Abstract
Transposable elements not only can change genomic positions and disperse across the gene pool, but also can jump to another species through horizontal transmission. Of late, the P element, a DNA transposon in insects, was shown to cross the genetic boundary from Drosophila melanogaster to D. simulans in Europe around 2006. To understand the dynamics of transposable elements, especially in the early stages of invasion, we examined 63 lines of D. simulans from 11 natural populations in Japan established in 1976–2015. Based on PCR analyses, P elements were demonstrated to exist in Japan in 2008 and later. One copy of the full‐length P element was identified and mapped to a site on chromosome 3 L in a genome. All of 18 copies of P elements examined shared “A” at the nucleotide position 2040, which is representative of the direct descendants of the original P element that invaded in D. simulans. We also found that some lines having P elements can induce intensive gonadal dysgenesis in D. simulans at 29°C. Our present results imply that P elements in D. simulans arrived at the east end of Asia just a few years later than or almost simultaneously to the initial invasion in Europe, Africa, and North America, suggesting a more astonishingly rapid spread than previously assumed.
Keywords: Drosophila, horizontal transmission, hybrid dysgenesis, P element, transposon
1. INTRODUCTION
Transposable elements (TEs) are one of the general components of genomes, especially in eukaryotes (Bergman, Quesneville, Anxolabéhère, & Ashburner, 2006; Craig, Craigie, Gellert, & Lambowitz, 2002; Kidwell & Lisch, 1997). TEs change genomic position, replicate their own copies in a genome, and, thus, can spread all across a gene pool via the usual inheritance. TEs also leap occasionally from one species to another through horizontal transmission (HT), by which TEs can move between organisms by means other than ordinary vertical gene transmission from parents to offspring (Kidwell, 1993; Le Rouzic & Capy, 2005). Molecular mechanisms of HT are almost unknown, but most probable molecular vehicles are bacteria and viruses that can shuttle TEs between organisms with ecological connections, for instance, prey–predator and host–parasite interactions (Drezen et al., 2017; Schaack, Gilbert, & Feschotte, 2010; Venner et al., 2017), although a mite, Proctolaelaps regalis, was proposed as a vector of the HT in Drosophila (Houck, Clark, Peterson, & Kidwell, 1991). The evolutionary relationship between TEs and the host genome remains controversial. TEs play important roles in genetic diversity of the hosts by insertion/deletion, illegitimate recombination, and domestication. However, their propagating behavior appear to have a selfish unconcern about the fitness of the hosts (Biemont & Vieira, 2006; Feschotte & Pritham, 2007; Hua‐Van, Le Rouzic, Boutin, Filee, & Capy, 2011; Orgel & Crick, 1980). For a successful invasion, TE needs a suitable cellular environment. Colonized once in a new gene pool, TEs can increase in copy number despite deleterious consequences for the host. The host, on the other hand, quickly develops regulatory systems for suppressing TEs’ transposition under a permissible level (Lee & Langley, 2012; Lewis et al., 2018; Senti, Jurczak, Sachidanandam, & Brennecke, 2015). As a consequence of such arm races, invaded TEs would spread first, but be totally inactive for a long time, and only their remnant sequences would remain. For bypassing this extinction scenario, another HT is the only escape route, except for domestication (Le Rouzic & Capy, 2009; Pinsker, Haring, Hagemann, & Miller, 2001; Schaack et al., 2010).
The P element, a DNA transposon in insects, is one of the most popular model systems of TEs (O'Hare & Rubin, 1983) and the causative factor of P‐M hybrid dysgenesis in Drosophila melanogaster (Bingham, Kidwell, & Rubin, 1982). A P strain carries many copies of P elements in genome, and an M strain has no copy. In the germline of F1 progeny of a cross between M females and P males, P elements transpose and lead to hybrid dysgenesis, including symptoms such as gonadal dysgenesis (GD) sterility, elevated mutations, segregation distortion, and male recombination (Ashburner, Golic, & Hawley, 2005; Kidwell, 1994). P elements recently invaded D. melanogaster by HT from another Drosophila species, probably D. willistoni, in the middle of the 20th century (Daniels, Peterson, Strausbaugh, Kidwell, & Chovnick, 1990; Kidwell, 1979). P elements dispersed worldwide and are currently virtually ubiquitous in wild populations of D. melanogaster (Anxolabéhère, Kidwell, & Periquet, 1988; Anxolabéhère, Nouaud, Periquet, & Tchen, 1985; Bonnivard & Higuet, 1999), Therefore, true M strains are limited to old laboratory lines that were established before the invasion of the P element (Kidwell, Kidwell, & Sved, 1977).
The fruit flies D. simulans and D. melanogaster (Figure 1) are closely related in the Drosophila melanogaster species subgroup of the genus Drosophila. They are highly similar in morphology, ecologically, and genetically (reviewed by Capy, Gibert, & Boussy, (2004)). P elements were known to invade only D. melanogaster, but not D. simulans (Brookfield, 1991). This was rather enigmatic, because the P element was repeatedly shown to be active in D. simulans if artificially introduced (Daniels, Strausbaugh, & Armstrong, 1985; Montchamp‐Moreau, 1990; Scavarda & Hartl, 1984) and able to increase in number with structural decay with time as in D. melanogaster (Daniels, Chovnick, & Kidwell, 1989; Higuet, Merçot, Allouis, & Montchamp‐Moreau, 1996; Kimura & Kidwell, 1994; Scavarda & Hartl, 1987). However, an occurrence of HT, of P elements transferring from D. melanogaster to D. simulans, was recently reported by Kofler, Hill, Nolte, Betancourt, & Schlötterer (2015), who demonstrated that isofemale lines of D. simulans established in North America in 2010 and South Africa in 2012 carried many P elements in their genomes. They indicated that some of P elements of D. simulans were 2,907 bp in length and thus autonomous. All P elements shared nucleotide “A” at the position 2,040. Considering that, in the single nucleotide polymorphism (SNP) at 2,040 of P elements in D. melanogaster, “G” is common and “A” is rare; they concluded that the P element of D. simulans was horizontally transmitted from D. melanogaster and the HT is likely to be a single event.
