Abstract
Biologists would be mistaken if they relegated living fossils to paleontological inquiry or assumed that the concept is dead. It is now used to describe entities ranging from viruses to higher taxa, despite recent warnings of misleading inferences. Current work on character evolution illustrates how analyzing living fossils and stasis in terms of parts (characters) and wholes (e.g., organisms and lineages) advances our understanding of prolonged stasis at many hierarchical levels. Instead of viewing the concept's task as categorizing living fossils, we show how its primary role is to mark out what is in need of explanation, accounting for the persistence of both molecular and morphological traits. Rethinking different conceptions of living fossils as specific hypotheses reveals novel avenues for research that integrate phylogenetics, ecological and evolutionary modeling, and evo-devo to produce a more unified theoretical outlook.
Keywords: character evolution, concepts, evolutionary rates, living fossil, stasis
Living fossils: contentious but necessary? Whether it is horseshoe crabs, coelacanths, or gingko trees, taxa that allegedly display extraordinary levels of morphological stasis over geological time have called out for a special explanation since Darwin (Lidgard and Hopkins 2015). Viewing extinct and extant representatives of these lineages side by side provokes an immediate judgment of similarity, regardless of whether it is justified. Why have these constellations of characters persisted for so long? What exactly is in need of explanation? Accounting for persistence over long periods of evolutionary time has encouraged researchers to apply the moniker living fossil to viruses, transposons, genomes, ribosomes, proteins, cell types, species, and higher taxa (Bell et al. 2008, Smith et al. 2012, Richardson et al. 2013, Schuldiner 2014, Werth and Shear 2014, Prangishvili 2015, Lupas and Alva 2017). Despite this proliferation of the label, there is widespread dissatisfaction with the concept (box 1). These concerns sometimes evoke the claim that the concept is not scientifically sensible (Vanschoenwinkel et al. 2012, Casane and Laurenti 2013, Mathers et al. 2013).
Box 1. Common criteria for and complaints about “living fossils” as a scientific concept.
Criteria often used to designate living fossils
• Prolonged geological duration relative to similar entities
• Slow evolutionary change relative to similar entities
• Gross similarity to an ancestral fossil
• Very low taxonomic richness today compared to the past
• Relic geographic range compared to the past
• Phylogenetic inference of specific characters as plesiomorphic
• Phylogenetic inference of a genealogical divergence between other groups that diverged in the distant past
• Known in the fossil record before being discovered alive
Complaints often lodged against the appropriateness of living fossil designation
• Ill-defined or cross-cutting definitional criteria
• Molecular genetic change despite apparent morphological stasis (and vice versa)
• Preservational or sampling biases in the fossil record
• Misclassification or faulty phylogenetic inference
• Confusion about what level of taxonomic hierarchy is in view (e.g., species versus higher taxa, or unrecognized cryptic species)
• Lineage originations and evolutionary rates not being reliably derived from fossils or molecular clocks
• Problematic expectations that morphological change occurs in concert with biotic and abiotic environmental change
Our aim in this article is to rethink the concept of a living fossil in a way that takes seriously both its routine use across disparate research questions in biology and the worries about its misleading inferences in order to create new paths for productive research that yield a more unified theoretical outlook. We first highlight the value of thinking in terms of parts (characters) and wholes (typically, organisms or lineages) to better understand stasis. Then, we move beyond concerns that concentrate on categorizing living fossils. As an alternative, we characterize a diverse array of questions associated with the biological research program that encompasses different conceptions of living fossils across hierarchical levels of organization. Our characterization of the rich space of questions surrounding slow or negligible rates of evolutionary change with respect to characters or groups thereof has several advantages, such as making sense of recent applications to molecular features, permitting the precise testing of specific hypotheses related to living fossils, and suggesting novel avenues for research. In addition, by focusing on explanatory goals and their relevant standards, we are able to provide increased conceptual unification to heterogeneous and relatively fragmented investigations into living fossils.
Living fossils: Parts and wholes
All scientific practice involves using proxies: measurements of particular properties that stand in for an entity or phenomenon. Biologists use subsets of characters (parts) and their differing states to discriminate among organisms or lineages (wholes). Suites of morphological or molecular characters also act as proxies in phylogenetic hypotheses. However, there is an ambiguity between the morphological and molecular parts of an organism and whole organisms or genomes of a lineage or clade when being evaluated with respect to stasis (box 2). Living fossil taxa such as coelacanths, limulids (horseshoe crabs), Lingula (lamp shell), Ginkgo (maidenhair tree), Ornithorhynchus (platypus), Sphenodon (tuatara), and Triops (tadpole shrimp) each exhibit a mix of ancient and derived characters. Fossils are seldom direct ancestors of living organisms. At best, the rock record yields only a small minority for certain abundant, readily fossilized, and well-studied living groups. Instead, fossils are typically instantiations of related lineages with their own histories of character evolution that help inform phylogenetic relationships. Even in well-studied taxa such as vertebrates, selections of morphological characters from different anatomical regions can imply different phylogenetic trees (Mounce et al. 2016).
