Abstract
Prostate cancer (PCa) is one of the most common malignancies in men globally. The aim of the present study was to identify the key genes and pathways involved in the occurrence of PCa. Gene expression profile (GSE55945) was downloaded from Gene Expression Omnibus, and the differentially expressed genes (DEGs) were identified. Subsequently, Gene ontology analysis, KEGG pathway analysis and protein-protein interaction (PPI) analysis of DEGs were performed. Finally, the identified key genes were confirmed by immunohistochemistry. The GO analysis results showed that the DEGs were mainly participated in cell cycle, cell division, cell development and cell junction. The KEGG pathway analysis showed that the DEGs were mainly enriched in proteoglycans in cancer, endocytosis, focal adhesion and hippo signaling pathway. The PPI analysis results showed that RPS21, FOXO1, BIRC5, POLR2H, RPL22L1 and NPM1 were the key genes involved in the occurrence of PCa, and the Module analysis indicated that the occurrence of PCa was associated with cell cycle, oocyte meiosis and ribosome biogenesis. IHC result showed that the expression of RPS21, BIRC5, POLR2H, RPL22L1 and NPM1 were significantly upregulated in PCa, while the expression of FOXO1 was significantly downregulated in PCa, matching with the bioinformatics analysis. Taken together, several key genes and pathways were identified involved in PCa, which might provide the potential biomarker for prognosis, diagnosis and drug targets.
Keywords: bioinformatics analysis, prostate cancer, differentially expressed gene, pathways, identification
Introduction
Prostate cancer (PCa) is one of the most common malignancies in men globally and the second leading cause of cancer associated mortality in developed countries (1,2). Like other cancers, PCa is considered to be a disease which caused by age, diet and gene aberrations (3). Accumulating evidences have demonstrated that a series of genes and pathways involved in the occurrence, progression and metastasis of PCa (4). At present, the underlying mechanism of PCa occurrence is still unclear, which limits the diagnosis and therapy. Therefore, it is urgent to identify the key genes and pathways involved in the occurrence of PCa (2,5).
Microarray is a useful tool for analysis of gene expression that can be applied to disease diagnosis and targeted therapy (6,7). During the past decade, hundreds of differentially expressed genes (DEGs) participated in biological processes, cell component, molecular functions and pathways of PCa were identified by microarray technology (8,9). However, previous studies of DEGs analysis have shown relative limitations, for example, no reliable biomarker was identified that could distinguish tumors from normal tissues (10). Therefore, gene expression in the occurrence of PCa needs to be further analyzed by microarray combining bioinformatics technology at present.
In the present study, Gene expression profile (GSE55945) was downloaded from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). The gene expression profile was analyzed, and the DEGs were identified between PCa group and normal group. Subsequently, gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and protein-protein interaction (PPI) analysis of DEGs were performed. Finally, the expression of screened key genes was verified by immunohistochemistry (IHC). The present study aimed to identify key genes and pathways which involved in the occurrence of PCa and explored the potential biomarker for prognosis, diagnosis and drug targets.
Materials and methods
Expression Profile Microarray
Gene expression profile (GSE55945) was downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/). GSE55945 was based on GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array), which was submitted by Arredouani et al, and contained 21 samples, including 18 PCa samples and 8 normal prostate samples (8).
Identification of DEGs
The gene expression profile was analyzed by Morpheus online tools (https://software.broadinstitute.org/morpheus/) as previous study (11). The DEGs were identified between PCa group and normal group. The significance of DEGs was identified by classical t-test. The change ≥twofold and P<0.05 was considered to indicate a statistically significant difference.
GO analysis and KEGG pathway analysis of DEGs
In order to analyse the function and pathway of the DEGs, DAVID database (https://david.ncifcrf.gov/) was used for GO analysis and KEGG pathway analysis as previous study (7). P<0.05 was considered to indicate a statistically significant difference.
PPI network and module analysis
Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org/cgi/input.pl) was a database that could assess the protein-protein interaction. The DEGs were mapped to STRING, and a score >0.4 was considered to be significant. Then, the Cytoscape software (version 3.3.0) was used to construct PPI networks. Finally, the modules of PPI network were screened by the plug-in Molecular Complex Detection (MCODE). In addition, the pathway analysis was performed in the modules. P<0.05 was considered to indicate a statistically significant difference.
