Abstract
Bladder cancer has the highest recurrence rate of all cancers due in part to inadequate transurethral resection. Inadequate resection is caused by the inability of cystoscopes to detect invisible lesions during the resection procedure. To improve detection and resection of nonmuscle invasive bladder cancer, we quantified the ability of a surface-enhanced Raman nanoparticle and endoscope system to classify bladder tissue as normal or cancerous. Both antibody-based (active) and tissue permeability-based (passive) targeting mechanisms were evaluated by topically applying nanoparticles to ex vivo human bladder tissue samples. Multiplexed molecular imaging of CD47 and Carbonic Anhydrase 9 tumor proteins gave a receiver operating characteristic area under the curve (ROC AUC of 0.93 (0.75, 1.00). Furthermore, passively targeted nanoparticles enabled tissue classification with an ROC AUC of 0.93 (0.73, 1.00). Passively targeted nanoparticles penetrated 5-fold deeper and bound to tumor tissue at 3.3-fold higher concentrations in cancer compared to normal bladder urothelium, suggesting the existence of an enhanced surface permeability and retention effect in human bladder cancer.
Keywords: bladder cancer, molecular imaging, surface-enhanced Raman scattering, passive targeting, multiplexing, Raman endoscopy
With a worldwide incidence and mortality of 330,000 and 123,000, respectively, bladder cancer is a major burden on global public health.1 75% of new cases are stage T1, Ta (non-invasive papillary carcinoma), or carcinoma in situ (Tis), and these stages are collectively referred to as nonmuscle invasive bladder cancer (NMIBC).2 The standard of care for NMIBC is transurethral resection (TUR) guided by white light cystoscopy (WLC).3 WLC has several limitations. First, WLC often results in incomplete resection of NMIBC. For example, one study showed 42% of T1 bladder cancer patients had residual disease in the resection region 6 weeks after an initial TUR procedure.4 Second, WLC is unable to detect some flat lesions, particularly high grade Tis lesions, which is an independent negative predictor of cancer progression.5−10 These limitations are partly responsible for the high recurrence rate of NMIBC of 60%–70%.3,10 Because improved detection of cancer could result in lower recurrence rates due to more complete disease resection,10−12 there is an unmet need for cystoscopy methods that can detect residual disease during WLC-guided resection.
We approached the problem of incomplete bladder cancer resection by developing intravesical surface-enhanced Raman scattering (SERS) nanoparticles that are targeted to bladder cancer. This approach was motivated by two potential advantages of intravesical SERS nanoparticles compared to other contrast agents. First, SERS nanoparticles produce very sharp (<1 nm) spectral lines, which enables multiplexed detection of up to eight nanoparticle channels.13−15 This multiplexing capability is in stark contrast with current cystoscopy systems, which can only detect one fluorescent dye. Given the molecular heterogeneity of cancer, we hypothesize that multiplexed imaging of many molecular targets with Raman may enable superior tissue classification compared to imaging only one target. Second, we anticipate that intravesical nanoparticles have a higher potential for clinical translation than systemically applied nanoparticles because of their more favorable pharmacokinetics.16 In particular, a major barrier to clinical translation of systemically administered SERS nanoparticles is the potential toxicity associated with long-term sequestration of nanoparticles in the spleen, bone-marrow, and liver.17 While the toxicity profile of intravesical gold-silica nanoparticles is currently unknown, quantum dots instilled into mouse bladder were only observed in extravesical organs in rare cases and showed no toxicity up to 7 days after instillation.16 We therefore expect that toxicity risks of gold-silica nanoparticles can be mitigated by intravesical administration, which has long been used by oncologists to limit systemic exposure of drugs and imaging contrast agents.16,18,19
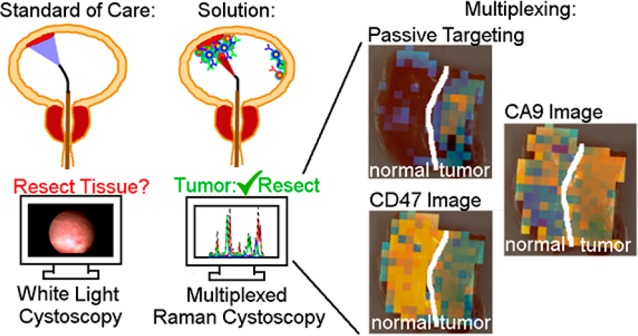
For context, Figure 1 describes the eventual clinical use of the SERS nanoparticles developed herein.20 A patient presents with a potential NMIBC, and if diagnosed, a physician recommends them for WLC-assisted transurethral resection (Figure 1a). Using the accessory channel of the cystoscope, the physician injects a co-mixture of SERS particles that are either nontargeted or actively targeted to cell surface targets (Figure 1b). Note that pre-instillation of nanoparticles does not alter the clinical routine because clinics performing photodynamic diagnosis pre-instill hexaminolevulinate for 1 h prior to cystoscopy. After pre-instillation, the unbound particles are washed out of the bladder (Figure 1c). Resection margins and ambiguous regions on the WLC image are identified and interrogated using a Raman endoscope (Figure 1d,e). Based on absolute and relative binding levels, we hypothesize that the nanoparticle-endoscope system can differentiate cancer (Figure 1d) and normal (Figure 1e) urothelium. The purpose of the current paper is to test that hypothesis and identify effective nanoparticle formulations and tissue classification algorithms that can discriminate normal and cancerous bladder tissue. We believe that accurate classification of bladder tissue with Raman cystoscopy would improve completeness of resection and prolong the time to recurrence.
Figure 1.
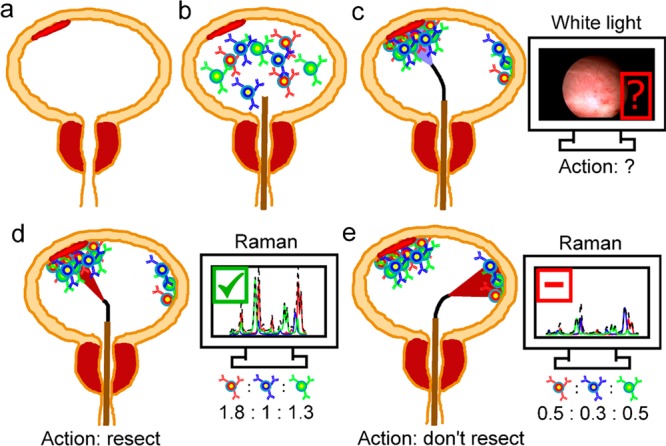
Proposed application of intraluminal SERS nanoparticles. (a) Patient presents with potential NMIBC (red tissue). (b) Before cystoscopy, intraluminal SERS nanoparticles are administered. Each nanoparticle color represents a different targeting mechanism (CA9, red; passive, blue; CD47, green). (c) Patient gets standard of care, which is transurethral resection guided by WLC. Regions ambiguous on WLC are subsequently interrogated with Raman endoscopy. (d, e) Based on absolute and relative binding levels of each channel, flat lesions that are identified, and cancerous tissue is resected.
Because NMIBC is localized to the bladder lumen, intravesical nanoparticles can only target tumor biomarkers expressed on the luminal surface. One such biomarker is the immunosuppressive protein CD47, which is upregulated in tumors and interacts with SIRP-α on macrophages to inhibit phagocytosis.21 A previous molecular cystoscopy study using quantum dots conjugated to anti-CD47 achieved cancer detection with a sensitivity and specificity of 82.9% and 90.5%, respectively.20 Another such biomarker, CA9, catalyzes hydration of carbon dioxide and is involved in pH regulation. CA9 was chosen as a biomarker because immunohistochemical analysis of human bladder cancer biopsies showed that CA9 was expressed on the luminal surface of 84% and 68% of Ta and T1 bladder cancers, respectively, but not on normal urothelium.22,23 The rationale behind also including the passively targeted nanoparticles was two-fold. First, binding of passively targeted nanoparticles to the bladder wall serves as an experimental control for nonspecific binding of actively targeted nanoparticles.24 Second, because cancer disrupts the urothelial layer that blocks intravesical compounds from entering the body,20 we hypothesized that passive binding of intravesical nanoparticles could be predictive of cancer.
The purpose of this work is to synthesize and characterize the SERS nanoparticles shown in Figure 1 and evaluate the relative strengths and challenges associated with active and passive targeting to human bladder cancer. Seven ex vivo human normal bladder and eight bladder tumor samples were imaged using the nanoparticle mixture and endoscope device shown in Figure 1. We compared the diagnostic performance of three post-processing techniques and active vs passive targeting in order to determine the optimal approach for discriminating ex vivo cancer from normal bladder samples. In this paper we show (1) evidence of passive targeting of intravesical nanoparticles, (2) an enhanced surface permeability and retention effect for topically applied nanoparticles, and (3) that bladder tissue can be classified as normal or tumor using multiplexed molecular Raman imaging. The enhanced surface permeability effect described herein represents a simple targeting mechanism that we believe will be useful for both diagnosis and therapy of bladder cancer.
Results/Discussion
SERS Nanoparticles Actively Target CD47 and CA9 in Cell Suspension
Figure 2 shows a schematic representation of the gold-silica nanoparticles used in this paper. The prefixes s420, s421, and s440 are used to differentiate SERS nanoparticles with different Raman dyes coating the gold surface. Each dye is paired with a different antibody, so that binding levels of each antibody-nanoparticle pair can be differentiated by spectrally unmixing the three dye signals (see below). The actively targeted nanoparticles are s420-CA9 and s440-CD47, and the passively targeted s421-IgG4 nanoparticle is coated with an isotype control IgG4 antibody that does not actively bind to human proteins. Physiochemical characterization of all three nanoparticle types is given in Supporting Figure 1.
