Abstract
Cell entry of anionic nano-objects has been observed in various types of viruses and self-assembled DNA nanostructures. Nevertheless, the physical mechanism underlying the internalization of these anionic particles across the negatively charged cell membrane remains poorly understood. Here, we report the use of virus-mimicking designer DNA nanostructures with near-atomic resolution to program “like-charge attraction” at the interface of cytoplasmic membranes. Single-particle tracking shows that cellular internalization of tetrahedral DNA nanostructures (TDNs) depends primarily on the lipid-raft-mediated pathway, where caveolin plays a key role in providing the short-range attraction at the membrane interface. Both simulation and experimental data establish that TDNs approach the membrane primarily with their corners to minimize electrostatic repulsion, and that they induce uneven charge redistribution in the membrane under the short-distance confinement by caveolin. We expect that the nanoscale like-charge attraction mechanism provides new clues for viral entry and general rules for rational design of anionic carriers for therapeutics.
Short abstract
Combined experiments and molecular simulations reveal the underlying mechanism of like-charge attraction during the cell entry of framework nucleic acids.
Introduction
The cell entry of exogenous materials has important implications for biomedical applications including bioimaging, drug delivery, and pathogen defense.1−3 Given the anionic nature of the cytoplasmic membrane, artificial delivery agents are often positively charged by design to overcome the electrostatic barrier. Nevertheless, various types of negatively charged viruses and nanoparticles have proven to be efficiently internalized by cells.4−9 For example, self-assembled DNA nanostructures can enter a variety of mammalian cell lines via receptor-mediated endocytosis,10−12 which has resulted in versatile therapeutic applications.10,11,13−20 Although like-charge attraction has been often seen in simple colloidal systems, which has been explained by the many-body effects at the micro-/nanoscale,21,22 the underlying physical mechanism for a similar effect at the cell-membrane interface has not been elucidated.
Structural DNA nanotechnology has enabled tailored design of arbitrarily shaped nanostructures with near-atomic resolution.23,24 In particular, the site-addressability of self-assembled DNA nanostructures holds great potential for developing framework nucleic acids (FNAs) harboring versatile functional molecules and nanoparticles with spatial order.13,25−29 More recent findings on the efficient cellular uptake of FNAs have aroused great interest.7,11,18,28 DNA molecules are intrinsically negatively charged in pH-neutral water solution and were traditionally regarded to be transfection inert. Hence, the obviation of transfection agents in delivering FNAs triggers the exploration of new mechanisms of cell entry as well as developing novel tailor-made FNA-based delivery agents.18−20
Results and Discussion
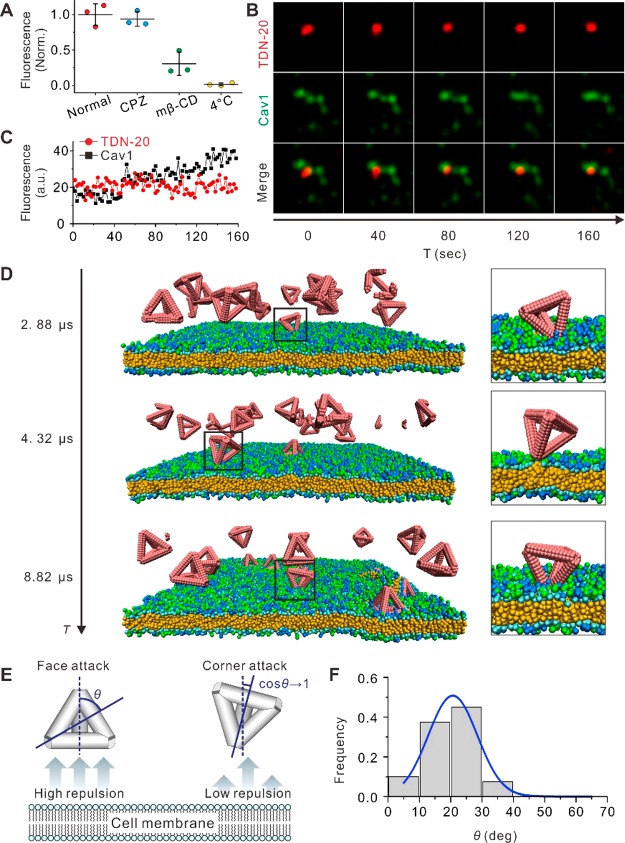
We first studied the cell entry pathway of a tetrahedral DNA nanostructure30 with edge length of 20 bp (TDN-20). To visualize the cellular uptake of TDN-20, we labeled it with a Cy3 fluorophore and incubated it with HeLa cells at 37 °C in the serum-free medium. Both confocal fluorescence microscopy and flow cytometry demonstrated that the fluorescence of Cy3 increased over time in the cytoplasm (Figure S1), suggesting efficient cell entry of TDN-20. The cellular uptake of TDN-20 relies on energy-dependent endocytosis since the incubation at 4 °C resulted in negligible fluorescence in cells. To examine the endocytic pathway of TDN-20, we employed pharmaceutical inhibitors to block the established pathways. The 3D confocal images showed that the presence of methyl-β-cyclodextrin (MβCD, an inhibitor of lipid-raft-/caveolin-mediated endocytosis) led to remarkably reduced fluorescence puncta in cells (Figure S1A). Quantitative analysis with flow cytometry revealed that the uptake of TDN-20 was suppressed by ∼54% by MβCD, while not by chlorpromazine (CPZ, an inhibitor of clathrin-mediated endocytosis) (Figure 1A), which was consistent with a previous observation that TDNs were internalized by cells via a lipid-raft-/caveolin-mediated endocytic pathway.11 To substantiate the role of caveolin in the cellular uptake of TDN-20, we employed time-resolved confocal imaging to real-time monitor the intracellular colocalization of TDN-20 with genetically encoded and endogenously expressed green-fluorescence-protein-fused caveolin-1 (GFP-Cav1) (Figure 1B). The observation of synchronized fluctuation of the two fluorophores confirmed that TDN-20 particles were internalized by the caveolar endocytic vesicles (Figure 1C and Movie S1).
