Abstract
Circulating microRNAs (miRNAs) are promising markers for cancer diagnosis and prognosis. Numerous studies evaluating miRNAs as markers for non-small cell lung cancer (NSCLC) have been conducted in recent years; however, the majority of candidate markers proposed via individual studies were inconsistent and no marker miRNAs for the diagnosis of early stage NSCLC have been established. In the present study, miR-145, miR-20a, miR-21 and miR-223, which were previously reported as candidate diagnostic markers of NSCLC, were re-evaluated. The serum levels of these miRNAs were quantified in 56 patients with stage I–IV NSCLC using the TaqMan microRNA assays and separately compared the levels at each stage with those in 26 control patients. The level of miR-145 was significantly reduced in patients with NSCLC, regardless of clinical stage, and its level increased following tumor resection in patients with stage I–II disease. These results indicate that miR-145 is relevant as a diagnostic marker for stages I–IV NSCLC. Additionally, the levels of miR-20a and miR-21 demonstrated notable differences among patients at different clinical stages. These miRNAs distinguished patients in a number of, but not all, stages of NSCLC from cancer-free control patients. These results indicated that it is essential to analyze miRNA levels at each stage separately in order to evaluate marker miRNAs for NSCLC diagnosis.
Keywords: circulating microRNAs, biomarkers, resection, TaqMan, reverse transcription-quantitative polymerase chain reaction
Introduction
In 2017, lung cancer was reported as the leading cause of cancer-associated mortalities globally (1). It has been reported that in Japan the 5-year survival rate in patients with stage IA lung cancer, who underwent resection for primary lung neoplasms in 2005, was 79.5% (2); however, it was as low as 20.0% in patients with stage IV lung cancer (2). Currently, the most effective treatment for early stage non-small cell lung cancer (NSCLC) remains surgical resection. However, the sensitivities of chest X-rays and sputum cytology are insufficient for early diagnosis of NSCLC, and there is no effective blood marker for its accurate detection (3). Therefore, the development of novel markers to screen for early stage NSCLC is essential to reduce the mortality rate associated with NSCLC.
microRNAs (miRNAs) are primarily 21–23 nucleotide-long noncoding RNAs, which negatively regulate target mRNAs via binding to their 3′-untranslated region. miRNAs that target the mRNAs of tumor suppressor genes and oncogenes are referred to as oncomiRs and anti-oncomiRs, respectively. miRNAs located in the blood have been regarded as promising non-invasive markers for diagnosing and predicting cancer prognosis. Although numerous studies have evaluated miRNAs as candidates for NSCLC markers in recent years, the majority of candidate markers proposed via individual studies are inconsistent (4,5).
There are two major possible causes for this discordance. The first is low reproducibility in the quantification of circulating miRNAs (6–9). Each study used different platforms to quantify notably short and scarce miRNAs in the blood (10). A major issue in quantification of circulating miRNA is that there is no internal control for serum and plasma samples (11). The second factor is that the circulating miRNAs are a mixture of various miRNAs and in the majority of cases, each type of miRNA is not tissue specific (12–16). Thus, these miRNAs may be derived from various normal cells, including leukocytes and vascular endothelial cells, as well as from tumor cells. During NSCLC progression, tumor cells develop a unique genetic profile. Thus, different cancer microenvironments and tumor progression stages may diversify the profiles of circulating miRNAs in each patient.
In the present study, the serum levels of four miRNAs previously reported as candidate markers of NSCLC in multiple studies were analyzed and were re-evaluated as diagnostic markers of NSCLC (17–19). Stem-loop reverse transcription (RT)-primers and a TaqMan® real-time polymerase chain reaction (PCR) system were used to amplify the miRNAs, and quantification was performed using cel-miR-39 as a spike-in control. A separate comparison between the serum levels in patients with stage I–II, III or IV NSCLC and the levels in the control group was performed. Furthermore, a comparison between the miRNA levels pre- and post-surgical resection in individual patients was performed.
Materials and methods
Patients and clinical specimens
The present study protocol was approved by the Ethical Committee of the Faculty of Medicine (approval no. H26-010) and the Ethical Committee of the Faculty of Health Sciences (approval no. 25-40) of Kyorin University (Tokyo, Japan). The serum samples used in the present study were collected between October 2014 and May 2016 at Kyorin University Hospital. Signed informed consent was obtained from all participants. Histological typing and staging of the tumors were performed according to the World Health Organization criteria (20) and the seventh edition of the Tumor-Node-Metastasis classification of malignant tumors (21), respectively. Serum samples from 26 cancer-free control group (healthy individuals or patients with cataract) and 56 patients with NSCLC were used. Table I summarizes the clinicopathological characteristics of the study subjects. The inclusion criteria for the patient sample collection were as follows: Presence of a pathological diagnosis of NSCLC and the absence of any previous lung cancer history, as well as other types of cancer. The blood samples were collected prior to any therapeutic procedures, including surgery, chemotherapy and radiotherapy. For the second examination of patients with stage I–II NSCLC, the samples were collected 6–12 months post-surgical resection. Peripheral blood was collected in VP-AS109K Vacutainer tubes (Terumo Corporation, Tokyo, Japan), incubated at room temperature for 30 min and then centrifuged at 1,500 × g for 10 min at 4°C to separate the serum. The serum was centrifuged again at 20,000 × g for 10 min at 4°C to remove cell debris, divided into 200 µl aliquots and stored at −80°C until use. Hemolyzed serum samples were excluded.
