Abstract
Liver cancer cells often exhibit mesenchymal phenotypes, a critical phenotypic alteration of cancer cells termed the epithelial-mesenchymal transition (EMT). To examine whether liver kinase B1 (LKB1) serves a potential role in EMT in liver carcinogenesis, in the present study, it was determined that the expression of LKB1 decreased in the hepatocellular carcinoma (HCC) cell line, compared with a normal liver cell line. LKB1 overexpression decreased cell motility and invasiveness. Furthermore, the loss of LKB1 induced the expression of several EMT marker proteins, including that of Zinc Finger E-Box Binding Homeobox 1 (ZEB1). Notably, the expression of Yes-associated protein (YAP) was positively associated with that of ZEB1 in LKB1-knockdown cells with a mesenchymal phenotype. Here, we describe the direct regulation of the Hippo pathway effector YAP by ZEB1. The findings of the present study demonstrate that ZEB1 regulates the expression of YAP and regulates the expression of downstream target genes to promote malignant progression.
Keywords: liver kinase B1, epithelial-mesenchymal transition, zinc finger E-box binding homeobox 1, yes-associated protein, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common primary hepatic malignancy and is the third leading cause of cancer-associated mortality globally (1). HCC is a remarkably heterogeneous entity, at the molecular and clinical level, with a variety of pathogenetic mechanisms that have not been completely elucidated (2). Therefore, it is necessary to identify the underlying mechanisms driving HCC, in order to identify appropriate treatment targets.
Numerous genetic alterations result in the dysfunction of cell growth and migration pathways and have been demonstrated to be associated with HCC progression (3). An increasing number of studies demonstrated that liver kinase B1 (LKB1) loss and mutations are identified in various cancer types, including lung, mammary gland and ovarian cancer, melanoma and HCC (4–6). However, the mechanism by which LKB1 functions is not completely understood.
Epithelial-mesenchymal transition (EMT) is a key process for cancer invasion and metastasis (7). EMT is a process that is activated by EMT-inducing transcription factors such as SNAIL, Twist and Zinc Finger E-Box Binding Homeobox (ZEB). Previous studies have demonstrated the potency of ZEB1 (8,9). ZEB1 is associated with aggressive behavior, invasion, drug resistance and poor prognosis in various cancer types, including lung, pancreatic and breast cancer (7,10).
The Yes-associated protein (YAP) is a transcriptional factor of the Hippo signaling pathway (11). A number of studies have supported a critical role for YAP in different types of cancer, including breast and lung cancer and pancreatic ductal adenocarcinoma (PDAC) (12,13). YAP has been demonstrated to be an essential promoter of the mutant KRAS oncogenic program, specifically inducing the expression of secreted factors such as CTGF and CYR61, and interacting with FOS, a proto-oncogene, to regulate the expression of EMT-associated genes such as E-cadherin, SLUG, SNAIL and Vimentin (13,14). These pieces of evidence suggest that YAP serves an important role in EMT. Recent studies have reported that YAP is activated in LKB1-mutant tumors (15–19). Further functional studies have reported that YAP as an oncogenic driver is often hyper activated in a wide number of human cancer types, including HCC and melanoma (14,19,20). However, the role of YAP in HCC progression in the context of LKB1-deficient HCC remains elusive.
The present study demonstrated that the overexpression of LKB1 in the HCC cell line Hep3B leads to the decreased motility and invasiveness of cells. Functional evidence is presented that suggests ZEB1 is an important mediator of LKB1-loss-induced EMT progression. Further study indicated YAP as a downstream coactivator of ZEB1 in LKB1-deficient HCC malignant progression. The present study indicated ZEB1 as a direct mediator of YAP and determined a critical role of YAP in HCC.
