Abstract
Introduction
Renal cell carcinoma (RCC) accounts for 3% of adult malignancies and more than 90% of kidney neoplasms. High rates of undiagnostic percutaneous kidney biopsies and difficulties in reliable pre-operative differentiation between malignant and benign renal tumors using contemporary imaging techniques result in large numbers of redundant surgeries. Absence of specific biomarkers for early detection and monitoring complicates on-time diagnosis of the disease and relapse. For the patients followed up after having a nephrectomy, a noninvasive and sensitive biomarker enabling early detection of disease relapse would be extremely useful.
Material and methods
The study is a review of recent knowledge regarding potential clinical applications of microRNAs (miRNAs) as biomarkers of RCC.
Results
MicroRNAs are essential regulators of various processes such as cell proliferation, differentiation, development and death; they have been implicated in diverse biological and pathological processes in RCC. There is a class of miRNAs that promote RCC development (oncomirs) and a class of miRNAs that negatively regulate oncogenes, suppress tumor growth and invasion, and thus could be considered treatment agents (anti-oncomirs). Separate miRNAs and specific miRNAs expression profiles have been identified, enabling early detection of the disease, prediction of response to systemic therapy, or prognostication of biological behavior of the disease.
Conclusions
The miRNA network analysis and gene profiling may help to identify the most sensible molecular signatures of RCC that can be used for diagnostic purposes, as well as poor prognosis signatures and poor therapeutic response signatures in patients who undergo systemic therapy.
Keywords: renal cell carcinoma, microRNA, diagnostics, prognosis, prediction, biomarker
INTRODUCTION
Renal cell carcinoma (RCC) is a relatively common pathology that is found in roughly 3% of all cases of malignant neoplasia in adults and approximately 90% of malignant kidney tumors. Nearly 1 in 69 men and 1 in 116 women will be diagnosed with RCC during their life. According to data of the U.S. National Cancer Institute, in 2016 the estimated number of new RCC cases was 62700, while the number of estimated deaths was 14240 (2.4% of all mortality due to oncological pathology). At the same time the 5-year survival rate of patients with RCC was 73.7% [1]. The most prevalent histological subtypes of renal cancer, are: clear-cell RCC (ccRCC, 60–80% of all patients), papillary RCC (pRCC, 10–15%), chromophobe RCC (chRCC, 5–10%) and other rare subtypes (<1%) [2]. Many molecular agents such as hypoxia-inducible factor (HIF), vascular endothelial growth factor (VEGF), carbonic anhydrase IX (CaIX), phosphatase and tensin homolog (PTEN), C-reactive protein (CRP), osteopontin, E-cadherin, CXCR4 (C-X-C chemokine receptor type 4), CD44, Ki67, p21, p53 and other potential RCC biomarkers have been investigated, however, not one of them demonstrated reliability in diagnostics, prediction of the treatment outcome or prognosis [3–6].
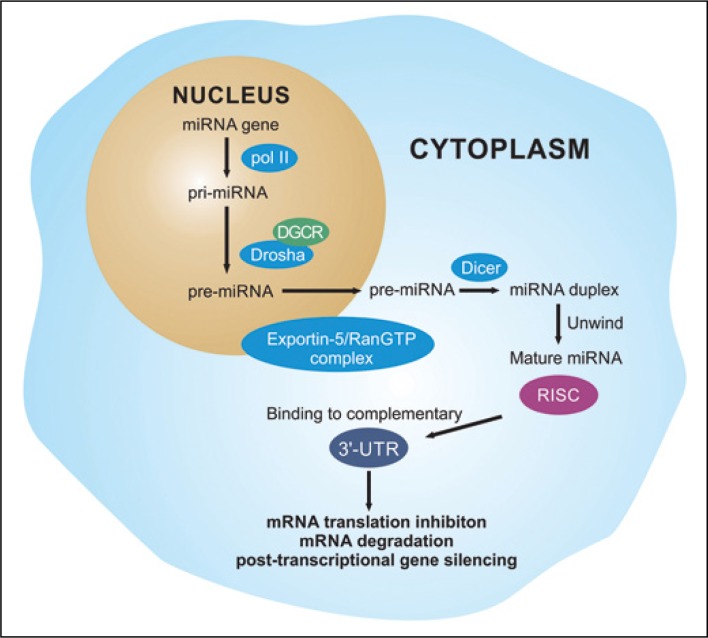
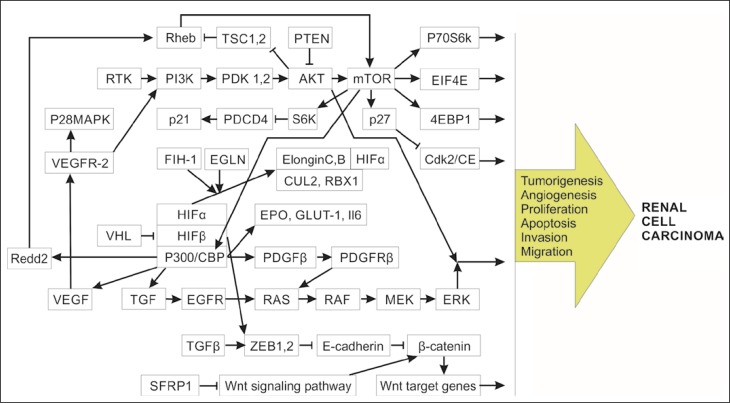
In the last decade the role of microRNAs (miRNA, miR) in the development of RCC in order to assess their potential diagnostic, predictive and prognostic value was intensively explored. The miRNAs are small non-coding RNAs that regulate the expression of a broad spectrum of genes by affecting the 3ˊ-untranslated regions (3ˊ-UTR) of complementary mRNAs (Figure 1). The miRNAs regulate cell growth and cell cycle, apoptosis, replicative potential, angiogenesis, tissue invasion and metastasizing in RCC development (Figure 2) [7]. There is a class of miRNAs that promote cancer development (oncomirs) and, conversely – a class of miRNAs that negatively regulate oncogenes, suppress tumor growth and invasion, and thus could be considered treatment agents for RCC (anti-oncomirs) [8]. Currently, no miRNAs are used in wide clinical practice, nevertheless the results of multiple studies suggest exceptional potential of miRNAs as RCC biomarkers.