Figure 1.

A picture of Drosophila melanogaster (left) and D. simulans (right). Males (upper) and females (lower). The photograph was taken with a digital microscope VHX‐6000 (KEYENCE, Japan). Bar: 1 mm
Hill, Schlötterer, & Betancourt (2016) demonstrated that crosses of D. simulans between the dysgenesis‐susceptible (DS) females and dysgenesis‐inducing (DI) males showed gonadal dysgenesis in the F1 progeny, but not in the reciprocal cross and that F1 females of dysgenesis‐resistant (DR) lines have normal ovaries from the crosses with DS females or DI males. Therefore, DS, DI, and DR strains of D. simulans are analogous to M, P, and Q strains of D. melanogaster, respectively. They implied that the P element causes hybrid dysgenesis in D. simulans and that its transposition is regulated by PIWI‐interacting RNAs (piRNA), which is the molecular basis of the cytotype for repressing P transposition in D. melanogaster (Brennecke et al., 2008; Kelleher, 2016; Khurana et al., 2011). More important, Hill et al. (2016) found that the oldest line of D. simulans having P elements was established in Portugal in 2006 and such lines emerged in Northeast America in 2008 and that P elements spread other regions in Africa and Northwest America from 2009 to 2014. Their results suggested that P elements invaded D. simulans earlier than 2006. This ongoing spreading of P elements serves an irreplaceable model for studying the early stage of TE's life cycle.
The aims of this research are to clarify the invasion of P elements in D. simulans in Asia. A survey of local populations showed that the D. simulans P element currently existed in Japanese populations. We also explored the abilities of P elements to cause hybrid dysgenesis and determined the full‐length sequence to identify the inserted chromosomal position as a first step to investigate the molecular nature of TEs in the future. Our results support the single event of HT from D. melanogaster to D. simulans and suggest a nearly simultaneous invasion in Japan, Africa, Europe, and North America.
2. MATERIALS AND METHODS
2.1. Fly lines
We used 63 isofemale lines of D. simulans from 11 localities in Japan collected from 1976 to 2015 (Supporting information Table S1). Three sets of isofemale lines, MSO12, FWU12, and Hikone15, were established with inseminated females collected by banana bait traps. The SPP, OGS, KMM, OEB, AM06, and TSM lines were provided by the Ehime Drosophila Stock Center. TN samples were caught in 2010 and ethanol‐preserved. Rakujuen (RM1) is one of the oldest laboratory stocks of D. simulans in Japan, established in 1976 (Kawanishi & Watanabe, 1977). Moreover, two lines of D. melanogaster, Harwich and Canton S, were used as P and M strains of the P‐M system of D. melanogaster, respectively. Harwich was provided Kyoto Drosophila Stock Center. Flies were maintained on standard food medium at 25°C except for the GD test (see below).
2.2. Gonadal dysgenesis tests
According to the standard method for the GD test in the P‐M system in D. melanogaster (Engels & Preston, 1980; Kidwell et al., 1977), a set of diagnostic crosses, cross A (DS females × tested males) and cross A* (tested females × DI males), were performed for the Hikone15 lines at 29°C (Hill et al., 2016). All F1 females were individually dissected in 5–7 days after eclosion and the GD score was calculated for each line as the percentage of undeveloped ovaries. We used RM1 and TSM31 as the control of DS and DI strains, respectively. The strain type was defined according to the previous studies; DI strains (more than 10% GD in cross A and less than 10% GD in cross A*), DS strains (less than 10% GD in cross A and more than 10% GD in cross A*), and DR strains (less than 10% GD in both crosses) (Hill et al., 2016; Kidwell, 1993). Similar set of diagnostic crosses were performed for the four Hikone15 lines (15‐5, 15‐10, 5‐14, and 15‐26) at 25°C.
Differences in ratio of dysgenic females between reciprocal cross were tested by Fisher's exact test (FET). Differences in GD% between two temperatures were also tested by FET. The statistical significant levels for multiple comparisons were corrected by the Bonferroni method.
2.3. PCR
Genomic DNA was extracted from four adult flies for each isofemale lines, or from one ethanol‐preserved fly, using the GenElute Mammalian Genomic DNA miniprep kit (Sigma‐Aldrich). After ethanol precipitation DNA was dissolved in 30 μl of sterile distilled water. Using the genomic DNA as templates, P elements of D. simulans were amplified with Ex Taq (Takara) by primer sets as follows: P176F (5′‐CAAAGCTGTGACTGGAGTAA‐3′) and P2812R (5′‐GTCGTATTGAGTCTGAGTGA‐3′), or P357F (5′‐AACGCAGATGCCGTACCTAG‐3′) and P2770R (5′‐AACCCTTAGCATGTCCGTGG‐3′). The PCR reaction conditions for 30 cycles were denaturing at 98°C for 10 s, annealing at 50°C for 30 s and polymerizing at 72°C for 2.5 min for the former set, and denaturing at 98°C for 10 s and annealing at 56°C for 30 s and polymerizing at 72°C for 2.5 min for the latter primer set.
2.4. Inverse PCR
Genomic DNA was extracted from ten flies of FWU12‐07 as above and was dissolved in 150 μl of sterile distilled water after ethanol precipitation. We digested the genomic DNA in 20 μl of solution with 15 units of EcoRI (Takara) at 37°C for 1 hr, because the canonical P elements should have only one EcoRI site at nucleotide 1711 (Kofler et al., 2015; O'Hare & Rubin, 1983). After heat denaturing EcoRI (65°C, 20 min), digested DNA was self‐ligated with 3 Weiss units of T4 DNA ligase (Promega) at 4°C for overnight. For convenience, hereafter, we call the part of P element 5′‐side of the EcoRI site “Left side” (~1.7 kb), and that the 3′‐side of the EcoRI site “Right side” (~1.2 kb). Inverse PCR was performed with the primer sets of Pinv HaeIII‐up (5′‐AAATTCGTCCGCACACAACC‐3′) and Pinv HaeIII‐down (5′‐AATCTTCACGGACACGCAGA‐3′) for the Left side with 0.4 units of Ex Taq (Takara 5 units/μl) in a total volume of 25 μl. PCR conditions were as follows: preheat at 95°C for 3 min, followed by 32 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 1 min, and additional extension at 72°C for 7 min.