Materials and methods
Plasmids
Plasmids were constructed using the following primers: LKB1 forward, 5′-GCGCTAGCATGGAGGTGGTGGACCCG-3′ and reverse, 5′-GCGCGGCCGCTCACTGCTGCTTGCAGGCC-3′; ZEB1 forward, 5′-GCGCTAGCATGGCGGATGGCCCCAGG-3′ and reverse, 5′-GCGCGGCCGCTTAGGCTTCATTTGTCTTTTCTTCAG-3′; The PCR product was purified and cloned into a pCDH lentivirus vector, and positive clones were sent for DNA sequencing to confirm there were no mutations. PCDH lentivirus vector was used as the empty vector control. For RNAi, the LKB1 RNAi sequence was as follows: 5′-UAGUUGAAUUUCCUUCUUCUU-3′; ZEB1 RNAi, 5′-GGUAGAUGGUAAUGUAAUA-3′; Scramble sequence, 5′-CATGATGATGGGCTCCATA-3′. For luciferase activity assay, the promoter (−2,000+ 5′UTR) of YAP was cloned into a pGL3-Basic vector (Promega Corporation, Madison, WI, USA). pGL3-SV40-Renilla plasmid (Promega Corporation) was used as the internal control.
Cell culture
The CRL-11233 cell line (American Type Culture Collection, Manassas, VA, USA) was cultured in bronchial epithelia growth medium (BEGM) supplemented with a BEGM Bullet kit (Lonza Group, Ltd., Basel, Switzerland). The human hepatocellular carcinoma cell line Hep3B (American Type Culture Collection) was cultured in Eagle's minimum essential medium (EMEM cat, no. 30-2003; American Type Culture Collection) +2 mM glutamine +1% Non-Essential Amino Acids (NEAA) +10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cultures were maintained in a humidified chamber at 5% CO2 and 37°C.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA of CRL-11233 and Hep3B cells treated as indicated was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). First strand cDNA was synthesized using a Reverse Transcription kit (iScript™ Reverse Transcription Supermix for RT-qPCR; Bio-Rad Laboratories, Inc., Hercules, CA, USA), according to the manufacturer's protocols. Gene expression of cells treated as indicated was detected with a QPCR kit (iQ™ SYBR®-Green Supermix; Bio-Rad Laboratories, Inc.), according to the manufacturer's protocols. All qPCR was performed at 95°C for 3 min, and then 40 cycles of 95°C for 15 sec and 60°C for 1 min using the CFX 96 realtime PCR machine (Bio-Rad Laboratories, Inc.). The specificity of the reaction was verified by melt curve analysis (21). The relative quantitation of each gene was performed using the comparative ∆∆Cq method (21). Primer sequences were as follows:
LKB1 forward, 5′-AGTCCAACATCACCATGCAG-3′ and reverse, 5′-TTCCCTTTCCTCGAACTGATTT-3′; SNAIL forward, 5′-ACAAGCACCAAGAGTCCG-3′ and reverse, 5′-ATGGCAGTGAGAAGGATGTG-3′; SLUG forward, 5′-ACTGCTCCAAAACCTTCTCC-3′ and reverse, 5′-TGTCATTTGGCTTCGGAGTG-3′; TWIST1 forward, 5′-CTCAGCTACGCCTTCTCG-3′ and reverse, 5′-ACTGTCCATTTTCTCCTTCTCTG-3′; ZEB1 forward, 5′-ACCCTTGAAAGTGATCCAGC-3′ and reverse, 5′-CATTCCATTTTCTGTCTTCCGC-3′; ZEB2 forward, 5′-GCCATCTGATCCGCTCTTATC-3′ and reverse, 5′-ACCTGTGTCCACTACATTGTC-3′; AXL forward, 5′-TTTATGACTATCTGCGCCAGG-3′ and reverse, 5′-TGTGTTCTCCAAATCTTCCCG-3′; CTGF forward, 5′-ACCAATGACAACGCCTCC-3′ and reverse, 5′-TTGGAGATTTTGGGAGTACGG-3′; SDPR forward, 5′-TCAAAGAGCGCATGGATAGG-3′ and reverse, 5′-TGGCAGGGATCTCATTTTCC-3′; YAP forward, 5′-AAGCTGCCCGACTCCTTCTTCAAG-3′ and reverse, 5′-TGAGCTCGAACATGCTGTGGAGTCAG-3′; GAPDH forward, 5′-AGGTGAAGGTCGGA-3′ and reverse, 5′-TTGAGGTCAATGAAG-3′.
