Abstract
Impairment of microglial functions, such as phagocytosis and/or dysregulation of immune responses, has been implicated as an underlying factor involved in the pathogenesis of various neurodegenerative disorders. Our previous studies have demonstrated that long intergenic noncoding RNA (lincRNA)-Cox2 expression is influenced by nuclear factor κB (NF-κB) signaling and serves as a coactivator of transcriptional factors to regulate the expression of a vast array of immune-related genes in microglia. Extracellular vesicles (EVs) have been recognized as primary facilitators of cell-to-cell communication and cellular regulation. Herein, we show that EVs derived from astrocytes exposed to morphine can be taken up by microglial endosomes, leading, in turn, to activation of Toll-like receptor 7 (TLR7) with a subsequent upregulation of lincRNA-Cox2 expression, ultimately resulting in impaired microglial phagocytosis. This was further validated in vivo, wherein inhibition of microglial phagocytic activity was also observed in brain slices isolated from morphine-administrated mice compared with control mice. Additionally, we also showed that intranasal delivery of EVs containing lincRNA-Cox2 siRNA (small interfering RNA) was able to restore microglial phagocytic activity in mice administered morphine. These findings have ramifications for the development of EV-loaded RNA-based therapeutics for the treatment of various disorders involving functional impairment of microglia.
Keywords: phagocytosis, drug abuse, morphine, extracellular vesicle, exosome, lincRNA, microglia, CNS
Introduction
Opioid analgesics, of which morphine is a classic example, are used extensively in the clinical setting for pain management owing to their beneficial effects. Paradoxically, however, there are also deleterious side effects associated with opiates such as opioid tolerance, addiction, and decline in cognitive performance.1, 2 Opiates exert dramatic effects on phagocytic cells including microglia, the resident phagocytic cells of the brain.3, 4, 5 Rapid phagocytic removal of dead, dying, infected cells or microbes is vital for brain homeostasis and for proper resolution of the insult by removing immunogens and limiting excessive immune responses.6 Phagocytic cells via their cell surface receptors can recognize pathogenic microorganisms and cellular debris that, in turn, trigger the internal cellular machinery resulting in the ingestion of the particles into the phagosomes, which can fuse with the lysosomes to be ultimately degraded in the phagolysosomes.7, 8 It has been well documented that the “find-me” and “eat-me” signals released by apoptotic cells serve to attract the phagocytes, resulting in the engulfment and clearance of apoptotic cells to maintain tissue homeostasis.9, 10 In the CNS, impairment of microglial phagocytosis renders the microglia unable to engulf and eliminate toxic pathogens and cellular debris, ultimately culminating in a toxic inflammatory milieu, a common feature underlying various neurodegenerative disorders.11, 12 Microglia express a wide array of phagocytic receptors that are involved in receptor-mediated phagocytosis in the CNS.13 Dysregulated expression of microglial phagocytic receptors has been shown to result in impaired microglial phagocytosis.14 The current study was undertaken to understand how morphine exposure impairs microglial phagocytosis involving the astrocyte-microglial crosstalk.
Extracellular vesicles (EVs) comprising microvesicles, exosomes, and apoptotic bodies play important roles as cargo-carrying vesicles mediating communication among diverse cells types and tissues, including the CNS.15, 16, 17, 18 Studies by others15 and us16 have demonstrated that EVs released from astrocytes, the most abundant cell type in the CNS, can be conduits for delivering the cargo of macromolecules to neighboring as well as distant cells, culminating in a wide spectrum of functional changes in the recipient cells. Exploiting this cargo delivery function of EVs has been critical in understanding how EVs can also be developed as vehicles for targeted delivery of therapeutic drugs to tissues.19 For example, an exciting study by Zhuang et al.20 has demonstrated that intranasal delivery of exosomes loaded with curcumin can target the microglia, resulting in reduction of lipopolysaccharide (LPS)-induced neuroinflammation.
Long intergenic noncoding RNAs (lincRNAs) are regulatory RNA molecules and play pivotal roles in the regulation of many cellular processes.21, 22 Our previous studies have demonstrated that the transcription of lincRNA-Cox2 is regulated by the nuclear factor κB (NF-κB) signaling pathway and is required for the transcription of late primary inflammatory-response genes.23, 24 In the current study, we demonstrate that morphine-stimulated astrocyte-derived EVs (morphine-ADEVs) can be taken up by microglial cells, leading, in turn, to impaired microglial phagocytosis via the Toll-like receptor 7 (TLR7)-NF-κB-lincRNA-Cox2 axis. Additionally, we also demonstrate that intranasal delivery of lincRNA-Cox2 small interfering RNA (siRNA) can restore microglial phagocytic activity in morphine-administered mice.
Results
Morphine Induces Release of EVs from Astrocytes
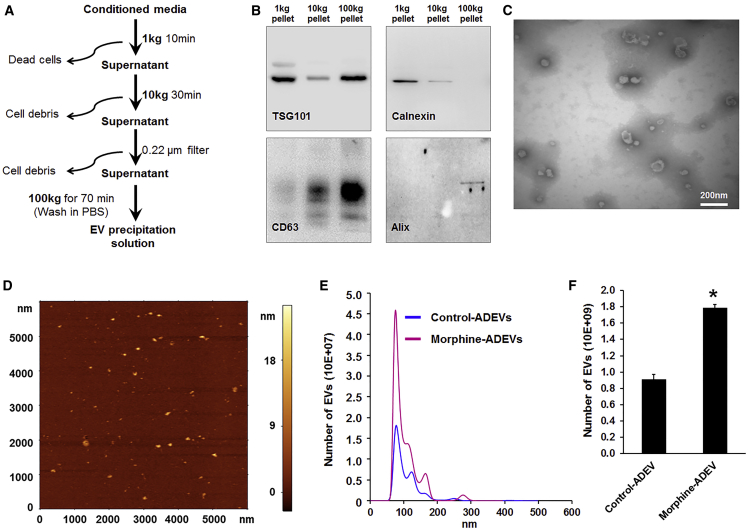
Astrocytes perform essential functions to sustain homeostasis within the CNS, including proper maintenance of water and ion balance, removal of extracellular glutamate, and release of neurotrophic factors.25 In addition, astrocytes have developed multiple avenues of intercellular communication and transfer of various factors including RNA. It is well known that astrocytes can communicate via direct cell-cell interactions and gap junctions; however, over the last few years, it is increasingly being appreciated that astrocytes also communicate with many, if not all, cells within the CNS, through the release and uptake of EVs. Herein we first sought to isolate and characterize EVs from conditioned media of human astrocyte A172 cells, as well as mouse and human primary astrocytes, using a differential ultracentrifugation procedure (Figure 1A).16, 26 Purified EVs were characterized by western blot for signature exosomal markers, as well as by transmission electron microscopy (TEM) and atomic force microscopy (AFM). As shown in Figure 1, immunoblotting of the EV lysates revealed the presence of exosomal markers TSG101, CD63, and Alix. Additionally, immunoblotting using calnexin was also done to demonstrate that isolated ADEVs were free of contamination with cell debris (Figure 1B). Further characterization of EVs using TEM and AFM demonstrated EVs ranging from 40 to 100 nm in diameter (Figures 1C and 1D). Given that astrocytes express high levels of mu opioid receptors and have been shown to respond to morphine, we next sought to examine the effect of morphine on the number and size of ADEVs using nanoparticle tracking analysis (NTA). As shown in Figures 1E and 1F, exposure of astrocytes to morphine (10 μM, 24 hr) resulted in significantly increased numbers of released EVs compared with the control cells. There was, however, no effect on the size distribution of EVs in the presence of morphine.
Figure 1.
Characterization of EVs Isolated from Astrocyte Culture
(A) Schematic representation of the major steps involved in the EV isolation from astrocyte culture. (B) Western blot characterization of astrocyte EVs. Protein isolated from astrocyte EVs was separated on SDS-PAGE and electroblotted onto nitrocellulose membrane. Blots were probed with exosome marker antibody against TSG101, CD63, and Alix. Calnexin was used as a control for cell debris contamination. (C) Representative electron micrograph of EVs isolated from A172 cells. Scale bar, 200 nm. (D) AFM image of EVs isolated from mouse primary astrocyte culture supernatants. (E) Size and particle distribution plots of isolated EVs from cell culture by NanoSight Tracking Analysis (NTA). The plot shows a peak size around 80 nm for the isolated EVs. (F) Number of EVs isolated from control and morphine-exposed human primary astrocytes. All experiments were done at least three independent times. Data are shown as mean ± SEM. *p < 0.05 versus control.
