Abstract
Upregulations of neuronal nitric oxide synthase (nNOS) in the rodent brain have been associated with neuronal aging. To address underlying mechanisms we generated SH-SY5Y neuronal cells constitutively expressing nNOS at a level similar to mouse brain (nNOS+ versus MOCK). Initial experiments revealed S-nitrosylations (SNO) of key players of protein homeostasis: heat shock cognate HSC70/HSPA8 within its nucleotide-binding site, and UBE2D ubiquitin conjugating enzymes at the catalytic site cysteine. HSPA8 is involved in protein folding, organelle import/export and chaperone-mediated LAMP2a-dependent autophagy (CMA). A set of deep redox and full proteome analyses, plus analysis of autophagy, CMA and ubiquitination with rapamycin and starvation as stimuli confirmed the initial observations and revealed a substantial increase of SNO modifications in nNOS+ cells, in particular targeting protein networks involved in protein catabolism, ubiquitination, carbohydrate metabolism and cell cycle control. Importantly, NO-independent reversible oxidations similarly occurred in both cell lines. Functionally, nNOS caused an accumulation of proteins, including CMA substrates and loss of LAMP2a. UBE2D activity and proteasome activity were impaired, resulting in dysregulations of cell cycle checkpoint proteins. The observed changes of protein degradation pathways caused an expansion of the cytoplasm, large lysosomes, slowing of the cell cycle and suppression of proliferation suggesting a switch of the phenotype towards aging, supported by downregulations of neuronal progenitor markers but increase of senescence-associated proteins. Hence, upregulation of nNOS in neuronal cells imposes aging by SNOing of key players of ubiquitination, chaperones and of substrate proteins leading to interference with crucial steps of protein homeostasis.
Abbreviations: BIAM, EZ-LInk Iodoacetyl-PEG2-Biotin; 2D-DIGE, Two-dimensional difference gel electrophoresis; CMA, Chaperone mediated autophagy; ERAD, Endoplasmic reticulum associated death; GO BP, GO CC, GO MF, Gene ontology for biological process, cellular component, molecular function; HSC70/HSPA8, Heat shock cognate of 70 kDa; nNOS/NOS1, Neuronal nitric oxide synthase; NO, Nitric oxide; ORA, Overrepresentation analysis; SILAC, Stable isotope labeling by amino acids in cell culture; SNO, S-nitrosylation; SNOSID, S-nitrosylation site identification; UBE2, Ubiquitin E2 ligase
Keywords: Redox modification, Nitric oxide, Autophagy, Ubiquitin, Chaperone, Lysosome, Posttranslational modification, Starvation, Rapamycin, Senescence
Graphical abstract
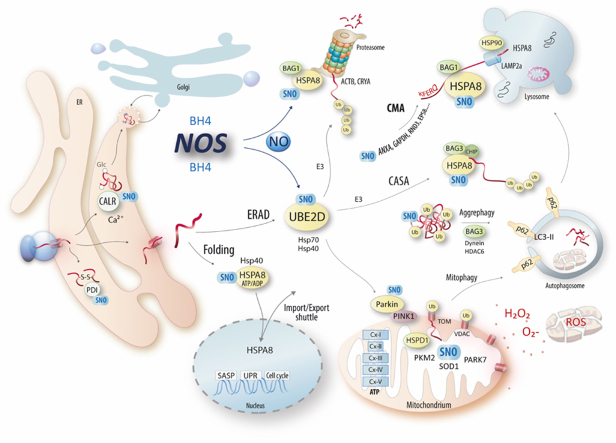
Illustration of direct protein S-nitrosylation (SNO) in protein folding and degradation pathways. Key SNO-targets identified and studied in the present study are HSPA8, and UBE2D isoenzymes. SNOing of Cys17 of HSPA8 likely compromises binding of ATP/ADP, which is essential for HSPA8's functions including protein folding, clathrin uncoating, protein shuttling to and from organelles, chaperone-mediated-autophagy (CMA) and chaperone assisted autophagy (CASA) and proteasomal degradation of specific proteins such as beta-actin. SNOing of UBE2D's catalytic site cysteine reduces its activity and interferes with the degradation of specific proteins, which rely on ubiquitination via UBE2D such as p53. Abbreviations, CMA, Chaperone mediated autophagy; CASA, Chaperone assisted autophagy; ERAD, ER associated degradation; UPS, Ubiquitin-Proteasome System; SASP, Senescence associated secretory phenotype; UPR, unfolded protein response; NOS, nitric oxide synthase; BH4, tetrahydrobiopterin
Highlights
-
•
Direct NO-mediated S-nitrosylation of HSAP8 interferes with chaperone-mediated autophagy. neuroblastoma cells.
-
•
Direct S-nitrosylation of UBE2D reduces ubiquitination and proteasomal degradation proteins.
-
•
NO-evoked disturbances of protein homeostasis lead to an aged and senescent phenotype.
-
•
NO-evoked pro-aging effects agree with NOS upregulations in the aging rodent brain.
1. Introduction
Nitric oxide is produced by nitric oxide synthases, and the neuronal isoform, nNOS/NOS1, is upregulated in the aging brain [1], [2], [3], [4] suggesting that NO-dependent posttranslational redox modifications such as S-nitrosylations (SNO) promote aging and interfere with neuronal functions and longevity. Indeed, protein S-nitrosylations precipitate protein misfolding [5], [6], contribute to the toxicity of beta amyloid protein or mutant Huntingtin [1], [3], [4], [7] and lead to disruptions of protein homeostasis [8], [9], [10], [11], [12], the latter a hallmark of a number of neurodegenerative diseases such as Alzheimer's and Parkinson's disease.
Protein degradation machineries can be direct targets of NO-evoked modifications, or these machineries are over-loaded with oxidized substrate proteins that are hard to digest [5], [8], [13], [14], particularly in the form of oxidized protein aggregates [15], [16]. The latter are normally not present in unstressed cells because endogenous quality control systems maintain protein homeostasis by coordinating protein synthesis and degradation [17], [18]. Likewise, SNO modifications are normally well balanced and constitute subtle transient regulations of protein functions [19], but prolonged cellular stresses such as starvation, radiation, hypoxia or ROS exposure increase the SNO and aggregate burden [20], [21], which is particularly detrimental for neurons [22].
Initial screening experiments revealed SNO modifications of key proteins involved in protein degradation, in particular the heat shock protein, HSC70/HSPA8, a master regulator of chaperone mediated autophagy (CMA) [23], [24], and ubiquitin 2 ligase, UBE2D suggesting that NO-dependent protein allostasis may be key to the understanding of its functions in neuronal aging. Hence, our study was centered on NO-evoked changes of proteostasis.
Eucaryotic cells utilize two major mechanistically distinct, complementary systems for protein degradation, the 26S proteasome, which recognizes client proteins labeled with ubiquitin, and the autophagolysosome [25], [26], [27], [28], [29]. The concerted actions ensure a specific and tightly regulated degradation process, which is highly sensitive to oxidative stress [30], [31], [32], [33], [34], [35]. Oxidized proteins are prone to form large aggregates due to covalent cross-linking or increased surface hydrophobicity and unless repaired or removed, these oxidized proteins are toxic [20], [36], [37], [38], [39].
Autophagy can be disrupted by SNO-modifications of upstream signaling molecules such as c-Jun N-terminal kinase (JNK1), and of the autophagosome assembly machinery [40]. Ubiquitination and proteasomal degradation is affected by S-nitrosylation of the substrate, of proteasome subunits or ubiquitin ligases [5], [6], [41], [42], [43]. Further SNO-ing may impair selective ubiquitin-dependent removal via the autophagosome [26], [44] by masking recognition sites for cargo adapter proteins like sequestosome 1 (SQSTM1/p62) [45], [46]. SQSTM1 accumulates itself if autophagy is impaired, which results in secondary deficits of proteasomal degradation [47], [48], [49].
A further link between the key degradation pathways is the very versatile HSPA8, which contributes to proteasomal degradation of some proteins such as beta actin [50], catalyzes protein folding and clathrin uncoating [51] and contributes to multiple transport and sorting mechanisms for endocytosis, organelle import and export [52], [53], [54] and ubiquitin dependent selective autophagy [55]. All of these functions require ATP binding. HSPA8 is the key carrier in chaperone-mediated autophagy, in which substrate proteins are recognized on the basis of a KFERQ-like motif, and then transferred into the lysosome via the receptor, LAMP2A [56], [57], [58].
CMA is activated in response to prolonged starvation [59], [60], exposure to toxic compounds, or oxidative and nitrosative stress [32], [61], [62], [63], [64], and a decline of CMA efficiency has been linked with aging processes [65]. Its substrates are heterogeneous, mostly cytoplasmic long-lived proteins including GAPDH, pyruvate kinases, annexins, RNAse A, RND3, EPS8 and HSC70/HSPA8 itself [66], [67], [68], [69], [70], which are all important for neuronal integrity.
Hence, NO-mediated redox changes are key to the understanding of proteostasis in the context of cellular stresses, aging and neurodegeneration [71], [72], [73], [74]. To further understand the implications of NO-mediated redox imbalances in this context we generated a cell model, in which the prototypical SH-SY5Y neuroblastoma cells constitutively express nNOS/NOS1 at a level similar to the mouse brain. These nNOS+ versus MOCK SH-SY5Y cells were exposed to "starvation"-evoked stresses and analyzed by a set of proteomic and functional analyses. The results show that NO-evoked protein allostasis leads to neuronal aging.
2. Results
2.1. 2D-SNO DIGE and SNOSID: S-nitrosylation upon rapamycin stimulation
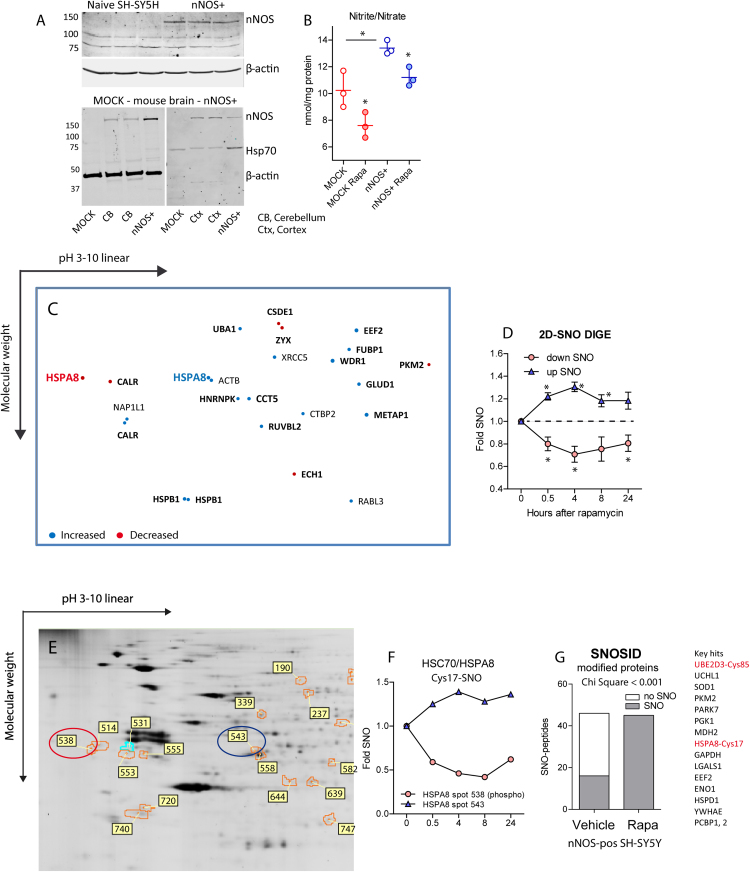
Naïve and MOCK-transduced SH-SY5Y cells did not express nNOS protein at a level strong enough to be detectable by standard infrared Western Blotting (Fig. 1A), but label-free proteomics revealed MOCK nNOS at a level comparable to low abundant proteins. Therefore, we generated SH-SY5Y cells by lentiviral transduction of NOS1, which constitutively express nNOS/NOS1 (nNOS+) at a level comparable to mouse cerebellum and cortex (Fig. 1A) and exceeding MOCK nNOS about 250fold (please see proteomics below), associated with an increase of nitrite/nitrate (Fig. 1B). Hence, nNOS+ cells replicate the situation of neurons, which express nNOS in the brain and upregulate it on aging or in the context of neurodegenerative diseases.
Fig. 1.
Expression of neuronal nitric oxide synthase (nNOS/NOS1), release of nitric oxide (NO) and NO-mediated S-nitrosylation in SH-SY5Y human neuroblastoma cells stimulated with rapamycin. A: Western Blot analysis of nNOS/NOS1 in naïve, MOCK and NOS1-transduced SH-SY5Y cells and mouse brain. Cells were transduced with a lentiviral vector for NOS1 (nNOS+) or backbone control vector (MOCK) followed by FACS cell sorting. β-actin or Hsp70 were used as loading controls. B: Nitric oxide production in MOCK and nNOS+ SH-SY5Y cells without/with rapamycin stimulation for 6 h. Nitrite/nitrate was determined with the Griess method. The scatter represent replicate cultures, the line is the mean, whiskers show the SD. Asterisks indicate significant changes versus vehicle treated MOCK cells (2-way ANOVA for "genotype" by "treatment", subsequent ttests with Šidák adjustment of alpha, adjusted P < 0.05). C: Exemplary 2D-SNO-DIGE spot map on stimulation of nNOS+ cells with rapamycin (1 µM) for 6 h. The size of the spots correlates with the extent of SNO-increase or decrease. Bold printed proteins were confirmed in subsequent assays. D: Fold change of S-nitrosylated proteins (SNO) in 2D-SNO-DIGE in nNOS+ SH-SY5Y cells stimulated with rapamycin (1 µM) (results of the individual proteins in DIB1_Table 1). Asterisks indicate significant time-dependent changes of S-nitrosylation across proteins (2-way-ANOVA within subject factor time, posthoc t-tests versus 0 h with Dunnett adjustment of alpha, P < 0.05). E, F: Exemplary partial detail view of a 2D-SNO DIGE gel showing regulated spots ofHSC70/HSPA8 in nNOS+ cells stimulated for 6 h with rapamycin (1 µM). The numbers indicate the regulated spots, picked and identified by ESI-MS/MS. The line graph shows the quantification of HSPA8 spots over time. G: Binominal frequency distribution of SNO modified proteins in nNOS+ cells on rapamycin stimulation as detected by SNO-site identification analysis (SNOSID, qualitative assay). Individual proteins are listed in DIB1_Table 2.
