Abstract
Filamin A (FlnA) is an actin cross-linking protein. Previous studies have demonstrated its role in tumor progression in a wide range of cancer types. It has been reported that FlnA interacts with the DNA damage response protein, breast cancer gene 1 (BRCA1), which is a tumor suppressor gene. However, to the best of our knowledge, there are no studies evaluating the association of these genes in human carcinomas. In the present study, the immunohistochemistry of a tissue microarray was used to investigate the clinical significance of FlnA and BRCA1 expression in pathological specimens collected from 424 patients treated for breast cancer. In addition, FlnA and BRCA1 expression was downregulated in the breast cancer cell line, MCF-7, through FlnA RNA interference. FlnA expression was exhibited by cancer tissues collected from 137 patients with breast cancer, which also exhibited high expression of BRCA1 and were associated with a relatively long survival time. A significant association was identified between FlnA protein expression and tumor size, and between FlnA protein expression and progesterone receptor expression. These results suggest that BRCA1 expression could be regulated by FlnA in the breast cancer cell line, MCF-7. Overall, the present study demonstrates that FlnA expression was associated with BRAC1 expression and tumor size in breast cancer, which provides important implications for future study of FlnA in the progression of human breast cancer.
Keywords: human breast cancer, filamin A, breast cancer gene 1, tumor progression
Introduction
Filamin A (FlnA) is an actin cross-linking protein (1), which serves a role in the organization of the orthogonal actin network, and is considered an essential component of the cytoskeleton networks that support various cellular functions (2). In addition to filamentous actin, FlnA interacts with >60 functionally diverse cellular proteins, including transmembrane receptors, signaling molecules and DNA damage repair proteins (1,3–5). These diverse interactions suggest that FlnA is essential for versatile cell signaling. Previous research has investigated the involvement of FlnA in different types of cancer (6,7). FlnA was originally identified as able to promote cancer progression. However, a recent study demonstrated that the effect of FlnA on cancer cells depended on its subcellular localization and corresponding binding partners (8).
Breast cancer type 1 susceptibility gene (BRCA1) is a DNA repair gene commonly associated with familial breast and ovarian cancers (9,10). Studies in BRCA1-knockout models have indicated that mice lacking BRCA1 have poor genomic integrity and cannot survive (11). It has been reported that FlnA may act as a binding partner of BRCA1, and that it was required for efficient regulation of early stages of DNA repair in a yeast two-hybrid screening model (12). However, to the best of our knowledge, the association of these genes in cancer patients and cell lines remains to be investigated.
In the present study, the expression pattern of FlnA and BRCA1 proteins were investigated in breast cancer tissues. The clinicopathological and prognostic value of the subcellular localizations of these proteins, and the association between FlnA/BRCA1 expression and clinicopathological characteristics of breast cancer patients were also investigated. Furthermore, the association between FlnA and BRCA1 genes was investigated through RNA interference in the breast cancer cell line, MCF-7. The results of the present study provide a basis for researching FlnA and BRCA1 function in tumor progression, as well as insight into the potential prognostic and therapeutic value of FlnA/BRCA1 expression.
Materials and methods
Patients
Tumor tissue specimens were collected from 424 women (mean age, 56.49 years; range, 26–94 years), who had been diagnosed with breast cancer. Patients who had received any preoperative chemotherapy or radiotherapy prior to surgery were excluded. A total of 88 tissues were collected at the Department of General Surgery of the Suzhou Municipal Hospital (Jiangsu, China) between January 2009 and December 2010. The remaining 336 tissues were collected from the Department of General Surgery of Shanghai Renji Hospital (Shanghai, China). All 424 cases were pathologically confirmed as breast cancer by hematoxylin and eosin (H&E) staining at the time of surgical resection. The clinical parameters of the patients are presented in Table I. Survival data were collected through patient follow-up each year for seven years. The present study was approved by the Ethics Review Board of Suzhou Vocational Health Technology College (Jiangsu, China), and written informed consent was obtained from each participant.
