Abstract
To investigate the effects of hypoxia on the features of cancer stem cells in the glioma cancer U87 cell line and underlying mechanism, stem cell markers and features in U87 were studied under the hypoxic and normoxic culture conditions by reverse transcription-quantitative polymerase chain reaction, western blot analysis, MTT, a colony formation test and flow cytometry. Compared to the normoxic group, the cluster of differentiation 133+ phenotype, clone formation rate and cell vitality were significantly elevated in U87 cells cultured in a hypoxic microenvironment. Also, the mRNA and protein expression of neurogenic locus notch homolog protein 1 (Notch1) and Oct3/4 were significantly elevated in U87 cells cultured in a hypoxic microenvironment, however, transcription factor SOX-2 expression was not significantly changed. These results indicate that hypoxia can promote the proliferation of glioma stem cells and maintain the characteristics of stem cells through the activation of Notch1 and Oct3/4 or Notch1 activation, affecting the biological characteristics of glioma cells.
Keywords: glioma, neurogenic locus notch homolog protein 1, Oct3/4
Introduction
Gliomas are neuroepithelial tumors and malignant gliomas are the most common type of primary tumors of the central nervous system (1). The prognosis is worse and the degree of malignancy is increased in tumors with a higher grade (1). Usually, the treatment for a glioma is surgery, complemented with radiotherapy and chemotherapy (2). Although, in recent years, the microsurgery technique, chemotherapy, immunotherapy, gene therapy, molecular targeted therapy and biological technology have progressed, the overall prognosis of patients with glioma has not improved (2). The mortality and recurrence rates are high, with the 5-year survival rate for the primary type of glioma, glioblastoma, being 5.1% and the one-year recurrence rate being ~70% (3). The main reasons for the high mortality of patients with glioma are resistance to treatment and local recurrence, which have been difficult problems to overcome in the treatment of gliomas (2).
Previous studies have revealed that cancer stem cells (CSCs) or CSC-like cells exist in a variety of tumors, and indicated that they serve an important role in the occurrence, development, invasion, metastasis and recurrence of tumors (4,5). The cluster of differentiation (CD)133 protein has been reported as a cell surface marker to human embryonic neural stem cells and cancer stem cells (6–9), and CD133 positive cells present stem cell properties when isolated from human brain tumors (6,7,10). In gliomas, glioma stem cells (GSCs) have also been reported to serve a role in the progression of glioma (11), which provides a novel theory, and may provide a breakthrough point for studying the mechanism of glioma treatment and improving the therapeutic effect.
The proliferation of normal stem cells is strictly regulated by the microenvironment, where cancer stem cells are also likely to specifically grow and exist (12). Internal tumors are often in hypoxic states during the growth process (13). Studies have demonstrated that a hypoxic microenvironment serves an important role in the occurrence and development of a tumor, changing the phenotype of tumor cells and increasing the malignant degree through hypoxic stress (14). The presence of hypoxia can promote tumor cell proliferation, promote angiogenesis, inhibit stem cell differentiation, increase resistance to radiotherapy and chemotherapy, and increase tumor invasion and metastasis (11,15–20). Cancer stem cells express several stem cell markers (21). Pluripotent stem cells are regulated by a series of transcription factors, including Oct3/4 and transcription factor SOX-2 (Sox2) (22,23). Studies demonstrated that changes in neurogenic locus notch homolog protein (Notch)1 signaling are associated with a number of human cancers and Notch1 has been speculated to be a prognostic marker for tumors (24–28). The present study established a hypoxic model in human U87 glioma cells, observed the effect of hypoxia on the expression of Oct3/4, Sox2 and Notch1 at mRNA and protein levels, and assessed the influence of the hypoxic environment on human U87 glioma cell physiology.
Materials and methods
Cell culture
The human glioma cell line, U87, was purchased from the Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in the incubator with 5% CO2 and 95% humidity. The medium was changed every 72 h. Previously, this widely used cell line was reported to be different from the original U87MG Uppsala cell line, but it is likely that the cells line is a bona fide human glioblastoma cell line of unknown origin (29). The authors of the current study posit that the possible difference between these two cell lines should not affect the conclusions of the present study.