Box 2. Recent paleontological work on character evolution.
Morphologies of fossil skeletons provide the most direct evidence for evaluating evolutionary modes in deep time. Attributions of living fossil status to modern organisms and to organismal parts with little or no fossil record rely on more indirect chains of inference, especially molecular phylogenetic hypotheses. However, there are hundreds of studies of fossil morphologies that bear on a central claim of the theory of punctuated equilibria: Most fossil species exhibit stasis (Lidgard and Hopkins 2015). A new approach analyzes this evidence comparatively using a consistent protocol based on model selection arbitrated by a measure of penalized likelihood (Hopkins and Lidgard 2012, Hunt et al. 2015). Two of the three canonical evolutionary modes—stasis and random walk (Brownian motion)—are about equally common. A third mode, gradual change, is comparatively rare. The relative commonness of stasis does not imply a total absence of character change (Voje 2016). Rather, nonaccumulating morphological fluctuations occur at scales that reflect neither large net evolutionary transformation nor speciation.
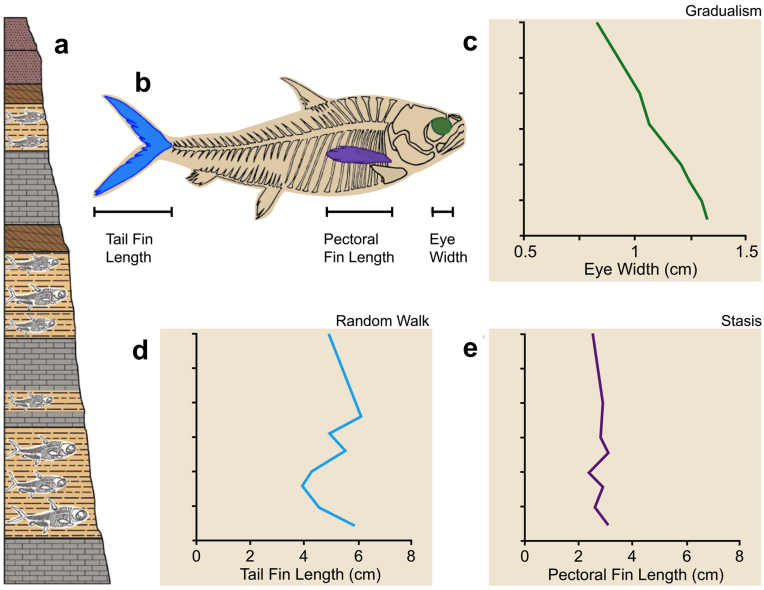
Single size and shape characters are taken to represent a species or lineage in most quantitative paleontological studies of evolutionary modes. Where multiple characters are recorded for the same species or lineage, analyses frequently distinguish separate character trajectories that correspond to different models of evolutionary change. Consider a hypothetical sequence of fossil fish populations of a given species or transitional lineage (figure 1). The fossil-bearing layers may not be spaced uniformly and the amount of absolute time represented can be variable. Many more characters are available than those selected to represent the lineage. Characters are measured in each sample population as successive steps in each character's evolutionary trajectory, which yields sample population measures of central tendency and variance. The best model fit for three canonical modes of evolution is chosen for each character trajectory: gradualism for the width of the eye, random walk for tail fin length, and stasis for pectoral fin length. This illustrates the importance of mosaic evolutionary patterns for different characters, which are revealed when relationships between parts and wholes are scrutinized explicitly. It implies that tracking stasis and change or comparing extant and extinct morphologies is a subtler endeavor than controversies about categorizing living fossils suggest.
The problem of part–whole ambiguity runs deep in biology. On a molecular level, it has long been recognized that mutation and amino-acid substitution rates are not constant across genes and taxa, or over long stretches of geological time (Szöllősi et al. 2014). Characters or character states are relatively more ancestral or derived, not whole organisms or lineages (Omland et al. 2008). Even when we consider a different reference point as a “whole,” such as a genome, this ambiguity applies. Genes of ancient prokaryote ancestors have persisted (in some form) through endosymbiotic evolutionary transitions into mitochondria and plastids in eukaryote clades. Mitochondria in mosses (Liu et al. 2014) and plastids in liverworts (Forrest et al. 2011) and cycads (Wu and Chaw 2015) show remarkable genetic stasis relative to other plant groups, especially angiosperms. Among metazoans, nonbilaterians show much lower rates of mitochondrial evolution than do bilaterians (Lavrov 2007), whereas metazoan mtDNA evolves rapidly compared to plant mtDNA. The DNA binding domain of transcription factors is remarkably conserved, but regions of protein–protein interaction exhibit dramatic changes (Lynch and Wagner, 2008, 2011). How do we understand what counts as parts and whole(s) through evolutionary stasis and change?
