Roe et al. examine the time-course of biomarker changes during conversion to Clinical Dementia Rating >0. The rate of change of cognitive and structural, but not molecular, biomarkers is greater in those who progress versus do not progress. Molecular biomarkers may become abnormal earlier, consistent with theoretical models.
Keywords: Alzheimer’s disease, biomarkers, APOE, amyloid imaging, cerebrospinal fluid
Abstract
Longer periods are needed to examine how biomarker changes occur relative to incident sporadic cognitive impairment. We evaluated molecular (CSF and imaging), structural, and cognitive biomarkers to predict incident cognitive impairment and examined longitudinal biomarker changes before and after symptomatic onset. Data from participants who were cognitively normal, underwent amyloid imaging using Pittsburgh compound B and/or CSF studies, and at least two clinical assessments were used. Stepwise Cox proportional hazards models tested associations of molecular (Pittsburgh compound B; CSF amyloid-β42, tau, ptau181, tau/amyloid-β42, ptau181/amyloid-β42), structural (normalized hippocampal volume, normalized whole brain volume), and cognitive (Animal Naming, Trail Making A, Trail Making B, Selective Reminding Test - Free Recall) biomarkers with time to Clinical Dementia Rating (CDR) > 0. Cognitively normal participants (n = 664), aged 42 to 90 years (mean ± standard deviation = 71.4 ± 9.2) were followed for up to 16.9 years (mean ± standard deviation = 6.2 ± 3.5 years). Of these, 145 (21.8%) participants developed a CDR > 0. At time of incident cognitive impairment, molecular, structural, and cognitive markers were abnormal for CDR > 0 compared to CDR = 0. Linear mixed models indicated rates of change in molecular biomarkers were similar for CDR = 0 and CDR > 0, suggesting that the separation in values between CDR = 0 and CDR > 0 must have occurred prior to the observation period. Rate of decline for structural and cognitive biomarkers was faster for CDR > 0 compared to CDR = 0 (P < 0.0001). Structural and cognitive biomarkers for CDR > 0 diverged from CDR 0 at 9 and 12 years before incident cognitive impairment, respectively. Within those who developed CDR > 0, a natural separation occurred for Pittsburgh compound B values. In particular, CDR > 0 who had at least one APOE ɛ4 allele had higher, and more rapid increase in Pittsburgh compound B, while APOE ɛ2 was observed to have slower increases in Pittsburgh compound B. Of molecular biomarker-positive participants followed for at least 10 years (n = 16–23), ∼70% remained CDR = 0 over the follow-up period. In conclusion, conversion from cognitively normal to CDR > 0 is characterized by not only the magnitude of molecular biomarkers but also rate of change in cognitive and structural biomarkers. Findings support theoretical models of biomarker changes seen during transition to cognitive impairment using longitudinal data and provide a potential time for changes seen during this transition. These findings support the use of molecular biomarkers for trial inclusion and cognitive/structural biomarkers for evaluating trial outcomes. Finally, results support a potential role for APOE ɛ in modulating amyloid accumulation in CDR > 0 with APOE ɛ4 being deleterious and APOE ɛ2 protective.
Introduction
Hypothesized models detailing molecular, structural, and cognitive changes before and after the onset of symptomatic cognitive impairment have been proposed (Sperling et al., 2011; Jack et al., 2013, 2015, 2018; Dubois et al., 2016). Development and refinement of these models has relied greatly on findings generated by cross-sectional studies, including examination of estimated age at symptomatic onset among persons with mutations causing autosomal dominant Alzheimer’s disease (Bateman et al., 2012). Within autosomal dominant Alzheimer’s disease studies, the estimated years to onset has allowed for evaluating changes in biomarkers in relation to a known event. Results from these studies suggest that asymptomatic mutation carriers in autosomal dominant Alzheimer’s disease families have molecular biomarker changes 15–20 years prior to estimated year of onset and changes in brain structural and cognitive symptoms 8–10 years prior to estimated year of onset (Bateman et al., 2012; Fagan et al., 2014).
More recent studies have demonstrated that biomarker abnormalities may be present during preclinical stages of sporadic Alzheimer’s disease (Stomrud et al., 2015; Sutphen et al., 2015; Toledo et al., 2015; Fletcher et al., 2016; Dumurgier et al., 2017; Insel et al., 2017). However, since there is no ‘estimated years to onset’ for sporadic Alzheimer’s disease, it has been difficult to elucidate when changes will occur as long follow-up periods are required. Relatively few longitudinal studies have been performed, and hypothesized models for sporadic Alzheimer’s disease have not defined specific time intervals. The few longitudinal studies that have been performed using a limited number of biomarkers suggest that the asymptomatic period for persons with preclinical Alzheimer’s disease may last for at least a decade (Buchhave et al., 2012; Roe et al., 2013; Stomrud et al., 2015).
A gap therefore exists with longitudinal studies examining multiple biomarkers (molecular, structural, and cognitive), such that longer periods are needed to examine how biomarker changes occur relative to incident sporadic cognitive impairment. In this study, we first examined molecular, structural, and cognitive biomarkers to predict incident cognitive impairment up to 16.9 years after initial evaluation. Symptomatic cognitive impairment was operationalized by a Clinical Dementia Rating (CDR) > 0 and encompassed both mild cognitive impairment and mild dementia. Our main question of interest, the magnitude and rate of change in biomarkers for persons who did, and did not, develop incident symptomatic cognitive impairment was then examined.
Materials and methods
Participant selection
Data were used from participants enrolled in longitudinal studies at the Knight Alzheimer’s Disease Research Center at Washington University in Saint Louis, USA, who were cognitively normal at their initial visit, underwent PET using Pittsburgh compound B (PIB) imaging and/or had CSF collected within 1 year of the baseline clinical assessment, and had at least one additional clinical assessment after their baseline visit. All procedures were reviewed and approved by the Washington University in St. Louis Human Research Protection Office and informed consent was obtained according to the Declaration of Helsinki from all participants.
Clinical assessment
Participants in Knight Alzheimer’s Disease Research Center longitudinal studies have annual clinical and psychometric assessments. During these assessments, cognitive normality was determined using the CDR (Morris, 1993). Experienced clinicians derive the CDR by integrating information obtained from interviews with the participant and separately with a collateral source who knows the participant well. These interviews rely on the principle of intra-individual change in cognitive and functional abilities, where the individual serves as his or her own control. The CDR is obtained via a standard scoring algorithm based on scores in six domains and indicates whether the participant has dementia, and if so, the severity of dementia (CDR 0 = cognitively normal, CDR 0.5 = very mild, CDR 1 = mild, CDR 2 = moderate, and CDR 3 = severe dementia) (Morris, 1993). Summation of the scores from the six domains results in the CDR Sum of Boxes, a continuous measure of cognitive impairment, which is often used as an outcome in clinical trials (Williams et al., 2009).
Design
We evaluated the ability of structural MRI and cognitive biomarkers together with the molecular biomarkers to predict time to a first diagnosis of cognitive impairment and evaluated the number of years that participants with ‘positive’ molecular biomarkers could remain cognitively normal. For participants who developed cognitive impairment over the follow-up period, changes in biomarkers before and after the date of first diagnosis of cognitive impairment were examined and compared to each other, and to biomarker changes for persons who remained cognitively normal. We also examined associations between APOE ɛ genotype and changes in molecular biomarkers across time.
Molecular biomarkers
Amyloid imaging
PIB imaging was used to determine brain amyloid burden (Klunk, 2011). Dynamic scans were used. Regional target-to-reference intensity ratio—standard uptake ratio—was estimated using 30 to 60 min post-injection as the time window for PIB and using the cerebellum cortex as the default reference region. Global amyloid-β burden was estimated using a set of regions of interest known to be sensitive to amyloid-β deposition (Su et al., 2013). PIB positivity was defined as having a standardized uptake value ratio (SUVR) ≥ 1.31 (Vlassenko et al., 2016). Partial volume correction was not applied.
CSF biomarkers
CSF analytes (Fagan et al., 2006) [amyloid-β42, tau and ptau181; Innotest, Fujirebio (formerly Innogenetics)] were measured using sensitive and quantitative enzyme-linked immunosorbent assays (ELISA). CSF was obtained using a 22-gauge Sprotte spinal needle to draw 20–30 ml of CSF at 8:00 am following an overnight fast. CSF samples were gently inverted and centrifuged at low speed to avoid possible gradient effects and frozen at −84°C after aliquoting into polypropylene tubes. Biomarker assays included a common reference standard, within-plate sample randomization and standardized protocol adherence. Samples were reanalysed if coefficients of variability exceed 25% [per Alzheimer’s Disease Neuroimaging Initiative (ADNI) criteria]; if there were ‘edge artefacts’; or if the pooled common CSF sample yielded widely discrepant values.