Hypoxic cell culture
U87 cells were cultured in DMEM containing fetal bovine serum, penicillin and streptomycin at 37°C in the closed incubation system (Xvivo system 300C; BioSpherix, Ltd., Parish, NY, USA) with 1% O2, 94% N2, 5% CO2 and 95% humidity. Cells were assessed following 2, 6, 12, 24, 48 and 72 h of culturing.
Cell viability
A total of 100 ml U87 cells at a density of 1.5×105 cells/ml were plated into each well of a 96-well plate and cultured under hypoxia with blank controls set up in the middle and at the edge of the 96-well plate. Cells were incubated for 2, 6, 12, 24, 48, and 72 h, with five replicates for each time point. Following incubation, the medium was replaced with fresh DMEM containing 20 ml MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and incubated further for 4 h. Subsequently, 150 ul/well DMSO was added into the 96-well plate, followed by agitating the plate for 10 min. Absorbance was measured by a microplate reader at 490 nm. The growth curves of U87 cells under low oxygen or normal conditions (37°C with 5% CO2) were drawn using the duration of the cells cultures and the mean value of absorbance of cells.
Apoptosis assay
A total of 2×106 U87 cells were suspended with DMEM containing 10% fetal bovine serum (both from Gibco), 100 U/ml penicillin and 100 U/ml streptomycin, and inoculated in the culture flask and cultured in the hypoxic environment or at normal conditions at 37°C for 48 h. Following culturing, the single cell suspension was fixed with 70% ethanol at 4°C overnight. Then cells were collected by centrifugation at 300 × g for 5 min, washed three times with PBS, resuspended with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) staining solutions using the FITC Annexin V Apoptosis Detection kit (BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer's protocols. The cells were subsequently mixed gently and incubated for 15 min at room temperature in the dark. Stained cells were analyzed using the BD FACSCalibur™ analyzer with FlowJo 7.6 software (BD Biosciences), which used FL1 and FL2 channels to produce scatter charts with PI and FITC as the parameters. The experiment was repeated three times.
Soft agar colony formation assay
Agar (BD Biosciences) plates were constructed with 0.5% agar forming the bottom layer and 0.3% agar forming the top layer. The plate contained a 1×104 cells/well single cell suspension and was cultured in hypoxic conditions or normal conditions for 2–3 weeks for colony formation. The colonies were subsequently stained by adding 0.2 ml/well of 1 mg/ml nitro blue tetrazolium chloride (ThermoFisher Scientific, Inc.) dissolved in PBS and incubated overnight at 37°C. Using a light microscope (magnification, 100×), colonies with a diameter >75 µm or colonies that contained >50 cells were counted. The rate of colony formation was calculated with the following formula: Colony formation rate (%)=number of colonies/number of inoculated cells ×100.
Flow cytometry analysis and sorting of CD133+ cells
Cells in the logarithmic growth phase were cultured under hypoxic conditions or normal conditions for 24 h at 37°C. They were subsequently washed three times with PBS, collected by centrifugation at 300 × g for 5 min at room temperature, and resuspended and dissociated into single cell suspension in the Pre-Sort Buffer (BD FACS™) by repeatedly pipetting. The cells were then centrifuged at 300 × g for 5 min at room temperature and the supernatant was discarded. The cells were washed with 8 ml PBS, blocked with 2 ml FcR Blocking Reagent (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) for 10 min at 4°C and incubated with 2 ml CD133/2 (293C3)-PE antibodies (dilution, 1:1,000; cat. no. 130-090-853; Miltenyi Biotec GmbH) for 10 min at 4°C in the dark. The cells were washed once with 2 ml PBS and then the proportion of CD133+ cells were detected using the BD FACSCalibur™ analyzer with FlowJo 7.6 software.
Reverse transcription-quantitative polymerase chain reaction analysis
Cells in the logarithmic growth phase at an appropriate density were seeded in culture flasks and cultured for 24 h, then cultured in normoxic or hypoxic environments. Total RNA was extracted using the TRIzol reagent kit (Invitrogen; Thermo Fisher Scientific Inc.) and reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. cDNA was used as a template for Oct3/4, Sox2, Notch1 and GAPDH gene amplification; GAPDH was used as the reference gene. Primer sequences for genes and reference gene are provided in Table I. The qPCR was run with SYBR Green (Takara Biotechnology Co., Ltd.) on 7500 fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific Inc) and the thermocycling conditions were as follows: Denaturation at 95°C for 5 min, and then 35 cycles of denaturation at 94°C for 20 sec, annealing at 60°C for 20 sec and extension at 72°C for 40 sec. The results were quantified with the 2−ΔΔCq method (30).
