | Cases (n=35) | High (n=15) | Low (n=20) | P-value |
| Age (years) | 0.2048 | |||
| >60 | 16 | 6 | 10 | |
| ≤60 | 19 | 9 | 10 | |
| Family history of cancer | 0.6257 | |||
| Yes | 18 | 7 | 11 | |
| No | 17 | 8 | 9 | |
| Tumor size (cm) | 0.0024 | |||
| ≥5.0 | 19 | 4 | 15 | |
| <5.0 | 16 | 11 | 5 | |
| TNM stage | 0.0039 | |||
| I–II | 17 | 11 | 6 | |
| III | 18 | 4 | 14 | |
| Lymph node metastasis | 0.0020 | |||
| Yes | 20 | 4 | 16 | |
| No | 15 | 11 | 4 | |
| Pausimenia | 0.4658 | |||
| Yes | 18 | 7 | 11 | |
| No | 17 | 8 | 9 | |
| FIGO stage | 0.0017 | |||
| I–II | 16 | 12 | 4 | |
| III–IV | 19 | 3 | 16 | |
| Distant metastasis | 0.5236 | |||
| Yes | 17 | 6 | 11 | |
| No | 18 | 9 | 9 | |
TNM, tumor-node-metastasis; FIGO, International Federation of Gynecology and Obstetrics.
Cell lines
Normal cervical epithelial cells (Ect1/E6E7) and two cervical cancer cell lines (SiHa and HeLa) were purchased from the American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI-1640 medium which contained 10% fetal bovine serum (FBS). The cells were incubated in an atmosphere containing 5% CO2 at 37°C.
Cell transfection
miRNA-143 mimics, miRNA-143 inhibitor or GOLM1 small interfering RNA (siRNA) as well as the corresponding controls were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and transfected into human cervical cancer cell lines by Lipofectamine 2000 (Invitrogen: Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) in line with the manufacturer's instructions. The cells were harvested 48 h post-transfection for further assays.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen: Thermo Fisher Scientific, Inc.) was used to isolate the total RNA from cervical cancer tissues and cells. cDNA was synthesized from 1 µg total RNA with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems: Thermo Fisher Scientific, Inc., Foster City, CA, USA). RT-qPCR was carried out on CFX96 real-time PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) with SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, China). The relative expression of mRNA was analysed using 2−ΔΔCq (16) (Table II).
Table II.
Primer sequences for RT-qPCR.
| Primers | Sequences | |
|---|---|---|
| miR-143 | F 5′-TGTAGTTTTCGGAGTTAGTGTCGCGC-3′ | |
| R 5′-CCTACGATCGAAAACGACGCGAACG-3′ | ||
| U6 | F 5′-GTTTTGTAGTTTTTGGAGTTAGTGTTGTGT-3′ | |
| R 5′-CTCAACCTACAATCAAAAACAACACAAACA-3′ | ||
| GOLM1 | F 5′-CGTGGAGCTGAAGAAGAACGAG-3′ | |
| R 5′-CCTGGTACAGCTTGTTGACGCT-3′ | ||
| GAPDH | F 5′-CACAATTGGGACCACAAGGG-3′ | |
| R 5′-AACTCATCACAGCACGTCACACC-3′ | ||
F, forward; R, reverse; U6: snRNA, small nuclear RNA; GOLM1, golgi membrane protein 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Migration and invasion assay
Transwell assays were conducted to value the cervical cancer cell invasion and migration capacities. A total of 1×105 cells with transfection of miR-143 mimics or inhibitor were detached, suspended and seeded in the upper chamber. In the meantime, DMEM containing 10% FBS was added to the bottom chambers. The difference between invasion and migration assays was that the Transwell upper chambers were precoated with Matrigel for invasion assays. After incubation for 24 h, the cells that remained on the upper chambers were removed with cotton swabs whereas the cells attached to the lower chambers were fixed and stained, respectively, using formaldehyde (4%) and crystal violet (0.1%) for the detection of the images under a microscope (Olympus Corp., Tokyo, Japan) in five randomly selected fields.