In initial experiments employing 2D-DIGE, SNOSID and SNO-SILAC, we used rapamycin to challenge proteostasis and impose a "starvation"-like stress. Rapamycin reduced free nitrite/nitrate levels (Fig. 1B), and in parallel, 2D-SNO-DIGE revealed rapamycin-evoked changes of SNOed proteins, which occurred with a maximum between 4 h and 8 h (Fig. 1C, D: spot map and time course; Fig. 1E exemplary gel). SNO-modified candidates are summarized in the accompanying Data-in-Brief article "Redox proteomics" in Table 1 (DIB1_Table 1). The most interesting hits were heat shock cognate of 70 kDa (HSC70/HSPA8), the small heat shock protein HSP27/HSPB1 and calreticulin (CALR), all chaperones and represented in more than one spot (Fig. 1E, F), possibly reflecting phospho-SNO switches. Bold printed hits (Fig. 1C) were confirmed in subsequent analyses.
SNOSID identified some crucial SNO-sites (DIB1_Table 2) in key candidates (Fig. 1G) including HSPA8 (Cys17) and the ubiquitin E2 ligase, UBE2D, the latter particularly interesting because the modified cysteine, Cys85, is the highly conserved catalytic site cysteine of all UBE2D isoforms essential for ubiquitin conjugation (mass spectrograms in Suppl. Figs. 1a, b). UBE2D ligases engage with a number of U-box or single Ring finger type E3 ligases, including CHIP/STUB1, SIAH1, RING1, SCF, XIAP, MDM2, TRAF6 and BRCA1, some of them additional SNO targets as revealed in subsequent analyses (CHIP/STUB1, RING1). Cysteine 17 of HSPA8 is localized within the nucleotide binding region (NBR, subdomain Ia), which is also involved in interactions with its cochaperone, BAG1 [54] and essential for ATP/ADP binding.
Some further SNO-sites were identified in Super SNO-SILAC experiments (DIB1_Table 3), and gene ontology of pooled SNOed proteins (72 individual proteins in 2D-DIGE, SNOSID and SNO Super SILAC) pointed to NO-mediated regulations of proteins involved in protein and carbohydrate metabolism, ubiquitination and Parkinson's disease (DIB1_Table 4).
2.2. Super SILAC: protein accumulation on rapamycin stimulation
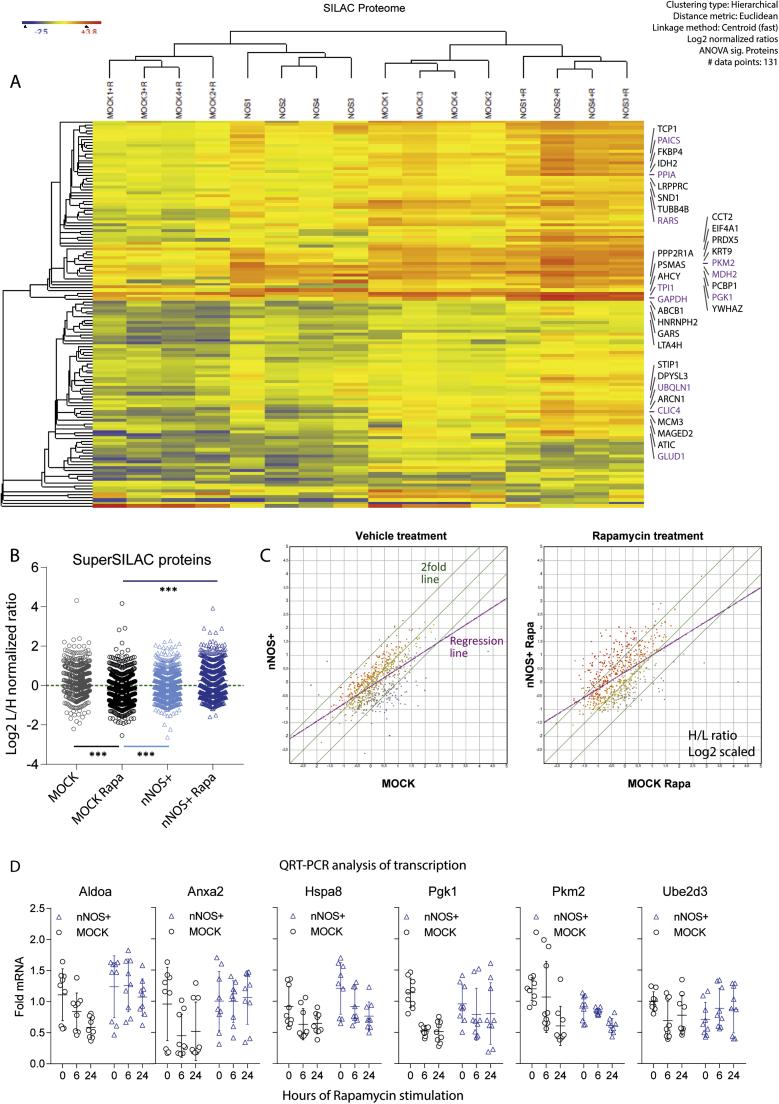
The initial screens suggested that nitric oxide by direct S-nitrosylation impacts on protein homeostasis under "starvation"-like stress evoked by mTOR inhibition. Indeed Super SILAC proteomics revealed higher protein levels in rapamycin-stimulated nNOS+ cells at 6 h as compared to MOCK cells, likely caused by protein accumulation (Fig. 2A-C, Data in Brief "Full Proteome" DIB2_Table 1) rather than transcription, because mRNA levels of candidate proteins were similar in nNOS+ and MOCK cells (Fig. 2D). Differentially expressed proteins (131 hits; ANOVA P and q < 0.05; DIB2_Table 1) encompassed 18 documented and putative CMA substrates, which carry one or more of 20 published KFERQ-like motifs [68], [69], [70], [75], [76], [77] (Fig. 2A, some highlighted in pink) including HSPA8, GAPDH, PGK1, PKM2, ANXA2, ALDOA and EEF1 and 2. Transcription of these candidates rather decreased on rapamycin stimulation or remained constant and did not differ between cell lines as far tested (Fig. 2D). Further CMA substrates were increased in nNOS+ cells on serum-free starvation including RND3 [68] and EPS8 [69] (Fig. 7C). Gene Ontology Overrepresentation analysis (ORA) revealed that differentially expressed proteins were involved in four major "biological processes" that can be captured with “protein folding”, “glycolysis”, “response to stress” and "protein complex assembly” (DIB2_Table 2).
Fig. 2.
Super SILAC analysis of protein expression in MOCK and nNOS+ SH-SY5Y neuroblastoma cells treated with vehicle or 1 µM rapamycin for 6 h A: Heatmap showing the Euclidean hierarchical clustering result of 131 significantly regulated proteins (ANOVA P and q < 0.05, Benjamini Hochberg adjusted) out of 669 valid proteins. The colour scale ranges from minus to plus 4 SD. Some exemplary CMA targets (with confirmed KFERQ-like motifs) are indicated. The list of all valid proteins (DIB2_Table 1, GO DIB2_Table 2) and of SNO-modified proteins (DIB1_Table 3, GO DIB1_Table 4) are presented in accompanying Data-in-Brief articles. B: Scatter plot showing Log2 transformed normalized Light/Heavy (L/H) ratios of valid proteins revealing an increase of protein levels in nNOS+ on stimulation with rapamycin (univariate ANOVA, posthoc t-tests with adjustment of alpha according to Šidák; adjusted P < 0.05). C: X-Y Scatter plots of normalized L/H ratios (log2 scaled) comparing genotypes on vehicle or rapamycin stimulation. The green lines show the 2fold range, the purple line is the regression line. B: Analysis of the mRNA by quantitative RT-PCR of candidate proteins, which accumulated in nNOS+ cells on rapamycin stimulation. There were no upregulations at the transcriptional level. Data are the ratios of normalized deltaCt values (2-ΔCt/2-ΔCt of MOCK at 0 h). Cycle numbers of two housekeeping genes, RPL37A and ACTB, were averaged for normalization. Each scatter is one culture, the line is the mean, error bars show the SD.
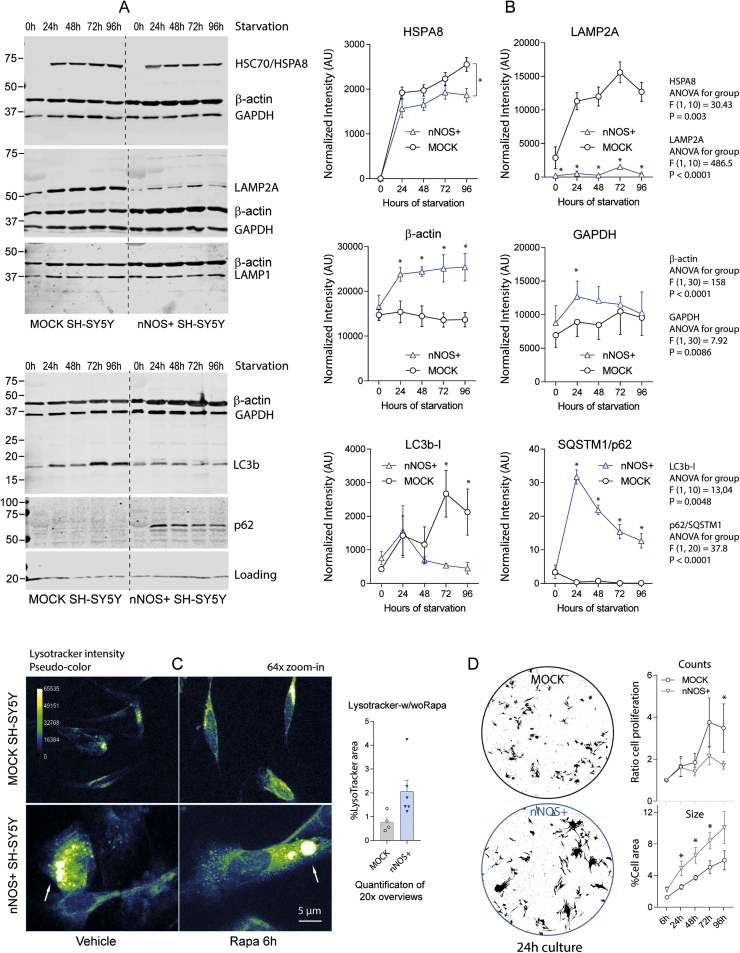
Fig. 7.
Western Blot analysis of chaperone-mediated and macro-autophagy on starvation in nNOS+ and MOCK SH-SY5Y cells and lysosome morphology. A, B: Exemplary blots and quantification (right panel line graphs) showing the time course of HSC70/HSPA8, LAMP2A, LAMP1, GAPDH, β-actin, LC3b-I/II and SQSTM1/p62. Blots were sequentially developed and infrared images superimposed. LC3b-II was not detected in some blots. p62 accumulated in nNOS+ cells but was not detected in MOCK cells. LAMP2A was almost absent in nNOS+ cells, whereas β-actin and GAPDH accumulated. The quantification shows the mean ± SD of ≥ 3 independent experiments. Intensities were normalized for blot loading. Asterisks show group differences at individual time points (2-way ANOVA, "genotype" X "time", posthoc t-tests with Šidák adjustment of alpha; adjusted P < 0.05). C: Lysotracker analysis of lysosome morphology in nNOS+ and MOCK SH-SY5Y cells treated with vehicle or 1 µM rapamycin for 6 h. Immunofluorescent lysotracker intensities are shown in pseudo-colour. nNOS+ cells have huge lysosomes (arrows). The normal lysosome size is 0.5–1 µm. D: Cell culture morphology of nNOS+ and MOCK SH-SY5Y cells at 24 h in culture. The line graph shows the ratio of cell proliferation versus baseline and the percentage area covered by cell bodies, the latter an indicator of cell size. Data are the mean and SD of 3 independent experiments. 2-way ANOVA as in B, * <0.05.
2.3. Deep redox proteome analyses on starvation
Motivated by the initial observations pointing to SNO-mediated changes of HSPA8 and in ubiquitination-dependent protein degradation pathways we performed deep proteome analyses to identify SNO-modified protein networks and pathways (SNO-BIAM Switch, Fig. 3), reversibly oxidized proteins (REDOX-BIAM switch, Fig. 4) and consequences for the full proteome (Fig. 5) on serum-free starvation. Starvation is considered to trigger CMA and links autophagy with ubiquitin-dependent proteasomal degradation [29], [60], [78].