After electrophoresis in a 0.8% agarose gel (Agarose S, Nippon gene), DNA fragments larger than 2.2‐kb were separately extracted and purified using Wizard SV Gel and PCR Clean‐Up System (Promega). The purified DNA was inserted into pGEM vector using pGEM‐T Easy Vector systems (Promega), and transformed to E. coli host, JM109. The flanking genomic sequence was determined using the Pinv HaeIII‐up or Pinv HaeIII‐down primer (see above). The obtained sequence was searched for D. simulans in FlyBase [http://flybase.org/] using Blast Search. Based on a concatenated sequence of determined and retrieved sequences from the database, a primer pair, 3L2ForNew (5′‐TGATGTGCGTCATTCAGCTT‐3′) and 3L2RevNew (5′‐CGACGAGAGGGAAATGAAAA‐3′), was designed to determine the size and structure of the inserted P element using Primer3Plus on the web [http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/]. The PCR condition was preheat at 95°C for 3 min, followed by for 32 cycles were denaturing at 95°C for 30 s, annealing at 50°C for 30 s and polymerizing at 72°C for 4 min.
2.5. Nucleotide sequencing
The DNA fragment was inserted into pGEM vector using pGEM‐T Easy Vector systems (Promega) and BigDye Terminator ver. 3.1 (Thermo Fisher Scientific) and Genetic Analyzer 3100 or 3500 (Thermo Fisher Scientific) were used for DNA sequencing. Cycling conditions were as follows: incubation at 96°C for 1 min followed by 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. We used eight primers as follows: P410F (5′‐AGAAGGCTATACCAGTGGGAG‐3′), P759F (5′‐ATTTCCTTTGCCCAGTCGTAC‐3′), P1227F (5′‐ACCTGGTTTAGCCATCCTGC‐3′), P1632F (5′‐GAGTGCTCGCAACCTTATGG‐3′), P1894F (5′‐TCGACCATCCCACTCCACTG‐3), P2250F (5′‐TGAGCCTGTCGATGATGAGC‐3′), M13M4, and M13reverse. The sequence of genomic P element obtained in this study was deposited in the DDBJ with the accession number LC274660.
3. RESULTS
3.1. P element sequence in D. simulans in Japan
To detect P element homologous sequences, we examined genomic DNAs from 63 isofemale lines of D. simulans from 11 Japanese local populations established from 1976 to 2015. PCR primer pairs, P176F and P2812R, were designed within the P element sequence. Thus, a full‐sized element will generate a product 2.6 kb long. All lines established after 2008 carried copies of P sequences containing full‐sized and internally truncated elements (Figure 2). The oldest lines carrying P elements were the TSM lines, established in Tsushima Island in 2008. As for the smaller elements, their contents, structure and number, seemed to vary from line to line, and there was no specific small element commonly existing across populations. The 15 lines established before 2006 amplified no P element homologous sequence. Similar results were obtained with another set of P primers, P257F and P2770R (Supporting information Figure S1). The RM1 showed a few faint signals in Figure 2, but nothing in another PCR (Supporting information Figure S1), suggesting that they were not specific products of P sequences.
Figure 2.
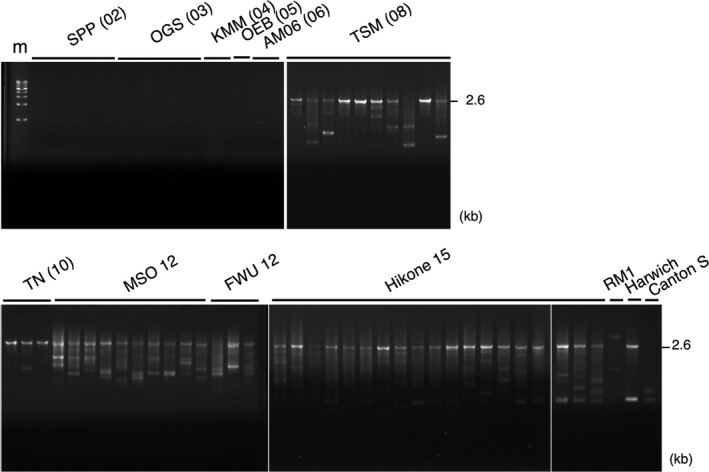
Genomic P elements in Drosophila simulans in Japan. The primer set of P176F and P2812R was used for PCR. The expected size of the full‐sized P elements is indicated as 2.6 kb. The samples were applied from left to right: SPP (02): ‐7, ‐8, ‐28, ‐29, and ‐30; OGS (03): ‐C27, ‐C30, ‐C31, ‐H3, and ‐H4; KMM (04): ‐1 and ‐2; OEB (05); AM06 (06): ‐3 and ‐4; TSM (08): ‐3, ‐27, ‐30, ‐31, ‐32, ‐36, ‐39, ‐58, ‐59, and ‐60; TN (10): ‐134, ‐136, and ‐140; MSO12: ten lines from ‐1 to ‐10; FWU12: ‐4, ‐6, and ‐7; Hikone15: ‐1 to ‐5, ‐7 to ‐11, and ‐13 to ‐21; and RM1 established in 1976. Harwich and Canton S were the standard P and M strains of the P‐M system of D. melanogaster. m: 1 kb DNA ladder marker (Takara)
Recent occurrence of P element does not appear limited in any small area, but suggests comparatively wide distribution in the Japanese archipelago. The local populations showed a monotonous result of with or without P sequence, hence no polymorphism in each population (Figure 3).
Figure 3.