Table I.
The nucleotide sequence of primers for reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence (5′-3′) | Product length (bp) |
|---|---|---|
| Oct3/4 | F: ACATGTGTAAGCTGCGGCC | 297 |
| R: GTTGTGCATAGTCGCTGCTTG | ||
| Transcription factor SOX-2 | F: TTGCTGCCTCTTTAAGACTAGGA | 75 |
| R: CTGGGGCTCAAACTTCTCTC | ||
| Neurogenic locus notch homolog protein 1 | F: CACTGTGGGCGGGTCC | 85 |
| R: GTTGTATTGGTTCGGCACCAT | ||
| GAPDH | F: CCTCAAGATCATCAGCAATGC | 101 |
| R: TGGTCATGAGTCCTTCCACG |
F, forward; R, reverse.
Western blot analysis
Cells in the logarithmic growth phase at an appropriate density were seeded in culture flasks and cultured for 24 h, then cultured in normoxic or hypoxic environments. Total protein was extracted with lysis buffer (pH 7.5) containing the following: 50 mM Tris-HCl; 100 mM NaCl; 1% TritonX-100; 1 mM EDTA; 2 mM sodium vanadate, and protease inhibitor. Protein concentration was determined using the bicinchoninic acid method. Protein (40 ug/lane) was loaded onto 12% SDS-PAGE gel to separate the proteins. The semi-dry method was used to transfer protein onto a polyvinylidene difluoride membrane (PVDF; 0.45 µm; EMD Millipore, Billerica, MA, USA). The PVDF membrane was incubated with 5% skim milk in TBST for 90 min at room temperature, followed by primary antibodies against Oct3/4 (cat. no. sc-9081), Sox2 (cat. no. sc-20088), Notch1 (cat. no. sc-9170) and GAPDH (cat. no. sc-25778) at a dilution of 1:1,000 (all from Santa Cruz Biotechnology Inc) at 4°C overnight. The membrane was then washed with TBST three times and incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibodies (dilution, 1:1,000; cat. no. sc-2004; Santa Cruz Biotechnology, Inc.) at 37°C for 2 h. Finally, the membrane was washed with TBST three times, developed with Clarity™ Western Electrochemiluminescence Substrate (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and detected by the ChemiDoc™ XRS + System with Image Lab™ Software (170–8265; Bio-Rad Laboratories, Inc.). GAPDH was used as the reference protein. The experiment was repeated three times.
Statistical analysis
The data are presented as mean ± standard deviation and analyzed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). A two-sample t-test or a one-way analysis of variance with the Tukey's Honest Significant Difference post-hoc test were used for statistical analysis. P<0.05 indicated that the difference between groups was statistically significant.
Results
Hypoxia increases U87 cell viability
To assess the effect of hypoxia on U87 cell viability, cells were cultured under hypoxic and normal conditions, and their viability were analyzed by the MTT assay. Hypoxic U87 cells exhibited a higher viability rate compared with the cells cultured under normal conditions when exposed to hypoxia for 48 and 72 h. Although the increased viability of hypoxic cells was slight, the difference between the two groups at 72 h was statistically significant (P<0.05; Fig. 1).
Figure 1.
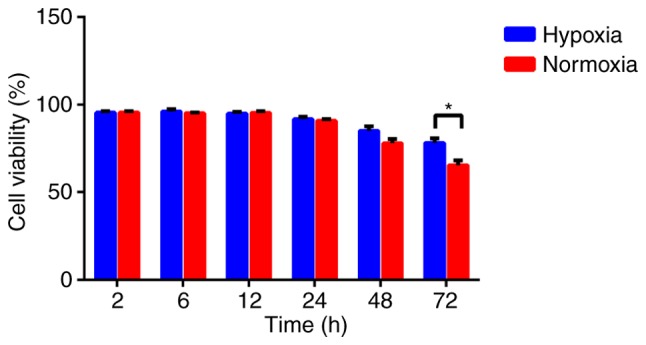
Hypoxia increases U87 cell viability. The cell viabilities of U87 cells cultured under hypoxic and normoxic conditions for 2, 6, 12, 24, 48 or 72 h. *P<0.05, hypoxic compared with normoxic.