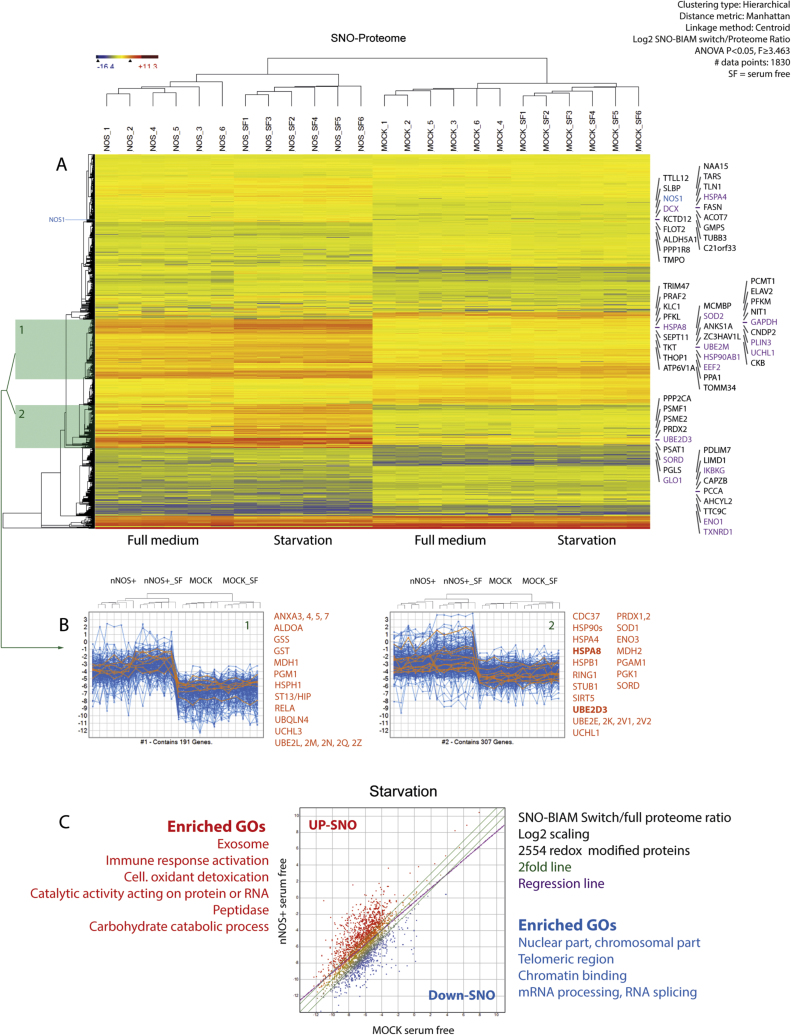
Fig. 3.
SNO-BIAM Switch analysis of protein S-nitrosylations in nNOS+ and MOCK SH-SY5Y neuroblastoma cells subjected to 24 h of serum free starvation. A: Heatmap of Euclidean hierarchical clustering of 1830 proteins with significant group-dependent differences of SNO-modifications (ANOVA P and q < 0.05, Benjamini Hochberg adjusted, 1830 displayed out of 2554 valid proteins). The data show LOG2-transformed ratios of SNO-BIAM Switch/full proteome to address changes in protein expression. The colour scale ranges from minus to plus 4 SD. B: Thumbnail line graph views of two major clusters (highlighted in green in 3A) with strong SNO upregulations under starvation in nNOS+ cells. The upper dendrograms show the experimental clusters (as in A). The orange lines show exemplary candidate proteins in part listed in orange. C: XY-Scatter plots of nNOS+ versus MOCK cells showing the SNO-BIAM Switch/Proteome LFQ intensity ratios (log2 scaled) on starvation. SNO-BIAM identified 2777 valid modified proteins out of which 2554 could be matched with the full proteome (DIB1_Table 5) and are presented. The green lines show the 2fold range, the purple line is the regression line.
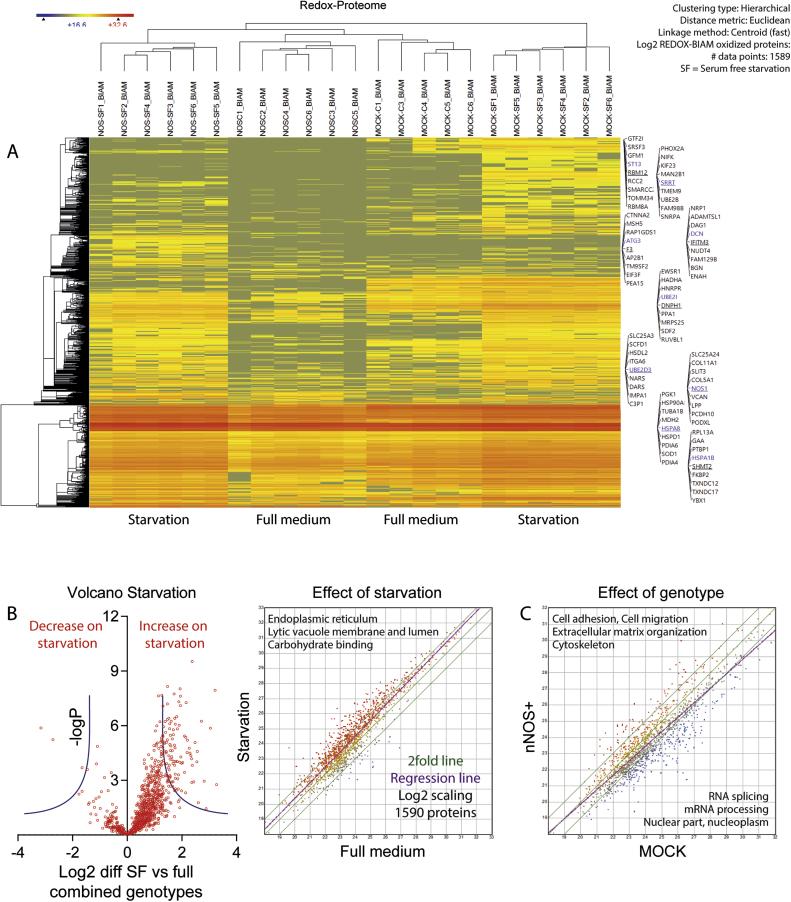
Fig. 4.
REDOX-BIAM switch analysis of reversible protein oxidations in nNOS+ and MOCK SH-SY5Y neuroblastoma cells subjected to 24 h of serum free starvation A: Heatmap of Euclidean clustering of 1589 identified valid proteins. "Zero" data were set to the minimum for clustering. The colour scale ranges from minus to plus 4 SD. B: Volcano plots of REDOX-BIAM switch proteins comparing starvation with full medium. nNOS+ and MOCK are combined. Proteins with decreased oxidation under starvation appear on the left side of the Y-axis. C: XY-Scatter plots of REDOX-BIAM switch LFQ intensities (log2 scaled) of 1589 valid proteins (DIB1_Table 6). The green lines show the 2fold range, the purple line is the regression line. Major GO terms associated with the regulated proteins are indicated in the upper left corner for increased proteins, in the lower right corner for decreased proteins. SF = serum free.
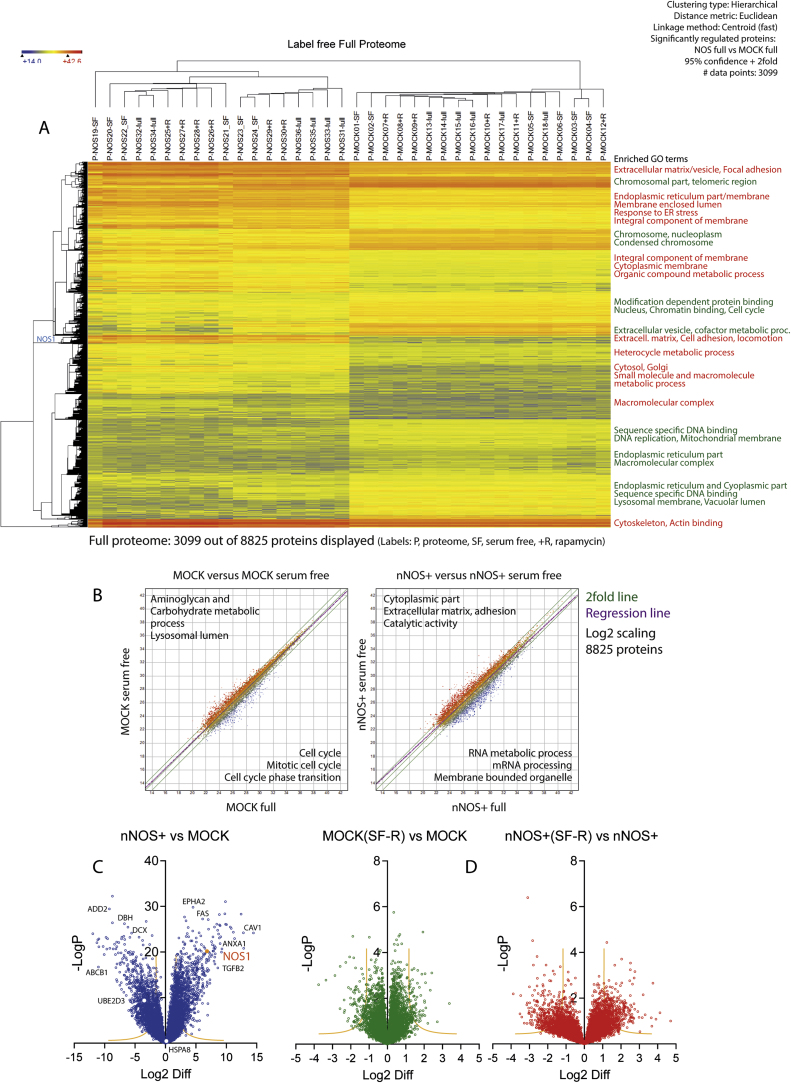
Fig. 5.
Full proteome analysis in nNOS+ and MOCK SH-SY5Y neuroblastoma cells subjected to 24 h of serum free starvation (SF) or rapamycin (+R, 1 µM) treatment versus full medium (full). A: Heatmap of Euclidean hierarchical clustering of 3099 significantly regulated proteins out of 8825 valid non-ambiguous proteins (DIB2_Table 3), which differed between nNOS+ and MOCK cells under full-nutrition conditions. The colour scale ranges from minus to plus 4 SD. Major GO terms (CC and BP) associated with the respective clusters are indicated in the right, red for proteins increased in nNOS+ , green for reduced proteins. On the left side, the position of NOS1/nNOS itself is shown. The experiments clustered into 2 major groups representing the genotypes. B: XY-Scatter plots of LFQ intensities of 8825 proteins (log2 scaled) show stronger effects of starvation in nNOS+ (right) than MOCK (left) cells. The green lines show the 2fold range, the purple line is the regression line. Major GO terms associated with the regulated proteins are indicated (upper left GO terms of increased proteins, lower right GO terms of decreased proteins). C, D: Volcano plots of the proteome (8825 proteins) comparing effects of the genotype (nNOS+ versus MOCK), and effects of combined "rapamycin or serum free starvation" versus "full medium" separately for MOCK and nNOS+. White dots in C indicate HSPA8 and UBE2D3. Proteins reduced in nNOS+ cells, or reduced under starvation appear on the left side of the Y-axis. nNOS itself was about 250-fold higher in nNOS+ than MOCK cells (log2 difference of 7.98, orange dots in C).
SNO-BIAM Switch and REDOX-BIAM Switch assays (DIB1_Table 5 and DIB1_Table 6) provided quantitative data of redox-modified proteins including low-abundant proteins, and the respective cysteine sites were identified in a number of proteins. Both Thiol-BIAM-switch approaches confirmed oxidation of Cys85 of UBE2D3 (Peo-Biotin site in DIB1-Table 6), and revealed Cys17 (also identified by SNOSID), Cys574 and Cys603 as redox sensitive sites in HSPA8 (DIB1-Table 5).
2.3.1. SNO-BIAM Switch assay for cysteine S-nitrosylations
In total, 2777 SNO-modified proteins were identified, out of which 2554 were matched with the full proteome and then transformed into the ratio to account for changes of protein levels. The proportion of nitrosylation of NOS1 (Fig. 3A, DIB1_Table 5) remained nearly constant despite its higher expression in nNOS+ cells (about 250fold according to the full proteome), showing that the SNO-BIAM Switch / Proteome ratio was well corrected for changes of protein expression. ANOVA of these ratios identified 1830 significant proteins (ANOVA P and q < 0.05, Benjamini Hochberg adjusted), which grouped into 10–12 major clusters (Fig. 3A) and revealed a clear separation of genotypes and treatments. Starvation for 24 h had minor effects in MOCK cells but increased levels of SNO modified proteins in nNOS+ cells (Fig. 3A–C). Thumbnail line graphs views of highlighted clusters (Fig. 3B) show key candidates, and scatter plots (Fig. 3C) reveal major gene ontology terms for "biological process" (GO BP) and "cellular compartment" (GO CC). In particular, proteins involved in protein and carbohydrate metabolism and redox control were increasingly SNO-modified in nNOS+ cells on starvation (Fig. 3B, C), whereas SNO modifications decreased in proteins involved in RNA processing (Fig. 3C).
2.3.2. REDOX-BIAM Switch Assay for oxidative protein modifications
The REDOX-BIAM switch assay primarily detects reversible oxidations including disulfides and sulfoxides and identified 1590 modified proteins in total (Fig. 4; DIB1_Table 6), out of which 1242 were significantly different between groups (ANOVA P and q < 0.05; 870 if only proteins were considered, which were detected in ≥2 samples of at least 2 groups). The number of oxidized proteins increased substantially on starvation, both in MOCK and in nNOS+ cells, particularly in proteins localized to the endoplasmic reticulum and the lysosome (GO CC) and involved in carbohydrate binding (GO BP) (Fig. 4B shows effects of starvation). Comparison of genotypes irrespective of treatments (Fig. 4C) reveals an nNOS+ mediated pro-oxidation of proteins involved in extracellular matrix organization, cytoskeleton and adhesion but down-oxidation of proteins involved in RNA processing. The similarity of starvation-evoked oxidation in MOCK and nNOS+ cells suggests that the presence of nNOS does not generally increase oxidation but rather specifically S-nitrosylation.
