Abstract
Background
In addition to its outstanding cellulase production ability, Trichoderma reesei produces a wide variety of valuable secondary metabolites, the production of which has not received much attention to date. Among them, sorbicillinoids, a large group of hexaketide secondary metabolites derived from polyketides, are drawing a growing interest from researchers because they exhibit a variety of important biological functions, including anticancer, antioxidant, antiviral, and antimicrobial properties. The development of fungi strains with constitutive, hyperproduction of sorbicillinoids is thus desired for future industry application but is not well-studied. Moreover, although T. reesei has been demonstrated to produce sorbicillinoids with the corresponding gene cluster and biosynthesis pathway proposed, the underlying molecular mechanism governing sorbicillinoid biosynthesis remains unknown.
Results
Recombinant T. reesei ZC121 was constructed from strain RUT-C30 by the insertion of the gene 12121-knockout cassette at the telomere of T. reesei chromosome IV in consideration of the off-target mutagenesis encountered during the unsuccessful deletion of gene 121121. Strain ZC121, when grown on cellulose, showed a sharp reduction of cellulase production, but yet a remarkable enhancement of sorbicillinoids production as compared to strain RUT-C30. The hyperproduction of sorbicillinoids is a constitutive process, independent of culture conditions such as carbon source, light, pH, and temperature. To the best of our knowledge, strain ZC121 displays record sorbicillinoid production levels when grown on both glucose and cellulose. Sorbicillinol and bisvertinolone are the two major sorbicillinoid compounds produced. ZC121 displayed a different morphology and markedly reduced sporulation compared to RUT-C30 but had a similar growth rate and biomass. Transcriptome analysis showed that most genes involved in cellulase production were downregulated significantly in ZC121 grown on cellulose, whereas remarkably all genes in the sorbicillinoid gene cluster were upregulated on both cellulose and glucose.
Conclusion
A constitutive sorbicillinoid-hyperproduction strain T. reesei ZC121 was obtained by off-target mutagenesis, displaying an overwhelming shift from cellulase production to sorbicillinoid production on cellulose, leading to a record for sorbicillinoid production. For the first time, T. reesei degraded cellulose to produce platform chemical compounds other than protein in high yield. We propose that the off-target mutagenesis occurring at the telomere region might cause chromosome remodeling and subsequently alter the cell structure and the global gene expression pattern of strain ZC121, as shown by phenotype profiling and comparative transcriptome analysis of ZC121. Overall, T. reesei ZC121 holds great promise for the industrial production of sorbicillinoids and serves as a good model to explore the regulation mechanism of sorbicillinoids’ biosynthesis.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1296-4) contains supplementary material, which is available to authorized users.
Keywords: Biosynthetic gene cluster, Secondary metabolites, Natural product, Yellow pigment, Sorbicillinoids, Trichoderma reesei
Background
Secondary metabolites are structurally heterogeneous, highly bioactive, and low-molecular weight compounds synthesized by bacteria, fungi, algae, plants, and animals [1–5]. Unlike primary metabolites, they are not directly essential for the growth of the corresponding production organism but can broaden the inhabitable environments or beat other competitive organisms in a given ecological niche [6]. Most secondary metabolites are derived from either nonribosomal peptides (NRPs) or polyketides (PKSs) or both mixed together, whereas others are derived from terpenes or fatty acids [7]. These molecules include important pharmaceuticals (e.g., penicillin, cyclosporin and statins), potent toxins (e.g., aflatoxins and trichothecenes) and Janus-faced compounds (e.g., ergot alkaloids), holding both economical and health implications for humans [8].
Sorbicillinoids, also termed “yellow pigment,” are derived from PKS and a large group of hexaketide secondary metabolites and include the cyclization on the carboxylate terminus. Sorbicillinoids are produced and secreted by both marine and terrestrial ascomycetes, including Trichoderma [9], Aspergillus [10], Penicillium [11], Streptomyces [1], Acremonium [12], Paecilomyces [13], and Eurotiomycete [14]. Most of these compounds possess the characteristic C1–C6 sorbyl sidechain and bi- or tri-cyclic frameworks that are extremely complex and highly oxygenated. Based on their structure, sorbicillinoids fall into four classes: monomeric sorbicillinoids, bisorbicillinoids, trisorbicillinoids and hybrid sorbicillinoids. They have a variety of biological activities, including anticancer [15], antioxidant [16], antiviral [17] and antimicrobial [18], showing promising applications in the agriculture, pharmaceutical, and food industries. Therefore, these yellow pigments have attracted considerable interest. Historically, however, most work was aimed at eliminating yellow pigments from fungal fermentation cultures for the production of products like β-lactams [19] and cellulase [9].
Studies of sorbicillinoids have been performed primarily with Trichaderma [9] and Penicillium species [11]. As a well-known industrial strain for the production of cellulases, hemicellulases, and recombinant proteins [20], T. reesei is also a rich source of secondary metabolites [21, 22], a point which often is overlooked. It has been observed that T. reesei forms yellow pigments during growth [23, 24], which have been identified as a mixture of sorbicillin, sorbicillinol and sorbicillinoids [9, 25, 26]. More recently, the biosynthesis pathway of sorbicillinoids in P. chrysogenum [27] and T. reesei [25] has been proposed, wherein all the related genes are clustered on the genome the same way as most genes involved generally in secondary metabolites production [9, 19, 25]. This cluster also contains two transcriptional factors, YPR1 and YRP2, and a transporter [9]. Still, the biosynthesis mechanism of sorbicillinoids remains obscure and strain improvement for sorbicillinoids’ hyperproduction has not been reported.
In this study, the recombinant T. reesei strain ZC121 was obtained by off-target mutagenesis resulting from the unsuccessful deletion of gene 121121. Strain ZC121 was found to produce only a small amount of cellulase and hemicellulase, but a record yield of sorbicillinoids. The effect of culture conditions on this sorbicillinoids’ hyperproduction was determined, including carbon source, light, pH and temperature. Furthermore, identification of the off-target mutagenesis, phenotype profiling and comparative transcriptional profiling were carried out to reveal the molecular mechanism(s) underlying sorbicillinoid hyperproduction of ZC121.
Materials and methods
Materials
Construction and propagation of plasmids were performed in Escherichia coli DH5α. Agrobacterium tumefaciens AGL-1 served as a T-DNA donor for transformation of T. reesei RUT-C30 (CICC 13052) [28]. E. coli DH5α and A. tumefaciens AGL-1 were cultured in Luria–Bertani (LB) with 220 rpm at 37 °C and 28 °C, respectively. T. reesei RUT-C30 and its derivatives were cultured on potato dextrose agar (PDA) plates at 28 °C with mixing at 200 rpm for conidia production and in Trichoderma minimal media (TMM) [29] with 2% (w/t) cellulose or other carbon sources (as indicated) for cellulase and sorbicillinoid production. Plasmid pXBthg was employed to construct plasmid pXBthg-121121 (Additional file 1: Fig. S1) [30]. The primers used in this study can be found in Additional file 1: Table S1. Fifty μg/mL of hygromycin B was utilized as the selection marker. All chemicals used in this study were purchased from Sigma-Aldrich, USA.
Strain construction
Genomic DNA was extracted from T. reesei cells grown in sabouraud dextrose broth (SDB) medium for 48 h at 28 °C using the E-Z 96 Fungal DNA Kit (Omega Bio-tek, Germany). The 1500-bp upstream or downstream region abutting gene 121121 (Additional file 1: Fig. S2) were amplified from the prepared DNA template, respectively. The 1500-bp downstream fragment was cloned into plasmid pXBthg at BamHI using ClonExpress™ II One Step Cloning Kit (Vazyme, China), which was followed by the cloning of the 1500-bp upstream one at XhoI, resulting in the plasmid pXBthg-121121 (Additional file 1: Fig. S1). pXBthg-121121 was then introduced into T. reesei RUT-C30 by the Agrobacterium tumefaciens-mediated transformation (AMT) method [28]. Four transformants ZC121-1, ZC121-2, ZC121-3, and ZC121-4 were obtained after selection on PDA plates containing 50 μg/mL hygromycin B and 200 μM cefotaxime.
Shake flask cultivation
Five percent (v/v) 107/mL conidia from T. reesei grown on PDA plates at 28 °C for 7 days were inoculated into 10 mL SDB and incubated at 28 °C with mixing at 200 rpm for 2 days. Ten percent (v/v) pre-grown mycelia were inoculated into 50 mL TMM media (pH 6) with 2% cellulose, lactose, glucose, galactose, or glycerol, and then incubated at 28 °C with mixing at 200 rpm for 5 days. A 0.5 mL culture sample was taken every 12 h. The samples were centrifuged at 14,000×g for 10 min at 4 °C and the supernatants were filtrated with 0.22 µm filter membranes and stored at − 80 °C for sorbicillinoid analysis and the cellulase activity assay.
The absorbance at 370 nm of the prepared supernatant as mentioned above was recorded using a UV spectrophotometer to determine the amount of sorbicillinoids present [31]. To determine the exact yield of sorbicillinoids, the prepared supernatant was dried at 40 °C overnight. Methanol was used to extract the sorbicillinoids from the dried supernatant powder several times until the powder became white. The supernatant containing sorbicillinoids in methanol was collected after centrifuging and methanol was removed by rotary evaporation. The crude sorbicillinoids product was then obtained and utilized to determine the standard curve (Additional file 1: Fig. S3) for the yield calculation of sorbicillinoids.