Patients and tissue samples
The tissue microarray was purchased from Alenabio Co., Ltd (Xian, China) including 60 PCa samples from patients and 10 normal prostate tissue samples from healthy donors. The procedures performed in this study involving human patients were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The present study was approved by the Research Ethics Committee of Weifang Medical University (Weifang, China).
IHC validation
IHC was performed as previous study (12). The tissue sample was blocked by 0.3% H2O2 and blocked by 10% bovine serum albumin (BSA) for 25 min. Then, the tissue sample was incubated overnight with anti-ribosomal protein S21 (RPS21), anti-forkhead box O1 (FOXO1), anti-baculoviral IAP repeat containing 5 (BIRC5), anti-RNA polymerase II subunit H (POLR2H), anti-ribosomal protein L22 like 1 (RPL22L1) and anti-nucleophosmin 1 (NPM1; 1:100 dilution; Proteintech, Wuhan, China) at 4°C and incubated with secondary antibody (1:1,000 dilution; Beyotime Institute of Biotechnology, Shanghai, China) for 1.5 h at 30°C. At last, 3,3-diaminobenzidine (DAB) was used for color visualization and hematoxylin was used for counterstained. In this study, eight pairs of normal and cancerous tissues were stained with each antibody. To evaluate the gene expression, a previously described scoring system was utilized (13). Briefly, the scores of two parameters were multiplied by the staining intensity (range, 0–3) and the percentage of positive cells [range, 0–4 (0, 0–10%; 1, 11–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%)]. The tissue sample with scores of 8 or higher was classified as positive staining. In addition, an independent sample t-test was applied to the results of staining.
Results
Identification of DEGs
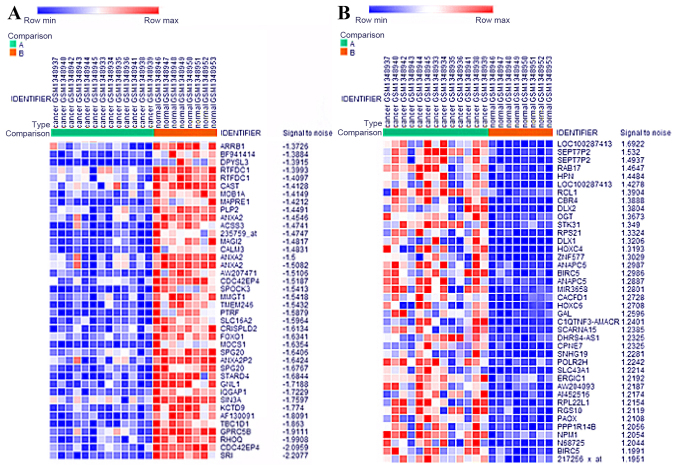
A total of 13 PCa samples and 8 normal samples were analyzed using Morpheus online tools (https://software.broadinstitute.org/morpheus/). A total of 2,000 DEGs were identified in PCa compared to normal group, including 1000 upregulated and 1,000 downregulated genes respectively. The heat map of DEGs expression (including top 40 upregulated and downregulated genes) was shown in Fig. 1.
Figure 1.
Heatmap of the top 40 upregulated genes and 40 downregulated genes. (A) blue, downregulated; (B) red, upregulated.
GO analysis of DEGs
All DEGs were uploaded to DAVID database (https://david.ncifcrf.gov/), and then GO analysis was conducted. The results showed that upregulated DEGs were enriched in biological processes, including cellular response to growth factor stimulus, phosphate metabolic process, cell development, cell cycle and cell division, while downregulated DEGs were enriched in biological processes, including signal transduction, cell communication, response to stimulus, cell projection organization and cell development (Table I). For cell component, upregulated DEGs were enriched in contractile fiber, actin cytoskeleton, anchoring junction, adherens junction and focal adhesion, while downregulated DEGs were enriched in membrane raft, actin cytoskeleton, adherens junction, anchoring junction and membrane-bounded vesicle (Table I). Furthermore, for molecular function, upregulated DEGs were enriched in enzyme binding, cytoskeletal protein binding, enzyme regulator activity, macromolecular complex binding and protein kinase binding, while downregulated DEGs were enriched in RNA binding, cytoskeletal protein binding, enzyme regulator activity, transcription factor binding and calcium ion binding (Table I).
Table I.