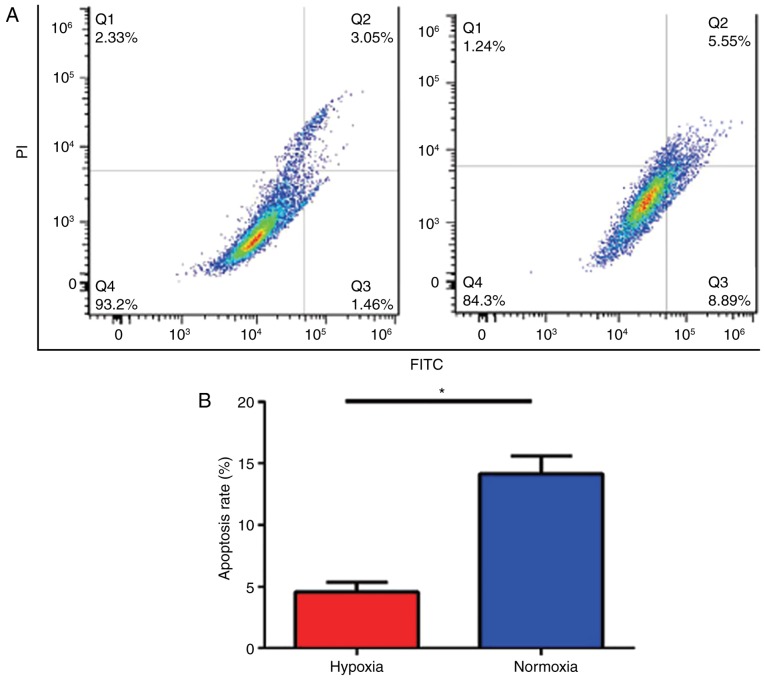
Hypoxia decreases U87 cell apoptosis
To assess if hypoxia induced apoptosis in U87 cells, cultured cells were analyzed using flow cytometry. The data demonstrated that the apoptosis rate in U87 cells cultured under hypoxic conditions was decreased compared with that in cells in normal conditions (Fig. 2A). The greatest difference in the apoptosis rate in U87 cells cultured under hypoxic or normoxic conditions was 48 h post treatment (4.55±0.46 and 14.15±0.84%, respectively); the difference in the apoptosis rate was statistically significant (P<0.05; Fig. 2B).
Figure 2.
Hypoxia decreases U87 cell apoptosis. (A) The effect of hypoxia on U87 cell apoptosis was detected by flow cytometry using the FL1 (FITC green) and FL2 (PI red fluorescence) channels. (B) The quantified apoptosis rate of hypoxic and normoxic cells after 48 h of culturing. *P<0.05. FITC, fluorescein isothiocyanate; PI, propidium iodide.
Hypoxia increases the colony formation ability of U87 cells
To assess the proliferation of U87 cells under hypoxic conditions, a colony formation assay was performed in U87 cells (Fig. 3A). U87 cells cultured under hypoxic conditions possessed a significantly greater clone formation ability (25.38±1.46%) compared with U87 cells cultured under normal conditions (7.25±1.56%; P<0.05; Fig. 3B).
Figure 3.