We examined the CSF variables of amyloid-β42, tau, ptau181, and the ratios of tau/amyloid-β42 and ptau181/amyloid-β42. In dichotomizing the CSF biomarkers, previously published cut-offs (Vos et al., 2013) were used for tau (339 pg/ml) and ptau181 (67 pg/ml). Because of concerns about upward drift in Innotest immunoassay amyloid-β42 values over the years (Schindler et al., 2018), positive and negative values of amyloid-β42 were assigned using assay- and date-specific cut-offs recommended by the Knight Alzheimer’s Disease Research Center Biomarker Core based on the work of Schindler et al. (2018). Using receiver operating curve (ROC) analyses, we determined the CSF amyloid-β42 cut-off that best distinguishes between amyloid PET positive and amyloid PET negative cases as a function of amyloid-β42 assay time period and assay type. Then, CSF amyloid-β42 levels were dichotomized as positive (if lower than the cut-off) or negative (if higher than the cut-off) according to the assay date and type (for more specific information, see Schindler et al., 2018). Adjusted values of amyloid-β42, tau/amyloid-β42, and ptau181/amyloid-β42 were constructed by calculating studentized residuals based on amyloid-β42 lot numbers and dates. Because cut-offs for the adjusted ratio variables are not yet available, we examined the frequency distributions of each variable and operationally defined the highest 30% of values as positive based on Alzheimer’s disease biomarker measurement and autopsy studies suggesting that roughly 30% of cognitively normal persons have preclinical Alzheimer’s disease (Price et al., 2009; Morris et al., 2010; Jansen et al., 2015).
Structural imaging
Scans were acquired using Siemens BioGraph mMR PET-MR 3T and Siemens TIM Trio 3 T MRI scanners.
To transition our cohort from the Siemens TIM Trio 3 T MRI to the Siemens Biograph 3 T molecular magnetic resonance (mMR), we performed direct correlations in a subset of our participants. Sixty-nine participants with a mean age of 65.9 years (CDR 0–0.5) received both the Trio and mMR MRI within 2 weeks; 67 participants were cognitively normal (CDR 0); two participants had a diagnosis of mild symptomatic Alzheimer’s disease (CDR 0.5). FreeSurfer v5.1 was used to segment the brain into various regions of interest for quantitative analysis.
For the left hippocampal volume as measured by Trio and the PET MRI, the estimated concordance correlation coefficient (CCC) on the raw data is 0.83 with a 95% confidence interval (CI) from 0.73 to 0.89, and after the standardization, the estimated CCC is 0.83 with a 95% CI from 0.74 to 0.90. For the right hippocampus volume as measured by Trio and the PET MRI, the estimated CCC on the raw data is 0.79 with a 95% CI from 0.67 to 0.87, and after the standardization, the estimated CCC is still 0.79 with a 95% CI from 0.67 to 0.87. Because of the two potential outliers in the scatter plot of the data on hippocampal volumes, we also performed rank-based CCC on these measures, the rank-based CCC for left hippocampal volume is 0.92 with a 95% CI from 0.86 to 0.95, and the rank-based CCC for right hippocampal volume is 0.91 with a 95% CI from 0.86 to 0.95, again both indicating excellent rank-based reproducibility of measuring hippocampal volumes. These findings are within the reported test-retest reliability range for repeat MRI visits on the same scanner (Han et al., 2006).
All MRI sessions were processed through the FreeSurfer image analysis suite using Dell PowerEdge 1950 servers with Intel Xeon processors running CentOS 5.5 Linux. FreeSurfer 5.3 (http://surfer.nmr.mgh.harvard.edu/) analyses involved cortical reconstruction and volumetric segmentation of T1-weighted images. The technical details of these procedures have been described previously (Dale et al., 1999; Fischl et al., 1999a, 2002). The cross-sectional processing pipeline included motion correction and segmentation of the subcortical white matter and deep grey matter volumetric structures on a T1-weighted image (Fischl et al., 2002), intensity normalized, registered to a spherical atlas, which used individual cortical folding patterns to match cortical geometry across participants (Fischl et al., 1999b), and parcellated into units based on gyral and sulcal structure (Desikan et al., 2006).
The structural biomarkers examined included: normalized whole brain volume and normalized hippocampal volume. Normalization was accomplished by computing the mean intracranial volume (ICV) for the current sample, performing a regression analysis using ICV as the independent variable and the raw volume as the dependent variable to obtain the b-weight, and then applied using the following equation: normalized volume = raw volume – [b-weight * (ICV for individual participant – mean sample ICV)] (Mathalon et al., 1993).
Psychometric battery
Independent of the CDR assessment, a psychometric test battery was administered to participants, typically within a few weeks of the CDR assessment. Psychometric tests common to all Knight Alzheimer’s Disease Research Center longitudinal protocols include Animal Naming (Goodglass and Kaplan, 1983), Trail Making A test (Armitage, 1946), Trail Making B test (Armitage, 1946), the Selective Reminding Test containing the Free Recall (SRTFREE) and Cued subtests (Grober et al., 1988), and the Mini-Mental State Examination (Folstein et al., 1975). Because baseline clinical and neuropsychometric assessments followed the National Alzheimer’s Disease Coordinating Center Uniform Data Set protocols (Morris et al., 2006; Beekly et al., 2007; Weintraub et al., 2009), data were available regarding behavioural changes, medications, and health history.
APOE ɛ genotyping
Briefly, all DNA samples underwent stringent quality control before genotyping with the Illumina 610 or the Omniexpress chip (Cruchaga et al., 2012). Complete information regarding APOE ɛ genotyping is available using previously described methods (Cruchaga et al., 2012).
Statistical analyses
Portions of the data were collected and managed using research electronic data capture (REDCap) tools (Harris et al., 2009). For all analyses, SAS statistical software version 9.4 (SAS Institute Inc.) was used, alpha = 0.05 was taken to indicate statistical significance, and all tests were two-tailed.
Biomarker prediction of incident CDR > 0
Six stepwise Cox proportional hazards models tested the association of each of the molecular biomarkers (PIB, adjusted CSF amyloid-β42, CSF tau, CSF ptau181, adjusted CSF tau/amyloid-β42, adjusted CSF ptau181/amyloid-β42) with time to incident CDR > 0. These models included terms for demographic [age, education, gender, race, number of APOE ɛ4 allele (0, 1, or 2) copies], psychometric (Animal Naming, Trail Making A, Trail Making B, SRTFREE, Selective Reminding Test – Cued Recall), and structural imaging (normalized hippocampal volume, normalized whole brain volume) measures for stepwise selection. Mini-Mental State Examination and CDR Sum of Boxes scores were not included as candidate variables because of extreme ceiling and floor effects. Because stepwise selection was used, data from participants with non-missing data on all candidate variables were used. Normalized hippocampal volume and normalized whole brain volume data from the MRI visit closest to the baseline clinical assessment were used. The significance level for entry and retention of each term was set at P = 0.05. Data from participants who did not become symptomatic over the follow-up period were censored at the date of last clinical assessment. We repeated the Cox proportional hazards models after first forcing age, gender, and education into the model and then allowed the stepwise selection method to select from the remaining variables those that met entry and retention criteria. Kaplan-Meyer survival curves and bubble plots were used to graphically illustrate survival findings.
Time remaining cognitively normal despite biomarker abnormality
For each molecular biomarker, we examined and reported the number and percentage of biomarker-positive individuals who remained cognitively normal among those followed at least 10 years. As exploratory analyses, we also examined differences in demographics and baseline cognitive scores for those who did and did not remain cognitively normal.
Comparison of biomarker changes across time for persons who did and did not develop incident CDR > 0
To examine longitudinal changes in biomarkers before and after CDR > 0 onset, spaghetti plots for individual biomarkers were constructed showing changes in that biomarker for persons who developed CDR > 0. A smoothed Loess curve illustrating biomarker changes before and after the first clinical assessment with CDR > 0 was fitted to the data. To compare biomarker changes in individuals who remained CDR 0 during an earlier and later period in their follow-up, we randomly chose an arbitrary clinical assessment date analogous to the date of incident CDR > 0 in the affected group. This allowed us to compare biomarker changes at an earlier and later period of follow-up within the same individual who remained CDR 0 (Fig. 1). A smoothed curve was then fitted to the data as described above. Linear mixed models were used to test whether there were differences between the slopes of biomarker change across time for the groups.