EVs Are Taken up by Microglia and Reach the Endosomes
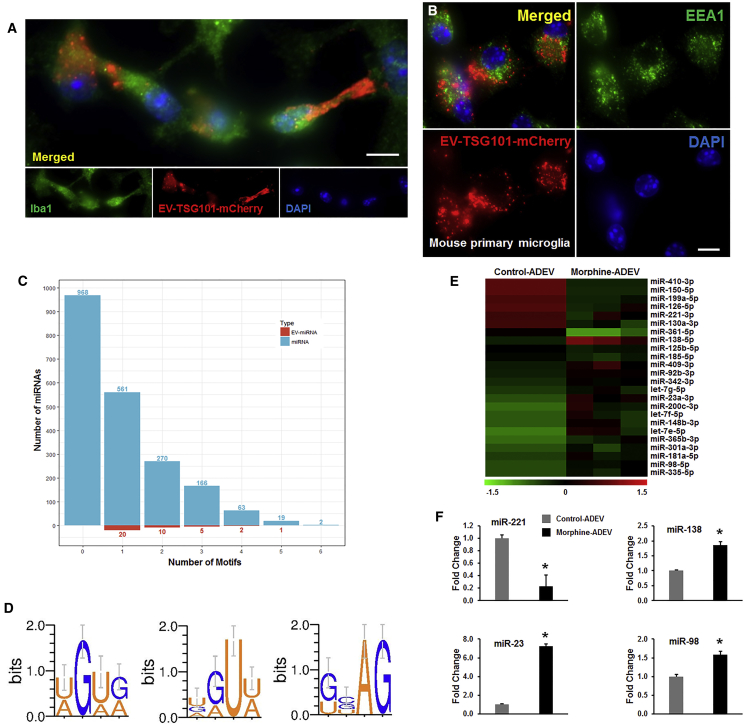
Because microglia are the primary phagocytic immune cells within the CNS, we next asked whether ADEVs could be taken up by microglia and, if so, could they reach the endosomes, the site for RNA-sensing TLRs. For this, we cultured mouse primary microglia with EVs isolated from mouse primary astrocytes that were transfected with a plasmid encoding the exosomal marker TSG101 fused with mCherry. As observed in Figure 2A, mouse primary microglia cultured with RFP-tagged EVs (30 min) demonstrated a fine granular fluorescent pattern within the cytoplasm, suggesting thereby that EVs could reach and release their contents into the microglial cells. We next sought to assess whether ADEVs taken up by microglia could reach the endosomes; mouse primary microglia were exposed to mouse primary astrocyte-derived RFP-tagged EVs for 30 min, followed by harvesting of cells for localization of EVs at the endosomes. Cells were immunostained for the endosomal marker EEA1 and also assessed for the RFP-tagged ADEVs by confocal microscopy. As shown in Figure 2B, microglia were able to take up mCherry-TSG101 ADEVs and, furthermore, these ADEVs were found to co-localize within the microglial endosomes.
Figure 2.
EVs Are Internalized by Microglial Cells and Localize to Late Endosomes
(A and B) Mouse primary microglial cells were incubated with EVs purified from pEF6.mCherry-TSG101-transfected mouse primary astrocytes for 20 min. Paraformaldehyde fixed microglial cells were permeabilized and stained for (A) Iba1 or (B) EEA1, followed by FITC-conjugated secondary antibody and visualized by fluorescence microscopy. Scale bars, 10 μm. (C) Barplot showing the number of miRNAs (light blue) and EV-miRNAs (red) that contain various numbers of AU- or GU-rich motifs. (D) Sequence logos AU- and GU-rich motifs of EV miRNAs. (E) Heatmap of expression profile of AU- and GU-rich miRNAs in EVs from control and morphine-stimulated human primary astrocytes. (F) qPCR validation of representative differentially expressed miRNAs identified from the RNA-seq. All experiments were done at least three independent times. *p < 0.05 versus control.
Endosomal receptors TLR7 and TLR8 are known to bind AU- or GU-rich single-stranded RNA ligands, leading to activation of the downstream NF-κB signaling pathway.27 For example, Forsbach et al.27 have identified 15 AU- or GU-containing 4-mers that function as TLR7/8 RNA agonists. Interestingly, EV microRNAs (miRNAs) containing the TLR7/8 binding motifs have also been shown to function as TLR7/8 agonists.18, 28, 29 Based on our findings that ADEVs taken up by the microglia could reach the endosomes, we next sought to investigate whether ADEV-miRNAs could also serve as TLR7/8 RNA agonists. For this, we performed an TLR7/8 binding elements search within the known mouse mature miRNA sequence database (miRBase,30 version 21). Moreover, the extracellular mouse miRNAs annotation was also obtained from the EV miRNA databases, ExoCarta31 and EVpedia.32 We found that almost 1,079 out of 2,049 mouse miRNAs contained at least one AU- or GU-rich 4-mers, as reported by Forsbach et al.,27 with 38 of them reported to be present in EVs (Figure 2C; Table S1). Based on these 38 reported EV-miRNAs, which are potential TLR7/8 RNA agonists, we next conducted motif prediction using MDS2,33 and three possible motifs were predicted (Figure 2D) covering 66.0% to 79.0% of the 38 EV-miRNAs as potential TLR7/8 RNA agonist. Next, we sought to examine whether morphine-ADEVs also contained the TLR7/8 motifs. For this, RNA was purified from control- or morphine-ADEVs isolated from human primary astrocytes and subjected to RNA-sequencing (RNA-seq) analyses. As shown in Figure 2E, 15 of the 4-mer containing miRNAs were significantly upregulated and, 9 of the 4-mer containing miRNAs were downregulated in morphine-ADEVs compared with control-ADEVs. Selective differentially expressed miRNAs identified from the RNA-seq analysis were further validated using qPCR (Figure 2F). Interestingly, the total reads from the RNA-seq data for miRNAs in morphine-ADEVs were significantly increased compared with that in control-ADEVs, indicating thereby that morphine-ADEV-miRNAs could serve as TLR7/8 RNA agonists in the recipient microglia.
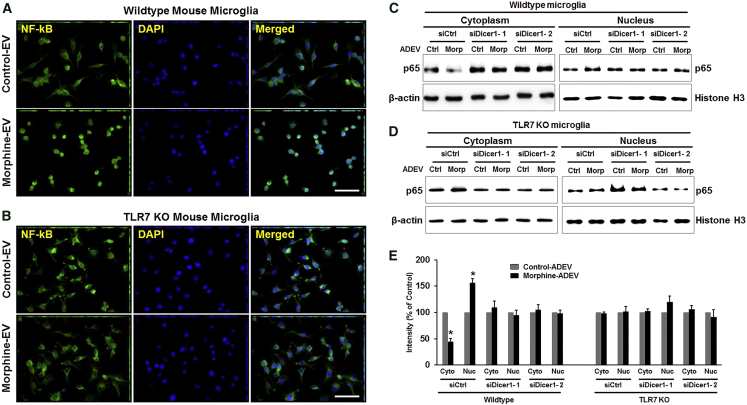
Morphine-ADEV-Induced Nuclear Translocation of Endogenous NF-κB p65 Involves Endosomal TLR7 Signaling in Microglia
Having determined that EVs can reach the endosomes and deliver AU- and GU-rich miRNAs, which could activate TLR7/8, the next logical step was to investigate the downstream signaling of the TLR7/8 pathway, in particular, nuclear translocation of NF-κB and microglial activation.34 To this end, we thus sought to examine whether morphine-induced EVs released from astrocytes could activate NF-κB in microglia. Mouse primary microglia were stimulated with either control- or morphine-ADEVs isolated from mouse primary astrocytes for 30 min followed by immunocytochemistry for translocation of p65. As shown in Figure 3A, microglial cells stimulated with morphine-ADEVs demonstrated significantly increased translocation of NF-κB p65 from the cytoplasm into the nucleus compared with cells exposed to control-ADEVs (Figure 3A). Intriguingly, and as expected, morphine-ADEVs failed to promote nuclear translocation of p65 in TLR7 knockout (KO) microglial cells (Figure 3B). These findings were further confirmed by immunoblotting. To further investigate whether morphine-ADEV-mediated translocation of NF-κB could be attributed to the miRNA cargo in the EVs, we transfected mouse primary astrocytes with either control or Dicer-siRNA for 24 hr, and subsequently exposed them to morphine for an additional 24 hr. EVs were then isolated from the conditioned media and exposed to either wild-type (WT) or TLR7 KO mouse microglial cells for 30 min, followed by detection of nuclear translocation of the NF-κB p65 subunit by western blotting and fluorescent immunostaining. As shown in Figures 3C–3E and Figure S1, EVs isolated from morphine-stimulated, Dicer KO astrocytes failed to induce nuclear translocation of NF-κB p65 in WT microglia. These data thus underpin the pivotal role of miRNA cargo of the morphine-ADEVs in activating the TLR7-NF-κB signaling pathway.
Figure 3.