Western blot analysis
CRL-11233 and Hep3B cells were washed in PBS and lysed in radioimmunoprecipitation assay buffer [25 mM Tris (pH 7.4), 150 mM NaCl, 0.5% sodium deoxycholate and 1% Triton X-100] with the addition of proteinase inhibitors (Pierce; Thermo Fisher Scientific, Inc.). The protein concentration determination was performed using bicinchoninic acid method (Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. Samples (30 µg) were loaded per well. Polyacrylamide gels (4–12%) were used and the proteins in the gels were transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked with 5% non-fat milk in TBS with 0.05% Tween-20 (TBST) at room temperature for 1 h, and the indicated primary antibodies were incubated at 4°C overnight whilst shaken. Membranes were washed in TBST for 10 min 3 times and incubated with secondary antibody at room temperature for 1 h, and then the membranes were washed in TBST for 10 min 3 times. The blots were detected using Pierce™ ECL Western Blotting Substrate (cat. no. 32106, Thermo Fisher Scientific, Inc.). Antibodies were as follows: Rabbit anti-LKB1 (ab199970; Abcam, Cambridge, UK), rabbit anti-ZEB1 (HPA027524; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), mouse anti-YAP (sc-101199), goat anti-CTGF (sc-14939), rabbit anti-ZEB2 (sc-48789), β-actin (sc-47778; all Santa Cruz Technology, Inc., Dallas, TX, USA), rabbit anti-AXL (cat. no. 8661), rabbit anti-SNAIL (cat. no. 3879), rabbit anti-SLUG (cat. no. 9585) and rabbit anti-Twist (cat. no. 46702; all Cell Signaling Technology, Inc., Danvers, MA, USA). The β-actin signal was used as a loading control. Secondary Antibodies were: Donkey anti-rabbit, horseradish peroxidase (HRP)-conjugate (sc-2313); donkey anti-mouse, HRP-conjugate (sc-2314); and donkey anti-goat, HRP-conjugate (sc-2020) (1:5,000; Santa Cruz Technology, Inc.). The densitometric analysis was conducted with ImageJ software (ImageJ bundled with 64-bit Java 1.8.0_112; National Institutes of Health, Bethesda, MD, USA).
In vitro Transwell™ assay
CRL-11233 and Hep3B cells were starved overnight and then seeded (1×105 cells/well) in upper 8-µm-pore membrane chambers coated with (invasion assay) or without (migration assay) Matrigel® (BD Biosciences, Franklin Lakes, NJ, USA). The complete medium (BEGM supplemented with a BEGM Bullet kit for CRL-11233 cells and EMEM +2 mM glutamine +1% NEAA +10% FBS for Hep3B cells, respectively) was loaded in the lower chamber. After 16 h of incubation in a humidified chamber at 5% CO2 and 37°C, cells that passed through the membrane were fixed in 4% paraformaldehyde at 4°C for 15 min, washed once with PBS, stained with 1% crystal violet at room temperature for 20 min, washed with PBS three times, and then the cell invasion was quantified by counting the number of cells passing through the pores from five different fields per sample at magnification, ×100 under a light microscope selected in a random manner.