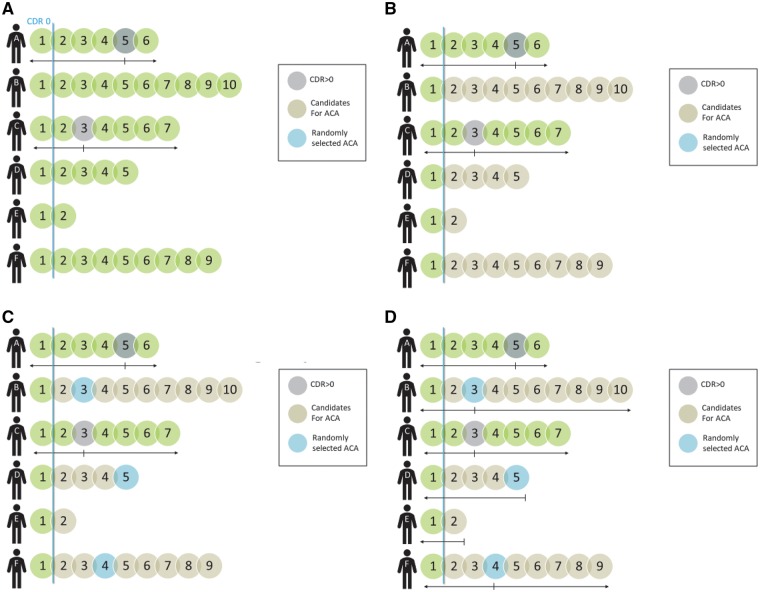
Figure 1.
Assignment of arbitrary clinical assessment (ACA) date. Only persons with a CDR of 0 at first assessment were included in analyses, so no participants could have developed CDR > 0 at that assessment (A). Participant A had a first CDR > 0 at Assessment 5, and Participant C at Assessment 3 (A). Biomarker behaviour before and/or after first assessment with a CDR > 0 was examined for Participants A and C, who developed cognitive impairment (B). For participants remaining CDR 0 across the follow-up period, all assessments following the first were candidates for ACA (B). An ACA was randomly assigned to one of the candidates (C). For candidates remaining CDR 0, biomarker behaviour before and/or after the assigned ACA was examined (D). Since Participant E only had one assessment after the first, Assessment 2 was assigned as his/her ACA (D).
Associations between APOE ɛ genotype and molecular biomarker cut-offs
Spaghetti plots illustrated changes in adjusted amyloid-β42, tau, and ptau181 values with time as they related to APOE ɛ genotype. Logistic regression was used to examine APOE ɛ genotypes as they relate to molecular biomarker cut-off values.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Data from n = 664 participants, ranging in age from 42 to 90 years at baseline (mean ± standard deviation = 67.6 ± 9.6 years), were available. Participants were followed for up to 16.9 years with a mean follow-up period of 6.2 ± 3.5 years. During follow-up, 145 (21.8%) participants became CDR > 0 and 519 remained cognitively normal. Table 1 presents the demographic characteristics of the sample.
Table 1.
Baseline demographics (n = 664)
| Developed CDR > 0 (n = 145) | Remained CDR 0 (n = 519) | Total | |||||
|---|---|---|---|---|---|---|---|
| n/Mean | %/SD | n/Mean | %/SD | P-value | n/Mean | %/SD | |
| Age, years | 73.9 | 7.6 | 65.9 | 9.3 | <0.0001 | 67.6 | 9.6 |
| Female, n | 83 | 57.2% | 307 | 59.2% | 0.680 | 390 | 58.7% |
| Minority race, n | 13 | 9.0% | 65 | 12.5% | 0.024 | 78 | 11.8% |
| APOE, n | 176 | 33.9% | 52 | 35.9% | 0.721 | ||
| ɛ2ɛ2 | 2 | 1.4% | 3 | 0.6% | 5 | 0.8% | |
| ɛ2ɛ3 | 15 | 10.3% | 62 | 12.0% | 77 | 11.6% | |
| ɛ2ɛ4 | 3 | 2.1% | 19 | 3.7% | 22 | 3.3% | |
| ɛ3ɛ3 | 76 | 52.4% | 278 | 53.6% | 354 | 53.3% | |
| ɛ3ɛ4 | 43 | 29.7% | 133 | 25.6% | 176 | 26.5% | |
| ɛ4ɛ4 | 6 | 4.1% | 24 | 4.6% | 30 | 4.5% | |
| Education, years | 15.7 | 3.2 | 16.0 | 2.6 | 0.175 | 15.9 | 2.7 |
| PIB, SUVR | 1.4 | 0.4 | 1.2 | 0.2 | <0.0001 | 1.2 | 0.3 |
| Adjusted CSF Aβ42 | −0.36 | .95 | 0.10 | 0.95 | <0.0001 | 0.0 | 1.0 |
| CSF tau, pg/ml | 371.0 | 206.0 | 276.3 | 151.9 | <0.0001 | 295.7 | 168.7 |
| CSF ptau181, pg/ml | 64.2 | 30.3 | 53.7 | 25.2 | <0.001 | 55.9 | 26.6 |
| Adjusted tau/Aβ42 | 0.62 | 1.48 | −0.16 | 0.77 | <0.0001 | 0.0 | 1.0 |
| Adjusted ptau181/Aβ42 | 0.56 | 1.40 | −0.14 | 0.82 | <0.0001 | 0.0 | 1.0 |
| SRTFREE | 26.9 | 6.2 | 31.3 | 5.6 | <0.0001 | 30.4 | 6.0 |
| Animal Naming | 19.0 | 5.4 | 22.0 | 5.6 | <0.0001 | 21.3 | 5.6 |
| Trail Making A | 36.8 | 15.2 | 31.6 | 12.3 | <0.001 | 32.8 | 13.1 |
| Trail Making B | 96.4 | 41.1 | 75.9 | 31.0 | <0.0001 | 80.4 | 34.5 |
| Normalized hippocampal volume, ml | 6.8 | 1.0 | 7.7 | 0.9 | <0.0001 | 7.6 | 1.0 |
| Normalized whole brain volume, ml | 963.7 | 63.2 | 1025.4 | 65.6 | <0.0001 | 10 164.4 | 68.7 |
| Follow-up time, years | 7.4 | 3.6 | 5.9 | 3.4 | <0.0001 | 6.2 | 3.5 |
Aβ = amyloid-β; SD = standard deviation.
Biomarker prediction of incident CDR > 0
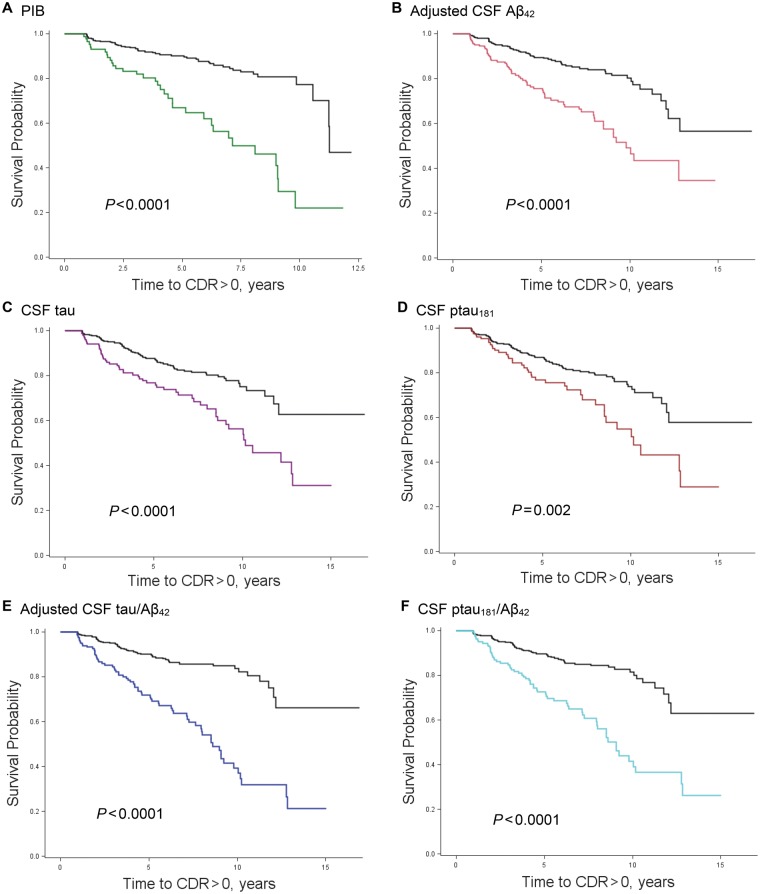
Figure 2 shows the survival curves for the unadjusted association between each of the molecular biomarkers and time to incident CDR > 0 for the 664 participants. Because non-missing data on all 13 candidate variables for stepwise selection were required, Cox proportional hazards analyses consisted of smaller subsamples (n = 286 for PIB and n = 302 for CSF). Forty-two participants in the PIB, and 39 in the CSF analyses developed CDR > 0. Molecular (PIB, CSF amyloid-β42, tau, tau/amyloid-β42, ptau/amyloid-β42), psychometric (SRTFREE), and structural (normalized hippocampal volume) biomarkers combined to predict first time to cognitive impairment in the models (Table 2). Trail Making B also independently contributed to prediction of CDR > 0 in the model testing PIB (P = 0.024). Age did not independently predict CDR > 0 once normalized hippocampal volume was included in the models. Supplementary Table 1 shows the results when age, gender and education were first forced into the models, and then the stepwise selection procedure was used to choose among the remaining candidate variables for model entry.