EVs Released from Morphine-Exposed Astrocytes Induce Nuclear Translocation of NF-κB
(A and B) Wild-type (A) and TLR7 KO (B) mouse primary microglial cells were treated with EVs (2 μg of EVs per 2 × 105 cells) isolated from morphine-exposed astrocytes for 30 min. The subcellular localization of endogenous p65 was visualized by indirect immunofluorescence using anti-p65 antibodies (green). Nuclear DNA was revealed by DAPI staining. Scale bars, 50 μm. (C and D) Primary mouse astrocytes were transfected with either control or Dicer-siRNA for 24 hr, followed by morphine exposure for 24 hr. Wild-type (C) and TLR7 KO (D) mouse microglial cells were exposed to EVs (30 min) isolated from astrocyte conditioned media followed by detection of NF-κB p65 in the nuclear fraction by western blot. (E) All experiments were done at least three independent times. Band intensities of the cytoplasmic and nuclear extracts were quantified and normalized against β-actin and histone H3, respectively, and compared with untreated cells. All data are presented as mean ± SD. *p < 0.05 versus control using Student’s t test.
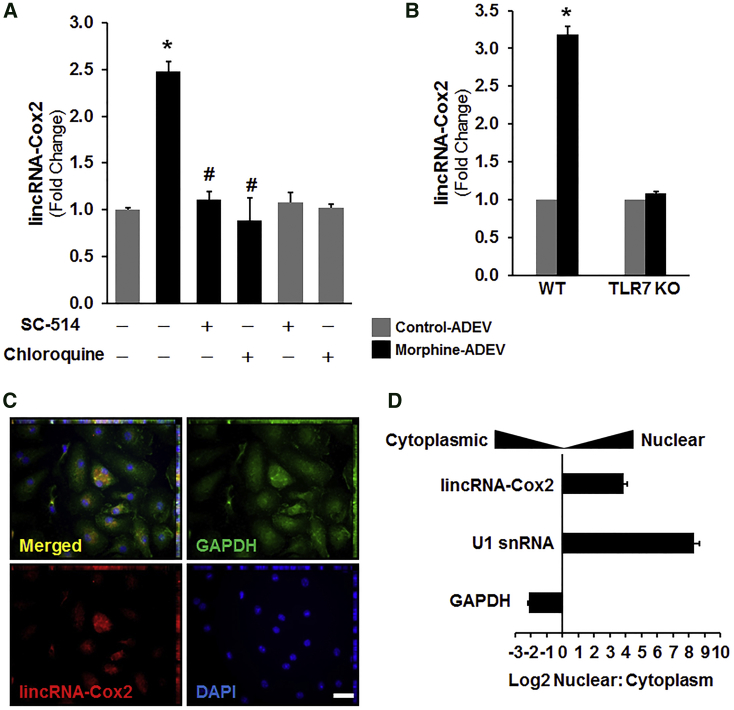
Morphine-ADEVs Induce lincRNA-Cox2 Expression in Microglia via the TLR7/NF-κB Signaling Pathway
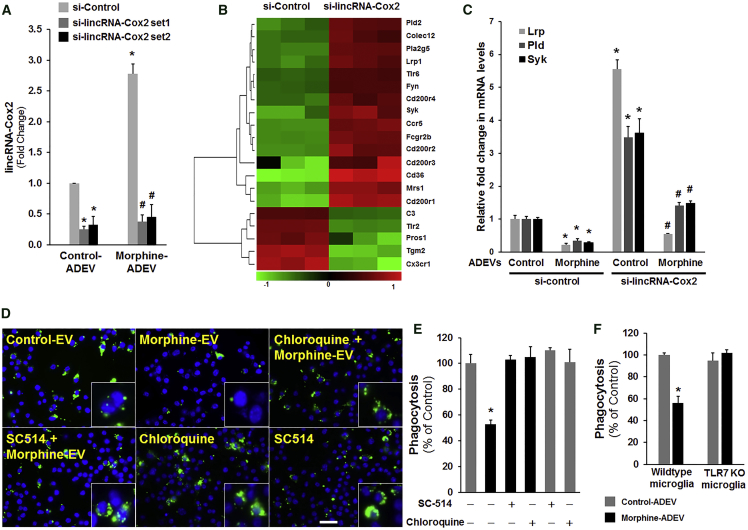
Studies from others35 and us24 have demonstrated that lincRNA-Cox2 is an early-primary gene regulated by NF-κB signaling in myeloid cells such as macrophages and microglia. Herein we sought to examine whether morphine-ADEVs that are taken up by microglial cells could, via the TLR7-NF-κB axis, lead to upregulation of lincRNA-Cox2 expression. To address this, we pretreated mouse primary microglia with either SC-514 (NF-κB inhibitor) or chloroquine (TLR7 inhibitor) followed by stimulation with either control- or morphine-ADEVs isolated from mouse primary astrocytes for 4 hr. Expression of lincRNA-Cox2 was analyzed by real-time PCR. As presented in Figure 4A, expression of lincRNA-Cox2 was significantly increased in microglial cells stimulated with morphine-ADEVs compared with its expression in microglia exposed to control-ADEVs. In microglia pretreated with either SC-514 or chloroquine, morphine-ADEVs failed to induce expression of lincRNA-Cox2 (Figure 4A). To further validate our results, we also assessed lincRNA-Cox2 expression in mouse primary microglia isolated from either C57BL/6N WT or TLR7 KO mice that were stimulated with either control- or morphine-ADEVs from mouse primary astrocytes. As shown in Figure 4B, exposure of WT microglia to morphine-ADEVs resulted in an upregulation of lincRNA-Cox2 expression, and in contrast and as expected, morphine-ADEVs failed to upregulate lincRNA-Cox2 expression in TLR7 KO microglial cells. Next, to determine the site of action of lincRNA-Cox2, we performed RNA fluorescent in situ hybridization (FISH) for lincRNA-Cox2 in microglia. As shown in Figure 4C, FISH analysis revealed nuclear, perinuclear, as well as cytoplasmic expression of lincRNA-Cox2 in microglial cells. Further validation of these findings was done by assessing the expression of lincRNA-Cox2 in subcellular fractions of microglia by real-time PCR. As shown in Figure 4D, lincRNA-Cox2 expression was predominantly localized to the nucleus in the microglia (Figure 4D), and as expected, GAPDH mRNA was localized to the cytosol, and U1 small nuclear RNA (snRNA) was confined to the nucleus. Consistent with previous studies,24 these results indicate that lincRNA-Cox2 could serve as a pivotal mediator regulating the expression of other genes at the level of transcription in microglia.
Figure 4.
Morphine Astrocyte EV-Mediated Induction of lincRNA-Cox2 Involves TLR7 and NF-κB
(A) Mouse primary microglial cells were pre-treated with either TLR7 inhibitor (chloroquine 25 μM) or NF-κB inhibitor (SC-514, 5 μM) for 1 hr, followed by exposure of cells for 4 hr to EVs (2 μg of EVs per 2 × 105 cells) isolated from astrocyte conditioned media. Total RNA isolated from cells was analyzed by real-time PCR for expression of lincRNA-Cox2. (B) Mouse primary microglial cells were isolated from wild-type and TLR7 KO pups, and exposed to EVs isolated from morphine-stimulated astrocyte conditioned media for 4 hr. Total RNA from cells was analyzed by real-time PCR for expression of lincRNA-Cox2. (C) Representative confocal images of ISH assay using a probe specific for lincRNA-Cox2 (red) combined with DAPI staining (blue) and immunostaining for GAPDH (green) in mouse primary microglia. Scale bar, 10 μm. (D) Subcellular fractionation followed by qPCR for lincRNA-Cox2, U1 snRNA, and GAPDH mRNA. All values are shown as the mean ± SEM. *p < 0.05 versus control; #p < 0.05 versus treatment.