Luciferase reporter assay
CRL-11233 and Hep3B cells (1×105 cells/well) were seeded in 24 wells overnight and transfected with indicated amounts of plasmids encoding ZEB1 or the corresponding empty vector controls using FuGENE HD transfection reagent (Promega Corporation), according to the manufacturer's protocol. Cells were collected after 48 h and luciferase activity was measured using a Dual-Luciferase Reporter assay system (Promega Corporation) according to the manufacturer's instruction. Relative expression values were normalized to the respective values of Renilla.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). For multiple comparisons, one-way ANOVA with Bonferroni's post-hoc test was applied. Data are presented as the mean ± standard deviation of the mean (SEM). All experiments were repeated at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
HCC cells show LKB1 downregulation and EMT progression
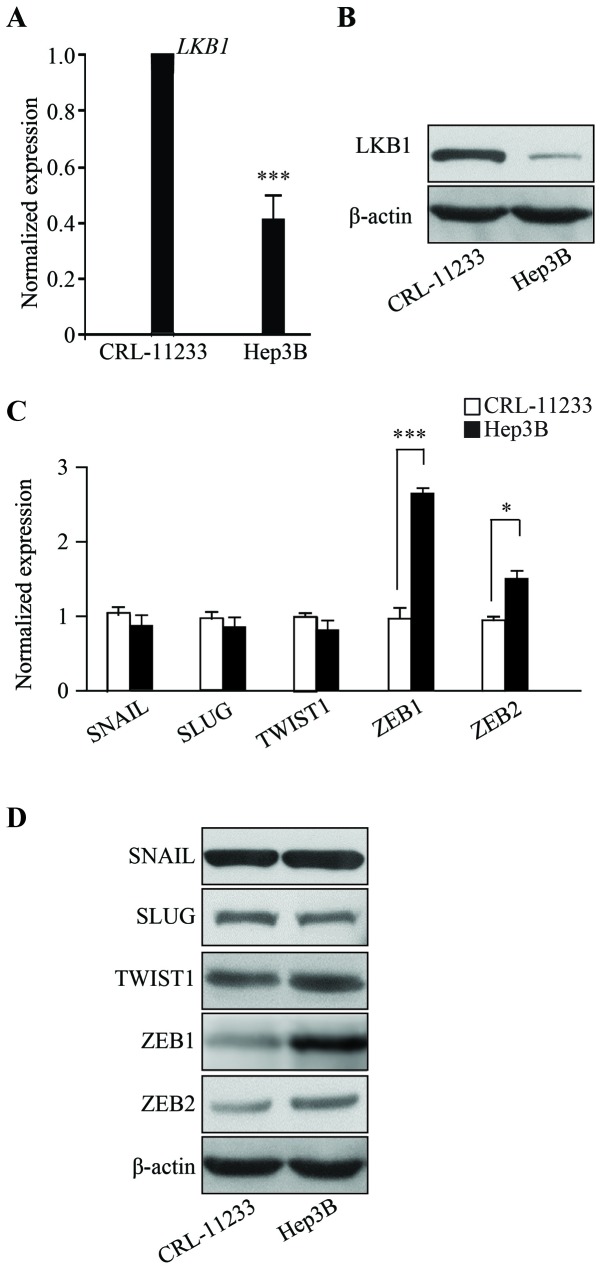
To characterize HCC, the present study examined the expression level of LKB1. RNAs from human the HCC line Hep3B were isolated and RT-qPCR was performed which demonstrated that, compared with the non-malignant normal liver cell line CRL-11233, the mRNA expression of LKB1 in Hep3B cells is significantly downregulated (Fig. 1A) (P<0.001). Consistent with the RT-qPCR data, LKB1 protein levels are decreased in Hep3B cells (Fig. 1B). The RT-qPCR and western blotting results demonstrated that the expression of EMT markers, namely SNAIL, SLUG, TWIST1, ZEB1 and ZEB2, are upregulated in Hep3B cells, when compared with the normal liver cell line CRL-11233 (Fig. 1C and D). These results demonstrated that LKB1 is downregulated in HCC and suggest that EMT may be involved in the progression of HCC.
Figure 1.
HCC cells exhibit LKB1 downregulation and EMT progression. Expression of LKB1 was demonstrated by (A) RT-qPCR and (B) western blot analysis. Expression of EMT markers was demonstrated by (C) RT-qPCR and (D) western blot analysis. *P<0.05, ***P<0.001. HCC, hepatocellular carcinoma; EMT, epithelial-mesenchymal transition; LKB1, liver kinase B1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SNAIL, Snail Family Transcriptional Repressor 1; SLUG, Snail family transcriptional repressor 2.