Western blot analysis
The cells were lysed in chilled RIPA lysis buffer on ice, and then centrifuged at 12,000 × g for 20 min at 4°C and collected the supernatants. Protein concentrations were assessed with a BCA Protein Assay kit (Bio-Rad Laboratories, Inc.). Protein lysate was separated by 10% SDS-PAGE, followed by being electroblotted onto a PVDF membrane (DuPont, Boston, MA, USA), which had been blocked with 5% non-fat milk in TBST for 1 h at room temperature. Next, the membrane was incubated with the primary antibodies: anti-GOLM1 antibody (1:300, cat. no. HPA010638; Atlas Antibodies, Stockholm, Sweden) and anti-GAPDH antibody (1:3,000; cat. no. ab9485) at 4°C overnight, and then rinsed the membrane and incubated it with anti-rabbit IgG (1:4,000; cat. no. ab191866) (both from Abcam, Cambridge, UK) for 2 h at room temperature. Chemiluminescence detection system (Beyotime Institute of Biotechnology, Shanghai, China) was used to detect the results and the protein expression was normalized to GAPDH.
Luciferase assay
The luciferase reporter assays were carried out to evaluate whether GOLM1 was targeted by miRNA-143. Cervical cancer cells were co-transfected with the wild-type or mutant GOLM1 3′UTR luciferase reporter vectors and miRNA-143 by Lipofectamine™ 2000 (Invitrogen: Thermo Fisher Scientific, Inc.). After transfection (48 h), Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI, USA) was used to measure the luciferase activities.
Statistical analysis
All the experiments were performed at least 3 times. The SPSS 18.0 version (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA) were used to evaluate the statistical analysis. Students t test and one way ANOVA followed by Tukeys post hoc test were used to analyze two or multiple groups, respectively. Spearmans correlation analysis was used to determnine the correlation between miR-143 and GOLM1 expressions in cervical cancer tissues. P<0.05 was considered to be a statistically significant difference.
Results
miRNA-143 is downregulated and GOLM1 is overexpressed in cervical cancer
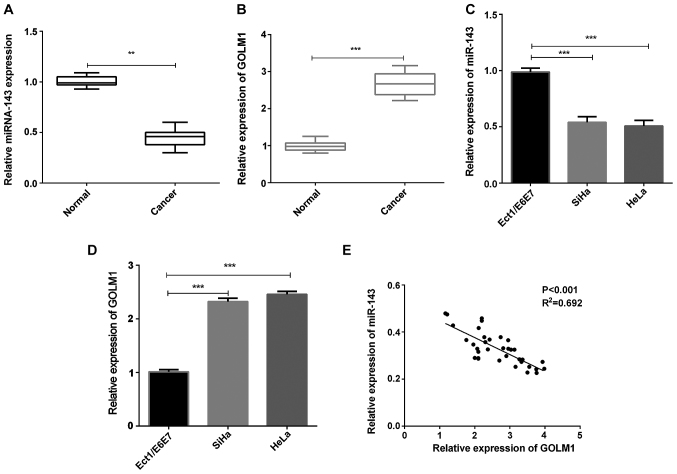
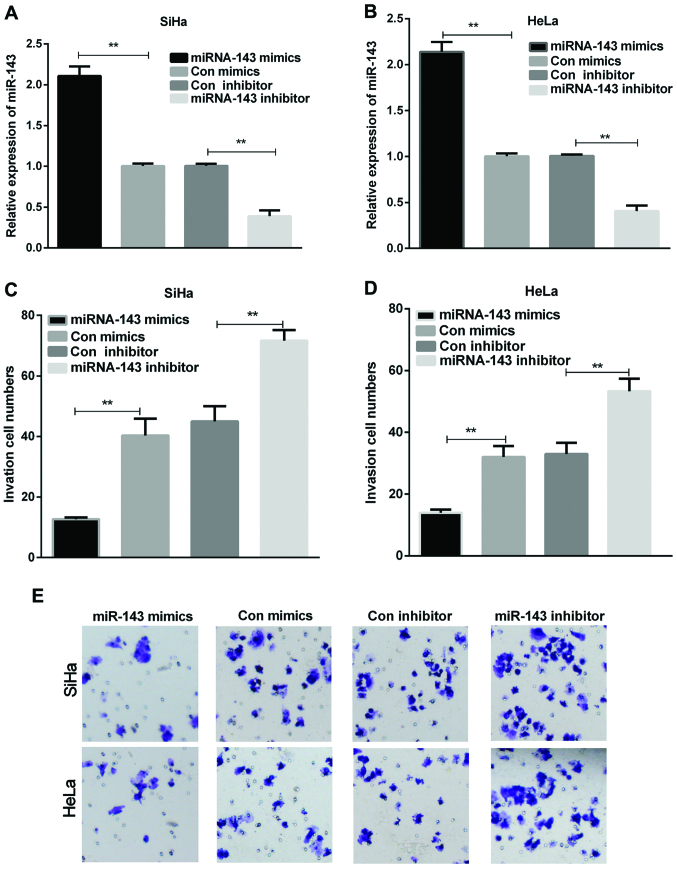