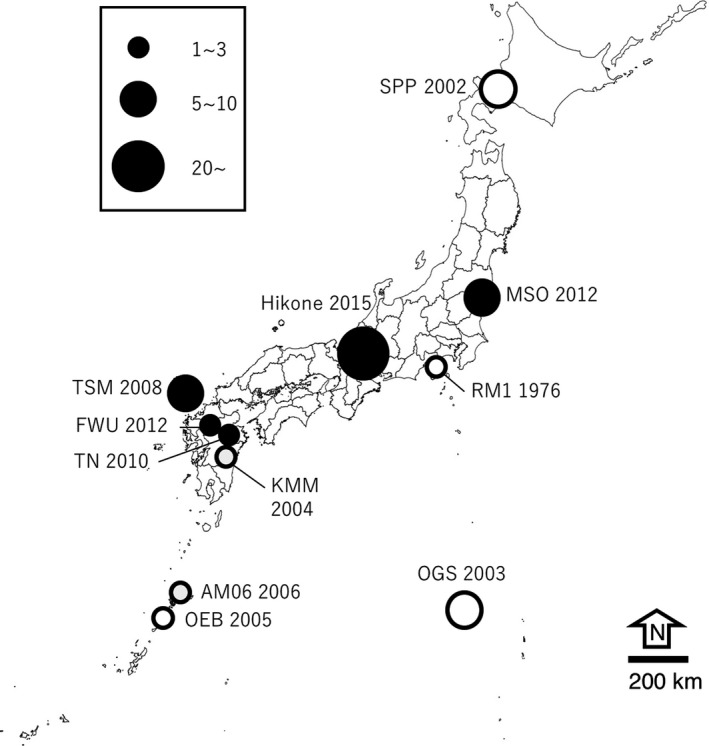
Geographic and chronological distribution of P elements in Drosophila simulans in Japan. A Closed circle indicates that all lines carried genomic P elements, and an open circle indicates no copy of P element. The number of lines examined is approximately depicted by the size of each circle. See Supporting information Table S1 and Figure 2 for detail
3.2. Full‐length P element in D. simulans
To obtain a complete sequence of the genomic P elements and identify the insertion site, we carried out inverse PCR with FWU12‐07, because it appeared to have some full‐sized and only a few other internally truncated smaller P elements in the genome (Figure 2, Supporting information Figure S1). We designed primer sets for 14 possible templates containing the flanking sequences of P element. We successfully obtained a 3.5‐kb fragment from a template, 3L#2. The nucleotide sequence was determined (3,447 nt). The inserted P element was the canonical complete element of 2,907 bp and the nucleotide at position 2,040 was “A”, thus the same as the P element that recently identified in D. simulans populations (Figure 4). Its upstream 154‐bp and downstream 385‐bp flanking sequences consistently indicated that the element is inserted in an 8 bp of GCACAGCC at 16,390,349–16,390,356 on the chromosome 3 L as the target. The insertion point is in the first intron of GD12441 gene, which is 19.6 kb in size and deduced to encode a glutamate synthase (FlyBase ID: FBgn0184173). Transcriptional direction of the P element was opposite to that of GD12441. The EcoRI sites presumed at the both ends in the initial digestion of inverse PCR were confirmed 1,231‐bp upstream and 904‐bp downstream of the target.
Figure 4.
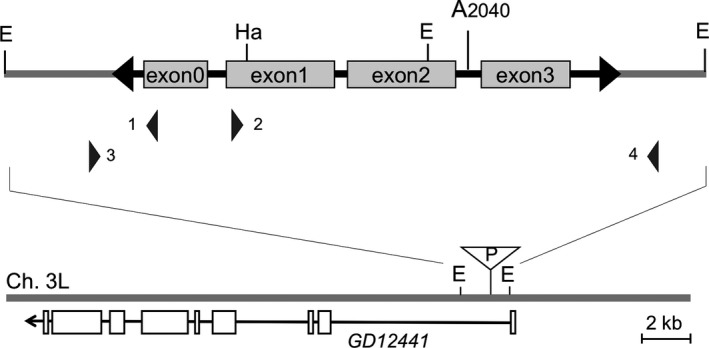
A complete P element in Drosophila simulans in FWU12‐7. A 2,907‐bp P element was located at GCACAGCC at 16,390,349‐16,390,356 of chromosome arm 3 L as the flanking target duplication. The nucleotide sequence of the P element was thoroughly identical, containing the A at the position 2,040, to that of the originally invaded element in D. simulans (Kofler et al., 2015). Approximate positions of the PCR primers are indicated; 1: Pinv HaeIII‐up, 2: Pinv HaeIII‐down, 3: 3L2RevNew, and 4: 3L2ForNew. The primer set 1&2 was used for inverse PCR and 3&4 for amplifying the region containing the P element. This element inserted in the first intron of GD12441 in the opposite direction. EcoRI sites (E) at 16,389,125 for the left and at 16,391,252 for the right side and HaeIII site (Ha) inside P element are indicated in the bottom map
In addition, we determined the nucleotide sequence of 17 DNA fragments obtained from MSO12, Hikone15s‐4, and TSM58 by PCR above. The nucleotide was “A” at the position 2,040 in each amplicon, with no exception (Supporting information Figure S2).
3.3. Hybrid dysgenesis of the flies in D. simulans in Japan
To know the phenotypic characteristics of the current D. simulans in Japan, we carried out the gonadal dysgenesis (GD) test. RM1 was tentatively used as a DS strain because it was free of the P element sequence in its genome (Figure 2, Supporting information Figure S1). First, each of three TSM lines was crossed to RM1 in both directions and F1 females were individually dissected in 5–7 days after eclosion to evaluate the development of ovaries. There was a large difference in the fraction of dysgenic females between the offspring in each reciprocal crosses at 29°C (FET, p < 0.01), but not at 25°C (Figure 5a). As a result, unidirectional gonadal dysgenesis was observed in all three lines examined at 29°C. Males of TSM lines induced significantly higher levels (more than 96%) of dysgenic F1 females when crossed to RM1 females. Thus, hereafter we tentatively used TSM31 as a control for the DI strain having both strong inducing ability and high repression potential of hybrid dysgenesis, and RM1 as a control of the DS strain having neither.
Figure 5.