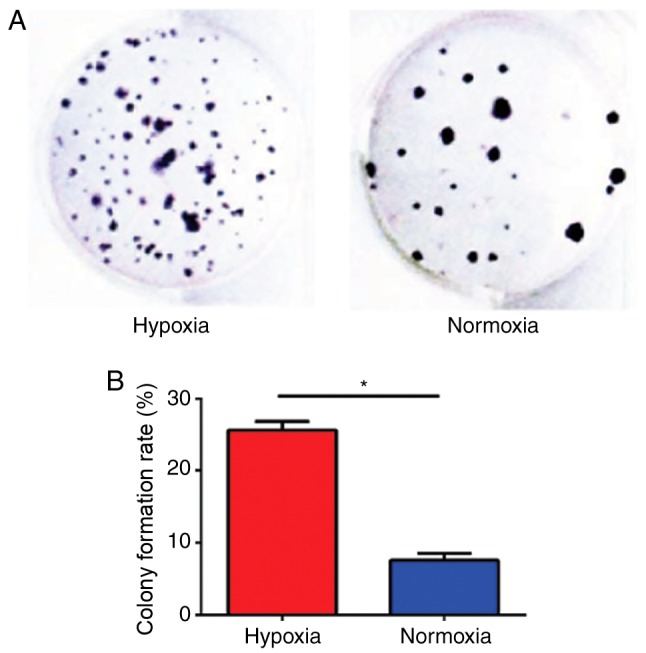
Hypoxia increases the colony formation rate of U87 cells. (A) Colony formation assays were performed in U87 cells cultured in hypoxia and normoxia. (B) The quantified colony formation rate of hypoxic and normoxic cells. *P<0.05, hypoxic compared with normoxic.
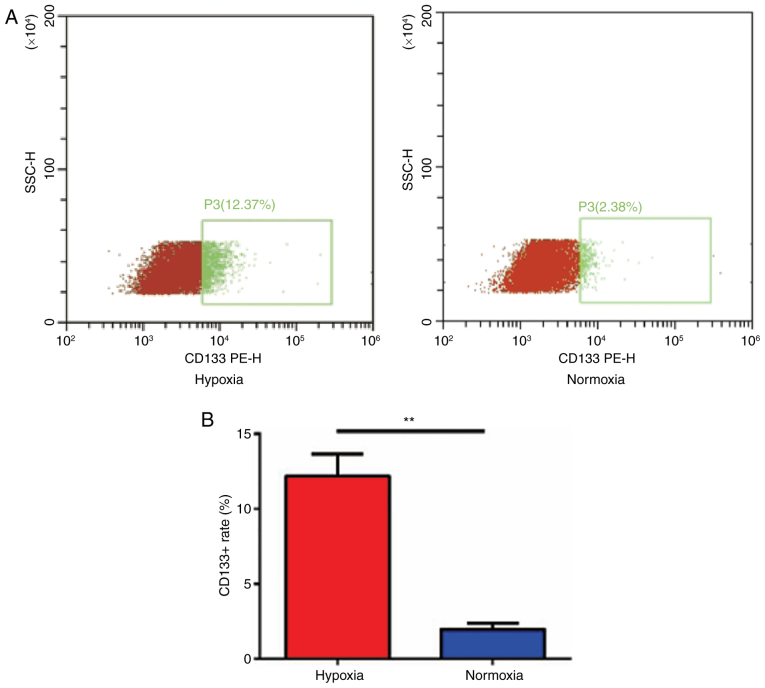
CD133 expression is increased in U87 cells cultured in a hypoxic environment
To assess if CD133 expression can be induced under hypoxic conditions, the expression of CD133 in U87 cells was detected using flow cytometry (Fig. 4A). The data demonstrated that the expression of CD133 in U87 cells cultured in a hypoxic environment was significantly higher (12.18±0.86%) compared with those cultured in a normal environment (1.96±0.23%; P<0.01; Fig. 4B).
Figure 4.
CD133 expression is increased in U87 cells cultured in a hypoxic environment. (A) Flow cytometry to detect the expression of CD133 of U87 cells under hypoxia and normoxia with the ratio of CD133+ cells shown in the red box. (B) Quantification of the CD133+ fraction of U87 cells. **P<0.01, hypoxic compared with normoxic. CD, cluster of differentiation; +, positive; SSC-H, side scatter height; PE-H, phycoerythrin height.