GO annotation of DEGs in PCa.
| A, Upregulated genes | |||
|---|---|---|---|
| Category | Term/gene function | Count | P-value |
| GOTERM_BP_FAT | Cellular response to growth factor stimulus | 27 | 4.7×10-6 |
| GOTERM_BP_FAT | Regulation of phosphate metabolic process | 25 | 6.2×10-6 |
| GOTERM_BP_FAT | Cell development | 54 | 6.3×10-5 |
| GOTERM_BP_FAT | Regulation of cell cycle | 28 | 3.1×10-3 |
| GOTERM_BP_FAT | Cell division | 19 | 3.7×10-3 |
| GOTERM_CC_FAT | Contractile fiber | 20 | 5.4×10-9 |
| GOTERM_CC_FAT | Actin cytoskeleton | 25 | 9.6×10-7 |
| GOTERM_CC_FAT | Anchoring junction | 31 | 2.5×10-6 |
| GOTERM_CC_FAT | Adherens junction | 30 | 4.6×10-6 |
| GOTERM_CC_FAT | Focal adhesion | 21 | 9.4×10-6 |
| GOTERM_MF_FAT | Enzyme binding | 53 | 6.1×10-6 |
| GOTERM_MF_FAT | Cytoskeletal protein binding | 31 | 9.0×10-3 |
| GOTERM_MF_FAT | Enzyme regulator activity | 30 | 9.5×10-4 |
| GOTERM_MF_FAT | Macromolecular complex binding | 34 | 3.9×10-3 |
| GOTERM_MF_FAT | Protein kinase binding | 18 | 4.4×10-3 |
| B, Downregulated genes | |||
| Category | Term/gene function | Count | P-value |
| GOTERM_BP_FAT | Regulation of signal transduction | 25 | 9.2×10-6 |
| GOTERM_BP_FAT | Regulation of cell communication | 25 | 3.9×10-5 |
| GOTERM_BP_FAT | Regulation of response to stimulus | 27 | 4.6×10-5 |
| GOTERM_BP_FAT | Cell projection organization | 26 | 5.4×10-5 |
| GOTERM_BP_FAT | Cell development | 33 | 1.0×10-4 |
| GOTERM_CC_FAT | Membrane raft | 11 | 2.6×10-4 |
| GOTERM_CC_FAT | Actin cytoskeleton | 12 | 2.6×10-3 |
| GOTERM_CC_FAT | Adherens junction | 14 | 7.5×10-3 |
| GOTERM_CC_FAT | Anchoring junction | 14 | 9.1×10-3 |
| GOTERM_CC_FAT | Membrane-bounded vesicle | 44 | 1.2×10-3 |
| GOTERM_MF_FAT | RNA binding | 43 | 9.3×10-6 |
| GOTERM_MF_FAT | Cytoskeletal protein binding | 17 | 1.1×10-4 |
| GOTERM_MF_FAT | Enzyme regulator activity | 18 | 2.3×10-3 |
| GOTERM_MF_FAT | Transcription factor binding | 11 | 7.7×10-3 |
| GOTERM_MF_FAT | Calcium ion binding | 12 | 2.9×10-3 |
GO, gene ontology; PCa, prostate cancer; DEGs, differentially expressed genes.
KEGG pathway analysis of DEGs
The KEGG pathway analysis results showed that upregulated DEGs were enriched in proteoglycans in cancer, endocytosis, focal adhesion, hippo signaling pathway and cGMP-PKG signaling pathway, whereas downregulated DEGs were enriched in proteoglycans in cancer, endocytosis, hippo signaling pathway, thyroid hormone signaling pathway and sulfur relay system (Table II).
Table II.
KEGG pathway analysis of DEGs in PCa.