Figure 2.
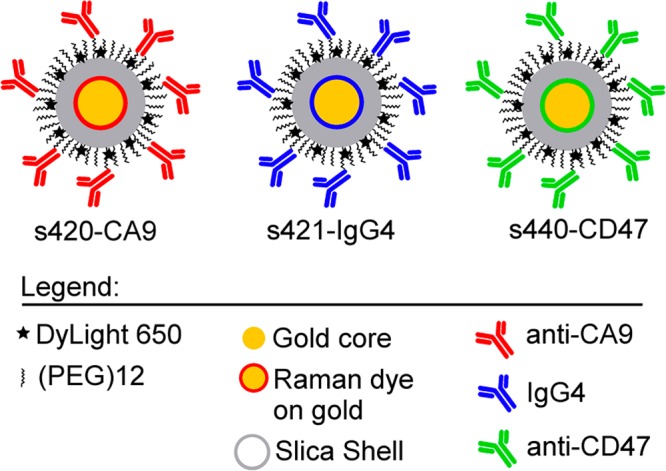
Schematic representation of the SERS nanoparticles developed herein. The Raman dyes were 4,4′-dipyridyl (s420, red dye), d8-4,4′-dipyridyl (s421, blue dye), and trans-1,2-bis(4-pyridyl)-ethylene (s440, green dye). The blue IgG4 nanoparticle is used as a negative experimental control for active binding of CA9- and CD47-targeted SERS nanoparticles.
We first used flow cytometry to validate the specific binding of fluorophore-labeled SERS nanoparticles to CD47 and CA9 in cell suspension. Figure 3a shows flow cytometry for cells labeled with s440-CD47, and Figure 3b shows median DL650 fluorescence for flow cytometry experiments ran in triplicate. As can be seen in Figure 3a,b, compared to CD47+ DLD-1 cells, CD47– DLD-1 cells exhibited 26-fold less nanoparticle binding (p < 0.005), labeling cells with s421-hIgG4 resulted in 25-fold less nanoparticle binding (p < 0.005), and epitope blocking resulted in 8-fold less nanoparticle binding (p < 0.0001). Similarly, Figure 3c shows flow cytometry data for HCT-116 cells labeled with s420-CA9 nanoparticles, and Figure 3d shows median DL650 fluorescence for flow cytometry experiments ran in triplicate. As can be seen in Figure 3c,d, compared to CA9+ HCT-116 cells, CA9– HCT-116 cells exhibited 6-fold less nanoparticle binding (p < 0.005), and labeling cells with s421-mIgG2b resulted in 8-fold less nanoparticle binding (p < 0.005). Furthermore, experiments that varied the available target antigen showed that the level of nanoparticle binding was proportional to antigen availability on the cell surface (Supporting Figure 2). These results suggest that the antibody-functionalized SERS nanoparticles actively target CD47 and CA9 in cell suspension.
Figure 3.
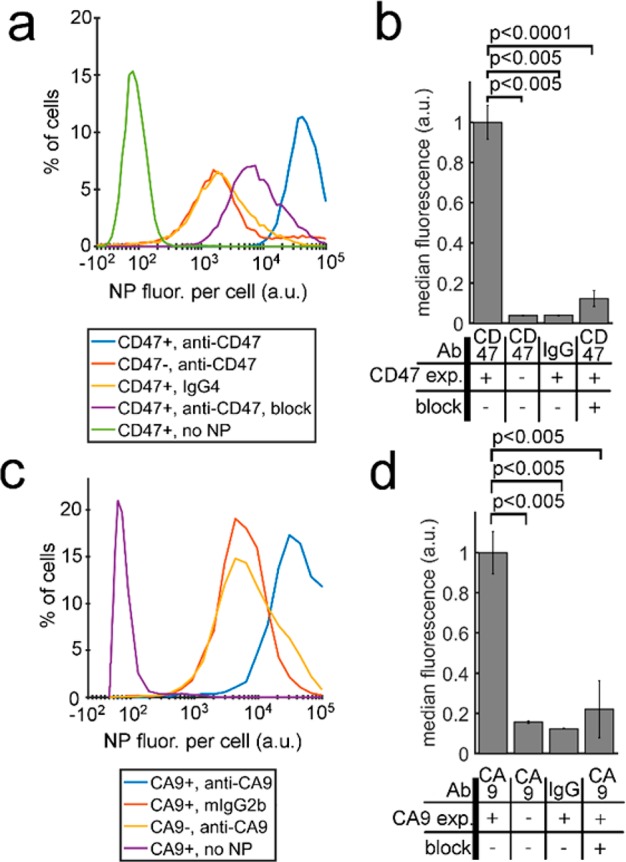
Validation of active targeting using flow cytometry. (a) Flow cytometry histogram showing binding of CD47-targeted SERS nanoparticles to cells with three negative controls. (b) Experiments from (a) were reported in triplicate and plotted in a bar graph. (c) Flow cytometry histogram showing binding of CA9-targeted SERS nanoparticles to cells with three negative controls. (d) Experiments from (c) were conducted in triplicate and plotted in a bar graph.
As can be seen in Figure 3a,c and Supporting Figure 3a, the isotype control nanoparticles had higher fluorescence than untreated cells, indicating that nanoparticles were binding nonspecifically to cells. Indeed, nonspecific binding is a common problem in the field of nanobiotechnology and is problematic during magnetic isolation of bacteria, cell lines, and circulating tumor cells.25−27Supporting Figure 3a shows that nonspecific binding varies by over an order of magnitude as the cross-linker length and number of antibodies are varied. When a mixture of nontargeted (mIgG2b) and anti-CD47 antibodies is conjugated to the nanoparticle surface with a (PEG)77 cross-linker, the ratio of specific to nonspecific binding increases 2.6-fold compared to anti-CD47 congugated with (PEG)12 cross-linker. The cause of this reduced nonspecific binding is currently unclear, but it is likely that nontargeting antibodies and a long cross-linker alter the surface–surface interaction between nanoparticles and cells in a way that reduces the nonspecific stickiness of the nanoparticles. These data suggest that coating nanoparticles with a mixture of targeted and nontargeted antibodies using a long cross-linker can reduce but not eliminate nonspecific binding of antibody-functionalized nanoparticles.
Raman Cystoscopy Enables Multiplexed Molecular Imaging of CD47 and CA9 in Cell Suspensions
Next we tested the multiplexing capabilities of the three-nanoparticle mixture (s420-CA9, s421-IgG4, s440-CD47) by applying the mixture to cells in suspension and imaging with a Raman endoscope (Figure 4). Four cell suspensions were made that did or did not have (in each possible combination) the CD47 or CA9 antigen available for nanoparticle binding. Cells were stained with a 1:1:1 mixture of the three nanoparticle types, washed, and then imaged.
Figure 4.
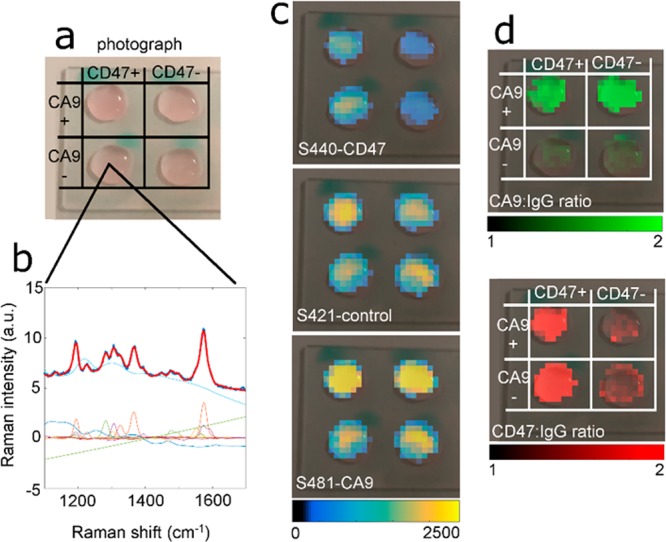
Multiplexed imaging of CD47 and CA9 on cells in suspension. (a) Photograph of cells positive and negative for CD47 and CA9. The top right sample in the 2 × 2 grid is positive for CA9 but negative for CD47. (b) Spectral unmixing of background, s420-CA9, s421-IgG4, s440-CD47. The thick red line represents the fit, and the underlying thick dark blue line is the raw data. Thin lines represent the SERS and background components of the signal. (c) Overlay of the s440, s421, and s420 channel images on the photograph. (d) Calculated CD47:IgG4 and CA9:IgG4 ratio images overlaid on the photograph.
Figure 4a shows a photograph of the cells with the four possible combinations of CD47 and CA9 expression. Figure 4b and Supporting Figure 4 show raw spectra, the least-squares fit (bold red line), and the decomposed components of the SERS and background spectra (thin colored lines). Figure 4c shows the decomposed images of each nanoparticle type. Figure 4d shows the active-to-passive ratios of s440-CD47 and s420-CA9 nanoparticles, both normalized on a pixel-by-pixel basis to s421-IgG4 binding. As can be seen in Figure 4d, the s440-CD47/s421-IgG4 ratio is >2 in CD47+ cells and close to 1 in CD47– cells, with a similar pattern for the CA9-targeted nanoparticles. These data suggest that our combined endoscope/nanoparticle system is capable of multiplexed molecular detection of both CD47 and CA9 in cell suspension.
