Abstract
Nucleic acids can form noncanonical four-stranded structures called G-quadruplexes. G-quadruplex-forming sequences are found in several genomes including human and viruses. Previous studies showed that the G-rich sequence located in the U3 promoter region of the HIV-1 long terminal repeat (LTR) folds into a set of dynamically interchangeable G-quadruplex structures. G-quadruplexes formed in the LTR could act as silencer elements to regulate viral transcription. Stabilization of LTR G-quadruplexes by G-quadruplex-specific ligands resulted in decreased viral production, suggesting the possibility of targeting viral G-quadruplex structures for antiviral purposes. Among all the G-quadruplexes formed in the LTR sequence, LTR-III was shown to be the major G-quadruplex conformation in vitro. Here we report the NMR structure of LTR-III in K+ solution, revealing the formation of a unique quadruplex–duplex hybrid consisting of a three-layer (3 + 1) G-quadruplex scaffold, a 12-nt diagonal loop containing a conserved duplex-stem, a 3-nt lateral loop, a 1-nt propeller loop, and a V-shaped loop. Our structure showed several distinct features including a quadruplex–duplex junction, representing an attractive motif for drug targeting. The structure solved in this study may be used as a promising target to selectively impair the viral cycle.
Introduction
G-quadruplexes are alternative secondary structures formed by guanine-rich nucleic acids. Four runs of at least two guanines linked by short mixed nucleotide sequences are prone to fold in a monomolecular G-quadruplex structure, built up from planar G-tetrads where four guanines interact through Hoogsteen hydrogen bonds.1 Different strand polarities and different loops and groove dimensions give rise to a large variety of G-quadruplex topologies.2,3 Physiological concentrations of potassium and sodium cations efficiently stabilize G-quadruplexes.4−6
Potential G-quadruplex forming sequences are widespread in the human genome and implicated in key genomic functions, such as transcription, replication, repair, and telomere maintenance.7−11 Particularly, overrepresented in the promoter regions of oncogenes, G-quadruplexes act as regulatory elements of gene expression.9,10 Targeting the G-quadruplex structures in the promoters of oncogenes c-myc, c-kit, and bcl-2 with G-quadruplex-stabilizing agents leads to gene transcription inhibition and decreased levels of gene expression,12 suggesting G-quadruplexes as promising anticancer targets.13,14
Besides the human genome, viral genomes also contain G-quadruplex-forming sequences, and emerging evidence suggests that they could be implicated in the regulation of key steps in the viral cycles.15 In Epstein–Barr virus (EBV) an RNA G-quadruplex regulates translation of EBNA1 mRNA.16,17 Multiple G-quadruplex-forming sequences located in the long control regions of some human papilloma virus (HPV) genomes suggest G-quadruplex involvement in transcriptional regulation.18 In herpes simplex virus-1 (HSV-1), G-quadruplexes that form in the virus DNA genome were visualized in infected cells and were shown to peak at the time of virus genome replication;19 in addition, the DNA replication step was affected by a G-quadruplex ligand, BRACO-19, inferring a regulatory role of G-quadruplexes in the viral replication.20
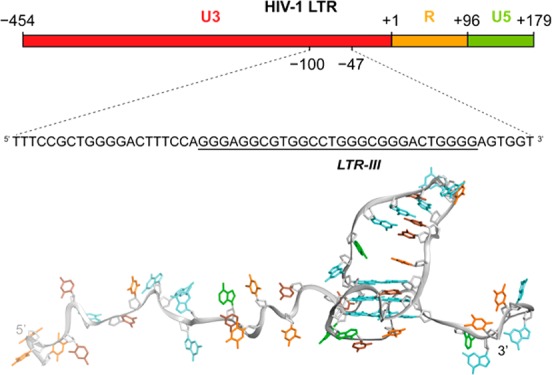
G-quadruplexes have also been described in the human immunodeficiency virus 1 (HIV-1), a lentivirus that is the etiological agent of the acquired immunodeficiency syndrome (AIDS). HIV-1 is characterized by a ssRNA genome that, once retrotranscribed by the viral reverse transcriptase enzyme, integrates into the host cell chromosome in the provirus form. The provirus can then undergo a productive replicative cycle or remain in a dormant state known as “latency”. Effective progression of the viral cycle relies on the proper function of the 5′-long terminal repeat (5′-LTR), which is characterized by transcription factor binding sites and serves as unique viral promoter.21 Formation of multiple G-quadruplexes in the viral and proviral genome,22,23 and in particular in the LTR promoter,22,24 has been reported. LTR G-quadruplexes act as repressor elements of viral transcription initiation: stabilization by G-quadruplex ligands intensifies this effect,25,26 while cellular proteins modulate viral transcription by inducing/unfolding LTR G-quadruplexes.27,28 The observation that 5′-LTR G-quadruplex forming sequences are conserved in all primate lentiviruses29 further validates viral G-quadruplexes as novel antiviral targets. However, selective targeting of viral G-quadruplexes with small molecules is challenging and very few compounds have been shown to recognize specific G-quadruplex structures.30 High-resolution structures of viral G-quadruplexes may give new insights to achieve higher level of selectivity and specificity.
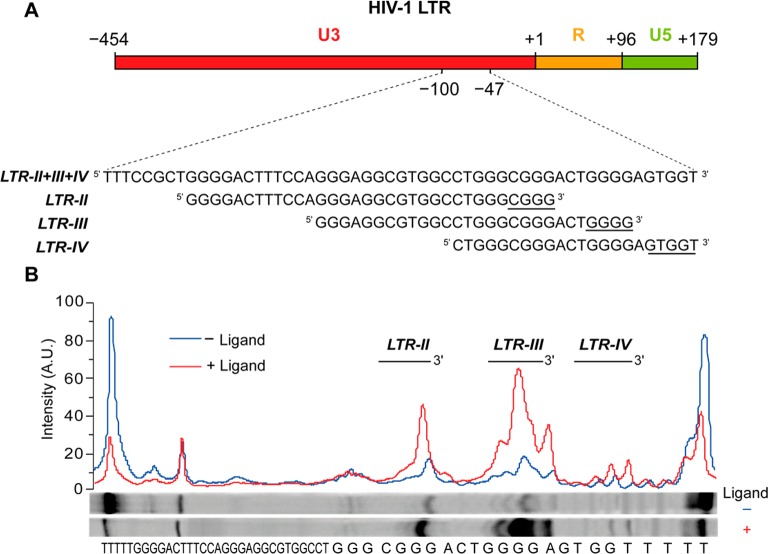
Within the LTR G-rich sequence, in the U3 region of the proviral genome, formation of multiple G-quadruplex conformations involving different G-tracts is possible. This sequence was divided into three main G-quadruplex-forming components, namely LTR-II, LTR-III, and LTR-IV (Figure 1A). In previous studies, the G-quadruplex formed by the LTR-III sequence showed the highest thermal stability in circular dichroism and FRET melting experiments. Moreover, Taq polymerase stop assay on the full-length LTR sequence, in K+ solution, revealed a stop site prevalently occurring at the LTR-III site and this effect was exacerbated with G-quadruplex ligands, such as BRACO-19 (Figure 1B).22,25
Figure 1.