Table I.
Characteristics of patients with NSCLC and control subjects.
| Characteristics | No. of patients with NSCLC (%) | No. of control subjects (%) | P-value |
|---|---|---|---|
| Total | 56 | 26 | |
| Age, years | 0.819 | ||
| ≤60 | 13 (23.2) | 8 (30.8) | |
| >60 | 43 (76.8) | 18 (69.2) | |
| Sex | 0.195 | ||
| Male | 38 (67.9) | 13 (50.0) | |
| Female | 18 (32.1) | 13 (50.0) | |
| Smoking status | <0.001 | ||
| Never | 15 (26.8) | 21 (80.8) | |
| Former | 13 (23.2) | 1 (3.8) | |
| Current | 28 (50.0) | 4 (15.4) | |
| Lung cancer stage | |||
| I | 10 (17.9) | 0 (0) | |
| II | 5 (8.9) | 0 (0) | |
| III | 9 (16.1) | 0 (0) | |
| IV | 32 (57.1) | 0 (0) | |
| Type of NSCLC | |||
| AC | 46 (82.1) | 0 (0) | |
| SQ | 10 (17.8) | 0 (0) |
Age (mean ± SD): NSCLC, 66.1±12.0; control, 65.8±14.0. AC, adenocarcinoma; SQ, squamous cell carcinoma; NSCLC, non-small cell lung cancer.
RNA extraction
The cel-miR-39 RNeasy Serum/Plasma Spike-In control (5.6×108 molecules; Qiagen GmbH, Hilden, Germany) was added to 200 µl serum sample following the addition of QIAzol (Qiagen GmbH), and RNA was then extracted using the miRNeasy Serum/Plasma kit (Qiagen GmbH), according to the manufacturer's protocol, with a minor modification: The volume of ultra-pure H2O used to elute the RNA was changed to 28 µl.
Reverse transcription-quantitative PCR (RT-qPCR)
The volume of the RNA eluent was fixed rather than the amount of total RNA used per RT (22–24). A total of 5 µl RNA eluent was used per each RT reaction. The TaqMan MicroRNA Reverse Transcription kit and RT primers in TaqMan MicroRNA assays [cat. no., 000200 (cel-miR-39); cat. no., 000580 (miR-20a-5p); cat. no., 000397 (miR-21-5p); cat. no., 002278 (miR-145-5p) and 002295 (miR-223-3p)] (Thermo Fisher Scientific, Inc., Waltham, MA, USA) for cel-miR-39, miR-20a-5p, miR-21-5p, miR-145-5p, and miR-223-3p were used for RT. RT was performed according to the manufacturer's protocols as follows: 16°C for 30 min; 42°C for 30 min; and 85°C for 5 min. TaqMan Universal Master mix II no UNG (Thermo Fisher Scientific, Inc.), and TaqMan probes and PCR primers in the TaqMan MicroRNA assays were used for qPCR. qPCR was performed in triplicate on a Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The 2−ΔΔCq method was used for relative quantification of miRNAs in each sample (25). ΔCq was determined as follows: Target miRNA Cq-cel-miR-39 Cq. The 2−ΔΔCq was used to determine the fold change (FC), where ΔΔCq was calculated as follows: (Median ΔCq of the patients with NSCLC)-(median ΔCq of the control group) or (median ΔCq of post-surgery)-(median ΔCq of pre-surgery).
Statistical analysis
The data of the present study were presented as the mean ± standard deviation. Nonparametric Mann-Whitney U or Kruskal-Wallis tests were performed to compare the demographic features between patients with NSCLC and the control group using Statcel3 software (Ohms Publishing Co., Ltd; Tokyo, Japan). A Mann-Whitney U test was used to compare miRNA levels between the patients with NSCLC and the control group. The Kruskal-Wallis and Steel-Dwass tests were used for analysis of overall group differences and for multiple comparisons, respectively. All P-values were two sided and P<0.05 was considered to indicate a statistically significant difference. Receiver operating characteristic (ROC) curves were constructed and the area under the ROC curve (AUC) was calculated to assess the performance of miR-145, miR-20a and miR-223 using JMP 13.0 software (SAS Institute, Inc., Cary, NC, USA).