2.4. Full Proteome under starvation and rapamycin stimulation
Full proteome analysis identified 8825 non-ambiguous proteins with ≥ 6 valid samples in total, out of which 3099 were differentially expressed between nNOS+ and MOCK cells on full nutrition (95% confidence, plus 2fold change) (Fig. 5A–D; DIB2_Table 3), with major differences between genotypes (5A, B, D) but mild effects of starvation or rapamycin at this time point. Treatment effects were stronger in nNOS+ than MOCK cells (Fig. 5B, D). NOS1/nNOS itself (indicated in 5A and C) was 250fold higher in nNOS+ as compared with MOCK cells. HSPA8a protein levels did not differ and UBE2D3 levels were lower in nNOS+ (Fig. 5C white dots). Overall, there was an even distribution of up- and downregulated proteins in nNOS+ versus MOCK cells (Fig. 5A, C), but GO analyses revealed different functions (Fig. 5A, B). Increased proteins in nNOS+ cells were involved in ER stress response, extracellular matrix organization, adhesion and metabolic processes (GO BP), whereas proteins localized to lysosomal membranes or chromatin (GO CC) or involved in cell cycle processes were reduced (Fig. 5A). Filtering proteins for those carrying known or predicted KFERQ-like motifs revealed a strong increase of cytoskeletal-associated proteins in nNOS+ cells.
2.5. Macroautophagy and chaperone mediated autophagy
2.5.1. Rapamycin
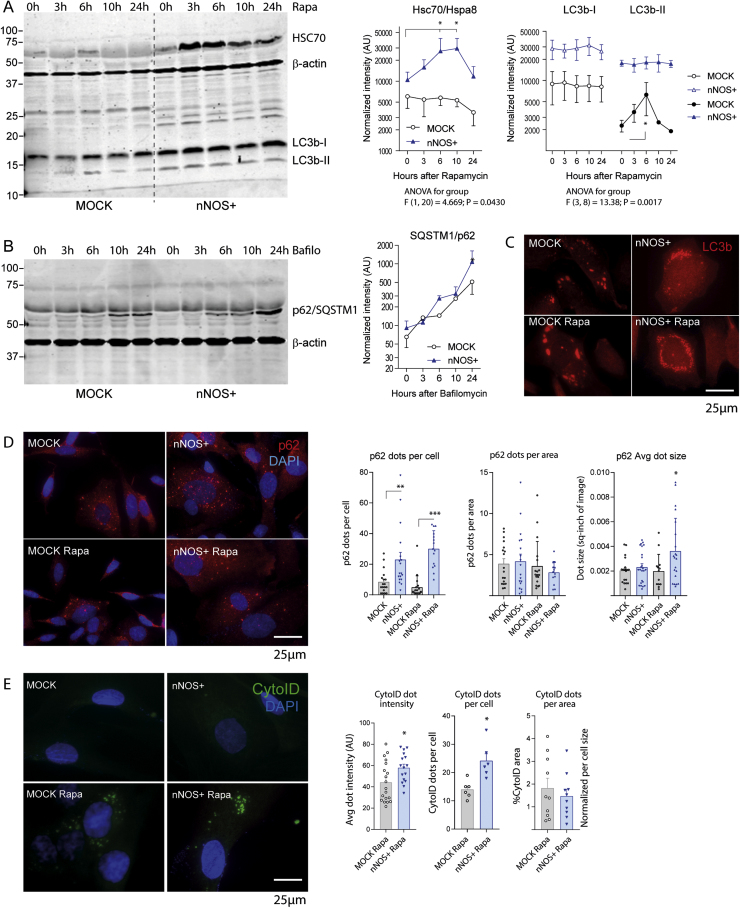
To further assess the impact of NO on specific protein degradation pathways, we analyzed standard markers of macroautophagy and chaperone mediated autophagy (CMA) using Western Blot and immunofluorescence analyses. On stimulation with rapamycin, we observed a time dependent increase of HSC70/HSPA8 in nNOS+ cells, but not in MOCK cells with a peak at 3–6 h (Fig. 6A; cell homogenate). The result agrees with the rapamycin-proteomes (Fig. 2, Fig. 5), i.e. increase at 6 h, but no difference at 24 h. LC3b-I (cytoplasm) and LC3b-II (autophagosome) levels were both higher in nNOS+ cells irrespective of the stimulation with rapamycin. LC3b-II mildly increased in MOCK cells at 6 h (Fig. 6A), which is the time of maximum effects of rapamycin. Rapamycin had no effect on p62 (not shown) but blocking the autophagic flux with bafilomycin resulted in a p62 accumulation in both cell lines (Fig. 6B), somewhat earlier/stronger in nNOS+ cells. The latter agrees with the proteomic results (SQSTM1/p62 9.9fold higher in nNOS+, -LogP 13.25). Immunofluorescent detection of LC3b-positive, p62-positive and CytoID-positive dots (Fig. 6C, D, E) revealed a higher number of p62-positive and CytoID-positive dots per cell in nNOS+ cultures, but similar numbers per area of the cytoplasm. In addition, the average p62 dot size was larger in nNOS+ cells. The abundance of LC3b-positive dots was similar in both lines. The results were somewhat inconclusive but pointed to lower efficiency of protein removal in nNOS+ cells.
Fig. 6.
Western Blot and immunofluorescence analysis of autophagy on rapamycin stimulation of nNOS+ and MOCK SH-SY5Y cells. A: Exemplary blots and quantification (right panel line graphs) showing the time course of HSC70/HSPA8, LC3b-I and LC3b-II. Blots were sequentially developed and infrared images superimposed. Intensities were normalized for protein loading and show the mean ± SD of ≥ 3 experiments. Asterisks indicate time-dependent significant changes within groups (2-way ANOVA, "genotype" X "time", posthoc t-tests versus baseline with Dunnett adjustment of alpha, adjusted P < 0.05). B: Exemplary blot and quantification showing accumulation of p62/SQSTM1 on stimulation with bafilomycin in both groups. The asterisk shows a significant difference between groups (2-sided unpaired ttests of Log10 transformed intensities at 24 h). C: Immunofluorescence of LC3b positive dots in nNOS+ and MOCK cells without/with stimulation with rapamycin for 6 h. The average number of LC3b dots per cell was higher in nNOS+ cells, but the average dot-size was higher in MOCK cells. D: Immunofluorescence analysis and quantification of SQSTM1/p62 with nuclear DAPI counterstain without/with rapamycin stimulation for 6 h. The average number of p62 immunoreactive dots per cell representing autophagolysosomes was higher in nNOS+ cells, and the average dot-size increased on rapamycin stimulation. The number of dots per area of the cytoplasm did not differ between groups. Asterisks indicate significant differences versus the respective control group as indicated (2-way ANOVA "genotype x treatment", subsequent t-tests versus specific controls using a Šídák-Bonferroni adjustment of alpha). E: CytoID® fluorescence of autopahosomes with nuclear DAPI counterstain on rapamycin stimulation for 6 h. The average intensity and average numbers of CytoID positive dots was higher in nNOS+ cells. Asterisks show significant differences between groups (2-sided, unpaired t-tests).
2.5.2. Starvation
The differences in HSPA8 levels pointed to changes in CMA, which is not normally affected by mTOR inhibition but may get constitutively active to remove toxic protein waste [79]. To further address CMA, cells were exposed to serum free starvation. HSC70/Hsp8 levels were similar in MOCK and nNOS+ cell homogenates (Fig. 7A). The most striking difference was a strong reduction of LAMP2A in nNOS+ cells and in parallel, accumulation of β-actin, GAPDH and p62 (Fig. 7A Western Blots, 7B quantification). GAPDH is a CMA substrate, while β-actin is supposed to undergo proteolytic cleavage on starvation [80] but its ubiquitin-dependent degradation via the proteasome depends on HSPA8 [50]. A search of the proteome for proteins carrying a motif highly similar to KFERQ retrieved 750 proteins, out of which 75 were significantly increased in nNOS+ cells (Suppl. Fig. 2). If they constitute CMA substrates depends on the exposure of the motif towards HSPA. Along with the accumulation of proteins, we observed enlarged lysosomes and an expansion of the cytoplasm or nNOS+ cells (Fig. 7C), whereas cell proliferation was reduced (Fig. 7D).
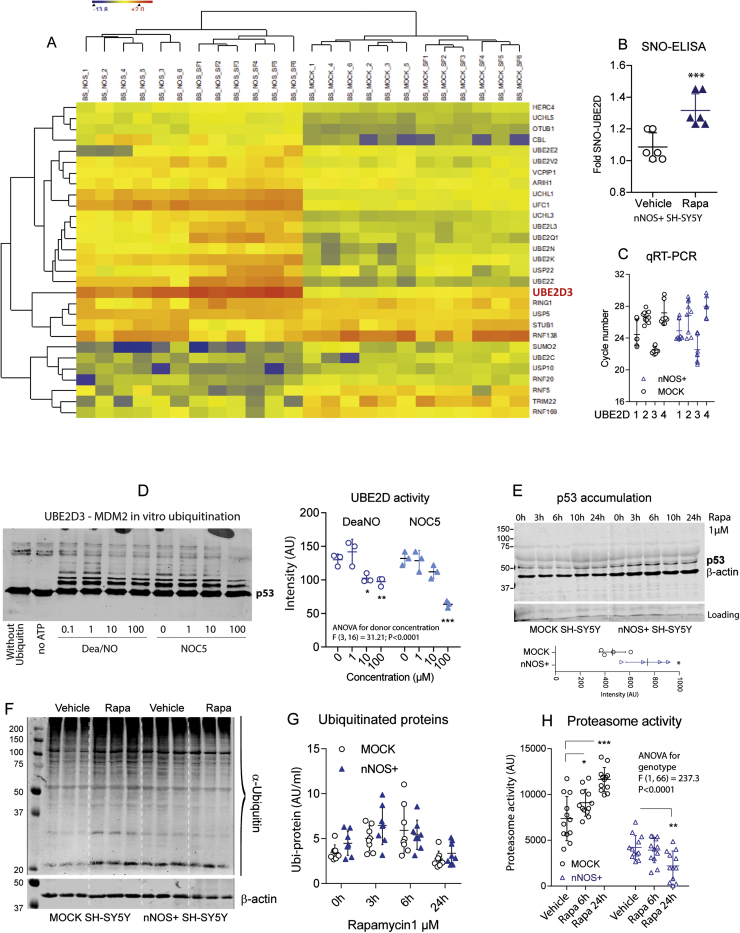
2.6. Ube2D activity and proteasome functions
Our initial experiments suggested that Cys85 of the catalytic site of UBE2D isoforms is a functionally relevant SNO-target in the ubiquitin proteasome system (UPS). Cys85, identified in SNOSID, SNO-SILAC and REDOX-BIAM, is highly conserved, and is required for the transfer of ubiquitin from E2 to E3 ligases. The SNO-BIAM Switch proteome revealed SNO modifications of further UBE2 enzymes (Fig. 8A). UBE2D3 was one of the strongest hits and its SNO-modification was further confirmed by SNO-immunoassay [81] (Fig. 8B). QRT-PCR analysis showed that SH-SY5Y cells primarily express UBE2D3 (Fig. 8C, lowest cycle number) in line with the proteomic data. Hence, we assessed effects of NO-donors on UBE2D3 enzyme activity in vitro. The test relies on in vitro ubiquitination of p53 by the E3 ligase MDM2, which is activated by UBE2D3. The NO donors DEA/NO and NOC-5 reduced the levels of ubiquitin-conjugated p53 in a dose dependent manner (Fig. 8D), and nNOS+ cells showed an increase of p53 levels upon rapamycin stimulation (Fig. 8E) suggesting reduced or slower ubiquitination-dependent removal of p53, which occurs mostly via the proteasome. General poly-ubiquitination as assessed per anti-ubiquitin Western Blotting and ELISA was not impaired in nNOS+ cells (Fig. 8F, G) but proteasome activity was reduced and further decreased on rapamycin stimulation (Fig. 8H), suggesting a profound impairment of UPS mediated protein degradation.
Fig. 8.
NO mediated redox modifications of UBE2D3 and functional consequences. A: Heatmap displaying 29 redox-modified significantly regulated proteins involved in ubiquitin conjugation and deconjugation (Euclidean hierarchical clustering, centroid linkage). The data show the SNO-BIAM Switch/Proteome ratio, Log2 transformed. The search criteria were "UBE", "UCL", "USP" in protein name or "E2", "E3" or "ubiquitin" in protein description. UBE2D3 was the strongest "up-oxidized" enzyme in this search. B: Confirmation of UBE2D S-nitrosylation using a SNO-BIAM Switch based sandwich ELISA with anti-UBE2D capture and anti-Biotin detection (*** P < 0.001; unpaired 2-sided t-test). C: Quantitative RT-PCR analysis of UBE2D isoenzymes in unstimulated nNOS+ and MOCK SH-SY5Y cells. UBE2D3 is the major isoform in both groups (lowest cycle number required for passing the threshold). D: Western Blot (left) and quantification (right) showing ubiquitination of His6-p53 (substrate) in an in vitro ubiquitination assay that uses recombinant proteins of E1 ligase, UBE2D3 (E2) and MDM2 (E3). The reaction was performed in the absence and presence of NO donors (DEA/NO and NOC-5) and quantified (scatter plot) via analysis of anti-p53 immunoblot. Asterisks indicate significant differences versus vehicle (2-way ANOVA, subsequent t-tests versus vehicle with Dunnett adjustment of alpha; adjusted P < 0.05). E: Western Blot and quantification showing p53 accumulation in nNOS+ cells as compared to MOCK cells on stimulation with rapamycin. F, G: Protein ubiquitination as assessed by Western Blot and ELISA using anti-ubiquitin detection. Rapamycin increased the abundance of ubiquitin-bound proteins in nNOS+ and MOCK cells without differences between genotypes. H: Proteasome activity as assessed by measuring the proteolytic activity of the 20 S core unit using a fluorogenic substrate. Proteasome activity increased on rapamycin stimulation in MOCK cells but decreased in nNOS+ cells. Asterisks indicate significant differences at the respective time points (2-way ANOVA for "genotype" by "treatment", subsequent t-tests with Šidák adjustment of alpha; adjusted P < 0.05). Scatter plots show results of individual experiments, the line is the mean and the whisker show the SD.