Figure 1.
The mechanisms of miRNA genesis and processing.
poll II – RNA polymerase II; pri-miRNA – primary miRNA; Drosha – RNase III enzyme; DGCR8 – Drosha cofactor (Pasha); pre-miRNA – precursor miRNA; RISC – RNA-induced silencing complex
Figure 2.
Mechanism of renal cell carcinoma pathogenesis based on dysregulated signaling pathways.
MicroRNAs in diagnostics of renal cell carcinoma
Currently, a high rate of undiagnostic percutaneous kidney biopsies (10–23%) and difficulties in reliable pre-operative differentiation between malignant and benign renal tumors (like oncocytoma and fat-poor angiomyolypoma) using contemporary imaging techniques result in the relatively high number of surgeries which might be considered as an overtreatment (7.5–33.6%) [9]. Absence of an accurate diagnostic biomarker for RCC promoted the interest of many researchers in studying miRNAs measured in tissues, serum or urine.
MiRNAs in tissues/serum
In 2007, Gottardo et al. reported that a composition of 4 miRs (miR-27, -28, -185, let-7f-2) was noticeably overexpressed in RCC specimens (p <0.05) in comparison to a healthy kidney [10]. Nakada and co-authors reported that 43 microRNAs were differently expressed in conventional RCC and in healthy kidney tissues: 37 miRs were significantly under-expressed in conventional RCC and the other 6 were overexpressed; the most significantly down-regulated miRs were microRNA-141 and microRNA -200c [11]. Another study validated in a multicenter cohort of 84 RCC patients (tissue, serum) and 93 healthy controls (serum) using quantitative real-time polymerase chain reaction (qRT PCR), showed that microRNA-1233 was significantly up-regulated in patients with RCC, enabling its detection with 77.4% sensitivity, 37.6%, specificity and area under the curve (AUC) of 0.588 [12]. Faragalla et al. in 2012 affirmed that miR-21 can be used as a diagnostic biomarker measured in RCCs tissues of different histologic subtypes, with the most significant expression levels in conventional and pRCCs. Measuring of microRNA-21 provided differentiation between ccRCC, pRCC, chRCC and oncocytoma with 90% specificity (95% confidence interval CI – 63.9–98.1%) and 83% sensitivity (95% CI, 53.5–97.6%) [13]. Redova et al. observed that microRNA-378 was up-regulated (AUC = 0.71, p = 0.0003) and microRNA-451 was down-regulated (AUC = 0.77, p <0.0001,) in the serum of patients with renal cancer in comparison to healthy controls. A composite use of microRNA-378 and microRNA-451 enabled diagnosis of RCC with the sensitivity, specificity and AUC of 81%, 83% and 0.86, respectively [14]. Zhao et al. reported that in primary ccRCC tissues the average microRNA-210 expression level was higher than in comparison to healthy controls (p = 0.004). In serum of patients with renal cancer, the average expression level of microRNA-210 was higher than in the control group (p <0.001), allowing for identification of RCC with 81.0% sensitivity, 79.4% specificity and an AUC of 0.874. Moreover, the average expression level of microRNA-210 in serum was noticeably decreased in patients with RCC following one week after surgical treatment (p = 0.001) [15]. In 2014, Chen et al. assessed the expression of microRNA-129-3p and microRNA-129-5p in 69 cases of paired renal tumors, healthy tissues and conventional renal cancer cell lines. Results showed that microRNA-129-3p instead of microRNA-129-5p was considerably under-expressed in ccRCC and chRCC; measuring of miR-129-3p expression in tissues allowed to differentiate conventional renal cancer from normal controls with 73.5% accuracy [16]. In another study, microRNA-210 serum expression levels were significantly higher in patients with ccRCC than in healthy controls (p = 0.001) – receiver operating characteristic (ROC) curve was 65% sensitivity, 83% specificity and AUC of 0.77 (95% CI, 0.65–0.89) [17]. In 2015, Fedorko et al. found that if analyzed in combination, serum levels of miR-210 and miR-378 enable identification of patients with RCC (significant overexpression) with 80% sensitivity and 78% specificity if (p <0.0001). Furthermore, miR-210 and miR-378 expression levels significantly diminished 3 months after radical nephrectomy (p <0.0001) [18].