Constellations of exoskeletal characters are proxies for stasis in the living fossil tadpole shrimps Triops (figure 2a, 2b) and Lepidurus. Characters including gross morphology of body segments, shape of the carapace, particular spines on the carapace, telson, and furca, and presence or absence of a supra-anal plate show mostly minor changes from appearances beginning in the Carboniferous (e.g., Kelber 1999). However, one or another of these diagnostic characters is often lacking in fossils. In addition, other morphological, reproductive, and ecological characters are extremely variable. When genetic characters are considered, the repeated detection of cryptic species in both genera (Vanschoenwinkel et al. 2012, Mathers et al. 2013) implies evolutionary diversification in living populations.
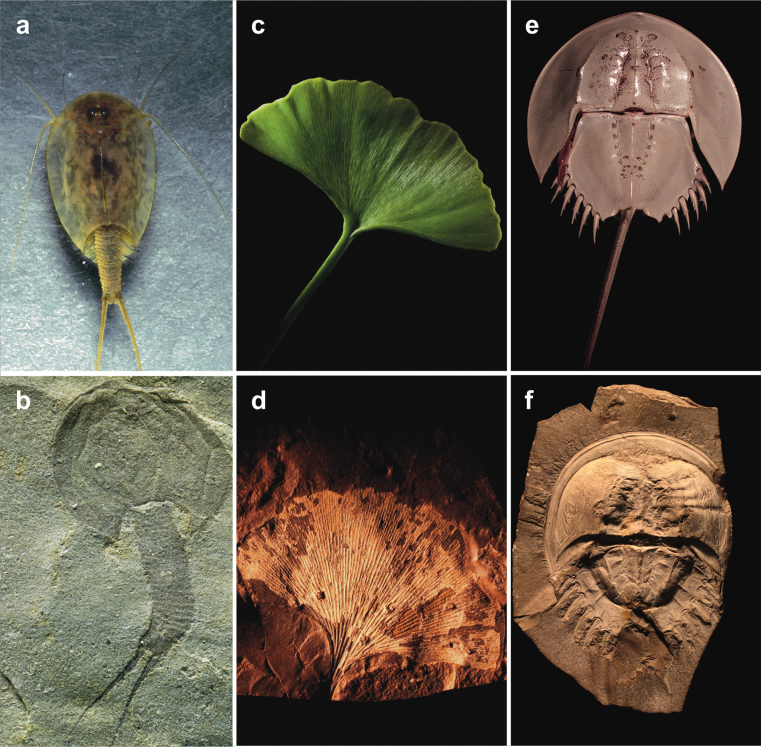
Figure 2.
Modern and ancient representatives of living fossils Triops, Ginkgo, and xiphosurans. (a) Triops cancriformis, recent, Hampshire, England (photograph: Roger Key). (b) T. cancriformis, Hassberge Fm., Triassic, Germany (PASS-074b; photograph: Klaus-Peter Kelber). (c) Ginkgo biloba leaf, recent. (d) G. cranei leaf, Sentinel Butte Fm., Paleocene, North Dakota (Field Museum FMNH pp34024). (e) Tachypleus, recent, Singapore (Yale University YPM IZ 055578). (f) Mesolimulus, Solnhofen Limestone, Late Jurassic, Germany (Yale University YPM IP 9011; both photographs: James Lamsdell).
Figure 1.
Evolutionary modes of different characters in a hypothetical fossil fish lineage exemplify results from hundreds of published studies (Hopkins and Lidgard 2012, Hunt et al. 2015). (a) Population samples are taken at successive intervals from sedimentary layers that contain fossils. (b) Characters are measured for each sample. Different evolutionary modes are seen in character trajectories plotted against stratigraphic positions for eye width (c), tail fin length (d), and pectoral fin length (e). Illustration: Monica Jurik.
Ginkgoalean fossils are commonly recognized from their leaves, sometimes indistinguishable from leaves of living Ginkgo biloba (figure 2c, 2d). Different developmental stages of modern leaves sometimes can be referred to separate fossil species or genera. Taxonomically important fructifications, seeds, wood, and leaf cuticle are seldom preserved together as whole plant fossils (this kind of ambiguity is widespread; species of incomplete mammalian fossils are sometimes determined only by the cusps of a few teeth). The genus may go back approximately 170 million years, but different morphological part proxies yield separate ages of first appearance (Zhou 2009). What combination of characters sufficiently represents a lineage as a whole?