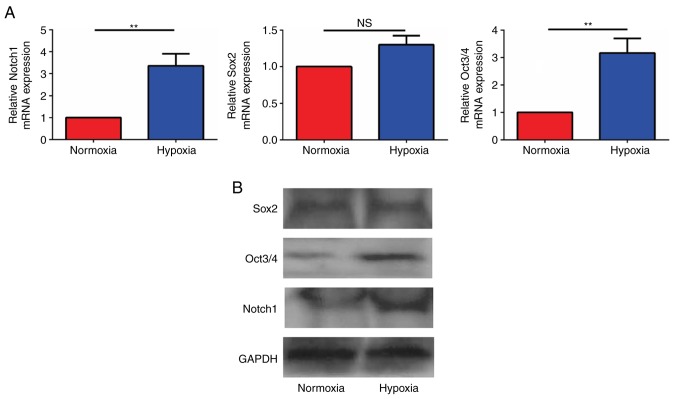
Hypoxia increases the mRNA expression of genes in the Notch signaling pathway
To detect mRNA expression of genes in the Notch signaling pathway in glioma cells, U87 cells cultured in hypoxia and normoxia for 24 h were collected, and the mRNA expression levels were detected by RT-qPCR. When the mRNA expression levels in the hypoxia group were compared with that of the normoxia group, the ratios were revealed to be 3.12±0.23 for Notch1, for 1.23±0.35 Sox2 and 3.17±0.30 for Oct3/4. The results demonstrated that the Notch1 and Oct3/4 mRNA levels in the hypoxia group were significantly increased compared with those in the normoxia group and that the difference was statistically significant (P<0.05; Fig. 5A). No significant difference in the expression of the stem cell associated gene, Sox2, was identified between the two groups.
Figure 5.
Hypoxia increases the expression of genes in the Notch signaling pathway. The (A) mRNA and (B) protein expression of Notch1, transcription factor SOX-2 and Oct3/4. Notch1, neurogenic locus notch homolog protein 1; NS, not significant. **P<0.01, hypoxia compared with normoxic.
Hypoxia increases the protein expression of genes in the Notch signaling pathway
To detect the protein expression of genes in the Notch signaling pathway in glioma cells, U87 cells cultured in hypoxia and normoxia environments for 24 h were collected, and the proteins were detected by western blot analysis. Notch1, Oct3/4 and Sox2 were identified in all groups, and the expression of Notch1 and Oct3/4 were higher in hypoxic cells compared with normoxic cells, while hypoxia did not increase Sox2 expression (Fig. 5B).
Discussion
Glioma is the most common type of primary tumor of the central nervous system (1) and recurrence, which is associated with the unlimited proliferation and invasive growth of tumor cells, is the greatest hindrance to successful treatment (2). Although differentiated tumor cells constituted the majority of the cell killed with the administration of current cancer treatments, a small amount of CSCs are resistant to the treatment (4,5). GSCs serve a very important role in the occurrence, development, invasion and recurrence of gliomas (6–9). GSCs have been demonstrated to cause tumors, maintain tumor growth, and are the root of tumor recurrence following surgery (6–8). In addition, there is a close association between tumor angiogenesis and resistance to chemotherapy or radiotherapy (10,11). Therefore, GSCs are a potential target for glioma therapy (31). Identifying a specific surface marker of CSCs is the key to further study of the occurrence, metastasis, recurrence and prognosis of the tumor. Multiple studies have revealed that CD133 is a specific surface marker of stem cells and CSCs (32–34). A hypoxic microenvironment is one of the important features of solid tumors as well as gliomas (35). Hypoxia can promote cell proliferation and angiogenesis in tumors, inhibit stem cell differentiation, increase tumor resistance to chemotherapy and radiotherapy, and make tumors more invasive (36).
The growth rate of tumors is usually greater than that of the blood vessels, which generates a hypoxic state inside the tumor (35). The hypoxic microenvironment has been revealed to be associated with the growth and metastasis of tumors (36). The results of the current study demonstrated that cell proliferation under hypoxia (1% O2) for >24 h was slightly higher compared with that under normoxia (20% O2), which suggested that the cell proliferation ability was increased following hypoxia induction. This paradoxical phenomenon may be attributed to changes in multiple signaling pathways that facilitate the de-differentiation and the acquisition of stem cell-like features in differentiated cancer cells (37). The ability of colony formation was also increased by hypoxia, which suggested that the microenvironment of hypoxia could promote stem cell characteristics in tumor cells. At the same time, it was demonstrated that CD133 expression in U87 cells increased 4–6-fold following hypoxia treatment, which is consistent with a previously reported 3–5-fold induction of CD133 positive fractions in glioblastoma-derived cells under hypoxia (38).