Table I.
Correlations between FlnA/BRCA1 expression and clinicopathological factors in breast cancer.
| FlnA expression | BRCA1 expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Factor | Number | Low (%) | High (%) | P-value | Number | Low (%) | High (%) | P-value |
| Age (years) | 0.719 | 0.307 | ||||||
| <60 | 259 | 177 (40.78) | 82 (18.8) | 259 | 145 (33.41) | 114 (26.27) | ||
| ≥60 | 175 | 120 (27.65) | 55 (12.67) | 175 | 89 (20.51) | 86 (19.82) | ||
| Tumor size | 0.027 | 0.064 | ||||||
| <2.5 cm | 218 | 137 (32.5) | 81 (19.2) | 172 | 127 (30.2) | 91 (21.6) | ||
| ≥2.5 cm | 203 | 148 (35.2) | 55 (13.1) | 163 | 100 (23.8) | 103 (24.5) | ||
| Lymph node metastasis | 0.869 | 0.535 | ||||||
| Negative | 72 | 47 (36.4) | 25 (19.4) | 72 | 24 (18.6) | 48 (37.2) | ||
| Positive | 57 | 38 (29.5) | 19 (14.7) | 57 | 22 (17.1) | 35 (27.1) | ||
| Differentiation | 0.392 | 0.099 | ||||||
| I | 3 | 3 (1.2) | 0 (0) | 3 | 3 (1.2) | 0 (0) | ||
| II | 171 | 118 (45.7) | 53 (20.5) | 171 | 96 (37.2.8) | 75 (29.1) | ||
| III | 84 | 62 (24) | 22 (8.5) | 84 | 56 (21.7) | 28 (10.9) | ||
| ER | 0.694 | 0.003 | ||||||
| ER (−~+) | 186 | 162 (38.3) | 74 (17.5) | 187 | 86 (20.3) | 101 (23.9) | ||
| ER (++~+++) | 236 | 125 (29.6) | 62 (14.7) | 236 | 143 (33.8) | 93 (22) | ||
| PR | 0.021 | 0.023 | ||||||
| PR (−~+) | 280 | 201 (47.5) | 79 (18.7) | 280 | 163 (38.5) | 117 (27.7) | ||
| PR (++~+++) | 143 | 86 (20.3) | 57 (13.5) | 143 | 66 (15.6) | 77 (18.2) | ||
| HER2 | 0.861 | 0.023 | ||||||
| HER2 (−~+) | 153 | 103 (24.3) | 50 (11.8) | 153 | 94 (22.2) | 59 (13.9) | ||
| HRE2 (++~+++) | 270 | 184 (43.5) | 86 (20.3) | 270 | 135 (31.9) | 135 (31.9) | ||
| Subtype | 0.521 | <0.001 | ||||||
| Lumin A | 83 | 57 (13.5) | 26 (6.2) | 83 | 49 (11.6) | 34 (8.1) | ||
| Lumin B | 116 | 76 (18.1) | 40 (9.5) | 116 | 44 (10.5) | 72 (17.1) | ||
| HER2 (+) | 147 | 103 (24.5) | 44 (10.5) | 147 | 85 (20.2) | 62 (14.7) | ||
| Basal like | 51 | 37 (8.8) | 14 (3.3) | 51 | 31 (7.4) | 20 (4.8) | ||
| Normal like | 24 | 13 (3.1) | 11 (2.6) | 24 | 19 (4.5) | 5 (1.2) | ||
P-values were calculated using Pearson Chi-square test or Chi-square test for trend in three or four levels of differentiation and subtype. ER (−~+) means the expression of ER was deficiency or weak. ER (++~+++) means the expression was moderate or strong. FlnA, filamin A; BRCA1, breast cancer gene 1; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor.