When we talk of a living fossil retaining an ancestral morphology, this retention concerns particular characters. However, if we ask whether the taxon in which these characters appear is geographically widespread, diverse in its extinct representatives, or a low-diversity relict population in the present, then we are expressly tracking whole organisms and their lineages. The carapaces of Jurassic Mesolimulus and modern Tachypleus (figure 2e, 2f) share numerous characters, but horseshoe crabs as a whole may have invaded land four separate times (Lamsdell 2016), with attendant evolutionary changes in ecology, physiology, and morphology among lineages. This ambiguity ramifies when we recognize that some taxa labeled as living fossils lack properties we might typically expect, such as geographic relict status, which is not applicable to taxa with extensive geographic ranges, such as modern horseshoe crabs.
Coelacanths are perhaps the epitome of living fossils, but patterns inferred from molecular characters as proxy parts are seldom uniform. Polymorphic genetic structure in living coelacanth populations suggests typical substitution rates (Casane and Laurenti 2013), but there is abundant evidence of slow genome evolution (Amemiya et al. 2010). The coelacanth genome has many active transposable elements (Naville et al. 2014). However, some elements are highly conserved (Smith et al. 2012) and Hox gene clusters appear to evolve slowly (Amemiya et al. 2010). Overall, when focused on either molecular or morphological characters that serve as proxies for species or lineages, there are rampant part–whole ambiguities in evaluating evolutionary stasis and change, many of which bear directly on controversies about categorizing living fossils.
Beyond categorizing living fossils
Divergent categorizations of species or supraspecific taxa have exposed multiple conceptions of living fossils (box 1). One conception, phylogenetic inference of specific characters as plesiomorphic or of a position intermediate between other groups that diverged in the distant past (even when fossil evidence is lacking), has parallels among conserved suborganismal entities. For instance, if a particular gene or gene family exhibits only minor sequence variation across numerous, distantly related living taxa, it is nearly always considered strongly conserved from an ancient divergence. Although highly conserved molecular sequences are not frequently labeled as living fossils, these parallels and similar part–whole ambiguities suggest an underlying unity for judgments of conserved molecular function and molecular living fossils.
Criteria for living fossil membership are often criticized as ill-defined or conflicting, compounded by the fact that most criteria rely on other contentious aspects that affect judgments, such as biases in the fossil record (box 1). This nurtures definitional debates about whether particular taxa should be considered living fossils (Nagalingum et al. 2011, Vanschoenwinkel et al. 2012, Casane and Laurenti 2013, Mathers et al. 2013, Cavin and Guinot 2014, Werth and Shear 2014, Bennett et al. 2017). Biologists often steer clear of definitional stalemates as unproductive. (Debate over species definitions is just one prominent example.) One reason for these stalemates is an assumption that the primary role of a concept is to categorize (i.e., figure out which set of entities should be classified by a particular term). In this sense, a concept is used to distinguish one thing from another (e.g., apples from oranges) or recognize commonalities (e.g., apples and oranges are both fruit).
However, concepts play many roles in human cognition generally and within scientific reasoning specifically. This provides an avenue out of definitional debates about what is (or is not) a living fossil; concepts also play a role in representing broad investigative domains (Brigandt and Love 2012). For example, the concept differentiation in developmental biology represents a research program aimed at understanding the causal factors and conditions that lead to the transformation of undifferentiated cells and tissues. This requires a variety of in-depth studies of the genomic architecture and genetic expression patterns of diverse cells in various states of differentiation and located in diverse microenvironmental conditions. Developmental biologists are seeking to discover many different things about differentiation (Love 2014), not merely whether something is or is not an example of differentiation. Similarly, the primary role of the living fossil concept is to mark out more precisely what requires explanation in a given instance for a particular entity in order to account for morphological and molecular stability or persistence over long periods of evolutionary time. From this perspective, we can take seriously the widespread invocation of living fossils across disparate biological research questions at different levels of organization and understand the legitimacy of divergent criteria used to isolate answers to these questions. This shift moves us away from semantics and toward both productive research and the possibility of a more unified conceptual framework for living fossils.