An accurate, but complex system of molecular classification of kidney cancer subtypes using the microRNA signature was proposed by Youssef et al.: the study enrolled 70 specimens – 20 conventional RCCs and 20 paired healthy tissues collected from the same patients, 10 papillary RCCs, 10 chromophobe RCCs and 10 oncocytomas. In result, 15 reliably differentially expressed miRs amongst RCC subtypes, oncocytomas, and healthy kidney tissues were detected. Sensitivity in differentiating healthy controls from RCC, ccRCC and pRCC was 97%, 100% and 97% respectively; accuracy to differentiate chRCC from oncocytoma was 100%. Moreover, the algorithm was cross-validated and demonstrated an accuracy of approximately 90% [19].
MiRNAs in urine
In contrast to a large number of studies involving measuring of miRNAs expression in tissues and serum of patients with RCC, only a few studies investigated the potential of miRNAs as urinary biomarkers. Brandenstein et al. in their work found that up-regulated miRNA-15a can be measured in urine from patients with ccRCC, but is barely detectable in cases of benign renal tumors (such as oncocytoma) and inflammation of the upper and lower urinary tract [20]. In our study, we assessed the expression of miRNA-15a in the urine of 67 adult patients with solid renal tumors before and after surgery (22 ccRCCs, 16 pRCCs, 14 chRCCs, 8 oncocytomas, 5 angiomyolipomas and 2 papillary adenomas) compared to 15 healthy controls using PCR. It was found that miRNA-15a expression was significantly up-regulated in RCC patients in comparison to benign tumors and healthy renal parenchyma (p <0.01). There was no significant difference in miR-15a expression levels between ccRCC, pRCC and chRCC. However, the presence of pathologically proven necrosis had an impact on miR-15a regulation in patients with RCC resulting in significantly (p <0.01) higher expression values in cases with necrosis in comparison with non-necrotic RCCs. Direct interconnection between RCC size and miR-15a expression value was registered: the Pearson correlation coefficient was 0.873. In differentiation between RCC and benign renal lesions we achieved 98.1% specificity and 100% sensitivity (95% CI 0.9–1.0) at a cut-off value of 5,00E-06 relative fluorescence units (RFU), with AUC of the ROC curve 0.955 [21].
Promising results in detection of RCC and identification of the most sensible biomarkers by means of microRNA profiling were presented in a number of works [22, 23]. However, further investigations with a larger number of patients of different stages, histologic subtypes and grades of differentiation between RCC and benign renal tumors and multicenter cross-validation are required for the implementation of the existing knowledge into routine clinical practice.
MicroRNAs in prediction of response to systemic therapy of renal cancer
In cases of advanced/metastasized RCC, when there are no indications for the surgical treatment, systemic therapy (ST) can be used as an alternative curative modality. A number of groups of agents for ST of RCC were proposed: chemotherapeutic, immunotherapeutic (interferon-α), targeted therapy agents (tyrosine kinase inhibitors, monoclonal antibody against circulating VEGF, mechanistic target of rapamycin inhibitors). Unfortunately, the treatment response rates are devastatingly low – 3–31% [24]. In this context, the prediction of RCC response to ST plays an essential role in treatment planning, enabling the avoidance in application of expensive treatment with side effects in cases with no potential benefit.
Chemotherapy
Chen and co-authors explored cell survival, cell cycle and programmed cell death in human kidney cells and 786-O cell line treated with chemotherapy using microRNA-381 and 5-fluorouracil. They observed that microRNA-381 enhances 786-O cells sensitiveness to 5-fluorouracil by mitosis inhibitor protein kinase WEE1 and of cyclin-dependent kinase 2 activation [25]. Sun and co-authors in 2017 found that overexpression of miR-451 strengthened drug resistance during chemotherapy with decreased cellular viability, and promoted cell apoptosis of GRC-1 cell line pretreated by adriamycin (ADM), while overexpressed activating transcription factor 2 (ATF-2) inverted the consequence induced by microRNA-451 increased expression. Moreover, miR-451 knockdown improved drug susceptibility, reduced programmed cell death rate, and improved cell viability of ACHN cell line induced by ADM; however, ATF-2 suppression reversed the low rate of cell apoptosis and the high rate of cell viability induced by miR-451 knockdown [26].
Immunotherapy
In 2015, Zhang et al. in their study that involved 82 patients with RCC, strived to determine a molecular biomarker that can predict the response of renal cancer cells to natural killer (NK) therapy. The results demonstrated that microRNA-183 expression in the serum of patients with RCC was significantly up-regulated compared with healthy controls; the expression levels were directly associated with the tumor grade of differentiation. Furthermore, Chromium-51 release assay demonstrated that the primary renal cancer cells with under-expressed microRNA-183 in serum were more responsive to the cytotoxic impact of natural killer cells [27].