Construction of tissue microarray (TMA)
Two specialized histopathologists (authors of this paper)isolated 2 1.6-mm sections from the centre and periphery of the tumors, and placed them into a paraffin block using a tissue puncher/arrayer (patent no. 200920350099.2). Sections of 4-µm were sliced from the paraffin block, deparaffinized in xylene for 10 min and conducted three times at room temperature and rehydrated using a descending ethanol series (100, 95, 80 and 70%) for immunohistochemical staining. The deparaffinized reagent was xylene and rehydrated reagent was ethanol (100, 95, 80, 70 and 0%).
Immunohistochemistry
A rabbit anti-human Filamin A antibody (dilution, 1:100; clone, PM61317; cat. no. MAB1678), and a mouse anti-human BRCA1 antibody (dilution, 1:150; clone, MS110; cat. no. OP92) were purchased from Abcam (Cambridge, UK). Other antibodies [rabbit anti-human ER antibody (dilution, 1:100; clone, SP1; cat. no. GT205611) rabbit anti-human PR antibody (dilution, 1:100; clone, SP2; cat. no. GT205711) rabbit anti-human HER2 antibody (dilution, 1:100; clone, SP3; cat. no. GT210011)] and the IHC kit (cat. no. KIT-5010) were purchased from Fuzhou Maixin Biotech, Co., Ltd. (Fuzhou, China). Immunohistochemical staining of FlnA and BRCA1 was performed according to the manufacturer's instructions.
Immunohistochemical staining of FlnA and BRCA-1 was examined by 2 independent pathologists (Suzhou Municipal Hospital, Jiangsu, China) who were blinded to the clinicopathological parameters of the breast cancer patients. A semi-quantitative scoring system was used, based on the distribution of tumor cells stained positively for FlnA and BRCA-1: 0, 0–10%; 1, 11–30%; 2, 31–60%, and 3, 61–100%. The distribution score was multiplied by the intensity factor: 1, staining intensity just exceeding background; 2, weak positive; 3, middle positive; 4, dark brown staining evident upon macroscopic inspection of the slide. The resulting score was divided by 4. For FlnA, final scores of 0–1 were considered to indicate low expression, and scores of 2–3 were considered to indicate high expression. For BRCA1, final scores of 0–2 were considered to indicate low expression, and a score of 3 was considered to indicate high expression.
Cell culture and transfection
The human breast cancer cell line, MCF-7, was obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). To acquire a better interference efficiency a total of 3 stealth RNA interference (RNAi) small interfering RNAs (siRNAs) target FlnA and 2 control RNAi were designed by Invitrogen (Thermo Fisher Scientific, Inc.) based on the FlnA GenBank accession no. NM_001456.3 (13). According to the FlnA sequence, the siRNAs sequences designed were: 5′-UGCAUUUGGCGGAAAGUGGGC-3′; 5′-UUUCUUCGGGUUCAGUUUGGG-3′; and 5′-UAUACUUUGACCUUGUUGGGG-3′. The control RNAi sequences were 5′-UUCUCCGAACGUGUCACGUTT-3′; and 5′-ACGUGACACGUUCGGAGAATT-3′. The RNAi with the greatest FlnA knockdown effect was also selected. MCF-7 cells were seeded at 1×105 in 24-well plates, and incubated for 24 h prior to transfection. The individual RNAi were transfected into MCF-7 cells at 10 µM using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). The cells were and cultured for 24 h prior to subsequent experimentation.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from MCF-7 cells using an RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) and cDNA was synthesized using a Primescript First cDNA synthesis kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturers' protocols. RT-qPCR was performed in triplicate using a Bio-Rad iCycler CFX96 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and a SYBR Green PCR Master mix (Takara Bio, Inc.), according to the manufacturer's protocol. The following primers were used: FlnA, forward, 5′-AGCCTCCACGAGACATCATC-3′ and reverse, 5′-CCAGTGTGTACTCCCCCTTG-3′; BRCA-1, forward, 5′-GGCTATCCTCTCAGAGTGACA-3′ and reverse, 5′-CTGATGTGCTTTGTTCTGGA-3′; GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′. The levels of gene expression were normalized to the expression of GAPDH. The thermocycling conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, with a final extension of 72°C for 3 min. The 2−∆∆Cq method was used for the quantification of FlnA and BRCA-1 gene expression (14).