| A, Upregulated genes | |||
|---|---|---|---|
| KEGG terms | Count | P-value | Genes |
| Proteoglycans in cancer | 13 | 5.0×10-5 | IQGAP1, ROCK2, ARHGEF12, TIMP3, CAV1, CAV2, FZD1, HOXD10, PAK1, PPP1R12A, PPP1R12B, SDC4, TGFB2 |
| Endocytosis | 12 | 1.9×10-4 | SH3GLB1, VPS37A, ARRB1, CAV1, CAV2, CHMP1B, CHMP7, SNX2, SPG20, TGFB2, TGFB3, VPS36 |
| Focal adhesion | 10 | 4.2×10-3 | ROCK2, CAV1, CAV2, COL4A6, CCND2, MYLK, PAK1, PPP1R12A, PPP1R12B, VCL |
| Hippo signaling pathway | 8 | 8.3×10-3 | MOB1A, WWTR1, YAP1, CCND2, FZD1, SNAI2, TGFB2, TGFB3 |
| cGMP-PKG signaling pathway | 8 | 1.4×10-3 | GNAI2, ROCK2, CALM1, CALM3, MEF2A, MYLK, NPR2, PPP1R12A |
| B, Downregulated genes | |||
| KEGG terms | Count | P-value | Genes |
| Proteoglycans in cancer | 8 | 1.6×10-3 | IQGAP1, ARHGEF12, TIMP3, CAV1, CAV2, FZD1, HOXD10, SDC4 |
| Endocytosis | 6 | 7.2×10-3 | SH3GLB1, VPS37A, ARRB1, CAV1, CAV2, SPG20 |
| Hippo signaling pathway | 5 | 4.2×10-2 | MOB1A, WWTR1, YAP1, FZD1, SNAI2 |
| Thyroid hormone signaling pathway | 4 | 7.6×10-2 | SIN3A, FOXO1, RXRA, SLC16A2 |
| Sulfur relay system | 2 | 8.4×10-2 | MOCS1, MOCS2 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; PCa, prostate cancer; DEGs, differentially expressed genes; FOXO1, forkhead box O1.
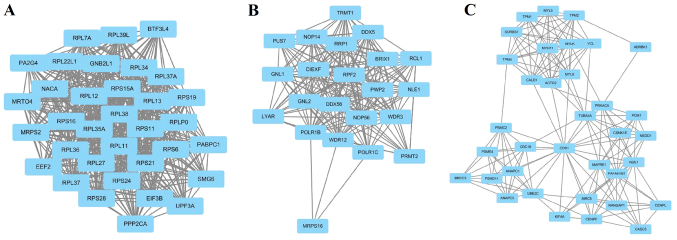
Module screening from the PPI network
The top 6 hub nodes with high degrees were screened by the STRING database. These hub genes included RPS21, FOXO1, BIRC5, POLR2H, RPL22L1 and NPM1. In addition, total nodes were analyzed by plug-ins MCODE, and the top three significant modules were selected (Fig. 2). The results showed that a total of 91 genes of functional annotation were involved in the modules that associated with cell cycle, oocyte meiosis and ribosome biogenesis in eukaryotes (data not shown).
Figure 2.
Top three modules from the PPI network. (A) module1, (B) module2, (C) module3. PPI, protein-protein interaction.
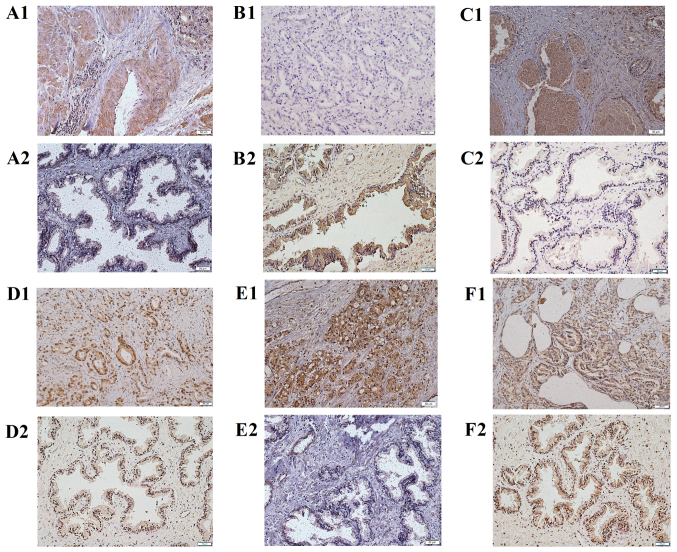
IHC validation of key genes in PCa samples
To verify the bioinformatics analysis data, the expression of key genes including RPS21, FOXO1, BIRC5, POLR2H, RPL22L1 and NPM1 were examined by IHC in PCa samples and normal samples. As shown in Fig. 3, compared with the normal tissues, the expression of RPS21, BIRC5, POLR2H, RPL22L1 and NPM1 were significantly upregulated in the cancer cells from tumor tissues, while the expression of FOXO1 was significantly downregulated in the cancer cells from tumor tissues (P<0.05). The IHC results were matched with the bioinformatics analysis.
Figure 3.