Figure 2.
Kaplan-Meier survival curves for molecular biomarkers. Time to first CDR > 0 for participants with abnormal (coloured line) and normal (black line) values of PIB (A), and CSF amyloid-β42 (B), tau (C), ptau181 (D), tau/amyloid-β42 (E), and ptau181/amyloid-β42 (F). Aβ = amyloid-β.
Table 2.
Results of Cox proportional hazards models
| P-value | HR | L95%CI | U95%CI | P-value | HR | L95%CI | U95%CI | |
|---|---|---|---|---|---|---|---|---|
| Pittsburgh Compound B | Adjusted CSF amyloid-β42 | |||||||
| Biomarker | 0.004 | 3.28 | 1.46 | 7.36 | 0.014 | 0.65 | 0.47 | 0.92 |
| SRTFREE | <0.0001 | 0.88 | 0.83 | 0.93 | 0.001 | 0.91 | 0.86 | 0.96 |
| Normalized hippocampal volume, ml | 0.004 | 0.59 | 0.41 | 0.84 | <0.0001 | 0.41 | 0.29 | 0.59 |
| Trail Making B | 0.024 | 1.01 | 1.001 | 1.02 | – | – | – | – |
| CSF tau | CSF ptau181 | |||||||
| Biomarker | 0.010 | 1.002 | 1.000 | 1.003 | – | – | – | – |
| SRTFREE | 0.002 | 0.92 | 0.87 | 0.97 | 0.002 | 0.91 | 0.86 | 0.97 |
| Normalized hippocampal volume, ml | <0.0001 | 0.40 | 0.28 | 0.57 | <0.0001 | 0.40 | 0.28 | 0.58 |
| Trail Making B | – | – | – | – | – | – | – | – |
| Adjusted CSF tau/amyloid-β42 | Adjusted CSF ptau181/amyloid-β42 | |||||||
| Biomarker | 0.000 | 1.54 | 1.22 | 1.96 | 0.003 | 1.44 | 1.13 | 1.83 |
| SRTFREE | 0.006 | 0.94 | 0.87 | 0.98 | 0.007 | 0.92 | 0.87 | 0.98 |
| Normalized hippocampal volume, ml | <0.0001 | 0.39 | 0.27 | 0.57 | <0.0001 | 0.38 | 0.26 | 0.55 |
| Trail Making B | – | – | – | – | – | – | – | – |
HR = hazard ratio; L95%CI = lower 95% confidence interval; U95%CI = upper 95% confidence interval.
Because normalized hippocampal volume was a consistent predictor in all models, bubble plots were used to graphically illustrate relationships between each of the molecular biomarkers and normalized hippocampal volume with regards to follow-up time, and CDR > 0 (Supplementary Fig. 1). As illustrated in those plots, none of the participants with a normalized hippocampal volume >8673 mm3 became CDR > 0 regardless of molecular biomarker values and length of follow-up. Similarly, lower SRTFREE baseline scores were associated with reduced time to symptomatic onset, such that only 1 of 34 persons (2.9%) with SRTFREE scores of 40 or above became CDR > 0 (data not shown).
Time remaining cognitively normal despite biomarker abnormality
Among participants followed for at least 10 years, the number and per cent of participants who were biomarker positive at baseline and who remained cognitively normal over the follow-up period were as follows: 15/18 (83.3%) for CSF amyloid-β42, 16/23 (69.6%) for CSF tau, 11/16 (68.8%) for CSF ptau181, 12/17 (70.6%) for CSF tau/amyloid-β42, and 13/17 (76.5%) for CSF ptau181/amyloid-β42. Only three participants with positive PIB values were followed for at least 10 years, and all three remained cognitively normal during this time. One participant with positive CSF biomarkers for tau, ptau181, CSF tau/amyloid-β42 and CSF ptau181/amyloid-β42 was followed for 15 years and remained cognitively normal. However, this participant had a normal baseline CSF amyloid-β42 value, and so may have not converted because he or she had a disease other than Alzheimer’s disease, such as suspected non-Alzheimer’s disease pathophysiology (SNAP) (Jack et al., 2016). Exploratory analyses indicated that persons who were resilient to underlying Alzheimer’s disease pathology tended to be younger and to have better performance on the Trail Making tests (Supplementary Table 2).
Comparison of biomarker changes across time for persons who did and did not develop incident CDR > 0
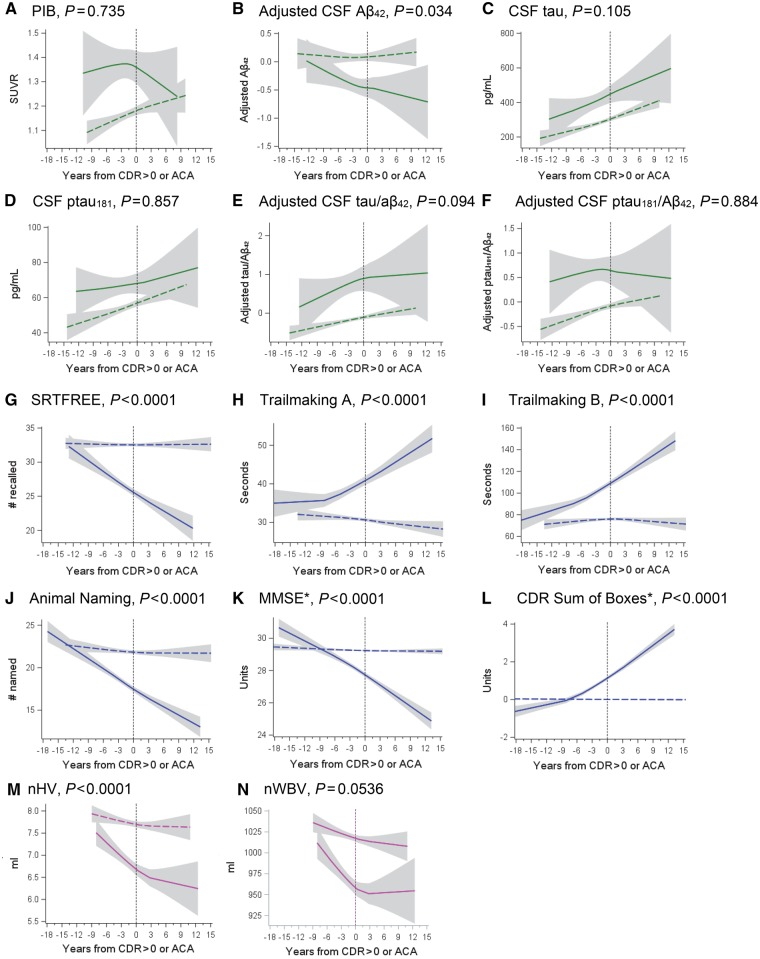
The length of biomarker data available for observation differed by biomarker and was dependent on how long each biomarker had been included in Knight Alzheimer’s Disease Research Center protocols. Including all biomarkers, data were collected from 8 May 1985 to 16 December 2016, a total of over 31 years. Specific information regarding when each biomarker was collected, and how much data were available per participant, is presented in Supplementary Table 3. As shown in Fig. 3, mixed linear models indicated that the rate of change of the molecular biomarkers, other than CSF amyloid-β42 (P = 0.034), did not differ for those who did and did not develop CDR > 0 (P > 0.094). Of note, Fig. 3A appears to show a downward curvature after the onset of CDR > 0 for that group. This is likely to be an artefact of the smaller number of data points following CDR 0.5 onset (see Fig. 4 for an illustration of how many data are available before and after CDR > 0 onset). However, highly significant group differences in rate of change were found for both cognitive biomarkers (Fig. 3G–L) and a structural measure—normalized hippocampal volume (Fig. 3M) (P < 0.0001). Group differences in the slope for the other structural measure—normalized whole brain volume—were marginally significant (P = 0.054). The overall magnitude of biomarker values, as reflected in the y-intercept (time 0), was more abnormal for the group that developed CDR > 0 for every biomarker examined compared to the group that remained cognitively normal (P < 0.0001). Supplementary Fig. 4 shows the Loess curves along with lines representing the linear fits.