The ability of Raman cystoscopy to detect more than one target directly addresses a limitation of fluorescence cystoscopes, which is that they can currently only detect one dye. Given the molecular and physiological heterogeneity of cancer, it is unlikely that a single biomarker will be able to identify cancer 100% of the time. One solution is to develop an imaging system that can detect multiple biomarkers. Currently available commercial fluorescence cystoscopes only provide 405 nm/635 nm excitation/emission wavelengths for photodynamic diagnosis. While in theory it is possible to modify endoscopes to detect a broader range of fluorescent dyes, the relatively broad emission spectra of fluorescent dyes and the background autofluorescence of tissue likely make simultaneous detection of more than 2–3 dyes problematic. SERS nanoparticles could be a solution to this limitation because the same Raman endoscope can image up to eight nanoparticles with a single excitation wavelength, and no hardware modifications are necessary when the number or combination of nanoparticle flavors is changed.15 Below, we test the hypothesis that multiplexed molecular imaging is superior to single-target imaging for classification of normal and cancerous bladder. Further studies will be needed to compare the effectiveness of fluorescent and Raman cystoscopy.
Classifying ex Vivo Human Bladder Tissue Using Multiplexed Imaging of CD47, CA9, and Passively Targeted Nanoparticles: Overview of Approach
Next, we topically applied passively and actively targeted nanoparticles to ex vivo bladder cancer specimens and quantified their ability to classify bladder tissue as cancerous or normal. Specifically, the 1:1:1 nanoparticle mixture and unmixing technique from Figure 4 was applied to N = 7 normal and N = 8 tumor human bladder tissue samples. Using this three-nanoparticle mixture, we evaluated three different approaches for classifying tissue as normal or cancer. The subsequent sections repeatedly refer to ratios and normalization approaches that are summarized in Table 1. The three approaches are analyzed in detail below, but here a brief qualitative summary is given for context.
Table 1. Summary of Signal Processing Approachesa.
| approach no. | N-plex | DoF | channel names | channel equations |
|---|---|---|---|---|
| Approach 1: No normalization | 3-plex | 3 | s420-CA9 | s420-CA9 |
| s421-IgG4 | s421-IgG4 | |||
| s440-CD47 | s440-CD47 | |||
| 1-plex | 1 | s421-IgG4 | s421-IgG4 | |
| Approach 2: Active-to-passive normalization | 3-plex | 2 | CA9:IgG4 ratio | (s420-CA9)/(s421-IgG4) |
| CD47:IgG4 ratio | (s440-CD47)/(s421-IgG4) | |||
| 2-plex | 1 | CA9:IgG4 ratio | (s420-CA9)/(s421-IgG4) | |
| 2-plex | 1 | CD47:IgG4 ratio | (s440-CD47)/(s421-IgG4) | |
| Approach 3: Active-to-sum normalization | 3-plex | 2 | CA9:3-sum ratio | (s420-CA9)/(s420-CA9 + s421-IgG4 + s440-CD47) |
| CD47:3-sum ratio | (s440-CD47)/(s420-CA9 + s421-IgG4 + s440-CD47) | |||
| 2-plex | 1 | CA9:2-sum ratio | (s420-CA9)/(s420-CA9 + s421-IgG4) | |
| 2-plex | 1 | CD47:2-sum ratio | (s440-CD47)/(s421-IgG4 + s440-CD47) |
N-plex: number of channels used in analysis. DoF: degrees of freedom.
For the first approach, we analyze the raw signal intensity levels of each nanoparticle type, finding that the passive binding levels of s421-IgG4 nanoparticles can accurately classify normal and cancerous bladder tissue (Figure 5). Passive targeting of intravesical nanoparticles has not been reported previously, and we hypothesize that passive targeting to cancer is caused by disruption of the urothelium by cancer, which enhances the tissue surface permeability to, and retention of, nanoparticles. That hypothesis was supported by experiments showing that the penetration depth of s421-IgG4 was on average 5-fold greater (Figure 6g), and the binding levels 3.3-fold greater, in tumor than normal tissue (Figure 5c). Those data suggested that cancer induces an enhanced surface permeability and retention effect to intravesical nanoparticles in the human bladder.
Figure 5.
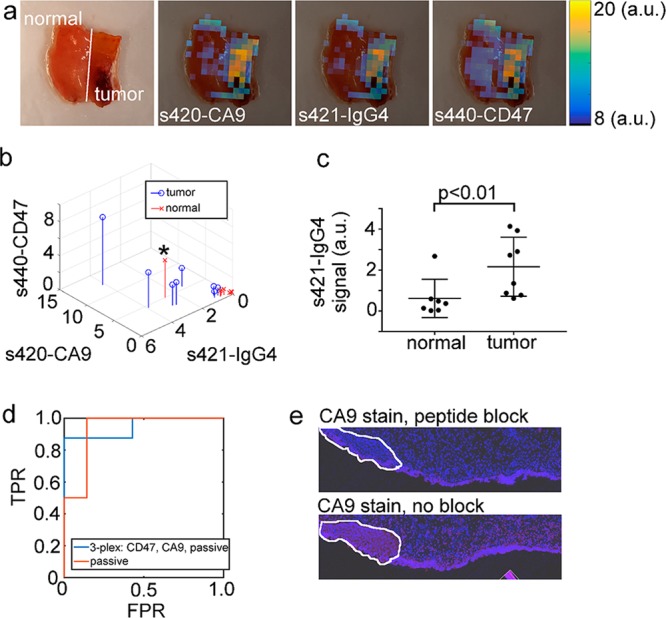
Approach 1: Tissue classification based on raw, non-normalized SERS signal intensity. (a) Photograph and overlays of unmixed s420-CA9, s421-IgG4, and s440-CD47 images. (b) Stem plot of the non-normalized signal in each channel for all N = 15 samples. (c) Statistical comparison of the binding levels of s421-IgG4 in normal and tumor samples. (d) ROC plot for tissue classification using either all three channels (3-plex) or only the s421-IgG4 channel (passive). (e) Histological analysis of the data point indicated by a black asterisk in frame (b). Blue is DAPI nuclear stain, and red is a CA9 stain.
Figure 6.
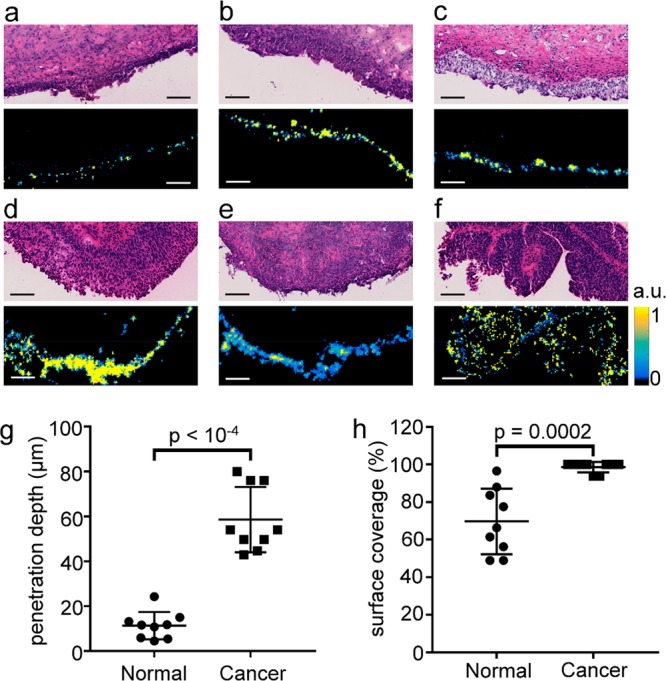
Penetration depth of SERS nanoparticles was 5-fold greater in tumor than normal tissue. (a–c) H&E stains (top) and s421-IgG4 Raman images (bottom) of normal bladder tissue from three separate patients. (d, e) H&E and s421-IgG4 Raman images for a low grade pNx (d), high grade T2 (e), and high grade pTa (f) bladder cancer tissues. All images are on the same scale, and all scale bars are 100 μm. All Raman images are on the same color scale, and the scale bar next to frame (f) applies to all Raman images. (g) Penetration depth in microns for normal and cancer tissue samples. (h) Percent of tissue surface with detectable Raman signal.
In order to overcome potential limitations with passive targeting, we next evaluated an approach commonly reported in the SERS literature which uses the ratio of actively targeted nanoparticles to passively targeted nanoparticles to classify tumor or normal tissue.24,28 The active-to-passive normalization technique exhibited a moderate classification accuracy that was hampered by experimental noise in the Raman signal (Figure 7). To overcome limitations imposed by noise, we designed an alternative approach where the signal from actively targeted nanoparticles was normalized to the sum of all three nanoparticles channels. This signal processing approach resulted in lower noise sensitivity and higher classification accuracy than the targeted-to-passive approach and a similar classification accuracy to passive targeting (Figure 8).
Figure 7.
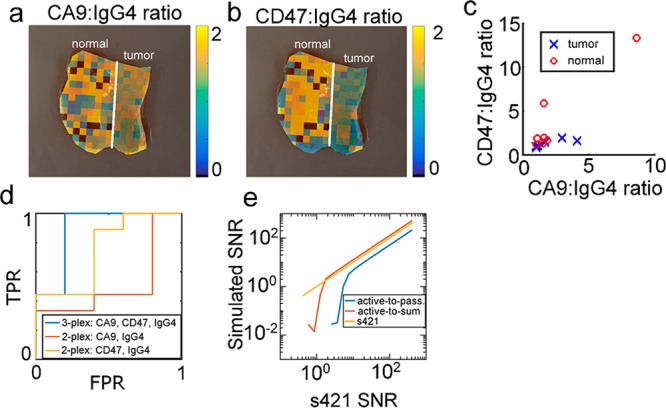
Approach 2: Tissue classification based on active-to-passive normalized data. (a) Overlay CA9:IgG4 ratio on same tissue as Figure 5. (b) CD47:IgG4 ratio. (c) scatterplot of CD47:IgG4 and CA9:IgG4 ratios for N = 15 bladder tissue samples. (d) ROC curve for classifying bladder tissue based on 2-plexed and 3-plexed data. (e) SNR simulations of active-to-passive (Approach 2) and active-to-sum (Approach 3) ratios.