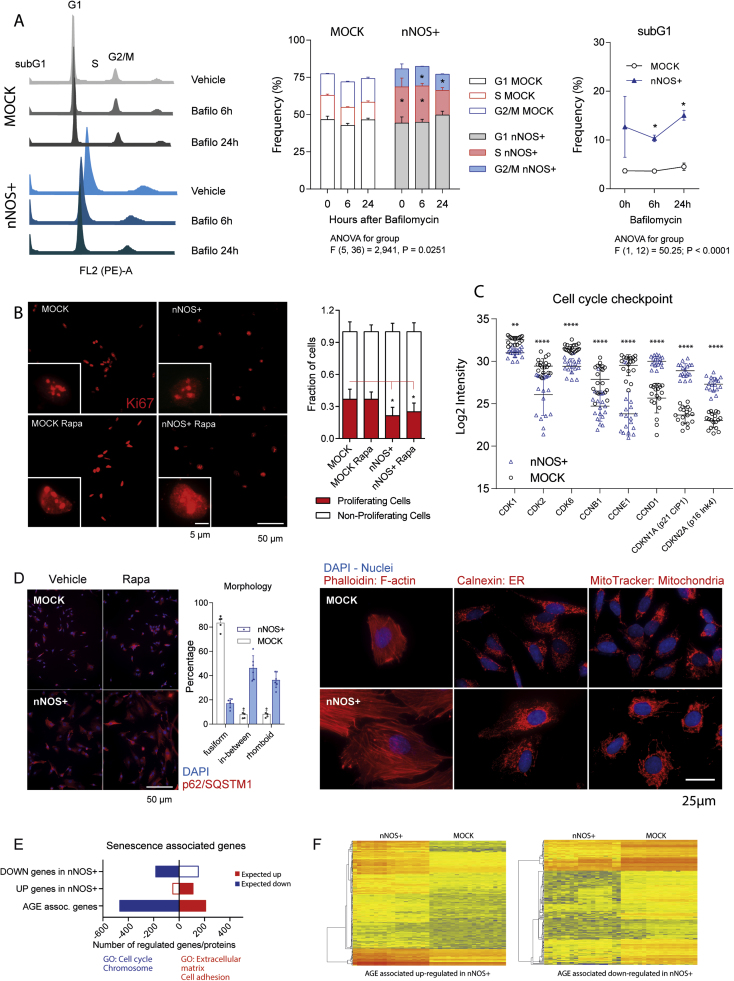
2.7. Cell cycle, proliferation and senescence
CMA and UPS are crucial for the regulation of the cell cycle [82], which was one of the GO terms associated with differentially expressed SNO-modified proteins. In addition HSPA8-BAG3 complexes are crucial for proper actin dynamics and spindle orientation during mitosis [83], [84]. Indeed, FACS analyses of the cell cycle revealed a higher fraction of subG1 cells in nNOS+ cultures, considered as apoptotic, and a lower fraction of G2/M cells (Fig. 9A) in line with the observed decrease of proteins involved in chromatin condensation and mitosis (proteome Fig. 5). Consequently, the number of Ki67 positive proliferating cells was reduced in nNOS+ cultures (Fig. 9B) and the proteome revealed a lower expression of cyclin dependent kinases but increase of cyclin D1, which is normally degraded via ubiquitin-dependent proteolysis at the end of the S-phase, allowing the entry into the M-phase. In addition, the CDK inhibitors p21-CIP1 (CDKN1A) and p16-Ink4 (CDKN2A) were strongly increased (Fig. 9C), in agreement with an "aged" phenotype of nNOS+ cells. Both are markers of senescent cells [85]. The change of the phenotype was reflected by morphometric changes of the cell architecture (Fig. 9D) showing an expansion of the cytoplasm and large distances to neighboring cells, the latter suggesting alterations of the secretome and extracellular matrix, reminiscent of senescent cells. Regulated genes were therefore compared with senescence-associated genes provided by the Human Senescence Gene Database (HSCGD; http://bioinfo.au.tsinghua.edu.cn/member/xwwang/HCSGD/index.html) [85], which is based on an integrative meta-analysis comprising 20 GEO datasets. Out of consistently age-associated genes (-logP > 3) 680 could be matched with the full proteome, and two-third were significantly regulated in nNOS+ cells (Fig. 9E, F). There was particularly high agreement with upregulated "pro-aging" candidates (Fig. 9F), whereas "anti-aging" genes that are supposed to be downregulated on senescence, were up or down-regulated in nNOS+ cells. The analysis agrees with a pro-aging phenotype but not/not yet with cellular senescence.
Fig. 9.
Cell cycle analysis, proliferation and morphology of nNOS+ and MOCK cells. A: FACS analysis of the cell cycle, based on propidium iodide staining in vehicle and bafilomycin stimulated cells. Stacked bar charts show the relative frequency of cells in G1, S and G2/M. The frequency of subG1 cells representing the apoptotic fraction is shown in the right line graph. Asterisks show significant differences between groups (2-way ANOVA for "genotype" by "treatment", subsequent t-tests with Šidák adjustment of alpha, adjusted P < 0.05). B: Exemplary immunofluorescence images of the proliferation marker, Ki67 and its quantification. The inserts show zoom-in images of individual cells. Asterisks show significant differences between groups as indicated (2-way ANOVA for "genotype" by "treatment", subsequent t-tests with Šidák adjustment of alpha, adjusted P < 0.05). C: Scatter plots showing the Log2 intensity of protein expression (full proteome data) of cyclins and cyclin dependent kinases and CDK-inhibitors in nNOS+ and MOCK cells. Asterisks show significant differences between groups (2-way ANOVA for "genotype" by "candidate protein", subsequent t-tests with Šidák adjustment of alpha, adjusted **P < 0.01; ****P < 0.0001). D: Immunofluorescence analysis of the cell and organelle morphology. The frequency of fusiform, rhomboid and "in-between" cells per image was counted with help of ImageJ by setting size inclusion/exclusion and circularity limits. Each scatter represents one culture. Frequencies differed significantly between groups (ChiSquare P < 0.005). E; F: Stacked bar charts and thumbnail heat maps showing the frequency and regulation of senescence-associated genes (HSCDB integrative meta-analysis, significant FDR adjusted genes), which were significantly regulated in nNOS+ versus MOCK cells (ANOVA P and q < 0.05, 2fold change). The agreement was high for upregulated genes. Genes downregulated on aging according to the Human Senescence Database, HSCDB, were mostly regulated in nNOS+ cells, but not unequivocally up or down.
3. Discussion
The present study identified protein S-nitrosylations and reversible oxidations on stimulation with rapamycin or starvation in nNOS+ SH-SY5Y human neuroblastoma cells, particularly targeting proteins involved in maintenance of protein homeostasis via chaperone mediated autophagy and ubiquitin-dependent protein disposal, with the prime candidates HSC70/HSPA8 and UBE2D isoenzymes. The oxidative modifications were associated with profound changes of the proteome pointing to nNOS-evoked pro-aging effects, which agrees with upregulations of nNOS in rodent brain on aging in association with a cognitive decline [1], [3], [4], [86]. The S-nitrosylations of key proteostasis pathways – particularly CMA and UPS - lead to accumulation of known or predicted CMA clients, of inhibitory cycle check-point proteins and of senescence-associated proteins, but decrease of neuronal precursor proteins and proteins involved in RNA processing and chromatin assembly, hence limiting renewal.
SH-SY5Y are the most popular model for research of aging and neurodegeneration, and various strategies have been developed to impose lineage differentiation, aging or disease-like phenotypes. The comparison of the proteome suggests that constitutive nNOS expression combines all of this, i.e. evokes a phenotypic switch that is known to occur upon retinoic acid treatment, sequential starvation [87], [88] or pro-aging genetic or pharmacologic manipulations [88], [89]. Expression of synaptic vesicle proteins (e.g. syntaxins) and differentiation or subtype markers (NOTCH1, 3, CRIM1, RND3, MOXD1) were increased in nNOS+ cells, whereas expression of neuronal progenitor makers (e.g. GAP43, DCX, NES, PHOX2A), proteins involved in neuron morphogenesis (NRCAM, MAP2, UNC5C) or neurotransmitter production and release (DBH, SLC18a3) were reduced. nNOS per se is no regulator of transcription, and transcription of candidate genes was unaltered. We therefore assume that the phenotypic change is secondary to the modification of protein disposal networks, which would agree with previous studies showing that oxidation-evoked modifications of the proteasome disrupt the UPS and consequently promote aging [16], [34], [90], [91], [92], [93], [94]. It is of note that signaling through retinoic acid alpha receptors leads to inhibition of CMA [64], and that CMA slowing also promotes aging [95] suggesting that retinoic acid evoked SH-SY5Y differentiation/aging works in part through accrual of CMA substrates. Reduced efficacy of protein degradation interferes with the cell cycle [82], [96], imposes ER stress [97] and if maintained, translation and protein folding will be impaired. Indeed, the rate of proliferation was reduced in nNOS+ cells. The cytoplasm was enlarged and was associated with alterations of extracellular proteins pointing to changes in adhesion, migration and a senescence associated secretory phenotype (SASP) [98]. It has to be considered that SH-SY5Y are slowly proliferating cells with doubling times of 2–3 days, which may explain why MOCK proteomes were only weakly affected by starvation for 24 h despite substantial oxidative modifications at this time point. Overall, effects of rapamycin or starvation were stronger in nNOS+ cells, but the predominant changes of the proteome depended on nNOS+ expression per se and were accentuated by the stimulation, suggesting that constitutive high or higher than normal levels of nNOS+ impose proteostasis stress. nNOS+ SH-SY5Y cells well replicate the situation of aging neurons, which upregulate nNOS in the aging brain.
Differences between nNOS+ and MOCK cells were stronger in the SNO-BIAM Switch assay than REDOX-BIAM switch assay. Although the principle of these assays is similar, the mild ascorbic acid evoked reduction in the SNO-BIAM Switch ensures that mainly SNO-modifications are detected, whereas REDOX-BIAM mainly detects other reversible oxidations including sulfoxidation and disulfide bridges. Hence, from the comparison of both assays we infer that nNOS expression does not generally increase oxidative events but rather specifically modifies proteins by S-nitrosylation, hence suggesting a specific pro-aging role of NO. Both assays are sensitive to detect low abundant proteins and therefore reveal redox modifications of protein networks.
Our initial experiments suggested that HSC70/HSPA8 and UBE2D isoenzymes are key candidates and the highly sensitive BIAM switch assay allowed us to describe the complex SNO-proteome under starvation encompassing roughly 2500 candidates and the ox-proteome with about 1600 candidates. The initial results were confirmed, and they showed the modifications in biological context of carbohydrate metabolism, protein ubiquitination and degradation, and redox processes. Oxidation of HSC70/HSPA8 is a key event that is accompanied by oxidative modifications of co-chaperones (e.g. BAG1–3, STUB1/CHIP, ST13, DNAJC12, CDC37) and further heat shock proteins including HSPB1, HSPD1 and HSP90S, that may contribute to changes of HSPA8-dependent mechanisms of protein homeostasis including protein folding, clathrin uncoating, assistance in UPS and chaperone mediated autophagy. The structure of HSPA8 is highly flexible and only partly known, and there is no prediction if redox modifications of HSPA8's Cys17, 574 or 603 affect its phosphorylation [99] or the ability to bind ATP/ADP, which is crucial to most of its functions [54]. Cysteine-17 is localized within the first lobe in subdomain Ia of the nucleotide binding site (NBR), which flexibly forms a pocket with the second lobe enabling the ATP/ADP cycling [100]. A Cys17 to lysine mutant (Cys17K) of HSPA8 retained only < 5% of ATPase activity [101], strongly suggesting that ATP hydrolysis and hence functions of this protein including its carrier services are subject to redox modifications. HSPA8 is supposed to be activated under cell stress such as starvation, oxidation or accumulation of unfolded proteins [55], [61], [63] and also binds proteins that are destined for selective ubiquitin-dependent autophagy [24], [102] including p62/SQSTM1 or subunits of inactive 26S proteasomes [29]. HSPA8a therefore links autophagy with UPS [103], [104], [105], [106], and it is of note that ubiquitination of p62 involves UBE2D3 [107], i.e. our second prime SNO candidate.
Oxidation of cysteine Cys85 in UBE2D will likely impair its function, because it is the catalytic site cysteine. In the ubiquitination cascade, a thioester-linked conjugate between the C-terminus of ubiquitin and the active site cysteine is formed, from which the E2 ligase shuttles the ubiquitin to the E3 ligase [108]. Hence, SNO-modification of this site presumably will interfere with the conjugation of ubiquitin [109]. UBE2D isoenzymes all share the Cys85 and interact with various Ring finger or U-box E3 ligases, including SIAH1 [110], which promotes ubiquitination and aggregation of alpha synuclein, and STUB1/CHIP [111], which acts in concert with HSPA8 in CMA. Hence, SNO-modification of Cys85 may alter the efficiency of protein ubiquitination and degradation through both the proteasomal and CMA pathways.
Inefficient disposal of protein waste is a major feature of neurodegenerative diseases [6], [112], [113], [114] and may also contribute to detrimental long-lasting neuronal adaptations after axonal injury [81]. In addition, previous studies showed that redox stresses increase the occurrence of aggregate prone proteins, which may be hard to digest by any of the alternative pathways [6], [115], [116], [117], [118]. In particular, SNO-modifications of GAPDH and SIAH are well described in the context of neuronal stress and neurodegeneration [66], [119], [120], [121] including Parkinson's disease, which is a bona fide disease linking pro-oxidation with dysfunctional protein degradation. SNO-modifications per se are a physiologic phenomenon and transiently or constitutively occur in several proteins. Our studies identified 2777, i.e. close to the number of approximately 3000, which have been collected in the dbSNO 2.0 (http://dbSNO.mbc.nctu.edu.tw; [19]) database including the C-terminal Cys574 and 603 of HSPA8, but so far not Cys17 or Cys85 of UBE2D. In contrast to irreversible oxidations, SNOing is a physiologic posttranslational modification but the proportion of the SNOed version of a protein or protein complex determines the functions or fate.