We first evaluated the expression of miR-143 and GOLM1 in cervical cancer tissue specimens. The results showed that the miRNA-143 expression in cervical cancer tissues was remarkably reduced in contrast with the normal tissue samples (Fig. 1A, P<0.01), whereas the GOLM1 mRNA expression in cervical cancer tissue samples was significantly increased (Fig. 1B, P<0.001). The expression of miRNA-143 and GOLM1 was then measured in cervical cancer cells (SiHa and HeLa). The results indicated that, compared to the control group, miRNA-143 expression in both SiHa and HeLa cells was significantly decreased (Fig. 1C, P<0.001), whereas the GOLM1 expression was significantly increased (Fig. 1D, P<0.001). Additionally, statistical analysis revealed that the miRNA-143 expression was negatively correlated with GOLM1 expression in the cervical cancer tissue samples (Fig. 1E). miRNA-143 suppresses cervical cancer cell invasion. HeLa and SiHa cells were transfected with miRNA-143 mimics or inhibitor, respectively. After 48 h, the transfection efficiency was measured. The results demonstrated that the miRNA-143 expression was dramatically upregulated in both SiHa and HeLa cells with transfection of miRNA-143 mimics, while the miRNA-143 expression level was downregulated in cervical cancer cells treated with miRNA-143 inhibitor (Fig. 2A and B, P<0.01). Additionally, cell invasion assay was performed using Transwell chambers. In the cell invasion assay, the overexpression of miRNA-143 decreased SiHa and HeLa cell invasion, whereas inhibition of the miRNA-143 expression enhanced SiHa and HeLa cell invasion (Fig. 2C-E, P<0.01). The results manifested that miR-143 inhibited cervical cancer cell invasion.
Figure 1.
miR-143 and GOLM1 expression in cervical cancer. (A and B) The miR-143 and GOLM1 mRNA expression in cervical cancer tissues (n=35) (**P<0.01, ***P<0.001). (C) miRNA-143 expression in cervical cancer cells was measured by RT-qPCR (***P<0.001). (D) The GOLM1 mRNA expression in cervical cancer cells was detected (***P<0.001). (E) Spearmans correlation analysis of miR-143 and GOLM1 expression. GOLM1, golgi membrane protein 1.