Figure 8.
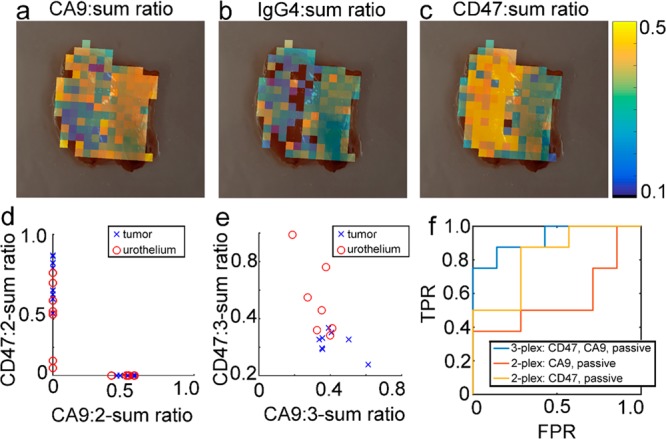
Approach 3: Classifying bladder tissue with active-to-sum normalized data. (a–c) CA9:sum, IgG4:sum, and CD47:sum overlays for the same tissue samples as in Figure 5a,b. (d) CD47:2-sum and CA9:2-sum ratios for all N = 15 tissue samples scanned on the endoscope. Because the 2-sum ratios only depend on either the s440-CD47 or s420-CA9 signal, the 2-sum ratios do not reflect correlations between s440-CD47 and s420-CA9 binding and thus fall directly on each axis. (e) CD47:3-sum and CA9:3-sum ratios for all N = 15 tissue samples. (f) ROC curves for classification of bladder tissue samples based on 3-plexed and 2-plexed active-to-sum ratios.
Approach 1: Classifying Tissue Based on Raw, Non-Normalized Signal Intensity
The first signal processing approach classified tissue by applying linear discriminant analysis to the raw intensity of each nanoparticle flavor. Figure 5a shows a white light image of a separate normal and tumor tissue sample positioned adjacent to each other and an overlay of each channel. As can be seen from Figure 5a, the raw intensity of each channel was higher on the tumor tissue relative to the normal tissue. Figure 5b shows a three-dimensional stem plot of the average signal intensity in each channel for all 15 tissue samples.
For the passively targeted s421-IgG4 nanoparticles, the average binding level was 3.3-fold higher on tumor than normal tissue (Figure 5c; mean/median/min/max was 0.62/0.37/0.023/2.7 for normal, 2.2/2.0/0.6/4.1 for tumor, Mann–Whitney U test p < 0.01), suggesting that passive binding of nanoparticles could be an accurate classifier of bladder tissue. ROC analysis gave an AUC of 0.95 (0.76, 1.0) and 0.93 (0.73, 1.0) for classifying tissue using all three channels or just s421-IgG4, respectively. The AUCs were not significantly different for the three-channel vs one-channel processing approach (p = 0.41). The leave-one-out cross-validation error rate (LOO-CV-ER) for the three- and one-channel approach were both 33%. These data suggest that the raw signal intensity of the s421-IgG4 channel was just as good at classifying bladder tissue as the three-nanoparticle mixture.
An interesting data point was the sample marked with a black asterisk at in Figure 5b, which was deemed normal by the surgeon but bound s421-IgG4 nanoparticles well above the level expected for normal tissue. During LOO-CV, the passive Raman classifier (Approach 1) assigned a likelihood of 93% that the tissue was tumor. Upon histological analysis of the tissue, there was clear signs of hyperplasia (outlined in white) and CA9 staining (red), suggesting that the sample may not have been actually normal (Figure 5e). CA9 is not expressed in normal bladder tissue,22,23 suggesting that the sample may contain cancer. While this histological data was not conclusive enough to reassign the entire tissue specimen to the tumor class, the data did indicate that the putatively normal tissue may have in fact contained cancer. This data point suggests that in some cases, passively targeted Raman nanoparticles could identify non-normal tissue in regions that appear normal on WLC.
Passive Targeting Was Mediated by Enhanced Permeability of SERS Nanoparticles in Cancer Relative to Normal Tissue
Our original hypothesis was that binding levels of the untargeted, non-normalized s421-IgG4 nanoparticles would be a poor classifier of cancer because there would be no mechanism targeting them to diseased cancer tissue. However, the data in Figure 5 show the unexpected result that s421-IgG4 binding levels were 3.3-fold higher in cancer than in normal and that s421-IgG4 binding levels classified tissue with an AUC ROC of 0.93 (0.73, 1.0). Higher binding to bladder cancer was unexpected because experiments in intact rat esophagus showed similar absolute binding levels in cancer and normal.29 In excised breast tissue, binding levels of SERS nanoparticles in tumor were 3-fold lower in tumor than normal.30 In ex vivo human bladders instilled with Qdot625-isotype control antibody conjugates, no nanoparticle uptake was observed in cancerous tissue.20 The comparison to quantum dots, which are 15–20 nm in diameter, suggests that the larger size of the SERS particles herein (120 nm) may contribute to their increased passive targeting.
Based on the data in Figure 5, we sought to better understand the mechanism behind passive targeting to bladder cancer by testing the hypothesis that bladder cancer exhibits higher permeability to SERS nanoparticles than normal bladder (Figure 6). This hypothesis was based on observations made in a previous study that noted that the surface layer of epithelial cells is disrupted in bladder cancer.20 It is well-known that the epithelial surface in the bladder exhibits cellular tight junctions and a GAG layer that protects the body by preventing the diffusion molecules and nanoparticles from the bladder lumen back into the bloodstream.16,20,31 We hypothesized that this protective layer partially blocks passive binding of SERS nanoparticles to normal bladder tissue, but that the disruption of the protective layer by cancer increases passive binding.
Tissue samples were stained and washed with the same method used for endoscope imaging, then frozen and sectioned orthogonally to the tissue surface (Supporting Figure 5a). H&E (top) and s421-IgG4 Raman images (bottom) of the resulting sections are shown in Figure 6 for normal (a–c) and cancerous (d–f) bladder tissues. As can be seen qualitatively from the Raman images, the nanoparticles penetrate deeper in tumor than normal tissue, suggesting that the cancer tissue was more permeable than normal to topically applied nanoparticles. The line profiles used for calculating penetration depth of nanoparticles are shown in Supporting Figure 5b,c (bottom). The resulting mean and standard deviation of penetration depth were 11 ± 6 μm and 58 ± 14 μm for normal and cancerous bladder tissues, respectively (Figure 6g, p < 10–4). The mean and standard deviation of the surface coverage were 70 ± 17% and 99 ± 3% for normal and cancerous bladder tissues, respectively (Figure 6h, p < 10–4).
In summary, s421-IgG4 exhibited higher penetration depth in bladder cancer compared to normal bladder, which was indicative of higher surface permeability in cancer. Furthermore, s421-IgG4 exhibited higher binding levels (Figure 5c) in bladder cancer than normal bladder, suggesting enhanced retention of IgG4-s421 in cancer. Had the retention of s421-IgG4 been equal between cancer and normal, enhanced penetration of s421-IgG4 in bladder cancer would have been negated by enhanced washout during the tissue washing step, and the cancer and normal tissue would have exhibited similar binding levels. Instead, the 3.3-fold higher binding levels of nanoparticles in the cancer samples suggest that nanoparticles in bladder cancer had higher retention during washing than did normal bladder. Thus, the combined data suggest the existence of an enhanced surface permeability and retention (ESPR) effect in bladder cancer, which appears to be a topical version of the well-known enhanced permeability and retention effect (EPR).32 The EPR effect, which refers to the accumulation of intraveneous macromolecules in tumors, is caused by fenestrations between defective vascular endothelial cells and poor lymphatic drainage in tumors. The ESPR effect reported herein refers to a similar but distinct concept where the normal bladder urothelium is disrupted by cancer, thus allowing improved penetration and retention of intravesical nanoparticles into the mucosa (Supporting Figure 6).
It is interesting to note that the passive targeting mechanism described above is related to physiological properties of the bladder tissue and thus may not be specific to gold-silica nanoparticles. Although further work is necessary, it is possible that similarly sized intravesical viral vectors,33 fluorescent nanoparticles such as quantum dots, and therapeutic nanoparticles34,35 could also exhibit passive targeting properties. Additionally, it is currently unclear how tumor stage and common stressors such as bacillus Calmette–Guerin immunotherapy, infection, and chemotherapy affect passive binding of nanoparticles to bladder tissue.
While the single-channel passive binding approach was effective at classifying normal and cancer tissues in our data set, it has two potential limitations. First, the single-channel approach does not use any normalization, which could be problematic in a clinical procedure; specifically, normalization controls for experimental variations in laser intensity and working distance, experimental variations in the staining and washing procedure, and tissue variations in passive targeting, allowing isolation of the component of signal due to active binding to the protein target. Second, molecular information inferred from active binding levels may be useful for discriminating subtle but important tissue differences like the difference between inflammation and cancer. Because of these potential advantages, we continued to explore additional processing techniques that employ normalization.
Approach 2: Active-to-Passive Normalization Exhibited Moderate Classification Accuracy, and Accuracy Was Limited by Experimental Noise
The active-to-passive normalization method, which is commonly used in the SERS literature and in Figure 5, normalizes the signal of s420-CA9 or s440-CD47 to s421-IgG4. Figure 7a,b shows the active-to-passive ratios overlaid on the same sample as in Figure 5a. Active-to-passive ratio images for all samples are shown in Supporting Figures 7 and 8. In Figure 7a,b, the calculated ratios in the urothelium appear noisy (discussed below), the s440-CD47 ratio appears higher in normal than in tumor tissue, and the CA9 ratio appears approximately the same between normal and tumor tissues.