Morphine-ADEVs Inhibited Microglial Phagocytosis
To determine the functional effects of induced lincRNA-Cox2 expression in microglia, we first designed lincRNA-Cox2 siRNAs, and the knockdown efficiency of our siRNAs was evaluated by real-time PCR in siRNA-transfected BV-2 cells. As shown in Figure 5A, both lincRNA-Cox2 siRNAs significantly downregulated the expression of lincRNA-Cox2 in microglia. We next performed microarray analysis to compare the gene expression profiles in both control and lincRNA-Cox2 knockdown BV-2 microglia. We have previously reported that lincRNA-Cox2 is a coregulator of transcription factor(s) controlling inflammatory responses in microglia and macrophages.24 The interrelationship between lincRNA-Cox2 and phagocytosis in microglia, however, remains largely unknown. Based on the microarray analysis, in lincRNA-Cox2 knocked-down microglia, we identified several phagocytic genes, including Lrp1, Syk, and Pld2, that were negatively regulated by lincRNA-Cox2 (Figure 5B). These findings were also validated in mouse primary microglia. As shown in Figure 5C, in mouse primary microglia exposed to morphine-ADEVs there was decreased expression of Lrp1, Pld2, and Syk compared with cells exposed to control-ADEVs. Interestingly, knocking down lincRNA-Cox2 significantly increased the expression of Lrp1, Pld2, and Syk in microglia exposed to either control- or morphine-ADEVs compared with microglia transfected with scrambled siRNA. To evaluate the effects of induced lincRNA-Cox2 on phagocytosis, we transfected BV-2 microglial cells with either control or lincRNA-Cox2-specific siRNA for 24 hr, followed by stimulation with either control- or morphine-ADEVs for another 24 hr. The cells were then allowed to internalize fluorescently labeled IgG-opsonized latex beads for 2 hr followed by extensive washes and fixing of cells. Fixed cells were then stained with DAPI and analyzed for phagocytic uptake of the beads using fluorescence microscopy (Figure S2A) and a fluorometric plate reader (Figure S2B). As shown in Figures S2A and S2B, microglia treated with morphine-ADEVs displayed significantly reduced internalization (≥40% inhibition) of IgG-opsonized latex beads compared with cells treated with control-ADEVs. Intriguingly, knockdown of lincRNA-Cox2 promoted internalization of IgG-opsonized latex beads in microglia following interaction with either control- or morphine-ADEVs compared with scrambled siRNA-transfected microglia (Figures S2A and S2B). Next, we sought to validate the role of morphine-ADEVs on microglial phagocytosis. For this, mouse primary microglial cells were transfected with either control- or lincRNA-Cox2 siRNA for 24 hr followed by exposure of cells to PKH26-labeled ADEVs (isolated from mouse primary astrocytes) for an additional 24 hr and subsequent incubation with fluorescent beads for 2 hr. As shown in Figure 2C, there was decreased internalization of the beads (green) in morphine-ADEV (red)-stimulated microglia compared with cells stimulated with control-ADEVs (red). In microglial cells transfected with lincRNA-Cox2 siRNA, however, there was increased internalization of beads (green) in morphine-ADEV (red)-stimulated microglia compared with cells transfected with control- siRNA and exposed to morphine-ADEVs (red). These findings thus underpinned the roles of morphine-ADEVs and lincRNA-Cox2 in microglial phagocytosis. We next sought to examine whether the TLR7-NF-κB signaling pathway was involved in phagocytosis. As presented in Figures 5D and 5E, in the presence of inhibitors of either TLR7 or NF-κB, morphine-ADEVs failed to abrogate microglial phagocytosis. These findings were further confirmed in TLR7 KO microglia. As shown in Figure 5F, morphine-ADEVs failed to inhibit phagocytosis in TLR7 KO microglia. Together, these findings demonstrated that morphine-ADEVs could be taken up by microglia, leading to activation of the TLR7-NF-κB signaling axis. This activation pathway, in turn, resulted in upregulation of lincRNA-Cox2 expression, culminating in impaired microglial phagocytosis.
Figure 5.
Knockdown of lincRNA-Cox2 Restores EV-Mediated Impairment of Microglial Phagocytosis
(A) Real-time PCR for lincRNA-Cox2 in microglia transfected with lincRNA-Cox2 siRNA. (B) Heatmap of differentially expressed phagocytosis-associated genes in lincRNA-Cox2 knockdown BV-2 cells compared with control cells. (C) Mouse primary microglial cells were transfected with lincRNA-Cox2 siRNA for 24 hr, followed by exposure of cells to astrocyte EVs for 4 hr, and subsequently assessed for the expression of Lrp, Pld2, and Syk by qPCR. (D and E) Mouse primary microglial cells were pre-treated with TLR7 inhibitor (chloroquine 25 μM) or NF-κB inhibitor (SC-514, 5 μM) for 1 hr, followed by exposure of cells to astrocyte EVs and subsequent exposure to fluorescent beads for 2 hr. Phagocytosis was measured using (D) fluorescence microscopy (scale bar, 20 μm; high-magnification images are shown in the bottom right corners) and (E) fluorometric plate reader. (F) TLR7 KO microglial cells were exposed to astrocyte-derived EVs and assessed for phagocytosis as described above. All data are presented as mean ± SD. *p < 0.05 versus control; #p < 0.05 versus treatment using Student’s t test.
Knockdown of lincRNA-Cox2 Restored Morphine-Impaired Phagocytic Activity In Vivo
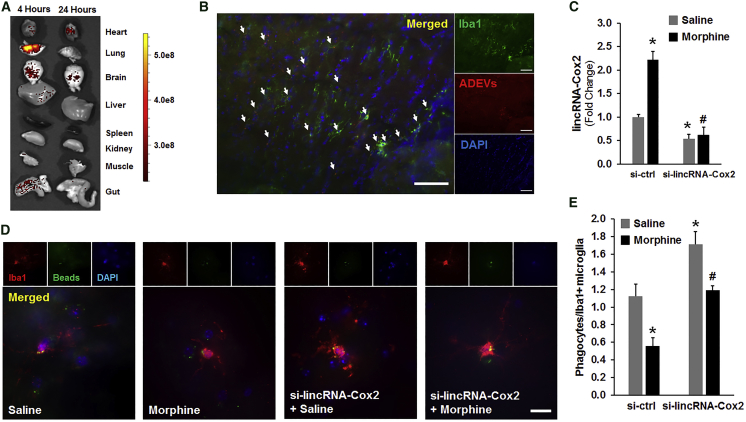
Having determined that knockdown of lincRNA-Cox2 in microglia was able to restore ADEV-mediated morphine-induced phagocytic impairment, we next sought to investigate whether knockdown of lincRNA-Cox2 could also restore microglial phagocytic activity in morphine-administrated mice. Targeting therapeutic drugs to the CNS is a major challenge in the field owing to the tight blood-brain barrier. Intranasal administration is a non-invasive route for drug delivery, which is widely used for the treatment of rhinitis and nasal polyposis.36, 37 In recent times, EVs have not only been recognized for their role as communication vesicles delivering cargo to neighboring or distant cells, but have also been implicated as conduits for efficient delivery of RNA drugs into the brain.20, 38 Before proceeding to test the efficacy of siRNA-loaded EVs in our system, it was first essential to assess the biodistribution of intranasally administered EVs. For this, WT mice were administered EVs labeled with the PKH26-dye intranasally and 4 and 24 hr later, animals were transcardially perfused with ice-cold PBS, followed by tissue harvesting and monitoring for delivery efficacy and biodistribution using the in vivo imaging system (IVIS). As shown in Figure 6A, labeled EVs were primarily localized in the lungs, brain, gut, liver, and heart following 4 hr of EV administration. Interestingly, in 24-hr post-EV administration mice, labeled EVs were found in the brain and heart, but were absent in the lungs and the gut. Next, we sought to examine the uptake of labeled EVs by microglia in sections of striatum using immunohistochemistry for PKH26+-labeled Iba1+ cells. As shown in Figure 6B, there was a significant and specific uptake of labeled EVs by the microglia. The next step was to assess the efficacy of the siRNA-loaded ADEVs in morphine-administered mice. For this, WT mice were given intraperitoneal injections of either saline or morphine (10 mg/kg) twice a day for 5 consecutive days. Simultaneously, we also initiated intranasal delivery of ADEVs (20 μg each time) loaded with either control or lincRNA-Cox2 siRNA to the two groups of mice 1 hr prior to morphine or saline injections. One hour after the last injection on day 5, mice were sacrificed followed by microglial isolation and assessment of lincRNA-Cox2 expression by real-time PCR. As shown in Figure 6C, the expression of lincRNA-Cox2 was significantly upregulated in microglia isolated from morphine-administrated mice compared with the saline-injected controls. Furthermore, intranasal delivery of lincRNA-Cox2 siRNA-loaded ADEVs resulted in a significant knockdown of lincRNA-Cox2 in microglia isolated from morphine-administered mice. The next step was to perform phagocytosis assay on the brain slices from the two groups of mice that were intranasally delivered loaded EVs. As shown in Figures 6D and 6E, there were fewer fluorescent beads phagocytosed in morphine-administrated microglia compared with saline-injected controls, suggesting thereby that morphine administration impaired microglial phagocytosis in the adult brain (Figures 6D and 6E). Intriguingly, intranasal delivery of lincRNA-Cox2 siRNA-loaded EVs restored microglial phagocytic activity in morphine-administrated mice compared with the morphine-administered mice that were treated with control siRNA-loaded EVs.
Figure 6.
Knockdown of lincRNA-Cox2 Ameliorates Morphine-Mediated Impairment of Phagocytic Activity of Microglia In Vivo
(A) PKH26-labeled EVs (20 μg) were intranasally delivered to WT mice (n = 3). Mice were sacrificed 4 and 24 hr posttreatment, and the harvested organs were imaged using a Xenogen IVIS 200 imager. A scale of the radiance efficiency is presented on the right. (B) Immunostaining of the brain sections of mice administered PKH26-labeled EVs for the microglial marker Iba1 (green). Representative micrographs are shown (original magnification ×20). Scale bar, 100 μm. (C) Real-time PCR for lincRNA-Cox2 in the CD11b+/CD45+ dim population of microglia sorted from the brains of various groups of mice. (D) Representative micrograph of brain slices (from 8-week-old mice) incubated with fluorescent beads (green). Scale bar, 20 μm. (E) Quantification of the number of Iba1-positive (red) microglia (with phagocytosed beads) normalized to the total number of Iba1-positive cells. n = 20 fields of view from three mice. All data are presented as mean ± SD; *p < 0.05 versus saline/si-control group; #p < 0.05 versus morphine/si-control group using Student’s t test.