Lessons from history can help us contextualize what researchers have argued is in need of (special) explanation. First, different conceptions and criteria of living fossils (box 1) derive from different explanatory expectations. Darwin never actually defined living fossil. He conveyed the idea with several examples. The logic and inferred criteria underlying his examples varied, and nearly all of his examples were genera or taxonomic groups of higher rank. By 1859, representatives of the living fossil brachiopod Lingula had been found alive and its morphologically simple shells also occurred in the oldest fossil-bearing geological layers. Then as now, simpler or unspecialized morphology has been related to longer geological durations, such as in crinoids (Liow 2004). In contrast, the platypus Ornithorhynchus had no known fossils in 1859 but was taxonomically intermediate in having the duck-like bill of a bird and fur of a mammal. From a modern phylogenetic perspective, the operational criteria involve a retention of plesiomorphic characters compared to sister groups. Living Ginkgo populations are tiny remnants of a broad ancient distribution. In contrast, living horseshoe crabs are geographically widespread. The multiple conceptions of living fossils represent distinct explanatory questions that require different operational definitions to gather the data required to test hypotheses. In these examples, criteria of adequacy diverge and range from when or where observations are made to what results are derived from a phylogenetic analysis. The category living fossil may be assigned or removed depending on which criteria are used and which proxies are measured to test particular hypotheses.
Second, increased sampling of the fossil record changes how different conceptions apply to perceived rates and status. Discoveries of soft tissue anatomy in Cambrian lingulid brachiopods show the evolution of dramatic changes toward modern species, despite retention of remarkably similar shell morphology (Zhang et al. 2005). Recent work on Crocodyliformes, classic living fossils, has uncovered diverse ecological adaptations, shifting evolutionary rates, and tangents from the generalized crocodylian form (Bronzati et al. 2015). Discoveries may also affect fossil age calibrations (moreso than rates of gene evolution alone), changing estimates of taxon ages, divergences, and rates in morphological and molecular phylogenetic results (Wagner and Marcot 2013). More and better-understood fossils in morphological phylogenetic analyses also can change relationships among stem groups and consequent molecular clock calibrations. For example, a new analysis of living fossil polypterid fishes may shift the radiation of crown group ray-finned fishes by 45 million years (Giles et al. 2017).
Finally, molecular phylogenetic methods have introduced new “parts” of lineages to evaluate for persistent evolutionary stasis. Molecular-oriented researchers pursue different part proxies than do morphological researchers. For instance, although genetically identified cryptic species are more common than once thought, their presence undercuts past assumptions of lineage stasis. More generally, gene sequences—as molecular proxies—show patterns that are often discordant with morphological evidence of living fossil status. This result coincides with complicated relationships that have been uncovered in genotype–phenotype maps (Wagner 2014) and G-matrix evolution (Jones et al. 2012), as well as between gene trees and species trees in phylogenetic reconstructions (Szöllősi et al. 2014). Cryptic species, rapid rates of change in some molecular characters, or substantial phylogenetic revisions are indicated for living fossil lingulid brachiopods (Luo et al. 2015), monoplacophorans (Kano et al. 2012), tuatara (Hay et al. 2008), tadpole shrimps (Vanschoenwinkel et al. 2012, Mathers et al. 2013), horseshoe crabs (Obst et al. 2012), bichirs (Near et al. 2014), and coelacanths (Casane and Laurenti 2013, Naville et al. 2014). This provokes new questions about relationships among molecular characters: Do constellations of molecular traits exhibit relative stasis through evolutionary time? If so, what is the nature of the links among different kinds of molecular characters, such as gene regulatory networks, transcription factors, signaling pathways, or conserved noncoding elements (Rebeiz et al. 2015, Polychronopoulos et al. 2017)? To what extent are developmental and morphological stasis reflected in genotype–phenotype maps or the evolving genome (Jones et al. 2012, Wagner 2014, Lowe et al. 2015, Niklas et al. 2015, Yao et al. 2016, Tschopp and Tabin 2017)?
Living fossils: A biological research program
What needs to be specified to understand how the living fossil concept plays the role of delineating what is in need of explanation and structuring a research program around different factors that are associated with long-term stasis? First, previous definitions of living fossils are critical to characterizing the research program, especially disentangling different questions that researchers have asked. Shared criteria of adequacy are a second element and comprise standards for evaluating descriptions of phenomena and putative explanations. A third element is explicit organizational structure, which pertains to how research questions of different kinds relate to one another. Having considered the links between different definitions and different questions above, we next briefly expand on the latter two elements.