In summary, our studies describe a nitric oxide dependent redox proteome that suggests SNO- mediated changes of protein disposal (i) by S-nitrosylation of HSPA8 and its co-chaperones affecting chaperone mediated autophagy, and (ii) by S-nitrosylation of UBE2D isoforms that impacts on ubiquitination of specific proteins and subsequent degradation in the proteasome or via ubiquitin-dependent autophagy. The observed changes of protein homeostasis change the phenotype of SH-SY5Y cells, from a neuroblast-like juvenile towards an adult or aged state, in which oxidation or starvation-evoked stress impose stronger imbalances than in control cells.
4. Methods
4.1. Cell culture
Human neuroblastoma cells (SH-SY5Y) were grown in RPMI 1640 medium supplemented with heat-inactivated 10% fetal bovine serum, 100 U/ml penicillin/streptomycin, and 2 mM glutamine at 37 °C in 5% CO2 atmosphere at 37 °C in a humidified tissue culture incubator. For SILAC experiments cells were grown in SILAC™ Protein ID & Quantitation Medium (Invitrogen) with SILAC stable isotopic [13C6]-L-arginine and [13C6] L-lysine and supplemented with 10% dialysed fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin/streptomycin. Cells were grown in SILAC medium for more than 10 passages before starting experiments.
4.2. Nitric oxide synthase (nNOS/Nos1) expressing neuroblastoma cells
A stable cell line of nNOS expressing SH-SY5Y cells was produced by lentiviral-mediated transduction of mouse nitric oxide synthase 1 (nNOS) using a lentiviral plasmid vector (GeneCopoeia, Mm04153 pReceiver-Lv). Control cells were transduced with the control lentivirus and are referred to as MOCK. Lentiviral particles were produced by transient co-transfection of HEK293T cells with 10 µg of vector DNA, together with three helper plasmids (5 µg of pMDL/RRE, 3 µg of RSV-Rev, and pMD2G-VSVG) using the calcium phosphate precipitation method. Cell supernatants containing lentivirus particles were harvested 48 h after transfection, passed through a 0.45 µm filter, and concentrated by centrifugation for 90 min at 40,000 rpm at 4 °C. The virus pellet was suspended in 25 µl cold 1x PBS and the titers were adjusted to 2–3 × 107 transduction units/ml. SH-SY5Y cells were transduced at 10 MOI in complete medium. After 3 days, the efficiency of virus infection was determined by flow cytometry of EGFP expression, and EGFP-positive cells were FACS sorted (FACS-Aria Cell sorter, Becton Dickinson, Germany).
4.3. Cell stimulation
To induce mTOR-dependent autophagy MOCK and nNOS+ SH-SY5Y cells (1 × 106) were grown to 60% confluence and were stimulated with 1 µM rapamycin (1 mM stock in ethanol or DMSO), which was added to the culture medium. An equal volume of vehicle was added to the control cells. Cells were supplemented with NOS cofactors including 10 µM NAD, 40 µM NADPH, and 100 µM tetrahydrobiopterin. For starvation, cells were cultured in serum free medium for 1–3 days.
4.4. Saville-Griess assay of NOS activity
The concentration of nitrite/nitrate was determined with the Saville-Griess assay [122] in a microtiter plate. A standard curve was prepared with serial dilutions of freshly prepared 40 μM NaNO2. One hundred micrograms of protein were added to a well and adjusted to 100 µl. 30 µl of 0.5% ammonium sulfamate (ASM; Aldrich) in water was added, followed by 30 µl of 2 N HCl, and incubation for 1–2 min. Subsequently, 40 µl of Griess reagent (sulphanilamide/N-1-napthylethylenediamine dihydrochloride (Promega) was added and incubated for 10 min at room temperature and the absorption at 595 nm was then measured using Spectra Fluor Plus® instrument and XFluor® software (Tecan, Crailsheim).
4.5. Quantitative RT PCR and Western blot
Total RNA was extracted from homogenized cells or tissue with the RNAeasy tissue Mini Kit (Qiagen), and reverse transcribed using poly-dT as a primer to obtain cDNA fragments. QRT-PCR was performed using an ABI prism 7700 TaqMan thermal cycler (Applied Biosystems) with the SybrGreen detection system and primer sets recommended by the manufacturer.
Whole cell protein extracts were prepared in RIPA lysis buffer (Sigma) or PhosphoSafe Buffer (Sigma Germany) containing a protease inhibitor cocktail (Roche) and PMSF 10 µg/ml, separated on a 10, 12 or 14% SDS-PAGE gel (30 µg/lane), transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Freiburg, Germany) by wet-blotting or electro-blotting. Blots were blocked and developed in Odyssee buffer or 5% skim milk in 1xPBS/Tween 20. For detection of specific proteins, blots were incubated with primary antibodies (Suppl. Table 1). β-actin, β-tubulin, GAPDH and Hsp70 were used as loading controls. Secondary antibodies conjugated with IRDye 680 or 800 (1:1000; LI-COR Biosciences, Bad Homburg, Germany) were used for detection. Blots were sequentially developed and analyzed on the Odyssey Infrared Imaging System (LI-COR Biosciences), superimposed and quantified using ImageStudio Light.
4.6. UBE2D activity
We assessed the concentration-dependent effects of nitric oxide on UBE2D3 activity employing an in vitro ubiquitination assay (MDM2 Ubiquitin Ligase Assay - p53 Substrate, BostonBiochem), in which UBE2D3 activates the Ring finger E3 ubiquitin ligase, MDM2 (murine double minute 2), which in turn conjugates ubiquitin with its substrate, His6-p53. Ubiquitination of the latter is subsequently quantified per Western Blot analysis with anti-His or anti-p53 antibody detection. The assay employs recombinant proteins for E1, UBE2D3, GST-MDM2 and the substrate, His6-p53. The reaction was prepared in 30 µl on ice by adding the components one by one in the absence and presence of 0.1–100 µM NO donors, DEA/NO and NOC-5 (both Sigma, added after UBE2D3), and finally started by adding ubiquitin. After 1 h incubation, the reaction was terminated by adding 8 µl 5X SDS-PAGE sample buffer and 2 µl 1 M DTT. Samples were directly submitted to standard 10% SDS-PAGE, electro-blotting onto nitrocellulose membranes and detection with anti-p53/anti mouse-IR-dye on an Odyssee infrared scanner.
4.7. Proteasome activity
Proteasome activity was analyzed according to the instructions for the Proteasome 20S Activity Assay (Sigma, # MAK172), which measures the chymotrypsin-like protease activity associated with the proteasome complex. The assay uses LLVY-R110 as a fluorogenic substrate. nNOS+ and MOCK cells were stimulated with 1 µM rapamycin for 6 h or 24 h, the last 2 h in the presence of the substrate, which was added to the culture medium. Cleavage of LLVY-R110 by the proteasome generates strongly green fluorescent R110, which was quantified on a SPECTRAFluor Plus 96-well plate reader at ex/em 492/535 nm.
4.8. Cell cycle analysis using flow cytometry
nNOS+ and MOCK cells were seeded in 10 cm dishes and starved or stimulated with 1 µM bafilomycin for 6 h and 24 h. Cells were harvested by trypsinization, fixed with 80% ethanol, washed, incubated for 5 min with 0.125% Triton X-100, washed again and stained with propidium iodide in PBS containing 0.2 mg/ml RNAse A. Stained cells were analyzed by flow cytometry (FACSCanto II, Becton Dickinson). The cell cycle distribution i.e. the percentage of cells in subG1, G0/G1, S and G2/M phase, was assessed using FlowJo V10.6.
4.9. Immunofluorescence analysis of autophagy and cell proliferation
Immunofluorescence studies in nNOS+ and MOCK cells assessed the cytoskeleton (F-actin labeling with phalloidin-Alexa-594), ER structure (calnexin), proliferation (Ki67), autophagy (CytoID, LC3b-II) and lysosomes (LysoTracker). Cultures were washed in PBS, fixed in 4% PFA in 1xPBS and immunostained or labeled with the respective dyes. Images were captured on an inverted AxioImager Z1 fluorescence microscope (Zeiss, Jena, Germany). FIJI ImageJ was used for counting Ki67 positive proliferating cells after threshold setting and generation of binary images. Nuclei were counter-stained with DAPI. Autophagy was estimated based on the strength and thickness of LC3b-II or CytoID® (CytoID® detection kit Enzo Life Sciences) positive dots. The CytoID® Autophagy Detection measures autophagic vacuoles and monitors autophagic flux in live cells using a cationic amphiphilic tracer dye that rapidly partitions into autophagic vacuoles where it shows bright fluorescence. The dye only weakly stains lysosomes. Quantification of immunofluorescence images was performed with ImageJ, using the particle counter. Images were split into the channels, transformed into binary images by setting the threshold according to the Isodata or Yen algorithm with minor adjustments. Size inclusion/exclusion thresholds and circularity limits were set for analysis of cells or subcellular structures.
4.10. Redox proteomics
The principle of the analyses of posttranslational redox modifications in proteins is similar for all techniques, which were employed in the present study and evolved over time. In the first step, non-oxidized sulfhydryl groups are masked with N-ethylmaleimide (NEM). Subsequently, originally oxidized residues are reduced, either mildly with ascorbic acid to reduce S-nitrosylated SH-groups (SNO sites) or generally with DTT to reduce reversible oxidations including sulfoxides and disulfides. Finally, the now newly generated SH-groups (i.e. the original oxidation sites) are labeled with biotinylated iodoacetamide or with Cy-dyes. Proteins are trypsinized, modified peptides are captured via streptavidin and analyzed per liquid chromatography/mass spectrometry or ELISA. The details of the different extraction procedures and LC/MS settings are described in accompanying Data in Brief articles DIB1 "Redox Proteomics" and DIB2 "Full Proteome". The raw MS proteomics data of the Full Proteome have been deposited to the ProteomeXchange Consortium via the PRIDE [123] partner repository with the dataset identifier PXD010538. Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD010538
4.11. Proteomic data analyses
For data analysis MaxQuant 1.6.1.0 [124], Perseus 1.6.0.2 [125] and ArrayStar (DNASTAR 15) were used. N-terminal acetylation (+42.01) and oxidation of methionine (+15.99), N-ethylmaleimide on cysteines (+125.05) and biotinylated iodoacetamide (+414.19) were selected as variable modifications for SNO/Redox-BIAM-Switch. The human reference proteome set (Redox-BIAM-Switch: Uniprot, July 2017, 701567 entries, SNO-BIAM Switch and Full proteome: April 2015, 68511 entries) was used as template to identify peptides and proteins with a false discovery rate (FDR) less than 1%. The minimal ratio count for label-free quantification (LFQ) was 1.
Proteins were quality filtered according to unique peptides, sequence coverage, putative contaminants and a minimum of 4 valid values or 6 valid samples in total. LFQ protein intensities were Log2 transformed and missing/zero values were imputed from the normal distribution (scattered missing values) or set to a minimum. ANOVA was performed before and after imputation, and consistent results considered as significant. Global averaging (i.e. adjustment to have a common mean, implemented in ArrayStar) was used as further adjustment to account for differences in cell morphology and hence relative differences of cytoplasm/cytoskeletal proteins and nuclear or organelle protein mass. Global averaging was used for SNO-BIAM Switch and Proteome before transforming the experiment into a BS/Proteome ratio, the latter to address changes of protein expression that may obscure or overestimate changes of protein oxidation. BS/Proteome ratios were then Log2-transformed and used for further analysis. Hierarchical clustering was employed to assess protein and treatment patterns using Euclidean distance metrics. Results are displayed as heat maps with dendrograms. For thumbnail line graph views, the Pearson correlation coefficient was used. Clusters were further analyzed for gene ontology annotation enrichments for “cellular component”, “biological process” and “molecular function", KEGG and reactome pathways, SMART domains and SP-PIR-Keywords to assess common localizations and functions. GO analyses were done with Perseus, ArrayStar or using the ”term enrichment analysis” and "functional gene clustering" tools of The Database for Annotation, Visualization and Integrated Discovery (DAVID, version 6.8) (http://david.abcc.ncifcrf.gov/home.jsp). STRING (https://string-db.org/) was used to generate protein networks and assess network GOs. Gene set enrichment analysis (GSEA) (http://www.gsea-msigdb.org) [126] was used to further assess functional implications, and regulated proteins were compared with genes listed in the Human Cellular Senescence Gene Database (HCSGD) (http://bioinfo.au.tsinghua.edu.cn/member/xwwang/HCSGD/) [85]. The P value was set at 0.05 adjusted according to Benjamini Hochberg. A batch peptide search (http://pir.georgetown.edu/pirwww/) for published/confirmed CMA-recognition sequences [75] was used to find known and putative CMA substrates [70], which were subsequently compared with significantly upregulated proteins.
4.12. Statistics
SPSS 24.0 and Graphpad Prism 6.0 were used for statistical evaluation for non-proteomic data. Data are presented as means ± SD. Data were analyzed using univariate or multivariate ANOVA and subsequent posthoc t-tests versus vehicle treated MOCK cells or baseline conditions according to Dunnett, or group-vise according to Sidak. Two groups were compared with 2-sided, unpaired Student's t-tests. Contingency tables and Chi Square tests were used to compare the number of modified proteins in SNOSID experiments. P was set at 0.05 for all statistical comparisons. VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html) and NetVenn (http://wheat.pw.usda.gov/NetVenn/) [127] were used to compare gene/protein lists.
Acknowledgements
We acknowledge the financial support of the Deutsche Forschungsgemeinschaft (SFB815 A12, SFB815 Z1 and CRC1080 A3). We thank Mirco Steger for operating mass spectrometry and Bela Zimmer for technical assistance and cell culture.