Figure 2.
miR-143 inhibits cell invasion in cervical cancer cells. (A and B) The miR-143 expression was examined using RT-qPCR in cervical cancer cells transfected with miR-143 mimics or inhibitor, respectively (**P<0.01). (C and D) The invasion cell numbers of cervical cancer cells were counted (**P<0.01). (E) Cell invasion was observed by the Transwell assay in cervical cancer cells treated with miR-143 mimics or inhibitor, respectively.
miRNA-143 inhibits cell migration in cervical cancer cells
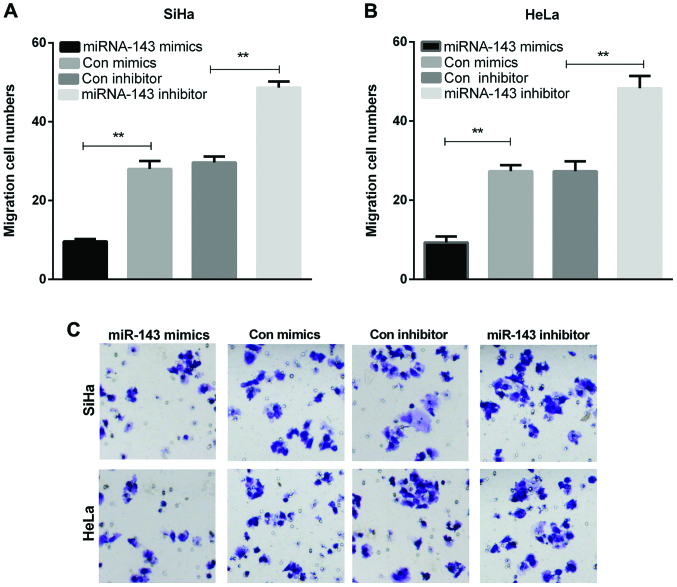
To explore the functions of miRNA-143 in metastasis, cell migration assay was also carried out by Transwell chambers. The overexpression of miRNA-143 significantly decreased SiHa and HeLa cell migration, whereas inhibiting the miRNA-143 expression increased SiHa and HeLa cell migration (Fig. 3A-C, P<0.01). This result indicated that miRNA-143 inhibited cervical cancer cell migration.
Figure 3.
miR-143 suppresses migration of cervical cancer cells. (A and B) The migration cell numbers of cervical cancer cells were counted (**P<0.01). (C) Cell migration was detected by the Transwell assay in cervical cancer cells with transfections of miR-143 mimics or inhibitor.
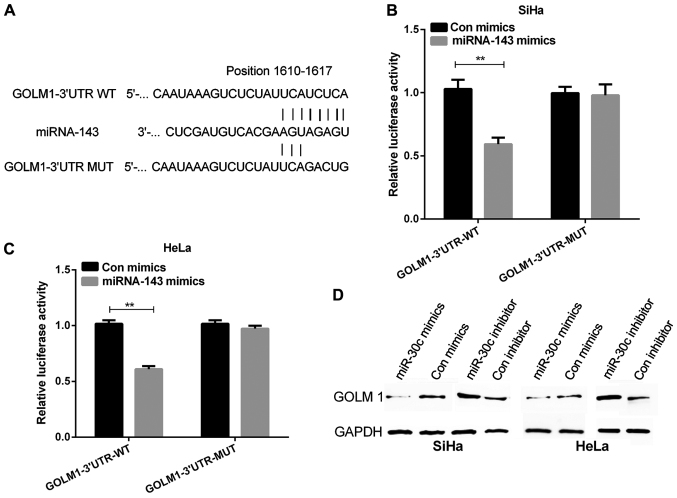
GOLM1 is a direct target of miRNA-143
To investigate the mechanism of miRNA-143 in inhibiting cervical cancer, we continued to explore the potential targets of miRNA-143. In the present study, TargetScan was used to find accurate potential target genes of miRNA-143, and it predicted GOLM1 as a potential target of miRNA-143. The wild-type and mutant GOLM1 3′UTRs were generated with sequences presented in Fig. 4A. Luciferase reporter assay was performed on cervical cancer cells which were transfected with miRNA-143 mimics. Subsequently, cells were co-transfected with wild or mutant GOLM1 3′UTR vectors. The results revealed that the miRNA-143 mimics remarkably declined luciferase activity of the GOLM1 3′UTR-wt vector. Moreover, the luciferase activity of GOLM1 3′UTR-mut vector was not affected by miRNA-143 mimics (Fig. 4B and C, P<0.01). Our findings suggested that GOLM1 was directly targeted by miRNA-143. In addition, the GOLM1 protein expression was remar-kably reduced in cells with the transfection of miRNA-143 mimics and obviously increased in cells with the transfection of miRNA-143 inhibitor compared with their respective controls (Fig. 4D).