Targeted therapy
In 2013, Berkers et al. found that miR-141 was significantly underexpressed in RCC patients with poor response to sunitinib in comparison to good responders, which was associated with epithelial-to-mesenchymal transition (EMT) in vivo. In vitro introduction of miR-141 inverted EMT and inhibited cellular viability in hypoxic conditions [28]. In another study, 673 microRNAs were screened using TaqMan Low Density Arrays (TLDA) in the setting of metastatic RCC (mRCC) in 41 patients with utmost phenotypes of assigned effectiveness and resistance to sunitinib. In a selected cohort of patients, 64 differentially expressed miRs were identified by TLDA; 7 of them were assessed by qRT PCR in an independent series. Among others, microRNA-942 allowed to predict efficacy of sunitinib with the highest accuracy (p = 0.0074). Furthermore, the new paracrine tract of up-regulation of matrix metallopeptidase 9 (MMP-9) and VEGF secretion through microRNA-942 expression and as a result enhancement in endothelial migration and resistance to sunitinib was depicted [29]. In 2015, Khella et al. analyzed miRNAs expression in patients with mRCC with a short and long (≤12 vs. >12 months) progression-free survival (PFS) in whom sunitinib was administered as a first-line therapy. In result, negative interconnection between the expression of microRNA-221 and its target VEGFR2 was evidenced. High levels of microRNA-221 were characteristic of patients with poor PFS, while VEGFR2 was associated with longer PFS. Gain-of-function studies demonstrated that microRNA-221 and microRNA-222 inhibited angiogenesis and cell proliferation in endothelial cells from the umbilical vein and promoted proliferation in ACHN cells [30].
In experimental work, Papadopoulos et al. assessed the cytotoxic effect of sunitinib and everolimus in Caki-1 renal cancer cells and the influence of the therapy on several BCL2-family and apoptosis-related miR clusters during and after treatment. It was found that both drugs had an inhibitive impact on time-dependent and dose-dependent cellular viability simultaneously promoting poly (ADP-ribose) polymerase cleavage. Significant shifts in expression of microRNA-15a, -16 and -145 under the impact of sunitinib and in expression levels of microRNA-15a, -145, BAX and BCL2 in everolimus application cohort were observed. Moreover, apoptosis in RCC cells was directly induced by both sunitinib and everolimus, at the same time affecting the regulation of BCL2 family members and apoptosis-related miRs [31].
Important data was published by Zheng el al.: in their study sorafenib was associated with autophagy activation in renal cancer cells (A489 and 786-0) that was interconnected with degradation of protein p62, upregulation of Beclin-1/autophagy protein 5 (ATG5) and conversion of light chain 3B-I/-II. Introducing of microRNA-30a in to A489/786-0 cells suppressed the expression of Beclin-1 and improved cytotoxicity induced by sorafenib. Conversely, a knockdown of microRNA-30a by means of exogenously expressed antagomiRNA-30a up-regulated expression of Beclin-1 and inhibited sorafenib-induced cytotoxicity in RCC cells [32].
In another recent work, an attempt was made to assign microRNA signature able to predict the therapeutic response to antiangiogenic tyrosine kinase inhibitor (TKI) treatment used as the first-line treatment in patients with RCC. As a result of the overseen analysis, it was found that miR-99b-5p was significantly down-regulated in patients with short progression free survivial (PFS) (<8 months) and TKI non-responders (progressive disease patients according to Response Evaluation Criteria in Solid Tumors) (p <0.0001, each) [33]. Such data demonstrates the potential of microRNAs as predictive biomarkers of solid tumors of RCC; however, further investigations are necessary.
MicroRNAs in renal cell carcinoma prognosis
Recurrence
In 2010, Hildebrandt et al. found that microRNA-9-1 and microRNA-9-3 methylation was more substantive in deoxyribonucleic acid (DNA) obtained from primary RCCs of recurrent patients (p-values 0.012 for miR-9-1 and 0.009 for miR-9-3) compared to patients with no recurrence. Moreover, miR-9-3 methy-lation was associated with a higher risk of recurrence (hazard ratio HR 5.85, 95% CI 1.30–26.35), increased levels of methylation of both microRNA-9-1 and microRNA-9-3 were characteristic of patients with decreased recurrence-free survival (RFS) time for about 30-month (p-values 0.034 for miR-9-1 and 0.007 for miR-9-3) [34]. In another study Nakata et al. noticed that miR-27a-3p levels (low vs. high HR, 2.33; 95% CI, 1.07–5.47, p = 0.0330) showed significant association with cancer progression, and miR-193a-3p levels (low vs. high HR, 1.93; 95% CI, 0.90–4.37, p = 0.0942) were associated with cancer progression [35]. In 2013, Gebauer et al. reported that higher relative methylation of microRNA-124-3 in ccRCCs tissues was associated with worse RFS (HR = 9.37, p = 0.0005) [36].
Metastasis
In a study accomplished by Slaby et al., it was observed that microRNA-106b expression levels were reliably lower in patients with RCCs in whom metastasis developed compared with non-metastatic cases (p = 0.030). Moreover, miR-106b expression level was predictive for early metastasis after nephrectomy in patients with renal cancer (long-rank p = 0.032) [37]. The scratch migration assays demonstrated that microRNA-506 mimics noteworthy suppressed migration of RCC cells in the Yang et al. study. Furthermore, the transwell invasion assay disclosed that the potential of renal cancer invasiveness transfected with microRNA-506 mimics was considerably decreased [38].