In embryonic stem cells, Oct3/4, Sox2 and other transcription factors may form a transcriptional network that regulates a large number of genes associated with differentiation of embryonic stem cell (39). Subtle changes in Oct3/4 protein expression have great influences on the differentiation of embryonic stem cells (40). Studies have demonstrated that Oct3/4 was only expressed in pluripotent cells, and that it maintained the pluripotency of embryonic stem cells, inhibited embryonic stem cell apoptosis and maintained their proliferative potential through the signal transduction pathway (40,41). The results of the current study supported the role of Oct3/4 in the proliferation of undifferentiated cells and stem cells that may be involved in tumor growth.
The Notch1 signaling pathway serves a major role in the maintenance of stem cell proliferation (42). In malignant tumors, the Notch1 signaling pathway is activated when the cells are transformed to malignant cells (43,44). The results of the current study demonstrated that the expression of Notch1 in U87 cells was affected by the hypoxia microenvironment. The Notch signaling pathway serves an important role in the self-renewal, proliferation and differentiation of neural stem cells or neural precursor cells (42). In addition, the Notch signaling pathway was also revealed to be highly activated in glioma, hematological and intestinal tumor, and other types of tumor, which suggested that the pathway can regulate the self-renewal and differentiation of CSCs, and serve an important role in malignant tumor formation and development (43–45). A previous study demonstrated that the Notch signaling pathway serves an essential role in hypoxia-mediated stem cell maintenance (37). Politi et al (44) revealed that the Notch signaling pathway is involved in the carcinogenesis of breast cancer, maintains the malignant phenotype of the mutant cells and is highly expressed in transplanted breast cancer tissue in nude mice. Jiang et al (45) demonstrated that compared with normal brain tissue, the expression of Notch1 was significantly higher in glioma tissues and increased with the grade of the glioma. Zhang et al (46) revealed that the activation of the Notch1 signaling pathway can stimulate the activation of RAC-α serine/threonine-protein kinase, activate the β-catenin and nuclear factor NF-κ-B signaling pathway, and promote the proliferation and invasion of glioma.
In the present study, it was demonstrated that in the hypoxic environment, Notch1 and its downstream target gene, Oct3/4, were markedly increased at mRNA and protein levels, while there appeared to be little change in Sox2 expression. These results indicated that hypoxia could promote the proliferation of GSCs and maintain the characteristics of stem cells by activating Notch1 and Oct3/4 or maintaining Notch1 activation.
Acknowledgements
Not applicable.
Funding
The present study was supported by Hubei Natural Science Foundation Project (grant no. 2016CFB575; Wuhan, China) and Wuhan University Renmin Hospital Guidance Fund (grant no. RMYD2018M09; Wuhan, China).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FZ designed the project, performed the assays and analyzed the data regarding U87 cell viability, cell apoptosis, CD 133 expression, and the Notch signaling pathway, and was a major contributor in writing the manuscript. HC performed the assays and analyzed the data regarding the U87 cell viability, colony formation, and Notch signaling pathway detection. ZZ, TY and GW performed the assays and analyzed the data regarding the U87 cell apoptosis, CD133 expression and Notch signaling pathway. QZ and SD contributed to the data acquisition and analysis regarding to the Notch signaling pathway detection. YZ was a major contributor to the project design, data analysis and manuscript editing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Davis ME. Glioblastoma: Overview of disease and treatment. Clin J Oncol Nurs. 2016;20(Suppl 5):S2–S8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. Central brain tumor registry of the united States (CBTRUS) statistical report: Primary brain and central nervous system tumors diagnosed in the united states in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 5.Soltanian S, Matin MM. Cancer stem cells and cancer therapy. Tumour Biol. 2011;32:425–440. doi: 10.1007/s13277-011-0155-8. [DOI] [PubMed] [Google Scholar]