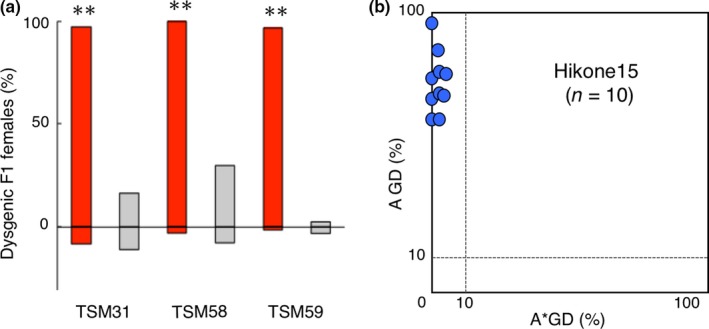
Hybrid dysgenesis in Drosophila simulans in Japan. (a) Fraction of dysgenic F1 females for TSM lines in both directions of each set of reciprocal crosses, with RM1 female (positive direction) and with RM1 males (negative direction). Crosses were performed at 29°C (red bar) and 25°C (gray bar). **Difference in the numbers of normal and dysgenic females was significant between the reciprocal crosses (FET, p < 0.01). See Supporting information Table S2 for detail. (b) A–A* graph of Hikone15 lines. Each dot indicates the dysgenic properties of one line. Dots gathered along the vertical axis, implying that they have higher inducibilities (more than 10% of the cross A GD%) and no, if any, susceptibilities (<10% of cross A* GD%), thus DI strains. See Supporting information Table S3 for detail. n: the number of lines examined
A series of GD tests was carried out for the Hikone15 lines at 29°C. GD% of the cross A (RM1 females × tested males) varied from 61.2 to 96.2%, suggesting considerable levels of inducing ability of P transposition in each line; GD% of cross A* (tested females x TSM31 males) were less than 4.7%, indicating almost full potential of repression (Figure 5b, Supporting information Table S3). To confirm the effects of high temperature of development in the GD test for D. simulans, we again performed GD test for the four lines, Hikone15‐5, 15‐10, 15‐14, and 15‐26, at 25°C. Significant differences were detected in GD% between temperatures, 25°C and 29°C, in the cross A (FET, p < 0.01), but not in the cross A* (Supporting information Table S3 for detail). Therefore, these ten Hikone lines were, from moderate to strong, DI strains. One line, Hikone15‐16, showed prominently strong inducibility, similar to TSM31.
4. DISCUSSION
4.1. P element invasion of D. simulans populations in Japan
The previous studies suggested that P elements invaded D. simulans probably somewhere in Europe, Africa, and North America earlier than 2006, and spread widely in many other areas investigated until 2014 (Hill et al., 2016; Kofler et al., 2015). These observations led us to assume that P elements would spread gradually, or from one population to the neighbor step by step, even if it proceeded much more quickly than in D. melanogaster. However, based on the present survey of D. simulans populations in Japan, P element copies were detected in all isofemale lines established after 2008, but no copy before 2006. P elements in Japanese populations are likely the descendants of the original P element, because they all shared “A” at the nucleotide position 2,040, as in European, African, and American populations. This supports the hypothesis of single event of HT in D. simulans (Kofler et al., 2015), and also implies that P elements reached to the east end of Asia almost simultaneously to other areas. Therefore, P elements dispersed worldwide in D. simulans in a curiously short time, and the spreading pattern is not as previously assumed. P elements are not likely to spread via a combination of the usual migration of individuals and vertical transmission in D. simulans. According to Kofler et al. (2015) human activity of transportation could help flies to move a long distance in a short time. If this is the case, P elements must have formed novel colonizing centers at many points of the earth and then dispersed in each area. To understand the dynamics of newly invaded TEs, further investigation of the current local populations for detail distribution of P elements is under study, because the numbers and localities of the lines examined in the present study were limited, partly due to the availability of the lines kept in lab and the stock centers.
4.2. Dysgenic properties associated with P elements in D. simulans
Our present GD test showed many strong DI strains, which can induce P element transposition, in the TSM and Hikone lines. This may be consistent with a unidirectional high‐temperature‐dependent ovarian dysgenesis in the reciprocal crosses (Figure 5a) and high GD% only in the cross A at 29°C, but not 25°C in the GD test (Supporting information Table S3). The results of PCR demonstrating apparently many copies of full‐sized P elements in the genomes also support this assumption. Four strong DI strains, TSM31, TSM58, TSM59, and Hikone15‐16, will be useful as a strong DI strain in the GD test, like Harwich as the strong P strain in D. melanogaster (Kidwell, 1979).
On the other hand, our present study found no DR strain in Japan, contrary to the observation in Africa and North America, where DR strains were frequently observed between 2008 and 2012 (Hill et al., 2016). Milder GD in D. simulans was hypothesized as one of the possible reasons of more swift spreading of P element than that in D. melanogaster, because the DI strains from Georgia showed <74% of inducing ability (Hill et al., 2016). Our present results of predominance of strong or moderate DI strains and no DR strain at least in two independent recent populations in Japan would not support this assumption, though. Rather, strong effect in GD and rapid spread may not be always exclusive.
In relation to the P element activity, it is noteworthy that one autonomous P element was identified in D. simulans in Japan and mapped to an 8‐bp on the third chromosome. Kofler et al. (2015) and Hill et al. (2016) showed the presence of P sequences in the genomes of D. simulans, many of which are full‐length, and mapped some P elements. However, no full‐length P element of D. simulans has yet been determined for its insertion position, because their results were mainly based on next‐generation sequencing (NGS). NGS serves a powerful tool for genomewide investigation by providing many short‐read sequences, but, even if a whole genome sequence is available as a reference, mapping TEs to the genome is not easy. Therefore, the full‐size P element was mapped for the first time in wild D. simulans in this study. Investigation of moving of individual P element is now under study using this element.