(A) LTR G-rich sequence in the U3 promoter region of HIV-1 proviral genome and the associated subsequences LTR-II, LTR-III, and LTR-IV. Underlines indicate most significant stop positions in the polymerase stop assay. (B) Gel quantification of a Taq polymerase stop assay in 100 mM potassium buffer on the LTR-II + III + IV template in the absence (blue) or presence of an excess (250 nM) of BRACO-19 (red).
The cellular protein nucleolin is involved in the regulation of viral promoter activity through binding to the LTR G-quadruplex structures.27 Specifically, the LTR G-quadruplex-stabilizing effect translates into the decrease of viral promoter activity. In contrast, the cellular protein hnRNP A2/B1 binds and unfolds the LTR G-quadruplexes, i.e. LTR-II and LTR-III, activating viral transcription.28 Interestingly, the activity of promoters with mutations totally or partially abolishing LTR-III G-quadruplex formation is not affected by nucleolin and hnRNP A2/B1 binding as compared to the wild-type sequence.
This evidence supports the key role of LTR-III G-quadruplex within the LTR G-quadruplex-folding motif in the regulatory events of HIV-1 transcription. Thus, selective targeting of the LTR-III G-quadruplex conformation with stabilizing ligands may represent an attractive strategy to inhibit virus production.
Here we report on the high-resolution NMR solution structure of the 28-nt LTR-III G-quadruplex 5′-GGGAGGCGTGGCCTGGGCGGGACTGGGG-3′, containing an interesting duplex–quadruplex junction that can potentially be specifically targeted. We also demonstrate that the LTR-III G-quadruplex structure persists in a longer LTR sequence, suggesting LTR-III as a major G-quadruplex structure formed in the HIV-1 LTR.
Materials and Methods
DNA Sample Preparation
Unlabeled and site-specific labeled DNA oligonucleotides were synthesized using reagents from Glen Research (Sterling, USA). Samples were deprotected in ammonium hydroxide solution, purified using Poly-Pak cartridges following Glen Research protocol, and then dialyzed overnight against 20 mM KCl solution. The excess of KCl was removed by dialysis against water for 2 h. Upon lyophilization DNA was obtained in powder form. DNA samples were dissolved in buffer containing 70 mM potassium chloride and 20 mM potassium phosphate (pH 7).
Gel Electrophoresis
DNA samples at 100 μM strand concentration in potassium phosphate buffer were loaded on 15% native polyacrylamide gel containing 10 mM KCl. An electrophoresis was run at 90 V for 30 min at room temperature in Tris-Borate-EDTA-KCl buffer, DNA bands were visualized by UV shadowing.
Circular Dichroism
CD spectra were recorded on a Jasco J-815 CD spectrometer at 20 °C using a quartz cuvette of 10-mm optical path length. The reported spectra of DNA samples at 5 μM concentration in potassium phosphate buffer (pH 7) were the average of 3 scans over the 220–320 nm wavelength range, at the scanning speed of 50 nm/min, baseline-corrected for buffer contribution.
Thermal Denaturing
Thermal denaturing experiments were performed on Jasco V-650 UV spectrometer. DNA samples at 5 μM or 100 μM strand concentration were initially heated to 95 °C for 5 min and cooled to 20 °C by a temperature ramping rate of 0.1 °C/min, followed by heating from 20 to 95 °C at the same rate. UV absorbance at 295 nm was measured every 0.5 °C. Obtained data were plotted as folded fraction against temperature and the melting temperature was determined as the value at which the folded fraction was 0.5.
NMR Spectroscopy
NMR experiments were performed on 600 and 800 MHz Bruker NMR spectrometers equipped with a cryoprobe. Unless otherwise stated, 1D NMR spectra were recorded at 25 °C. 2D JR-HMBC, TOCSY, and 13C-HSQC experiments were recorded at 35 °C. NOESY experiments in H2O were performed at 7 °C (mixing time, 75 ms) and 25 °C (mixing time, 200 ms). NOESY experiments in D2O were performed at 35 °C with two different mixing times, 100 and 300 ms.
Structure Calculation
LTR-III G-quadruplex structures were calculated using a routine simulated annealing procedure with the XPLOR-NIH program31,32 based on NMR-derived distance and dihedral constraints. Distance constraints extracted from NOESY experiments were manually classified using 5 types of distances (2.4 ± 0.6, 3.0 ± 0.75, 3.8 ± 0.95, 4.8 ± 1.2, 5.2 ± 1.8 Å). Dihedral angles were constrained based on intraresidue H1′-H8 NOE peak intensities and the canonical B-DNA backbone conformation for the stem-loop.32
Data Deposition
The NMR chemical shifts of LTR-III have been deposited in the Biological Magnetic Resonance Bank (accession code 34302) and the coordinates of LTR-III have been deposited in the Protein Data Bank (accession code 6H1K).
