Abstract
Correlation between microRNA (miRNA)-206 and miRNA-145 expression and prognosis in breast cancer was investigated. Breast cancer specimens and paracancerous tissues of 372 breast cancer patients who underwent surgical resection in the First Affiliated Hospital of Shantou University Medical College from September 2010 to September 2014 were included. qRT-PCR was used to detect the expression of miR-206 and miR-145 in breast cancer and paracancerous tissues, and patients were divided into high and low expression groups according to the median expression level to plot survival curve. Expression levels of miR-145 and miR-206 in breast cancer tissues were 2.24±1.23 and 0.76±0.24, respectively. Expression level of miR-145 was significantly lower, while expression level of miR-206 was significantly higher in tumor tissues than in paracancerous tissues (p<0.05). The 3-year survival rates of miR-145 low expression group and miR-206 high expression group were also lower than that of miR-145 high expression group and miR-206 low expression group, respectively (p<0.05). Expression of miR-206 is upregulated and expression of miR-145 is downregulated in breast cancer, which may have an impact on the prognosis of patients. miR-206 and miR-145 may serve as important indicators to predict prognosis of patients with breast cancer in the future.
Keywords: breast cancer, miR-206, miR-145, prognosis
Introduction
Breast cancer is one of the most common malignant tumors in women, accounting for 8–12% of all malignancies (1). Coates et al (2) showed that in 2015 breast cancer affected approximately 1.4 million new cases, and the incidence is rising. Breast cancer frequently occurs in developed countries in Europe and North America, and incidence is highest in the United States (3). Turner et al (4) predict that the incidence of breast cancer will exceed 50% within the next 50 years and will become the second most common malignant tumor after gastric cancer. In addition, breast cancer at early stage usually shows no obvious symptoms, and can be easily ignored, leading to the high mortality rate. Hindié and Groheux (5) showed that the 5-year survival rate of breast cancer patients was only 62.4%. Because of its high incidence and mortality, breast cancer has long been a hot clinical research topic. MicroRNAs (miRNAs) have been proven to participate in the development of many types of tumors (6–8). Among them, miRNA-206 and miRNA-145 were proved by Sun et al (9) to be associated with female ovarian cancer. Therefore, we speculate that miRNA-206 and miRNA-145 may also show unique expression in breast cancer. Our study investigated the application values of miRNA-206 and miRNA-145 as prognostic or therapeutic indicators for breast cancer.
Patients and methods
Patient data
Breast cancer specimens and paracancerous tissues (within 5 cm around the tumor) of 372 breast cancer patients who underwent surgical resection in the First Affiliated Hospital of Shantou University Medical College (Shantou, China) from September 2010 to September 2014 were included. Patients were aged 30–75 years with an average age of 45.32±7.21 years (Table I). Pathological classification and staging were based on the 2007 International Breast Cancer Typing Guidelines (10).
Table I.
Basic information of the patients.