Figure 3.
Biomarker changes for persons who did and did not develop CDR > 0. Comparison of selected biomarker, structural, and clinical changes for persons who did and did not develop CDR > 0. Solid lines represent those who developed dementia and dotted lines indicate persons who remained cognitively normal. P-values indicate whether there was a significant difference in the mean slope of each group. Aβ = amyloid-β; ACA = arbitrary clinical assessment; MMSE = Mini-Mental State Examination; nHV = normalized hippocampal volume; nWBV = normalized whole brain volume. *No persons had MMSE scores >30, nor CDR Sum of Boxes scores < 0. Extension of lines across the x-axis for these measures is an artefact of the curve-fitting process.
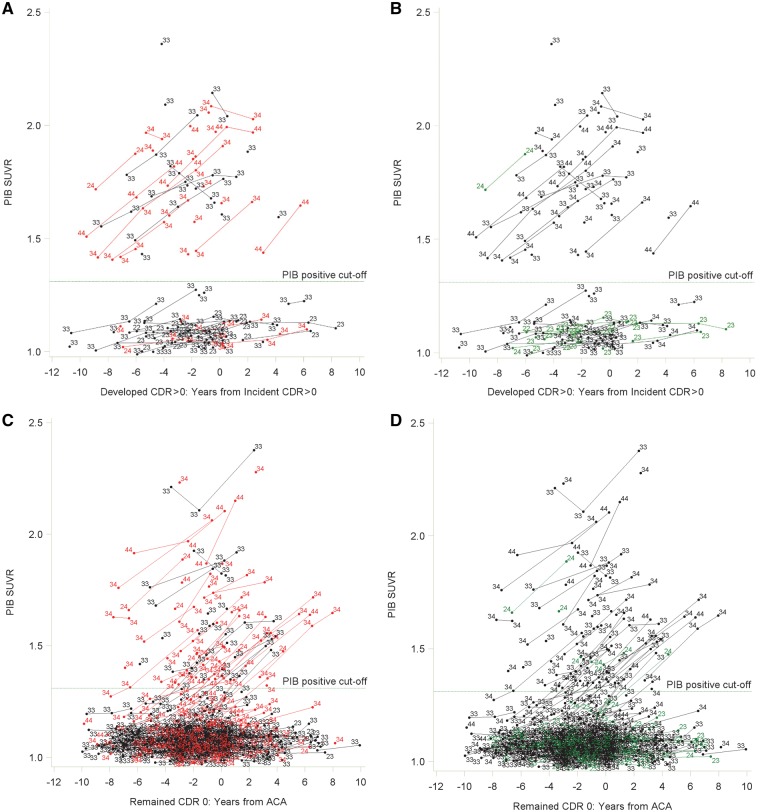
Figure 4.
Spaghetti plots for PIB accumulators and non-accumulators. Spaghetti plots for PIB accumulators and non-accumulators who developed CDR > 0 showing the relationship between having at least one APOE ɛ4 allele (in red) (A), having at least one APOE ɛ2 allele (in green) (B), and magnitude and changes in PIB SUVR values with time. Also shown are spaghetti plots illustrating relationships between having at least one APOE ɛ4 allele (C), having at least one APOE ɛ2 allele (D), and magnitude and changes in PIB SUVR values with time for individuals who remained cognitively normal over the follow-up period. Data points are labelled with the specific APOE ɛ genotype for that individual. ACA = arbitrary clinical assessment; APOE ɛ4 = at least one APOE ɛ4 allele; APOE ɛ2 = at least one APOE ɛ2 allele; SUVR = standard uptake value ratio.
Associations between APOE ɛ genotype and molecular biomarker values
Figure 4A and B shows that among those who developed CDR > 0, there was a clear separation of participants according to the amount and rate of amyloid accumulation. Individuals with high initial PIB values showed increased accumulation over the follow-up period, whereas those with low initial values generally did not show increased accumulation of fibrillar amyloid with time. For ease of discussion, and based on where the separation of the data occurred, individuals with PIB values above the PIB-positive cut-off of 1.31 SUVR (Vlassenko et al., 2016) were assigned to an ‘accumulator’ group, and those with PIB values below this cut-off were assigned to a ‘non-accumulator’ group. However, as can be seen in Fig. 4, any cut-off value within the approximate range of 1.3–1.4 SUVR could have been used. Linear mixed models confirmed that the rate of change in PIB values across the study period differed significantly for the accumulators and non-accumulators (P = 0.044).
Further, as shown in Fig. 4A, the majority of accumulators had one or more APOE ɛ4 alleles whereas the majority of non-accumulators did not. Figure 4B shows the same data, only now highlighting APOE ɛ2 individuals. APOE ɛ2 has a protective effect on amyloid accumulation, such that almost all participants with an APOE ɛ2 allele were non-accumulators. Only one participant in the accumulator group had an APOE ɛ2 allele, but that person also had an APOE ɛ4 allele. Logistic regression analyses confirmed the graphic information, indicating that being an accumulator was highly associated with having an APOE ɛ4 allele [odds ratio (OR) = 9.73, 95% CI = 2.87–32.94, P < 0.001], and that accumulators were less likely to have an APOE ɛ2 allele (OR = 0.10, 95% CI = 0.01–0.90, P = 0.040), and were older (OR = 1.07, 95% CI = 1.004–1.15, P = 0.038) than non-accumulators.
Among participants who remained cognitively normal (Fig. 4C and D), there was no clear separation into accumulator and non-accumulator groups. To explore the proportion of CDR 0 participants who showed this rapid increase in PIB, analogous to the CDR > 0 accumulators, we first found the 25th percentile of the slopes of the CDR > 0 accumulators. We then operationally defined an ‘accumulator’ in the CDR 0 group as persons with slopes above that 25th percentile. Of the 208 persons who remained cognitively normal and had at least two PIB measurements, 64 (30.8%) could be considered to be accumulators. In the CDR 0 group, APOE ɛ genotype was again associated with PIB behaviour across time. The majority of cognitively normal persons with high and rising PIB values had an APOE ɛ4 allele (Fig. 4C), whereas most persons with APOE ɛ2 values had low, stable values of PIB across the follow-up period (Fig. 4D). All cognitively normal persons with APOE ɛ2 who showed high and rising PIB values also had an APOE ɛ4 allele (Fig. 4D).
T-tests indicated that among those who developed CDR > 0, PIB accumulators were found to have lower mean values of adjusted CSF amyloid-β42, higher values of CSF ptau181, smaller normalized whole brain volume, and similar cognition (Supplementary Table 4).
In contrast to the PIB results, CSF biomarkers did not show a clear pattern of separation observed for participants who developed CDR > 0. The association between APOE ɛ genotype and CSF biomarkers was also different from those observed for PIB accumulators and non-accumulators (Fig. 5 and Supplementary Figs 2 and 3). Here, many individuals who became CDR > 0 and who were APOE ɛ4 had CSF amyloid-β42, tau, and ptau181 values in the normal range, and a few CDR > 0 and who were APOE ɛ2 had CSF biomarker values in the abnormal range. However, logistic regression indicated that generally, APOE ɛ genotypes were associated with lower CSF amyloid-β42 values for persons who developed CDR > 0 [APOE ɛ4: OR (95% CI) = 7.68 (2.17–27.26), P = 0.002; APOE ɛ2: OR (95% CI) = 0.14 (0.03–0.81), P = 0.028] and those who did not [APOE ɛ4: OR (95% CI) = 3.17 (2.04–4.94), P < 0.0001; APOE ɛ2: OR (95% CI) = 0.53 (0.27–1.02), P = 0.055]. No significant relationships were found between APOE ɛ and tau and ptau181 values (P > 0.096).
Figure 5.