Results
LTR-III Forms a Stable Intramolecular G-Quadruplex Structure
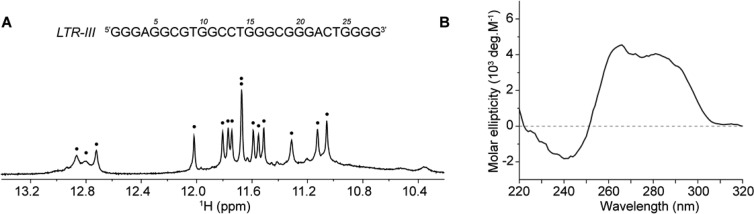
The 28-nt LTR-III sequence d[GGGAGGCGTGGCCTGGGCGGGACTGGGG] contains six tracts of 2–4 guanines (underlined). Using UV, CD, and NMR spectroscopy we investigated the G-quadruplex formation of LTR-III in K+ solution. The NMR spectrum of LTR-III showed 12 well-resolved peaks from 10.5 to 12.5 ppm, suggesting the formation of three G-tetrads, and three peaks from 12.5 to 13.5 ppm, suggesting the formation of Watson–Crick base pairs (Figure 2A). The CD spectrum of LTR-III showed a maximum peak at 260 nm and a shoulder peak around 285 nm, suggesting the formation of a nonparallel G-quadruplex topology (Figure 2B).33,34
Figure 2.
Characterization of the LTR-III sequence in K+ solution. (A) NMR imino proton spectrum of LTR-III. Imino protons are labeled with black dots. (B) CD spectrum of LTR-III.
The melting temperature of LTR-III, measured by UV absorption (Figure S1A) in ∼100 mM K+, was found to be 65.5 °C and independent of the DNA strand concentration (5 to 100 μM), consistent with the formation of a monomeric G-quadruplex structure. Additionally, on a native gel the migration of LTR-III was similar to that of a monomeric three-layered G-quadruplex structure35 (Figure S1B). Overall, these data support the formation of an intramolecular monomeric G-quadruplex structure.
LTR-III G-Quadruplex Adopts a (3 + 1) Folding Topology Containing a Diagonal Stem-Loop
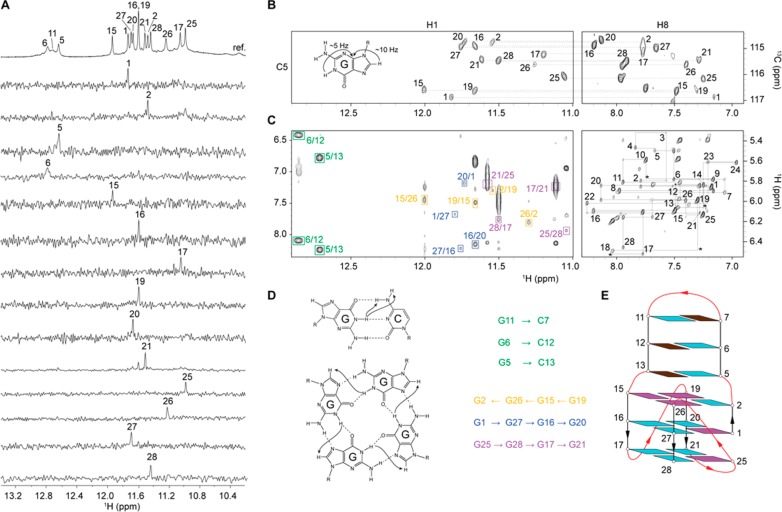
To elucidate the folding topology of LTR-III, NMR spectral assignment was performed using well-established protocols.36 Imino protons (H1) involved in base-pairing formation were assigned using site-specific low-enrichment (2–4%) 15N-labeling (Figure 3A),37 except for G11 for which H1 was assigned using NOE connectivities observed at low temperature (10 °C) (Figure S2). Subsequently, imino protons of guanines were correlated to their corresponding aromatic protons (H8) using through-bond JR-HMBC experiment38 (Figure 3B). Other aromatic protons were assigned or confirmed using H-to-D site-specific labeling and correlations through bond and space (TOCSY, 13C-HSQC, and NOESY experiments).
Figure 3.
NMR spectral assignments and folding topology of LTR-III. (A) Assignment of imino (H1) protons from 15N-filtered spectra of samples containing 2–4% of 15N-enriched isotope at the indicated position; the reference spectrum is shown on the top. (B) Assignment of H8 protons using H1–H8 through-bond correlation in JR-HMBC experiment. (C) Right panel, H8/H6–H1′ NOE sequential connectivities in NOESY spectrum in D2O at 35 °C (mixing time 300 ms). Intraresidue cross-peaks are labeled with residue number. Cross-peaks marked with asterisks are seen at lower threshold. Left panel, H1–H8 NOE cyclical connectivities in NOESY spectrum in H2O at 25 °C (mixing time, 200 ms). Cross-peaks used for G-tetrads determination are framed and colored based on the G-tetrad participation. Cross-peaks between guanines H1 protons and cytosine amino protons involved in Watson–Crick base pairs are framed and labeled in green. (D) Folding topology of LTR-III. Guanines in anti and syn conformations are cyan and magenta, respectively; cytosines are brown.
Strong intensity of the intraresidue H1′-H8 NOE cross-peaks observed in NOESY experiment (mixing time 100 ms) indicated a syn glycosidic conformation for G1, G15, G19, G25 and G26.
H1–H8 NOE connectivities (Figure 3C) allowed to establish the formation of a three-layered G-quadruplex core composed of G2•G26•G15•G19, G1•G27•G16•G20 and G25•G28•G17•G21; the hydrogen-bond directionality of the first G-tetrad is in opposite direction compared to that of the two other G-tetrads. NOEs between guanine imino protons and cytosine amino protons established three Watson–Crick base pairs G5•C13, G6•C12, and C7•G11 (Figure 3C, Figure S2). The folding topology of LTR-III is consistent with the slow exchange of imino protons of guanines in the central G-tetrad (G1, G27, G16, and G20) in a solvent exchange experiment (Figure S3).
LTR-III forms a (3 + 1) G-quadruplex folding topology with three strands (G15–G17, G19–G21, and G26–G28) pointing down and one strand (G1–G2) pointing up; the G-tetrad core has two medium grooves, a wide and a narrow groove (Figure 3E). Four loops connect the tetrads: a 1-nt propeller loop ( residue 18), a 3-nt lateral loop (from residue 22 to residue 24), a V-shaped loop (between residue 25 and residue 26), and a 12-nt diagonal loop (from residue 3 to residue 14) containing three Watson–Crick base pair (Figure 3E).
The V-shaped loop is formed between G25 and G26 residues with structural features similar to those observed in a G-quadruplex formed by an intronic human sequence.39
Within the long 12-nt diagonal loop, six nucleotides are interacting by Watson–Crick hydrogen bonds to form a hairpin (or stem-loop) structure with a capping G8-T9-G10 loop (Figure 3E). A possible additional base pair (A4•T14 or G3•T14) at the junction bridging the large distance (>20 Å) between the diagonal corners of the G-tetrads32 was not observed in our experiments, even at low pH and temperature (Figure S4).