| Cases (n=372) | No. | % |
|---|---|---|
| Age (years) | ||
| <50 | 152 | 40.9 |
| ≥50 | 220 | 59.1 |
| Body weight (kg) | ||
| <60 | 164 | 44.1 |
| ≥60 | 208 | 55.9 |
| Residence | ||
| City | 254 | 68.3 |
| Countryside | 118 | 31.7 |
| Ethnicity | ||
| Han nationality | 364 | 97.8 |
| Minority | 8 | 2.2 |
| Types | ||
| Non-invasive cancer | 97 | 26.1 |
| Early invasive cancer | 134 | 36.0 |
| Special type invasive cancer | 51 | 13.7 |
| Non-special type invasive cancer | 90 | 24.2 |
| Pathological staging | ||
| I | 46 | 12.4 |
| II | 96 | 25.8 |
| III | 136 | 36.6 |
| IV | 94 | 25.3 |
| T staging | ||
| Tis | 35 | 9.4 |
| T1 | 94 | 25.3 |
| T2 | 127 | 34.1 |
| T3 | 62 | 16.7 |
| T4 | 54 | 14.5 |
| N staging | ||
| N1 | 86 | 23.1 |
| N2 | 174 | 46.8 |
| N3 | 112 | 30.1 |
| Distant metastasis | ||
| Yes | 243 | 65.3 |
| No | 129 | 34.7 |
Inclusion and exclusion criteria
Inclusion criteria were: Patients confirmed with breast cancer by pathological biopsy in the First Affiliated Hospital of Shantou University Medical College. Cancerous tissue was placed in liquid nitrogen and stored at −80°C immediately after surgical resection. Before the operation, patients were not treated with radiotherapy or chemotherapy and patients with complete medical record. Exclusion criteria were: Combined with other cardiovascular and cerebrovascular diseases, respiratory tract and gastrointestinal disease patients, pregnant women, long-term bedridden patients, patients with physical disabilities, surgery-intolerant patients, patients transferred to other hospitals during treatment and patients who received unauthorized treatment. The study was approved by the Ethics Committee of the First Affiliated Hospital of Shantou University Medical College. Signed informed consents were obtained from the patients or guardians.
Main instruments and reagents
LightCycler real-time PCR instrument (Roche Diagnostics, Basel, Switzerland), total RNA extraction TRIzol kit (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA), M-MLV reverse transcriptase kit was from Vazyme Biotech Co., Ltd., (Nanjing, China). miR-206, miR-145 and real-time PCR kit from Biomiga China (Shanghai, China). Primers of miR-206, miR-145 and U6 (endogenous control) used in PCR reaction were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Table II).
Table II.
Primers of miR-206, miR-145 and U6.
| Primer sequences | ||
|---|---|---|
| miR-206 | F | 5′-ATCCAGTGCGTGTCGTG-3′ |
| R | 5′-TGCTTGGAATGTAAGGAAG-3′ | |
| miR-145 | F | 5′-ACACTCCAGCTGGGCAGGTCAAAAGGGTCC-3′ |
| R | 5′-GGTGTCGTGGAGTCG-3′ | |
| U6 | F | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| R | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
F, forward; R, reverse.
Detection methods
Breast cancer tissue (80 mg) was ground in liquid nitrogen. TRIzol reagent was added and mixed, and the mixture was kept at room temperature for 30 min. Total RNA was extracted in strict accordance with the manufacturer's instructions. The extracted RNA was tested by ultraviolet spectrophotometer (Bio Rad, Hercules, CA, USA) and electrophoresis to determine the concentration and purity. Total RNA was then reverse-transcribed according to the instructions of reverse transcription kit, and cDNA samples were stored at −20°C. PCR reaction system was prepared according to the manufacturer's instructions (10.5 µl), and DEPC water was added to make a 20 µl volume. PCR reaction conditions: 94°C for 10 min, followed by 40 cycles of 94°C for 45 sec, 60°C for 45 sec and 72°C for 45 sec. Data were analyzed using the software provided by the manufacturer by 2−ΔΔCq method (11). U6 was used as endogenous control. The average of three replicates was used as the final result.
Statistical analysis
SPSS 22.0 statistical software was used for data analysis. Measurement data are expressed as mean × standard deviation (SD), comparisons between two groups was performed by t-test. Enumeration data were expressed as rate. Survival curves were plotted using Kaplan-Meier method and compared by log-rank test. P<0.05 indicated that the difference was statistically significant.
Results
miR-145 expression
Expression level of miR-145 in breast cancer tissues was 2.24±1.23, and in paracancerous tissues was 6.24±1.51. Expression level of miR-145 in breast cancer tissues was significantly lower than that in paracancerous tissues (t=39.61, p<0.001) (Fig. 1).
Figure 1.