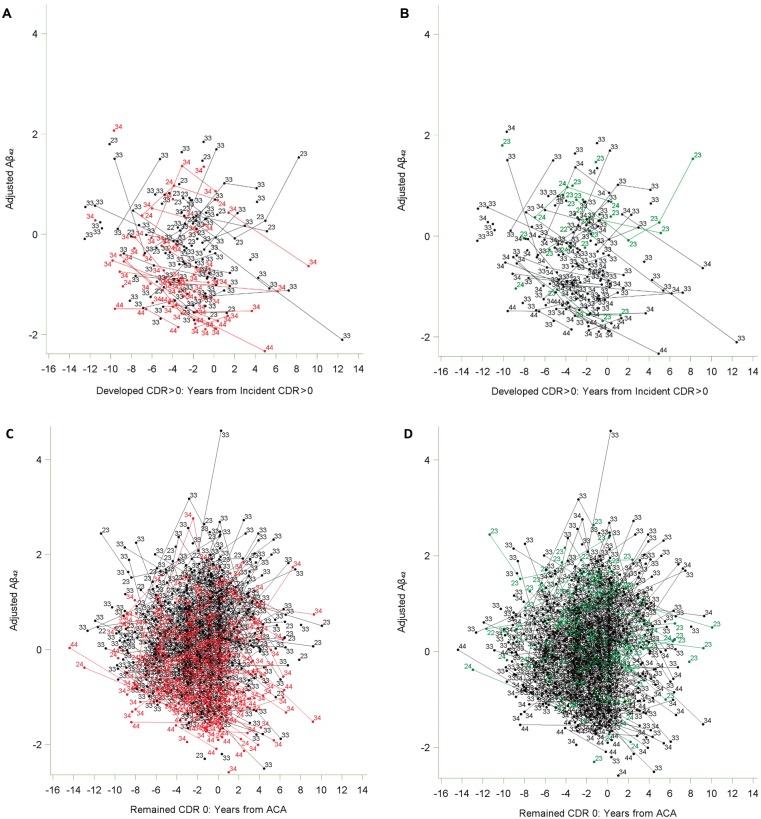
Spaghetti plots for adjusted CSF amyloid-β42. Spaghetti plots for CSF adjusted amyloid-β42 showing the relationship between having at least one APOE ɛ4 allele (in red), having at least one APOE ɛ2 allele (in green), and magnitude and changes in CSF amyloid-β42 with time for participants who did (A and B) and did not (C and D) develop CDR > 0. Data points are labelled with the specific APOE ɛ genotype for that individual. Because appropriate cut-off values for amyloid-β42 depended on date and lot number, no reference line is presented. Aβ = amyloid-β; ACA = arbitrary clinical assessment.
Discussion
We explored how molecular, psychometric, and structural biomarkers predict time to CDR > 0, and how they change close to the time of CDR > 0 onset. Results from our first set of analyses indicate that molecular (amyloid imaging and CSF), cognitive (free recall), and structural (normalized hippocampal volume) biomarkers independently contribute to prediction of the onset of symptomatic sporadic Alzheimer’s disease. Although past work has demonstrated that these biomarkers individually are predictive of incident dementia (Grober et al., 2000; Fagan et al., 2007; Morris et al., 2009; Wolz et al., 2011), our results indicate that given a wide variety of biomarkers to choose from, these three biomarker types provide different, non-redundant information and combine to predict onset of CDR > 0.
As expected from previous longitudinal studies (Fagan et al., 2007; Morris et al., 2009; Stomrud et al., 2015; Baker et al., 2017; Donohue et al., 2017; Dumurgier et al., 2017), all molecular biomarkers, with the exception of CSF ptau181, predicted time to onset of incident cognitive impairment. However, around 70% of cognitively normal persons with abnormal Alzheimer’s disease molecular biomarkers remained cognitively normal for at least the next 10 years. Potential reasons why participants with abnormal molecular biomarkers may remain cognitively normal include greater cognitive reserve (Stern, 2012) and fewer other risk factors for dementia (e.g. head trauma, diabetes, hypertension, cerebrovascular disease). Although based on small numbers of participants, our results suggest that persons resilient to the effects of underlying Alzheimer’s disease pathology may be younger and have better scores on the Trail Making tests. One participant remained cognitively normal 15 years after positive tau and ptau181 measurements. However, because this participant had a normal CSF amyloid-β42 value, he or she may have had a non-Alzheimer’s disease disorder, such as SNAP (Jack et al., 2016).
Better structural (as measured by larger normalized hippocampal volume) and cognitive performance (higher SRTFREE scores) values at baseline were associated with later onset of CDR > 0. Persons with normalized hippocampal volume >8673 mm3 did not become CDR > 0 regardless of length of follow-up or baseline molecular biomarker values. Although these results are consistent with the brain reserve hypothesis (Stern, 2014), which would suggest that large hippocampal volumes at baseline may protect against the development of incident symptomatic Alzheimer’s disease, it is possible that smaller hippocampi resulted from a cumulative neurodegenerative process. As seen in Fig. 3M, participants who subsequently developed CDR > 0 had similar hippocampal volumes as those who remained CDR = 0 at 10 or more years prior to symptomatic onset. Some of the observed decreases that may start to develop at 10 years before clinical onset of symptoms may reflect tau-mediated changes within the hippocampus; however, further studies using tau PET are needed. Likewise, only one (2.9%) participant with a SRTFREE score above 39 developed CDR > 0 regardless of molecular biomarker values and length of follow-up. Of note, neither structural (normalized hippocampal volume) nor free recall performance (SRTFREE) are considered when calculating CDR, yet they are often performed in the clinical evaluation of patients.
Others have reported that smaller normalized hippocampal volumes are associated with ageing (Knopman et al., 2016). Both normalized hippocampal volume (r = −0.563) and whole brain volume (r = −0.666) were highly correlated with age (P ≤ 0.0001), and with each other (r = 0.700, P < 0.0001) in our sample, indicating that they share variance. Variables that are highly related can be thought of as proxies of each other, and therefore, once normalized hippocampal volume was included in stepwise models, age contributed little additional independent predictive information, and so failed to enter the models predicting time to incident CDR > 0. Indeed, when age was first forced into each model before stepwise selection of other variables, age was not significant in the CSF tau and ptau181 models upon stepwise entry of normalized hippocampal volume.
In our second, and main, set of analyses examining the behaviour of molecular biomarkers surrounding transition to CDR > 0, we found no differences in the rate of change for participants who did and did not develop cognitive impairment (with the exception of CSF amyloid-β42). However, individuals who developed CDR > 0 had abnormal intercept values for each of the biomarkers. Assuming that participants had similar levels of molecular biomarkers early in life, the separation between the groups, reflected in the magnitudes of the intercepts, must have occurred years prior to our observation period.
However, the assumption that participants had similar levels of molecular biomarkers at an earlier time point may not hold true for all molecular biomarkers. Studies in autosomal dominant Alzheimer’s disease have suggested that initial amyloid-β42 levels at younger ages before symptom onset are higher among mutation carriers compared to mutation non-carriers but that there are no differences in CSF tau and ptau181 values (Bateman et al., 2012; Reiman et al., 2012). Later, amyloid-β42 values decrease for mutation carriers compared to non-carriers (Bateman et al., 2012). These studies have also demonstrated overproduction of CSF amyloid-β42 in mutation carriers compared to controls in vivo (Potter et al., 2013). Results have been interpreted as consistent with a model of autosomal dominant Alzheimer’s disease development whereby increased, abnormal amyloid-β42 production occurs, followed by a reduction in CSF amyloid-β42 levels as amyloid-β is sequestered into amyloid plaques (Potter et al., 2013). It has been suggested that a similar process occurs in sporadic Alzheimer’s disease (Selkoe and Hardy, 2016). Thus, if changes in CSF amyloid-β42 occur first in the pathological process, such that amyloid-β42 levels are higher initially (i.e. before the period of observation in this study) in persons who will develop Alzheimer’s disease, but levels of other biomarkers are similar, as suggested by autosomal dominant Alzheimer’s disease research, our results do not imply that changes in CSF amyloid-β42 occur before those of other molecular biomarkers. CSF amyloid-β42 data collected many years before onset of cognitive impairment are required to address this possibility. Further, CSF amyloid-β42 values are thought to reflect an ongoing pathological state, rather than accumulation of neuropathological load (Jack et al., 2018). Finally, the P-value indicating a difference in slope of CSF amyloid-β42 for participants who did and did not progress was relatively large (P = 0.034) compared to the P-values indicating slope differences for normalized hippocampal volume and the cognitive biomarkers, suggesting that replication of these results is necessary.
Persons who developed CDR > 0 had significantly greater decline in structural (normalized hippocampal volume) and cognitive (SRTFREE, Trail Making A and B) values compared to those who remained cognitively normal. Overall these results complement existing hypothesized models and suggest that molecular biomarker changes occur prior to structural or cognitive markers in the Alzheimer’s disease pathological process (Jack et al., 2013).
These results have implications for clinical trials in preclinical Alzheimer’s disease if replicated in other cohorts. Research into disease-modifying drug treatment relies on assessing change in the slope of decline as an outcome to demonstrate efficacy (Aisen, 2015). Therefore, desirable outcomes for clinical trials in preclinical Alzheimer’s disease are those that show clear, abnormal decline among persons who will develop symptomatic Alzheimer’s disease compared to those who will not, and where the decline occurs relatively close to the time of symptomatic onset.