Statistical analysis
All data are presented as the mean ± standard deviation. The χ2 test was used to analyze the association between FlnA/BRCA1 expression and clinicopathological features. Unpaired Student's t-test was used for all other comparisons. All tests were performed using software SPSS 17.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference. Kaplan-Meier survival plots were generated, and comparisons between the survival curves were made with the log-rank test.
Results
FlnA protein expression levels in breast cancer tissue
FlnA protein expression was assessed in breast cancer and normal breast tissue specimens (Fig. 1A and B). FlnA protein was mainly detected in the myoepithelial and basal cell cytoplasm or intercellularly in breast cancer tissue. FlnA protein expression was detected in 52.6% breast cancer tissues, whereas FlnA was rarely detectable in normal breast tissues or benign tumor ductal epithelium sections. The rate of positive expression in normal breast epithelium was 5.9%.
Figure 1.

Immunohistochemical analysis of FlnA expression in breast tissues. (A) Low FlnA expression in breast cancer tissue. (B) High FlnA expression in breast cancer tissue. Magnification, ×400. FlnA, filamin A.
Association of FlnA protein expression with clinicopathological and patient prognosis features
The associations between FlnA protein expression level and various clinicopathological features of the patients with breast cancer were investigated. The protein expression level of FlnA was negatively associated with tumor size (P<0.05; Table I). The rate of positive FlnA staining increased with progesterone receptor expression (P<0.05). No significant association was identified between FlnA protein expression level and age, lymph node metastasis, differentiation, estrogen receptor status or Her2/neu protein expression (P>0.05; Table I).
Only 84 cases in Suzhou Municipal Hospital were investigated for survival. The mean duration of follow-up was 84 months after surgery and 75th percentile of duration was 74 months. During the follow-up period, 14 mortalities occurred (16.67%). The overall survival rate of patients exhibiting FlnA overexpression was determined by the by the Kaplan-Meier survival analysis and log-rank test, and was significantly increased compared with those who did not exhibit FlnA overexpression (P=0.025; Fig. 2).
Figure 2.
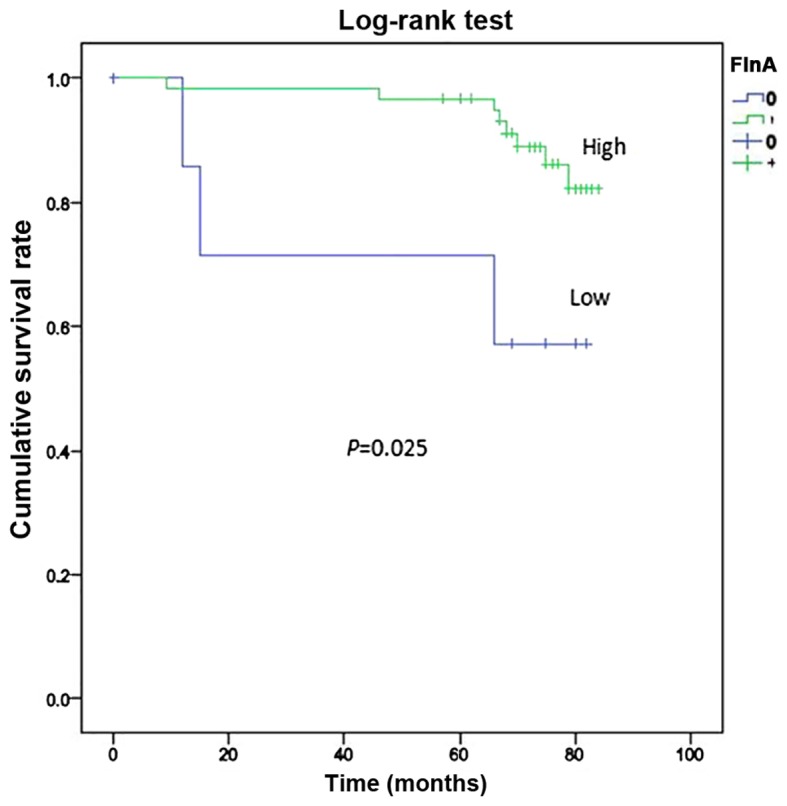
Kaplan-Meier survival analysis of the association between survival rate and FlnA expression. The overall survival rate of patients with high FlnA expression, was significantly increased compared with the rate of those with low FlnA expression, as determined by the log-rank test (P=0.025). FlnA, filamin A.