Figure 1.
TDN internalization via corner attack. (A) Flow cytometric fluorescence analysis of cells fed with TDN-20 under different inhibitor treatments. (B) Time-resolved confocal images of a TDN-20 particle (red) colocalized with GFP-Cav1 (Cav1, green) in a living HeLa cell at different time points. (C) Time-resolved fluorescence intensity fluctuation of the particles from part B. (D) Time evolution of simulation of TDN-20 interacting with the cell membrane and representative snapshots of the TDNs attacking the membrane. (E) Schematic illustration of the attack modes of TDN-20. (F) Simulated attacking angle distribution of TDN-20. Blue curve, distribution fitting.
Having elucidated the biochemical mechanism of TDN uptake, we next explore the underlying physical mechanism for the like-charge attraction that breaks down the electrostatic barrier at the membrane interface. To mimic the lipid-raft-mediated endocytosis, we constructed a piece of cell-membrane model filled with negatively charged or neutral lipids, with an average surface charge density of −0.04 e/nm2 (within the reported physiological range from −0.02 to −0.2 e/nm2),31,32 which provides electrostatic repulsion to the anionic DNA nanostructures (Figure S2).33,34 This membrane was enriched with simplified models of lipid-raft proteins (i.e., caveolin-1, with a molar ratio to lipids of 0.5, detailed in the Supporting Information) to imitate short-range attraction to DNA nanostructures (with an attraction strength of 5 kBT referring to the reported physiological range35) in the lipid raft region. Then, we performed dissipative particle dynamics (DPD) simulations to investigate the interactions between TDN-20 and the modeled cell membrane by placing 16 TDN-20 model structures above the membrane (initial distance Z = 10.0 nm). From the calculated consequences of their interactions, we found that short-range attraction from the caveolin-1 was essential for the entry of TDN-20 (Figure S3). None of the anionic TDN-20 structures could reach the cell membrane without such an attractive force, which agrees well with the experimental observation under energy-deprived conditions (i.e., incubation at 4 °C, Figure 1A). In the presence of the short-range attraction mimicking lipid-raft-mediated endocytosis, 7 of 16 TDN-20 structures reached the membrane within 15 μs in a typical simulation, with time-resolved snapshots shown in Figure S4. Of note, the overall electric potential of the membrane was still below zero.
Further analysis of the simulation data revealed that TDN-20 structures predominantly attacked the membrane with their corners (Figure 1D and Movie S2). Although we initially placed TDN-20 structures such that one of their facets faced up with the membrane surface, most TDN-20 particles rotated during the approaching time of 15 μs to realize a “corner attack” (Figure S4), which might have minimized the repulsion between the negatively charged lipids and TDN-20 (Figure 1A). To verify this hypothesis, we conducted a control simulation by fixing the facet orientation of TDN-20 throughout the 15 μs approaching time. We found that only 2 of the 16 “frozen” TDN-20 structures (Figure S5) could reach the membrane, implying the significance of orientation adjustment of TDN-20 during their cell entry.
To quantify the frequency of the corner attack, we next investigated the distribution of attack angles from multiple simulated TDN-20 attacking events. The angle θ, between the line vertical to the cell membrane and the line ligating the TDN’s attacking corner, and its geometric center are illustrated in Figure 1E. From 40 independent simulations (Figure 1F), we found that the majority (∼92.5%) of the TDN-20 structures attacked the cell membrane with θ < 30°, indicating the corner attack mode.