Figure 7c compares the distribution of the CA9:IgG4 and CD47:IgG4 ratios in 15 ex vivo human tissue samples. Qualitatively, there was an overlap between the tumor and normal distributions. We quantitatively compared the ability of 2-plexed and 3-plexed active-to-passive data to classify bladder tissue by calculating the area under the receiver operating characteristic curve (AUC ROC, Figure 7d) and LOO-CV-ER for each case. For the 2-plexed cases, the receiver operating characteristic area under the curve (ROC AUC) was 0.57 (0.24, 0.86) and the LOO-CV-ER was 60% for the CA9:IgG4 ratio, and the ROC AUC was 0.82 (0.52, 1.0) and the LOO-CV error rate was 33% for the CD47:IgG4 ratio. When the CD47:IgG4 and CA9:IgG4 were both used to classify tissue based on their linear discriminant analysis (LDA) score (3-plexed case), the ROC AUC was 0.82 (0.55, 1.0) and the LOO-CV-ER was 27%. For the active-to-passive targeting approach, the CD47 and IgG4 2-plex AUC was not significantly different from the 3-plex AUC (p = 0.5), indicating that with active-to-passive targeting, addition of the CA9 channel is did not improve tissue classification.
We hypothesized that the cause of the wide ROC AUC confidence intervals and LOO-CV error rates reported above are caused by high noise in the active-to-passive ratios on the urothelium (see for example urothelium of Figure 7a,b). To test this, we simulated the experimental error of the calculated active-to-passive binding ratios (Figure 7d). The signal-to-noise ratio (SNR) of the s421-IgG4 channel in Figure 7a was 1.7, and simulations show that this results in a very low (≪1) SNR of the s440/s421 ratio. The low SNR of the ratio is probably due to noise bringing the denominator of the ratio close to zero in some pixels and not others, causing the pixel-to-pixel variance of the ratio to be very large. Experimentally, the CD47:IgG4 ratio for the image in Figure 7b had a standard error of the mean (SEM) of 41%. Notably, the median CD47:IgG4 ratios for tumor and urothelium were 1.3 and 1.9, respectively, corresponding to an effect size of 33%. Because the effect size and SEM are comparable, it is not surprising that experimental noise degrades the classifier performance when using the active-to-passive normalization approach. In the future, this problem could be solved using brighter SERS particles36−38 or a more sensitive endoscope setup, but for the current study we developed the active-to-sum normalization method, which had a simulated SNR 24% higher than the s421-IgG4 channel (Figure 7d). The active-to-sum metric for the urothelium in Figure 7a had a SEM of 4.6%, which is 9-fold lower than the SEM of the active-to-passive metric, indicating that active-to-sum normalization exhibits lower noise sensitivity than active-to-passive normalization.
Approach 3: Active-to-Sum Normalization Exhibited Lower Noise and Higher Classification Accuracy Compared to Targeted-to-Passive Normalization
For the active-to-sum normalization method, each channel is normalized to the sum of all three channels. Overlays of active-to-sum normalized data are shown in Figure 8a–c and Supporting Figures 7 and 8. Qualitatively, the CA9-to-sum ratio appeared higher in the tumor than normal, and the CD47-to-sum ratio looked lower in the tumor than normal. Figure 8b,c is analogous to Figure 7b and shows how the clustering of data changes as additional molecular information is added. As can be seen in the two-channel active-to-sum normalization cases (Figure 8d), there was overlap between the tumor and urothelium distributions. A possible cause of this overlap is that the CA9:2-sum and CD47:2-sum ratios only contain information from one molecular target and as a result are less able to differentiate between the normal and cancer tissue. For the CA9:2-sum ratio, the mean and standard deviation for tumor and normal tissues were 0.6 ± 0.1 and 0.6 ± 0.1, respectively (t test p = 0.70). For the CD47:2-sum ratio the mean and standard deviation for tumor and normal tissues were 0.5 ± 0.1 and 0.7 ± 0.1, respectively (t test p = 0.04). The ROC AUC and LOO-CV-ER were 0.57 (0.24, 0.87) and 73% for the CA9:2-sum ratio, while for the CD47:2-sum ratio, they were 0.82 (0.55, 1.0) and 33%. These data suggest that the CD47:2-sum ratio is the best classifier if only two channels are used.
The active-to-sum normalization technique which uses all three channels has two degrees of freedom, which are shown for normal and tumor samples in Figure 8e. The resulting ROC AUC was 0.93 (0.75, 1.0) and the LOO-CV-ER was 20%. The ROC AUC for the three channel case was statistically significantly greater than the s420-CA9:2-sum ROC (p = 0.01) but not the s440-CD47 AUC (p = 0.14). Thus, the tighter confidence intervals, higher AUC, and lower LOO-CV-ER suggest a trend toward higher classification accuracy with three channels compared to two channels, although the trend was not statistically significant compared to the CD47:2-sum ratio. We believe that this trend justifies further development of multiplexed molecular imaging strategies because multiplexed detection of two targets could outperform detection of a single target.
Summary: Both Passively and Actively Targeted Nanoparticles Accurately Classified Bladder Cancer with an AUC ROC > 0.9
Of all of the tissue approaches investigated herein, the two that most warrant further study are the non-normalized s421-IgG4 nanoparticles and 3-plexed active-to-sum normalized nanoparticles. These two approaches both resulted in a ROC AUC of 0.93, and the AUCs were not statistically different (p = 0.50). For clinical translation, the decision to use passive or active targeting is influenced by a trade-off between nanoparticle formulation simplicity and measurement robustness. In particular, the passive targeting approach requires a simple formulation with only one nanoparticle flavor that may not require an antibody or biomarker. Furthermore, with the passive targeting, the signal processing is simpler because cancer is identified by positive contrast. We anticipate that the simpler formulation behind passive targeting will facilitate regulatory approval and adoption by physicians. However, the actively targeted approach also has some clear advantages. First, active targeting uses normalization during post-processing, which helps control the experimental variations listed above. Second, as more bladder cancer surface biomarkers continue to be discovered, additional Raman channels can be included which could improve classification accuracy. Furthermore, it is possible that the molecular approach can discriminate tissue injury like inflammation from cancer, which would reduce the time physicians spend resecting inflamed but otherwise healthy tissue.
Limitations
While the data herein suggest that intravesical nanoparticles could be a promising approach for improving tissue resection of NMIBC, there are a few limitations of this study worth mentioning. First, all data in this study were carried out in ex vivo tissue specimens and not whole bladders. Because both the biological (e.g., nanoparticle endocytosis) and physical (e.g., motion of nanoparticle solution) behaviors of the system are different ex vivo, we currently do not know how the classification performance and staining procedure shown herein will generalize to direct instillation in whole bladders. Second, the active targeting approach is limited by the availability of cell surface bladder biomarkers. As more biomarkers continue to be discovered and validated, we believe that the diagnostic accuracy of the active targeting approach will increase further. Finally, it is unclear if Raman cystoscopy or fluorescence cystoscopy will provide the most clinical utility. Fluorescence cystoscopy has the advantages of high imaging speed and commercially available hardware, while Raman is preferable for multiplexing multiple nanoparticle flavors.13
Conclusions
This article evaluates the tissue classification performance of a Raman nanoparticle-endoscope system designed to supplement WLC. We found that 1-plexed imaging of passively targeted nanoparticles and 3-plexed imaging of CD47, CA9, and passively targeted nanoparticles classified human tissue with a ROC AUC of 0.93 (0.73, 1.0) and 0.93 (0.75, 1.0), respectively. Tissue penetration depth was on average 5-fold higher in tumors than normal, which suggests an ESPR effect in bladder cancer. Although the trend was not statistically significant, the 3-plexed active-to-sum approach classified tissue more accurately than the 2-plexed active-to-sum approaches, indicating that multiplexed detection of multiple molecular targets with Raman endoscopy could improve identification of residual disease during WLC. Furthermore, the nanoparticle-endoscope system identified one sample that appeared normal on WLC as tumor, and upon subsequent histological analysis, that sample exhibited hyperplasia and positive CA9 staining. These data suggest that Raman endoscopy may outperform the gold standard in some cases. Collectively, the data herein support Raman endoscopy and intravesical SERS nanoparticles as a promising approach for improving resection of NMIBC.
Methods
Antibodies
For SERS nanoparticle functionalization, Hu5F9-G439 was obtained courtesy of the Irving Weissman Lab and used for targeting to CD47. For targeting to CA9, mouse monoclonal antibody (GT12) was obtained additive-free from GeneTex. hIgG4 isotype control antibody was obtained from antibodiesonline.com. For tissue immunofluorescence clone B6H12.2 (Novus Biologicals) was used for CD47 detection and NB100-417 (Novus Biologicals) was used for CA9 detection. For negative controls the CA9 deactivating peptide (NB100-417PEP, Novus Biologicals) and a mouse IgG1 isotype (product no. MAB002, R&D Biosystems) for CD47 were used.