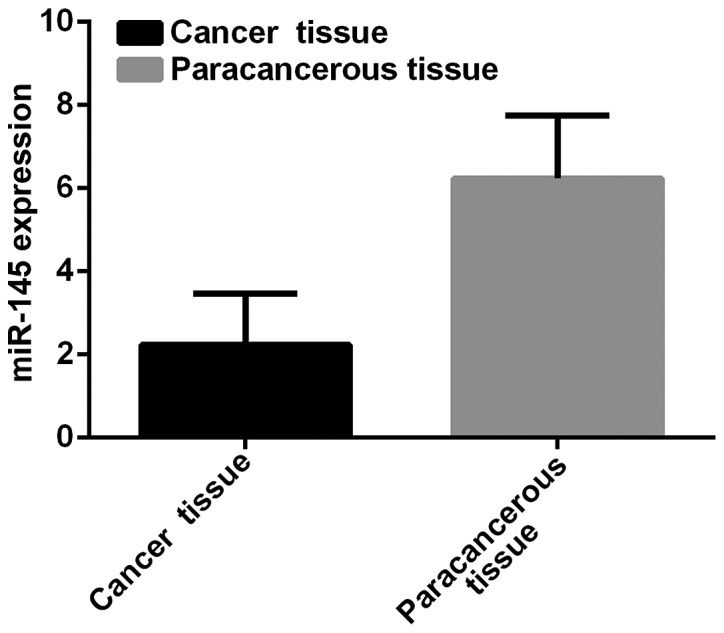
miR-145 expression in breast cancer and paracancerous tissues. Expression level of miR-145 in breast cancer tissues was 2.24±1.23 and that in paracancerous tissues was 6.24±1.51.
miR-206 expression
Expression level of miR-206 in breast cancer tissues was 0.76±0.24, and in paracancerous tissues was 0.12±0.08. Expression level of miR-206 in breast cancer tissues was significantly higher than that in paracancerous tissues (t=48.79, p<0.001) (Fig. 2).
Figure 2.
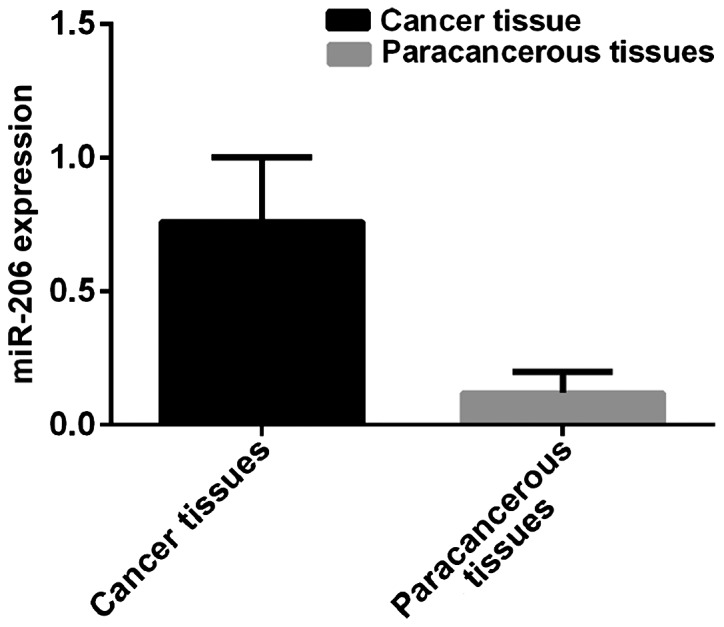
miR-206 expression in breast cancer and paracancerous tissues. Expression level of miR-206 in breast cancer tissues was 0.76±0.24 and that in paracancerous tissues was 0.12±0.08.