Solution Structure of LTR-III G-Quadruplex
NMR solution structures of LTR-III were calculated based on restraints obtained from NMR experiments (Table 1). Ten lowest-energy structures were superimposed using heavy atoms in the G-tetrad core and represented in Figure 4A. Both the G-tetrad core and the stem-loop are well-converged individually (Figure 4, Figure S5, Table 1), however the orientations between them vary (Figure 4), mainly due to the lack of constraints involving G3 and A4 residues where few inter-residue NOEs were detected. In addition, peak broadening was observed for G3 indicating a possible flexible linker between the G-tetrad core and the stem loop.
Table 1. Statistics of the Computed Structures of LTR-III.
| NMR Restraints | ||
|---|---|---|
| distance restraints | D2O | H2O |
| intraresidue | 179 | 0 |
| sequential (i, i + 1) | 165 | 11 |
| long-range (i, ≥ i + 2) | 16 | 47 |
| other restraints | ||
| hydrogen bond | 24 | |
| dihedral angle | 35 | |
| Structure Statistics | |
|---|---|
| NOE violations | |
| number (>0.2 Å) | 0.000 ± 0.000 |
| maximum violation (Å) | 0.098 ± 0.048 |
| RMSD of violations (Å) | 0.007 ± 0.004 |
| deviations from ideal covalent geometry | |
| bond lengths (Å) | 0.003 ± 0.000 |
| bond angles (deg) | 0.716 ± 0.013 |
| improper dihedrals (deg) | 0.358 ± 0.012 |
| pair-wise all heavy atom RMSD values (Å) | |
| all heavy atoms of G-tetrad core | 0.63 |
| all heavy atoms including residues G5 to C13 | 1.74 |
Figure 4.
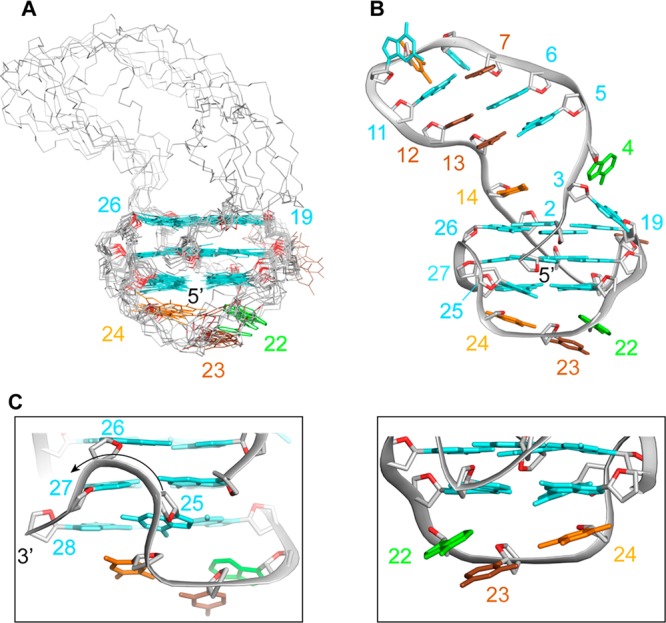
NMR solution structure of LTR-III. (A) Superposition of the ten lowest-energy structures based on the G-quadruplex-core. Bases of the stem-loop are omitted for clarity. (B) Ribbon view of the lowest energy structure. (C) Zoom-in on the V-shaped loop and the 3′-end-capping. Backbones are gray. O4′ atoms are red. Guanine bases are cyan, adenine are green, thymine are orange, and cytosine are brown.
The stem-loop is composed of three Watson–Crick base pairs (G5•C13, G6•C12, and C7•G11) showing regular B-DNA-like features. In our calculated model, T14 is stacked on top of the G2•G26•G15•G19 tetrad, as seen by numerous NOEs (Figure S6), while the G3 and A4 residues are pointing outside. In the lateral loop A22-C23-T24, A22 and T24 stack below G21 and G25, respectively while C23 is positioned below A22 and T24. The V-shaped loop between G25 and G26 is bridging the last and first G-tetrads with both syn G25 and G26 residues.
LTR-III Sequence Mutations: Probing the Stem-Loop and Quadruplex-Duplex Junction
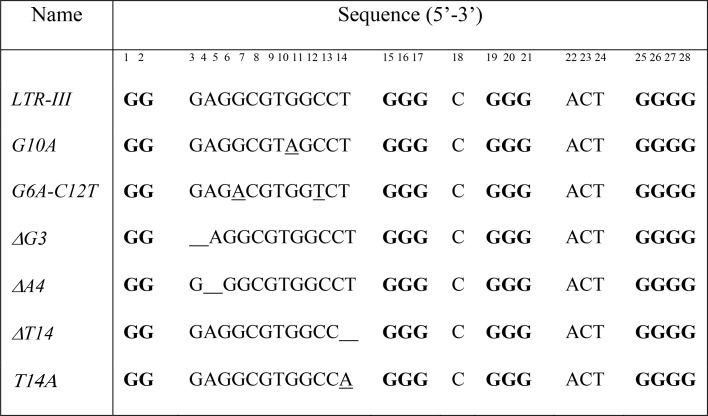
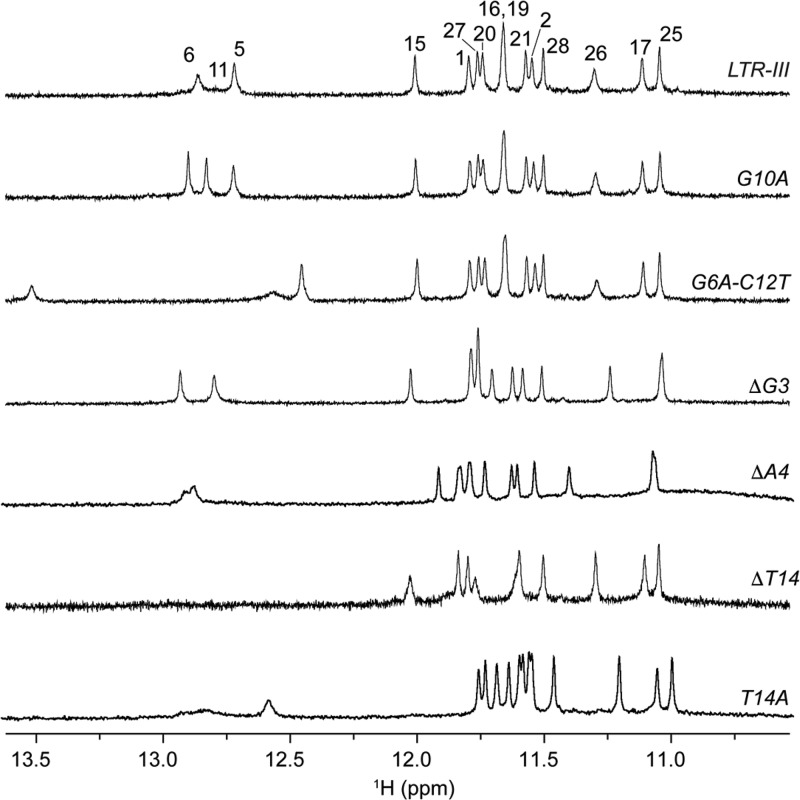
We investigated the effects of different sequence mutations in the LTR-III G-quadruplex structure (Table 2). In particular, we mutated residues in the diagonal stem-loop and at the quadruplex–duplex junction. The diagonal stem loop of the LTR-III G-quadruplex is composed of Watson–Crick base pairs and a capping GTG loop. Previous studies on stem-loop duplexes showed that a GCA or GTA loop in the stem-loop structure could favor hairpin formation.32,40 GTG loop of LTR-III was mutated to a GTA loop (G10A sequence) (Figure 5, Figure S7). Imino proton spectrum of the G10A sequence showed three peaks in the 12.5 to 13.0 ppm region significantly sharper than those of LTR-III, suggesting a more stable hairpin formation. To replace the G6•C12 base pair by an A•T base pair, G6 and C12 were substituted by A6 and T12, respectively, in the G6A-C12T sequence. NMR imino proton spectrum of G6A-C12T showed one significant downfield-shifted peak at ∼13.5 ppm (Figure 5, Figure S7), supporting the formation of an A•T base pair.