Discussion
EVs play an important role in cell-cell communication among diverse cells types and tissues, including the CNS. The primary function of these vesicles is the delivery of their cargo comprising cellular miRNAs, proteins, and DNA to neighboring as well as distant cells, resulting in impairment of cellular functions.16, 39, 40, 41 For example, in our previous study, we have shown that both morphine and HIV Trans-Activator of Transcription (Tat) protein upregulated miR-29b in ADEVs, which, in turn, resulted in neuronal death. Along these lines, studies by Yelamanchili et al.18 also demonstrated that HIV infection resulting in significant upregulation of miR-21 in blood macrophage-derived EVs in turn directly activated the TLR7-dependent downstream necroptosis pathway. Our miRNA-seq profiling of morphine-ADEVs demonstrating enrichment of miRNAs with an AU- or GU-rich motif critical for direct activation of TLR7 is in keeping with published reports on the role of EV miRNAs in this process.28, 29 The process by which EV-miRNA-mediated activation of TLR7 leads to upregulation of lincRNAs that regulate cellular function(s), however, remains less understood. In the current study, we demonstrate that exposure of mouse microglial cells to morphine-ADEVs resulted in impaired phagocytic function involving the TLR7-NF-κB-lincRNA-Cox2 axis. To determine whether morphine-ADEVs increased lincRNA-Cox2 in vivo, mice were intranasally administered ADEVs isolated from either control- or morphine-stimulated mouse primary astrocytes, and brain sections were assessed for the expression of lincRNA-Cox2 using double in situ hybridization (ISH) and immunostaining for Iba1. Additionally, microglia were also isolated ex vivo by flow cytometry from the mice brains and assessed for the expression of lincRNA-Cox2 by qPCR, and as shown in Figures S3A and S3B, intranasal delivery of morphine-ADEVs increased the expression of lincRNA-Cox2 in microglia. Future studies using human astrocytes and microglia are warranted to provide additional insights in the underlying mechanism(s). Recently, Abud et al.42 have demonstrated that human microglial-like cells (iMGLs) can be generated from iPSCs (induced pluripotent stem cells) and can be used for investigation of microglial functions in various CNS disorders, including substance use disorder. Another advantage of such a model system is that it enables investigating the effect of ADEVs on microglia from the same donor. It must be noted that the miRNA cargo in EVs can also affect the functioning of the recipient cells via miRNA-mRNA interactions. Future IPA (Ingenuity Pathway Analysis) bioinformatic analysis will be essential to understand the complex regulatory networks regulated by the dysregulated miRNAs in morphine-ADEVs. Future efforts will be aimed at downregulating the dysregulated miRs in the microarray screen, which, in turn, could further help establish a link between the EV-TLR7/8 and linc-RNA-Cox2 pathway in the context of microglial phagocytosis.
Phagocytes play a vital role in the maintenance of tissue homeostasis by promoting rapid clearance of apoptotic cells, pathogens, and other insults, leading, in turn, to the elimination of immunogens and prevention or resolution of inflammation.43 Elegant studies by Ninković et al.44 have previously demonstrated impairment of bacterial phagocytosis by macrophages following morphine exposure, and that this effect was mediated by opioid receptors and the subsequent downstream inhibition of actin polymerization and pathogen internalization. These findings thus suggested that the inhibitory effects of morphine on macrophage phagocytosis and pathogen clearance could lead to exacerbation of disease pathogenesis and recovery from a wide range of diseases and injuries. Controversial in vitro studies demonstrating morphine-mediated enhancement of Cryptococcus neoformans and Mycobacterium tuberculosis phagocytosis by human microglia, involving the complement or G protein-coupled opiate receptors, respectively, have also been reported.3, 4 It has been shown that chronic morphine administration can mediate both the activation and repression of phagocytosis-related genes in the striatum of C57BL/6N mice45 (data obtained from NCBI-GEO database accession code GEO: GSE7762, analyzed with GEO2R tool, and presented in Table S2). In the current study, we identified 20 phagocytosis-related genes regulated by lincRNA-Cox2, and out of these, 15 are negatively regulated. We also demonstrate yet another novel function of lincRNA-Cox2, that of negatively regulating microglial phagocytosis using both in vitro and in vivo model systems. Our previous studies shed light on the role of lincRNA-Cox2 as a critical mediator for the development of inflammatory responses in both microglia and macrophages. The current finding of its inhibitory role in microglial phagocytosis has ramifications for therapeutics. Knockdown of lincRNA-Cox2 could thus be considered as a therapeutic strategy aimed at restoring both microglial phagocytic function and mitigating neuroinflammation associated with opiate abuse.
Gene therapy is becoming a promising tool for the treatment of human diseases that cannot be cured by conventional therapies. In fact, RNA-based approaches such as siRNA, antisense RNA, and miRNAs—potent sequence-selective inhibitors of transcription—are rapidly being developed as therapeutics. Elegant studies have demonstrated the use of EVs to successfully deliver siRNAs to specific cell types in vivo in rodents.46 Intranasal administration of exosomes is considered a noninvasive method for rapid delivery of exosome-encapsulated drug(s) to the brain with selective uptake by microglial cells.20, 36, 47, 48 Manipulating EVs with miRNAs, anti-miRNAs, or siRNAs in vivo could thus be envisioned as an efficient means to deliver these molecules to the target cells or organs. Our data clearly demonstrate that exposure of astrocytes to morphine leads to increased release of ADEVs, which upon uptake by the microglia result in upregulated expression of a NF-κB-regulated gene, lincRNA-Cox2, in the recipient cells, ultimately leading to impaired microglial phagocytosis. Furthermore, our in vivo study, for the first time, also demonstrates that intranasal delivery of lincRNA-Cox2 siRNA restored microglial phagocytic activity of morphine-administered mice.
In conclusion, our findings suggest that exposure of microglial cells to morphine-ADEVs results in impaired phagocytic function via the TLR7-NF-κB-lincRNA-Cox2 axis. We also demonstrate that knockdown of lincRNA-Cox2 restores morphine-impaired phagocytic activity in microglia in vivo. These findings have ramifications for the development of EV-loaded RNA drug target(s) as therapeutics for a multitude of neurodegenerative disorders including those associated with opiate abuse.
Materials and Methods
Animals
All animal procedures were performed in strict accordance with the protocols approved by the Institutional Animal Care and Use Committee at the University of Nebraska Medical Center (UNMC) and the NIH. C57BL/6N WT mice were purchased from Charles River Laboratories (Wilmington, MA, USA) and housed under conditions of constant temperature and humidity on a 12-hr light and 12-hr dark cycle, with lights on at 7:00 a.m. Food and water were available ad libitum. Tlr7−/− mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and bred in the UNMC animal facility. Pregnant WT mice were purchased from Charles River Laboratories.
Intranasal Delivery of EVs in Mice
For intranasal administration of EVs, we anesthetized C57BL/6N mice and placed them in a supine position in an anesthesia chamber. EVs (20 μg/100 μL) in saline were administered intranasally as drops with a small pipette every 2 min into alternating sides of the nasal cavity for a total of 10 min. To determine tissue distribution of EVs in vivo, we harvested various tissues and monitored for efficiency of delivery and biodistribution using a Xenogen IVIS 200 imager.
Cell Culture and Cell Lines
The human astrocytic cell line A172 (no. CRL-1620; American Type Culture Collection [ATCC]) was cultured as described previously49 and maintained in DMEM with high glucose containing 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/mL), streptomycin (100 μg/mL), essential amino acids, and vitamins. In this study, A172 cells were used within 30 passages. Human primary astrocytes were obtained from ScienCell Research Laboratories (Carlsbad, CA, USA) and were cultured in DMEM/F12 medium (Invitrogen Life Technologies, Carlsbad, CA, USA) containing 10% heat-inactivated FBS, 2 mM glutamine, sodium bicarbonate, gentamicin, non-essential amino acids, and vitamins.
Mouse primary astrocytes were prepared from whole brains of post-natal (1- to 3-day-old) C57BL/6N mice and plated on poly-D-lysine pre-coated cell culture flasks containing DMEM (10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin). The cells were grown in a humidified atmosphere of 5% CO2/95% air at 37°C. When the astrocytes reached confluence, they were passaged by trypsinization and plated at a density of 106 cells/well on 24-well culture plates in a final volume of 1 mL of DMEM and grown in a humidified atmosphere of 5% CO2/95% air at 37°C. Two days later, the astrocytes were used for experimentation. Immunocytochemical analyses demonstrated that the cultures were comprised of >95% glial fibrillary acidic protein (GFAP)-positive astrocytes.