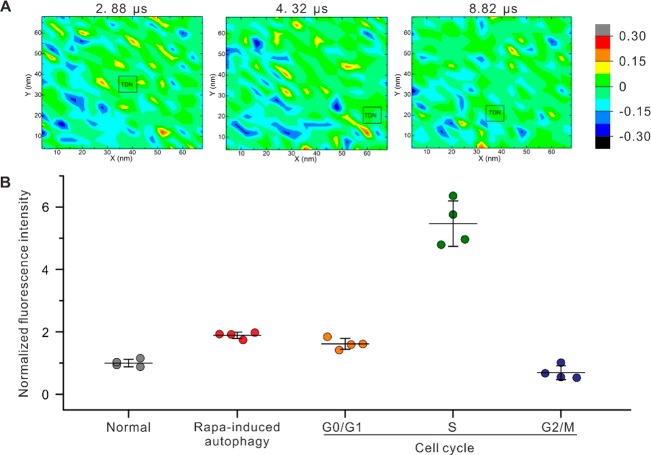
Next, we examined the effects of TDN-20 exerted on the cell membrane. Although a cell membrane is negatively charged in general, the semifluidic membrane is filled with unevenly distributed neutral, negatively, and positively charged lipids. Indeed, our simulation revealed that most TDN-20 particles attacked the membrane on hot spots with slight negative charge or even charge reversal (Figure 2A). Interestingly, confocal fluorescence imaging demonstrated that the fluorescence of TDN-20 was condensed into bright spots scattering on the membrane, suggesting the presence of hot spots on the membrane for efficient cell entry (Figure S1). When the negatively charged TDN-20 approached the membrane, the electrostatic repulsion also exerted the lipid spots, driving charge redistribution in the semifluidic membrane. Thus, negatively charged lipids were pushed away while the positively charged molecules were drawn close to the hot spots, resulting in “image charges” that are often observed in many-body physical systems with like-charge attraction.21 Control simulation studies showed that a frozen membrane with fixed lipids only allowed the entry of few TDN-20 structures (3 out of 16, Figure S6), suggesting that the fluidity of the membrane is necessary for the cell entry of TDN-20.
Figure 2.
Charge redistribution of the cell membrane. (A) Representative landing spots of TDN-20 on the membrane at different time points in a typical simulation. (B) Cytometric measurements of TDN-20 uptake by HeLa cells in rapamycin-induced autophagy or in different phases. Fluorescence intensities are normalized to that from normal cells.
The findings on the TDN-driven charge redistribution of the cell membrane motivated us to explore the cellular uptake of TDN-20 in different statuses of cells. Cells at different physiological status or with external stimuli may exhibit varied membrane potential, mechanical tension, and density of membrane proteins, etc.,36−38 which lead to varied membrane fluidity and charge distribution, and thus different uptake efficiency. Hence, we studied the cell entry of TDN-20 at different phases of a cell cycle and in autophagic cells triggered by rapamycin (methods of cell synchronization and rapamycin treatment are detailed in the SI). Flow cytometry revealed that the uptake efficiency varied greatly in these statuses (Figure 2B). In particular, the S-phase cell could uptake ∼5-fold more TDN-20 than the G0/G1-phase ones (Figure 2B). These results suggest that the cell status has prominent effects on the cell entry of DNA nanostructures, which might be attributed to the variation of membrane fluidity and charge distribution as observed in the simulation (Figures S7 and S8).
We further investigated whether the variation of the attraction parameters has an effect on the cell entry of TDN-20. We found that (Figures S9 and S10) the TDN-20 attacks the membrane with their corners and adsorbs on the hot spots of the membrane, regardless of how the attraction strength changes (increasing to 10.0 kBT or decreasing to 2.5 kBT, within the reported physiological range35). The engulfment after the adsorption showed difference. When the attraction strength increased (to 10.0 kBT), most of the adsorbed TDN-20 structures were engulfed by the cell membrane within the 15 μs simulation. On the contrary, when the attraction strength was weak (2.5 kBT), the TDN-20 structures remained attached on the cell membrane because the attraction force could not overcome the bending energy of the membrane deformation.39 A similar phenomenon was observed when the density of caveolin-1 in the membrane was reduced (i.e., the receptor–lipid ratio set to 0.25, Figure S11). In general, although these variations of attraction parameters affect the engulfment efficiency of TDN-20, the like-charge attractions (i.e., the corner attack mode and the hot spot adsorption) always occur, suggesting its robustness and universality.