The composition of the crude sorbicillinoids product from T. reesei ZC121 and RUT-C30 were analyzed using LC–MS system (G2-XS QTof, Waters) coupled with a UPLC column (2.1 × 100 mm ACQUITY UPLC BEH C18 column containing 1.7 μm particles). Reserpine was used as the internal standard [32]. Two microlitre samples dissolved in methanol were injected onto the C18 column at a flow rate of 0.4 mL/min. The gradient for elution was 2% buffer B (0.1% formic acid in water) for 0.5 min, 2–20% buffer B for 5 min, 20–95% buffer B for 6 min, and finally 95% buffer B for 2 min. Mass spectrometry was operated using the electrospray source in positive ion mode with MSe acquisition according to the following settings: the selected mass ranged from 50 to 1200 m/z and leucine enkephalin (m/z 556.2771) was used as the “lock mass option” for recalibration. The ionization parameters were set as follows: capillary voltage was 2.5 kV, collision energy was 40 eV, source temperature was 120 °C, and the desolvation gas temperature was 400 °C. Data acquisition and processing were performed using Masslynx 4.1.
Analysis methods
For cellulase activity assay, biomass measurement, RT-qPCR analysis and whole genome resequencing, please refer to our previous studies [33, 34]. DNA content was determined according to our previous study [34].
Microscopy observation
Mycelia of T. reesei RUT-C30 and ZC121 were placed onto a glass slide, covered with a cover glass, and observed with an inverted confocal laser scanning microscope SP8 (Leica, Germany) equipped with a 100 × 1.4 NA oil-immersion objective. The excitation wavelength used was 405 nm and the emission was checked in the range of 415–495 nm.
Sporulation assay
Trichoderma reesei was cultivated on PDA plates at 28 °C for 5 days to obtain mature mycelia. A portion of the mature mycelia was inoculated onto another PDA plate and incubated at 28 °C for 5 days. The spores were then collected using 4 mL of sterilized 0.02% Tween 80. The spores were counted using a hemocytometer and light microscope.
RNA-seq analysis
The total RNA of T. reesei RUT-C30 was extracted with the RNA extraction Kit (Omega Bio-tek, Germany). The purity, concentration and integrity of the isolated RNA was assayed by the NanoDrop® spectrophotometer (Thermo Fisher, USA), the Qubit® RNA assay kit on Qubit® 3.0 Fluorometer (Life Technologies, USA) and the RNA Nano 6000 assay kit with the Bioanalyzer 2100 system (Agilent Technologies, USA), respectively.
Illumina RNA sequencing was carried out with two duplicates by Genscript following their standard analysis method. Gene FPKMs were computed by summing the FPKMs of the transcripts in each gene group with Cuffdiff (v1.3.0). FPKM represents “fragments per kilobase of exon per million fragments mapped,” which is calculated from the length of the fragments and the reads count mapped to each fragment. Genes with the absolute value of log2 (fold change) ≥ 1 and the corrected p values less than 0.05 were assigned as significantly differentially expressed genes (DEGs) by Cuffdiff (v2.2.1). GO enrichment analysis of DEGs was implemented with Perl module (GO::TermFinder) [35]. GO terms with corrected p value less than 0.05 were significantly enriched among the differentially expressed genes. R functions (phyper and q value) were used to test for the statistical enrichment of the differentially expressed genes among the KEGG pathways. KEGG pathways with corrected p value less than 0.05 were considered to be significantly enriched among the differentially expressed genes.
Results
Trichoderma reesei recombinant strains with off-target mutagenesis displayed markedly reduced cellulase production, but significantly enhanced sorbicillinoids production
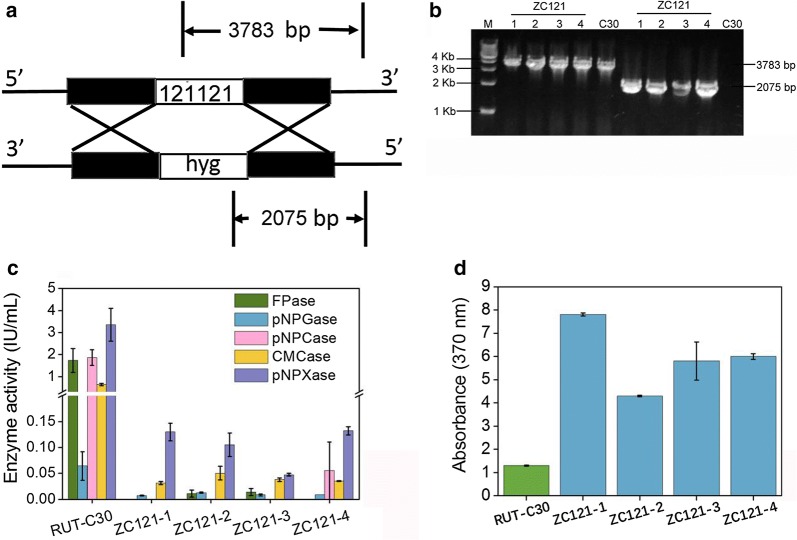
Gene 121121 encodes a candidate fungal regulatory protein that contains a Zn(2)Cys(6) fungal-type DNA binding domain. Gene 121121 is located next to β-glucosidase gene, cel3d [46]. Both genes belong to a tightly co-expressed genome region as identified by analysis of transcriptional data of T. reesei producing cellulases [46]. Moreover, gene cel3d was considered to be significantly involved in cellulase production by T. reesei [33]. Based on these previous findings, we presumed that gene 121121 might play a role in T. reesei cellulase production. Gene 121121 did not affect cellulase production when cells were grown on lactose [46], although no study was found which explored the effect of deletion of gene 121121 on cellulase synthesis. Therefore, we attempted to knockout gene 121121 in T. reesei RUT-C30 using homologous recombination mediated by AMT (Fig. 1a and Additional file 1: Fig. S1).
Fig. 1.
a Schematic description of the knockout of gene 121121 by homologous recombination using plasmid pXBthg-121121 (Additional file 1: Fig. S1). b PCR cloning of the gene fragments 3783 bp and 2075 bp as shown in a from T. reesei RUT-C30 and the recombinant strains ZC121-1, ZC121-2, ZC121-3, and ZC121-4. c Cellulase and hemicellulase activities in the culture supernatant of T. reesei RUT-C30, ZC121-1, ZC121-2, ZC121-3, and ZC121-4 grown on 2% cellulose were measured on day 5, including the activities of FPase (the filter paper activity), pNPGase (the BGL activity), pNPCase (the CBH activity), CMCase (the CMC activity), and pNPXase (the β-xylosidase activity). Meanwhile, d the corresponding yellow pigment production was determined by measuring the absorbance at 370 nm. The error bars indicate the standard deviations of three biological replicates
Four transformants, ZC121-1, -2, -3, -4, were obtained after selection using the marker, hygromycin. Unfortunately, the deletion of 121121 was not successful as shown by PCR result that gene 121121 was successfully cloned from all of four transformants, individually (Fig. 1b). This unsuccessful deletion was further confirmed by whole genome resequencing using NGS sequencing (Additional file 1: Fig. S4). However, we have successfully cloned the gene, hyg, in the four transformants mentioned above, demonstrating that the knockout cassette was randomly inserted into the chromosome of these strains by off-target mutagenesis [36].
Cellulase production in the four recombinant strains induced by cellulose on day 5 was assayed as described (Fig. 1c). The FPase, pNPCase, CMCase, pNPGase, and pNPXase activities of all four mutants were in the range of 0–1.9 IU/mL, 0.007–0.21 IU/mL, 0–0.37 IU/mL, 0.03–1.2 IU/mL, and 0.1–2.9 IU/mL, respectively, showing noticeably reduced cellulase activities when compared to that of RUT-C30, which was 6.1 IU/mL FPase, 0.5 IU/mL pNPCase, 6.8 IU/mL CMCase, 2.1 IU/mL pNPGase, and 4.6 IU/mL pNPXase. Obviously, the off-target mutagenesis caused serious inhibition of cellulase and hemicellulase production in T. reesei.
Surprisingly, we observed that these mutants displayed a much greater yellow color production than RUT-C30 both in liquid culture (Additional file 1: Fig. S4) and on the PDA plates (Additional file 1: Fig. S5), indicating far more sorbicillinoids were produced. For quantitative comparison, the absorbance of liquid cultures of T. reesei at 370 nm was used to measure the sorbicillinoids’ production (Fig. 1d) on cellulose [31]. The sorbicillinoids’ production in strains ZC121-1, ZC121-2, ZC121-3, and ZC121-4 was increased by 6, 3.3, 4.5, 4.6-fold, respectively, when compared to RUT-C30 production. In contrast to the dramatic decrease of cellulase and hemicellulase activities on cellulose, a significantly enhancement of yellow pigment production was observed by these recombinant strains. This finding demonstrated that off-target mutagenesis in strain ZC121 has switched cellulase production to sorbicillinoids’ production on cellulose. The recombinant strain ZC121-1 was selected for further study and was referred to as ZC121 because it displayed the highest yellow pigment production found in this study.