4.3. Evolutionary aspect of P regulatory systems in Drosophila hosts
P elements were shown to have changed the host species via HT, from D. willistoni to D. melanogaster in the 20th century (Daniels et al., 1990), and very recently, from D. melanogaster to D. simulans (Kofler et al., 2015). The host species invaded by TEs needs to develop regulatory systems against selfish jumping of TEs, but the evolutionary process of establishing suppression is not always clear. In D. melanogaster, in addition to the major repression by the piRNAs (Brennecke et al., 2008) and the 66 kDa repressor produced by the full‐length element (O'Hare & Rubin, 1983), some internally deleted variants are thought to play important roles in suppressing P transposition, for instance, A12 (Andrews & Gloor, 1995; Gloor et al., 1993), D50 (Rasmusson, Raymond, & Simmons, 1993), SP (Rasmusson et al., 1993), SR (Corish, Black, Featherston, Merriam, & Dover, 1996) and KP (Black, Jackson, Kidwell, & Dover, 1987; Jackson, Black, & Dover, 1988; Simmons, Grimes, & Czora, 2016) elements, each of which has a specific size. In particular, KP elements of D. melanogaster increased in number (Andrews & Gloor, 1995; Black et al., 1987) and became predominant together with full‐length elements (FP + KP predominance) worldwide (Itoh & Boussy, 2002; Itoh, Sasai, Inoue, & Watada, 2001; Itoh, Takeuchi, Yamaguchi, Yamamoto, & Boussy, 2007; Itoh, Woodruff, Leone, & Boussy, 1999; Itoh et al., 2004; Ogura, Woodruff, Itoh, & Boussy, 2007). FP + KP predominance may support an advantageous effect of KP elements by preventing P elements from transposing. An alternative plausible hypothesis was suggested that KP elements have more ability of transposition than other P elements (Fukui, Inoue, Yamaguchi, & Itoh, 2008). Based on our PCR analyses in D. simulans, all lines examined carried some copies of full‐length and smaller derivative elements. However, in striking contrast to D. melanogaster, no specific smaller elements were frequently observed (Figure 1, Supporting information Figure S1).
Only several years may be too short for a unique repressor variant, like KP elements, to evolve de novo in D. simulans. Such smaller repressor elements, however, were not found in D. willistoni (Regner, Pereira, Alonso, Abdelhay, & Valente, 1996), although D. willistoni should have the longest evolutionary history with P elements among the three species (Powell & Gleason, 1996). On the other hand, repression by the piRNAs was shown to work in D. simulans (Hill et al., 2016) and D. melanogaster (Brennecke et al., 2008; Khurana et al., 2011), but still unknown in D. willistoni. From the view of evolution of repression systems, careful investigation is required, for instance, comparing expression, fine structure, and insertion sites of the genomic P elements both between populations and between species.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
MI conceived and designed the study. MI, YY, and NI collected some wild flies and established the isofemale lines. YY, NI, MS, and YK contributed to data acquisition and analysis. MI, YY, and NI drafted the first manuscript. MI, NI, YK, YY, and MS contributed to the revision. MI, NI, YK, YY, and MS approved final submission.
DATA ACCESSIBILITY
DNA sequences DDBJ accession number: LC274660.
Supporting information
ACKNOWLEDGMENTS
We thank Y. Tamiaki and Y. Usami for providing technical support and I. A. Boussy for careful reading of the manuscript and valuable comments. We also thank Ehime Drosophila Stock Center and Kyoto Drosophila Stock Center for providing fly lines.
Yoshitake Y, Inomata N, Sano M, Kato Y, Itoh M. The P element invaded rapidly and caused hybrid dysgenesis in natural populations of Drosophila simulans in Japan. Ecol Evol. 2018;8:9590–9599. 10.1002/ece3.4239
REFERENCES
- Andrews, J. D. , & Gloor, G. B. (1995). A role for the KP leucine zipper in regulating P element transposition in Drosophila melanogaster . Genetics, 141, 587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anxolabéhère, D. , Kidwell, M. G. , & Periquet, G. (1988). Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Molecular Biology and Evolution, 5, 252–69. [DOI] [PubMed] [Google Scholar]
- Anxolabéhère, D. , Nouaud, D. , Periquet, G. , & Tchen, P. (1985). P‐element distribution in Eurasian populations of Drosophila melanogaster: A genetic and molecular analysis. Proceedings of the National Academy of Sciences of the United States of America, 82, 5418–22. 10.1073/pnas.82.16.5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. , Golic, K. G. , & Hawley, R. (2005). Drosophila: A laboratory handbook. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, xxviii. [Google Scholar]
- Bergman, C. M. , Quesneville, H. , Anxolabéhère, D. , & Ashburner, M. (2006). Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biology, 7, R112 10.1186/gb-2006-7-11-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemont, C. , & Vieira, C. (2006). Genetics: junk DNA as an evolutionary force. Nature, 443, 521–4. 10.1038/443521a [DOI] [PubMed] [Google Scholar]
- Bingham, P. M. , Kidwell, M. G. , & Rubin, G. M. (1982). The molecular basis of PM hybrid dysgenesis: The role of the P element, a P‐strain‐specific transposon family. Cell, 29, 995–1004. 10.1016/0092-8674(82)90463-9 [DOI] [PubMed] [Google Scholar]
- Black, D. M. , Jackson, M. S. , Kidwell, M. G. , & Dover, G. A. (1987). KP elements repress P‐induced hybrid dysgenesis in Drosophila melanogaster . EMBO Journal, 6, 4125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnivard, E. , & Higuet, D. (1999). Stability of European natural populations of Drosophila melanogaster with regard to the PM system: A buffer zone made up of Q populations. Journal of Evolutionary Biology, 12, 633–647. 10.1046/j.1420-9101.1999.00063.x [DOI] [Google Scholar]