Survival
Faragalla et al. found that RCCs with higher stage and grade were associated with significantly higher microRNA-21 levels in tissue samples. MicroRNA-21-positive patients had a reliably shorter disease-free survival (HR 2.15, 95% CI 1.16-3.98, p = 0.014) [14]. According to Goto et al., microRNA 486 expression in RCC samples was about 2.7 fold higher when compared to healthy kidney tissues (P <0.0001). In 46 cases of RCCs of stage III and IV overexpressed microRNA 486, which was associated with poor cancer specific mortality (CSM), independent of other covariates and TNM staging (P = 0.0064). Besides, according to the Kaplan Meier analysis, microRNA 486 expression was associated with CSM in 14 patients with RCC (of III and IV stages) that were not treated with interferon α (P = 0.0574) [39]. According to our unpublished data, we observed poor cancer specific survival (CSS) in patients with RCC and overexpressed miR-15a in tumor tissues. Patients with renal cancer and miRNA-15a expression ≤0.10 RFU 3-year and 5-year CSS was 100% and 97.0% accordingly, the mean overall survival (OS) was 59.88 ±0.12 months (95% CI 59.66–60.11); 3-year and 5-year CSS in patients with miR-15a expression >0.10 RFU was 83.9% and 54.8% respectively; the mean OS was 49.74 ±2.16 months (95% CI – 59.66–60.11). Optimistic results were described in studies where microRNA profiling was executed in order to identify a molecular signature of a poor prognosis in patients with RCC [40]. We summarized the available data on miRs impact on RCC pathogenesis and oncological characteristics of the tumor which may play an important role in predicting disease outcome (Tables 1 and 2). A list of miRNAs that are known to be directly associated with renal cell carcinoma prognosis is presented in Table 3.
Table 1.
Oncomirs in pathogenesis of renal cell carcinoma [1–40]
| MicroRNA | Target | Pathway/mechanism |
|---|---|---|
| miR-7 | Cell migration, proliferation and apoptosis | |
| miR-15a-5p | Cell proliferation, migration, invasion and apoptosis | |
| miR-17-5p | VEGF-A, EGLN3 | Cell cycle, migration, proliferation, and invasion, modulation of the differentiation of mesenchymal stem cells |
| miR-21 | TORC1, FasL, TIMP3, TCF21, PDCD4, TPM1. | Akt/TORC1/KISS1/ PTEN/Akt/IKKβ and NFκB-dependent cyclin D1 expression/Activation of caspase pathway/ cell proliferation and cell apoptosis |
| miR-23b | POX | HIF/apoptosis |
| miR-28-5p | Mad2 | VHL/mitotic checkpoint function/chromosomal instability |
| miR-29b | KIF1B | Apoptosis, proliferation and invasion ability |
| miR-30b | Cell proliferation, invasion, migration and apoptosis | |
| miR-106b | Cell proliferation, migration and apoptosis | |
| miR-122 | mTOR, OCLN, Sprouty2 | PI3K/Akt/Cell proliferation, invasion and migration |
| miR-142-3p | Cellular migration, proliferation and apoptosis | |
| miR-155 | BACH1, E2F2 | Cell proliferation, migratory activity and apoptosis |
| miR-195-3p | Cell proliferation, migration, invasion and apoptosis | |
| miR-203a | GSK-3β | Cell proliferation, migration, and apoptosis |
| miR-210 | ISCU1/2 | VHL/HIF1α/centrosome amplification/ migratory and invasive potential of ACHN metastatic RCC cells |
| miR-217 | HIF-1α/AXL/ LncRNA HOTAIR/ proliferation, migration, EMT process and apoptosis | |
| miR-224 | VHL, SMAD4, SMAD5, DIO1 | VHL/ HIF1α/Tissue |
| hypothyroidism in RCC | ||
| miR-590-5p | PBRM1 | Inhibition of G1/S transition /cell proliferation and invasion |
Table 2.