Hyperproduction of sorbicillinoids in strain ZC121 is a constitutive process
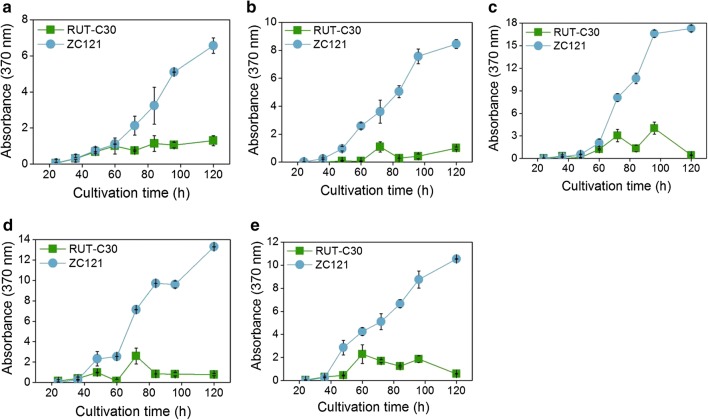
In previous studies, the ability of T. reesei to produce sorbicillinoids was reported to vary with different carbon sources [19, 37–39]. To see whether the superior sorbicillinoids’ production of strain ZC121 compared to strain RUT-C30 is dependent on carbon source, both the sorbicillinoids and cellulase production of strains ZC121 were measured during the time course of growth on TMM containing cellulose, lactose, glucose, galactose or glycerol as the individual carbon source (Fig. 2 and Additional file 1: Fig. S6). The maximal absorbance at 370 nm of the culture supernatant of strain ZC121 was 6.6, 8.4, 17.3, 13.3 and 10.5 for cellulose, lactose, glucose, galactose and glycerol, respectively. Note that these values are 5.1, 8.4, 4.3, 5.1 and 4.5-fold that of RUT-C30 (Fig. 2). Obviously, strain ZC121 produced remarkably increased sorbicillinoids on all tested carbon sources. The highest sorbicillinoids’ production was found with glucose as the carbon source at 120 h of growth, followed by galactose, glycerol, lactose, and cellulose—in descending order. An absorbance of OD370 = 17.3 for the culture supernatant from growth on glucose corresponds to a concentration of 627 μg/mL yellow pigments (see “Materials and methods” section). Except for growth on cellulose, both ZC121 and RUT-C30 display very low cellulase production on lactose, glucose, galactose, and glycerol (Additional file 1: Fig. S5), which is reasonable because these four carbon sources have been shown to be inefficient inducers cellulases.
Fig. 2.
The sorbicillinoids’ production in the culture supernatant of T. reesei RUT-C30 and ZC121 grown on TMM medium containing a cellulose, b lactose, c glucose, d galactose and e glycerol, respectively, was assayed by measuring the absorbance at 370 nm at the indicated time points. Error bars indicate SDs from three independently grown cultures
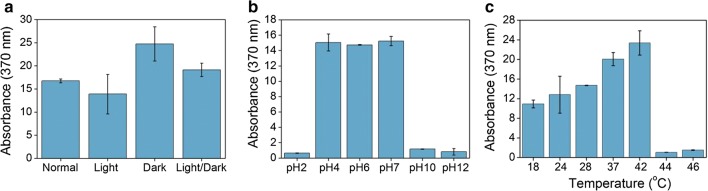
Using TMM + glucose as the culture medium, we also tested the effects of other culture conditions on the sorbicillinoids’ production ability of ZC121, including light, pH and temperature. Strain ZC121 was grown under normal lab light condition, constant light, constant darkness, or cycles of 12 h light–12 h dark (light–dark cycles) for 120 h. The highest absorbance (OD370 = 24.7) was observed for cell growth under constant darkness, whereas the absorbance under the other three light conditions were comparable internally (Fig. 3). It seems that the lighting condition only affects the production of sorbicillinoids in strain ZC121 to some limited extent, with the highest sorbicillinoids’ production observed for growth under constant darkness (Fig. 3a). However, strain ZC121 displayed excellent sorbicillinoids’ production ability under all tested light conditions. Strain ZC121 exhibited the hyperproduction of yellow pigment at pH 4, 6 and 7, but displayed sharply reduced pigment production at extreme pH values (pH 2, 10 and 12), which was probably related to the poor growth of strain ZC121 under these extreme pH conditions (Fig. 3b). The sorbicillinoids’ production declined somewhat when the culture temperature was decreased to 18 °C, but growth at 18 °C and 24 °C still maintained 74% and 87% of growth at 28 °C, respectively (Fig. 3c). In contrast, yellow pigment production was enhanced along with the increased temperature up to 42 °C (Fig. 3c). Beyond 42 °C, the sorbicillinoids’ production was reduced sharply, because strain ZC121 did not grown at all (Fig. 3c). Obviously, the high production performance of ZC121 was maintained over a limited range as the culture temperature was varied from the optimum temperature of 28 °C. In summary, the hyperproduction of sorbicillinoids in T. reesei ZC121 was a constitutive process, independent of carbon source, light, pH, and temperature.
Fig. 3.
The effect of light (a), pH (b) and temperature (c) on the sorbicillinoids’ production ability of T. reesei ZC121. Unless otherwise indicated, T. reesei was cultivated on TMM + glucose (pH 6) at 28 °C with 200 rpm for 5 days. Error bars indicate SDs from three independently grown cultures
Identification of the off-target mutagenesis in recombinant strain ZC121
The random insertion of the knockout cassette of pxBthg-121121 (Additional file 1: Fig. S2) into the chromosome of T. reesei ZC121 could cause collateral mutations, which might contribute to the outstanding yellow pigment production shown by ZC121. Therefore, the identification of the insertional site might help us further understand the regulatory mechanism of sorbicillinoids’ biosynthesis. To this end, the whole genome resequencing using NGS sequencing was employed to find the random insertional sites. The whole genome resequencing of T. reesei ZC121 resulted in a total of 16,078,934 150-bp paired-end reads with mean depth coverage of 89.78% and Q30 percentage of 92.42%. These clean reads covered 89.78% of the reference genome of RUT-C30. The NGS sequencing result (NCBI Accession Number: SRR6906202) show that one copy of the cassette was probably inserted at 1–60 bp of T. reesei RUT-C30 genome KI911238.1 (https://www.ncbi.nlm.nih.gov/nuccore/572281258/). The insertion site was at the telomere of chromosome IV of T. reesei. No coding genes were found in the neighboring sequence of 3 kb.
Identification of sorbicillinoids in the culture supernatant of strain ZC121
To determine which types of sorbicillinoids were produced by strain ZC121, LC–MS analysis of the culture supernatant of strain ZC121 cultivated on glucose for 5 days was performed (Table 1). A total of seven known sorbicillinoid-related compounds were identified, including sorbicillin, sorbicillinol, bisorbicillinol, dihydrosorbicillinol, oxosorbicillinol, bisvertinolone, and dihydrobisvertinolone. Furthermore, five structure-unknown compounds were detected. These compounds have been reported to be sorbicillinoid-related but are not named [19]. Peak area was utilized to roughly assess the abundance of these compounds (Table 1). Sorbicillinol was the most abundant product; bivertinolone was second. Sorbicillinol has been found to be the major sorbicillinoid-related product in several studies [13, 19, 25] and considered to be the building block for the other sorbicillinoids [19]. Bivertinolone can inhibit the biosynthesis of β-1,6-glucan and is an effective 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenger [40]. Its direct precursor, oxosorbicillinol, was also abundantly accumulated in the supernatant of strain ZC121. Only a trace amount of sorbicillin was detected, which agreed with the early findings [41].
Table 1.
Metabolite profiling for sorbicillinoids in the culture broth of strain ZC121 grown on glucose for 5 days
| Compound | Name | Formula | Acquired [M+H]+ | RT (min) | Peak area |
|---|---|---|---|---|---|
| 1 | Sorbicillinol | C14H16O4 | 249.1135 | 6.94 | 333,191 |
| 2 | Bisvertinolone | C28H32O9 | 513.2125 | 12.08 | 300,654 |
| 3 | Oxosorbicillinol | C14H16O5 | 265.108 | 9.03 | 111,253 |
| 4 | Bisorbicillinol | C28H32O8 | 497.2175 | 11.89 | 99,103 |
| 5 | Dihydrobisvertinolone | C28H34O9 | 515.228 | 12.57 | 40,780 |
| 6 | Dihydrosorbicillinol | C14H17O4 | 251.13 | 7.89 | 22,754 |
| 7 | Sorbicillin | C14H16O3 | 233.1178 | 5.63 | 7002 |
| 8 | Unknown | C12H17ON | 192.1388 | 5.26 | 36,046 |
| 9 | Unknown | C11H12O3 | 193.0865 | 6.67 | 35,210 |
| 10 | Unknown | C15H20O4N2 | 293.1501 | 4.56 | 9430 |
| 11 | Unknown | C12H14O3 | 207.1021 | 6.94 | 5678 |
| 12 | Unknown | C15H20O5N2 | 309.145 | 4.58 | 1021 |
Characterization of T. reesei ZC121
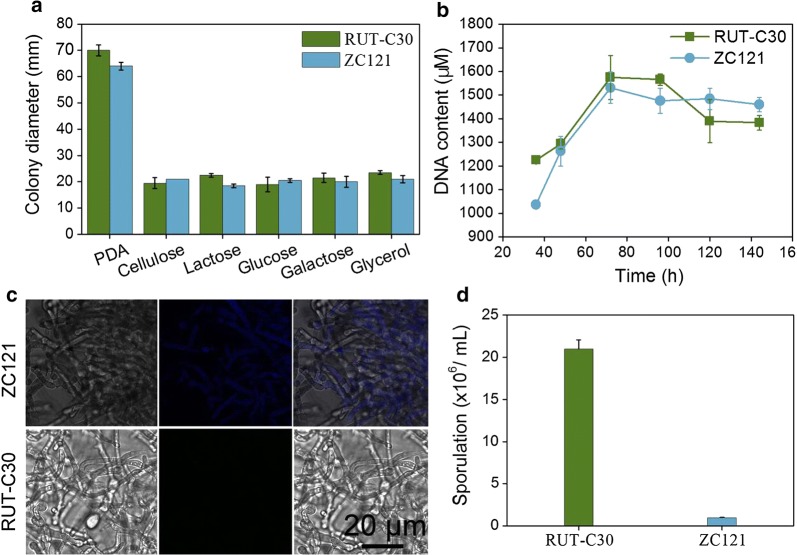
Growth of strains ZC121 and RUT-C30 was studied by measuring colony diameters on PDA plates and TMM plates with different carbon sources (i.e., cellulose, lactose, glucose, galactose or glycerol) (Fig. 4a and Additional file 1: Fig. S6). The colony diameters of these two strains were nearly the same regardless the carbon sources (Fig. 4a). Furthermore, the growth of strain ZC121 and RUT-C30 in TMM + 2% cellulose was assayed by DNA content measurement (Fig. 4b), as we were unable to accurately measure the dry T. reesei biomass due to the interference from insoluble cellulose. No significant difference of growth was observed between strain ZC121 and RUT-C30 (Fig. 4b). These results suggest that off-target mutagenesis did not affect the growth of T. reesei on different carbon sources.