Mouse primary microglia cells were obtained from 1- to 3-day-old C57BL/6N or TLR7−/− newborn pups, as described previously.50, 51 After digestion and dissociation of the dissected brain cortices in Hank’s buffered salt solution supplemented with trypsin (0.25%), mixed glial cultures were prepared by resuspending the cell suspension in DMEM supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. Cells were plated at 20 × 106 cells/flask density onto 75-cm2 cell culture flasks. The cell medium was replaced every 5 days, and after the first medium change, macrophage colony-stimulating factor (M-CSF; 0.25 ng/mL; PeproTech, Rocky Hill, NJ, USA) was added to the flasks to promote microglial proliferation.
The BV-2 microglial cell line was generously provided by Dr. Sanjay Maggirwar (University of Rochester Medical Center, Rochester, NY, USA). Cells were grown and routinely maintained at 37°C and 5% CO2 in DMEM supplemented with 10% heat-inactivated FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin.
EV Isolation
EVs were prepared from the supernatant of primary astrocytes and A172 cells by differential centrifugations as previously described.16, 26 In brief, conditioned media were harvested, centrifuged at 1,000 × g for 10 min to eliminate cells, and again spun at 10,000 × g for 30 min, followed by filtration through a 0.22-μm filter to remove cell debris. EVs were pelleted by ultracentrifugation (Beckman Ti70 rotor; Beckman Coulter, Brea, CA, USA) at 100,000 × g for 70 min. EVs were assessed for their protein content using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). TSG101 and CD63 were detected by western blot as exosome markers. EVs were further quantified by NTA using a NanoSight (model NS300), as previously reported.26
Cell and EV Transfection
Plasmid and siRNA transfections were performed using Lipofectamine 2000 (catalog no. 11668027; Life Technologies) according to the manufacturer’s instructions. In brief, cells were transfected with plasmid (500 ng) or targeted siRNA (20 pM) mixed with 2 μL of Lipofectamine 2000 diluted in 100 μL of Opti-MEM (catalog no. 31985062; Life Technologies). The resulting siRNA-lipid complexes were added to the cells, incubated for 6 hr, and the medium changed into fresh DMEM. Next, the medium was changed to 10% FBS-containing medium for 20-hr incubation. The transfected cells were then ready for use in experiments. pEF6.mCherry-TSG101 was a gift from Quan Lu (Addgene plasmid 38318).52 CD63-pEGFP was a gift from Paul Luzio (Addgene plasmid 62964). Sequences of mouse Dicer1 siRNA oligonucleotides used in this study were: mouse Dicer1-siS1, 5′-GrUrGrUrCrArUrCrUrUrGrCrGrArUrUrCrUrArUrUr-3′; mouse Dicer1-si AS1, 5′-UrArGrArArUrCrGrCrArArGrArUrGrArCrArCrUrUr-3′; mouse Dicer1-siS1, 5′-CrCrArArCrUrArCrCrUrCrArUrArUrCrCrCrArUrUr-3′; mouse Dicer1-si AS2, 5′-UrGrGrGrArUrArUrGrArGrGrUrArGrUrUrGrGrUrUr-3′.
EVs were transfected with siRNA using Exo-Fect Exosome Transfection Reagent (SBI; System Biosciences) according to the manufacturer’s instructions. Sequences of mouse lincRNA-Cox2 siRNA oligonucleotides used in this study were: Control-(si)S1, 5′-rUrArArGrGrCrUrArUrGrArArGrArGrArUrArCrUrU-3′; Control-(si)AS1, 5′-rGrUrArUrCrUrCrUrUrCrArUrArGrCrCrUrUrArUrU-3′; lincRNA-Cox2-(si)S1, 5′-rGrCrCrCrUrArArUrArArGrUrGrGrGrUrUrGrUrUrU-3′; lincRNA-Cox2-(si)AS1, 5′-rArCrArArCrCrCrArCrUrUrArUrUrArGrGrGrCrUrU-3′; lincRNA-Cox2-(si)S2, 5′-rGrArGrArGrArGrArGrUrGrUrArCrUrUrArArArUrU-3′; lincRNA-Cox2-(si)AS2, 5′-rUrUrUrArArGrUrArCrArCrUrCrUrCrUrCrUrCrUrU-3′.
Western Blotting
Treated cells or EVs were lysed using the Mammalian Cell Lysis kit (Sigma, St. Louis, MO, USA) and quantified using the micro BCA Protein Assay kit (Pierce, Rockford, IL, USA). Equal amounts of the corresponding proteins were electrophoresed in an SDS-polyacrylamide gel (10%–12%) under reducing conditions followed by transfer to polyvinylidene fluoride (PVDF) membranes. The blots were blocked with 5% non-fat dry milk in PBS. Western blots were then probed with antibodies recognizing the CD63 antibody (1:1,000; ab216130; Abcam, Cambridge, MA, USA), TSG101 (1:1,000; ab125011; Abcam, Cambridge, MA, USA), NF-κB p65 (1:2,000; ab16502; Abcam, Cambridge, MA, USA), histone H3 (1:1,000; catalog no. 9715S; Cell Signaling Technology, Danvers, MA, USA), and β-actin (1:4,000; A5316; Sigma, St. Louis, MO, USA). The secondary antibodies were alkaline phosphatase-conjugated to goat anti-mouse/rabbit IgG (1:5,000). Signals were detected by chemiluminescence and imaged on the FLA-5100 (Fujifilm, Valhalla, NY, USA) digital image scanner; densitometry was performed utilizing ImageJ software (NIH).53
Electron Microscopy
EV pellets were prepared for negative staining employing a slightly modified procedure. Using wide-bore tips, 3 μL of EV pellet was gently placed on 200-mesh Formvar-coated copper grids, allowed to adsorb for 4–5 min, and processed for standard uranyl acetate staining. In the last step, the grid was washed with three changes of PBS and allowed to semi-dry at room temperature before observation in TEM (Hitachi H7500 TEM; Hitachi, Tokyo, Japan).
Atomic Force Microscopy
The 1-(3-aminopropyl) silatrane (APS) mica functionalization procedure was used, in which freshly cleaved mica is treated with APS, as previously described.54, 55 Twenty microliters (8.3 × 109 EVs/mL) of EV sample was deposited on APS mica for 20 min at room temperature. Then 200 μL of the PBS buffer was added to the sample. The sample was then subjected to AFM imaging. AFM imaging of EVs was carried out using the Asylum Research MFP3D (Santa Barbara, CA, USA) instrument. Imaging was performed in tapping mode at room temperature. An MSNL probe with cantilever “E” (Bruker Corporation) was employed for imaging. The nominal spring constant of the MSNL “E” cantilevers was ∼0.1 N/m.
RNA Isolation and Sequencing
RNA was extracted from EVs, which were isolated from conditioned media of human primary astrocytes of three donors. RNA samples were then shipped on dry ice to LC Sciences (Houston, TX, USA) for miRNA sequencing.
Real-Time PCR
For quantitative analysis of mRNA expression, comparative real-time PCR was performed with the use of the SYBR Green PCR Master Mix (Applied Biosystems). The sequences for the amplification of lincRNA-Cox2 were: 5′-AGTATGGGATAACCAGCTGAGGT-3′ (forward) and 5′- GAATGCTGAGAGTGGGAGAAATAG-3′ (reverse); the primer sequences for the amplification of U1 were as follows: 5′-CCATGATCACGAAGGTGGTTT-3′ (forward); 5′-TATGCAGTCGAGTTTCCCGC-3′ (reverse); and the primer sequences for the amplification of GAPDH were as follows: 5′-TGCACCACCAACTGCTTAGC-3′ (forward); 5′-ATGCCAGTGAGCTTCCCGTT-3′ (reverse). All reactions were run in triplicate.
Immunofluorescence
Cells grown on sterile coverslips were incubated with TSG101-mCherry/CD63-EGFP-labeled EVs at 40% confluency. After 20–30 min, cells were washed extensively with PBS and then fixed with 4% paraformaldehyde for 20 min at room temperature followed by permeabilization with 0.3% Triton X-100 in PBS for 30 min. The samples were incubated with a blocking buffer containing 10% goat serum (NGS) in PBS for 2 hr at room temperature followed by addition of rabbit anti-Iba1 antibody (1:1,000; Wako Laboratory Chemicals) or rabbit anti-NF-κB p65 antibody (1:1,000; Abcam) or rabbit anti-EEA1 antibody (1:200; Cell Signaling Technology) and incubated overnight at 4°C. Primary Abs were labeled with secondary anti-rabbit Abs conjugated to the fluorescent probes Alexa Fluor 488 or Alexa Fluor 594, and nuclei were labeled with DAPI.