Nucleic BRCA1 expression in breast cancer tissue and its association with clinicopathological parameters
Adjacent normal breast tissue stained strongly positive for BRCA1 in the cell nuclei, and no cytoplasmic or membranous staining was observed (Fig. 3A). In contrast, the protein expression level of BRCA1 in breast cancer tissue was demonstrated to be either nucleic or cytoplasmic (Fig. 3B). Of the 424 breast cancer tissues stained for BRCA1, complete loss of nucleic expression was detected in 207 (48.82%) cases. Absent nucleic staining was considered to indicate low expression.
Figure 3.
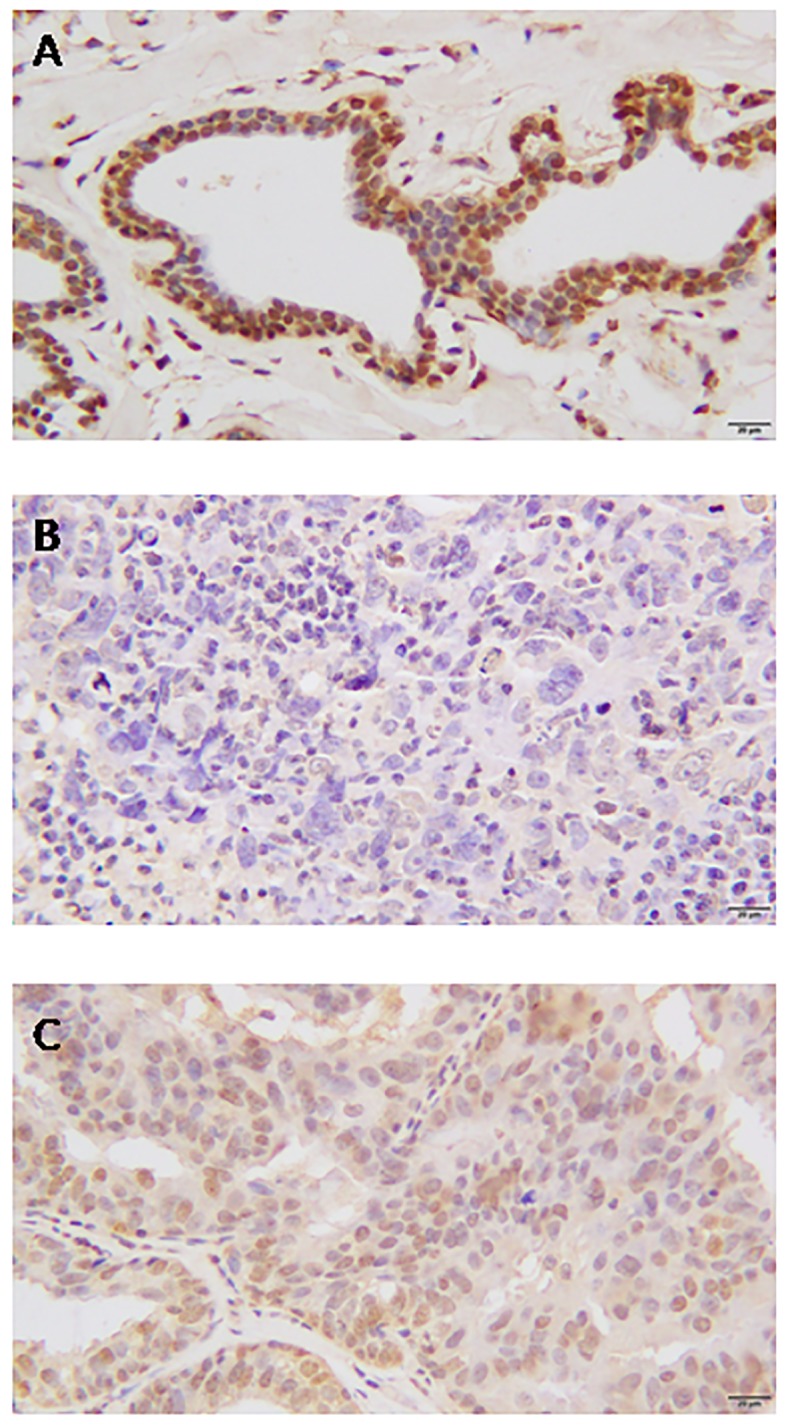
Immunohistochemical analysis of BRCA1 expression in breast tissues. (A) Normal breast tissue. (B) Low BRCA1 expression in breast cancer tissue. (C) High BRCA1 expression in breast cancer tissue. Magnification, ×400. BRCA1, breast cancer gene 1.
High nucleic BRCA1 protein expression level was associated with clinicopathological features, including breast cancer molecular subtype, phenotype, and estrogen and progesterone receptor expression (P<0.05; Table I). However, no association was detected between nucleic BRCA1 protein expression and lymph node metastasis, age, tumor size, differentiation or Her2/neu expression (P>0.05; Table I).
Correlation of FlnA and BRCA1 expression
A positive correlation between FlnA expression and BRCA1 nucleic expression was identified, indicating that high expression of BRCA1 was associated with high expression of FlnA (P<0.001; Table II).
Table II.
Correlation between FlnA and BRCA1 protein expression in patients with breast cancer (χ2=0.203; P<0.001).
| FlnA expression | |||
|---|---|---|---|
| BRCA1 expression | Negative | Positive | Total |