Fig. 4.
Characterization of T. reesei ZC121. Diameters of the colonies of T. reesei RUT-C30 and ZC121 grown on PDA plates, and TMM plates with 2% cellulose, 2% lactose, 2% glucose, 2% galactose and 2% glycerol, respectively, were measured (a). Meanwhile, the growth of RUT-C30 and ZC121 cultured in TMM + 2% cellulose for cellulase production was determined by measuring the cellular DNA content (b), while their hyphae were profiled by fluorescence microscopy at Ex/Em = 405/415–495 nm (c). The sporulation of T. reesei RUT-C30 and ZC121 on PDA plates was counted on day 5 (d)
The morphology of ZC121 was different from that of RUT-C30. The number of the branched hyphae of ZC121 decreased and its hyphae became thinner and longer when compared to RUT-C30 (Fig. 4c). Also, the sorbicillinoids were not only secreted out of the cells, but also filled the fungal cell resulting in blue fluorescence emission at 405 nm excitation—as observed under fluorescence confocal microscopy (Fig. 4c). Furthermore, the spore amount of strain ZC121 was 1 × 106/mL, only 4.8% of that of RUT-C30 (2.1 × 107/mL) (Fig. 4d), demonstrating that the off-target mutagenesis leads to noticeable reduction of sporulation in T. reesei ZC121. This negative link between secondary metabolite production and conidiation in fungi was frequently found in early studies [20]. For example, when deleting the velvet protein VeA in A. nidulans, the abolishment of penicillin and aflatoxin secondary metabolite production was accompanied with the enhancement in asexual conidiation. The low level expression of sorbicillinoid-related genes during asexual growth was also reported [12]. Overall, with the off-target mutagenesis, the growth rate and biomass of T. reesei ZC121 was not affected significantly, but its morphology changed with marked sporulation reduction.
Transcription patterns of strain ZC121
RNA-seq analysis was performed to understand how the off-target mutagenesis influences the transcriptional level of strain ZC121. The sequences of the total reads were mapped to the reference genome of T. reesei RUT-C30 (https://www.ncbi.nlm.nih.gov/genome/323?genomeassembly_id=49799) with coverage of 93.24–95.63%. A total of 9544 unique transcripts were detected. Genes were differentially expressed between the two strains when the average reads of the corresponding transcripts differed with |log2Ratio| ≥ 1 and adjusted p values ≤ 0.05. By comparing strain ZC121 to RUT-C30, we obtained 638 and 1006 differentially expressed genes (DEGs) under cellulose and glucose growth conditions, respectively (Additional file 1: Table S4). Among these, 341 DEGs were found under both glucose and cellulose growth conditions, of which 327 DEGs expression changing trends were identical between these two carbon sources, whereas 14 DEGs expression changing trends were opposite (Additional file 1: Table S4).
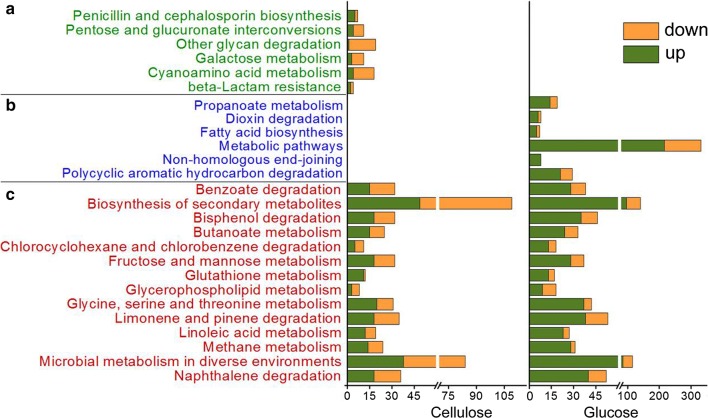
KEGG pathway enrichment analysis showed that 14 out of the top 20 enriched pathways were shared by growth on both glucose and cellulose conditions (Fig. 5c). This result implies that the impact of the off-target mutagenesis in ZC121 at the transcription level share a lot in common between growth on different carbon sources. The “other glycan degradation” was enriched only on cellulose (Fig. 5a). Most genes in this pathway were related to cellulase production induced by cellulose and were down-regulated, notably. This result is in line with reports of the inhibited cellulase production in strain ZC121 for growth on cellulose in comparison with RUT-C30.
Fig. 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs in strain ZC121 in comparison with the parental strain RUT-C30 on cellulose or glucose. The y axis represents the name of the top 20 enriched pathways, and the x axis shows the number of DEGs in the enriched pathways. The enriched pathways were categorized into pathways enriched only under glucose condition (a), only under cellulose condition (b), and under both glucose and cellulose conditions (c)
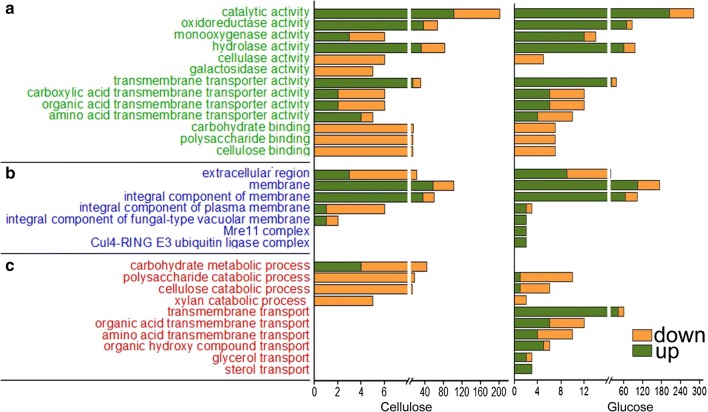
The enriched molecular functions “catalytic activity” and “cellulose binding” were observed for growth on both glucose and cellulose (Fig. 6a), including two enriched subcategories “oxidoreductase activity” and “hydrolase activity, acting on glycosyl bonds”. “Transmembrane transporter activity” was enriched only under glucose growth conditions (Fig. 6a).
Fig. 6.
Gene ontology (GO) functional enrichment analysis of DEGs of strain ZC121 compared to the parental strain RUT-C30 grown on cellulose and glucose as the sole carbon source, respectively. The y axis represents the name of the most enriched GOs that belong to different ontologies: (a) the molecular function, (b) the cellular component and (c) the biological process, while the x axis represents the number of DEGs in each enriched GO
For the enriched cellular components, both “membrane” and “extracellular region” were enriched in the presence of glucose or cellulose (Fig. 6b), which is reasonable given that both cellulases and sorbicillinoids are secreted outside of the fungal cells. Under the “membrane” category, DEGs are mainly involved in the intrinsic component of membrane (Fig. 6b), including the plasma membrane, vacuolar membrane, nuclear membrane, and mitochondrial membrane. Deletion of gene 121121 might pose a significant change on cell structure of T. reesei, which might be responsible for the morphology change mentioned above (Fig. 4c); as well as the condition defect (Fig. 4d).
In the presence of cellulose, the most enriched DEGs in the category of biological process belong to “carbohydrate metabolic process” which mainly included “xylan catabolic process” and “cellulose catabolic process” (Fig. 6c). Most of these DEGs were downregulated, which is consistent with the markedly reduced cellulase production and the result of the KEGG pathway enrichment analysis. Under glucose growth conditions, the most enriched biological process changed to “transmembrane transport” and “oxidation–reduction process” (Fig. 6c).
Most DEGs involved in the cellulase production were downregulated in T. reesei ZC121 on cellulose
There are 74 genes known or predicted to be related to cellulase/hemicellulase production in T. reesei, of which 43 were DEGs with notably downregulated mRNA levels, except gene xdh1 (Additional file 1: Table S5). Specifically, the transcriptional levels of cellulase/hemicellulase genes, such as cellobiohydrolase cel6a (CBH II), endoglucanases egl1, egl2, egl3, egl4, and egl5, β-glucosidases cel3a (bgl1), cel3d, cel3e, cel1a, and cel1b, β-xylosidase bxl1, and xylanases xyn1, xyn2 and xyn3 were all reduced markedly in strain ZC121 (Table 2), matching well with the nearly abolished cellulase/hemicellulase activities found in the supernatant of ZC121 culture. The auxiliary proteins, such as swollenin (encoded by swo1) [42], the glycoside hydrolase Family 61 (GH61 s) GH61b and GH61a [43] and the cellulose-induced proteins CIP-1 and CIP-2 [44], which have been shown to improve the hydrolysis of cellulose, were also downregulated significantly in ZC121 grown on cellulose. In addition, three well-known cellulase transcription activators xyr1 [45], ace3 [46], and crt1 [47] were significantly downregulated in strain ZC121 grown on cellulose, whereas cellulase transcription repressor, cre1 [48], was downregulated under both cellulose and glucose growth conditions.