Results
Comparison of miRNA serum levels between patients with stage I–II, III or IV NSCLC and control group
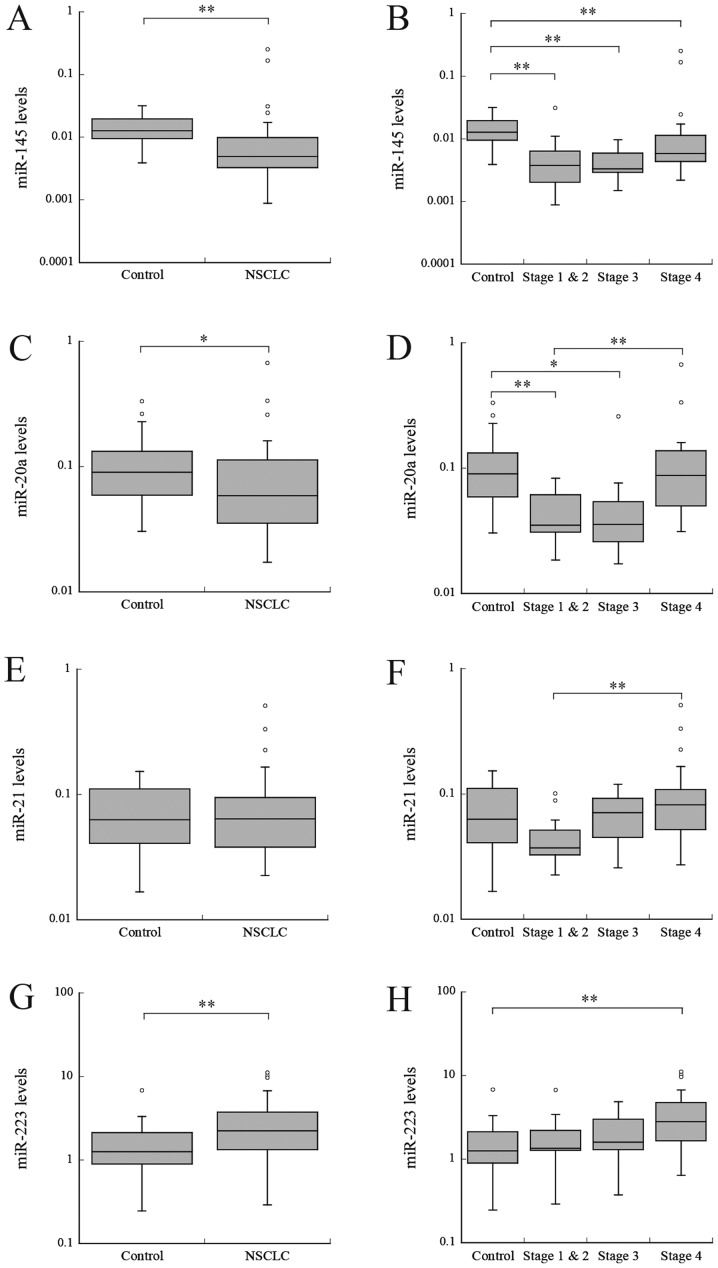
miR-145. Firstly, the miRNA serum levels of patients with NSCLC were compared with the levels in the control group. The miR-145 serum level was significantly reduced in patients with NSCLC, compared with the control group (P<0.001; Fig. 1A). Subsequently, the levels in patients with stage I–II, III or IV NSCLC were compared with levels in the control group. The serum levels in the patients in each group were significantly reduced, compared with the levels in the control group (P<0.01; Fig. 1B). The fold change (FC) in stage I–II, III and IV NSCLC were 0.31, 0.23, and 0.46, respectively (Table II). No significant difference in the serum levels between NSCLC stages was identified.
Figure 1.
Relative levels of serum miRNAs in patients with NSCLC and cancer-free control group. (A) miR-145 expression between control and NSCLC, and (B) among different NSCLC stages. (C) miR-20a expression between control and NSCLC, and (D) among different NSCLC stages. (E) miR-21 expression between control and NSCLC, and (F) among different NSCLC stages. (G) miR-223 expression between control and NSCLC, and (H) among different NSCLC stages. The upper and lower limits of the boxes and lines inside the boxes represent the 75th and 25th percentiles and the median, respectively. *P<0.05 and **P<0.01. miR/miRNA, microRNA; NSCLC, non-small cell lung cancer.
Table II.
Serum miRNAs differentially expressed in patients with NSCLC and control subjects.
| ΔCq median (median relative level) | ||||
|---|---|---|---|---|
| miRNA | Stage (n) | NSCLC | Control | FC |
| miR-145 | I–II (15) | 8.06 (0.004) | 6.31 (0.013) | 0.31 |
| miR-20a | I–II (15) | 4.83 (0.035) | 3.48 (0.090) | 0.39 |
| miR-145 | III (9) | 8.24 (0.003) | 6.31 (0.013) | 0.23 |
| miR-20a | III (9) | 4.82 (0.035) | 3.48 (0.090) | 0.39 |
| miR-145 | IV (32) | 7.43 (0.006) | 6.31 (0.013) | 0.46 |
| miR-223 | IV (32) | −1.49 (2.818) | −0.317 (1.246) | 2.26 |
FC, fold change; miR/miRNA, microRNA; NSCLC, non-small cell lung cancer.
miR-20a
In patients with NSCLC, the levels of miR-20a were significantly reduced, compared with the control group (P<0.05; Fig. 1C). The miR-20a levels were significantly different among different NSCLC stages (P<0.001; Fig. 1D). The levels were significantly reduced in patients with stages I–II and III NSCLC, compared with the control group (P<0.01 and P<0.05, respectively); however, no significant difference was identified between patients with stage IV NSCLC and the control group (Fig. 1D). The FC in stage I–II and III NSCLC were 0.39 and 0.39, respectively (Table II).
miR-21
No significant difference in miR-21 levels were identified between patients with NSCLC and the control group (P=0.968; Fig. 1E). However, the levels were significantly increased in patients with stage IV NSCLC, compared with patients with stage I–II NSCLC (P<0.01; Fig. 1F).
miR-223
Levels of miR-223 were significantly increased in patients with NSCLC, compared with the control group (P<0.01; Fig. 1G). No significant difference was identified between the levels in patients with stages I–II or III NSCLC and the control group; however, the levels were significantly increased in patients with stage IV NSCLC, compared with the control group. (P<0.01; Fig. 1H). The FC in stage IV NSCLC was 2.26 (Table II). No significant difference was observed in the serum levels between NSCLC stages.