Figure 4.
miR-143 regulates GOLM1 expression via targeting its 3′UTR. (A) According to the TargetScan, the miR-143 target sites in the sequence of GOLM1 were predicted. (B and C) The fluorescence activity of the GOLM1 3′UTR was measured by the luciferase reporter gene assay in cervical cancer cells that were co-transfected with wild-type or mutational type GOLM1 3′UTR and miR-143, respectively (**P<0.01). (D) Western blot analysis detected the GOLM1 protein expression in cervical cancer cells with transfection of miR-143 mimics or inhibitor. GOLM1, golgi membrane protein 1.
The effects of GOLM1 on regulating the effects of miR-143 on cervical cancer cell invasion and migration
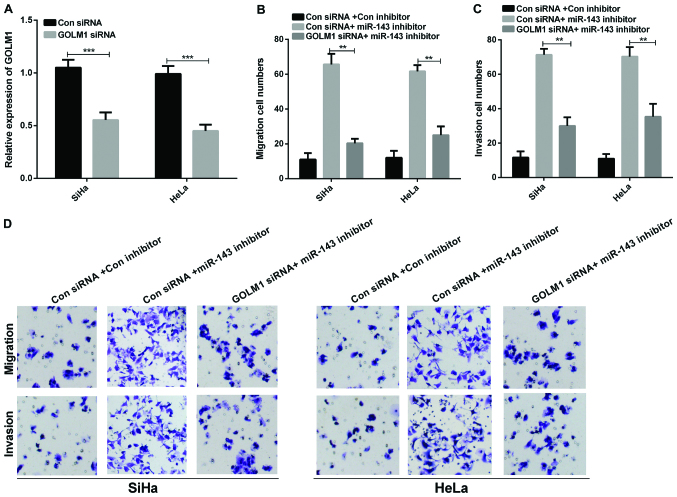
We sequentially studied whether GOLM1 was needed in regulating the miR-143 ability in cell invasion and migration of cervical cancer. GOLM1 siRNAs were transfected into SiHa and HeLa cells to knockdown endogenous GOLM1 and RT-qPCR was conducted to examine the GOLM1 mRNA expression. The results revealed that the GOLM1 expression in cervical cancer cells transfected with GOLM1 siRNAs was significantly reduced compared to the control group (Fig. 5A, P<0.001). Furthermore, Transwell assays were conducted to measure the invasion and migration of cervical cancer cells co-transfected with GOLM1 siRNA and miR-143 inhibitor. Our above results indicate that deletion of GOLM1 markedly reversed miR-143-medicated suppression of cell migration and invasion in cervical cancer cells (Fig. 5B-D, P<0.01).
Figure 5.
GOLM1 may regulate miR-143 function in cervical cancer cell migration and invasion. (A) The GOLM1 mRNA expression in cervical cancer cells with transfection of GOLM1 siRNA (***P<0.001). (B) The migration and (C) the invasion cell numbers of SiHa and HeLa transfcted with GOLM1 siRNA and control siRNA were counted (**P<0.01). (D) Transwell assay showed cell invasion and migration in cervical cancer cells co-transfected with GOLM1 siRNA and miR-143 inhibitor. GOLM1, golgi membrane protein 1; siRNA, small interfering RNA.