The expression of DEGs in PCa compared to normal tissues. (A1) RPS21 of PCa; (A2) RPS21 of control; (B1) FOXO1 of PCa; (B2) FOXO1 of control; (C1) BIRC5 of PCa; (C2) BIRC5 of control; (D1) POLR2H of PCa; (D2) POLR2H of control; (E1) RPL22L1 of PCa; (E2) RPL22L1 of control; (F1) NPM1 of PCa; (F2) NPM1 of control. IHC, magnification, ×400. Scale bar, 50 µm. Control: Normal tissues. DEGs, differentially expressed genes; PCa, prostate cancer; IHC, immunohistochemistry; FOXO1, forkhead box O1; BIRC5, baculoviral IAP repeat containing 5; RPS21, ribosomal protein S21; POLR2H, RNA polymerase II subunit H; RPL22L1, ribosomal protein L22 like 1; NPM1, nucleophosmin 1.
Discussion
In the present study, we uploaded GSE21815 and identify 2,000 DEGs (upregulated and downregulated) between PCa and normal tissues by bioinformatics analysis. Go analysis and KEGG pathway analysis showed that the DEGs were mainly involved in cell cycle, cell division, cell development and cell junction. The results of PPI analysis showed that some key genes might play an important role in the occurrence, progression and metastasis of PCa that could provide the potential biomarker for prognosis, diagnosis and drug targets.
In this study, the gene expression profile of GSE55945 was downloaded from GEO which including 18 PCa samples and 8 normal prostate samples. The results showed that a total of 2,000 DEGs were identified in PCa compared to normal group, including 1,000 upregulated and 1,000 downregulated genes respectively. Previous studies have demonstrated that co-expression genes were frequently involved in similar biological function and signal pathway (2,9). Therefore, GO analysis and KEGG pathway analysis was further performed.
The GO analysis showed that the DEGs were mainly participated in cell cycle, cell division, cell development and cell junction (Table I). This result was consistent with other studies, and the dysregulation of cell function lead to the occurrence, progression and metastasis of PCa (2,8). The KEGG pathway analysis showed that the DEGs were mainly enriched in proteoglycans in cancer, endocytosis, focal adhesion and hippo signaling pathway (Table II). Previous study demonstrated that dysregulation of the hippo pathway exerts a significant impact on cancer development. For example, activated hippo signaling pathway was observed in many types of cancers, including colon, liver, breast, lung and ovary (14). Recent study implied that proteoglycans exert diverse functions in the occurrence of cancer. For instance, proteoglycans contributed to the formation of provisional matrix for tumor growth affecting cell-cell and cell-matrix interactions and signal transduction of tumor cells. Proteoglycans also regulated the phenotype of tumor cells and tumor stroma angiogenesis (15). In addition, other studies showed that endocytosis and focal adhesion were closely related to tumorigenesis (16,17). Bibens-Laulan et al demonstrated that the high expression of galectin-7 in ovarian and breast cancer cells was due to the endocytosis (16). Kanteti et al indicated that focal adhesion kinase play an important role in tumor cell phenotype such as survival, proliferation, migration and invasion (17).
Finally, PPI analysis was performed and the key genes were identified (Fig. 2). Subsequently, the result was confirmed by IHC. As shown in Fig. 3, the expression of RPS21, BIRC5, POLR2H, RPL22L1 and NPM1 were significantly upregulated in PCa, while the expression of FOXO1 was significantly downregulated in PCa. The IHC results were matched with the bioinformatics analysis.