Anti-oncomirs in pathogenesis of renal cell carcinoma [1–40]
| MicroRNA | Target | Pathway/mechanism |
|---|---|---|
| miR-1 | TAGLN2 | Cell proliferation, invasion, apoptosis and cell cycle arrest |
| miR-20b-5p | VEGFA, PAR-1, MALAT1 | Cellular proliferation, migration and apoptosis |
| miR-22 | PTEN | Cell growth, migration and invasion |
| miR-23b | POX | HIF/apoptosis |
| miR-30c | Slug | VHL/HIF/epithelial-mesenchymal transition, cell migration |
| miR-30d | Cyclin E2 | Cyclin E2/cell proliferation and colony formation, G1 phase arrest |
| miR-34a | Notch1, GAS1 | Cell growth, cell cycle arrest |
| miR-99a | mTOR, IGF-1R | IGF-1R/G1-phase cell cycle arrest, cells growth, clonability, migration and invasion |
| miR-133a | TAGLN2 | Cell proliferation, invasion, apoptosis and cell cycle arrest |
| miR-133b | MMP-9 | Cell proliferation, migration and invasion |
| miR-133b miR-135a |
Bcl-2 | JAK2/STAT3/cell apoptosis |
| miR-135a | c-MYC | Cell cycle, pathways in cancer, DNA replication, and focal adhesion |
| miR-138 | HIF-1α, vimentin, EZH2 | HIF/ apoptosis and cell migration/cell senescence |
| miR-143 miR-145 |
HK2 | Cell proliferation and invasion |
| miR-145 | ADAM17, ANGPT2, NEDD9 | HIF2α/VEGF/MMP9/CCND1/ ARE/Cell proliferation and migration, G2-phase arrest |
| miR-148a | AKT2 | Cell proliferation, colony formation, migration and invasion |
| miR-182-5p | FLOT1 | AKT/FOXO3a/ proliferation, tumorigenicity, G1-phase arrest |
| miR-192 miR-194 |
MDM2, TYMS, ZEB2 | Cell proliferation and invasion |
| miR-199a-3p | HGF/c-Met | HGF/c-Met/cell proliferation and caused G1 phase arrest |
| miR-200c | ZEB1 | Akt /epithelial-to-mesenchymal transition modulation, metastatic ability |
| miR-205 | SFK, ZEB2 | Phospho-Src–regulated ERK1/2 pathway/ cell proliferation, colony formation, migration, and invasion |
| miR-206 | GAK | Cell proliferation, migration and invasion, cell cycle arrest |
| miR-215 | MDM2, TYMS, ZEB2 | Cell proliferation and invasion |
| miR-218 | Caveolin-2, CXCR7 | Focal Adhesion Pathway/cell viability, migration and invasion ability |
| miR-490-5p | PIK3CA | Tumourigenicity |
| miR-497 | VEGFR-2, ACHN | MEK/ERK, p38 MAPK/cell viability, migration and invasion |
| miR-508-3p miR-509-3p |
Cell invasion, migration and apoptosis | |
| miR-509-5p | FIGN, SFRS11, HMGA2, GOLGA1 | Cell migration, proliferation and anti-apoptosis |
| miR-584 | ROCK-1 | 3′UTR luciferase activity of ROCK-1/cell motility inhibition |
| miR-708 | ZEB2, BMI1 | Cell growth, clonability, invasion, migration and apoptosis |
| miR-1285 | TGM2 | Cell proliferation and invasion |
| miR-1291 | SLC2A1/GLUT1 | Cell proliferation, migration and invasion |
| miR-1826 | CTNNB1, MEK1 | Cell proliferation, invasion and migration, apoptosis and G 1 arrest in VHL-inactivated renal cancer cells |
Table 3.
MicroRNAs associated with renal cell carcinoma prognosis [1–40]
| MicroRNA | Pathway/mechanism |
|---|---|
| miR-9 | Cancer development and metastatic recurrence |
| miR-19a | Poor prognosis |
| miR-21 | Disease-free and overall survival rates, stage and grade, advanced clinic-pathological features and poor prognosis |
| miR-21/10b ratio | Disease severity and survival, poor prognosis in metastasis-free patients |
| miR-23b/27b cluster | Good overall survival |
| miR-27a-3p | Predictive factor for recurrence |
| miR-100 | Advanced tumor T stage, presence of metastasis, overall and tumor-specific survival |
| miR-106 | Early metastasis after nephrectomy |
| miR-124-3 | Advanced tumors and disease recurrence |
| miR-126 | Cancer-specific survival |
| miR-155 | Poor clinical outcomes |
| miR-187 | Lower survival rates |
| miR-217 | Lower survival rates |
| miR-221 | Poor overall survival |
| miR-321 | Tumor proliferation and survival rates |
| miR-424 | Tumor proliferation and survival rates |
| miR-429 | Linked to metastasis and poor prognosis |
| miR-486 | Cancerspecific mortality after nephrectomy |
| miR-497 | Poor prognosis |
| miR-630 | Lower overall survival |
| miR-1236 | Favorable survival |
Despite the potential value of microRNAs in the disease outcome prognosis, currently none of them supplemented existing RCC prognostic nomograms like the The University of California in Los Angeles integrated Staging System (UISS), stage-size-grade-necrosis (SSIGN) score, Karakiewicz's nomogram or the Memorial Sloan-Kettering Cancer Center (MSKCC) prognostic system.