Prognosis of patients
According to the median value of miR-145 and miR-206 expression, patients were divided into miR-145 high expression group (≥2.24, 219 cases), miR-145 low expression group (<2.24, 153 cases), miR-206 high expression group (≥0.76, 194 cases) and miR-206 low expression group (<0.76, 178 cases). Patients were followed up for 3 years by telephone, review and mail. Follow-up was performed until December 2017 or death of the patients. A total of 354 patients finished the follow-up, and follow-up success rate was 95.2%. Survival rates at 1, 2 and 3 years in miR-145 low expression group were 83.1, 71.2 and 59.8%, respectively, while survival rates at 1, 2 and 3 years in miR-145 high expression group were 89.5, 79.1 and 70.6%, which were significantly better than those in miR-145 low expression group (p=0.028). Survival rates at 1, 2 and 3 years in miR-206 high expression group were 77.3, 67.5 and 55.7%, respectively, while survival rates at 1, 2 and 3 years in miR-206 low expression group were 89.9, 81.5 and 75.8%, respectively, which were significantly higher than those in miR-206 high expression group (p=0.034) (Figs. 3 and 4).
Figure 3.
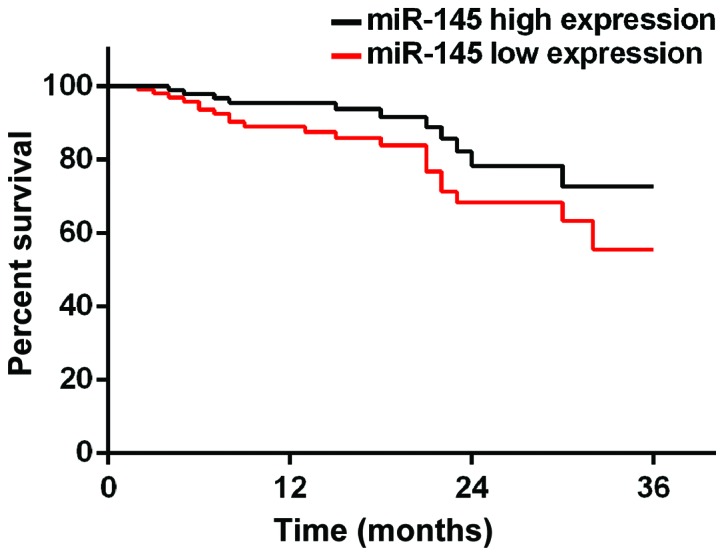
Three-year survival curve of patients with high and low expression level of miR-145. Survival rates at 1, 2 and 3 years in miR-145 low expression group were 83.1, 71.2 and 59.8%, respectively. Survival rates at 1, 2 and 3 years in miR-145 high expression group were 89.5, 79.1 and 70.6%, respectively.
Figure 4.
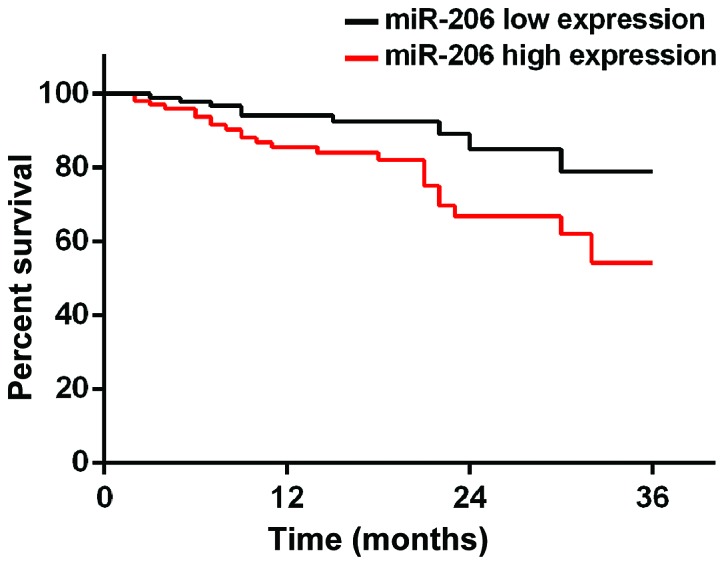
Three-year survival curve of patients with high and low expression level of miR-206. Survival rates at 1, 2 and 3 years in miR-206 high expression group were 77.3, 67.5 and 55.7%, respectively. Survival rates at 1, 2 and 3 years in miR-206 low expression group were 89.9, 81.5 and 75.8%, respectively.