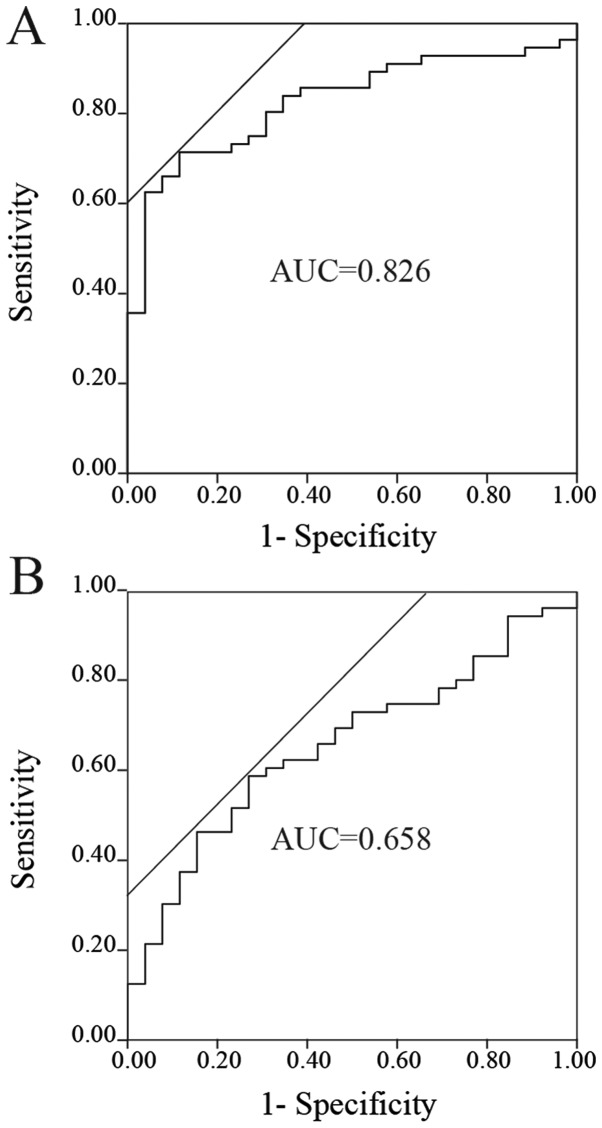
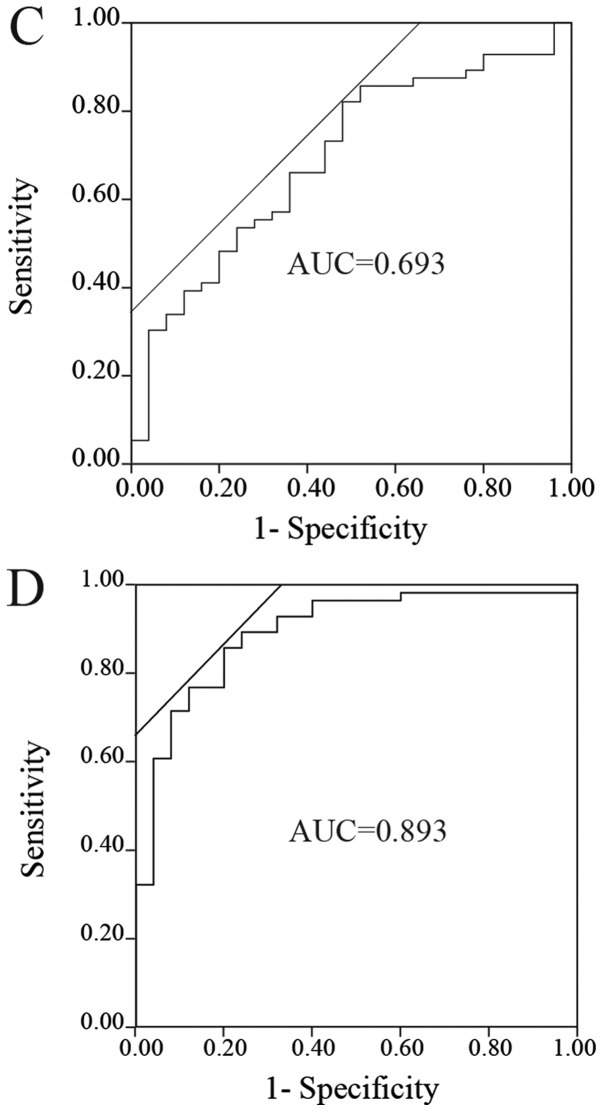
ROC analyses of the miRNAs to distinguish patients with NSCLC from the control group
The ROC curves of miR-145, miR-20a and miR-223 for evaluation as diagnostic markers are depicted in Fig. 2A-C. The AUCs for miR-145, miR-20a and miR-223 were 0.826 (sensitivity, 0.714, and specificity, 0.885, at the optimal cutoff point of 0.00764; Fig. 2A), 0.658 (sensitivity, 0.589, and specificity, 0.731, at the optimal cutoff point of 0.0665; Fig. 2B) and 0.693 (sensitivity, 0.821, and specificity, 0.520, at the optimal cutoff point of 1.304; Fig. 2C), respectively. Furthermore, ROC analysis was performed for combinations of these miRNAs. The combination of miR-145 and miR-223 yielded the highest AUC (AUC, 0.893; sensitivity, 0.857; and specificity, 0.800; Fig. 2D). The other combinations, including miR-145 and miR-20a, miR-20a and miR-223, and miR-145, miR-20a and miR-223, had AUCs of 0.815, 0.787 and 0.876, respectively.