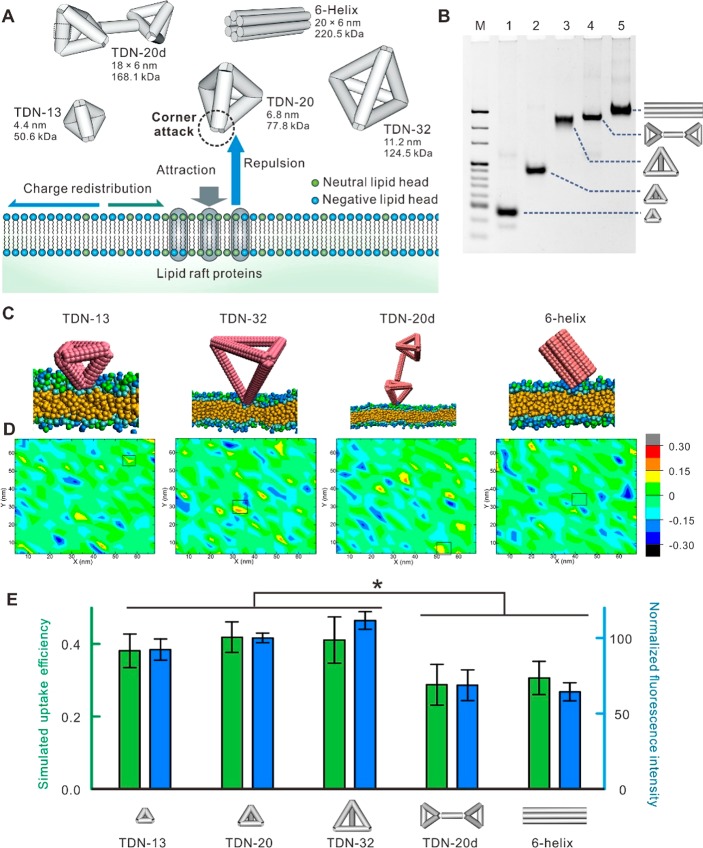
To examine the generality of like-charge attraction at the membrane interface, we synthesized four new DNA nanostructures with defined sizes and shapes (Figure 3A), including two differently sized TDNs (with edge length of 13 and 32 bp, referred to as TDN-13 and TDN-32, respectively), a TDN-20 dimer (TDN-20d), and a 6-helix DNA tubular nanostructure (6-helix). The polyacrylamide gel electrophoresis (PAGE) image (Figure 3B) presents clear bands with migration rates corresponding to the expected molecular weights, which verified the formation of these nanostructures. Next, we performed a simulation for the cell entry of these DNA nanostructures. We observed that all these nanostructures generally adopt the corner attack mode (Figure 3C and Figure S12) to approach the hot spots on the membrane (Figure 3D), consistent with our findings on TDN-20. Both experimental observation (confocal images shown in Figure S13 and cytometric analysis shown in Figure 3E) and simulation data (Figure 3E) revealed that all TDNs showed similar efficiency of cell entry, regardless of their sizes within the range ∼4–11 nm, suggesting the size-independence of lipid-raft-/caveolin-mediated endocytosis. However, TDN-20d and 6-helix showed ∼30% reduction in cell entry (Figure 3E). Hence, the cell entry is size-independent in the range of several tens of nanometers whereas dependent on the shape of DNA nanostructures.
Figure 3.
Generality of like-charge attraction at the membrane interface. (A) Schematic of the cell entry of structures including TDN-13, TDN-20, TDN-32, TDN-20d, and 6-helix. (B) PAGE electrophoresis characterization of the structures. (C) Representative simulation snapshots of DNA nanostructures attacking the cell membrane. (D) Representative landing spots of the structures on the cell membrane in typical simulations. (E) Cell entry efficiency of DNA nanostructures (green, simulation; blue, cytometric measurements. *, p < 0.05).
Given that the cellular uptake efficiency of TDN-20d was reduced by ∼30% as compared to that of TDN-20 (Figures 3E and 4A), possibly because of the presence of the rigid double-stranded DNA linker in TDN-20d that restricts its rotational freedom, we reason that studies on the difference in cellular uptake efficiency between TDN-20 and TDN-20d provide a possible means to experimentally verify the “corner attack” mechanism. Biochemical studies revealed that the cellular uptake of TDN-20d exploited the same lipid-raft-/caveolin-mediated endocytic pathway (Figure S14). However, confocal imaging showed reduced cellular uptake efficiency of TDN-20d as compared to that of TDN-20 (Figure 4A). Single-particle tracking with the total internal reflection fluorescence (TIRF) microscopy provides dynamic information on the cell entry (Figure 4B,C). Given that TIRF senses a few hundred nanometers above the glass slide, TDN-20/TDN-20d particles were visible only when they were within the illuminated layer at the cell bottom (Figure 4B). The observed fluorescence vanishing suggested that they left the illuminated layer by moving upward and entering the cell, providing single-particle tracking of TDN-20/TDN-20d in real time. We found that the fluorescence spots of TDN-20 disappeared more rapidly than those of TDN-20d (Figure 4C), consistent with the ensemble flow cytometric quantification and the simulation. The rate of cellular uptake for TDN-20 was ∼6% faster than that for TDN-20d, suggesting the difference in their rotation freedom that possibly accounts for the observed difference in cell entry.
Figure 4.