Table 2.
The main DEGs related to (hemi) cellulase in strain ZC121 (C121)
| Gene IDa | Gene name | Description | log2 (C121/CC30) | p value | Category |
|---|---|---|---|---|---|
| 122470 | cel6a | Exoglucanase 2 | − 6.43 | 5.00E−05 | Cellulase |
| 5304 | egl1 | Endoglucanase I precursor | − 5.75 | 5.00E−05 | Cellulase |
| 124931 | egl2 | Endoxylanase II | − 2.63 | 0.0001 | Cellulase |
| 72489 | egl3 | Endoglucanase III | − 6.74 | 5.00E−05 | Cellulase |
| 139633 | egl4 | Endoglucanase-4 | − 5.70 | 5.00E−05 | Cellulase |
| 25940 | egl5 | Endo-1,4-beta-glucanase V | − 2.50 | 5.00E−05 | Cellulase |
| 136547 | cel3a | Beta-d-glucoside glucohydrolase I | − 4.35 | 5.00E−05 | Cellulase |
| 122639 | cel3d | Hypothetical protein | − 1.94 | 5.00E−05 | Cellulase |
| 74305 | cel3e | Hypothetical protein | − 1.88 | 5.00E−05 | Cellulase |
| 127115 | cel1a | Beta-glucosidase | − 3.13 | 5.00E−05 | Cellulase |
| 77989 | cel1b | Glycoside hydrolase | − 2.90 | 5.00E−05 | Cellulase |
| 122518 | cel61b | Endoglucanase VII | − 6.84 | 5.00E−05 | Cellulase |
| 77521 | bxl1 | Family 43 glycoside hydrolase | − 1.32 | 0.0017 | TF |
| 38418 | xyn1 | Endo-1,4-β-xylanase 1 | − 1.45 | 0.0009 | Hemicellulases |
| 124931 | xyn2 | Endo-1,4-β-xylanase 2 | − 2.63 | 0.0001 | Hemicellulases |
| 23616 | xyn3 | Xylanase III | − 6.17 | 5.00E−05 | Hemicellulases |
| 104220 | swo1 | Swollenin | − 3.52 | 5.00E−05 | Nonenzymatic cellulose attacking enzymes |
| 121449 | cip1 | Hypothetical protein | − 6.30 | 5.00E−05 | Nonenzymatic cellulose attacking enzymes |
| 125575 | cip2 | Hypothetical protein | − 5.19 | 5.00E−05 | Nonenzymatic cellulose attacking enzymes |
| 98788 | xyr1 | Xylanase regulator 1 | − 2.28 | 5.00E−05 | TF |
| 98455 | ace3 | Hypothetical protein | − 2.29 | 5.00E−05 | TF |
| 109243 | crt1 | General substrate transporter | − 3.04 | 5.00E−05 | TF |
| 23706 | cre1 | Hypothetical protein | − 1.63 | 0.0001 | TF |
aGene ID was assigned based on the T. reesei RUT-C30 genome database (https://www.ncbi.nlm.nih.gov/genome/323?genome_assembly_id=49799)
DEGs involved in sorbicillinoid biosynthesis and other secondary metabolism
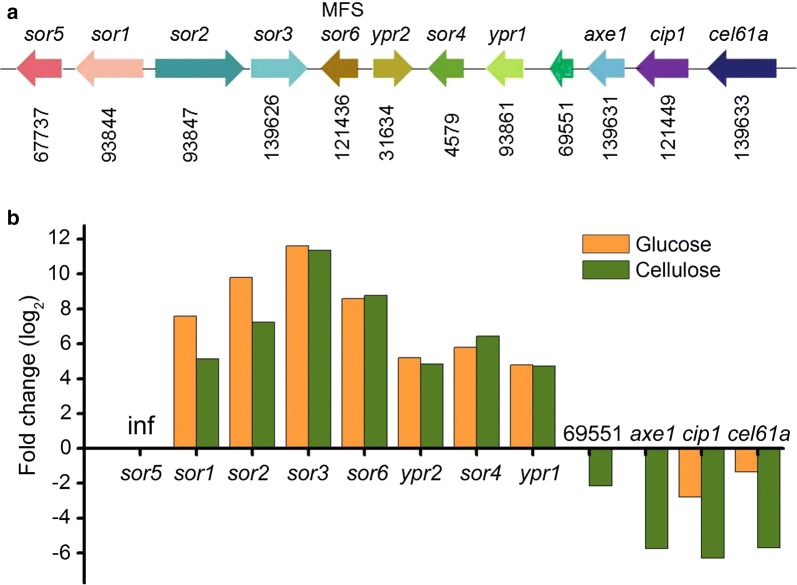
Genes involved in the biosynthesis of sorbicillinoids were clustered in the T. reesei genome and named the “sorbicillinoid gene cluster” [14, 25]. This T. reesei gene cluster contains a highly reducing PKS Sor 1, a non-reducing PKS Sor 2, a flavin adenine dinucleotide (FAD)-dependent monooxygenase Sor3, a FAD/flavin mononucleotide-containing dehydrogenase Sor4, two transcription factors YPR1 and YPR2 [3], a MFS transporter Sor6, and a short-chain dehydrogenase/reductase Sor 5 (Fig. 7 and Additional file 1: Table S2). We provisionally named genes in the sorbicillinoid gene cluster in this study, because the naming of this cluster was inconsistent in previous studies [25, 26]. It is worth noting that all genes in the sorbicillinoid cluster were upregulated significantly in strain ZC121 when using glucose or cellulose as the sole carbon source (Fig. 7b and Additional file 1: Table S2). This result agrees well with the excellent overproduction of sorbicillinoids in strain ZC121 (Fig. 2). Under both conditions tested, the expression of Sor5 was detected in strain ZC121, but not in RUT-C30 (Fig. 7b and Additional file 1: Table S2). Gene sor5 in this study, which is designated as sor7 in the literature, encodes a short-chain dehydrogenase/reductase [14]. Its ortholog from P. rubens was not located in the sorbicillinoid gene cluster. Moreover, gene sor5 was not found in most other fungi. It seems that the off-target mutagenesis activates the expression of gene sor5, which might play a role in the notable overexpression of genes in the sorbicillinoid cluster. However, how the activation of gene Sor5 functions in T. reesei ZC121 is unclear, because studies performed on gene sor5 are rare.
Fig. 7.
a Genomic organization of the sorbicillinoid gene cluster and its neighboring cellulase-related genes in T. reesei. The relative transcription levels of these genes in strain ZC121 to RUT-C30 are shown under both cellulose and glucose conditions b. Gene 102500 and axe1 were not DEGs under glucose condition, so their fold change was not presented. Inf: the transcript of sor5 was not detected in RUT-C30, but in ZC121
In the previous study, an extensive search for PKS-, NRPS- and hybrid synthetase genes using basic local alignment search tool (BLAST) revealed the 11 PKS-, 11 NRPS- and 4 hybrid synthetase genes in the T. reesei genome (http://genome.jgipsf.org/pages/blast.jsf?db=Trire2), which could be appointed to 23 distinct gene clusters [21]. Also, a total of 31 predicted transcription factor (TF) genes were found in a 50 kb radius of these synthase genes. Among them, five synthases and four TFs were significantly upregulated in strain ZC121 under both tested conditions (Additional file 1: Table S6), of which two synthases and two TFs (Additional file 1: Table S2) are involved in the sorbicillin biosynthesis pathway, as we discussed above. The transcription level of pks8 (90904) were increased under both conditions (Table 3). Its homologues, adaA from A. niger [10] or vrtA from Penicillium aethiopicum [11], were involved in anthracenone and naphthacenedione biosynthesis, respectively. A hybrid PKS-NRPS synthase (128011) and its neighboring TF (74475) was significantly upregulated (Table 3), whose closest annotated hit was fusaproliferin synthase [21]. Fusaproliferin, a mycotoxin from Fusarium spp. PKS 77957, is a nonribosomal peptide synthetase, whose homologue in Aspergillus fumigatus is essential for fumigaclavine C production. Fusaproliferin has been studied extensively [49]. This outcome shows that other secondary metabolites, other than sorbicillinoids, might also be produced in strain ZC121 and thus it is worth studying them in the future.
Table 3.