RPS21 was the first identified key gene, belonging to ribosomal proteins (RPS) family. RPS are the pivotal components of ribosome, which associated with proliferation, differentiation, DNA repair and apoptosis of cell (18). Arthurs et al reported that RPS21 was upregulated in PCa and might serve to be a possible biomarker (19), which was correspond to our study. To our knowledge, this was the only study of RPS21 in cancer up to now. Moreover, Huang et al reported that RPS27L may be a useful index for predicting prognoses in colorectal cancer (20). Li et al also reported that knockdown of RPSL26 or RPSL29 significantly inhibits cell proliferation in pancreatic cancer (21). The second key gene was FOXO1, belonging to the FOXOs family, which involved in cell proliferation, differentiation and apoptosis by the regulation of multiple genes (22). Consistent with our study, previous studies demonstrated the expression of FOXO1 was downregulated in PCa (23,24). Moreover, other studies further proposed that FOXO1 was a key tumor suppressor in cancer, including PCa (22,25). The third identified key gene was BIRC5, which play an important role in the occurrence and progression of cancer (26). Wang et al reported that BIRC5 was involved in the tumorigenesis of colorectal cancer (27). In addition, BIRC5 was reported to be associated with microtubule-kinetochore attachment, interacting with cell adhesion (28,29). Therefore, BIRC5 might play a critical role in metastasis of PCa. The fourth identified key gene was POLR2H, which was the necessary subunit of RNA polymerase II, which was essential for transcription of DNA (30). To our knowledge, the related study of POLR2H was extremely rare. This study indicated that POLR2H was involved in the occurrence and progression of PCa for the first time, and the mechanism need to be further explored. The fifth key gene was RPL22L1, which was identified as a trace component of ribosome (31,32). Wu et al reported that RPL22L1 could promote ovarian cancer metastasis by inducing epithelial-to-mesenchymal transition (33). Moreover, recent study also demonstrated that RPL22L1 could play vital and definite roles in hematopoietic development (34). The sixth key gene was NPM1, which play an crucial role in cell growth and proliferation (35). Leotoing ea al reported that NPM1 was significantly upregulated in prostate tumour cells, indicating that NPM1 might be an enhancer in progression of PCa (36,37). Further study showed that NPM1 was critical for migration and invasion of PCa, and knockdown of NPM1 resulted in a decrease in the growth of the tumor cell (35).
Module analysis of PPI indicated that the occurrence of PCa was associated with cell cycle, oocyte meiosis and ribosome biogenesis. It is well known that cell cycle is involved in the occurrence, progression and metastasis of cancer (38–40). Moreover, previous studies demonstrated that ribosome biogenesis was closely related to tumorigenesis, and suppressing ribosome biogenesis could inhibit cancer development (41,42). Therefore, drugs targeted ribosome biogenesis was proposed recently for cancer therapy, which could repress tumor cell proliferation without genotoxic activity (43).
In conclusion, our study identified some key genes and pathways by bioinformatics analysis, which might be involved in the occurrence of PCa. The molecular mechanism of these key genes in the occurrence of PCa need to be further studied.
Acknowledgements
Not applicable.
Funding
The present study was supported by Natural Science Foundation of Shandong Province (grant nos. ZR2013CM032, ZR2014CL034, ZR2015HM028 and ZR2017MH103), Science and Technology Development Plan of Shandong Province (grant no. 2015GSF118178) and Medical and Health Development Plan of Shandong Province (grant no. 2017WS058).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GSE55945 repository, https://www.ncbi.nlm.nih.gov/geo/.
Authors' contributions
WF and ZFP designed the study. SF and ZL performed the data analysis and wrote the paper. ZG and ZWP identified the DEGs. SH and XL carried out the GO analysis, KEGG pathway analysis and PPI analysis. CZ and WY carried out immunohistochemical analysis. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