Figure 2.
ROC analyses of the miRNAs. ROC curves for (A) miR-145, (B) miR-20a, (C) miR-223 and (D) the combination of miR-145 and miR-223 to distinguish patients with non-small-cell lung cancer from control groups. miR/miRNA, microRNA; ROC, receiver operating curve; AUC, area under the curve.
Comparison of miRNA serum levels pre- and post-surgical resection
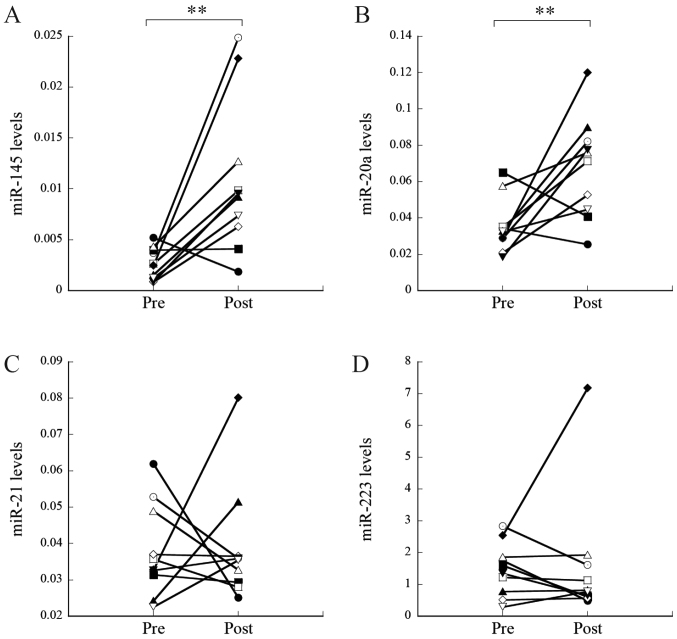
Subsequently, the miRNA levels pre- and post-tumor resection were compared in 10 patients with stage I–II NSCLC who underwent surgery. The levels of miR-145 and miR-20a were significantly increased post-resection, compared with levels pre-resection (P=0.002 and P=0.007, respectively; Fig. 3A and B, respectively). The FC of miR-145 and miR-20a were 3.00 and 2.24, respectively (Table III). As a result, the levels of miR-145 and miR-20a post-resection were similar to the levels in the control group (P=0.120 and P=0.077, respectively). In contrast, no significant changes were observed for miR-21 and miR-223 (P=0.88 and P=0.45, respectively; Fig. 3C and D, respectively).
Figure 3.
Relative levels of serum miRNAs pre- and post-tumor resection. Serum levels of (A) miR-145, (B) miR-20a, (C) miR-21 and (D) miR-223 pre- and post-tumor resection. **P<0.01. miR/miRNA, microRNA; pre, pre-tumor resection; post, post-tumor resection.
Table III.
Serum miRNAs differentially expressed pre- and post-surgical resection.
| ΔCq median (median relative level) | ||||
|---|---|---|---|---|
| miRNA | Stage (n) | Post-surgery | Pre-surgery | FC |
| miR-145 | I–II (10) | 6.73 (0.009) | 8.60 (0.003) | 3.00 |
| miR-20a | I–II (10) | 3.76 (0.074) | 4.94 (0.033) | 2.24 |
FC, fold change; miR/miRNA, microRNA; NSCLC, non-small cell lung cancer.
Discussion
In the present study, four serum miRNAs were evaluated as markers for early diagnosis of NSCLC. The serum level of miR-145 was significantly reduced in patients with NSCLC at all stages, compared with the control group (FC, 0.23–0.46) (Table II). These results demonstrated that serum miR-145 distinguishes patients in all stages of NSCLC from cancer-free control group, with high sensitivity and specificity. ROC analysis revealed that miR-145 demonstrated a notable AUC, indicating that among the miRNAs examined, it was the most suitable diagnostic marker for NSCLC. The decline in miR-145 levels in NSCLC observed in the present study is in agreement with a previous study, in which miR-145 expression was reduced in a number of tumor cell lines or tumor tissues, including NSCLC, and acted as an anti-oncomiR (26).