| Negative | 176 | 54 | 230 |
| Positive | 111 | 83 | 194 |
| Total | 287 | 137 | 424 |
FlnA, filamin A; BRCA1, breast cancer gene 1.
Silencing FlnA expression downregulates BRCA1 expression in MCF-7 cells
To assess whether FlnA regulates BRCA1 expression, RNAi technology was used to knock down FlnA expression in MCF-7 cells. RT-qPCR analysis revealed effective and specific knockdown of FlnA after 48 h of transfection (Fig. 4A and B). FlnA was decreased by ~50% following transfection of MCF-7 cells by FlnA-specific stealth RNAi siRNAs, and BRCA-1 expression was also decreased (Fig. 4A and C).
Figure 4.
Analysis of the knockdown efficiency of FlnA by stealth RNAi siRNA, and the regulation of BRCA1 expression in MCF-7 cells. (A) FlnA and BRCA1 mRNA expression was measured by reverse transcription-quantitative polymerase chain reaction. Expression of (B) FlnA and (C) BRCA1 mRNA was measured 48 h after FlnA siRNA transfection. Data are presented as the mean ± standard deviation of triplicate samples. FlnA, filamin A; BRCA1, breast cancer gene 1; RNAi, RNA interference; siRNA, small interfering RNA.
Discussion
FlnA was first identified as a non-muscle actin filament cross-linking protein, or gelation factor, in 1975 (15). FlnA serves as a scaffold protein for >90 binding partners, including channels, receptors, intracellular signaling molecules, transcription factors, transmembrane receptors and DNA damage repair proteins (16). FlnA has been associated with cancer metastasis and progression. Originally it was suggested to be a cancer-promoting protein (17), however, it has been more recently established that FlnA serves a dual role in cancers, depending on its subcellular localization and the corresponding binding partners (8).