Shared criteria of adequacy are evaluative standards for deciding whether descriptions or explanations meet the aims of a research community and constitute a central element of conceptual unity in these communities. Living fossil and stasis are relative terms established by comparison: What's changing (or not)? How fast? In relation to what? By what assumptions or theoretical model? Making the metric(s) of comparison overt and interpretable helps to define a domain within which specific questions can be asked by establishing the relationships among theoretical contexts, measurements, and reality (Houle et al. 2011). Comparisons should initially focus on entities of the same hierarchical level or kind: species with species, proteins with proteins, a skull bone with “the same” skull bone. This typically demands an inference of homology. Morphological or molecular entities must have properties that can be measured or estimated in order to determine degrees of variation in evolutionary patterns or rates. A specified temporal interval of comparison with a robust chronology is often required because scaling affects evolutionary patterns and theoretical models (Uyeda et al. 2011, Hunt et al. 2015). Within an observed or inferred interval, entities must be phylogenetically unambiguous, and the boundaries of a comparison should be set relative to some inclusive phylogenetic group.
Another element of conceptual unity derives from organizational structure, which makes explicit the need to distinguish different types of questions (e.g., geographical distribution of extant and extinct taxa versus temporal duration of particular lineages or suborganismal parts) and articulate thematic and dependency relationships between these questions (e.g., connections between questions about ecological stability and morphological stability, or answers to questions about organismal stasis relying on answers to questions about stasis for proxy characters). How many characters count as a constellation? How is “negligible rate of evolutionary change” operationalized? What do “geographically widespread,” “diverse,” “relict,” and the “same” lineage or clade mean? Different criteria need to be made explicit in the contexts of what a scientist is focused on and in relation to other allied questions in order to flesh out the broader research program of living fossils—explaining why there appears to be a slow or negligible rate of evolutionary change (or stasis) with respect to constellations of molecular or morphological characters in genealogical lineages.
Retention of some phenotypic (traditionally morphological) characters does not adequately explain change or the lack thereof in other phenotypic characters. The same can be said for molecular characters. The role of the living fossil concept can be understood as setting an integrated agenda for research—interrelated suites of questions about patterns in need of explanation and processes relevant to specific character constellations and wholes—that advances our understanding of evolutionary stasis across hierarchical levels of organization. The relations among questions provide a means to navigate the complex architecture of the living fossils problem agenda and can be observed in abstract formulations of these questions.
What mechanisms are responsible for the retention of particular groups of ancestral morphological characters over long periods of time within a lineage? Constraints on evolutionary change—structural, physiological, functional, developmental, and genetic regulatory—can affect groups of characters at all levels of biological organization and are becoming more amenable to comparative phylogenetic and experimental analyses (Pyron 2015, Tschopp and Tabin 2017). It is just as important to consider the role of stabilizing selection in model theoretic and empirical analyses (Jones et al. 2012, Voje 2016). Although these intrinsic and extrinsic influences remain difficult to disentangle, it is possible to see the roles of both within an integrated explanatory framework (Love 2015).
For slow rates of change, how are suites of morphological and molecular characters related? Although much commotion surrounds discordance of morphological and molecular rates of change in categorizing living fossils, less attention has focused on congruent patterns, and still less on links of genetic or developmental pathways and the expression of suites of putatively static morphological characters (Pyron 2015, Rebeiz et al. 2015). Conservation of gene regulatory networks that link transcription factors to gene expression, and ultimately the development of different components of phenotypes, is now widely appreciated, with consequences for both evolvability and constraint. The extent to which such molecular networks resist alteration and also act as determinants contributing to morphological stasis is not yet resolved (Wagner 2014, Niklas et al. 2015, Rebeiz et al. 2015). Such approaches are uncovering relationships among suites of characters that may help to integrate genotypic and phenotypic components of prolonged stasis.
Why do some but not all constellations of characters exhibit apparent stasis over long periods of time in the same lineage? Addressing this question involves evaluating developmental entrenchment, morphological modularity, and functional integration of character constellations in conjunction with phylogenetic inferences of evolutionary persistence (Goswami et al. 2015, Hunt and Slater 2016). Because of the hierarchical organization of gene regulatory networks, different components exhibit distinct grades of conservation or stability in relation to their internal organization, developmental role, and context-dependent functionality (Niklas et al. 2015, Rebeiz et al. 2015). Differences in patterns and strength of modularity and integration among different character constellations could identify the explanatory roles of these phenomena in comprehending patterns of evolutionary stasis and change, as in a recent study on lanternfishes (Denton and Adams 2015).