- 6.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 7.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 8.Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 10.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–436. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 11.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 12.Costa FF, Seftor EA, Bischof JM, Kirschmann DA, Strizzi L, Arndt K, Bonaldo Mde F, Soares MB, Hendrix MJ. Epigenetically reprogramming metastatic tumor cells with an embryonic microenvironment. Epigenomics. 2009;1:387–398. doi: 10.2217/epi.09.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eales KL, Hollinshead KE, Tennant DA. Hypoxia and metabolic adaptation of cancer cells. Oncogenesis. 2016;5:e190. doi: 10.1038/oncsis.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen RL. Brain tumor hypoxia: Tumorigenesis, angiogenesis, imaging, pseudoprogression, and as a therapeutic target. J Neurooncol. 2009;92:317–335. doi: 10.1007/s11060-009-9827-2. [DOI] [PubMed] [Google Scholar]
- 15.Birner P, Piribauer M, Fischer I, Gatterbauer B, Marosi C, Ambros PF, Ambros IM, Bredel M, Oberhuber G, Rössler K, et al. Vascular patterns in glioblastoma influence clinical outcome and associate with variable expression of angiogenic proteins: Evidence for distinct angiogenic subtypes. Brain Pathol. 2003;13:133–143. doi: 10.1111/j.1750-3639.2003.tb00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Børresen-Dale AL, et al. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathornsumetee S, Cao Y, Marcello JE, Herndon JE, II, McLendon RE, Desjardins A, Friedman HS, Dewhirst MW, Vredenburgh JJ, Rich JN. Tumor angiogenic and hypoxic profiles predict radiographic response and survival in malignant astrocytoma patients treated with bevacizumab and irinotecan. J Clin Oncol. 2008;26:271–278. doi: 10.1200/JCO.2007.13.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL. Intratumoral hypoxia, radiation resistance, and HIF-1. Cancer Cell. 2004;5:405–406. doi: 10.1016/S1535-6108(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 20.Vaupel P, Mayer A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134:505–512. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez F, Huangfu D. Mechanisms underlying the formation of induced pluripotent stem cells. Wiley Interdiscip Rev Dev Biol. 2016;5:39–65. doi: 10.1002/wdev.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–673. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 24.Fazio C, Ricciardiello L. Inflammation and notch signaling: A crosstalk with opposite effects on tumorigenesis. Cell Death Dis. 2016;7:e2515. doi: 10.1038/cddis.2016.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jundt F, Pröbsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, Bargou RC, Manz R, Stein H, Dörken B. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103:3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- 26.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human ras-transformed cells. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 27.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amaya-Chanaga CI, Rassenti LZ. Biomarkers in chronic lymphocytic leukemia: Clinical applications and prognostic markers. Best Pract Res Clin Haematol. 2016;29:79–89. doi: 10.1016/j.beha.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Evers P, Lee PP, DeMarco J, Agazaryan N, Sayre JW, Selch M, Pajonk F. Irradiation of the potential cancer stem cell niches in the adult brain improves progression-free survival of patients with malignant glioma. BMC Cancer. 2010;10:384. doi: 10.1186/1471-2407-10-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- 33.Wu Y, Wu PY. CD133 as a marker for cancer stem cells: Progresses and concerns. Stem Cells Dev. 2009;18:1127–1134. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 34.Li Z. CD133: A stem cell biomarker and beyond. Exp Hematol Oncol. 2013;2:17. doi: 10.1186/2162-3619-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 36.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Eberhart, hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177:1491–1502. doi: 10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizzino A. Concise review: The Sox2-Oct4 connection: Critical players in a much larger interdependent network integrated at multiple levels. Stem Cells. 2013;31:1033–1039. doi: 10.1002/stem.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 41.Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: Evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 43.Li JL, Harris AL. Notch signaling from tumor cells: A new mechanism of angiogenesis. Cancer Cell. 2005;8:1–3. doi: 10.1016/j.ccr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 44.Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14:341–347. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Jiang L, Wu J, Chen Q, Hu X, Li W, Hu G. Notch1 expression is upregulated in glioma and is associated with tumor progression. J Clin Neurosci. 2011;18:387–390. doi: 10.1016/j.jocn.2010.07.131. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Chen T, Zhang J, Mao Q, Li S, Xiong W, Qiu Y, Xie Q, Ge J. Notch1 promotes glioma cell migration and invasion by stimulating β-catenin and NF-κB signaling via AKT activation. Cancer Sci. 2012;103:181–190. doi: 10.1111/j.1349-7006.2011.02154.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.