The procedures performed in this study involving human patients were in accordance with the ethics standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The present study was approved by the Research Ethics Committee of Weifang Medical University. Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Djulbegovic M, Beyth RJ, Neuberger MM, Stoffs TL, Vieweg J, Djulbegovic B, Dahm P. Screening for prostate cancer: Systematic review and meta-analysis of randomised controlled trials. BMJ. 2010;341:c4543. doi: 10.1136/bmj.c4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, Guo K, Liang Z, Li F, Wang H. Identification of candidate genes that may contribute to the metastasis of prostate cancer by bioinformatics analysis. Oncol Lett. 2018;15:1220–1228. doi: 10.3892/ol.2017.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 4.Cancer genome atlas research network: The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hafeez BB, Zhong W, Fischer JW, Mustafa A, Shi X, Meske L, Hong H, Cai W, Havighurst T, Kim K, Verma AK. Plumbagin, a medicinal plant (Plumbago zeylanica)-derived 1,4-naphthoquinone, inhibits growth and metastasis of human prostate cancer PC-3M-luciferase cells in an orthotopic xenograft mouse model. Mol Oncol. 2013;7:428–439. doi: 10.1016/j.molonc.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profiling in colorectal cancer using microarray technologies: Results and perspectives. Cancer Treat Rev. 2009;35:201–209. doi: 10.1016/j.ctrv.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Liang B, Li C, Zhao J. Identification of key pathways and genes in colorectal cancer using bioinformatics analysis. Med Oncol. 2016;33:111. doi: 10.1007/s12032-016-0829-6. [DOI] [PubMed] [Google Scholar]
- 8.Arredouani MS, Lu B, Bhasin M, Eljanne M, Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA, Sanda MG. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794–5802. doi: 10.1158/1078-0432.CCR-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marín-Aguilera M, Codony-Servat J, Kalko SG, Fernández PL, Bermudo R, Buxo E, Ribal MJ, Gascón P, Mellado B. Identification of docetaxel resistance genes in castration-resistant prostate cancer. Mol Cancer Ther. 2012;11:329–339. doi: 10.1158/1535-7163.MCT-11-0289. [DOI] [PubMed] [Google Scholar]
- 10.Lascorz J, Hemminki K, Försti A. Systematic enrichment analysis of gene expression profiling studies identifies consensus pathways implicated in colorectal cancer development. J Carcinog. 2011;10:7. doi: 10.4103/1477-3163.78268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi B, Liu G, Zhou W, Lv H, Liu Y, Liu J. Identification of genes and pathways in the synovia of women with osteoarthritis by bioinformatics analysis. Mol Med Rep. 2018;17:4467–4473. doi: 10.3892/mmr.2018.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng W, Zhou D, Meng W, Li G, Zhuang P, Pan Z, Wang G, Cheng Z. Growth retardation induced by avian leukosis virus subgroup J associated with down-regulated Wnt/β-catenin pathway. Microb Pathog. 2017;104:48–55. doi: 10.1016/j.micpath.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Zhang J, Zhan X, Lin T, Yang M, Hu J, Han B, Hu S. SOX4 is associated with poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;452:614–621. doi: 10.1016/j.bbrc.2014.08.124. [DOI] [PubMed] [Google Scholar]
- 14.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theocharis AD, Karamanos NK. Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biol pii: S0945-053X. 2017;30313-X doi: 10.1016/j.matbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Bibens-Laulan N, St-Pierre Y. Intracellular galectin-7 expression in cancer cells results from an autocrine transcriptional mechanism and endocytosis of extracellular galectin-7. PLoS One. 2017;12:e0187194. doi: 10.1371/journal.pone.0187194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanteti R, Mirzapoiazova T, Riehm JJ, Dhanasingh I, Mambetsariev B, Wang J, Kulkarni P, Kaushik G, Seshacharyulu P, Ponnusamy MP, et al. Focal adhesion kinase a potential therapeutic target for pancreatic cancer and malignant pleural mesothelioma. Cancer Biol Ther. 2018;19:316–327. doi: 10.1080/15384047.2017.1416937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Nag S, Zhang X, Wang MH, Wang H, Zhou J, Zhang R. Ribosomal proteins and human diseases: Pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015;35:225–285. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arthurs C, Murtaza BN, Thomson C, Dickens K, Henrique R, Patel HRH, Beltran M, Millar M, Thrasivoulou C, Ahmed A. Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS One. 2017;12:e0186047. doi: 10.1371/journal.pone.0186047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang CJ, Yang SH, Lee CL, Cheng YC, Tai SY, Chien CC. Ribosomal protein S27-like in colorectal cancer: A candidate for predicting prognoses. PLoS One. 2013;8:e67043. doi: 10.1371/journal.pone.0067043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, Ge M, Yin Y, Luo M, Chen D. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol Cell Biochem. 2012;370:127–139. doi: 10.1007/s11010-012-1404-x. [DOI] [PubMed] [Google Scholar]