Why do constellations of characters that represent defining features of species (or supraspecific taxa) persist over long durations and exhibit little net evolutionary change when compared to other lineages? This question requires exploring hypotheses of niche conservatism, habitat tracking, and functional specialization of parts, in addition to genotypic and phenotypic components of stasis. Increasingly sophisticated modeling and comparative analyses are synthesizing evidence from molecular and morphological characters, and functional or ecological parameters, relevant to rates of character evolution (Denton and Adams 2015, Pyron 2015, Hunt and Slater 2016, Lloyd 2016, Price and Schmitz 2016, Lamsdell et al. 2017). Similarity in niche-related trait values sustained in closely related groups of lineages is consistent with hypotheses of phylogenetic niche conservatism. Long-term tracking of paleoecological conditions may perpetuate the survival of certain lineages, and niche modeling can help to distinguish different survival scenarios (Stigall 2012). Parts of organisms relevant to particular functions such as feeding (Herrera-Flores et al. 2017) or collaborative components of transcriptional machinery (Lynch and Wagner, 2008, 2011) can be analyzed by focusing on the coadaptation of traits or convergence among sets of characters recognized phylogenetically in more distantly related lineages. These approaches bring extrinsic factors—biogeography, ecology, adaptation, and independent phylogenetic contrasts with other groups of organisms—into the foreground of the research program, and not only for organisms and lineages. Niche conservatism, habitat tracking, and functional specialization of parts all apply equally in molecular environments.
How are perceived declines in living fossil groups (from previous high levels of taxonomic diversity to low levels today) related to patterns in phylogenetic sister groups, and to origination and extinction dynamics? When comparing species-rich and species-poor sister groups, the former sometimes have been interpreted as derived or advanced and the latter as basal living fossils. However, the underlying patterns may be more complicated or the explanatory reasoning flawed (Omland et al. 2008, Nagalingum et al. 2011, Lamsdell et al. 2017). One might hypothesize, for example, that differences in the diversity of sister groups are simply due to chance variations, or alternatively that reduced diversity represents some sort of replacement. Monotremes have been called survivors, sister taxa to placental and therian mammals, conceivably discounting the similar ages of these groups and the fact that all are a mix of symplesiomorphic and independently evolved characters. Assuming that sustained extinctions and slow evolutionary rates are uniformly characteristic of living fossil groups is also problematic. Modern crown group gymnosperms (living fossils more abundant in the Early Mesozoic) are considerably younger—not more ancient—than living crown group angiosperms (Crisp and Cook 2011). Extinction and radiation surges as late as 5 million years ago also have been discovered in living fossil cycads, a gymnosperm subclade (Nagalingum et al. 2011). Together with the evolutionary dynamics in bichir fishes (Near et al. 2014), these studies point to a more nuanced understanding attainable from combined fossil and phylogenetic analyses that include both molecular and morphological characters.
Why do some living fossils exhibit “relict” geographic distribution (i.e., distribution that is significantly more restricted than in the geologic past)?
Ginkgo, Cercidiphyllum, and related genera, once widespread in the Northern Hemisphere, became confined to Eastern Asia near the end of the Cenozoic. Distribution range modeling linked with climatic fluctuations indicates that seasonal aridity more than temperature was a constraining factor (Huang et al. 2015). Cenozoic co-occurrence of Ginkgo and Cercidiphyllum in disturbed streamside environments suggests prolonged conservation of their habitats (Zhao et al. 2016). Molecular phylogeography, niche modeling, paleoclimatology, and life-history traits reveal an even more dynamic geographic history through glaciations. Finer-scale studies reveal incongruent episodes of retreat, colonization, and expansion linked to changes in preferred habitats, as well as regional temperature and aridity fluctuations, topography, and hydrology. G. biloba's more restrictive, disjoint distribution relative to C. japonicum may relate to longer generation times, more climatically vulnerable reproductive periods, or more limited dispersal of its fleshy seeds compared to wind dispersal (Zhao et al. 2016), implicating population demographic and life-history traits as potential explanations.
How is molecular or morphological stasis for constellations of characters affected by their manifestation at different junctures in a life history? Most studies of evolutionary tempo and mode have focused on changes in adult morphology, but part–whole patterns of stasis also are manifested in different life-history stages, conserved genetic mechanisms, and cellular–physical mechanisms or morphogenetic patterns. Developmentally circumscribed examples of stasis can be observed in both living and fossil forms, such as similar larval forms across taxa with radically different adult forms (Wray 1995, Marlow et al. 2014), cell lineage morphogenesis (Hunt and Yasuhara 2010), or pollen formation and structure (Matamoro-Vidal et al. 2016). Molecular genetic and embryological body-axis patterning studies are reinforcing ancient, stable relationships across deuterostomes (Lowe et al. 2015), certain gene sequence motifs and functionally conserved enhancers appear to predate the evolution of chordates (Yao et al. 2016), and studies of rare fossil embryos are integrating data from developmental genetics and paleontology (Organ et al. 2015).
What are our null expectations concerning living fossils?