DEGs that are synthetases and transcriptional factors involved in secondary metabolism, which were found under both cellulose and glucose conditions
| Gene IDa | Gene name | Description | log2 (C121/CC30) | p value | Category | ||
|---|---|---|---|---|---|---|---|
| Cellulose | Glucose | Cellulose | Glucose | ||||
| 90904 | 90904 | Putative TPA: polyketide synthase | 4.98278 | 5.88729 | 5.00E−05 | 5.00E−05 | Synthase |
| 128011 | 128011 | Hypothetical protein | 3.33221 | 2.94805 | 5.00E−05 | 0.0001 | TF |
| 74475 | 74475 | Hypothetical protein | 2.96889 | 3.35194 | 5.00E−05 | 0.0115 | TF |
| 77957 | 77957 | Putative nonribosomal peptide synthase GliP-like protein | 4.56203 | 2.51002 | 5.00E−05 | 5.00E−05 | Synthase |
| 77229 | 77229 | Hypothetical protein | 2.27 | 2.09 | 5.00E−05 | 5.00E−05 | TF |
aGene ID was assigned based on the T. reesei RUT-C30 genome database (https://www.ncbi.nlm.nih.gov/genome/323?genome_assembly_id=49799)
Discussion
Sorbicillinoids have potential pharmaceutical value as antimicrobial, antivirus, and anticancer agents [50]. Moreover, they could be utilized as pigments and food colorants (yellow pigments) as well. Since they were first discovered in 1948 from Penicillium notatum [51], studies related to sorbicillinoids have focused on finding new compounds with similar structure [52], elucidating chemical structures and biological activities [16, 50], establishing the complex biosynthetic pathway [41] and developing chemical synthesis methods [53]. However, both strain engineering to increase sorbicillinoids’ production and the relevant regulatory mechanism is less studied. T. reesei is well-known for its prominent enzyme-secreted ability using cellulose as the efficient inducer and widely utilized in industry as a work horse for the production of both cellulase and heterologous recombinant proteins [54]. For the first time, an overwhelming switch from cellulase production to sorbicillinoids’ production using cellulose as carbon source was reported in recombinant T. reesei strain ZC121. Coincidentally, an expression shift from cellulase-related genes to genes in the sorbicillinoid gene cluster was found by transcriptional profiling with steep downregulation of 42 genes involved in cellulase production and significant upregulation of all genes in the sorbicillinoid cluster. This switch would enable T. reesei to contribute more energy and metabolites to sorbicillinoids’ production, considering that cellulase production is a heightened energy-efficient process. This shift, at the same time, would allow the easier separation of sorbicillinoids from a culture supernatant that contains only low titers of cellulases. More importantly, T. reesei was first engineered to directly degrade cellulose to produce other platform chemical compounds, rather than proteins. The routine strategy for cellulose-based chemical production is the combination of T. reesei for cellulose degradation and other model industrial microorganisms for desired compound production via co-culturing [29] and consolidated bioprocessing [55], achieving the degradation of cellulose to produce platform compounds, such as isobutanol [29] and ethanol [56]. These strategies using multiple microorganisms often require complicated and stringent operations, with the compatibility between two or more working microorganisms as another major challenge [55]. Therefore, it is of great value to have T. reesei mutants that can produce just the necessary amount of cellulases/hemicellulases to support the host cells in the production of valuable products in high yield.
In most studies related to sorbicillinoids, glucose is usually used as the carbon source with the highest sorbicillinoids’ production of OD370 = 13 reported in the literature [31], which is lower than OD370 = 17.3 of strain ZC121 in this study. Upon growth on cellulose, T. reesei cultures with the deletion of gene XPP1 can produce a small amount of yellow pigment with OD370 ≈ 0.32 [31], much less than the OD370 = 6.6 we observed in strain ZC121. Strain ZC121 generated the highest reported amounts of sorbicillinoids when grown on cellulose or glucose, as far as we know. Other carbon sources, such as lactose, glycerol, and galactose have been reported to be utilized by T. reesei to produce remarkable amounts of sorbicillinoids. Furthermore, the sorbicillinoids’ production in fungi has been reported to be generally impacted by culture conditions, such as carbon source [9, 37], light exposure time [39], temperature, and pH. By contrast, the recombinant strain ZC121 displayed hyperproduction of sorbicillinoids regardless the culture conditions, so long as T. reesei can grow well. This constitutive hyperproduction would benefit the future industry application of strain ZC121 for sorbicillinoids’ production with great flexibility and easy fermentation operation.
Currently, sorbicillinoids are not produced by industry. Various efforts have been made to access these compounds by chemical synthesis in the laboratory and only for research purposes [41]. However, most of the chemical synthesis routes are cumbersome, as the structure of sorbicillinoids is complex [53]. Whether or not these chemical methods are applicable in the industry is still unknown. Moreover, the chemical method is less environment-friendly than the biosynthesis. Therefore, it is highly desired to have recombinant microorganisms that can produce constantly sorbicillinoids with high yield.
The global regulator laeA is well-known for being involved in the secondary metabolism by regulating some polyketides [57]. Protein LaeA directly interacts in the nucleus with transcription factors of the trimeric Velvet complex, consisting of the Velvet domain proteins VeA and VelB [57]. The heterotrimeric velvet complex VelB/VeA/LaeA correlates fungal development and secondary metabolism in response to light. Moreover, it is found that both the knockout and overexpression of gene laeA affected the expression of some sor genes [14]. Nevertheless, the mRNA level of all these three genes remained unchanged in strain ZC121 in comparison with RUT-C30. Moreover, VosA, a recently identified regulator of fungal sporogenesis [58], another binding partner of VelB [57], was also not affected in strain ZC121. This finding is in line with the observation that T. reesei ZC121 exhibited high sorbicillinoids’ production as a function of light exposure time as we showed above.
The major types of sorbicillinoids generated in strain ZC121 from growth on glucose were sorbicillinol and bisvertinolone as analyzed by LC–MS (Table 1). It is worth noting that bisvertinolone is a potential anticancer agent (acts by inhibiting β-1,6-glucan biosynthesis or serving as an effective 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical scavenger [40]. Currently, it is only available by intricate chemical methods [53]. Bisvertinolone has never been reported to be the major product in T. reesei. Furthermore, the precursor of bisvertinolone, oxosorbicillinol was the third most abundant compound in the culture medium (Table 1). Thus, it seems that T. reesei ZC121 might serve as a good starting strain to obtain mutants that primarily produce bisvertinolone by further strain optimization.
Interestingly, four genes related to cellulase production are in close vicinity to the sorbicillinoid gene cluster, including gene 102500, axe1, cip1 and cel61a, forming a “sorbicillinoid-cellulase” supercluster (Fig. 7a). In contrast to the remarkably upregulated expression of genes in the sorbicillinoid gene cluster, these four genes were all DEGs steeply downregulated in ZC121 on cellulose growth, while only two genes were DEGs markedly reduced on glucose growth (Fig. 7b). Genes encoding candidate carbohydrate-active enzymes (CAZy) for polysaccharide degradation are prone to clustering in the T. reesei genome, leading to 25 genomic regions of high CAZy gene density [59]. These regions usually possess genes participating in secondary metabolism as well, forming “superclusters”. These kinds of superclusters have also been observed for other secondary metabolites in other fungi, playing a key role in the corresponding secondary metabolite production [60–62]. However, whether and how the “sorbicillinoid–cellulase” supercluster impacts the secondary metabolism for sorbicillinoids and cellulase is still unknown and is worth exploring in future studies.
We have initially aimed to specifically knockout gene 121121 in T. reesei RUT-C30 to study its function on cellulase production by AMT utilizing homologous recombination; however, this approach unfortunately failed. Continued work gave rise to the unexpected off-target mutagenesis at the telomere region of chromosome IV. The insertion of T-DNA into the genome of the recipient cell is random [63]. This randomness of T-DNA integration leads to inefficient targeting, resulting in off-target mutagenesis. It has been shown that T-DNA can insert into the telomeric region by non-homologous recombination (NHR) [64]. Therefore, although the similarity between the flanking 1500-bp regions of gene 121121 and the integration site at the telomere of chromosome IV of T. reesei is not high (40.4% and 39.0% for the upstream and downstream flanking regions, respectively), off-target integration was observed at the telomere of chromosome IV of T. reesei. The inefficient gene targeting by AMT has been a long-standing issue in plant and fungi [36]. Strategies like generation of double strand breaks at genomic positions of interest, downregulation of enzymes involved in NHEJ pathway, and concomitant translocation of the homing endonuclease I-SceI, have been explored to improve targeted integration of AMT [36]. Nevertheless, target mutagenesis can be embraced as a potent mutagenesis strategy for strain engineering as we showed here, together with the traditional methods using nitrosoguanidine (NTG) or UV irradiation. The insertion of T-DNA into the telomeric region induced gross chromosome rearrangement [64]. Chromosome remodeling plays an important role in cellulase production [33, 34, 65]. It is tempting to speculate that the off-target mutagenesis probably gave rise to the excellent sorbicillinoids’ production ability of strain ZC121, which came about by chromosome rearrangement and subsequently affected gene expression as we found using transcriptome analyses.
Conclusion
We constructed recombinant T. reesei strain ZC121 from T. reesei RUT-C30 by off-target mutagenesis during the failed knockout of gene 121121. The knockout cassette for gene 121121 was found to insert at the telomere of chromosome IV of T. reesei. Strain ZC121 exhibited constitutive hyperproduction of sorbicillinoids under all tested culture conditions, including varied carbon source, light, pH, and temperature. Particularly, an overwhelming switch from cellulase production to sorbicillinoids production was observed in strain ZC121 on cellulose. Coincidently, a similar shift was also observed at the transcriptional level in ZC121 cultivated on cellulose, with steep downregulation of 42 genes involved in cellulase production and significant upregulation of all genes in the sorbicillinoid cluster. For the first time, T. reesei alone can degrade cellulose to directly produce valuable compounds (sorbicillinoids in this case) other than proteins, paving the way for the industrial production of cellulase-based chemical compounds.