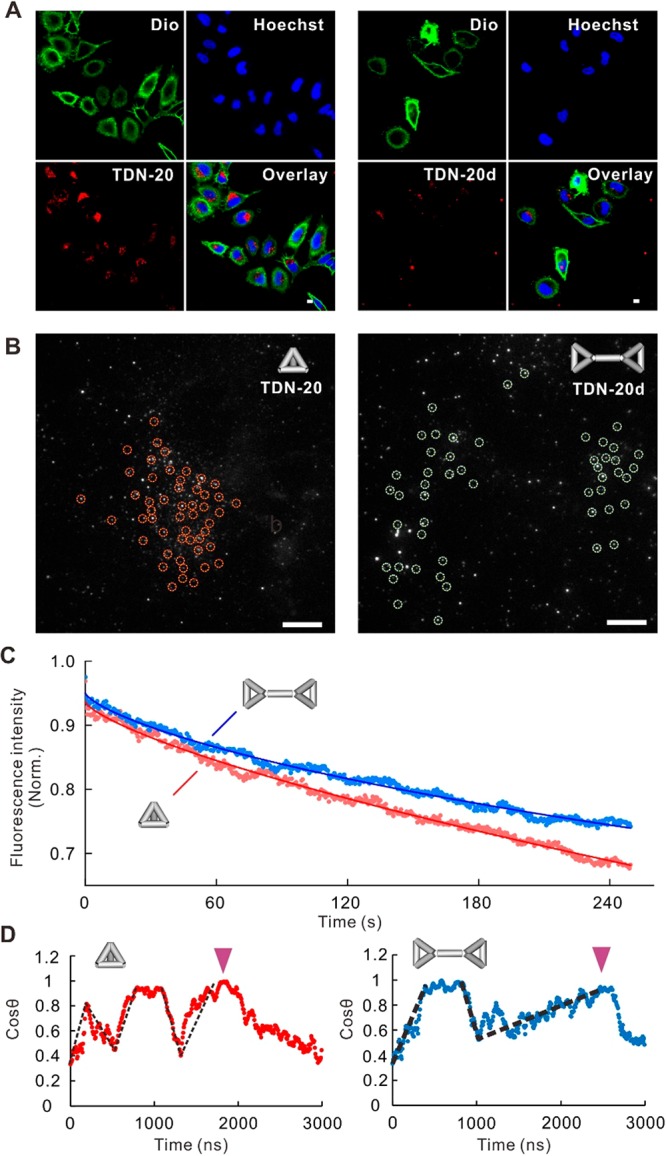
Comparison of the attack between TDN monomer and dimer. (A) Confocal fluorescence images of cells fed with TDN-20 (left) or TDN-20d (right). Green, cell membrane stained with Dio; blue, cell nucleus stained with Hoechst 33258; red, Cy3-labeled TDN-20 or TDN-20d. Scale bar, 1 μm. (B) TIRF imaging of DNA nanostructures (TDN20 and L-TDN20, marked with dashed circles, respectively) at the bottom of the cells. Scale bar, 1 μm. (C) Dynamics of fluorescence intensity decrease reflecting the cell entry kinetics of the two structures. (D) Representative dynamics of attacking angles of a TDN-20 (left) and a TDN20-d (right) during the approaching process. Black dashed lines indicate the rotation of the structures; purple arrows mark the time points when the structures touch the membrane.
To further verify this hypothesis, we investigated the rotational diffusion rates of TDN-20 and TDN-20d during their approaching processes via simulation. Representative results shown in Figure 4D revealed that the rotational frequency of a TDN-20 structure (5 turns before touching) was higher than that of a TDN-20d (3 turns before touching). Moreover, it took TDN-20 1.8 μs to reach the membrane in a corner attack manner, which was faster than TDN-20d (∼2.5 μs), indicating that the TDN monomer possesses higher freedom to adopt the corner attack mode than the dimer. Thus, the presence of the rigid dsDNA linker in the TDN-20d structure restricted the rotational diffusion rate, which renders TDN-20d biochemically identical to TDN-20 whereas kinetically different for cell entry.
In addition, we simulated the cellular uptake of another two types of anionic nanoparticles (i.e., spherical nanoparticle and triangular pyramid nanoparticle without holes on its faces) with size and charge similar to that of TDN-20. We found that the triangular pyramid nanoparticles also adopt the corner attack mode and land on the hot spot regions of the membrane (Figure S15). The spherical nanoparticles also tend to approach the hot spots on the membrane (Figure S16), but the corner attack mode cannot be observed because of the structural isotropy. Thus, fewer spherical particles entered the cell (4 out of 16, Figure S16) compared to TDN-20, suggesting the important role of corner attack in cell entry.
Having substantiated the mechanism for the cell entry of FNAs, we next studied the thermodynamics of TDN-20–membrane interactions. The endocytosis process of freely diffusive TDN-20 (as defined by Z > 10 nm) could be divided into three stages (Figure 5A): (i) approaching (Z > 8 nm); (ii) adsorption (4 < Z < 8 nm); and (iii) engulfment (Z < 4 nm). At the first stage, freely diffusive TDN-20 minimally interacted with the membrane lipids, as characterized by the little variation of the potential of mean force (PMF) during the Brownian diffusion of TDN-20. Here, TDN-20 was controlled by the long-range Coulomb repulsion from the charged lipids in the membrane (Figure 5B), whereas the TDN–receptor attraction stayed approximately 0 (Figure 5C), leading to a positively increasing PMF (Figure 5D) that formed an energy barrier for the approach of TDN-20. However, when TDN-20 rotated to the orientation for corner attack at the end of this stage, the reduced electrostatic repulsion allowed TDN-20 to overcome the energy barrier to approach the membrane. At the second stage, the short-range attraction from the lipid-raft proteins dominated the motions of the TDN-20; i.e., the decrease of TDN–receptor interaction energy (Figure 5C) overtook the increase of TDN–lipid interaction energy (Figure 5B), which pushed TDN-20 toward the membrane with rapidly decreased PMF. At the last stage, the TDN-20 particles were engulfed and entered the cell via caveolin-mediated vesicles. Hence, the confinement provided by caveolin-1 and the rotation freedom of TDN-20 lead to the apparent like-charge attraction for its effective cell entry.
Figure 5.