Discussion
Despite regular cancer screening, cervical cancer still remains a common tumor in women (17). It is important to elucidate the underlying mechanism of cervical cancer development. Increasing number of research has indicated that the aberrant miRNA expression has important functions in tumor progression. Previous studies also demonstrated that the dysregulation of miRNAs potentially serves as a therapeutic target in various cancers including cervical cancer (18,19). Therefore, the study of the miRNA expression and functions in cervical cancer may help to improve the treatment strategy for refractory cervical cancer.
miRNA-143 is one of the most crucial miRNAs which has been implicated in tumor progression. Previously, it has been verified that miR-143 is downregulated in various tumors and can inhibit the development of multiple cancers including leukemia (20), breast cancer (21), and osteosarcoma (22). The miRNA-143 expression has been found to be significantly downregulated in many tumors (23,24), involving human cervical cancer tissues (25), thus, demonstrating a potentially suppressive effect of miRNA-143 on tumor development. Nevertheless, limited efforts have been made to elucidate the mechanism of miRNA-143 in cervical cancer. The role of miRNA-143 in the regulation of cell migration and invasion remains unclear.
GOLM1 has two variants which encode the same protein (26). Previous studies have demonstrated that the GOLM1 expression is overexpressed in human PCa tissues (27), HCC (28), and lung adenocarcinoma (29). Nevertheless, little is known about the possible role of GOLM1 in cervical cancer. The present study aimed to reveal the relationship between the miRNA-143 and GOLM1 expression in cervical cancer. Data revealed that the miRNA-143 expression was downregulated dramatically in cervical cancer tissue samples in contrast to normal samples while GOLM1 was overexpressed in cervical cancer tissue samples. Moreover, the low expression of miRNA-143 and GOLM1 overexpression were also found in cervical cancer cells. In addition, the miRNA-143 expression negatively correlated with the GOLM1 expression in cervical cancer tissues. We also found that miRNA-143 suppressed cell invasion and migration by modulating GOLM1 in cervical cancer.
In summary, our study demonstrated that the miRNA-143 expression is low in cervical cancer and it inhibited cell invasion and migration via targeting GOLM1. These data provided new insights into the tumor-suppressive effects of miRNA-143 in cervical cancer, suggesting that miRNA-143 may be a therapeutic strategy and an effective biomarker for cervical cancer treatment in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MZ and XC contributed to the conception of the study. JW contributed significantly to the analysis and manuscript preparation. XH performed the data analyses and contributed to the writing of the manuscript. RR helped to perform the statistical analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Gansu Provincial Hospital (Lanzhou, China). Each patient enrolled in this study provided a signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li XL, Liu XX, Cao GS, Ju DD, Jiang H. Narrowing resection of parametrial tissues is feasible in low-risk cases of stage IA2-IB1 cervical cancer. J Cancer. 2016;7:1481–1486. doi: 10.7150/jca.15466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Tjalma WAA. Diagnostic performance of dual-staining cytology for cervical cancer screening: A systematic literature review. Eur J Obstet Gynecol Reprod Biol. 2017;210:275–280. doi: 10.1016/j.ejogrb.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Malo TL, Perkins RB, Lee JH, Vadaparampil ST. Primary care physicians' adherence to expert recommendations for cervical cancer screening and prevention in the context of human papillomavirus vaccination. Sex Transm Dis. 2016;43:438–444. doi: 10.1097/OLQ.0000000000000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, Zhang Y, Zhang S. MicroRNA-92 regulates cervical tumorigenesis and its expression is upregulated by human papillomavirus-16 E6 in cervical cancer cells. Oncol Lett. 2013;6:468–474. doi: 10.3892/ol.2013.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiffman M, Wentzensen N. Human papillomavirus infection and the multistage carcinogenesis of cervical cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:553–560. doi: 10.1158/1055-9965.EPI-12-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du J, Wang L, Li C, Yang H, Li Y, Hu H, Li H, Zhang Z. MicroRNA-221 targets PTEN to reduce the sensitivity of cervical cancer cells to gefitinib through the PI3K/Akt signaling pathway. Tumour Biol. 2016;37:3939–3947. doi: 10.1007/s13277-015-4247-8. [DOI] [PubMed] [Google Scholar]