Table 2. LTR-III and Mutated Sequencesa.
Mutations are underlined. Guanines participating in the G-tetrad core are in boldface. Residue numbers are shown on top.
Figure 5.
LTR-III sequence mutational analysis. NMR spectra of the LTR-III sequence (on the top) and LTR-III mutated sequences. Imino protons of LTR-III are labeled by corresponding residue numbers.
The junction between the stem-loop and the G-tetrad core is an important structural feature. Deletion of the G3 base in the ΔG3 sequence led to 1D NMR and CD spectra with features similar to those of LTR-III (Figure 5, Figure S7): peaks at 12.5–13.0 ppm remained sharp and slight variations were observed for peaks from 10.8 to 12.2 ppm. This indicates that the G3 base is not crucial for the formation of the G-tetrad core or the stem-loop, consistent with NOE data and our calculated structure. In contrast, mutation/deletion of A4 and T14 resulted in the disappearance or broadening of the resonances in the 12.5–13.0 ppm region, while 10–12 resonances in the 10.8–12.2 ppm region were still observed despite a pronounced chemical shift variation. These data suggest a possible role of A4 and T14 in the quadruplex–duplex junction and the stabilization of the duplex stem.
Similar results were also observed for mutated sequences containing both improved cap, as in the G10A sequence, and mutations at the quadruplex–duplex junction (Figure S8).
Whereas in most of our calculated models T14 is stacked on the top G-tetrad and the G3 and A4 are pointing outside, in some models the A4 base is close to T14. To test the hypothesis on the formation of a transient Watson–Crick base pair between A4 and T14, we ran structure calculation with additional Watson–Crick A4•T14 base pair constraints. The formation of an A•T Watson–Crick base pair was compatible with the structure and our collected NOEs (Figure S9). We also tested the formation of a possible G3•T14 base pair in our structural calculation by adding hydrogen-bond constraints, but no stable base-pair could be observed without a large NOE violation or high increase in energy penalty.
LTR-III G-Quadruplex Structure Persists in a Longer LTR Sequence
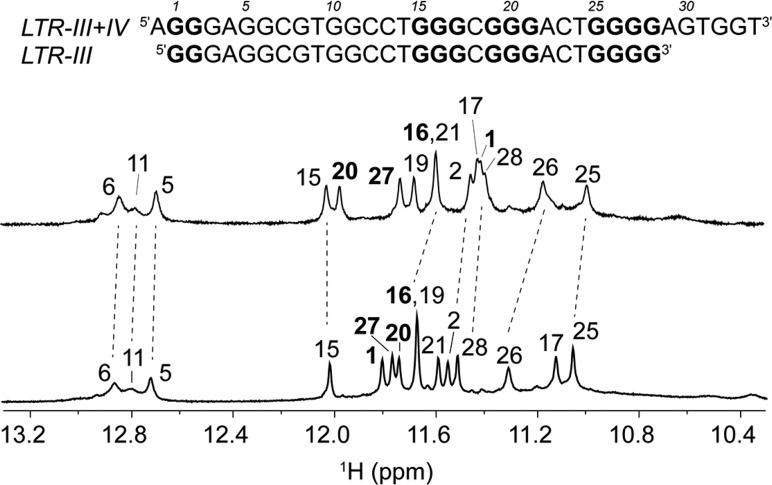
Formation of LTR-III G-quadruplex was assessed in a longer sequence containing LTR-III and LTR-IV sequences.27 In principle, the LTR-III+IV sequence is able to alternatively form both LTR-III and LTR-IV G-quadruplexes. However, NMR spectrum of LTR-III+IV displayed 12 well-resolved peaks at 10–12.5 ppm and 3 broad peaks at 12.5–13.5 ppm in the imino proton region, which shared many similarities with the 1D NMR spectrum of LTR-III sequence (Figure 6), suggesting that LTR-III+IV might form a G-quadruplex fold containing a stem-loop similarly to LTR-III.
Figure 6.