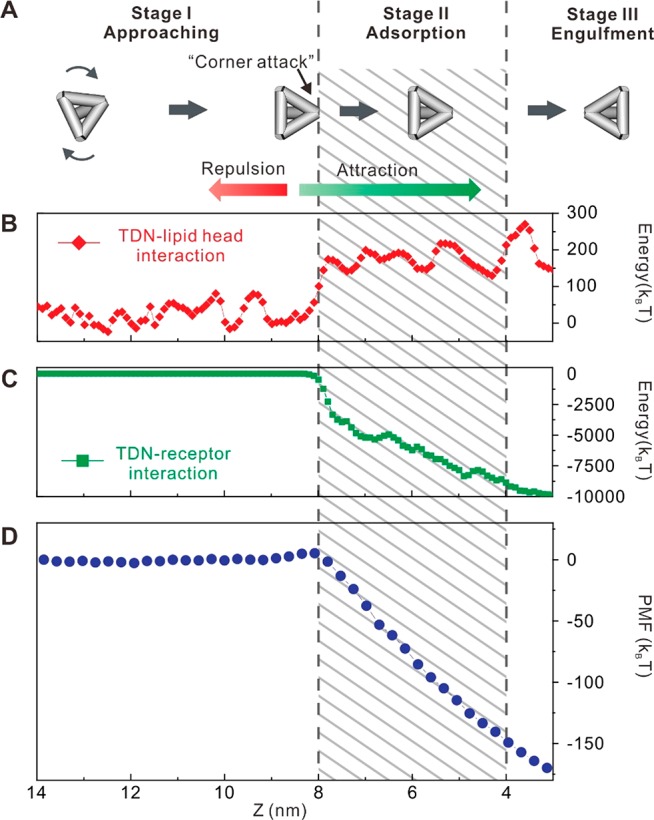
Cell entry stages of TDN-20. (A) Schematic illustration of the three-stage cell entry process of TDN-20. (B) Simulated interaction energy between a TDN and the lipid heads on the cell membrane as functions of distance Z between the center of mass (COM) of the TDN and the central plane of the cell membrane. (C) Simulated interaction energy between the TDN and lipid-raft proteins (receptors) on the cell membrane as functions of Z. (D) Potential of mean force (PMF) between the TDN and the membrane as a function of Z.
Conclusions
Recent studies have experimentally investigated the correlation between the geometric parameters of DNA nanostructures and their cellular uptake efficiency. For example, Wang et al. and Bastings et al. studied the cellular uptake of DNA origami structures and found that size, shape, and compactness play crucial roles in determining their uptake efficiency.40,41 Meanwhile, Rahman et al.42 studied DNA brick nanostructures with varied aspect ratio and observed no significant difference among them in cellular uptake. However, experimental studies in the complex biological environments tend to be interfered by a lot of uncontrolled variables, leading to contradictory conclusions. Here, this study offers a physics perspective with well-controlled parameters, and reveals the “corner attack” mode that can hardly be observed directly but whose effects can be verified by experiments.
Because of the limitation of present computing abilities, this work employed the coarse-grained (CG) modeling strategy, which omitted the atomic detail of the DNA structures and the receptor proteins, and may cause some distortions between the simulation and the reality. For the same reason, the size of the modeled FNA structures in this work is limited within ∼10 nm. In future work, with the improvement of computing power, we aspire to develop simulation methods with atomic/molecular accuracy, and investigate a wider spectrum of FNA structures (e.g., DNA origami with larger sizes and more shapes).
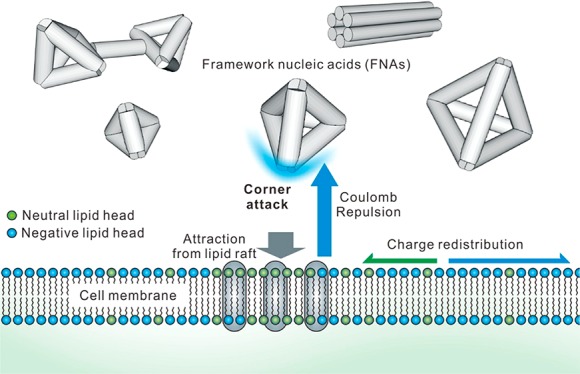
In summary, we have established the like-charge attraction mechanism in FNA-programmed cell entry. We find that the confinement of the lipid-raft proteins leads to the like-charge attraction between FNAs and the cytoplasmic membrane. The corner attack of FNAs and the charge redistribution of the membrane play important roles in this process. By designing FNAs with varied size and distinct shape, we observe that the like-charge attraction is shape-dependent whereas minimally size-dependent. Hence, FNAs provide a designer platform for studying structural parameters governing cell entry.40,43 In particular, the resemblance of TDNs to polyhedral viruses may have implications in pathogen infection.2,44 We also envision that the like-charge attraction mechanism sheds light on designing novel cellular delivery strategies.
Materials and Methods
All oligonucleotides purified with high-performance liquid chromatography (HPLC) were obtained from Invitrogen (Shanghai, China), and the sequences are shown in Table S1. TDN-13, TDN-20, and TDN-32 were synthesized using the previously reported method. HeLa cells were obtained from Shanghai Institute of Biological Sciences, and were cultured in a 5% CO2 environment at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) and supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibico), 100 international units/mL penicillin, 100 mg/mL streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen). No unexpected or unusually high safety hazards were encountered in this work.
The detailed procedures for cell synchronization and cellular uptake studies are provided in the Supporting Information. Confocal images were obtained using a Leica TCS SP8 confocal microscope. Total internal reflection fluorescence (TIRF) microscopic imaging was conducted with a Leica AM TIRF MC total internal reflection fluorescence microscope. Flow cytometric analysis was performed with a FACS Calibur flow cytometer (BD Biosciences).