Discussion
Breast cancer is a malignant tumor that seriously affects life and health of females. Incidence and mortality of breast cancer rank in the top area among all malignancies (12). Breast cancer at early stage shows no obvious symptoms and most patients are diagnosed at advanced stages, and thus missing the best treatment time, leading to poor prognosis. At present, pathogenesis of breast cancer is still unclear. Poortmans et al (13) believed that the occurrence of breast cancer is mainly caused by genetic factors, while Tutt et al (14) showed that the occurrence of breast cancer is closely related to cancer stem cells. miRNAs as a group of endogenous non-protein-coding RNA (15,16) have been proved to be directly involved in tumorigenesis and development. Most miRNAs are highly conserved, cell-specific, and have a strong ability to regulate cell proliferation and apoptosis (17). Among them, miR-206 plays a key role in the regulation of cell proliferation, apoptosis, invasion and migration. miR-206 may play a role as a tumor suppressor gene and may also have oncogenic functions and has been proven to promote muscle differentiation by downregulating the P180 subunit of DNA polymerase and muscle transcription factors (18). miR-145 is expressed in many eukaryotic organisms and plays a role in regulating gene expression and has multiple targets that are associated with oncogenes (19). With the deepening of research, miR-206 and miR-145 have been proven to be closely correlated with breast cancer. Therefore, in this study, expression levels of miR-206 and miR-145 in breast cancer and paracancerous tissues were measured, and the correlation with prognosis were analyzed with an expectation of providing references for diagnosis and treatment of breast cancer.
The results of this study indicate that miR-206 is upregulated in breast cancer tissues and miR-145 is downregulated in breast cancer tissues. This is in agreement with the finding by Kim et al (20) and Oksuz et al (21) on the role of miR-206 and miR-145 in ovarian and uterine cancer, suggesting that miR-206 and miR-145 may be involved in the occurrence and development of breast cancer. Expression of miR-206 and miR-145 may be related to the severity of breast cancer, the degree of differentiation, lymph node metastasis and the depth of invasion. miR-206 and miR-145 can be used as tumor markers in the early diagnosis of breast cancer because they can downregulate mRNA expression of ERα and its coregulatory proteins, inhibit the proliferation of breast cancer cells and participate in the development of breast cancer. However, the mechanism of miR-206 and miR-145 in breast cancer remains unclear.
miR-206 and miR-145 may also inhibit the development of breast cancer. Because miR-206 is upregulated in breast cancer and miR-145 is downregulated, and high level of miR-206 expression and level of miR-145 expression was correlated with poor prognosis, suggesting that miR-206 and miR-145 can be used as a prognostic indicator for patients with breast cancer.
There are still some shortcomings in this study. For example, the sample size was small, and it is not ruled out that there may be differences in expression levels of miR-206 and miR-145 among different age groups. We will conduct a longer follow-up survey of patients in this study to confirm our conclusions.