Comparison between LTR-III and LTR-III+IV sequences. Imino proton NMR spectra of LTR-III+IV (top) and LRT-III (bottom) G-quadruplexes with the spectral assignments of guanines participating in the formation of three G-tetrads and the stem-loop, unambiguously obtained from site-specific isotopic labeling studies (Figure S10). Residues in the middle G-tetrad layer are shown boldface as observed by solvent exchange experiments.
Using site-specific labeling strategy, we demonstrated that the 12 well-resolved peaks in the imino proton region of the LTR-III+IV sequence originate from the LTR-III part of the sequence, while the guanines involved only in LTR-IV G-quadruplex structure (G30, G32, and G33) are not engaged in Hoogsteen hydrogen bond formation (Figure 6, Figure S10).
Moreover, solvent exchange experiments showed that the guanines involved in the central tetrad of LTR-III+IV G-quadruplex (G1, G16, G20, and G27) exactly correspond to the guanines in the same position of LTR-III (Figure 6, Figure S11).
According to these data, it is clear that the longer sequence favors the single conformation of LTR-III G-quadruplex, conserving its unique features. This fact suggests that the LTR-III G-quadruplex, previously demonstrated to be the major and most stable form of the considered region, is prevalent in the longer and more dynamic context.
Discussion
In this work, we demonstrated that LTR-III folds in a hybrid quadruplex–duplex conformation with a three-layered G-tetrad core arranged in a (3 + 1) topology and a long 12-nt loop forming a hairpin structure. NMR analysis of the sequence named LTR-III+IV and able to form both LTR-III and LTR-IV G-quadruplexes showed that the folding topology of LTR-III is still conserved, suggesting the preferential folding of LTR-III G-quadruplex within the dynamic context of multiple conformations.
Hybrid quadruplex–duplex structures have been described previously as artificial constructs with different relative quadruplex–duplex orientations, exploring junction and connection varieties.32 Our structure of LTR-III G-quadruplex containing a duplex hairpin across a diagonal loop reveals a significant tilting between the helical axis of the duplex and that of the quadruplex, contrasting the feature observed for an artificial hybrid quadruplex–duplex also containing a duplex hairpin across a diagonal loop (PDB code 2M91) (Figure S12). This difference arises from the difference in the junction composition: in the 2M91 structure an adaptor G•A base pair suitably bridges the large distance (>20 Å) across the diagonal corners of a G-tetrad, while in LTR-III the junction structure formed with T14 on one strand and G3-A4 on the other strand of the duplex might be more floppy and dynamic, providing an opportunity for targeting. Even though bioinformatic studies on the human genome showed the potential of over 80 000 sequences prone to fold in such a structure,41 so far high-resolution structures of naturally occurring and biologically relevant hybrid quadruplex–duplex topologies have not been reported.41−45
The guanine content in the HIV-1 G-quadruplex forming region is highly conserved.22 Mutations in the sequence that forms the stem-loop may disrupt Watson–Crick base pairing in the duplex component of the structure. Therefore, the conservation of the nucleotides participating in the stem-loop formation has also been assessed, revealing high percentages of conservation (70–99%) for all the nucleotides with the exception of cytosine in position 7, which displayed around 50% of probability for thymine mutation.
Conserved multiple G-quadruplex structures in the LTR promoter region of HIV-1 and primate lentiviruses have been proposed as regulatory elements of viral transcription22,29 and therefore as promising targets for viral cycle inhibition. Stabilization of viral G-quadruplexes by the well-known G-quadruplex ligand BRACO-19 resulted in the inhibition of viral production.25 Recently, newly synthesized naphthalene diimide (NDI) compounds with an extended core were found to act as antiviral agents with a G-quadruplex related mechanism, selectively targeting viral over telomeric G-quadruplexes.26 Moreover, a novel NDI Cu(II) complex was found to act as DNA-cleaving agent, targeting the LTR-III G-quadruplex with high selectivity.61 In particular, the binding geometry of the NDI Cu(II) derivative to the LTR-III structure resolved here defined the proximity of the Cu catalytic site to the nearby regions and helped explain the sharp cleavage observed at two main sites of the LTR-III sequence.
Interestingly, we found that mutations in the LTR-IV G-quadruplex component do not abolish the inhibitory effect on viral transcription probably due to the stable presence of LTR-III conformation.46 Therefore, selective targeting of the major LTR-III G-quadruplex component may be a promising strategy for viral transcription inhibition.
Such a singular structure of LTR-III G-quadruplex opens the possibility of improving selectivity by targeting the quadruplex-duplex junction. Examples of compounds targeting this feature may come from DHFBI fluorogens intercalating on the junction between the G-quadruplex and hairpin of RNA light-up aptamers,47 or from recently published molecules that can simultaneously bind G-quadruplex and a proximal duplex.48,49
Conclusion
The emerging importance of the LTR G-quadruplexes as antiviral targets opened the possibility of exploring novel G-quadruplex ligands as anti-HIV-1 agents. Considering the high G-quadruplex content in human cells, one of the main challenges is to achieve selectivity toward viral G-quadruplexes. We provided here a starting point to the rational drug design approach by defining the solution structure of LTR-III G-quadruplex, the major component within the LTR G-quadruplex-forming motif. Given the fact that the majority of G-quadruplex binding ligands tested so far display structural features directed to target G-tetrads prevalently by stacking interaction50−59 and high selectivity can be achieved at duplex region, our findings open new perspectives to the possibility of discriminating among different G-quadruplex conformations. The future approach may thus be directed to the development of small molecules with structural features compatible with unique loop sequences and arrangements. This strategy toward the LTR-III structure presented in this work may provide new selective anti-HIV-1 agents with a G-quadruplex-mediated mechanism of action.
Acknowledgments
This work was supported by the Singapore National Research Foundation Investigatorship (NRF-NRFI2017-09) and Nanyang Technological University (NTU Singapore) grants to A.T.P.; Bill and Melinda Gates Foundation (GCE grants OPP1035881 and OPP1097238), and the European Research Council (ERC Consolidator grant 615879) grants to S.N.R. The authors acknowledge the use of NMR facilities at the NTU Institute of Structural Biology.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b05332.