Acknowledgments
This work was financially supported by the National Key R&D Program of China (2016YFA0400900), the National Natural Science Foundation of China (NSFC 21604060, 11474155, 11774147, 21675167, 31571014, U1532119, 21775157, 11575278, 21505148, 21390414), and the Chinese Academy of Sciences (Instrument Developing Project, the Open Large Infrastructure Research and the Key Research Program of Frontier Sciences QYZDJ-SSW-SLH031).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00383.
DNA sequences, synthesis of FNAs, cell experimental details, simulation methods, supplementary cell imaging and cytometric results, and supplementary simulation results (PDF)
Movie S1: time-resolved confocal animation of a TDN-20 particle colocalized with GFP-Cav1 in a living HeLa cell (red, TDN-20 particle; green, Cav1) (AVI)
Movie S2: simulated animation of a TDN-20 particle attacking the cell membrane (AVI)
Author Contributions
⊥ H.D. and J.L. contributed equally to this study.
The authors declare no competing financial interest.
Supplementary Material
References
- Peer D.; Karp J. M.; Hong S.; Farokhzad O. C.; Margalit R.; Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- Marsh M.; Helenius A. Virus entry: Open sesame. Cell 2006, 124, 729–740. 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfbeis O. S. An overview of nanoparticles commonly used in fluorescent bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. 10.1039/C4CS00392F. [DOI] [PubMed] [Google Scholar]
- Yoo P. J.; Nam K. T.; Qi J.; Lee S. K.; Park J.; Belcher A. M.; Hammond P. T. Spontaneous assembly of viruses on multilayered polymer surfaces. Nat. Mater. 2006, 5, 234–240. 10.1038/nmat1596. [DOI] [PubMed] [Google Scholar]
- Sun X.; Liu Z.; Welsher K.; Robinson J. T.; Goodwin A.; Zaric S.; Dai H. Nano-graphene oxide for cellular imaging and drug delivery. Nano Res. 2008, 1, 203–212. 10.1007/s12274-008-8021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostiainen M. A.; Kasyutich O.; Cornelissen J. J.; Nolte R. J. Self-assembly and optically triggered disassembly of hierarchical dendron-virus complexes. Nat. Chem. 2010, 2, 394–399. 10.1038/nchem.592. [DOI] [PubMed] [Google Scholar]
- Walsh A. S.; Yin H.; Erben C. M.; Wood M. J.; Turberfield A. J. DNA cage delivery to mammalian cells. ACS Nano 2011, 5, 5427–5432. 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- Mout R.; Moyano D. F.; Rana S.; Rotello V. M. Surface functionalization of nanoparticles for nanomedicine. Chem. Soc. Rev. 2012, 41, 2539–2544. 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chithrani B. D.; Ghazani A. A.; Chan W. C. W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- Bhatia D.; Surana S.; Chakraborty S.; Koushika S. P.; Krishnan Y. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat. Commun. 2011, 2, 339. 10.1038/ncomms1337. [DOI] [PubMed] [Google Scholar]
- Liang L.; Li J.; Li Q.; Huang Q.; Shi J.; Yan H.; Fan C. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem., Int. Ed. 2014, 53, 7745–7750. 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- Surana S.; Bhat J. M.; Koushika S. P.; Krishnan Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat. Commun. 2011, 2, 340. 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- Veetil A. T.; Chakraborty K.; Xiao K.; Minter M. R.; Sisodia S. S.; Krishnan Y. Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol. 2017, 12, 1183–1189. 10.1038/nnano.2017.159. [DOI] [PubMed] [Google Scholar]
- Modi S.; Nizak C.; Surana S.; Halder S.; Krishnan Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 2013, 8, 459–467. 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- Modi S.; Swetha M. G.; Goswami D.; Gupta G. D.; Mayor S.; Krishnan Y. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009, 4, 325–330. 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- Halo T. L.; McMahon K. M.; Angeloni N. L.; Xu Y.; Wang W.; Chinen A. B.; Malin D.; Strekalova E.; Cryns V. L.; Cheng C.; Mirkin C. A.; Thaxton C. S. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 17104–17109. 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H.; Hao L.; Narayan S. P.; Auyeung E.; Mirkin C. A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 7625–7630. 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Fan C.; Pei H.; Shi J.; Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 2013, 25, 4386–4396. 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- Chen Y. J.; Groves B.; Muscat R. A.; Seelig G. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 2015, 10, 748–760. 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- Li J.; Green A. A.; Yan H.; Fan C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 2017, 9, 1056–1067. 10.1038/nchem.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A. E.; Grier D. G. Like-charge attractions in metastable colloidal crystallites. Nature 1997, 385, 230–233. 10.1038/385230a0. [DOI] [Google Scholar]