LKB1 downregulates the expression of the EMT marker ZEB1 in Hep3B cells
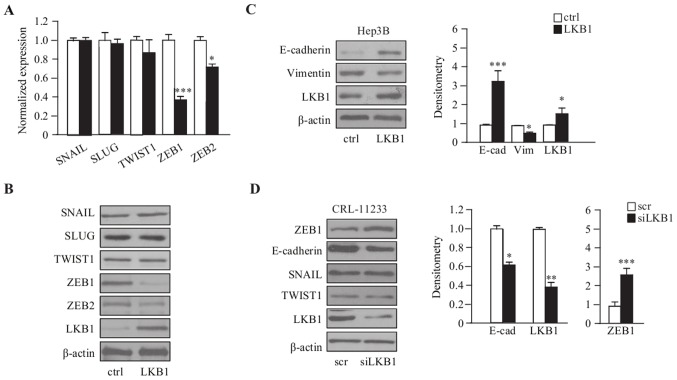
To illustrate the correlation of EMT and LKB1, LKB1 was transiently overexpressed in Hep3B cells and the expression of EMT markers examined; for SNAIL, SLUG, TWIST1, ZEB1 and ZEB2, the RT-qPCR and immunoblot results demonstrated that the expression of ZEB1 and ZEB2 were significantly downregulated in LKB1-overexpressing Hep3B cells, compared with the empty vector control cells (Fig. 2A and B). Consistently, the epithelial marker E-cadherin was upregulated, and the mesenchymal marker Vimentin was downregulated, as demonstrated by western blot analysis (Fig. 2C). Furthermore, LKB1 depletion by siRNA in the normal liver cell line CRL-11233, exhibited an upregulation of ZEB1; however, this was not observed with additional EMT markers such as SNAIL and TWIST1, and the downregulation of E-cadherin (Fig. 2D). Taken together, these results suggest that LKB1 primarily regulates the expression of ZEB1 in order to regulate EMT process in HCC cells.
Figure 2.
LKB1 downregulates ZEB1 expression in Hep3B cells. (A) RT-qPCR and (B) western blotting (B) results demonstrated that overexpression of LKB1 inhibited the expression of ZEB1 but no other EMT-inducing genes in Hep3B cells. (C) Western blot analysis demonstrated that overexpression of LKB1 increased the expression of E-cadherin and decreased the expression of vimentin in Hep3B cells. (D) Western blot analysis demonstrated that the knockdown of LKB1 increased the expression of ZEB1 and decreased the expression of E-cadherin in CRL-11233 cells. *P<0.05; **P<0.01; ***P<0.001. LKB1, liver kinase B1; EMT, epithelial-mesenchymal transition; ZEB1, Zinc Finger E-Box Binding Homeobox 1; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
EMT induced by LKB1 loss promotes HCC migration and invasion through ZEB1
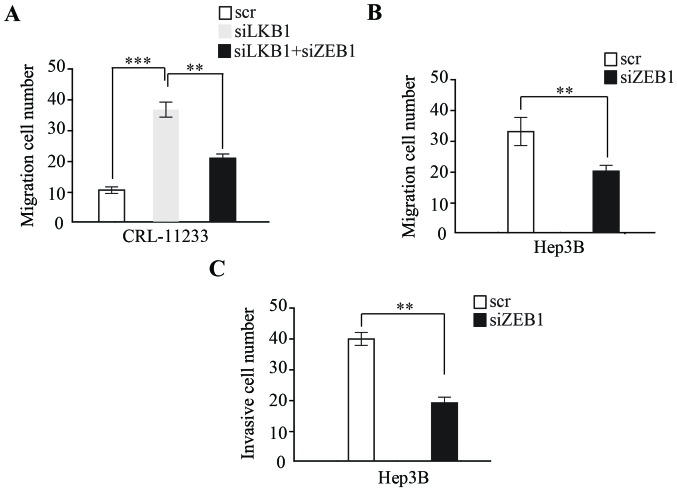
To examine whether EMT induced by LKB1 regulates HCC migration and invasion, the present study examined LKB1-depleted CRL-11233 cells, which demonstrated a high migration capability; additionally, the depletion of ZEB1 by siRNA significantly inhibited the migration capability of CRL-11233 cells (Fig. 3A). Furthermore, the expression of ZEB1 by siRNA was depleted in Hep3B cells, with a migration and invasion assay demonstrating that, compared with a scramble RNA control, ZEB1 knockdown significantly inhibited the migration and invasion capability of HCC cells (Fig. 3B and C).