CONCLUSIONS
Data described in many investigations displays the prominent potential of microRNAs as diagnostic, predictive and prognostic biomarkers of renal cell carcinoma. MiRNA network analysis and gene profiling may help to identify the most sensible molecular signatures of RCC that can be used for diagnostic purpose, as well as poor prognosis signatures and poor therapeutic response signatures in patients who undergo systemic therapy. However, application of such novel biomarkers in routine clinical practice still requires further research, a larger number of patients and multicenter cross-validation.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
References
- 1.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review 1975-2016. Bethesda, MD: National Cancer Institute; 2016. [Google Scholar]
- 2.Capitanio U, Cloutier V, Zini L, et al. A critical assessment of the prognostic value of clear cell, papillary and chromophobe histological subtypes in renal cell carcinoma: a population-based study. BJU Int. 2009;103:1496. doi: 10.1111/j.1464-410X.2008.08259.x. [DOI] [PubMed] [Google Scholar]
- 3.Sim SH, Messenger MP, Gregory WM, et al. Prognostic utility of pre-operative circulating osteopontin, carbonic anhydrase IX and CRP in renal cell carcinoma. Br J Cancer. 2012;107:1131. doi: 10.1038/bjc.2012.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatino M, Kim-Schulze S, Panelli MC, et al. Serum vascular endothelial growth factor and fibronectin predict clinical response to high-dose interleukin-2 therapy. J Clin Oncol. 2009;27:2645. doi: 10.1200/JCO.2008.19.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, Feng G, Gentil-Perret A, Genin C, Tostain J. Serum carbonic anhydrase 9 level is associated with postoperative recurrence of conventional renal cell cancer. J Urol. 2008;180:510. doi: 10.1016/j.juro.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Zhao H, Ljungberg B, Grankvist K, Rasmuson T, Tibshirani R, Brooks JD. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goga A, Benz C. Anti-Oncomir Suppression of Tumor Phenotypes. Mol Interv. 2007;4:199–202. doi: 10.1124/mi.7.4.6. [DOI] [PubMed] [Google Scholar]
- 9.Volpe A, Terrone C, Scarpa RM. The Current Role of Percutaneous Needle Biopsies of Renal Tumours. Arch Ital Urol Androl. 2009;81:107–112. [PubMed] [Google Scholar]
- 10.Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA Profiling in Kidney and Bladder Cancers. Urol Oncol. 2007;5:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Nakada C, Matsuura K, Tsukamoto Y, et al. Genome-Wide microRNA Expression Profiling in Renal Cell Carcinoma: Significant down-Regulation of miR-141 and miR-200c. J Pathol. 2008;4:418–427. doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]
- 12.Wulfken LM, Moritz R, Ohlmann C, et al. MicroRNAs in Renal Cell Carcinoma: Diagnostic Implications of Serum miR-1233 Levels. PloS One. 2011;9:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faragalla H, Youssef M, Scorilas A, et al. The Clinical Utility of miR-21 as a Diagnostic and Prognostic Marker for Renal Cell Carcinoma. J Mol Diagn. 2012;4:385–392. doi: 10.1016/j.jmoldx.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Redova M, Poprach A, Nekvindova J, et al. Circulating miR-378 and miR-451 in Serum Are Potential Biomarkers for Renal Cell Carcinoma. J Transl Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao A, Li G, Péoc'h M, Genin C, Gigante M. Serum miR-210 as a Novel Biomarker for Molecular Diagnosis of Clear Cell Renal Cell Carcinoma. Exp Mol Pathol. 2013;1:115–120. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Ruan A, Wang X, et al. MiR-129-3p, as a Diagnostic and Prognostic Biomarker for Renal Cell Carcinoma, Attenuates Cell Migration and Invasion via Downregulating Multiple Metastasis- Related Genes. J Can Res Clin Oncol. 2014;8:1295–1304. doi: 10.1007/s00432-014-1690-7. [DOI] [PubMed] [Google Scholar]
- 17.Iwamoto H, Kanda Y, Sejima T, Osaki M, Okada F, Takenaka A Serum miR-210 as a Potential Biomarker of Early Clear Cell Renal Cell Carcinoma. Int J Oncol. 2014;1:53–58. doi: 10.3892/ijo.2013.2169. [DOI] [PubMed] [Google Scholar]
- 18.Fedorko M, Stanik M, Iliev R, et al. Combination of MiR-378 and MiR-210 Serum Levels Enables Sensitive Detection of Renal Cell Carcinoma. Int J Mol Sci. 2015;10:23382–23389. doi: 10.3390/ijms161023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youssef M, White NM, Grigull J, et al. Accurate Molecular Classification of Kidney Cancer Subtypes Using microRNA Signature. Eur Urol. 2011;5:721–730. doi: 10.1016/j.eururo.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Brandenstein M, Pandarakalam JJ, Kroon L, et al. MicroRNA 15a, Inversely Correlated to PKCα, Is a Potential Marker to Differentiate between Benign and Malignant Renal Tumors in Biopsy and Urine Samples. Am J Pathol. 2012;5:1787–1797. doi: 10.1016/j.ajpath.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Mytsyk Yu, Dosenko V, Borys Yu, et al. The Possibility of Application of Detected in Urine MicroRNA-15a for Diagnostics of Renal Cell Carcinoma. Exp Clin Physiol Biochy. 2017;1:49–53. [Google Scholar]