Acknowledgments
Conflict of interest
The authors have no conflict of interest
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.10.002.
Appendix A. Supplementary material
Supplementary material
References
- 1.Liu P., Smith P.F., Appleton I., Darlington C.L., Bilkey D.K. Hippocampal nitric oxide synthase and arginase and age-associated behavioral deficits. Hippocampus. 2005;15(5):642–655. doi: 10.1002/hipo.20085. [DOI] [PubMed] [Google Scholar]
- 2.Liu P., Smith P.F., Appleton I., Darlington C.L., Bilkey D.K. Regional variations and age-related changes in nitric oxide synthase and arginase in the sub-regions of the hippocampus. Neuroscience. 2003;119(3):679–687. doi: 10.1016/s0306-4522(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 3.de la Torre J.C., Pappas B.A., Prevot V., Emmerling M.R., Mantione K., Fortin T., Watson M.D., Stefano G.B. Hippocampal nitric oxide upregulation precedes memory loss and A beta 1-40 accumulation after chronic brain hypoperfusion in rats. Neurol. Res. 2003;25(6):635–641. doi: 10.1179/016164103101201931. [DOI] [PubMed] [Google Scholar]
- 4.Liu P., Smith P.F., Appleton I., Darlington C.L., Bilkey D.K. Nitric oxide synthase and arginase in the rat hippocampus and the entorhinal, perirhinal, postrhinal, and temporal cortices: regional variations and age-related changes. Hippocampus. 2003;13(7):859–867. doi: 10.1002/hipo.10138. [DOI] [PubMed] [Google Scholar]
- 5.Uehara T., Nakamura T., Yao D., Shi Z.Q., Gu Z., Ma Y., Masliah E., Nomura Y., Lipton S.A. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441(7092):513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura T., Lipton S.A. Emerging roles of S-nitrosylation in protein misfolding and neurodegenerative diseases. Antioxid. Redox Signal. 2008;10(1):87–101. doi: 10.1089/ars.2007.1858. [DOI] [PubMed] [Google Scholar]
- 7.Haun F., Nakamura T., Shiu A.D., Cho D.H., Tsunemi T., Holland E.A., La Spada A.R., Lipton S.A. S-nitrosylation of dynamin-related protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington's disease. Antioxid. Redox Signal. 2013;19(11):1173–1184. doi: 10.1089/ars.2012.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tegeder I., Scheving R., Wittig I., Geisslinger G. SNO-ing at the nociceptive synapse? Pharmacol. Rev. 2011;63(2):366–389. doi: 10.1124/pr.110.004200. [DOI] [PubMed] [Google Scholar]
- 9.Benhar M., Stamler J.S. A central role for S-nitrosylation in apoptosis. Nat. Cell Biol. 2005;7(7):645–646. doi: 10.1038/ncb0705-645. [DOI] [PubMed] [Google Scholar]
- 10.Lipton S.A., Choi Y.B., Pan Z.H., Lei S.Z., Chen H.S., Sucher N.J., Loscalzo J., Singel D.J., Stamler J.S. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364(6438):626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 11.Lane P., Hao G., Gross S.S. S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci. STKE. 2001;2001(86) doi: 10.1126/stke.2001.86.re1. (re1) [DOI] [PubMed] [Google Scholar]
- 12.Foster M.W., Hess D.T., Stamler J.S. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 2009;15(9):391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butterfield D.A., Poon H.F., Clair D., St, Keller J.N., Pierce W.M., Klein J.B., Markesbery W.R. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol. Dis. 2006;22(2):223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Gu Z., Nakamura T., Lipton S.A. Redox reactions induced by nitrosative stress mediate protein misfolding and mitochondrial dysfunction in neurodegenerative diseases. Mol. Neurobiol. 2010;41(2–3):55–72. doi: 10.1007/s12035-010-8113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies K.J., Shringarpure R. Preferential degradation of oxidized proteins by the 20S proteasome may be inhibited in aging and in inflammatory neuromuscular diseases. Neurology. 2006;66(2 Suppl 1):S93–S96. doi: 10.1212/01.wnl.0000192308.43151.63. [DOI] [PubMed] [Google Scholar]
- 16.Grune T., Merker K., Sandig G., Davies K.J. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003;305(3):709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 17.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 18.Powers E.T., Balch W.E. Costly mistakes: translational infidelity and protein homeostasis. Cell. 2008;134(2):204–206. doi: 10.1016/j.cell.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y.J., Lu C.T., Su M.G., Huang K.Y., Ching W.C., Yang H.H., Liao Y.C., Lee T.Y. dbSNO 2.0: a resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Res. 2015;43:D503–D511. doi: 10.1093/nar/gku1176. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grune T., Reinheckel T., Davies K.J. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11(7):526–534. [PubMed] [Google Scholar]
- 21.Kriegenburg F., Poulsen E.G., Koch A., Kruger E., Hartmann-Petersen R. Redox control of the ubiquitin-proteasome system: from molecular mechanisms to functional significance. Antioxid. Redox Signal. 2011;15(8):2265–2299. doi: 10.1089/ars.2010.3590. [DOI] [PubMed] [Google Scholar]
- 22.Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9(11):826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 23.Tekirdag K.A., Cuervo A.M. Chaperone-mediated autophagy and endosomal microautophagy: joint by a chaperone. J. Biol. Chem. 2017 doi: 10.1074/jbc.R117.818237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Yang Q., Mao Z. Chaperone-mediated autophagy: machinery, regulation and biological consequences. Cell. Mol. Life Sci. 2011;68(5):749–763. doi: 10.1007/s00018-010-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinsztein D.C. Autophagy induction rescues toxicity mediated by proteasome inhibition. Neuron. 2007;54(6):854–856. doi: 10.1016/j.neuron.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Ebrahimi-Fakhari D., Cantuti-Castelvetri I., Fan Z., Rockenstein E., Masliah E., Hyman B.T., McLean P.J., Unni V.K. Distinct roles in vivo for the ubiquitin-proteasome system and the autophagy-lysosomal pathway in the degradation of alpha-synuclein. J. Neurosci. 2011;31(41):14508–14520. doi: 10.1523/JNEUROSCI.1560-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kageyama S., Sou Y.S., Uemura T., Kametaka S., Saito T., Ishimura R., Kouno T., Bedford L., Mayer R.J., Lee M.S., Yamamoto M., Waguri S., Tanaka K., Komatsu M. Proteasome dysfunction activates autophagy and the Keap1-Nrf2 pathway. J. Biol. Chem. 2014;289(36):24944–24955. doi: 10.1074/jbc.M114.580357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall R.S., Vierstra R.D. Eat or be eaten: the autophagic plight of inactive 26S proteasomes. Autophagy. 2015;11(10):1927–1928. doi: 10.1080/15548627.2015.1078961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishii T., Sakurai T., Usami H., Uchida K. Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26 S proteasome. Biochemistry. 2005;44(42):13893–13901. doi: 10.1021/bi051336u. [DOI] [PubMed] [Google Scholar]
- 31.Aiken C.T., Kaake R.M., Wang X., Huang L. Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteom. 2011;10(5) doi: 10.1074/mcp.M110.006924. (R110 006924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niforou K., Cheimonidou C., Trougakos I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014;2:323–332. doi: 10.1016/j.redox.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiffin R., Bandyopadhyay U., Cuervo A.M. Oxidative stress and autophagy. Antioxid. Redox Signal. 2006;8(1–2):152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- 34.Korovila I., Hugo M., Castro J.P., Weber D., Hohn A., Grune T., Jung T. Proteostasis, oxidative stress and aging. Redox Biol. 2017;13:550–567. doi: 10.1016/j.redox.2017.07.008. (Epub 2017 Jul 12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hohn T.J., Grune T. The proteasome and the degradation of oxidized proteins: part III-Redox regulation of the proteasomal system. Redox Biol. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. (eCollection 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giordano S., Darley-Usmar V., Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biol. 2014;2:82–90. doi: 10.1016/j.redox.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakajima H., Amano W., Kubo T., Fukuhara A., Ihara H., Azuma Y.T., Tajima H., Inui T., Sawa A., Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J. Biol. Chem. 2009;284(49):34331–34341. doi: 10.1074/jbc.M109.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reeg S., Jung T., Castro J.P., Davies K.J.A., Henze A., Grune T. The molecular chaperone Hsp70 promotes the proteolytic removal of oxidatively damaged proteins by the proteasome. Free Radic. Biol. Med. 2016;99:153–166. doi: 10.1016/j.freeradbiomed.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poppek D., Grune T. Proteasomal defense of oxidative protein modifications. Antioxid. Redox Signal. 2006;8(1–2):173–184. doi: 10.1089/ars.2006.8.173. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar S., Korolchuk V.I., Renna M., Imarisio S., Fleming A., Williams A., Garcia-Arencibia M., Rose C., Luo S., Underwood B.R., Kroemer G., O'Kane C.J., Rubinsztein D.C. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell. 2011;43(1):19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Azad N., Vallyathan V., Wang L., Tantishaiyakul V., Stehlik C., Leonard S.S., Rojanasakul Y. S-nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel antiapoptotic mechanism that suppresses apoptosis. J. Biol. Chem. 2006;281(45):34124–34134. doi: 10.1074/jbc.M602551200. [DOI] [PubMed] [Google Scholar]
- 42.Kapadia M.R., Eng J.W., Jiang Q., Stoyanovsky D.A., Kibbe M.R. Nitric oxide regulates the 26S proteasome in vascular smooth muscle cells. Nitric Oxide. 2009;20(4):279–288. doi: 10.1016/j.niox.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Yao D., Gu Z., Nakamura T., Shi Z.Q., Ma Y., Gaston B., Palmer L.A., Rockenstein E.M., Zhang Z., Masliah E., Uehara T., Lipton S.A. Nitrosative stress linked to sporadic Parkinson's disease: s-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA. 2004;101(29):10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamark T., Kirkin V., Dikic I., Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8(13):1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 46.Kirkin V., Lamark T., Sou Y.S., Bjorkoy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., Bilusic I., Theurillat J.P., Overvatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 47.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 48.Inami Y., Waguri S., Sakamoto A., Kouno T., Nakada K., Hino O., Watanabe S., Ando J., Iwadate M., Yamamoto M., Lee M.S., Tanaka K., Komatsu M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011;193(2):275–284. doi: 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riley B.E., Kaiser S.E., Shaler T.A., Ng A.C., Hara T., Hipp M.S., Lage K., Xavier R.J., Ryu K.Y., Taguchi K., Yamamoto M., Tanaka K., Mizushima N., Komatsu M., Kopito R.R. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 2010;191(3):537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bercovich B., Stancovski I., Mayer A., Blumenfeld N., Laszlo A., Schwartz A.L., Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 1997;272(14):9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- 51.Newmyer S.L., Schmid S.L. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J. Cell Biol. 2001;152(3):607–620. doi: 10.1083/jcb.152.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y., Thomas J.O. The transport of proteins into the nucleus requires the 70-kilodalton heat shock protein or its cytosolic cognate. Mol. Cell. Biol. 1992;12(5):2186–2192. doi: 10.1128/mcb.12.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terada K., Ueda I., Ohtsuka K., Oda T., Ichiyama A., Mori M. The requirement of heat shock cognate 70 protein for mitochondrial import varies among precursor proteins and depends on precursor length. Mol. Cell Biol. 1996;16(11):6103–6109. doi: 10.1128/mcb.16.11.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stricher F., Macri C., Ruff M., Muller S. HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting. Autophagy. 2013;9(12):1937–1954. doi: 10.4161/auto.26448. (Epub 2013 Oct 8) [DOI] [PubMed] [Google Scholar]
- 55.Behl C. BAG3 and friends: co-chaperones in selective autophagy during aging and disease. Autophagy. 2011;7(7):795–798. doi: 10.4161/auto.7.7.15844. [DOI] [PubMed] [Google Scholar]
- 56.Cuervo A.M., Dice J.F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273(5274):501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 57.Bandyopadhyay U., Cuervo A.M. Entering the lysosome through a transient gate by chaperone-mediated autophagy. Autophagy. 2008;4(8):1101–1103. doi: 10.4161/auto.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bandyopadhyay U., Kaushik S., Varticovski L., Cuervo A.M. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 2008;28(18):5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn P.F., Mesires N.T., Vine M., Dice J.F. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1(3):141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 60.Kaushik S., Cuervo A.M. Chaperone-mediated autophagy. Methods Mol. Biol. 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dohi E., Tanaka S., Seki T., Miyagi T., Hide I., Takahashi T., Matsumoto M., Sakai N. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem. Int. 2012;60(4):431–442. doi: 10.1016/j.neuint.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 62.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic. Biol. Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiffin R., Christian C., Knecht E., Cuervo A.M. Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell. 2004;15(11):4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anguiano J., Garner T.P., Mahalingam M., Das B.C., Gavathiotis E., Cuervo A.M. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 2013;9(6):374–382. doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moreno-Blas D., Gorostieta-Salas E., Castro-Obregon S. Connecting chaperone-mediated autophagy dysfunction to cellular senescence. Ageing Res. Rev. 2017;41:34–41. doi: 10.1016/j.arr.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Hara M.R., Agrawal N., Kim S.F., Cascio M.B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J.H., Tankou S.K., Hester L.D., Ferris C.D., Hayward S.D., Snyder S.H., Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005;7(7):665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 67.Sen N., Hara M.R., Ahmad A.S., Cascio M.B., Kamiya A., Ehmsen J.T., Agrawal N., Hester L., Dore S., Snyder S.H., Sawa A. GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63(1):81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J., Yang J., Fan X., Hu S., Zhou F., Dong J., Zhang S., Shang Y., Jiang X., Guo H., Chen N., Xiao X., Sheng J., Wu K., Nie Y., Fan D. Chaperone-mediated autophagy regulates proliferation by targeting RND3 in gastric cancer. Autophagy. 2016 doi: 10.1080/15548627.2015.1136770. (0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welsch T., Younsi A., Disanza A., Rodriguez J.A., Cuervo A.M., Scita G., Schmidt J. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp. Cell Res. 2010;316(12):1914–1924. doi: 10.1016/j.yexcr.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cuervo A.M., Gomes A.V., Barnes J.A., Dice J.F. Selective degradation of annexins by chaperone-mediated autophagy. J. Biol. Chem. 2000;275(43):33329–33335. doi: 10.1074/jbc.M005655200. [DOI] [PubMed] [Google Scholar]