Figure 3.
EMT induced by LKB1 loss promotes HCC migration and invasion through ZEB1. (A) Knockdown of LKB1 promotes the motility of CRL-11233 cells compared with the scramble control RNA. Knockdown of ZEB1 in LKB1 depleted CRL-11233 cells partially inhibits motility of CRL-11233. (B) Knockdown of ZEB1 inhibits the migration of Hep3B cells. (C) Knockdown of ZEB1 inhibits the invasiveness of Hep3B cells. Data are expressed as the mean numbers of independent triplicate experiments. ***P<0.001; **P<0.01. scr, scramble; ZEB1, Zinc Finger E-Box Binding Homeobox 1; LKB1, liver kinase B1; si, small interfering.
ZEB1 regulates the expression of YAP to promote YAP signaling
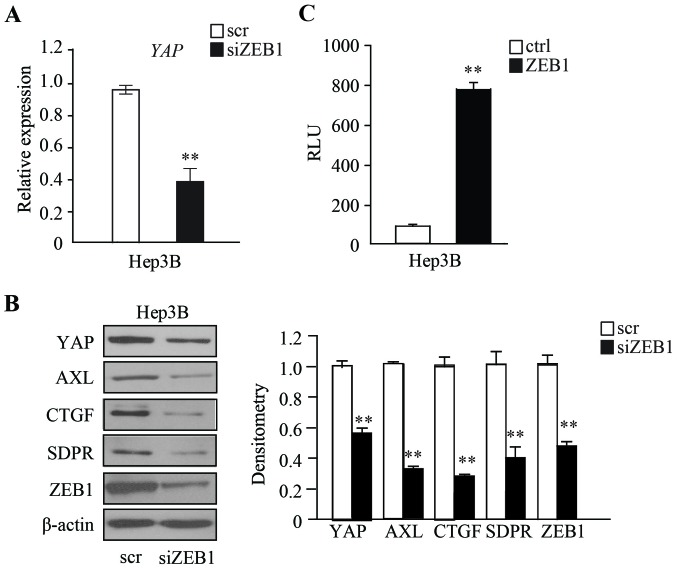
The present study then attempted to further determine the mechanism by which ZEB1 functions in HCC cells. It has been reported that upregulation of Hippo signaling regulates tumor progression in various cancers, such as NSCLC, breast, pancreatic and colon cancer (12). In order to examine whether ZEB1 regulates the expression of YAP, the expression of ZEB1 was transiently depleted in Hep3B cells and the expression of YAP was examined. The RT-qPCR and western blot analysis results demonstrated that the expression of YAP is significantly downregulated in ZEB1-depleted Hep3B cells, compared with scramble RNA control cells (Fig. 4A and B). YAP and ZEB1 have been reported to co-localize in the nucleus (10); therefore, the present study investigated whether the expression of YAP is directly regulated by ZEB1, by performing a luciferase reporter assay. The promoter and 5′UTR region of YAP were cloned into the promoter region of luciferase genes using a pGL3-Basic plasmid. The plasmid was then co-transfected with ZEB1-expressing plasmids into Hep3B cells, and the luciferase reporter assay was performed after 48 h. Relative Luminometer Units (RLU) results demonstrated that Hep3B cells expressing ZEB1 had a markedly higher RLU when compared with cells transfected with an empty vector, indicating that ZEB1 could directly bind to the promoter and 5′UTR region of YAP gene (**P<0.01) (Fig. 4C). The expressions of downstream target genes that are regulated by YAP were then examined. Western blot analysis demonstrated that AXL, CTGF, and SDPR are downregulated in ZEB1-depleted Hep3B cells, compared with in scramble RNA control cells (Fig. 4B).