Because changes in molecular biomarkers, other than CSF amyloid-β42, were similar for those who did and did not develop CDR > 0 around the time of onset or arbitrary clinical assessment, little to no change in trajectory would be expected for these measures regardless of the efficacy of the treatment. Instead, our results support the idea that changes in cognition should be considered in preclinical Alzheimer’s disease populations (Henley et al., 2015). Separation between the Mini-Mental State Examination and CDR Sum of Boxes mean scores occurs at around 8 years prior to symptomatic onset in our sample (Fig. 3K and L). The separation between the groups who did and did not develop symptomatic Alzheimer’s disease occurred even earlier, around 12 years, on the other cognitive tests examined here: SRTFREE, Trail Making A and B, and Animal Naming (Fig. 3G–J). These tests do not have ceiling effects among cognitively normal persons, unlike the Mini-Mental State Examination. Our results also suggest that structural change in normalized hippocampal volume (Fig. 3M) shows dramatic decline prior to symptomatic onset and may also be a suitable choice to investigate the effects of disease-modifying treatments. On the other hand, the molecular biomarkers that show overall separation in magnitude between those who do and do not go on to develop Alzheimer’s disease, but show little slope differences across the groups, may be most useful in screening for inclusion in clinical trials rather than as outcomes.
The finding of slope differences in one measure of amyloid, CSF amyloid-β42, but not in another, PIB, may be associated with differences in what each measure represents. It is possible that CSF amyloid-β42 may reflect what is occurring at the time of study, while PIB may reflect not only changes at the time of scan, but also total accumulation up to that point.
A surprising finding was the stark division of PIB values among persons who developed CDR > 0. Among these participants (Fig. 4A and 4B), PIB longitudinal behaviour separated into two distinct groups. One group had high (above the 1.31 SUVR criterion for PIB positivity, Vlassenko et al., 2016) and rising values for PIB as symptomatic onset approached, the other group had initial PIB values below the cut-off and maintained PIB negativity across the observation period, despite developing CDR > 0. No other molecular biomarkers examined here, including CSF amyloid-β42 showed this same pattern of separation (Fig. 4 and Supplementary Figs 2 and 3). These results are consistent with work by others suggesting that there are PIB ‘accumulators’ and ‘non-accumulators’ (Villain et al., 2012). Accumulators who developed CDR > 0 tended to have more abnormal CSF amyloid-β42 and ptau181 biomarker values, but similar hippocampal volumes and cognitive scores (Supplementary Table 4). Considered in the context of the recently published National Institute on Aging – Alzheimer Association guidelines (Jack et al., 2018), these results suggest that accumulators may be further along on the Alzheimer’s disease pathological continuum compared to non-accumulators, or that non-accumulators may have developed a CDR > 0 due, at least in part, to factors that may independently increase the risk of Alzheimer’s disease (e.g. vascular factors, lower cognitive reserve), or their cognitive impairment might be non-Alzheimer’s disease in nature (e.g. Lewy body dementia, vascular dementia).
In contrast, among those who remained cognitively normal, there was no clear separation point of PIB behaviour into two groups. As would be expected, some people showed increasing PIB levels with time, rising from the group with low, stable values, suggesting that these people may be transitioning to CDR > 0 due to Alzheimer’s disease, whereas others show little increase in PIB levels across the follow-up period (Fig. 4A and B). Based on the slopes of accumulators in the CDR > 0 group, we found that 30.8% of those who remained CDR 0 could be considered accumulators suggesting that these individuals may be transitioning to preclinical Alzheimer’s disease.
Analyses indicated that having an APOE ɛ4 allele is strongly associated with being a PIB accumulator, and that having an APOE ɛ2 allele is associated with being a non-accumulator. No clear separation into accumulator and non-accumulator groups was shown for CSF molecular biomarkers. CSF amyloid-β42, but not tau or ptau181, was also linked to APOE ɛ genotype, consistent with previous cross-sectional findings (Morris et al., 2010).
In addition to the possibility, mentioned earlier, that cognitive impairment may be due to a condition other than Alzheimer’s disease, our research has other limitations. We used a sample of research volunteers willing to take part in cognitive, imaging, and molecular biomarker assessments. Therefore, the degree to which the results of this study will generalize to the larger population is unknown. The amount of time during which particular biomarkers were collected varied over the course of data collection, such that more information was available for some biomarkers (e.g. CDR Sum of Boxes) than for others (e.g. PIB). Greater statistical power and stability of findings is yielded by biomarkers with more data available. Both the confidence intervals and number of participants contributing data (Supplementary Fig. 5) indicate that for all biomarkers, greatest confidence in the stability of findings occurs approximately 9 years before, and 6 years after, incident cognitive impairment. As noted earlier, hippocampal volume is related to age. Hippocampal volume is also susceptible to TDP-43 pathology (Josephs et al., 2017) and hippocampal sclerosis (Leverenz et al., 2002); therefore, the extent to which hippocampal atrophy is due to Alzheimer’s disease or another pathology in this study is unknown. As noted earlier, CSF amyloid-β42 values in our cohort have exhibited upward drift over the years (Schindler et al., 2018) although we attempted to control for drift statistically by adjusting values by lot number and date. As in previous work, we examined time to first CDR > 0 (Fagan et al., 2007; Roe et al., 2013). However, it is possible that persons who developed CDR > 0 may receive a CDR 0 on a subsequent assessment, but in our experience, often these individuals will eventually progress to Alzheimer’s disease. The analysis of those individuals who oscillate is outside the realm of the current study and will be investigated in future manuscripts. Also, the high number of participants with SNAP in this cohort limits the ability to make conclusions regarding Alzheimer’s disease biomarkers. Given these limitations, replication of our results in additional samples is needed.
Despite these limitations, these results generally support the pathological sequence of biomarker events proposed in current theoretical models of Alzheimer’s disease and help to provide specific time points as to when biomarker changes begin to occur in the sequence of Alzheimer’s disease development.
Supplementary Material
Acknowledgements
The authors thank the participants, investigators, and staff of the Knight Alzheimer’s disease Research Center Clinical Core for participant assessments, Genetics Core for APOE ɛ genotyping, Biomarker Core for cerebrospinal fluid analysis, and the Imaging Core for amyloid and structural imaging. Imaging facilities were supported by the Washington University Institute of Clinical and Translational Sciences grant UL1TR000448 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). Imaging analyses used the services of the Neuroimaging Informatics and Analysis Center, supported by NIH grant 5P30NS048056.
Glossary
Abbreviations
- CDR
Clinical Dementia Rating
- PIB
Pittsburgh compound B
- SRTFREE
Selective Reminding Test – Free Recall
Funding
Funding for this study was provided by the National Institute on Aging [R01AG056466, R01AG043434 P50AG005681, P01AG003991, and P01AG026276]; Fred Simmons and Olga Mohan, the Farrell Family Research Fund, the Paula and Rodger O. Riney Fund, the Daniel J Brennan MD Fund, and the Charles and Joanne Knight Alzheimer’s Research Initiative of the Washington University Knight Alzheimer’s disease Research Center. The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Competing interests
The authors report no competing interests.