A number of studies have reported abnormal expression of FlnA in various types of cancer. Overexpression of FlnA has been observed in multiple malignancies, including hepatocellular carcinoma, breast cancer, colon cancer, melanoma and prostate cancer (17–19). In the present study, it was demonstrated that FlnA was overexpressed in breast cancer tissue compared with distant normal breast tissue, which is consistent with previous reports (20–22). Furthermore, it was demonstrated that FlnA was highly expressed in myoepithelial cells of normal breast tissues.
BRCA1 is a tumor suppressor gene, which is expressed in all cells. BRCA1 mutations lead to an increased predisposition to breast and ovarian cancer (23). BRCA1 has been implicated in several aspects of the DNA damage response (DDR) and FlnA has been reported to function in DNA repair (12,24). A previous study identified FlnA as a partner of BRCA1, and demonstrated that FlnA is required for the efficient regulation of the early stages of DNA repair (12). However, to the best of our knowledge, no further research regarding the association between FlnA and BRCA1 has been performed.
In the present study, the expression of BRCA1 was analyzed in 424 breast cancer tissue samples using immunochemistry, and a positive association between FlnA and BRCA1 was identified (P<0.001). Knockdown of FlnA expression using stealth RNAi siRNA in MCF-7 cells resulted in downregulation of BRCA1 expression. It is speculated that FlnA regulates BRCA1 expression in breast cancer, which was verified in another cell line (data not shown). However, further research is required to determine the mechanism by which FlnA regulates BRCA1 expression.
Overall, the present study indicates that increased FlnA expression is exhibited in breast cancer originating from epithelial cells, and that FlnA expression is associated with tumor progression. A positive association between FlnA and BRCA1 expression was also demonstrated, and it is hypothesized that FlnA regulates the expression of BRCA1. The precise mechanism, and whether FlnA may be used as a prognostic indictor of breast cancer, remains to be determined.
Acknowledgements
The authors would like to thank Dr Yumin Hu (Soochow University) for providing the breast cancer cell line. The authors would like to also thank the staff in the Pathology Department of Suzhou Municipal Hospital for collating the specimens of breast cancer.
Funding
The present study was supported by grants from Jiangsu Province University Outstanding Science and Technology Innovation Team (grant no. 2015023) and Jiangsu Provincial Medical Youth Talent.
Availability of data and materials
The data used to support the findings of this study are available from the corresponding author upon request.
Authors' contributions
Research was designed by JS. Patient samples and data were provided by ML and GB. IHC was performed by XL, LW and YG. Cell experiments in vitro were performed by ZS and JY. Statistical analysis of data was performed by YG. The manuscript was critically reviewed by all authors. The study was supervised by JS.
Ethics approval and consent to participate
Prior to commencing this study the written informed consent was obtained from each patient accepting the surgical treatment. The present study was approved by the Ethics Review Board of Suzhou Vocational Health Technology College.