- 22.Duan X, Kong Z, Liu Y, Zeng Z, Li S, Wu W, Ji W, Yang B, Zhao Z, Zeng G. β-arrestin2 contributes to cell viability and proliferation via the down-regulation of FOXO1 in castration-resistant prostate cancer. J Cell Physiol. 2015;230:2371–2381. doi: 10.1002/jcp.24963. [DOI] [PubMed] [Google Scholar]
- 23.Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, Chung LW, Zhou W, Dong JT. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting androgen receptor signaling in prostate cancer. Cancer Res. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 24.Yu JJ, Wu YX, Zhao FJ, Xia SJ. miR-96 promotes cell proliferation and clonogenicity by down-regulating of FOXO1 in prostate cancer cells. Med Oncol. 2014;31:910. doi: 10.1007/s12032-014-0910-y. [DOI] [PubMed] [Google Scholar]
- 25.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Zhang X, Wang L, Zheng G, Du L, Yang Y, Dong Z, Liu Y, Qu A, Wang C. Investigation of cell free BIRC5 mRNA as a serum diagnostic and prognostic biomarker for colorectal cancer. J Surg Oncol. 2014;109:574–579. doi: 10.1002/jso.23526. [DOI] [PubMed] [Google Scholar]
- 28.Vitale I, Galluzzi L, Senovilla L, Criollo A, Jemaà M, Castedo M, Kroemer G. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ. 2011;18:1403–1413. doi: 10.1038/cdd.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du YJ, Hou YL, Hou WR. Molecular characterization of a gene POLR2H encoded an essential subunit for RNA polymerase II from the Giant Panda (Ailuropoda Melanoleuca) Mol Biol Rep. 2013;40:1495–1498. doi: 10.1007/s11033-012-2192-9. [DOI] [PubMed] [Google Scholar]
- 31.Sugihara Y, Honda H, Iida T, Morinaga T, Hino S, Okajima T, Matsuda T, Nadano D. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J Proteome Res. 2010;9:1351–1366. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- 32.O'Leary MN, Schreiber KH, Zhang Y, Duc AC, Rao S, Hale JS, Academia EC, Shah SR, Morton JF, Holstein CA, et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 2013;9:e1003708. doi: 10.1371/journal.pgen.1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu N, Wei J, Wang Y, Yan J, Qin Y, Tong D, Pang B, Sun D, Sun H, Yu Y, et al. Ribosomal L22-like1 (RPL22L1) promotes ovarian cancer metastasis by inducing epithelial-to-mesenchymal transition. PLoS One. 2015;10:e0143659. doi: 10.1371/journal.pone.0143659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Duc AC, Rao S, Sun XL, Bilbee AN, Rhodes M, Li Q, Kappes DJ, Rhodes J, Wiest DL. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell. 2013;24:411–425. doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loubeau G, Boudra R, Maquaire S, Lours-Calet C, Beaudoin C, Verrelle P, Morel L. NPM1 silencing reduces tumour growth and MAPK signalling in prostate cancer cells. PLoS One. 2014;9:e96293. doi: 10.1371/journal.pone.0096293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Léotoing L, Meunier L, Manin M, Mauduit C, Decaussin M, Verrijdt G, Claessens F, Benahmed M, Veyssière G, Morel L, Beaudoin C. Influence of nucleophosmin/B23 on DNA binding and transcriptional activity of the androgen receptor in prostate cancer cell. Oncogene. 2008;27:2858–2867. doi: 10.1038/sj.onc.1210942. [DOI] [PubMed] [Google Scholar]
- 37.Dai L, Li J, Xing M, Sanchez TW, Casiano CA, Zhang JY. Using serological proteome analysis to identify serum anti-nucleophosmin 1 autoantibody as a potential biomarker in european-american and african-american patients with prostate cancer. Prostate. 2016;76:1375–1386. doi: 10.1002/pros.23217. [DOI] [PubMed] [Google Scholar]
- 38.Yun SJ, Moon SK, Kim WJ. Investigational cell cycle inhibitors in clinical trials for bladder cancer. Expert Opin Investig Drugs. 2013;22:369–377. doi: 10.1517/13543784.2013.751097. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Z, Liu J, Wang C, Wang Y, Jiang Y, Guo M. MicroRNA-25 regulates small cell lung cancer cell development and cell cycle through cyclin E2. Int J Clin Exp Pathol. 2014;7:7726–7734. [PMC free article] [PubMed] [Google Scholar]
- 40.Soták M, Sumová A, Pácha J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann Med. 2014;46:221–232. doi: 10.3109/07853890.2014.892296. [DOI] [PubMed] [Google Scholar]
- 41.Gentilella A, Kozma SC, Thomas G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta. 2015;1849:812–820. doi: 10.1016/j.bbagrm.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Yin C, Wang L, Zhang S, Qian Y, Ma J, Zhang Z, Xu Y, Liu S. HSPC111 governs breast cancer growth by regulating ribosomal biogenesis. Mol Cancer Res. 2014;12:583–594. doi: 10.1158/1541-7786.MCR-13-0168. [DOI] [PubMed] [Google Scholar]
- 43.Brighenti E, Treré D, Derenzini M. Targeted cancer therapy with ribosome biogenesis inhibitors: A real possibility? Oncotarget. 2015;6:38617–38627. doi: 10.18632/oncotarget.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GSE55945 repository, https://www.ncbi.nlm.nih.gov/geo/.