Functionalization of Raman Particles
SERS particles were obtained from Cabot Security Materials (formally Oxonica Materials). SERS particles were functionalized with antibodies using a procedure similar to the one outlined in ref (24). First, surface disulfides on SERS particles were reduced by suspending the SERS nanoparticles in freshly prepared solution of 10 mM dithiothreitol (Sigma-Aldrich) and 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS, Sigma-Aldrich) at pH 7.3 for 30 min. After washing the reduced nanoparticles three times in 10 mM MOPS, nanoparticles were mixed with Dylight 650 (DL650)-maleimide (Thermo Fisher) at a molar ratio of 60,000 dye molecules per nanoparticle in pH 7.3 MOPS buffer and allowed to incubate for 3 h, resulting in 3000 dye molecules per nanoparticle. Dye-labeled nanoparticles were then washed five times in 10 mM MOPS buffer to remove unbound dye. Next, dye-labeled nanoparticles were suspended at 400 pM, followed by addition of antibody and SM-(PEG)12 cross-linker (NHS-PEG12-maleimide, Thermo Fisher part 22112). The reaction contained 3000 cross-linkers per nanoparticle and 200 antibodies per nanoparticle, and the reaction was allowed to proceed at room temperature for 3 h. Next, remaining sulfhydryl groups on the nanoparticle surface were PEGylated by incubating the nanoparticle solution with 600,000 molar excess of MM-(PEG)12 (methyl-PEG12-maleimide, Thermo Fisher part 22711) at room temperature for 3 h. Finally, the functionalized nanoparticles were washed three times in nanoparticle storage solution (sterile 1% bovine serum albumin in 10 mM MOPS buffer at pH 7.3) and stored at 3 nM in nanoparticle storage solution with 0.03% sodium azide. Herein, functionalized nanoparticles will be referred to by their flavor and target. For example, s440 nanoparticles targeted to CD47 are called s440-CD47.
Flow Cytometry of Functionalized Nanoparticles Bound to DLD-1 and HCT-116 Cells
Binding specificity of antibody-targeted gold-silica nanoparticles to their target cell surface protein was evaluated using flow cytometry as described previously.24 Samples were performed by preparing aliquots of 200,000 cells at 2 million cells/mL (100 μL volume) and adding 5 μL of 3 nM Oxonica-DL650-antibody-(PEG)12 to achieve a final nanoparticle concentration of 150 pM. Nanoparticle/cell mixtures were incubated for 15 min at 4 °C, washed once in 2 mL PBS + 1% BSA, and resuspended in 200 μL PBS + 1% BSA for analysis. For epitope blocking experiments, prior to adding nanoparticles, DLD-1 cells were suspended in anti-CD47 ranging from 1 ng/mL to 100 μg/mL anti-CD47 for 30 min on ice. For experiments testing binding to CD47, wild-type and CD47 knockout DLD1 cells were provided courtesy of D. Kurtz and I. Weissman. For experiments testing binding to CA9, CA9 expression was induced in HCT-116 cells with transient transfection as described below. CA9 epitope blocking was performed on HCT-116 cells transfected with the CA9 plasmid with 35 ug/mL anti-CA9 for 1 h on ice. Cell sorting/flow cytometry analysis for this project was done on instruments in the Stanford Shared FACS Facility.
Transfection of CA9 into HCT116 Cells
A plasmid encoding the eGFP gene fused to the intracellular C-terminus of hCA9 (NM_001216.2) was obtained from VectorBuilder (Cyagen Biosciences Inc., Santa Clara, CA). The human protein portion of the plasmid was sequenced by Sequetech (Mountain View, CA) and had a >99% match to hCA9 according to BLAST (blast.ncbi.nlm.nih.gov). Transient transfections of hCA9 into HCT116 cells were generated in 6 well plates using the standard Lipofectamine 3000 (Thermo Fisher) protocol. Transfected cells were allowed to grow for 4 days before they were used for FACS experiments.
Spectral Unmixing and Background Subtraction of Raw Raman Spectra
All signal processing codes can be found at https://github.com/ryanmdavis/MolecularEndoscopy and has been described previously.14 Spectral unmixing and tissue background signal removal of the three nanoparticle flavors (s440, s421, s481) were performed by representing the problem as a linear system of equations given by
| 1 |
where b is a N × 1 matrix that contains one raw Raman spectrum obtained by the endoscope at a single location on tissue, A is a N × M matrix composed of M different N × 1 reference spectra (described below) that specify the forward problem, and x is a M × 1 matrix that quantifies the contribution of each reference spectrum to the raw signal b. The M reference spectra were categorized as either SERS signal, tissue background signal, or a linear background signal. Reference SERS signals were the background-subtracted SERS signal of s440, s421, and s481. Tissue background signal spectra were the first six principal components of 100 spectra obtained from fresh mouse DLD1 xenograft tumors (no nanoparticles). For Raman microscopy, two additional background spectra were added into the model: the spectra of glass and optimal cutting temperature (OCT) compound (Fisher Scientific part 23-730-571).
Once A was constructed from the M reference spectra, the inverse problem was solved by taking the Moore–Penrose pseudoinverse of A (A+) in MATLAB (Mathworks, Natick, MA, USA). This allows one to obtain the least-squares fit of the M reference spectra to the raw spectrum b using a matrix multiplication:
| 2 |
where the M elements of x̂ are the contribution of each reference spectrum to b.14 For endoscope spectra, unmixing was performed using data in the range 1100–1700 cm–1. For microscope spectra, unmixing was performed using data in the range 900–1700 cm–1.
Each experiment also had a concentration reference that controlled deviations from a 1:1:1 concentration ratio in the nanoparticle solution used to stain the tissue. This concentration reference was achieved by applying 5 μL of the nanoparticle mixture used to stain tissue onto a 10 mm2 section of filter paper. The stained filter paper was then placed next to the stained tissue sample, and both were scanned in the same experiment. During image reconstruction, on a pixel-by-pixel basis, the Raman intensity in each channel was divided by the average intensity of the same channel in the concentration reference filter paper. This procedure controlled deviations from the nominal 1:1:1 concentration ratio in the nanoparticle mixture.
Raman Endoscope and Imaging
All imaging was performed with a Raman endoscope that was built in-house.14,15,40 The laser wavelength was 785 nm. Ex vivo tissue samples were stained with a 1:1:1 mixture of s420-CA9:s421-IgG4:s440-CD47 nanoparticles, with each nanoparticle flavor at 300 pm. Staining was performed at room temperature and placed on a quartz microscope slide which in turn was placed on a movable stage. A forward-facing endoscope probe was positioned perpendicular to the surface of tissue at a distance of <1 cm. The laser power was measured with a laser power meter and adjusted to 40 mW at the beginning of each experiment. The sample was raster scanned under the endoscope probe with a step size of 1 mm and a signal integration time of 1 s per image pixel. Raw spectra were unmixed and displayed as an image using Matlab as described in the Spectral Unmixing and Background Subtraction of Raw Raman Spectra section.
The ability of the nanoparticle-endoscope system to discriminate
normal and cancerous bladder was assessed using region of interest
(ROI) analysis of Raman images. Maps of nanoparticle levels (x̂ from eq 2) were manually registered to photographs of the tissue samples.
Regions of interest were manually drawn on the images. Care was taken
to exclude the margin surrounding the edges of the samples, as the
edges had exposed submucosa that exhibited strong passive binding
of the nanoparticles. The mean signal in each ROI was assigned to
the conditions “normal” or “cancer” based
on the surgeon’s diagnosis and pathology report. The standard
error of the mean signal ratio 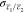 of two channels c1 and c2 of different
tissue class
(i.e., tumor or normal) was calculated
using the standard error propagation formula:
of two channels c1 and c2 of different
tissue class
(i.e., tumor or normal) was calculated
using the standard error propagation formula:
where 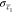 ,
, 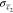 , c̅1,
and c̅2 are the standard error of
the means and the means of the signal in channel c1 and c2. When c1 and c2 came from the same
tissue class, the standard error of the mean was calculated directly
from the distribution of ratios across different tissue samples.
, c̅1,
and c̅2 are the standard error of
the means and the means of the signal in channel c1 and c2. When c1 and c2 came from the same
tissue class, the standard error of the mean was calculated directly
from the distribution of ratios across different tissue samples.
Tissue Procurement and Staining
Nineteen patients who were treated with TURBT or radical cystectomy at the Stanford University hospital or Palo Alto VA consented to their tissue being used in this study. All patient’s tissue samples were stained with the 3-nanoparticle mixture and imaged on the microscope (6 samples) or endoscope (15 samples). Tissue was harvested under the approval of the institutional review boards (IRBs). Cystectomy-derived samples were harvested after bladder removal and placed on ice within 15 min of bladder removal. TURBT-derived samples were placed on ice within 15 min of resection.
Samples were then stained with SERS nanoparticles at room temperature by dabbing the luminal surface of the tissue in 20 μL of nanoparticle mixture every 20 s for 5 min. Stained samples were then washed by submerging the sample for 15 s in 50 mL PBS. Excess PBS was removed from the tissue sample by placing the edge of the sample (not the luminal surface) in contact with a task wipe (Kimwipe, Kimtech Sciences). Stained and washed samples were then imaged with a Raman microscope or frozen in OCT compound for histological analysis. Table S1 contains the clinical pathology results and a summary of the allocation of tissue samples to different experiments.
Tissue Classification Approaches
Three signal processing approaches were evaluated for classification accuracy (Table 1). In Table 1, the values s420-CA9, s421-IgG4, and s440-CD47 refer to the signal intensity of each channel in a given pixel. For all approaches, Matlab’s LDA routine fitcdiscr was used to determine the posterior probability of a tissue sample being tumor. ROC curves were generated by feeding the posterior tumor probability of each sample into Matlab’s perfcurve routine. ROC AUC error bars were calculated with Matlab’s perfcurve using the percentile method. ROC curves were compared using Delong’s method for paired ROCs in the pROC package41 in R (R Core Team, Vienna, Austria).