- Brennecke, J. , Malone, C. D. , Aravin, A. A. , Sachidanandam, R. , Stark, A. , & Hannon, G. J. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science, 322, 1387–1392. 10.1126/science.1165171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J. F. (1991). Models of repression of transposition in P‐M hybrid dysgenesis by P cytotype and by zygotically encoded repressor proteins. Genetics, 128, 471–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy, P. , Gibert, P. , & Boussy, I. A. (2004). Drosophila melanogaster, Drosophila simulans: So similar, so different. Dordrecht, the Netherlands: Springer; 10.1007/978-94-007-0965-2 [DOI] [PubMed] [Google Scholar]
- Corish, P. , Black, D. M. , Featherston, D. W. , Merriam, J. , & Dover, G. A. (1996). Natural repressors of P‐induced hybrid dysgenesis in Drosophila melanogaster: A model for repressor evolution. Genetical Research, 67, 109–21. 10.1017/S0016672300033577 [DOI] [PubMed] [Google Scholar]
- Craig, N. L. , Craigie, R. , Gellert, M. , & Lambowitz, A. M. (2002). Mobile DNA II. Washington, DC: American Society for Microbiology Press; 10.1128/9781555817954 [DOI] [Google Scholar]
- Daniels, S. B. , Chovnick, A. , & Kidwell, M. G. (1989). Hybrid dysgenesis in Drosophila simulans lines transformed with autonomous P elements. Genetics, 121, 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, S. B. , Peterson, K. R. , Strausbaugh, L. D. , Kidwell, M. G. , & Chovnick, A. (1990). Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics, 124, 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, S. B. , Strausbaugh, L. D. , & Armstrong, R. A. (1985). Molecular analysis of P element behavior in Drosophila simulans transformants. Molecular and General Genetics, 200, 258–65. 10.1007/BF00425433 [DOI] [PubMed] [Google Scholar]
- Drezen, J. M. , Gauthier, J. , Josse, T. , Bezier, A. , Herniou, E. , & Huguet, E. (2017). Foreign DNA acquisition by invertebrate genomes. Journal of Invertebrate Pathology, 147, 157–168. 10.1016/j.jip.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Engels, W. R. , & Preston, C. R. (1980). Components of hybrid dysgenesis in a wild population of Drosophila melanogaster . Genetics, 95, 111–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C. , & Pritham, E. J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics, 41, 331–68. 10.1146/annurev.genet.40.110405.090448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui, T. , Inoue, Y. , Yamaguchi, M. , & Itoh, M. (2008). Genomic P elements content of a wild M’ strain of Drosophila melanogaster: KP elements do not always function as type II repressor elements. Genes & Genetic Systems, 83, 67–75. 10.1266/ggs.83.67 [DOI] [PubMed] [Google Scholar]
- Gloor, G. B. , Preston, C. R. , Johnson‐Schlitz, D. M. , Nassif, N. A. , Phillis, R. W. , Benz, W. K. , … Engels, W. R. (1993). Type I repressors of P element mobility. Genetics, 135, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuet, D. , Merçot, H. , Allouis, S. , & Montchamp‐Moreau, C. (1996). The relationship between structural variation and dysgenic properties of P elements in long‐established P‐transformed lines of Drosophila simulans . Heredity (Edinb), 77(Pt 1), 9–15. 10.1038/hdy.1996.102 [DOI] [PubMed] [Google Scholar]
- Hill, T. , Schlötterer, C. , & Betancourt, A. J. (2016). Hybrid dysgenesis in Drosophila simulans associated with a rapid invasion of the P‐Element. PLoS Genetics, 12, e1005920 10.1371/journal.pgen.1005920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck, M. A. , Clark, J. B. , Peterson, K. R. , & Kidwell, M. G. (1991). Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science, 253, 1125–8. 10.1126/science.1653453 [DOI] [PubMed] [Google Scholar]
- Hua‐Van, A. , Le Rouzic, A. , Boutin, T. S. , Filee, J. , & Capy, P. (2011). The struggle for life of the genome's selfish architects. Biology Direct, 6, 19 10.1186/1745-6150-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M. , & Boussy, I. A. (2002). Full‐size P and KP elements predominate in wild Drosophila melanogaster . Genes & Genetic Systems, 77, 259–267. 10.1266/ggs.77.259 [DOI] [PubMed] [Google Scholar]
- Itoh, M. , Fukui, T. , Kitamura, M. , Uenoyama, T. , Watada, M. , & Yamaguchi, M. (2004). Phenotypic stability of the P‐M system in wild populations of Drosophila melanogaster . Genes & Genetic Systems, 79, 9–18. 10.1266/ggs.79.9 [DOI] [PubMed] [Google Scholar]
- Itoh, M. , Sasai, N. , Inoue, Y. , & Watada, M. (2001). P elements and PM characteristics in natural populations of Drosophila melanogaster in the southernmost islands of Japan and in Taiwan. Heredity, 86, 206–212. 10.1046/j.1365-2540.2001.00817.x [DOI] [PubMed] [Google Scholar]
- Itoh, M. , Takeuchi, N. , Yamaguchi, M. , Yamamoto, M. T. , & Boussy, I. A. (2007). Prevalence of full‐size P and KP elements in North American populations of Drosophila melanogaster . Genetica, 131, 21–8. 10.1007/s10709-006-9109-2 [DOI] [PubMed] [Google Scholar]
- Itoh, M. , Woodruff, R. C. , Leone, M. A. , & Boussy, I. A. (1999). Genomic P elements and P‐M characteristics of eastern Australian populations of Drosophila melanogaster . Genetica, 106, 231–245. 10.1023/A:1003918417012 [DOI] [PubMed] [Google Scholar]
- Jackson, M. S. , Black, D. M. , & Dover, G. A. (1988). Amplification of KP elements associated with the repression of hybrid dysgenesis in Drosophila melanogaster . Genetics, 120, 1003–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanishi, M. , & Watanabe, T. (1977). Ecological factors controlling the coexistence of the sibling species Drosophila simulans and D. melanogaster . Japanese Journal of Ecology, 27, 279–283. [Google Scholar]
- Kelleher, E. S. (2016). Reexamining the P‐Element Invasion of Drosophila melanogaster Through the Lens of piRNA Silencing. Genetics, 203, 1513–31. 10.1534/genetics.115.184119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana, J. S. , Wang, J. , Xu, J. , Koppetsch, B. S. , Thomson, T. C. , Nowosielska, A. , … Theurkauf, W. E. (2011). Adaptation to P element transposon invasion in Drosophila melanogaster . Cell, 147, 1551–63. 10.1016/j.cell.2011.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G. (1979). Hybrid dysgenesis in Drosophila melanogaster: The relationship between the P– M and I– R interaction systems. Genetical Research, 33, 205 10.1017/S0016672300018358 [DOI] [Google Scholar]