Figure 4.
ZEB1 regulates the expression of YAP to promote YAP signaling. (A) RT-qPCR showed the effect of ZEB1 on YAP expression. Data are expressed as the mean numbers of independent triplicate experiments. **P<0.01. (B) Western blotting results demonstrated that knockdown of ZEB1 inhibited the expression of YAP and YAP target genes in the Hep3B cell line. (C) A Luciferase reporter assay demonstrated that ZEB1 expression increased the reporter activity of the YAP promoter in Hep3B cells. Data are expressed as the mean numbers of independent triplicate experiments. **P<0.01. ZEB1, Zinc Finger E-Box Binding Homeobox 1; YAP, Yes-associated protein; RT-qPCR, reverse transcription quantitative polymerase chain reaction; scr, scramble; si, small interfering.
Taken together, these results demonstrated that the loss of LKB1 promotes the expression of EMT marker ZEB1, which in turn promotes EMT and the Hippo signaling pathway by interacting with YAP, resulting in HCC progression.
Discussion
The results of the present study demonstrate that ZEB1, an activator of EMT induced by LKB1-loss, promotes HCC progression by activating YAP signaling. LKB1 is a frequently mutated tumor suppressor in HCC (22). It is indicated that LKB1 may regulate cell growth, metabolism, polarity and mobility. The loss of LKB1 contributes to a more aggressive phenotype in tumor cells, including EMT (14). LKB1 inactivation triggers EMT in lung cancer cells through the induction of ZEB1 (4).
ZEB1 is a tumor-specific molecule, and the elevated expression of ZEB1 has been observed in numerous types of cancer, including lung, breast and pancreatic head cancer (23,24). ZEB1 has been reported as a LKB1 target, due to a marked increase in ZEB1 levels observed upon LKB1 depletion in mice (24,25).
A previous study demonstrated that deletion of YAP in liver cells suppresses hepatomegaly and hepatocyte hyperplasia induced by acute deletion of LKB1 in mice (26). YAP is a well-studied oncogene in the liver that drives liver enlargement as well as liver tumor formation; a previous study revealing that ectopic expression of YAP resulted in accelerated liver tumor progression (14). In addition, depletion of YAP attenuates the metastasis potential of cells (27). The results of the present study demonstrated that YAP activation promotes liver cancer progression. Notably, ZEB1 was identified as a crucial regulator of YAP in LKB1-deficient HCC progression; however, the present study only examined the function of ZEB1 and YAP in vitro, and future studies should use a mouse model is required in order to characterize the function and association of ZEB1 with HCC in vivo.