- Squires T. M.; Brenner M. P. Like-charge attraction and hydrodynamic interaction. Phys. Rev. Lett. 2000, 85, 4976–4979. 10.1103/PhysRevLett.85.4976. [DOI] [PubMed] [Google Scholar]
- Funke J. J.; Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 2016, 11, 47–52. 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- Seeman N. C.; Sleiman H. F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Yang Y.; Wang J.; Shigematsu H.; Xu W.; Shih W. M.; Rothman J. E.; Lin C. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 2016, 8, 476–483. 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel A.; Lill P.; Stegemann P.; Bravo-Rodriguez K.; Schöneweiß E.-C.; Merdanovic M.; Gudnason D.; Aznauryan M.; Gamrad L.; Barcikowski S.; Sanchez-Garcia E.; Birkedal V.; Gatsogiannis C.; Ehrmann M.; Saccà B. Tailored protein encapsulation into a DNA host using geometrically organized supramolecular interactions. Nat. Commun. 2017, 8, 14472. 10.1038/ncomms14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N.; Pei H.; Ge Z. L.; Simmons C. R.; Yan H.; Fan C. H. Charge transport within a three-dimensional DNA nanostructure framework. J. Am. Chem. Soc. 2012, 134, 13148–13151. 10.1021/ja302447r. [DOI] [PubMed] [Google Scholar]
- Li J.; Pei H.; Zhu B.; Liang L.; Wei M.; He Y.; Chen N.; Li D.; Huang Q.; Fan C. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano 2011, 5, 8783–8789. 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Wang T.; Liu W.; Xin H. L.; Li H.; Ke Y.; Shih W. M.; Gang O. Prescribed nanoparticle cluster architectures and low-dimensional arrays built using octahedral DNA origami frames. Nat. Nanotechnol. 2015, 10, 637–644. 10.1038/nnano.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R. P.; Schaap I. A.; Tardin C. F.; Erben C. M.; Berry R. M.; Schmidt C. F.; Turberfield A. J. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 2005, 310, 1661–1665. 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- Dobrzynska I.; Szachowicz-Petelska B.; Sulkowski S.; Figaszewski Z. Changes in electric charge and phospholipids composition in human colorectal cancer cells. Mol. Cell. Biochem. 2005, 276, 113–119. 10.1007/s11010-005-3557-3. [DOI] [PubMed] [Google Scholar]
- Szachowicz-Petelska B.; Dobrzynska I.; Skrodzka M.; Darewicz B.; Figaszewski Z. A.; Kudelski J. Phospholipid composition and electric charge in healthy and cancerous parts of human kidneys. J. Membr. Biol. 2013, 246, 421–425. 10.1007/s00232-013-9554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. M.; Tian W. D.; Ma Y. Q. Designing nanoparticle translocation through membranes by computer simulations. ACS Nano 2012, 6, 1230–1238. 10.1021/nn2038862. [DOI] [PubMed] [Google Scholar]
- Ding H. M.; Ma Y. Q. Theoretical and computational investigations of nanoparticle-biomembrane interactions in cellular delivery. Small 2015, 11, 1055–1071. 10.1002/smll.201401943. [DOI] [PubMed] [Google Scholar]
- Bongrand P. Ligand-receptor interactions. Rep. Prog. Phys. 1999, 62, 921. 10.1088/0034-4885/62/6/202. [DOI] [Google Scholar]
- Kim J. A.; Aberg C.; Salvati A.; Dawson K. A. Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population. Nat. Nanotechnol. 2012, 7, 62–68. 10.1038/nnano.2011.191. [DOI] [PubMed] [Google Scholar]
- Boucrot E.; Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 7939–7944. 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonderlin W. F.; Woodfork K. A.; Strobl J. S. Changes in membrane potential during the progression of MCF-7 human mammary tumor cells through the cell cycle. J. Cell. Physiol. 1995, 165, 177–185. 10.1002/jcp.1041650121. [DOI] [PubMed] [Google Scholar]
- Deserno M. Elastic deformation of a fluid membrane upon colloid binding. Phys. Rev. E 2004, 69, 031903. 10.1103/PhysRevE.69.031903. [DOI] [PubMed] [Google Scholar]
- Wang P. F.; Rahman M. A.; Zhao Z. X.; Weiss K.; Zhang C.; Chen Z. J.; Hurwitz S. J.; Chen Z. G.; Shin D. M.; Ke Y. G. Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J. Am. Chem. Soc. 2018, 140, 2478–2484. 10.1021/jacs.7b09024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastings M. M. C.; Anastassacos F. M.; Ponnuswamy N.; Leifer F. G.; Cuneo G.; Lin C.; Ingber D. E.; Ryu J. H.; Shih W. M. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett. 2018, 18, 3557–3564. 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- Rahman M. A.; Wang P. F.; Zhao Z. X.; Wang D. S.; Nannapaneni S.; Zhang C.; Chen Z. J.; Griffith C. C.; Hurwitz S. J.; Chen Z. G.; Ke Y. G.; Shin D. M. Systemic delivery of Bc12-targeting siRNA by DNA nanoparticles suppresses cancer cell growth. Angew. Chem., Int. Ed. 2017, 56, 16023–16027. 10.1002/anie.201709485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langecker M.; Arnaut V.; List J.; Simmel F. C. DNA nanostructures interacting with lipid bilayer membranes. Acc. Chem. Res. 2014, 47, 1807–1815. 10.1021/ar500051r. [DOI] [PubMed] [Google Scholar]
- Lakadamyali M.; Rust M. J.; Babcock H. P.; Zhuang X. W. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 9280–9285. 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.