Serum miR-20a level was significantly reduced in patients with stages I–II and III NSCLC, compared with the control group, although the difference was not significant in patients with stage IV NSCLC. These results indicated that decreased levels of miR-20a were able to distinguish patients with stages I–II and III NSCLC from cancer-free control group. The majority of circulating miRNAs, except for a number of miRNAs, including miR-122, which has hepatocyte-specific expression, are broadly expressed in various normal cells (27,28). It is possible that miR-145 and miR-20a are released from a number of normal cells in cancer-free patients and that this release is suppressed by tumorigenesis. Similar downregulation of miRNA expression in patients with NSCLC has been observed in other studies, including miR-125a-5p, miR-25, miR-126 (23), miR-16-5p, miR-17b-5p, miR-19-3p, miR-20a-5p, miR-92-3p (29), miR-328-3p, miR-375, miR-139, miR-486, miR-191, miR-200b, miR-183 and miR-145 (30). In contrast, the levels of miR-20a in patients with stage IV NSCLC were similar to that in the control group. Additionally, serum levels were significantly increased in patients with stage IV NSCLC, compared with patients with stage I–II NSCLC, for two miRNAs (miR-20a and miR-21; P<0.01). In these cases, the release of miRNAs from normal cells and/or tumor cells may be accelerated in the advanced stage by unknown mechanisms. It has been reported that tumor cells release excessive amounts of extracellular vesicles (EVs), which contain non-coding RNAs and DNA fragments, and function in intercellular communication between tumor cells and cells in metastatic niches (31,32). Therefore, increases in these miRNAs in advanced stages, compared with early stages, may be associated with the increase in EVs in circulation. Another probable cause is apoptosis or necrosis in cancerous lesions, whereby cellular RNAs may be released into circulation (14,16). Regardless of the reason, the results for miR-20a and miR-21 notably indicate that it is essential to analyze miRNA levels at each stage separately when evaluating candidate miRNAs as diagnostic markers.
In contrast, the serum level of miR-223 was significantly increased in patients with NSCLC, compared with the control group. miR-223, alone may not be suitable as a diagnostic marker of NSCLC considering its relatively low AUC (0.693); however, it yielded a notable AUC (0.893) when used in combination with miR-145.
Comparison of miRNA levels pre- and post-surgical resection in an individual patient is an effective approach to evaluate candidate marker miRNAs (33–35). The use of this approach in the present study confirmed the relevance of using miR-145 and miR-20a as markers for stage I–II NSCLC. Additionally, serum miR-145 and miR-20a increased following tumor removal in the majority of patients with stage I–II NSCLC (FC, 3.00 and 2.24, respectively) (Table III). These results indicated that these miRNAs may be beneficial as tumor markers in the follow-up of patients with NSCLC, at least for those with stage I–II NSCLC. Notably, Leidinger et al (36) contradicted the idea of a general decrease in circulating miRNAs post-surgical resection of tumors. They reported that the levels of a number of plasma miRNAs peaked at 2 weeks post-tumor resection. These changes in miRNA levels post-surgery may be caused by inflammation at the surgical sites. To avoid this inflammatory effect, a second examination at 6–12 months post-resection was performed, rather than immediately following resection.
The four circulating miRNAs examined in the present study have been previously evaluated as NSCLC markers in a number of other studies (18,23,24,29,30,37–44). The results of some of these studies are inconsistent with those of the present study. For example, a number of studies reported that circulating miR-21 increased in patients with NSCLC (6,43,44), whereas other studies reported that it did not change (23,34). Table IV summarizes the results of associated studies with NSCLC, in which relative quantification using spike-in control or absolute quantification was performed (18,23,24,29,30,40,42,44). For example, the results from Arab et al (30) regarding miR-145 in stage I–IV and those of Fan et al (29) for miR-20a in stage I–IIIB are consistent with the present results. In contrast, the results of Arab et al (30) for miR-20a in stage I–IIIA, Lv et al (42) for miR-223 in stage I–III, Wang et al (24) for miR-145 and Chen et al (18) for miR-145 are inconsistent with the present data. One possible explanation for the discrepancy is the difference in patient cohort included in each study. In numerous studies, miRNA levels were compared between all patients with NSCLC and control subjects, and the distribution of stages was not taken into consideration (19,40). As aforementioned, changes in the levels of a number of miRNAs determined in the early stages of NSCLC may not be evident in the advanced stages. Therefore, the distribution of patients with each stage may notably affect the results when evaluating miRNAs as diagnostic markers. There may also be other factors that affect serum miRNA levels. In the present study, the percentage of smokers was significantly different between the patients with NSCLC and the control group. In our preliminary study, the serum level of miR-21 decreased in healthy passive smokers (unpublished data). Therefore, patient conditions, including smoking status, may affect miRNA levels, thereby causing inconsistency among studies. In future studies, sufficient numbers of patients with uniform conditions in each stage of NSCLC are required to accurately demonstrate the clinical relevance of serum miRNAs as diagnostic markers.
Table IV.
Previous studies performed with similar platforms to the present study.