- Kidwell, M. G. (1993). Lateral transfer in natural populations of eukaryotes. Annual Review of Genetics, 27, 235–56. 10.1146/annurev.ge.27.120193.001315 [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G. (1994). The Wilhelmine E. Key 1991 Invitational Lecture. The evolutionary history of the P family of transposable elements. Journal of Heredity, 85, 339–46. 10.1093/oxfordjournals.jhered.a111478 [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G. , Kidwell, J. F. , & Sved, J. A. (1977). Hybrid Dysgenesis in Drosophila melanogaster: A syndrome of aberrant traits including mutation, sterility and male recombination. Genetics, 86, 813–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G. , & Lisch, D. (1997). Transposable elements as sources of variation in animals and plants. Proceedings of the National Academy of Sciences of the United States of America, 94, 7704–7711. 10.1073/pnas.94.15.7704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K. , & Kidwell, M. G. (1994). Differences in P element population dynamics between the sibling species Drosophila melanogaster and Drosophila simulans . Genetical Research, 63, 27–38. 10.1017/S0016672300032055 [DOI] [PubMed] [Google Scholar]
- Kofler, R. , Hill, T. , Nolte, V. , Betancourt, A. J. , & Schlötterer, C. (2015). The recent invasion of natural Drosophila simulans populations by the P‐element. Proceedings of the National Academy of Sciences of the United States of America, 112, 6659–63. 10.1073/pnas.1500758112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic, A. , & Capy, P. (2005). The first steps of transposable elements invasion. Genetics, 169, 1033–1043. 10.1534/genetics.104.031211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic, A. , & Capy, P. (2009). Theoretical approaches to the dynamics of transposable elements in genomes, populations, and species. In: Lankenau D.‐H. & Volff J.‐N. (Ed.), Transposons and the Dynamic Genome, (pp. 1‐19). Springer‐Verlag: Berlin, Heidelberg: ISBN: 978‐3‐642‐02004‐9. [Google Scholar]
- Lee, Y. C. G. , & Langley, C. H. (2012). Long‐term and short‐term evolutionary impacts of transposable elements on Drosophila . Genetics, 192, 1411–1432. 10.1534/genetics.112.145714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S. H. , Quarles, K. A. , Yang, Y. , Tanguy, M. , Fr Zal, L. , Smith, S. A. , … Giraud, I. (2018). Pan‐arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nature Ecology & Evolution, 2, 174 10.1038/s41559-017-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp‐Moreau, C. (1990). Dynamics of P‐M hybrid dysgenesis in P‐transformed lines of Drosophila simulans . Evolution, 44, 194–203. [DOI] [PubMed] [Google Scholar]
- Ogura, K. , Woodruff, R. C. , Itoh, M. , & Boussy, I. A. (2007). Long‐term patterns of genomic P element content and P‐M characteristics of Drosophila melanogaster in eastern Australia. Genes & Genetic Systems, 82, 479–87. 10.1266/ggs.82.479 [DOI] [PubMed] [Google Scholar]
- O'Hare, K. , & Rubin, G. M. (1983). Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell, 34, 25–35. 10.1016/0092-8674(83)90133-2 [DOI] [PubMed] [Google Scholar]
- Orgel, L. E. , & Crick, F. H. C. (1980). Selfish DNA: The ultimate parasite. Nature, 284, 604–607. 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- Pinsker, W. , Haring, E. , Hagemann, S. , & Miller, W. J. (2001). The evolutionary life history of P transposons: From horizontal invaders to domesticated neogenes. Chromosoma, 110, 148–158. 10.1007/s004120100144 [DOI] [PubMed] [Google Scholar]
- Powell, J. R. , & Gleason, J. M. (1996). Codon usage and the origin of P elements. Molecular Biology and Evolution, 13, 278–279. 10.1093/oxfordjournals.molbev.a025564 [DOI] [PubMed] [Google Scholar]
- Rasmusson, K. E. , Raymond, J. D. , & Simmons, M. J. (1993). Repression of hybrid dysgenesis in Drosophila melanogaster by individual naturally occurring P elements. Genetics, 133, 605–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regner, L. P. , Pereira, M. S. , Alonso, C. E. , Abdelhay, E. , & Valente, V. L. (1996). Genomic distribution of P elements in Drosophila willistoni and a search for their relationship with chromosomal inversions. Journal of Heredity, 87, 191–8. 10.1093/oxfordjournals.jhered.a022984 [DOI] [PubMed] [Google Scholar]
- Scavarda, N. J. , & Hartl, D. L. (1984). Interspecific DNA transformation in Drosophila . Proceedings of the National Academy of Sciences of the United States of America, 81, 7515–7519. 10.1073/pnas.81.23.7515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavarda, N. J. , & Hartl, D. L. (1987). Germ line abnormalities in Drosophila simulans transfected with the transposable P element. Journal of Genetics, 66, 1–15. [Google Scholar]
- Schaack, S. , Gilbert, C. , & Feschotte, C. (2010). Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends in Ecology & Evolution, 25, 537–546. 10.1016/j.tree.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti, K.‐A. , Jurczak, D. , Sachidanandam, R. , & Brennecke, J. (2015). piRNA‐guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes & Development, 29, 1747–1762. 10.1101/gad.267252.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, M. J. , Grimes, C. D. , & Czora, C. S. (2016). Cytotype regulation facilitates repression of hybrid dysgenesis by naturally occurring KP elements in Drosophila melanogaster . G3 (Bethesda), 6, 1891–1897. 10.1534/g3.116.028597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner, S. , Miele, V. , Terzian, C. , Biemont, C. , Daubin, V. , Feschotte, C. , & Pontier, D. (2017). Ecological networks to unravel the routes to horizontal transposon transfers. PLoS Biology, 15, e2001536 10.1371/journal.pbio.2001536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences DDBJ accession number: LC274660.