- 22.Li J, Huang JH, Qu QH, et al. Evaluating the microRNA-Target Gene Regulatory Network in Renal Cell Carcinomas, Identification for Potential Biomarkers and Critical Pathways. Int J Clin Exp Med. 2015;8:7209–7219. [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav S, Khandelwal M, Seth A, Saini AK, Dogra PN, Sharma A. Serum microRNA Expression Profiling: Potential Diagnostic Implications of a Panel of Serum microRNAs for Clear Cell Renal Cell Cancer. Urology. 2017;104:64–69. doi: 10.1016/j.urology.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Choueiri TK, Motzer RJ. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N Engl J Med. 2017;376:354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 25.Chen B, Duan L, Yin G, Tan J, Jiang X. MiR-381, a Novel Intrinsic WEE1 Inhibitor, Sensitizes Renal Cancer Cells to 5-FU by up-Regulation of Cdc2 Activities in 786-O. J Chemother. 2013;25:229–238. doi: 10.1179/1973947813Y.0000000092. [DOI] [PubMed] [Google Scholar]
- 26.Sun X, Lou L, Zhong K, Wan L. MicroRNA-451 Regulates Chemoresistance in Renal Cell Carcinoma by Targeting ATF-2 Gene. Exp Biol Med. 2017;242:1299–1305. doi: 10.1177/1535370217701625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Di W, Dong Y, et al. High Serum miR-183 Level Is Associated with Poor Responsiveness of Renal Cancer to Natural Killer Cells. Tumour Biol. 2015;12:9245–9249. doi: 10.1007/s13277-015-3604-y. [DOI] [PubMed] [Google Scholar]
- 28.Berkers J, Govaere O, Wolter P, et al. A Possible Role for MicroRNA-141 Down-Regulation in Sunitinib Resistant Metastatic Clear Cell Renal Cell Carcinoma Through Induction of Epithelial-to- Mesenchymal Transition and Hypoxia Resistance. J Urol. 2013;189:1930–1938. doi: 10.1016/j.juro.2012.11.133. [DOI] [PubMed] [Google Scholar]
- 29.Prior C, Perez-Gracia L, Garcia-Donas J, et al. Identification of Tissue microRNAs Predictive of Sunitinib Activity in Patients with Metastatic Renal Cell Carcinoma. PloS One. 2014;9:e86263. doi: 10.1371/journal.pone.0086263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khella HW, Butz H, Ding Q, et al. MiR-221/222 Are Involved in Response to Sunitinib Treatment in Metastatic Renal Cell Carcinoma. Molecular Therapy: Mol Ther. 2015;23:1748–1758. doi: 10.1038/mt.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papadopoulos EI, Yousef G.M, Scorilas A. Cytotoxic Activity of Sunitinib and Everolimus in Caki-1 Renal Cancer Cells Is Accompanied by Modulations in the Expression of Apoptosis-Related microRNA Clusters and BCL2 Family Genes. Biomed Pharmacother. 2015;70:33–40. doi: 10.1016/j.biopha.2014.12.043. [DOI] [PubMed] [Google Scholar]
- 32.Zheng B, Zhu H, Gu D, et al. MiRNA-30a-Mediated Autophagy Inhibition Sensitizes Renal Cell Carcinoma Cells to Sorafenib. Biochem Biophys Res Commun. 2015;459:234–239. doi: 10.1016/j.bbrc.2015.02.084. [DOI] [PubMed] [Google Scholar]
- 33.Lukamowicz-Rajska M, Mittmann C, Prummer M, et al. MiR-99b-5p Expression and Response to Tyrosine Kinase Inhibitor Treatment in Clear Cell Renal Cell Carcinoma Patients. Oncotarget. 2016;7:78433–7447. doi: 10.18632/oncotarget.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildebrandt MA, Gu J, Lin J, et al. Hsa-miR-9 Methylation Status Is Associated with Cancer Development and Metastatic Recurrence in Patients with Clear Cell Renal Cell Carcinoma. Oncogene. 2010;29:5724–5728. doi: 10.1038/onc.2010.305. [DOI] [PubMed] [Google Scholar]
- 35.Nakata W, Uemura M, Sato M, et al. Expression of miR-27a-3p Is an Independent Predictive Factor for Recurrence in Clear Cell Renal Cell Carcinoma. Oncotarget. 2015;6:21645–21654. doi: 10.18632/oncotarget.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebauer K, Peters I, Dubrowinskaja N, et al. Hsa-Mir-124-3 CpG Island Methylation Is Associated with Advanced Tumours and Disease Recurrence of Patients with Clear Cell Renal Cell Carcinoma. B J Cancer. 2013;108:131–138. doi: 10.1038/bjc.2012.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slaby O, Jancovicova J, Lakomy R, et al. Expression of miRNA-106b in Conventional Renal Cell Carcinoma Is a Potential Marker for Prediction of Early Metastasis after Nephrectomy. J Exp Clin Cancer Res. 2010;29:90. doi: 10.1186/1756-9966-29-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Zhang H, Zhang H, Chen S, Yan Y, Zheng J. MiR-506 Is down-Regulated in Clear Cell Renal Cell Carcinoma and Inhibits Cell Growth and Metastasis via Targeting FLOT1. PloS One. 2015;10:e0120258. doi: 10.1371/journal.pone.0120258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goto K, Oue N, Shinmei S, et al. Expression of miR-486 Is a Potential Prognostic Factor after Nephrectomy in Advanced Renal Cell Carcinoma. Mol Clin Oncol. 2013;1:235–240. doi: 10.3892/mco.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang K, Xu H. Prognostic Value of Meta-Signature miRNAs in Renal Cell Carcinoma: An Integrated miRNA Expression Profiling Analysis. Sci Rep. 2015;5:10272. doi: 10.1038/srep10272. [DOI] [PMC free article] [PubMed] [Google Scholar]