In conclusion, miR-206 is upregulated and miR-145 is downregulated in breast cancer tissues, which may affect the prognosis of patients. miR-206 and miR-145 may be used as important prognostic indicators for patients with breast cancer in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YQ wrote the manuscript and was responsible for collecting the tissues. XH contributed to the extraction of RNA. XQ performed PCR. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the First Affiliated Hospital of Shantou University Medical College (Shantou, China). Signed informed consents were obtained from the patients or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Etti J, Patel R, Pinter T, Schmidt M, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 2.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn HJ. Panel Members: Tailoring therapies - improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swain SM, Baselga J, Kim SB, Ro J, Semiglazov VM, Ciruelos E, Ferrero JM, Schneeweiss A, Heeson S, et al. CLEOPATRA Study Group: Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372:724–734. doi: 10.1056/NEJMoa1413513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner NC, Ro J, André F, Loi S, Verma S, Iwata HN, Loibl S, Bartlett Huang C, Zhang K, et al. PALOMA3 Study Group: Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 5.Hindié E, Groheux D. Regional nodal irradiation in early-stage breast cancer. N Engl J Med. 2015;373:1877–1878. doi: 10.1056/NEJMc1510505. [DOI] [PubMed] [Google Scholar]
- 6.Narimatsu R, Patterson BK. High-throughput cervical cancer screening using intracellular human papillomavirus E6 and E7 mRNA quantification by flow cytometry. Am J Clin Pathol. 2005;123:716–723. doi: 10.1309/FE70AVNY75TDDJUH. [DOI] [PubMed] [Google Scholar]
- 7.Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, Huang MD, Shu YQ. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 8.Kim KT, Lee HW, Lee HO, Kim SC, Seo YJ, Chung W, Eum HH, Nam DH, Kim J, Joo KM, et al. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 2015;16:127. doi: 10.1186/s13059-015-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y, Wang L, Wang S, He Q, Huang J, et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6:25533–25574. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 11.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Jr, Dees EC, Perez EA, Olson JA, Jr, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373:2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Poortmans PM, Collette S, Kirkove C, Van Limbergen E, Budach V, Struikmans H, Collette L, Fourquet A, Maingon P, Valli M, et al. EORTC Radiation Oncology and Breast Cancer Groups: Internal mammary and medial supraclavicular irradiation in breast cancer. N Engl J Med. 2015;373:317–327. doi: 10.1056/NEJMoa1415369. [DOI] [PubMed] [Google Scholar]
- 14.Tutt A, Ellis P, Kilburn L, Gilett C, Pinder S, Abraham J, Barrett S, Barrett-Lee P, Chan S, Cheang M. The TNT trial: A randomized phase III trial of carboplatin (C) compared with docetaxel (D) for patients with metastatic or recurrent locally advanced triple negative or BRCA1/2 breast cancer (CRUK/07/012) Cancer Res. 2015;75(9 Suppl) doi: 10.1158/1538-7445.SABCS14-S3-01. Abst S3-01. [DOI] [Google Scholar]
- 15.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zargar H, Espiritu PN, Fairey AS, Mertens LS, Dinney CP, Mir MC, Krabbe LM, Cookson MS, Jacobsen NE, Gandhi NM, et al. Multicenter assessment of neoadjuvant chemotherapy for muscle-invasive bladder cancer. Eur Urol. 2015;67:241–249. doi: 10.1016/j.eururo.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta Y, Möller S, Witte M, Belheouane M, Sezin T, Hirose M, Vorobyev A, Niesar F, Bischof J, Ludwig RJ, et al. Dissecting genetics of cutaneous miRNA in a mouse model of an autoimmune blistering disease. BMC Genomics. 2016;17:112. doi: 10.1186/s12864-016-2455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge X, Lyu P, Cao Z, Li J, Guo G, Xia W, Gu Y. Overexpression of miR-206 suppresses glycolysis, proliferation and migration in breast cancer cells via PFKFB3 targeting. Biochem Biophys Res Commun. 2015;463:1115–1121. doi: 10.1016/j.bbrc.2015.06.068. [DOI] [PubMed] [Google Scholar]
- 19.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TH, Song JY, Park H, Jeong JY, Kwon AY, Heo JH, Kang H, Kim G, An HJ. miR-145, targeting high-mobility group A2, is a powerful predictor of patient outcome in ovarian carcinoma. Cancer Lett. 2015;356:1–945. doi: 10.1016/j.canlet.2014.11.011. (2 Pt B) [DOI] [PubMed] [Google Scholar]
- 21.Oksuz Z, Serin MS, Kaplan E, Dogen A, Tezcan S, Aslan G, Emekdas G, Sezgin O, Altintas E, Tiftik EN. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015;42:713–720. doi: 10.1007/s11033-014-3819-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.