Patient consent for publication
Prior to commencing this study, the approval from the Ethics Review Board of Suzhou Vocational Health College was granted.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Feng Y, Walsh CA. The many faces of filamin: A versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 2.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: Promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Meng X, Yuan Y, Maestas A, Shen Z. Recovery from DNA damage-induced G2 arrest requires actin-binding protein filamin-A/actin-binding protein 280. J Biol Chem. 2004;279:6098–6105. doi: 10.1074/jbc.M306794200. [DOI] [PubMed] [Google Scholar]
- 4.Duval D, Lardeux A, Le Tourneau T, Norris RA, Markwald RR, Sauzeau V, Probst V, Le Marec H, Levine R, Schott JJ, Merot J. Valvular dystrophy associated filamin A mutations reveal a new role of its first repeats in small-GTPase regulation. Biochim Biophys Acta. 2014;1843:234–244. doi: 10.1016/j.bbamcr.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpeläinen I, Munday AD, Hartwig JH, Stossel TP, Ylänne J. The structure of the GPIb-filamin A complex. Blood. 2016;107:1925–3192. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao QQ, Zhang TP, Zhao WJ, Liu ZW, You L, Zhou L, Guo JC, Zhao YP. Filamin A: Insights into its exact role in cancers. Pathol Oncol Res. 2016;22:245–252. doi: 10.1007/s12253-015-9980-1. [DOI] [PubMed] [Google Scholar]
- 7.Yue J, Lu H, Liu J, Berwick M, Shen Z. Filamin-A as a marker and target for DNA damage based cancer therapy. DNA Repair (Amst) 2012;11:192–200. doi: 10.1016/j.dnarep.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savoy RM, Ghosh PM. The dual role of filamin A in cancer: Can't live with (too much of) it, can't live without it. Endocr Relat Cancer. 2013;20:R341–R356. doi: 10.1530/ERC-13-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Breast cancer linkage consortium. Lancet. 1994;343:692–695. doi: 10.1016/S0140-6736(94)91578-4. [DOI] [PubMed] [Google Scholar]
- 10.Easton DF, Bishop DT, Ford D, Crockford GP. Genetic linkage analysis in familial breast and ovarian cancer: Results from 214 families. The breast cancer linkage consortium. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- 11.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 12.Velkova A, Carvalho MA, Johnson JO, Tavtigian SV, Monteiro AN. Identification of Filamin A as a BRCA1-interacting protein required for efficient DNA repair. Cell Cycle. 2010;9:1421–1433. doi: 10.4161/cc.9.7.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Hartwig JH, Stossel TP. Isolation and properties of actin, myosin, and a new actinbinding protein in rabbit alveolar macrophages. J Biol Chem. 1975;250:5696–5705. [PubMed] [Google Scholar]
- 16.Nakamura F, Stossel TP, Hartwig JH. The filamins: Organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanagan LA, Chou J, Falet H, Neujahr R, Hartwig JH, Stossel TP. Filamin A the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J Cell Biol. 2001;155:511–517. doi: 10.1083/jcb.200105148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ai J, Huang H, Lv X, Tang Z, Chen M, Chen T, Duan W, Sun H, Li Q, Tan R, et al. FLNA and PGK1 are two potential markers for progression in hepatocellular carcinoma. Cell Physiol Biochem. 2011;27:207–216. doi: 10.1159/000327946. [DOI] [PubMed] [Google Scholar]
- 19.Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu Q, Xu GJ, Song JD, Zhao FK. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121:2596–2605. doi: 10.1002/ijc.23016. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X, Yue J, Lu H, Campbell N, Yang Q, Lan S, Haffty BG, Yuan C, Shen Z. Inhibition of filamin-A reduces cancer metastatic potential. Int J Biol Sci. 2013;9:67–77. doi: 10.7150/ijbs.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alper O, Stetler-Stevenson WG, Harris LN, Leitner WW, Ozdemirli M, Hartmann D, Raffeld M, Abu-Asab M, Byers S, Zhuang Z, et al. Novel anti-filamin-A antibody detects a secreted variant of filamin-A in plasma from patients with breast carcinoma and high-grade astrocytoma. Cancer Sci. 2009;100:1748–1756. doi: 10.1111/j.1349-7006.2009.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian HM, Liu XH, Han W, Zhao LL, Yuan B, Yuan CJ. Differential expression of filamin A and its clinical significance in breast cancer. Oncol Lett. 2013;6:681–686. doi: 10.3892/ol.2013.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 24.Kim KM, Moon YJ, Park SH, Park HJ, Wang SI, Park HS, Lee H, Kwon KS, Moon WS, Lee DG, et al. Individual and combined expression of DNA damage response molecules PARP1, gammaH2AX, BRCA1, and BRCA2 predict shorter survival of soft tissue sarcoma patients. PLoS One. 2016;11:e0163193. doi: 10.1371/journal.pone.0163193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.