Supplementary Figures S1–S12 (PDF)
Author Contributions
# Authors E.B. and B.H. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Gellert M.; Lipsett M. N.; Davies D. R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A. 1962, 48, 2013–8. 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J.; Phan A. T.; Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007, 35 (22), 7429–55. 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem., Int. Ed. 2004, 43 (6), 668–98. 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- Sen D.; Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990, 344 (6265), 410–4. 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Lane A. N.; Chaires J. B.; Gray R. D.; Trent J. O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008, 36 (17), 5482–515. 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud N. V.; Plavec J.. The role of cations in determining quadruplex structure and stability. In Quadruplex Nucleic Acids, Neidle S.; Balasubramanian S., Eds. Royal Society of Chemistry, 2006; pp 100–130. [Google Scholar]
- Maizels N.; Gray L. T. The G4 genome. PLoS Genet. 2013, 9 (4), e1003468. 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel-Hertsch R.; Beraldi D.; Lensing S. V.; Marsico G.; Zyner K.; Parry A.; Di Antonio M.; Pike J.; Kimura H.; Narita M.; Tannahill D.; Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48 (10), 1267–72. 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- Rhodes D.; Lipps H. J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43 (18), 8627–37. 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrat A.; Lacroix L.; Mergny J. L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44 (4), 1746–59. 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M.; Ding Y.; Burrows C. J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (10), 2604–2609. 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Hurley L. H.; Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discovery 2011, 10 (4), 261–75. 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino-Reale G.; Zaffaroni N.; Folini M. Emerging role of G-quadruplex DNA as target in anticancer therapy. Curr. Pharm. Des. 2017, 22 (44), 6612–6624. 10.2174/1381612822666160831101031. [DOI] [PubMed] [Google Scholar]
- Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1 (5), 0041. 10.1038/s41570-017-0041. [DOI] [Google Scholar]
- Ruggiero E.; Richter S. N. G-quadruplexes and G-quadruplex ligands: targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46 (7), 3270–3283. 10.1093/nar/gky187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat P.; Zhong J.; Lekieffre L.; Cowieson N. P.; Clancy J. L.; Preiss T.; Balasubramanian S.; Khanna R.; Tellam J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10 (5), 358–64. 10.1038/nchembio.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista M. J.; Martins R. P.; Billant O.; Contesse M. A.; Findakly S.; Pochard P.; Daskalogianni C.; Beauvineau C.; Guetta C.; Jamin C.; Teulade-Fichou M. P.; Fahraeus R.; Voisset C.; Blondel M. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8, 16043. 10.1038/ncomms16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tluckova K.; Marusic M.; Tothova P.; Bauer L.; Sket P.; Plavec J.; Viglasky V. Human papillomavirus G-quadruplexes. Biochemistry 2013, 52 (41), 7207–16. 10.1021/bi400897g. [DOI] [PubMed] [Google Scholar]
- Artusi S.; Perrone R.; Lago S.; Raffa P.; Di Iorio E.; Palu G.; Richter S. N. Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res. 2016, 44 (21), 10343–10353. 10.1093/nar/gkw968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artusi S.; Nadai M.; Perrone R.; Biasolo M. A.; Palu G.; Flamand L.; Calistri A.; Richter S. N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 2015, 118, 123–31. 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L. A.; Bentley K.; Peeters A.; Churchill M. J.; Deacon N. J. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000, 28 (3), 663–668. 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Nadai M.; Frasson I.; Poe J. A.; Butovskaya E.; Smithgall T. E.; Palumbo M.; Palu G.; Richter S. N. A dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013, 56 (16), 6521–30. 10.1021/jm400914r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D.; Sullivan M. A.; Sharma G.; Bambara R. A. U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry 2014, 53 (16), 2581–93. 10.1021/bi4016692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrane S.; Kerkour A.; Bedrat A.; Vialet B.; Andreola M. L.; Mergny J. L. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: a potential target for anti-HIV drug development. J. Am. Chem. Soc. 2014, 136 (14), 5249–52. 10.1021/ja501500c. [DOI] [PubMed] [Google Scholar]
- Perrone R.; Butovskaya E.; Daelemans D.; Palu G.; Pannecouque C.; Richter S. N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014, 69 (12), 3248–58. 10.1093/jac/dku280. [DOI] [PubMed] [Google Scholar]
- Perrone R.; Doria F.; Butovskaya E.; Frasson I.; Botti S.; Scalabrin M.; Lago S.; Grande V.; Nadai M.; Freccero M.; Richter S. N. Synthesis, binding and antiviral properties of potent core-extended naphthalene diimides targeting the HIV-1 long terminal repeat promoter G-quadruplexes. J. Med. Chem. 2015, 58 (24), 9639–52. 10.1021/acs.jmedchem.5b01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni E.; Frasson I.; Scalabrin M.; Perrone R.; Butovskaya E.; Nadai M.; Palu G.; Fabris D.; Richter S. N. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res. 2015, 43 (18), 8884–97. 10.1093/nar/gkv897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalabrin M.; Frasson I.; Ruggiero E.; Perrone R.; Tosoni E.; Lago S.; Tassinari M.; Palu G.; Richter S. N. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 2017, 7, 45244. 10.1038/srep45244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R.; Lavezzo E.; Palu G.; Richter S. N. Conserved presence of G-quadruplex forming sequences in the long terminal repeat promoter of Lentiviruses. Sci. Rep. 2017, 7 (1), 2018. 10.1038/s41598-017-02291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M. H.; Wang Y. Q.; Yu Z. Y.; Hu L. N.; Ou T. M.; Chen S. B.; Huang Z. S.; Tan J. H. Discovery of a new four-leaf clover-like ligand as a potent c-MYC transcription inhibitor specifically targeting the promoter G-quadruplex. J. Med. Chem. 2018, 61 (6), 2447–2459. 10.1021/acs.jmedchem.7b01697. [DOI] [PubMed] [Google Scholar]