Tissue Immunofluorescence and Raman Microscopy
All tissues were embedded in OCT compound, frozen while fresh, and sectioned into 10 μm sections. For CA9 staining, samples were fixed in PFA, stained with 0.2 μg/mL rabbit polyclonal anti-CA9 (Novus Biologicals NB100–147), and detected with anti-rabbit F(ab′)2-Alexa Fluor 647 conjugate (Cell signaling Technology, 4414S). As a negative control, these stained images were compared to images obtained by pre-incubating the CA9 antibody with the immunizing peptide (Novus Biologicals NB100-417PEP). For CD47 staining, samples were fixed in acetone, stained with 0.2 μg/mL mouse monoclonal B6H12.2 (Novus Biologicals NBP2-31106), and detected with anti-mouse F(ab′)2-Alexa Fluor 647 conjugate (Cell signaling Technology 4410S). As a negative control, tissue was treated with 0.2 μg/mL mouse IgG1 (Novus Biologicals, MAB002) instead of B6H12.2. All tissue immunofluorescence images were acquired on a Hamamatsu Nanozoomer (Hamamatsu Photonics, Hamamatsu City, Japan).
For Raman microscopy, 50 μm sections that were adjacent to immunofluorescence sections were adhered to a quartz slide. Then Permount Mounting Medium (Fischer Scientific, SP15–100) was placed on top of the sample, and a glass coverslip was quickly pressed on top of the sample and allowed to harden. Raman signal was measured on a InVia microscope (Renishaw, Wotton-under-Edge, England) with a 50× objective on standard mapping mode (5 μm step size in both directions). Images were reconstructed in Matlab using the same procedure described in the Spectral Unmixing and Background Subtraction of Raw Raman Spectra section.
Quantification of Penetration Depth and Surface Coverage
The position of the bladder tissue surface was estimated by manually drawing a line on the s421-IgG4 Raman image of the tissue (red lines in Supporting Figure 5b,c, bottom). The surface was then smoothed with a median filter with a window width of 10 pixels. Next, for each pixel in the surface line (red) image, profiles were generated normal to the estimated tissue surface (white lines in Supporting Figure 5b,c, bottom). For clarity, Supporting Figure 5b,c only shows every tenth image profile (white lines). The penetration depth was calculated separately for each profile normal to the tissue surface as the number of pixels with s421-IgG4 signal five standard deviations above the mean background signal. The surface coverage percent was calculated as the percent of profiles with at least one pixel five standard deviations above the mean background signal. Three normal and three tumor samples were subjected to this analysis, and for each sample, three separate 100 μm sections of surface were analyzed resulting in 9 total data points for each tissue type.
Acknowledgments
CD47 antibody and DLD1 cells obtained from I. Weissman lab. We acknowledge Z. Qiu for instructions on and assembly of Raman endoscope. We thank C. Zavaleta for advice on staining tissue with Raman nanoparticles. We thank J. Rosenberg for advice on statistical comparison of paired ROC curves.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b03217.
Physiochemical characterization of SERS nanoparticles, nanoparticle binding vs antigen availability, nonspecific binding of SERS nanoparticles to cells in suspension and tissue specimens, high-resolution image of decomposed Raman spectrum, details of passive penetration experiments, illustration of the enhanced surface permeability and retention effect, full Raman image data set (PDF)
Research reported in this publication was supported by (1) the Center for Cancer Nanotechnology Excellence for Translational Diagnostics (CCNE-TD) at Stanford University through award number U54 CA199075 from the National Cancer Institute (NCI) of the National Institutes of Health (NIH), (2) the NIH grant R01 CA160986, and (3) the Cancer-Translational Nanotechnology Training (Cancer-TNT) Program of the National Cancer Institute under award number: T32 CA196585.
The authors declare no competing financial interest.
Supplementary Material
References
- Antoni S.; Ferlay J.; Soerjomataram I.; Znaor A.; Jemal A.; Bray F.; Catto J.; Asia S.; Asia S. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 2017, 71, 96–108. 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Babjuk M.; Burger M.; Zigeuner R.; Shariat S. F.; Sylvester R. J.; Kaasinen E.; van Rhijn B. W. G.; Comperat E.; Palou J.; Roupre M.; Bohle A.; et al. EAU Guidelines on Non – Muscle-Invasive Urothelial Carcinoma of the Bladder: Update 2013. Eur. Urol. 2013, 64, 639–653. 10.1016/j.eururo.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Aldousari S.; Kassouf W. Update on the Management of Non-Muscle Invasive Bladder Cancer. Can. Urol. Assoc. J. 2010, 4, 56–64. 10.5489/cuaj.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaibold H. E.; Sivalingam S.; May F.; Hartung R. The Value of a Second Transurethral Resection for T1 Bladder Cancer. BJU Int. 2006, 97, 1199–1201. 10.1111/j.1464-410X.2006.06144.x. [DOI] [PubMed] [Google Scholar]
- Lerner S. P.; Liu H.; Wu M. F.; Thomas Y. K.; Witjes J. A. Fluorescence and White Light Cystoscopy for Detection of Carcinoma in Situ of the Urinary Bladder. Urol. Oncol. Semin. Orig. Investig 2012, 30, 285–289. 10.1016/j.urolonc.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Schmidbauer J.; Witjes F.; Schmeller N.; Donat R.; Susani M.; Marberger M. Improved Detection of Urothelial Carcinoma In Situ With Hexaminolevulinate Fluorescence Cystoscopy. J. Urol. 2004, 171, 135–138. 10.1097/01.ju.0000100480.70769.0e. [DOI] [PubMed] [Google Scholar]
- Grossman H. B.; Gomella L.; Fradet Y.; Morales A.; Presti J.; Ritenour C.; Nseyo U.; Droller M. J. A Phase III, Multicenter Comparison of Hexaminolevulinate Fluorescence Cystoscopy and White Light Cystoscopy for the Detection of Superficial Papillary Lesions in Patients With Bladder Cancer. J. Urol. 2007, 178, 62–67. 10.1016/j.juro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Jocham D.; Witjes F.; Wagner S.; Zeylemaker B.; van Moorselaar J.; Grimm M.; Muschter R.; Popken G.; Onig F. K.; Uchel R. K. N.; Kurth K.; et al. Improved Detection and Treatment of Bladder Cancer Using Hexaminolevulinate Imaging: A Prospective, Phase Iii Multicenter Study. J. Urol. 2005, 174, 862–866. 10.1097/01.ju.0000169257.19841.2a. [DOI] [PubMed] [Google Scholar]
- Fradet Y.; Grossman H. B.; Gomella L.; Lerner S.; Cookson M.; Albala D.; Droller M. J. A Comparison of Hexaminolevulinate Fluorescence Cystoscopy and White Light Cystoscopy for the Detection of Carcinoma In Situ in Patients With Bladder Cancer: A Phase III, Multicenter Study. J. Urol. 2007, 178, 68–73. 10.1016/j.juro.2007.03.028. [DOI] [PubMed] [Google Scholar]
- Chang T. C.; Marcq G.; Kiss B.; Trivedi D. R.; Mach K. E.; Liao J. C. Image-Guided Transurethral Resection of Bladder Tumors – Current Practice and Future Outlooks. Bladder Cancer 2017, 3, 149–159. 10.3233/BLC-170119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babjuk M.; Soukup V.; Petrík R.; Jirsa M.; Dvorácek J. 5-Aminolaevulinic Acid-Induced Fluorescence Cystoscopy during Transurethral Resection Reduces the Risk of Recurrence in Stage Ta/T1 Bladder Cancer. BJU Int. 2005, 96, 798–802. 10.1111/j.1464-410X.2004.05715.x. [DOI] [PubMed] [Google Scholar]
- Daniltchenko D. I.; Riedl C. R.; Sachs M. D.; Koenig F.; Daha K. L.; Pflueger H.; Loening S. A.; Schnorr D. Long-Term Benefit of 5-Aminolevulinic Acid Fluorescence Assisted Transurethral Resection of Superficial Bladder Cancer: 5-Year Results of a Prospective Randomized Study. J. Urol. 2005, 174, 2129–2133. 10.1097/01.ju.0000181814.73466.14. [DOI] [PubMed] [Google Scholar]