Additional file
Additional file 1: Fig. S1. Schematic illustration of the plasmid pXBthg-121121. hyg: homomycin resistance; LB, left border of binary vector; RB, right border of binary vector; Kan: kanamycin resistance; 121121-up: the 1500 bp upstream of gene 121121; 121121-down: the 1500 bp downstream of gene 121121. Fig. S2. The 1500-bp upstream and downstream regions of gene 121121. Fig. S3. The standard curve of sorbicillinoids. Fig. S4. The whole genome resequencing of strain ZC121 shows the unsuccessful deletion of gene 121121. Fig. S5. The color of the culture supernatant of T. reesei RUT-C30 (a) and ZC121 (b) grown on glucose. Fig. S6. Cellulolytic enzyme activities in the culture supernatant of T. reesei ZC121 and RUT-C30 grown on 2% cellulose, lactose, glucose, galactose and glycerol were assayed on day 5, including the activities of FPase (the filter paper activity) (a), pNPGase (the BGL activity) (b), pNPCase (the CBH activity) (c), CMCase (the CMC activity) (d) ahd pNPXase (the β-xylosidase activity) (e). The error bars indicate the standard deviations of three biological replicates. Table S1. Primers used in this study. Table S2. Comparative transcription levels of the sorbicillinoid gene cluster and its neighboring cellulase-related genes in T. reesei ZC121on cellulose and glucose.
Authors’ contributions
CL and FL conceived and designed the study. CL carried out the majority of the experiments. WS conducted part of enzyme activity measurement experiments and the extraction of sorbicillionoids. ZZ provided the plasmid pXBthg and helped us to design and construct the plasmid pXBthg-121121 used in this study.SY helped us to analyze part of the RNA sequencing results. CL, FL and ZC analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Authors thank the National Natural Science Foundation of China (31700040), the Fundamental Research Funds for the Central Universities, and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. ZC thanks the University of Michigan for supporting his sabbatical leave.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (31700040), the Fundamental Research Funds for the Central Universities, and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. ZC acknowledges the support from University of Michigan.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- pNPGase
the β-glucosidase activity
- pNPCase
the CBH activity
- CMCase
the CMC activity
- FPase
the filter paper activity
- pNPXase
the β-xylosidase activity
- AMT
agrobacterium tumefaciens-mediated transformation
- PDA
potato dextrose agar
- SDB
sabouraud dextrose broth
- TMM
Trichoderma minimal medium
- MFS
major facilitator superfamily sugar transporter
- PKSs
polyketides
- NRPs
non-ribosomal peptides
- DEGs
differentially expressed genes
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NGS
next-generation sequencing
- FPKMs
fragments per kilobase of transcript sequence per millions base pairs sequenced
- RT-qPCR
quantitative real-time PCR
Contributor Information
Chengcheng Li, Email: lichengcheng1162@126.com.
Fengming Lin, Phone: +86-83791810, Email: linfengming@seu.edu.cn.
Wei Sun, Email: sunnysoon@seu.edu.cn.
Shaoxun Yuan, Email: 230159460@seu.edu.cn.
Zhihua Zhou, Email: zhouzhihua@sippe.ac.cn.
Fu-Gen Wu, Email: wufg@seu.edu.cn.
Zhan Chen, Email: zhanc2010@gmail.com.
References
- 1.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Kikuchi H. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz B, Boyl C, Draeger S, Römmert AK, Krohn K. Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol Res. 2002;106:996–1004. doi: 10.1017/S0953756202006342. [DOI] [Google Scholar]
- 3.Leflaive J, Ten-Hage L. Algal and cyanobacterial secondary metabolites in freshwaters: a comparison of allelopathic compounds and toxins. Freshw Biol. 2007;52:199–214. doi: 10.1111/j.1365-2427.2006.01689.x. [DOI] [Google Scholar]
- 4.Pichersky E, Gang DR. Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci. 2000;5:439–445. doi: 10.1016/S1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- 5.Forbey JS, Harvey AL, Huffman MA, Provenza FD, Sullivan R, Tasdemir D. Exploitation of secondary metabolites by animals: a response to homeostatic challenges. Integr Comp Biol. 2009;49:314–328. doi: 10.1093/icb/icp046. [DOI] [PubMed] [Google Scholar]
- 6.Wallace RJ. Antimicrobial properties of plant secondary metabolites. Proc Nutr Soc. 2004;63:621–629. doi: 10.1079/PNS2004393. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 8.Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3:408. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- 9.Derntl C, Rassinger A, Srebotnik E, Mach RL, Mach-Aigner AR. Identification of the main regulator responsible for synthesis of the typical yellow pigment produced by Trichoderma reesei. Appl Environ Microbiol. 2016;82:6247–6257. doi: 10.1128/AEM.01408-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aerts D, Hauer EE, Ohm RA, Arentshorst M, Teertstra WR, Phippen C, Wösten HA. The FlbA-regulated predicted transcription factor Fum21 of Aspergillus niger is involved in fumonisin production. Antonie Van Leeuwenhoek. 2018;111:311–322. doi: 10.1007/s10482-017-0952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chooi YH, Cacho R, Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kontani M, Sakagami Y, Marumo S. First β-1,6-glucan biosynthesis inhibitor, bisvertinolone isolated from fungus, Acremonium strictum and its absolute stereochemistry. Tetrahedron Lett. 1994;35:2577–2580. doi: 10.1016/S0040-4039(00)77175-9. [DOI] [Google Scholar]
- 13.Sugaya K, Koshino H, Hongo Y, Yasunaga K, Onose JI, Yoshikawa K, Abe N. The biosynthesis of sorbicillinoids in Trichoderma sp. USF-2690: prospect for the existence of a common precursor to sorbicillinol and 5-epihydroxyvertinolide, a new sorbicillinoid member. Tetrahedron Lett. 2008;49:654–657. doi: 10.1016/j.tetlet.2007.11.121. [DOI] [Google Scholar]
- 14.Druzhinina IS, Kubicek EM, Kubicek CP. Several steps of lateral gene transfer followed by events of ‘birth-and-death’ evolution shaped a fungal sorbicillinoid biosynthetic gene cluster. BMC Evol Biol. 2016;16:269. doi: 10.1186/s12862-016-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balde EHS, Andolfi A, Bruyère C, Cimmino A, Lamoral-Theys D, Vurro M, Damme MV, Altomare C, Mathieu V, Kiss R, Evidente A. Investigations of fungal secondary metabolites with potential anticancer activity. J Nat Prod. 2010;73:969–971. doi: 10.1021/np900731p. [DOI] [PubMed] [Google Scholar]
- 16.Abe N, Hirota A. Chemical studies of the radical scavenging mechanism of bisorbicillinol using the 1,1-diphenyl-2-picrylhydrazyl radical. Chem Commun. 2002;2002:662–663. doi: 10.1039/b200039n. [DOI] [PubMed] [Google Scholar]
- 17.Peng J, Zhang X, Du L, Wang W, Zhu T, Gu Q, Li D. Sorbi catechols A and B, antiviral sorbicillinoids from the marine-derived fungus Penicillium chrysogenum PJX-17. J Nat Prod. 2014;77:424–428. doi: 10.1021/np400977e. [DOI] [PubMed] [Google Scholar]
- 18.Maskey RP, Grün-Wollny I, Laatsch H. Sorbicillin analogues and related dimeric compounds from Penicillium notatum. J Nat Prod. 2005;68:865–870. doi: 10.1021/np040137t. [DOI] [PubMed] [Google Scholar]
- 19.Salo O, Guzmán-Chávez F, Ries MI, Lankhorst PP, Bovenberg RA, Vreeken RJ, Driessen AJ. Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl Environ Microbiol. 2016;82:3971–3978. doi: 10.1128/AEM.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta VK, Steindorff AS, de Paula RG, Silva-Rocha R, Mach-Aigner AR, Mach RL, Silva RN. The post-genomic Era of Trichoderma reesei: what’s next? Trends Biotechnol. 2016;3(4):970–982. doi: 10.1016/j.tibtech.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee PK, Horwitz BA, Kenerley CM. Secondary metabolism in Trichoderma—a genomic perspective. Microbiology. 2012;158:35–45. doi: 10.1099/mic.0.053629-0. [DOI] [PubMed] [Google Scholar]
- 22.Zeilinger S, Gruber S, Bansal R, Mukherjee PK. Secondary metabolism in Trichoderma—chemistry meets genomics. Fungal Biol Rev. 2016;30:74–90. doi: 10.1016/j.fbr.2016.05.001. [DOI] [Google Scholar]
- 23.Nakari-Setälä T, Aro N, Kalkkinen N, Alatalo E, Penttilä M. Genetic and biochemical characterization of the Trichoderma reesei hydrophobin HFBI. Eur J Biochem. 1996;235:248–255. doi: 10.1111/j.1432-1033.1996.00248.x. [DOI] [PubMed] [Google Scholar]
- 24.Seiboth B, Karimi RA, Phatale PA, Link R, Hartl L, Sauer DG, Kristina M, Smith KM, Baker SE, Freitag M, Kubicek CP. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol. 2012;84:1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Derntl C, Guzmán Chávez F, Mello-de-Sousa TM, Busse HJ, Driessen AJ, Mach RL, Mach-Aigner AR. In vivo study of the sorbicillinoid gene cluster in Trichoderma reesei. Front Microbiol. 2017;8:2037. doi: 10.3389/fmicb.2017.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jørgensen MS. Unraveling the secondary metabolism of the biotechnological important filamentous fungus Trichoderma reesei (Teleomorph Hypocrea jecorina). Technical University of Denmark. 2013. p. 164.