Candidate TLR7/8 RNA Agonists miRNA Search and Motif Prediction
To identify the potential mouse miRNAs that are capable of functioning as the TLR7/8 RNA agonists, we performed an unbiased search as follows: first, all known mouse mature miRNAs were collected from miRBase30 (Version 21), and the mouse EV-miRNAs were also obtained from two EV databases: ExoCarta31 and EVpedia.32 Next, previously reported 15 AU- and GU-rich 4-mers from the Forsbach et al.27 study were scanned for all known mouse miRNAs. Finally, the EV-associated miRNAs that contained the reported AU- and GU-rich 4-mers were considered as TLR7/8 RNA agonists. Furthermore, sequence motif analysis was performed based on the selected candidates using the tool MDS2.33
Cell Fractionation
Cells were fractionated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit to generate cytosolic and nuclear pools as described by the manufacturer (Thermo Fisher).
lincRNA In Situ Hybridization
In situ hybridization for lincRNA was performed as described previously.56 Biotin-labeled antisense DNA probes (5′-catggtttctcagaatgtcc-3′, 5′-gcaacagtttcattcacagc-3′, 5′-aactgaagctttcactctcc-3′, 5′-gcaaatcacagtcagttgct-3′, 5′-gactacttcctgctgattag-3′, and 5′-ctggataggactatttctcc-3′) were designed using the online probe designer at http://www.singlemoleculefish.com and synthesized by Integrated DNA Technologies. The cells were fixed in 4% paraformaldehyde for 10 min, and washed and permeabilized with 0.25% Triton X-100 in PBS for 15 min. Next, the slides were washed in PBS three times, followed by incubation in hybridization buffer for 1 hr. Subsequently, the DNA probes in hybridization buffer were added and incubated in a humid chamber overnight at 55°C. The next day, the slides were washed in 2 × saline-sodium citrate (SSC) three times for 2 min each at 42°C and 0.2 × SSC three times for 2 min each at 42°C. Blocking was performed with blocking solution (3% NGS + 1% BSA in PBS) at room temperature for 2 hr and then incubated with horseradish peroxidase (HRP)-labeled reagent (SA-HRP) and cell-type marker at 4°C overnight in a humidified chamber. On the third day, slides were washed three times for 5 min each in TBS at room temperature with agitation. Slides were incubated in streptavidin-HRP for 30 min at room temperature, then washed three times for 5 min each in TBS buffer at room temperature. After that the slides were incubated with the appropriate labeled secondary antibody in 1% BSA, 1% NGS diluted in PBS at room temperature for 1 hr. Then the slides were washed in 0.1% Tween 20 in TBS three times for 5 min each at room temperature. The slides were then mounted using Prolong gold anti-fade reagent with DAPI (Invitrogen).
Adult Microglia Isolation
Microglia were isolated from whole-brain homogenates by Percoll gradient centrifugation according to previous reports50, 51 with slight modifications. In brief, the brains were homogenized in PBS (pH 7.4) by passing through a 70-μm nylon cell strainer. Resulting homogenates were centrifuged at 600 × g for 6 min. Supernatants were removed and cell pellets were resuspended in 70% isotonic Percoll (GE Healthcare, Uppsala, Sweden) at room temperature. A discontinuous Percoll density gradient was layered as follows: 70%, 50%, 35%, and 0% isotonic Percoll. The gradient was centrifuged for 20 min at 2,000 × g, and microglia were collected from the interphase between the 70% and 50% Percoll layers. Cells were washed and then resuspended in sterile PBS followed by flow cytometry analysis by gating the myeloid cells for the CD11b+/CD45dim population.
Phagocytosis Assay
The phagocytic activity of adult microglia of live brain slices was analyzed as described previously.57, 58 Brains from 8-week-old mice were washed in carbogen-saturated (95% O2 and 5% CO2) artificial cerebrospinal fluid (ACSF) containing (in mM): NaCl 126; KCl 2.5; MgSO4 1.3; CaCl2 2.5; NaH2PO4 1.25; NaHCO3 26; and D-glucose 10 (pH 7.4; all from Sigma). Coronal slices (130 μm) were prepared using a Vibratome (Microm, Walldorf, Germany) at 4°C and allowed to rest in ACSF buffer at room temperature for 1 hr before incubation with fluorescein isothiocyanate (FITC)-labeled rabbit IgG-coated latex beads 0.1 μm mean particle size (1:100; Cayman Chemical) for 60 min at 37°C. The slices were washed and fixed with 4% paraformaldehyde. To visualize microglia, slices were permeabilized (0.25% Triton X-100, 1% BSA, and 10% goat serum in PBS) and incubated with anti-Iba1 (1:750; Wako), followed by goat anti-rabbit Alexa 488 (1:250; Invitrogen) and Hoechst 33258 (1:10,000; Sigma-Aldrich). Fluorescent images were acquired at room temperature on a Zeiss Observer Z1 microscope (Carl Zeiss, Germany); images were processed using AxioVs 40 4.8.0.0 software (Carl Zeiss MicroImaging). Photographs were acquired using an AxioCam MRm digital camera (Carl Zeiss, Germany).
To investigate the ability of microglia to internalize particles, microglial cells (2 × 105 cells/mL) were plated on a 24-well plate and allowed to adhere overnight, and then incubated with FITC-labeled rabbit IgG-coated latex beads as described above, intensively washed, and finally visualized at ×20 magnification with a microscope.
Statistical Analysis
Statistical analysis was performed using Student’s t test or one-way ANOVA followed by Holm-Sidak test (SigmaPlot 11.0). The appropriate test was clarified in the figure legends. Results were judged statistically significant if p < 0.05 by ANOVA.
Author Contributions
G.H. and S.B. designed the experiments and wrote the manuscript. K.L., F.N., L.Y., B.W.D., C.T., Z.S., and M.K. performed the experiments. J.S., J.C., and G.H. performed bioinformatics analysis. S.C., Y.L.L., X.-M.C., G.H., and S.B. analyzed data.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This work was supported by grants DA041751, DA043164, MH112848, DA040397, DA043138 (to S.B.), and DA042704 and DA046831 (to G.H.) from the NIH. The support of the Nebraska Center for Substance Abuse Research is acknowledged. The project described was also supported by the NIH, National Institute of Mental Health (grant 2P30MH062261). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.019.
Contributor Information
Guoku Hu, Email: guoku.hu@unmc.edu.
Shilpa Buch, Email: sbuch@unmc.edu.
Supplemental Information
References
- 1.Fields H.L. The doctor’s dilemma: opiate analgesics and chronic pain. Neuron. 2011;69:591–594. doi: 10.1016/j.neuron.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fields H.L., Margolis E.B. Understanding opioid reward. Trends Neurosci. 2015;38:217–225. doi: 10.1016/j.tins.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipovsky M.M., Gekker G., Hu S., Hoepelman A.I., Peterson P.K. Morphine enhances complement receptor-mediated phagocytosis of Cryptococcus neoformans by human microglia. Clin. Immunol. Immunopathol. 1998;87:163–167. doi: 10.1006/clin.1998.4518. [DOI] [PubMed] [Google Scholar]
- 4.Peterson P.K., Gekker G., Hu S., Sheng W.S., Molitor T.W., Chao C.C. Morphine stimulates phagocytosis of Mycobacterium tuberculosis by human microglial cells: involvement of a G protein-coupled opiate receptor. Adv. Neuroimmunol. 1995;5:299–309. doi: 10.1016/0960-5428(95)00020-3. [DOI] [PubMed] [Google Scholar]
- 5.Roy S., Ninkovic J., Banerjee S., Charboneau R.G., Das S., Dutta R., Kirchner V.A., Koodie L., Ma J., Meng J., Barke R.A. Opioid drug abuse and modulation of immune function: consequences in the susceptibility to opportunistic infections. J. Neuroimmune Pharmacol. 2011;6:442–465. doi: 10.1007/s11481-011-9292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergsbaken T., Fink S.L., Cookson B.T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadok V.A., Chimini G. The phagocytosis of apoptotic cells. Semin. Immunol. 2001;13:365–372. doi: 10.1006/smim.2001.0333. [DOI] [PubMed] [Google Scholar]