- 71.DiAntonio A., Hicke L. Ubiquitin-dependent regulation of the synapse. Annu. Rev. Neurosci. 2004;27:223–246. doi: 10.1146/annurev.neuro.27.070203.144317. [DOI] [PubMed] [Google Scholar]
- 72.Bingol B., Schuman E.M. Synaptic protein degradation by the ubiquitin proteasome system. Curr. Opin. Neurobiol. 2005;15(5):536–541. doi: 10.1016/j.conb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 73.Sutton M.A., Schuman E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127(1):49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 74.Li F., Sonveaux P., Rabbani Z.N., Liu S., Yan B., Huang Q., Vujaskovic Z., Dewhirst M.W., Li C.Y. Regulation of HIF-1alpha stability through S-nitrosylation. Mol. Cell. 2007;26(1):63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaushik S., Bandyopadhyay U., Sridhar S., Kiffin R., Martinez-Vicente M., Kon M., Orenstein S.J., Wong E., Cuervo A.M. Chaperone-mediated autophagy at a glance. J. Cell Sci. 2011;124(Pt 4):495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li P., Ji M., Lu F., Zhang J., Li H., Cui T., Li Wang X., Tang D., Ji C. Degradation of AF1Q by chaperone-mediated autophagy. Exp. Cell Res. 2014;327(1):48–56. doi: 10.1016/j.yexcr.2014.05.013. (Epub 2014 May 29) [DOI] [PubMed] [Google Scholar]
- 77.Park J.S., Kim D.H., Yoon S.Y. Regulation of amyloid precursor protein processing by its KFERQ motif. BMB Rep. 2016;49(6):337–342. doi: 10.5483/BMBRep.2016.49.6.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirkin V., Lamark T., Johansen T., Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5(5):732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 79.Koga H., Martinez-Vicente M., Arias E., Kaushik S., Sulzer D., Cuervo A.M. Constitutive upregulation of chaperone-mediated autophagy in Huntington's disease. J. Neurosci. 2011;31(50):18492–18505. doi: 10.1523/JNEUROSCI.3219-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song Q., Wei T., Lees-Miller S., Alnemri E., Watters D., Lavin M.F. Resistance of actin to cleavage during apoptosis. Proc. Natl. Acad. Sci. USA. 1997;94(1):157–162. doi: 10.1073/pnas.94.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scheving R., Wittig I., Heide H., Albuquerque B., Steger M., Brandt U., Tegeder I. Protein S-nitrosylation and denitrosylation in the mouse spinal cord upon injury of the sciatic nerve. J. Proteom. 2012;75(13):3987–4004. doi: 10.1016/j.jprot.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 82.Hubbi M.E., Semenza G.L. An essential role for chaperone-mediated autophagy in cell cycle progression. Autophagy. 2015;11(5):850–851. doi: 10.1080/15548627.2015.1037063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fuchs M., Luthold C., Guilbert S.M., Varlet A.A., Lambert H., Jette A., Elowe S., Landry J., Lavoie J.N. A role for the chaperone complex BAG3-HSPB8 in actin dynamics, spindle orientation and proper chromosome segregation during mitosis. PLoS Genet. 2015;11(10):e1005582. doi: 10.1371/journal.pgen.1005582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varlet A.A., Fuchs M., Luthold C., Lambert H., Landry J., Lavoie J.N. Fine-tuning of actin dynamics by the HSPB8-BAG3 chaperone complex facilitates cytokinesis and contributes to its impact on cell division. Cell Stress Chaperon. 2017;22(4):553–567. doi: 10.1007/s12192-017-0780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dong Q., Han H., Liu X., Wei L., Zhang W., Zhao Z., Zhang M.Q., Wang X. HCSGD: an integrated database of human cellular senescence genes. J. Genet. Genom. = Yi chuan xue bao. 2017;44(5):227–234. doi: 10.1016/j.jgg.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 86.Adachi M., Abe M., Sasaki T., Kato H., Kasahara J., Araki T. Role of inducible or neuronal nitric oxide synthase in neurogenesis of the dentate gyrus in aged mice. Metab. Brain Dis. 2010;25(4):419–424. doi: 10.1007/s11011-010-9224-8. [DOI] [PubMed] [Google Scholar]
- 87.Shipley M.M., Mangold C.A., Szpara M.L. Differentiation of the SH-SY5Y human neuroblastoma cell line. J. Vis. Exp. 2016;108:53193. doi: 10.3791/53193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xicoy H., Wieringa B., Martens G.J. The SH-SY5Y cell line in Parkinson's disease research: a systematic review. Mol. Neurodegener. 2017;12(1):10. doi: 10.1186/s13024-017-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wernersson J., Johansson I., Larsson U., Minth-Worby C., Pahlman S., Andersson G. Activated transcription of the human neuropeptide Y gene in differentiating SH-SY5Y neuroblastoma cells is dependent on transcription factors AP-1, AP-2alpha, and NGFI. J. Neurochem. 1998;70(5):1887–1897. doi: 10.1046/j.1471-4159.1998.70051887.x. [DOI] [PubMed] [Google Scholar]
- 90.Friguet B., Bulteau A.L., Chondrogianni N., Conconi M., Petropoulos I. Protein degradation by the proteasome and its implications in aging. Ann. N.Y. Acad. Sci. 2000;908:143–154. doi: 10.1111/j.1749-6632.2000.tb06643.x. [DOI] [PubMed] [Google Scholar]
- 91.Ding Q., Dimayuga E., Keller J.N. Proteasome regulation of oxidative stress in aging and age-related diseases of the CNS. Antioxid. Redox Signal. 2006;8(1–2):163–172. doi: 10.1089/ars.2006.8.163. [DOI] [PubMed] [Google Scholar]
- 92.Reeg S., Grune T. Protein oxidation in aging: does it play a role in aging progression? Antioxid. Redox Signal. 2015;23(3):239–255. doi: 10.1089/ars.2014.6062. (Epub 2014 Oct 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung T., Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60(11):743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 94.Hohn A., Konig J., Grune T. Protein oxidation in aging and the removal of oxidized proteins. J. Proteom. 2013;92:132–159. doi: 10.1016/j.jprot.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 95.Cuervo A.M., Dice J.F. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 2000;275(40):31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 96.Pathania A.S., Guru S.K., Kumar S., Kumar A., Ahmad M., Bhushan S., Sharma P.R., Mahajan P., Shah B.A., Sharma S., Nargotra A., Vishwakarma R., Korkaya H., Malik F. Interplay between cell cycle and autophagy induced by boswellic acid analog. Sci. Rep. 2016;6:33146. doi: 10.1038/srep33146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cybulsky A.V. The intersecting roles of endoplasmic reticulum stress, ubiquitin- proteasome system, and autophagy in the pathogenesis of proteinuric kidney disease. Kidney Int. 2013;84(1):25–33. doi: 10.1038/ki.2012.390. [DOI] [PubMed] [Google Scholar]
- 98.Ozcan S., Alessio N., Acar M.B., Mert E., Omerli F., Peluso G., Galderisi U. Unbiased analysis of senescence associated secretory phenotype (SASP) to identify common components following different genotoxic stresses. Aging. 2016;8(7):1316–1329. doi: 10.18632/aging.100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsukahara F., Maru Y. Identification of novel nuclear export and nuclear localization-related signals in human heat shock cognate protein 70. J. Biol. Chem. 2004;279(10):8867–8872. doi: 10.1074/jbc.M308848200. [DOI] [PubMed] [Google Scholar]
- 100.Flaherty K.M., DeLuca-Flaherty C., McKay D.B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346(6285):623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 101.Wilbanks S.M., McKay D.B. Structural replacement of active site monovalent cations by the epsilon-amino group of lysine in the ATPase fragment of bovine Hsc70. Biochemistry. 1998;37(20):7456–7462. doi: 10.1021/bi973046m. [DOI] [PubMed] [Google Scholar]
- 102.Koga H., Cuervo A.M. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol. Dis. 2011;43(1):29–37. doi: 10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Myeku N., Figueiredo-Pereira M.E. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. J. Biol. Chem. 2011;286(25):22426–22440. doi: 10.1074/jbc.M110.149252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cohen-Kaplan V., Ciechanover A., Livneh I. Stress-induced polyubiquitination of proteasomal ubiquitin receptors targets the proteolytic complex for autophagic degradation. Autophagy. 2017;13(4):759–760. doi: 10.1080/15548627.2016.1278327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang J., Peng H., Xu Y., Xie X., Hu R. SQSTM1/p62 (sequestosome 1) senses cellular ubiquitin stress through E2-mediated ubiquitination. Autophagy. 2017:1–2. doi: 10.1080/15548627.2017.1332566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cohen-Kaplan V., Livneh I., Avni N., Fabre B., Ziv T., Kwon Y.T., Ciechanover A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA. 2016;113(47):E7490–e7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peng H., Yang J., Li G., You Q., Han W., Li T., Gao D., Xie X., Lee B.H., Du J., Hou J., Zhang T., Rao H., Huang Y., Li Q., Zeng R., Hui L., Wang H., Xia Q., Zhang X., He Y., Komatsu M., Dikic I., Finley D., Hu R. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res. 2017;27(5):657–674. doi: 10.1038/cr.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Page R.C., Pruneda J.N., Amick J., Klevit R.E., Misra S. Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates. Biochemistry. 2012;51(20):4175–4187. doi: 10.1021/bi300058m. (Epub 2012 May 14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakata E., Satoh T., Yamamoto S., Yamaguchi Y., Yagi-Utsumi M., Kurimoto E., Tanaka K., Wakatsuki S., Kato K. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18(1):138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 110.Lee J.T., Wheeler T.C., Li L., Chin L.S. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum. Mol. Genet. 2008;17(6):906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 111.Graf C., Stankiewicz M., Nikolay R., Mayer M.P. Insights into the conformational dynamics of the E3 ubiquitin ligase CHIP in complex with chaperones and E2 enzymes. Biochemistry. 2010;49(10):2121–2129. doi: 10.1021/bi901829f. [DOI] [PubMed] [Google Scholar]
- 112.Friedman L.G., Lachenmayer M.L., Wang J., He L., Poulose S.M., Komatsu M., Holstein G.R., Yue Z. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J. Neurosci. 2012;32(22):7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Komatsu M., Wang Q.J., Holstein G.R., Friedrich V.L., Jr., Iwata J., Kominami E., Chait B.T., Tanaka K., Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA. 2007;104(36):14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caballero B., Wang Y., Diaz A., Tasset I., Juste Y.R., Stiller B., Mandelkow E.M., Mandelkow E., Cuervo A.M. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell. 2017 doi: 10.1111/acel.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Benhar M., Forrester M.T., Stamler J.S. Nitrosative stress in the ER: a new role for S-nitrosylation in neurodegenerative diseases. ACS Chem. Biol. 2006;1(6):355–358. doi: 10.1021/cb600244c. [DOI] [PubMed] [Google Scholar]
- 116.Nakamura T., Cieplak P., Cho D.H., Godzik A., Lipton S.A. S-Nitrosylation of Drp1 links excessive mitochondrial fission to neuronal injury in neurodegeneration. Mitochondrion. 2010 doi: 10.1016/j.mito.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chung K.K., David K.K. Emerging roles of nitric oxide in neurodegeneration. Nitric Oxide. 2010;22(4):290–295. doi: 10.1016/j.niox.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 118.Cannizzo E.S., Clement C.C., Morozova K., Valdor R., Kaushik S., Almeida L.N., Follo C., Sahu R., Cuervo A.M., Macian F., Santambrogio L. Age-related oxidative stress compromises endosomal proteostasis. Cell Rep. 2012;2(1):136–149. doi: 10.1016/j.celrep.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hara M.R., Snyder S.H. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell. Mol. Neurobiol. 2006;26(4–6):527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rott R., Szargel R., Haskin J., Shani V., Shainskaya A., Manov I., Liani E., Avraham E., Engelender S. Monoubiquitylation of alpha-synuclein by seven in absentia homolog (SIAH) promotes its aggregation in dopaminergic cells. J. Biol. Chem. 2008;283(6):3316–3328. doi: 10.1074/jbc.M704809200. [DOI] [PubMed] [Google Scholar]
- 121.Szargel R., Rott R., Eyal A., Haskin J., Shani V., Balan L., Wolosker H., Engelender S. Synphilin-1A inhibits seven in absentia homolog (SIAH) and modulates alpha-synuclein monoubiquitylation and inclusion formation. J. Biol. Chem. 2009;284(17):11706–11716. doi: 10.1074/jbc.M805990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Griess P. Bemerkungen zu der abhandlung der H.H. Weselsky und Benedikt, “Ueber einige azoverbindungen.”. Chem. Ber. 1879;12:426–428. [Google Scholar]
- 123.Vizcaino J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J.A., Sun Z., Farrah T., Bandeira N., Binz P.A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R.J., Kraus H.J., Albar J.P., Martinez-Bartolome S., Apweiler R., Omenn G.S., Martens L., Jones A.R., Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 125.Tyanova S., Cox J. Perseus: a bioinformatics platform for Integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 1711;2018:133–148. doi: 10.1007/978-1-4939-7493-1_7. [DOI] [PubMed] [Google Scholar]
- 126.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y., Thilmony R., Gu Y.Q. NetVenn: an integrated network analysis web platform for gene lists. Nucleic Acids Res. 2014;42:W161–W166. doi: 10.1093/nar/gku331. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material