In conclusion, the present study determined a functional link between YAP and ZEB1 in the context of LKB1-deficient HepB3 cells. Therefore, these findings demonstrated that ZEB1 not only regulates the expression of YAP, but also serves as a co-activator of YAP in regulating the expression of downstream target genes to promote malignant progression. Furthermore, considering the prevalence of LKB1 mutations in various cancer types, the data may also suggest a novel target for the treatment of other LKB1-deficient cancer cases.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BQ conducted the experiment and collected the data, WW designed the project. JZ, GF and DL analyzed the data and drafted the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2011;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy BC, Kohno T, Iwakawa R, Moriguchi T, Kiyono T, Morishita K, Sanchez-Cespedes M, Akiyama T, Yokota J. Involvement of LKB1 in epithelial-mesenchymal transition (EMT) of human lung cancer cells. Lung Cancer. 2010;70:136–145. doi: 10.1016/j.lungcan.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Rhodes LV, Tate CR, Hoang VT, Burks HE, Gilliam D, Martin EC, Elliott S, Miller DB, Buechlein A, Rusch D, et al. Regulation of triple-negative breast cancer cell metastasis by the tumor-suppressor liver kinase B1. Oncogenesis. 2015;4:e168. doi: 10.1038/oncsis.2015.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: Control of metabolism and cell growth. Future Oncol. 2010;6:457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez-Tilló E, Siles L, de Barrios O, Cuatrecasas M, Vaquero EC, Castells A, Postigo A. Expanding roles of ZEB factors in tumorigenesis and tumor progression. Am J Cancer Res. 2011;1:897–912. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, Sun Y, Ma L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann W, Mossmann D, Kleemann J, Mock K, Meisinger C, Brummer T, Herr R, Brabletz S, Stemmler MP, Brabletz T. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. Nat Commun. 2016;7:10498. doi: 10.1038/ncomms10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 13.Thongon N, Castiglioni I, Zucal C, Latorre E, D'Agostino V, Bauer I, Pancher M, Ballestrero A, Feldmann G, Nencioni A, et al. The GSK3β inhibitor BIS I reverts YAP-dependent EMT signature in PDAC cell lines by decreasing SMADs expression level. Oncotarget. 2016;7:26551–26566. doi: 10.18632/oncotarget.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohseni M, Sun J, Lau A, Curtis S, Goldsmith J, Fox VL, Wei C, Frazier M, Samson O, Wong KK, et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014;16:108–117. doi: 10.1038/ncb2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q, Li J, Sun S, Chen X, Zhang H, Li B, Sun S. YAP/TAZ-mediated activation of serine metabolism and methylation regulation is critical for LKB1-deficient breast cancer progression. Biosci Rep. 2017;37:BSR20171072. doi: 10.1042/BSR20171072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Gao Y, Li F, Tong X, Ren Y, Han X, Yao S, Long F, Yang Z, Fan H, et al. YAP promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of survivin. Cancer Res. 2015;75:4450–4457. doi: 10.1158/0008-5472.CAN-14-3396. [DOI] [PubMed] [Google Scholar]
- 18.Gao Y, Zhang W, Han X, Li F, Wang X, Wang R, Fang Z, Tong X, Yao S, Li F, et al. YAP inhibits squamous transdifferentiation of Lkb1-deficient lung adenocarcinoma through ZEB2-dependent DNp63 repression. Nat Commun. 2014;5:4629. doi: 10.1038/ncomms5629. [DOI] [PubMed] [Google Scholar]
- 19.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher ML, Grun D, Adhikary G, Xu W, Eckert RL. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget. 2017;8:110257–110272. doi: 10.18632/oncotarget.22628. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Barbier-Torres L, Delgado TC, García-Rodríguez JL, Zubiete-Franco I, Fernández-Ramos D, Buqué X, Cano A, Gutiérrez-de Juan V, Fernández-Domínguez I, Lopitz-Otsoa F, et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 2015;6:2509–2523. doi: 10.18632/oncotarget.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Lu X, Huang L, Wang W, Jiang G, Dean KC, Clem B, Telang S, Jenson AB, Cuatrecasas M, et al. Different thresholds of ZEB1 are required for Ras-mediated tumour initiation and metastasis. Nat Commun. 2014;5:5660. doi: 10.1038/ncomms6660. [DOI] [PubMed] [Google Scholar]
- 24.Bronsert P, Kohler I, Timme S, Kiefer S, Werner M, Schilling O, Vashist Y, Makowiec F, Brabletz T, Hopt UT, et al. Prognostic significance of Zinc finger E-box binding homeobox 1 (ZEB1) expression in cancer cells and cancer-associated fibroblasts in pancreatic head cancer. Surgery. 2014;156:97–108. doi: 10.1016/j.surg.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 25.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]
- 26.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J, Wu Y, Yang L, Du J, Gong K, Chen W, Dai J, Li X, Xi S. Repression of YAP by NCTD disrupts NSCLC progression. Oncotarget. 2017;8:2307–2319. doi: 10.18632/oncotarget.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.