| Author, date | miRNA | Stage | Sample | RT primer | qPCR | Normalization | Quantification | (Refs.) |
|---|---|---|---|---|---|---|---|---|
| Chen et al, 2012 | 20a↑, 145↑ and 223↑ | I–IV | Serum | Stem-Loopa | TaqMan | – | Absolute | (18) |
| Wang et al, 2015 | 20a→ and 21→ | I–II | Serum | Stem-Loopa | TaqMan | Spike-in | 2−ΔΔCq | (23) |
| Wang et al, 2015 | 145↑ | nd | Serum | Stem-Loopa | TaqMan | Spike-in | 2−ΔΔCq | (24) |
| Fan et al, 2016 | 20a↓ | I–IIIB | Serum | Stem-Loopa | TaqMan | – | Absolute | (29) |
| Arab et al, 2017 | 21↑, 20a↑ and 145↓ | I–IIIA | Plasma | Universalb | SYBR | Spike-in | 2−ΔΔCq | (30) |
| Arab et al, 2017 | 21↑, 20a↑ and 145↓ | IIIB-IV | Plasma | Universalb | SYBR | Spike-in | 2−ΔΔCq | (30) |
| Yu et al, 2014 | 20a→ | I–IV | Plasma | nd | SYBR | Spike-in | 2−ΔΔCq | (40) |
| Lv et al, 2017 | 223↑ | I–III | Serum | Stem-Loop | SYBR | – | Absolute | (42) |
| Zhou et al, 2017 | 21↑ | I–IV | Plasma | Stem-Loopc | SYBR | Spike-in | Absolute | (44) |
| The present study | 20a↓ and 145↓ | I–II | Serum | Stem-Loopa | TaqMan | Spike-in | 2−ΔΔCq | |
| The present study | 20a↓ and 145↓ | III | Serum | Stem-Loopa | TaqMan | Spike-in | 2−ΔΔCq | |
| The present study | 145↓ and 223↑ | IV | Serum | Stem-Loopa | TaqMan | Spike-in | 2−ΔΔCq |
Thermo Fisher Scientific, Inc. Waltham, MA, USA
Qiagen GmbH, Hilden, Germany
Guangzhou RiboBio Co., Ltd. Guangzhou, China. ↓, downregulated; ↑, upregulated; →, no change, nd, not determined; miRNA, microRNA; RT, Reverse transcription; qPCR, quantitative polymerase chain reaction.
Another possible cause for the discrepancy may have resulted from differences in the assays used in individual studies. RT-qPCR using the TaqMan miRNA assays, which is a gold standard in miRNA quantification, was used in the present study. Additionally, fixed volume-RNA elution was used rather than fixed weight-total RNA samples in RT due to the concentration of total RNA in the serum being too low to measure accurately and the concentration of total RNA was significantly increased in patients with NSCLC (7,45,46). Furthermore, a spike-in control was used to normalize the variation in RNA extraction and as a reference for the relative quantification instead of internal controls, including U6 RNA or miR-16, due to U6 RNA being unstable in serum (47), and miR-16 levels being significantly increased in the plasma of patients with NSCLC, compared with healthy control group (48). However, spike-in controls cannot normalize variations caused by factors prior to RNA isolation (6,11). These differences in the conditions of miRNA quantification may have caused the aforementioned inconsistencies.
Recently, the usefulness of serum miRNA levels as diagnostic markers for cancer has been questioned due to numerous studies demonstrating inconsistent results in various types of cancer, including NSCLC and breast cancer (4–9). Therefore, the study of circulating miRNAs as cancer markers requires further validation to advance into clinical practice. Standardization in the quantification of circulating miRNAs and clarification of individual or environmental factors affecting circulating miRNA levels are required to exploit their potential (6–8). Furthermore, the results of the present study indicated that it is essential to take care when evaluating circulating miRNAs as diagnostic markers for NSCLC due to the potentiation variation in their levels with tumor progression.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by JSPS KAKENHI (grant no. 25460698).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TA and HO designed the study. HK, HT, AO, TW, SK, TY, ST and NM made substantial contributions to the conception and to the design of the present study, and collected clinical samples. TA conducted the experiments and wrote the manuscript. TA, KO, SK and HO interpreted the experimental results. HO, HK, HT, AO and TW revised the manuscript critically for important intellectual content. TA and MU performed statistical analysis. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study protocol was approved by the Ethical Committee of the Faculty of Medicine (approval no. H26-010) and the Ethical Committee of the Faculty of Health Sciences (approval no. 25-40) of Kyorin University (Tokyo, Japan). Signed informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Goya T, Asamura H, Yoshimura H, Kato H, Shimokata K, Tsuchiy R, Sohara Y, Miya T, Miyaoka E, Japanese Joint Committee of Lung Cancer Registry Prognosis of 6644 resected non-small cell lung cancers in Japan: A Japanese lung cancer registry study. Lung Cancer. 2005;50:227–234. doi: 10.1016/j.lungcan.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 3.Wu GX, Raz DJ. Lung cancer screening. Cancer Treat Res. 2016;170:1–23. doi: 10.1007/978-3-319-40389-2_1. [DOI] [PubMed] [Google Scholar]
- 4.Witwer KW. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin Chem. 2015;61:56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 5.Moretti F, D'Antona P, Finardi E, Barbetta M, Dominioni L, Poli A, Gini E, Noonan DM, Imperatori A, Rotolo N, et al. Systematic review and critique of circulating miRNAs as biomarkers of stage I–II non-small cell lung cancer. Oncotarget. 2017;8:94980–94996. doi: 10.18632/oncotarget.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, Etheridge A, Luo Y, Ding Y, Wang K. Current state of circulating microRNAs as cancer biomarkers. Clin Chem. 2015;61:1138–1155. doi: 10.1373/clinchem.2015.241190. [DOI] [PubMed] [Google Scholar]