- 27.Guzmán-Chávez F, Salo O, Nygård Y, Lankhorst PP, Bovenberg RA, Driessen AJ. Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum. Microb Biotechnol. 2017;10:958–968. doi: 10.1111/1751-7915.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong YH, Wang XL, Wang TH, Jiang Q. Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Appl Microbiol Biotechnol. 2007;73:1348–1354. doi: 10.1007/s00253-006-0603-3. [DOI] [PubMed] [Google Scholar]
- 29.Minty JJ, Singer ME, Scholz SA, Bae CH, Ahn JH, Foster CE, Liao JC, Lin XN. Design and characterization of synthetic fungal–bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci USA. 2013;110:14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L, Zhang J, Zou G, Wang C, Zhou Z. Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta-glucosidase gene from Penicillium decumbens. Enzyme Microb Technol. 2011;49:366–371. doi: 10.1016/j.enzmictec.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Derntl C, Kluger B, Bueschl C, Schuhmacher R, Mach RL, Mach-Aigner AR. Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc Natl Acad Sci USA. 2017;114:E560–E569. doi: 10.1073/pnas.1609348114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali H, Ries MI, Nijland JG, Lankhorst PP, Hankemeier T, Bovenberg RA, Vreeken RJ, Driessen AJ. A branched biosynthetic pathway is involved in production of roquefortine and related compounds in Penicillium chrysogenum. PLoS ONE. 2013;8:e65328. doi: 10.1371/journal.pone.0065328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C, Lin F, Li Y, Wei W, Wang H, Qin L, Zhou ZH, Li BZ, Wu F-G, Chen Z. A β-glucosidase hyper-production Trichoderma reesei mutant reveals a potential role of cel3D in cellulase production. Microb Cell Fact. 2016;15:151. doi: 10.1186/s12934-016-0550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Lin F, Zhou L, Qin L, Li B, Zhou ZH, Jin MJ, Chen Z. Cellulase hyper-production by Trichoderma reesei mutant SEU-7 on lactose. Biotechnol Biofuels. 2017;10:228. doi: 10.1186/s13068-017-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolloos M, Hooykaas Minty JJPJ, Van Der Zaal BJ. Enhanced targeted integration mediated by translocated I-SceI during the agrobacterium mediated transformation of yeast. Sci Rep. 2015;5:8345. doi: 10.1038/srep08345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van den Berg MA. Penicillium chrysogenum: genomics of an antibiotics producer. In: Genomics of soil-and plant-associated fungi. Berlin: Springer; 2013. p. 229–54.
- 38.Atanasova L, Knox BP, Kubicek CP, Druzhinina IS, Baker SE. The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot Cell. 2013;12:1499–1508. doi: 10.1128/EC.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monroy AA, Stappler E, Schuster A, Sulyok M, Schmoll M. A CRE1-regulated cluster is responsible for light dependent production of dihydrotrichotetronin in Trichoderma reesei. PLoS ONE. 2017;12:e0182530. doi: 10.1371/journal.pone.0182530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe N, Arakawa T, Hirota A. The biosynthesis of bisvertinolone: evidence for oxosorbicillinol as a direct precursor. Chem Commun. 2002;3:204–205. doi: 10.1039/b109505f. [DOI] [PubMed] [Google Scholar]
- 41.Harned AM, Volp KA. The sorbicillinoid family of natural products: isolation, biosynthesis, and synthetic studies. Nat Prod Rep. 2011;28:1790–1810. doi: 10.1039/c1np00039j. [DOI] [PubMed] [Google Scholar]
- 42.Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssönen E, Bhatia A, Ward M, Penttilä M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- 43.Merino ST, Cherry J. Progress and challenges in enzyme development for biomass utilization. Adv Biochem Eng Biotechnol. 2007;108:95–120. doi: 10.1007/10_2007_066. [DOI] [PubMed] [Google Scholar]
- 44.Kuhad RC, Deswal D, Sharma S, Bhattacharya A, Jain KK, Kaur A, Pletschke BI, Singh A, Karp M. Revisiting cellulase production and redefining current strategies based on major challenges. Renew Sust Energy Rev. 2016;55:249–272. doi: 10.1016/j.rser.2015.10.132. [DOI] [Google Scholar]
- 45.Mello-de-Sousa TM, Rassinger A, Derntl C, Poças-Fonseca MJ, Mach RL, Mach-Aigner AR. The relation between promoter chromatin status, xyr1 and cellulase expression in Trichoderma reesei. Curr Genomics. 2016;17:145–152. doi: 10.2174/1389202917666151116211812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Häkkinen M, Valkonen MJ, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, Pakula TM. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7:14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, Kou Y, Xu J, Cao Y, Zhao G, Shao J, Liu W. Two major facilitator superfamily sugar transporters from Trichoderma reesei and their roles in induction of cellulase biosynthesis. J Biol Chem. 2013;288:32861–32872. doi: 10.1074/jbc.M113.505826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Portnoy T, Margeot A, Linke R, Atanasova L, Fekete E, Sándor E, Hartl L, Karaffa L, Druzhinina IS, Seiboth B, Crom SL, Kubicek CP. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genomics. 2011;12:269. doi: 10.1186/1471-2164-12-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Hanlon KA, Gallagher L, Schrettl M, Jöchl C, Kavanagh K, Larsen TO, Doyle S. Nonribosomal peptide synthetase genes pesL and pes1 are essential for fumigaclavine C production in Aspergillus fumigatus. Appl Environ Microbiol. 2012;78:3166–3176. doi: 10.1128/AEM.07249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng JJ, Wang XH, Xu D, Fu XX, Zhang XP, Lai DW, Zhou LG, Zhang GZ. Sorbicillinoids from fungi and their bioactivities. Molecules. 2016;21:715. doi: 10.3390/molecules21060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cram DJ. Mold metabolites. II. The structure of sorbicillin, a pigment produced by the mold Penicillium notatum. J Am Chem Soc. 1948;70:4240–4243. doi: 10.1021/ja01192a077. [DOI] [PubMed] [Google Scholar]
- 52.Ma L, Liu W, Huang Y, Rong X. Two acid sorbicillin analogues from saline lands-derived fungus Trichoderma sp. J Antibiot. 2011;64:645. doi: 10.1038/ja.2011.54. [DOI] [PubMed] [Google Scholar]
- 53.Nicolaou KC, Simonsen KB, Vassilikogiannakis G, Baran PS, Vidali VP, Pitsinos EN, Couladouros EA. Biomimetic explorations towards the bisorbicillinoids: total synthesis of bisorbicillinol, bisorbibutenolide, and trichodimerol. Angew Chem Int Ed. 1999;38:3555–3559. doi: 10.1002/(SICI)1521-3773(19991203)38:23<3555::AID-ANIE3555>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 54.Xue X, Wu Y, Qin X, Ma R, Luo H, Su X, Yao B. Revisiting overexpression of a heterologous β-glucosidase in Trichoderma reesei: fusion expression of the Neosartorya fischeri Bgl3A to cbh1 enhances the overall as well as individual cellulase activities. Microb Cell Fact. 2016;15:122. doi: 10.1186/s12934-016-0520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 56.Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ Sci. 2014;7:1446–1453. doi: 10.1039/c3ee41753k. [DOI] [Google Scholar]
- 57.Bayram Ö, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon N-J, Keller NP, Yu J-H, Braus GH. VelB/VeA/LaeA complex coordinates light signal wit h fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 58.Ni M, Yu JH. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS ONE. 2007;2:e970. doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 60.Wiemann P, Sieber CM, von Bargen KW, Studt L, Niehaus EM, Espino JJ, et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santamarta I, López-García MT, Kurt A, Nárdiz N, Álvarez-Álvarez R, Pérez-Redondo R, Martín JF, Liras P. Characterization of DNA-binding sequences for CcaR in the cephamycin–clavulanic acid supercluster of Streptomyces clavuligerus. Mol Microbiol. 2011;81:968–981. doi: 10.1111/j.1365-2958.2011.07743.x. [DOI] [PubMed] [Google Scholar]
- 62.Mast Y, Weber T, Gölz M, Ort-Winklbauer R, Gondran A, Wohlleben W, Schinko E. Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralis. Microb Biotechnol. 2011;4:192–206. doi: 10.1111/j.1751-7915.2010.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SI, Veena, Gelvin SB. Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 2007;51:779–791. doi: 10.1111/j.1365-313X.2007.03183.x. [DOI] [PubMed] [Google Scholar]
- 64.van Hattikum H, Bunduck P, Hooykaas PJJ. Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 2001;22:6550–6558. doi: 10.1093/emboj/20.22.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin H, Wang Q, Shen Q, Ma J, Fu J, Zhao Y. Engineering Aspergillus oryzae A-4 through the chromosomal insertion of foreign cellulase expression cassette to improve conversion of cellulosic biomass into lipids. PLoS ONE. 2014;9:e108442. doi: 10.1371/journal.pone.0108442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Schematic illustration of the plasmid pXBthg-121121. hyg: homomycin resistance; LB, left border of binary vector; RB, right border of binary vector; Kan: kanamycin resistance; 121121-up: the 1500 bp upstream of gene 121121; 121121-down: the 1500 bp downstream of gene 121121. Fig. S2. The 1500-bp upstream and downstream regions of gene 121121. Fig. S3. The standard curve of sorbicillinoids. Fig. S4. The whole genome resequencing of strain ZC121 shows the unsuccessful deletion of gene 121121. Fig. S5. The color of the culture supernatant of T. reesei RUT-C30 (a) and ZC121 (b) grown on glucose. Fig. S6. Cellulolytic enzyme activities in the culture supernatant of T. reesei ZC121 and RUT-C30 grown on 2% cellulose, lactose, glucose, galactose and glycerol were assayed on day 5, including the activities of FPase (the filter paper activity) (a), pNPGase (the BGL activity) (b), pNPCase (the CBH activity) (c), CMCase (the CMC activity) (d) ahd pNPXase (the β-xylosidase activity) (e). The error bars indicate the standard deviations of three biological replicates. Table S1. Primers used in this study. Table S2. Comparative transcription levels of the sorbicillinoid gene cluster and its neighboring cellulase-related genes in T. reesei ZC121on cellulose and glucose.