- 8.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li WH, Wu HJ, Li YX, Pan HG, Meng T, Wang X. MicroRNA-143 promotes apoptosis of osteosarcoma cells by caspase-3 activation via targeting Bcl-2. Biomed Pharmacother. 2016;80:8–15. doi: 10.1016/j.biopha.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Wu D, Huang P, Wang L, Zhou Y, Pan H, Qu P. MicroRNA-143 inhibits cell migration and invasion by targeting matrix metalloproteinase 13 in prostate cancer. Mol Med Rep. 2013;8:626–630. doi: 10.3892/mmr.2013.1501. [DOI] [PubMed] [Google Scholar]
- 13.Kladney RD, Bulla GA, Guo L, Mason AL, Tollefson AE, Simon DJ, Koutoubi Z, Fimmel CJ. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. doi: 10.1016/S0378-1119(00)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Li L, Hu L, Peng T. Golgi phosphoprotein 2 (GOLPH2/GP73/GOLM1) interacts with secretory clusterin. Mol Biol Rep. 2011;38:1457–1462. doi: 10.1007/s11033-010-0251-7. [DOI] [PubMed] [Google Scholar]
- 15.Mao Y, Yang H, Xu H, Lu X, Sang X, Du S, Zhao H, Chen W, Xu Y, Chi T, et al. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut. 2010;59:1687–1693. doi: 10.1136/gut.2010.214916. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Janicek MF, Averette HE. Cervical cancer: Prevention, diagnosis, and therapeutics. CA Cancer J Clin. 2001;51:92–114. doi: 10.3322/canjclin.51.2.92. [DOI] [PubMed] [Google Scholar]
- 18.Shi C, Zhang Z. MicroRNA-362 is downregulated in cervical cancer and inhibits cell proliferation, migration and invasion by directly targeting SIX1. Oncol Rep. 2017;37:501–509. doi: 10.3892/or.2016.5242. [DOI] [PubMed] [Google Scholar]
- 19.Lin M, Xue XY, Liang SZ, Li YX, Lv YY, He LH, Xu KC, Zhang LF, Chen JB, Niu LZ. MiR-187 overexpression inhibits cervical cancer progression by targeting HPV16 E6. Oncotarget. 2017;8:62914–62926. doi: 10.18632/oncotarget.17516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and −145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH, Chang YC, Lin WC, Shen CY, Huang CS, Hsieh FJ, et al. Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol Cancer. 2015;14:36. doi: 10.1186/s12943-015-0301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T, et al. MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther. 2011;19:1123–1130. doi: 10.1038/mt.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232. doi: 10.1186/1471-2407-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q, Dan ZL, Tian DA, Zhang P. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol. 2013;19:7758–7765. doi: 10.3748/wjg.v19.i43.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sültmann H, Scheffner M, Hoppe-Seyler K, Hoppe-Seyler F. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015;11:e1004712. doi: 10.1371/journal.ppat.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HJ, Lv D, Zhang Y, Peng T, Ma X. Golgi phosphoprotein 2 in physiology and in diseases. Cell Biosci. 2012;2:31. doi: 10.1186/2045-3701-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Wang X, Li B, Lu J, Chen G. Diagnostic significance of overexpression of Golgi membrane protein 1 in prostate cancer. Urology. 2012;80:952.e1–7. doi: 10.1016/j.urology.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Bachert C, Fimmel C, Linstedt AD. Endosomal trafficking and proprotein convertase cleavage of cis Golgi protein GP73 produces marker for hepatocellular carcinoma. Traffic. 2007;8:1415–1423. doi: 10.1111/j.1600-0854.2007.00621.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F, Gu Y, Li X, Wang W, He J, Peng T. Up-regulated Golgi phosphoprotein 2 (GOLPH2) expression in lung adenocarcinoma tissue. Clin Biochem. 2010;43:983–991. doi: 10.1016/j.clinbiochem.2010.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.