- 7.Tiberio P, Callari M, Angeloni V, Daidone MG, Appierto V. Challenges in using circulating miRNAs as cancer biomarkers. Biomed Res Int. 2015;2015:731479. doi: 10.1155/2015/731479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8:61–81. doi: 10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 9.Larrea E, Sole C, Manterola L, Goicoechea I, Armesto M, Arestin M, Caffarel MM, Araujo AM, Araiz M, Fernandez-Mercado M, Lawrie CH. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int J Mol Sci. 2016;17:pii: E627. doi: 10.3390/ijms17050627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D'Andrade P, DeMayo M, Dennis L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 11.Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data normalization strategies for microRNA quantification. Clin Chem. 2015;61:1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem Sci. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi F, Nicassio F, Marzi M, Belloni E, Dall'olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G, Di Fiore PP. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 19.Markou A, Sourvinou I, Vorkas PA, Yousef GM, Lianidou E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer. 2013;81:388–396. doi: 10.1016/j.lungcan.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 20.The World Health Organization histological typing of lung tumours. Second edition. Am J Clin Pathol. 1982;77:123–136. doi: 10.1093/ajcp/77.2.123. [DOI] [PubMed] [Google Scholar]
- 21.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L, International Association for the Study of Lung Cancer International Staging Committee. Participating Institutions The IASLC lung cancer staging project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 22.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Yang D, Zhang H, Wei X, Ma T, Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, et al. Early detection of lung cancer in serum by a panel of MicroRNA biomarkers. Clin Lung Cancer. 2015;16:313–319.e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Wang RJ, Zheng YH, Wang P, Zhang JZ. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:765–771. [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Das AV, Pillai RM. Implications of miR cluster 143/145 as universal anti-oncomiRs and their dysregulation during tumorigenesis. Cancer Cell Int. 2015;15:92. doi: 10.1186/s12935-015-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Danielson LS, Menendez S, Attolini CS, Guijarro MV, Bisogna M, Wei J, Socci ND, Levine DA, Michor F, Hernando E. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am J Pathol. 2010;177:908–917. doi: 10.2353/ajpath.2010.091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM, et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol. 2017;35:872–878. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan L, Qi H, Teng J, Su B, Chen H, Wang C, Xia Q. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol. 2016;37:7777–7784. doi: 10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 30.Arab A, Karimipoor M, Irani S, Kiani A, Zeinali S, Tafsiri E, Sheikhy K. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet. 2017;216–217:1–158. doi: 10.1016/j.cancergen.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Taverna S, Giallombardo M, Gil-Bazo I, Carreca AP, Castiglia M, Chacártegui J, Araujo A, Alessandro R, Pauwels P, Peeters M, Rolfo C. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: Critical analysis of evidence and potential role in clinical practice. Oncotar. 2016;7:28748–28760. doi: 10.18632/oncotarget.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopatina T, Gai C, Deregibus MC, Kholia S, Camussi G. Cross talk between cancer and mesenchymal stem cells through extracellular vesicles carrying nucleic acids. Front Oncol. 2016;6:125. doi: 10.3389/fonc.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, Zhang YK. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190–3197. doi: 10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 34.Aushev VN, Zborovskaya IB, Laktionov KK, Girard N, Cros MP, Herceg Z, Krutovskikh V. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS One. 2013;8:e78649. doi: 10.1371/journal.pone.0078649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Liu Y, Wang C, Deng T, Liang H, Wang Y, Huang D, Fan Q, Wang X, Ning T, et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci Rep. 2015;5:12921. doi: 10.1038/srep12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leidinger P, Keller A, Backes C, Huwer H, Meese E. MicroRNA expression changes after lung cancer resection: A follow-up study. RNA Biol. 2012;9:900–910. doi: 10.4161/rna.20107. [DOI] [PubMed] [Google Scholar]
- 37.Sanfiorenzo C, Ilie MI, Belaid A, Barlési F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang D, Shen Y, Wang M, Yang R, Wang Z, Sui A, Jiao W, Wang Y. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22:540–548. doi: 10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 39.Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Jiang L, Sun C, Li Guo L, Lin M, Huang J, Zhu L. Decreased circulating miR-375: A potential biomarker for patients with non-small-cell lung cancer. Gene. 2014;534:60–65. doi: 10.1016/j.gene.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 41.Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu RX, Liu SH, Yi QT, Li J, Song CH. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035–3042. doi: 10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 42.Lv S, Xue J, Wu C, Wang L, Wu J, Xu S, Liang X, Lou J. Identification of a panel of serum microRNAs as biomarkers for early detection of lung adenocarcinoma. J Cancer. 2017;8:48–56. doi: 10.7150/jca.16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Mao F, Shen T, Luo Q, Ding Z, Qian L, Huang J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, Cheng W, Wang F, Qi LW, Chen Y, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Driscoll L. Extracellular nucleic acids and their potential as diagnostic, prognostic and predictive biomarkers. Anticancer Res. 2007;27:1257–1265. [PubMed] [Google Scholar]
- 46.Zampetaki A, Mayr M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb Haemost. 2012;108:592–598. doi: 10.1160/TH12-02-0097. [DOI] [PubMed] [Google Scholar]
- 47.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, Xie H, Xu Y, Zeng X. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454:210–214. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 48.Sromek M, Glogowski M, Chechlinska M, Kulinczak M, Szafron L, Zakrzewska K, Owczarek J, Wisniewski P, Wlodarczyk R, Talarek L, et al. Changes in plasma miR-9, miR-16, miR-205 and miR-486 levels after non-small cell lung cancer resection. Cell Oncol (Dordr) 2017;40:529–536. doi: 10.1007/s13402-017-0334-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.