- Schwieters C. D.; Kuszewski J. J.; Tjandra N.; Marius Clore G. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160 (1), 65–73. 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Lim K. W.; Phan A. T. Structural basis of DNA quadruplex-duplex junction formation. Angew. Chem., Int. Ed. 2013, 52 (33), 8566–9. 10.1002/anie.201302995. [DOI] [PubMed] [Google Scholar]
- Vorlickova M.; Kejnovska I.; Bednarova K.; Renciuk D.; Kypr J. Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes. Chirality 2012, 24 (9), 691–8. 10.1002/chir.22064. [DOI] [PubMed] [Google Scholar]
- Del Villar-Guerra R.; Trent J. O.; Chaires J. B. G-quadruplex secondary structure obtained from circular dichroism spectroscopy. Angew. Chem., Int. Ed. 2018, 57 (24), 7171–7175. 10.1002/anie.201709184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do N. Q.; Phan A. T. Monomer-dimer equilibrium for the 5′-5′ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chem. - Eur. J. 2012, 18 (46), 14752–9. 10.1002/chem.201103295. [DOI] [PubMed] [Google Scholar]
- Adrian M.; Heddi B.; Phan A. T. NMR spectroscopy of G-quadruplexes. Methods 2012, 57 (1), 11–24. 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Phan A. T.; Patel D. J. A site-specific low-enrichment (15)N,(13)C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002, 124 (7), 1160–1. 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- Phan A. T. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR 2000, 16 (2), 175–8. 10.1023/A:1008355231085. [DOI] [PubMed] [Google Scholar]
- Kuryavyi V.; Patel D. J. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure 2010, 18 (1), 73–82. 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.; Chou S. H.; Xu J.; Reid B. R. Structure of a single-cytidine hairpin loop formed by the DNA triplet GCA. Nat. Struct. Mol. Biol. 1995, 2 (11), 1012–7. 10.1038/nsb1195-1012. [DOI] [PubMed] [Google Scholar]
- Lim K. W.; Jenjaroenpun P.; Low Z. J.; Khong Z. J.; Ng Y. S.; Kuznetsov V. A.; Phan A. T. Duplex stem-loop-containing quadruplex motifs in the human genome: a combined genomic and structural study. Nucleic Acids Res. 2015, 43 (11), 5630–46. 10.1093/nar/gkv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.; Gaerig V.; Cui Y.; Kang H.; Gokhale V.; Zhao Y.; Hurley L. H.; Mao H. Tertiary DNA structure in the single-stranded hTERT promoter fragment unfolds and refolds by parallel pathways via cooperative or sequential events. J. Am. Chem. Soc. 2012, 134 (11), 5157–64. 10.1021/ja210399h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onel B.; Carver M.; Wu G.; Timonina D.; Kalarn S.; Larriva M.; Yang D. A New G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc. 2016, 138 (8), 2563–70. 10.1021/jacs.5b08596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. J.; Cui Y.; Yin H.; Scheid A.; Hendricks W. P.; Schmidt J.; Sekulic A.; Kong D.; Trent J. M.; Gokhale V.; Mao H.; Hurley L. H. A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J. Am. Chem. Soc. 2016, 138 (41), 13673–13692. 10.1021/jacs.6b07598. [DOI] [PubMed] [Google Scholar]
- Greco M. L.; Kotar A.; Rigo R.; Cristofari C.; Plavec J.; Sissi C. Coexistence of two main folded G-quadruplexes within a single G-rich domain in the EGFR promoter. Nucleic Acids Res. 2017, 45 (17), 10132–10142. 10.1093/nar/gkx678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadai M.; Doria F.; Scalabrin M.; Pirota V.; Grande V.; Bergamaschi G.; Amendola V.; Winnerdy F. R.; Phan A. T.; Richter S. N.; Freccero M.. A Catalytic and Selective Scissoring Molecular Tool for Quadruplex Nucleic Acids. J. Am. Chem. Soc. 2018, submitted [DOI] [PMC free article] [PubMed]
- De Nicola B.; Lech C. J.; Heddi B.; Regmi S.; Frasson I.; Perrone R.; Richter S. N.; Phan A. T. Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res. 2016, 44 (13), 6442–51. 10.1093/nar/gkw432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige J. S.; Wu K. Y.; Jaffrey S. R. RNA mimics of green fluorescent protein. Science 2011, 333 (6042), 642–6. 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. Q. N.; Lim K. W.; Phan A. T. A dual-specific targeting approach based on the simultaneous recognition of duplex and quadruplex motifs. Sci. Rep. 2017, 7 (1), 11969. 10.1038/s41598-017-10583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamitsu S.; Obata S.; Phan A. T.; Hashiya K.; Bando T.; Sugiyama H. Simultaneous binding of hybrid molecules constructed with dual DNA-binding components to a G-quadruplex and its proximal duplex. Chem. - Eur. J. 2018, 24 (17), 4428–4435. 10.1002/chem.201705945. [DOI] [PubMed] [Google Scholar]
- Haider S. M.; Parkinson G. N.; Neidle S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003, 326 (1), 117–25. 10.1016/S0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Gaw H. Y.; Patel D. J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005, 1 (3), 167–73. 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell N. H.; Parkinson G. N.; Reszka A. P.; Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130 (21), 6722–4. 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- Dai J.; Carver M.; Hurley L. H.; Yang D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011, 133 (44), 17673–80. 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzicalupi C.; Ferraroni M.; Bilia A. R.; Scheggi F.; Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013, 41 (1), 632–8. 10.1093/nar/gks1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoludis J. M.; Miller S. T.; Jeffrey P. D.; Barrett S. P.; Rablen P. R.; Lawton T. J.; Yatsunyk L. A. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012, 134 (50), 20446–56. 10.1021/ja3088746. [DOI] [PubMed] [Google Scholar]
- Chung W. J.; Heddi B.; Tera M.; Iida K.; Nagasawa K.; Phan A. T. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J. Am. Chem. Soc. 2013, 135 (36), 13495–501. 10.1021/ja405843r. [DOI] [PubMed] [Google Scholar]
- Chung W. J.; Heddi B.; Hamon F.; Teulade-Fichou M. P.; Phan A. T. Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC3. Angew. Chem., Int. Ed. 2014, 53 (4), 999–1002. 10.1002/anie.201308063. [DOI] [PubMed] [Google Scholar]
- Kotar A.; Wang B.; Shivalingam A.; Gonzalez-Garcia J.; Vilar R.; Plavec J. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem., Int. Ed. 2016, 55 (40), 12508–11. 10.1002/anie.201606877. [DOI] [PubMed] [Google Scholar]
- Wirmer-Bartoschek J.; Bendel L. E.; Jonker H. R. A.; Grun J. T.; Papi F.; Bazzicalupi C.; Messori L.; Gratteri P.; Schwalbe H. Solution NMR structure of a ligand/hybrid-2-G-quadruplex complex reveals rearrangements that affect ligand binding. Angew. Chem., Int. Ed. 2017, 56 (25), 7102–7106. 10.1002/anie.201702135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.