- Zavaleta C. L.; Smith B. R.; Walton I.; Doering W.; Davis G.; Shojaei B.; Natan M. J.; Gambhir S. S. Multiplexed Imaging of Surface Enhanced Raman Scattering Nanotags in Living Mice Using Noninvasive Raman Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 13511–13516. 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai E.; Sensarn S.; Zavaleta C. L.; Van de Sompel D.; Loewke N. O.; Mandella M. J.; Gambhir S. S.; Contag C. H. High-Sensitivity, Real-Time, Ratiometric Imaging of Surface-Enhanced Raman Scattering Nanoparticles with a Clinically Translatable Raman Endoscope Device. J. Biomed. Opt. 2013, 18, 096008. 10.1117/1.JBO.18.9.096008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta C. L.; Garai E.; Liu J. T. C.; Sensarn S.; Mandella M. J.; Van de Sompel D.; Friedland S.; Van Dam J.; Contag C. H.; Gambhir S. S. A Raman-Based Endoscopic Strategy for Multiplexed Molecular Imaging. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E2288–E2297. 10.1073/pnas.1211309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Chang T.; Marcq G.; Liu C.; Kiss B.; Rouse R.; Mach K. E.; Cheng Z.; Liao J. C. In Vivo Biodistribution and Toxicity of Intravesical Administration of Quantum Dots for Optical Molecular Imaging of Bladder Cancer. Sci. Rep. 2017, 7, 9309. 10.1038/s41598-017-08591-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakor A. S.; Luong R.; Paulmurugan R.; Lin F. I.; Kempen P.; Zavaleta C.; Chu P.; Massoud T. F.; Sinclair R.; Gambhir S. S. The Fate and Toxicity of Raman-Active Silica-Gold Nanoparticles in Mice. Sci. Transl. Med. 2011, 3, 79ra33. 10.1126/scitranslmed.3001963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-J.; Droller M. J.; Liao J. C. New Optical Imaging Technologies for Bladder Cancer: Considerations and Perspectives. J. Urol. 2012, 188, 361–368. 10.1016/j.juro.2012.03.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highley M. S.; van Oosterom A. T.; Maes R. A.; De Bruijn E. A. Intravesical Drug Delivery Pharmacokinetic and Clinical Considerations. Clin. Pharmacokinet. 1999, 37, 59–73. 10.2165/00003088-199937010-00004. [DOI] [PubMed] [Google Scholar]
- Pan Y.; Volkmer J.-P.; Mach K. E.; Rouse R. V.; Liu J.-J.; Sahoo D.; Chang T. C.; Metzner T. J.; Kang L.; van de Rijn M.; Skinner E. C.; Gambhir S. S.; Weissman I. L.; Liao J. C. Endoscopic Molecular Imaging of Human Bladder Cancer Using a CD47 Antibody. Sci. Transl. Med. 2014, 6, 260ra148. 10.1126/scitranslmed.3009457. [DOI] [PubMed] [Google Scholar]
- Willingham S. B.; Volkmer J.-P.; Gentles A. J.; Sahoo D.; Dalerba P.; Mitra S. S.; Wang J.; Contreras-Trujillo H.; Martin R.; Cohen J. D.; Lovelace P.; Scheeren F. A.; Chao M. P.; Weiskopf K.; Tang C.; Volkmer A. K.; Naik T. J.; Storm T. A.; Mosley A. R.; Edris B.; et al. The CD47-Signal Regulatory Protein Alpha (SIRPa) Interaction Is a Therapeutic Target for Human Solid Tumors. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 6662–6667. 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte T.; Seligson D. B.; Rao J. Y.; Yu H.; De Martino M.; Kawaoka K.; Wong S. G.; Belldegrun A. S.; Pantuck A. J. Carbonic Anhydrase IX in Bladder Cancer: A Diagnostic, Prognostic, and Therapeutic Molecular Marker. Cancer 2009, 115, 1448–1458. 10.1002/cncr.24163. [DOI] [PubMed] [Google Scholar]
- Klatte T.; Belldegrun A. S.; Pantuck A. J. The Role of Carbonic Anhydrase IX as a Molecular Marker for Transitional Cell Carcinoma of the Bladder. BJU Int. 2008, 101 (Suppl), 45–48. 10.1111/j.1464-410X.2008.07650.x. [DOI] [PubMed] [Google Scholar]
- Wang Y. W.; Khan A.; Som M.; Wang D.; Chen Y.; Leigh S. Y.; Meza D.; McVeigh P. Z.; Wilson B. C.; Liu J. T. C. Rapid Ratiometric Biomarker Detection with Topically Applied SERS Nanoparticles. Technology 2014, 2, 118–132. 10.1142/S2339547814500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foddai A.; Elliott C. T.; Grant I. R. Maximizing Capture Efficiency and Specificity of Magnetic Separation for Mycobacterium Avium Subsp. Paratuberculosis Cells. Appl. Environ. Microbiol. 2010, 76, 7550–7558. 10.1128/AEM.01432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers J. J.; Xiong Y.; Jin X.; Shao M.; Tong X.; Farag S.; Zborowski M. Quantification of Non-Specific Binding of Magnetic Micro- and Nanoparticles Using Cell Tracking Velocimetry: Implication for Magnetic Cell Separation and Detection. Biotechnol. Bioeng. 2010, 105, 1078–1093. 10.1002/bit.22635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J. H.; Tam K. A.; Park S.-j.; Cha A.; Hong S. Recent Advances in Nanotechnology-Based Detection and Separation of Circulating Tumor Cells. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2016, 8, 223–239. 10.1002/wnan.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. W.; Doerksen J. D.; Kang S.; Walsh D.; Yang Q.; Hong D.; Liu J. T. C. Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection-Enhanced Topical Staining with SERS-Coded Nanoparticles. Small 2016, 12, 5612–5621. 10.1002/smll.201601829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. W.; Kang S.; Khan A.; Bao P. Q.; Liu J. T. C. In Vivo Multiplexed Molecular Imaging of Esophageal Cancer via Spectral Endoscopy of Topically Applied SERS Nanoparticles. Biomed. Opt. Express 2015, 6, 3714–3723. 10.1364/BOE.6.003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Kang S.; Khan A.; Ruttner G.; Leigh S. Y.; Murray M.; Abeytunge S.; Peterson G.; Rajadhyaksha M.; Dintzis S.; Javid S.; Liu J. T. C. Quantitative Molecular Phenotyping with Topically Applied SERS Nanoparticles for Intraoperative Guidance of Breast Cancer Lumpectomy. Sci. Rep. 2016, 6, 21242. 10.1038/srep21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhasarkar S.; Banerjee R. Intravesical Drug Delivery: Challenges, Current Status, Opportunities and Novel Strategies. J. Controlled Release 2010, 148, 147–159. 10.1016/j.jconrel.2010.08.031. [DOI] [PubMed] [Google Scholar]
- Maeda H.; Wu J.; Sawa T.; Matsumura Y.; Hori K. Tumor Vascular Permeability and the EPR Effect in Macromolecular Therapeutics: A Review. J. Controlled Release 2000, 65, 271–284. 10.1016/S0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- Shore N. D.; Boorjian S. A.; Canter D. J.; Ogan K.; Karsh L. I.; Downs T. M.; Gomella L. G.; Kamat A. M.; Lotan Y.; Svatek R. S.; Bivalacqua T. J.; Grubb R. L.; Krupski T. L.; Lerner S. P.; Woods M. E.; Inman B. A.; Milowsky M. I.; Boyd A.; Treasure F. P.; Gregory G.; Sawutz D. G.; Yla-Herttuala S.; Parker N. R.; Dinney C. P. N. Intravesical RAd–IFNa/Syn3 for Patients with High-Grade, Bacillus Calmette-Guerin–refractory or Relapsed Non–muscle-Invasive Bladder Cancer: A Phase II Randomized Study. J. Clin. Oncol. 2017, 35, 3410–3416. 10.1200/JCO.2017.72.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.; Wang Y.; Ke H.; Wang Q.; Lv X.; Wu H.; Tang Y.; Yang X.; Chen C.; Zhao Y.; Chen H. Protein-Nanoreactor-Assisted Synthesis of Semiconductor Nanocrystals for Efficient Cancer Theranostics. Adv. Mater. 2016, 28, 5923–5930. 10.1002/adma.201506119. [DOI] [PubMed] [Google Scholar]
- Yang T.; Tang Y.; Liu L.; Lv X.; Wang Q.; Ke H.; Deng Y.; Yang H.; Yang X.; Liu G.; Zhao Y.; Chen H. Size-Dependent Ag2S Nanodots for Second Near-Infrared Fluorescence/Photoacoustics Imaging and Simultaneous Photothermal Therapy. ACS Nano 2017, 11, 1848–1857. 10.1021/acsnano.6b07866. [DOI] [PubMed] [Google Scholar]
- Huang J.; Guo M.; Ke H.; Zong C.; Ren B.; Liu G.; Shen H.; Ma Y.; Wang X.; Zhang H.; Deng Z.; Chen H.; Zhang Z. Rational Design and Synthesis of Γfe < inf > 2</Inf > O < inf > 3</Inf>@Au Magnetic Gold Nanoflowers for Efficient Cancer Theranostics. Adv. Mater. 2015, 27, 5049–5056. 10.1002/adma.201501942. [DOI] [PubMed] [Google Scholar]
- Harmsen S.; Bedics M. a.; Wall M. a.; Huang R.; Detty M. R.; Kircher M. F. Rational Design of a Chalcogenopyrylium-Based Surface-Enhanced Resonance Raman Scattering Nanoprobe with Attomolar Sensitivity. Nat. Commun. 2015, 6, 6570. 10.1038/ncomms7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen S.; Huang R.; Wall M. a; Karabeber H.; Samii J. M.; Spaliviero M.; White J. R.; Monette S.; O’Connor R.; Pitter K. L.; Sastra S. a; Saborowski M.; Holland E. C.; Singer S.; Olive K. P.; Lowe S. W.; Blasberg R. G.; Kircher M. F. Surface-Enhanced Resonance Raman Scattering Nanostars for High-Precision Cancer Imaging. Sci. Transl. Med. 2015, 7, 271ra7. 10.1126/scitranslmed.3010633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Wang L.; Zhao F.; Tseng S.; Narayanan C.; Shura L.; Willingham S.; Howard M.; Prohaska S.; Volkmer J.; Chao M.; Weissman I. L.; Majeti R. Pre-Clinical Development of a Humanized Anti-CD47 Antibody with Anti-Cancer Therapeutic Potential. PLoS One 2015, 10, e0137345. 10.1371/journal.pone.0137345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garai E.; Sensarn S.; Zavaleta C. L.; Loewke N. O.; Rogalla S.; Mandella M. J.; Felt S. A.; Friedland S.; Liu J. T. C.; Gambhir S. S.; Contag C. H. A Real-Time Clinical Endoscopic System for Intraluminal, Multiplexed Imaging of Surface-Enhanced Raman Scattering Nanoparticles. PLoS One 2015, 10, e0123185. 10.1371/journal.pone.0123185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X.; Turck N.; Hainard A.; Tiberti N.; Lisacek F.; Sanchez J. C.; Müller M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinf. 2011, 12, 77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