References
- Aisen PS. Cognitive/clinical endpoints for pre-dementia AD trials. J Prev Alzheimers Dis 2015; 2: 82–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage SG. An analysis of certain psychological tests used in the evaluation of brain injury. Psychological Monographs 1946; 60: 1–48. [Google Scholar]
- Baker JE, Lim YY, Pietrzak RH, Hassenstab J, Snyder PJ, Masters CL et al. Cognitive impairment and decline in cognitively normal older adults with high amyloid-β: a meta-analysis. Alzheimer’s Dement 2017; 6: 108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 2012; 367: 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekly DL, Ramos EM, Lee WW, Deitrich WD, Jacka ME, Hubbard JL et al. The National Alzheimer’s Coordinating Center (NACC) database: the uniform data set. Alzheimers Dis Assoc Disord 2007; 21: 249–58. [DOI] [PubMed] [Google Scholar]
- Buchhave P, Minthon L, Zetterberg H, Wallin ÅK, Blennow K, Hansson O. Cerebrospinal fluid levels of β-amyloid 1–42, but not of tau, are fully changed already 5 to 10 years before the onset of Alzheimer dementia. Arch Gen Psychiatry 2012; 69: 98–106. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Nowotny P, Bales K, Pickering EH, Mayo K et al. Cerebrospinal fluid APOE levels: An endophenotype for genetic studies for Alzheimer’s disease. Hum Mol Genet 2012; 21: 4558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 1999; 9: 179–94. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006; 31: 968–80. [DOI] [PubMed] [Google Scholar]
- Donohue MC, Sperling RA, Petersen R, Sun CK, Weiner M, Aisen PS. Association between elevated brain amyloid and subsequent cognitive decline among cognitively normal persons. JAMA 2017; 317: 2305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Hampel H, Feldman HH, Scheltens P, Aisen P, Andrieu S et al. Preclinical Alzheimer’s disease: definition, natural history, and diagnostic criteria. Alzheimers Dementia 2016; 12: 292–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumurgier J, Hanseeuw BJ, Hatling FB, Judge KA, Schultz AP, Chhatwal JP et al. Alzheimer’s disease biomarkers and future decline in cognitive normal older adults. J Alzheimers Dis 2017; 60: 1451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Aá42 in humans. Ann Neurol 2006; 59: 512–19. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Roe CM, Xiong C, Morris JC, Holtzman DM. Cerebrospinal fluid tau/á-amyloid 42 ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol 2007; 64: 343–9. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Xiong C, Jasielec MS, Bateman RJ, Goate AM, Benzinger TLS et al. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci Transl Med 2014; 6; 226ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33: 341–55. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system. Neuroimage 1999a; 9: 195–207. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RBH, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp 1999b; 8: 272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher E, Villeneuve S, Maillard P, Harvey D, Reed B, Jagust W et al. beta-amyloid, hippocampal atrophy and their relation to longitudinal brain change in cognitively normal individuals. Neurobiol Aging 2016; 40: 173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. Mini-mental State: a practical method for grading the cognitive state of patients for the clinicians. J Psychiatr Res 1975; 12: 189–98. [DOI] [PubMed] [Google Scholar]
- Goodglass H, Kaplan E. Boston diagnostic aphasia examination booklet. Philadelphiam, PA: Lea & Febiger; 1983. [Google Scholar]
- Grober E, Buschke H, Crystal H, Bang S, Dresner R. Screening for dementia by memory testing. Neurology 1988; 38: 900–3. [DOI] [PubMed] [Google Scholar]
- Grober E, Lipton RB, Hall C, Crystal H. Memory impairment on free and cued selective reminding predicts dementia. Neurology 2000; 54: 827–32. [DOI] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage 2006; 32: 180–94. [DOI] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42: 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley DB, Dowsett SA, Chen YF, Liu-Seifert H, Grill JD, Doody RS et al. Alzheimer’s disease progression by geographical region in a clinical trial setting. Alzheimers Res Ther 2015; 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel PS, Ossenkoppele R, Gessert D, Jagust W, Landau S, Hansson O et al. Time to amyloid positivity and preclinical changes in brain metabolism, atrophy, and cognition: evidence for emerging amyloid pathology in Alzheimer’s disease. Front Neurosci 2017; 11: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018; 14: 535–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Wiste HJ, Weigand SD, Knopman DS, Mielke MM, Vemuri P et al. Different definitions of neurodegeneration produce similar amyloid/neurodegeneration biomarker group findings. Brain 2015; 138 (Pt 12): 3747–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Chételat G, Dickson D, Fagan AM, Frisoni GB et al. Suspected non-Alzheimer disease pathophysiology-concept and controversy. Nat Rev Neurol 2016; 12: 117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013; 12: 207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen WJ, Ossenkoppele R, Knol DL, Tijms BM, Scheltens P, Verhey FRJ et al. Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA 2015; 313: 1924–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, Dickson DW, Tosakulwong N, Weigand SD, Murray ME, Petrucelli L et al. Rates of hippocampal atrophy and presence of post-mortem TDP-43 in patients with Alzheimer’s disease: a longitudinal retrospective study. Lancet Neurol 2017; 16: 917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE. Amyloid imaging as a biomarker for cerebral β-amyloidosis and risk prediction for Alzheimer dementia. Neurobiol Aging 2011; 32(Suppl 1): S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopman DS, Jack CR, Wiste HJ, Weigand SD, Vemuri P, Lowe VJ et al. Age and neurodegeneration imaging biomarkers in persons with Alzheimer disease dementia. Neurology 2016; 87: 691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverenz JB, Agustin CM, Tsuang D, Peskind ER, Edland SD, Nochlin D et al. Clinical and neuropathological characteristics of hippocampal sclerosis: a community-based study. Arch Neurol 2002; 59: 1099–106. [DOI] [PubMed] [Google Scholar]
- Mathalon DH, Sullivan EV, Rawles JM, Pfefferbaum A. Correction for head size in brain-imaging measurements. Psychiatry Res 1993; 50: 121–39. [DOI] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 1993; 43: 2412–4. [DOI] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Grant EA, Head D, Storandt M, Goate AM et al. Pittsburgh Compound B imaging and prediction of progression from cognitive normality to symptomatic Alzheimer’s disease. Arch Neurol 2009; 66: 1469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol 2010; 67: 122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Weintraub S, Chui HC, Cummings J, DeCarli C, Ferris S et al. The Uniform Data Set (UDS): clinical and cognitive variables and descriptive data from Alzheimer Disease Centers. Alzheimers Dis Assoc Disord 2006; 20: 210–16. [DOI] [PubMed] [Google Scholar]
- Potter R, Patterson BW, Elbert DL, Ovod V, Kasten T, Sigurdson W et al. Increased in vivo amyloid-beta42 production, exchange, and loss in presenilin mutation carriers. Sci Transl Med 2013; 5: 189ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, McKeel DW, Buckles VD, Roe CM, Xiong C, Grundman M et al. Neuropathology of nondemented aging: presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging 2009; 30: 1026–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Quiroz YT, Fleisher AS, Chen K, Velez-Pardo C, Jimenez-Del-Rio M et al. Brain imaging and fluid biomarker analysis in young adults at genetic risk for autosomal dominant Alzheimer’s disease in the presenilin 1 E280A kindred: a case-control study. Lancet Neurol 2012; 11: 1048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe CM, Fagan AM, Grant EA, Hassenstab J, Moulder KL, Maue Dreyfus D et al. Amyloid imaging and CSF biomarkers in predicting cognitive impairment up to 7.5 years later. Neurology 2013; 80: 1784–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler SE, Sutphen CL, Teunissen C, McCue LM, Morris JC, Holtzman DM et al. Upward drift in cerebrospinal fluid amyloid β 42 assay values for more than 10 years. Alzheimers Dement 2018; 14: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 2016; 8: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7: 280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. Lancet Neurol 2012; 11: 1006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern Y. Cognitive reserve: implications for assessment and intervention. Folia Phoniatr Logop 2014; 65: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomrud E, Minthon L, Zetterberg H, Blennow K, Hansson O. Longitudinal cerebrospinal fluid biomarker measurements in preclinical sporadic Alzheimer’s disease: a prospective 9-year study. Alzheimers Dement 2015; 1: 403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, D’Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS et al. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One 2013; 8: e73377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphen CL, Jasielec MS, Shah AR, Macy EM, Xiong C, Vlassenko AG et al. Longitudinal cerebrospinal fluid biomarker changes in preclinical alzheimer disease during middle age. JAMA Neurol 2015; 72: 1029–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Zetterberg H, van Harten AC, Glodzik L, Martinez-Lage P, Bocchio-Chiavetto L et al. Alzheimer’s disease cerebrospinal fluid biomarker in cognitively normal subjects. Brain 2015; 138 (Pt 9): 2701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain N, Chételat G, Grassiot B, Bourgeat P, Jones G, Ellis KA et al. Regional dynamics of amyloid-β deposition in healthy elderly, mild cognitive impairment and Alzheimer’s disease: a voxelwise PiB-PET longitudinal study. Brain 2012; 135: 2126–39. [DOI] [PubMed] [Google Scholar]
- Vlassenko AG, McCue L, Jasielec MS, Su Y, Gordon BA, Xiong C et al. Imaging and cerebrospinal fluid biomarkers in early preclinical alzheimer disease. Ann Neurol 2016; 80: 379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SJB, Xiong C, Visser PJ, Jasielec MS, Hassenstab J, Grant EA et al. Preclinical Alzheimer’s disease and its outcome: a longitudinal cohort study. Lancet Neurol 2013; 12: 957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub S, Salmon D, Mercaldo N, Ferris S, Graff-Radford NR, Chui H et al. The Alzheimer’s Disease Centers’ Uniform Data Set (UDS): the neuropsychological test battery. Alzheimers Dis Assoc Disord 2009; 23: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MM, Roe CM, Morris JC. Stability of the clinical dementia rating: 1979–2007. Arch Neurol 2009; 66: 773–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D et al. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS One 2011; 6: e25446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.