- 8.Uribe-Querol E., Rosales C. Control of phagocytosis by microbial pathogens. Front. Immunol. 2017;8:1368. doi: 10.3389/fimmu.2017.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segawa K., Nagata S. An apoptotic ‘eat me’ signal: phosphatidylserine exposure. Trends Cell Biol. 2015;25:639–650. doi: 10.1016/j.tcb.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Zagórska A., Través P.G., Lew E.D., Dransfield I., Lemke G. Diversification of TAM receptor tyrosine kinase function. Nat. Immunol. 2014;15:920–928. doi: 10.1038/ni.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amor S., Puentes F., Baker D., van der Valk P. Inflammation in neurodegenerative diseases. Immunology. 2010;129:154–169. doi: 10.1111/j.1365-2567.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass C.K., Saijo K., Winner B., Marchetto M.C., Gage F.H. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu R., Shen Q., Xu P., Luo J.J., Tang Y. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 2014;49:1422–1434. doi: 10.1007/s12035-013-8620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abiega O., Beccari S., Diaz-Aparicio I., Nadjar A., Layé S., Leyrolle Q., Gómez-Nicola D., Domercq M., Pérez-Samartín A., Sánchez-Zafra V. Neuronal hyperactivity disturbs ATP microgradients, impairs microglial motility, and reduces phagocytic receptor expression triggering apoptosis/microglial phagocytosis uncoupling. PLoS Biol. 2016;14:e1002466. doi: 10.1371/journal.pbio.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickens A.M., Tovar-Y-Romo L.B., Yoo S.W., Trout A.L., Bae M., Kanmogne M., Megra B., Williams D.W., Witwer K.W., Gacias M. Astrocyte-shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions. Sci. Signal. 2017;10 doi: 10.1126/scisignal.aai7696. eaai7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu G., Yao H., Chaudhuri A.D., Duan M., Yelamanchili S.V., Wen H., Cheney P.D., Fox H.S., Buch S. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis. 2012;3:e381. doi: 10.1038/cddis.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu G., Yang L., Cai Y., Niu F., Mezzacappa F., Callen S., Fox H.S., Buch S. Emerging roles of extracellular vesicles in neurodegenerative disorders: focus on HIV-associated neurological complications. Cell Death Dis. 2016;7:e2481. doi: 10.1038/cddis.2016.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yelamanchili S.V., Lamberty B.G., Rennard D.A., Morsey B.M., Hochfelder C.G., Meays B.M., Levy E., Fox H.S. miR-21 in extracellular vesicles leads to neurotoxicity via TLR7 signaling in SIV neurological disease. PLoS Pathog. 2015;11:e1005032. doi: 10.1371/journal.ppat.1005032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krämer-Albers E.M. Ticket to ride: targeting proteins to exosomes for brain delivery. Mol. Ther. 2017;25:1264–1266. doi: 10.1016/j.ymthe.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang X., Xiang X., Grizzle W., Sun D., Zhang S., Axtell R.C., Ju S., Mu J., Zhang L., Steinman L. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011;19:1769–1779. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs J.A., Wolvetang E.J., Mattick J.S., Rinn J.L., Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–877. doi: 10.1016/j.neuron.2015.09.045. [DOI] [PubMed] [Google Scholar]
- 22.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong Q., Gong A.Y., Zhang X.T., Lin C., Ma S., Chen J., Hu G., Chen X.M. LincRNA-Cox2 modulates TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 2016;30:1187–1197. doi: 10.1096/fj.15-279166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu G., Gong A.Y., Wang Y., Ma S., Chen X., Chen J., Su C.J., Shibata A., Strauss-Soukup J.K., Drescher K.M., Chen X.M. LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J. Immunol. 2016;196:2799–2808. doi: 10.4049/jimmunol.1502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sofroniew M.V., Vinters H.V. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu G., Gong A.Y., Roth A.L., Huang B.Q., Ward H.D., Zhu G., Larusso N.F., Hanson N.D., Chen X.M. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog. 2013;9:e1003261. doi: 10.1371/journal.ppat.1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forsbach A., Nemorin J.G., Montino C., Müller C., Samulowitz U., Vicari A.P., Jurk M., Mutwiri G.K., Krieg A.M., Lipford G.B., Vollmer J. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann S.M., Krüger C., Park B., Derkow K., Rosenberger K., Baumgart J., Trimbuch T., Eom G., Hinz M., Kaul D. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 29.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathivanan S., Fahner C.J., Reid G.E., Simpson R.J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D.K., Lee J., Kim S.R., Choi D.S., Yoon Y.J., Kim J.H., Go G., Nhung D., Hong K., Jang S.C. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31:933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao T., Shu J., Cui J. A systematic approach to RNA-associated motif discovery. BMC Genomics. 2018;19:146. doi: 10.1186/s12864-018-4528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawai T., Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grassin-Delyle S., Buenestado A., Naline E., Faisy C., Blouquit-Laye S., Couderc L.J., Le Guen M., Fischler M., Devillier P. Intranasal drug delivery: an efficient and non-invasive route for systemic administration: focus on opioids. Pharmacol. Ther. 2012;134:366–379. doi: 10.1016/j.pharmthera.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Bitter C., Suter-Zimmermann K., Surber C. Nasal drug delivery in humans. Curr. Probl. Dermatol. 2011;40:20–35. doi: 10.1159/000321044. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 39.Roth W.W., Huang M.B., Addae Konadu K., Powell M.D., Bond V.C. Micro RNA in exosomes from HIV-infected macrophages. Int. J. Environ. Res. Public Health. 2015;13:h13010032. doi: 10.3390/ijerph13010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konadu K.A., Chu J., Huang M.B., Amancha P.K., Armstrong W., Powell M.D., Villinger F., Bond V.C. Association of cytokines with exosomes in the plasma of HIV-1-seropositive individuals. J. Infect. Dis. 2015;211:1712–1716. doi: 10.1093/infdis/jiu676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang N., Sun B., Gupta A., Rempel H., Pulliam L. Monocyte exosomes induce adhesion molecules and cytokines via activation of NF-κB in endothelial cells. FASEB J. 2016;30:3097–3106. doi: 10.1096/fj.201600368RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abud E.M., Ramirez R.N., Martinez E.S., Healy L.M., Nguyen C.H.H., Newman S.A., Yeromin A.V., Scarfone V.M., Marsh S.E., Fimbres C. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arandjelovic S., Ravichandran K.S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 2015;16:907–917. doi: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ninković J., Roy S. Morphine decreases bacterial phagocytosis by inhibiting actin polymerization through cAMP-, Rac-1-, and p38 MAPK-dependent mechanisms. Am. J. Pathol. 2012;180:1068–1079. doi: 10.1016/j.ajpath.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korostynski M., Piechota M., Kaminska D., Solecki W., Przewlocki R. Morphine effects on striatal transcriptome in mice. Genome Biol. 2007;8:R128. doi: 10.1186/gb-2007-8-6-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Boorn J.G., Schlee M., Coch C., Hartmann G. SiRNA delivery with exosome nanoparticles. Nat. Biotechnol. 2011;29:325–326. doi: 10.1038/nbt.1830. [DOI] [PubMed] [Google Scholar]
- 47.Lakhal S., Wood M.J. Intranasal exosomes for treatment of neuroinflammation? Prospects and limitations. Mol. Ther. 2011;19:1754–1756. doi: 10.1038/mt.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visweswaraiah A., Novotny L.A., Hjemdahl-Monsen E.J., Bakaletz L.O., Thanavala Y. Tracking the tissue distribution of marker dye following intranasal delivery in mice and chinchillas: a multifactorial analysis of parameters affecting nasal retention. Vaccine. 2002;20:3209–3220. doi: 10.1016/s0264-410x(02)00247-5. [DOI] [PubMed] [Google Scholar]
- 49.Bethel-Brown C., Yao H., Hu G., Buch S. Platelet-derived growth factor (PDGF)-BB-mediated induction of monocyte chemoattractant protein 1 in human astrocytes: implications for HIV-associated neuroinflammation. J. Neuroinflammation. 2012;9:262. doi: 10.1186/1742-2094-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao K., Guo M., Niu F., Yang L., Callen S.E., Buch S. Cocaine-mediated induction of microglial activation involves the ER stress-TLR2 axis. J. Neuroinflammation. 2016;13:33. doi: 10.1186/s12974-016-0501-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao H., Ma R., Yang L., Hu G., Chen X., Duan M., Kook Y., Niu F., Liao K., Fu M. MiR-9 promotes microglial activation by targeting MCPIP1. Nat. Commun. 2014;5:4386. doi: 10.1038/ncomms5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nabhan J.F., Hu R., Oh R.S., Cohen S.N., Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banerjee S., Sun Z., Hayden E.Y., Teplow D.B., Lyubchenko Y.L. Nanoscale dynamics of amyloid β-42 oligomers as revealed by high-speed atomic force microscopy. ACS Nano. 2017;11:12202–12209. doi: 10.1021/acsnano.7b05434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyubchenko Y.L., Shlyakhtenko L.S., Ando T. Imaging of nucleic acids with atomic force microscopy. Methods. 2011;54:274–283. doi: 10.1016/j.ymeth.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaudhuri A.D., Yelamanchili S.V., Fox H.S. Combined fluorescent in situ hybridization for detection of microRNAs and immunofluorescent labeling for cell-type markers. Front. Cell. Neurosci. 2013;7:160. doi: 10.3389/fncel.2013.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minami S.S., Min S.W., Krabbe G., Wang C., Zhou Y., Asgarov R., Li Y., Martens L.H., Elia L.P., Ward M.E. Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med. 2014;20:1157–1164. doi: 10.1038/nm.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krabbe G., Halle A., Matyash V., Rinnenthal J.L., Eom G.D., Bernhardt U., Miller K.R., Prokop S., Kettenmann H., Heppner F.L. Functional impairment of microglia coincides with Beta-amyloid deposition in mice with Alzheimer-like pathology. PLoS ONE. 2013;8:e60921. doi: 10.1371/journal.pone.0060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.