Although this question has seldom been asked of suborganismal entities, there is precedence for the use of null models of morphological disparity and taxonomic diversification in paleobiology. Should we expect very long-lived lineages to be common or rare, morphologically distant or average compared to related lineages? Simple null models of morphological evolution and lineage diversification in which characters are not subject to selection and do not affect diversification predict greater numbers of ancestral characters than derived ones over time, increasing character-level stasis (and the probability of living fossils) in a relative sense as diversification occurs (Raup et al. 1973). This also can be an expectation of rate-homogeneous models in which characters become integrated or correlated in some way (Wagner and Estabrook 2015). In addition, there is evidence that morphological distribution is related to lineage longevity; very-long-duration lineages often tend to be more average than expected when compared with others in their clade or paraclade (Liow, 2004, 2007). Things get more complicated or even reversed in models that incorporate parameters related to extinction, logistic or character-dependent diversification rates, and species selection. For example, Wagner and Estabrook (2014) evaluated a range of such models using several hundred morphological and stratigraphic data sets and found (in most cases) that groups containing ancestral characters experienced early loss of diversity and frequent shifts in the rate of character-dependent diversification, with the latter likely being determined by elevated net extinction—in short, these models suggest we should expect living fossils to be uncommon. Framing null expectations of this kind for highly conserved, hypothetically “living fossil” molecules and genetic mechanisms are more complicated still (Lynch and Wagner 2008, Wagner 2014, Niklas et al. 2015, Rebeiz et al. 2015, Polychronopoulos et al. 2017). However, answers to other questions within the research program, such as how suites of molecular characters are related or why some but not all constellations of molecular characters exhibit apparent stasis over long periods of time, will contribute to more robust null models that yield increasingly precise predictions for both molecular and morphological characters. This is a reminder that all of these research questions are interrelated, and therefore better characterizations of different patterns of stasis and better accounts of the mechanisms underlying them fosters conceptual unification for heterogeneous investigations of living fossils.
Conclusions
The research questions detailed above correspond to several of the cross-cutting membership criteria that make the living fossil concept contentious. Empirical advances in quantitatively evaluating evolutionary modes among different morphological characters in fossil lineages (box 2; Hopkins and Lidgard 2012, Hunt et al. 2015) open a path for investigating those membership criteria by generating rigorous measurements of stability, which then can be used to explore stasis in bundles of molecular and morphological characters. A central insight of this work is that thinking explicitly about relationships between parts (characters or character constellations) and wholes (e.g., organisms or lineages) requires revisions in how we understand stereotypical living fossils. Our synthesis suggests a parallel interpretation of highly conserved molecules and genetic mechanisms: the relative stability of particular bundles of molecular characters represents an ongoing explanatory task for contemporary biologists. More generally, our synthesis consolidates a broad array of heterogeneous and relatively fragmented investigations into living fossils, providing a more unified conceptual framework that steers clear of semantic debates and facilitates interdisciplinary research on fundamental evolutionary questions surrounding molecular and morphological stasis.
Instead of viewing the living fossil concept in terms of categorization—what criteria should define living fossils—it is better to understand its role as setting a unified agenda for research. This brings to the foreground hypotheses about patterns in need of explanation and processes relevant to specific character constellations and wholes for which they may serve in proxy roles. Interrelated research questions motivating these hypotheses differ and require combinations of criteria and methods to achieve explanatory adequacy. Increasingly powerful phylogenetic and fossil sampling models are discerning different rates of change among characters and lineages (Wagner and Marcot 2013, Denton and Adams 2015, Hunt and Slater 2016, Lloyd 2016, Price and Schmitz 2016, Tschopp and Tabin 2017). Integrative models are analyzing connections between prolonged stasis and timescale (Uyeda et al. 2011, Near et al. 2014, Pyron 2015, Price and Schmitz 2016), ecology (Stigall 2012, Price and Schmitz 2016, Lamsdell et al. 2017), and biogeography (Stigall 2012, Huang et al. 2015). Stable genetic or developmental pathways as character constellations can be analyzed in light of developmental and functional integration and modularity (Wagner 2014, Denton and Adams 2015, Rebeiz et al. 2015, Tschopp and Tabin 2017), with integration and modularity also becoming amenable to quantitative study in paleontology (Wilson 2013, Goswami et al. 2015). In summary, different disciplinary perspectives, data, and methods can be integrated within an organized structure of problems to achieve more comprehensive and unified answers to questions about slow or negligible rates of evolutionary change—stasis—for diverse kinds of parts and wholes in living systems.
Acknowledgments
Comments from Lee-Hsiang Liow, Jeremiah Smith, and Peter Wagner improved the manuscript, as did images from Klaus-Peter Kelber, Roger Key, and James Lamsdell, and drafting by Monica Jurik. We are grateful to all. ACL’s research was supported in part by a grant from the John Templeton Foundation (“From Biological Practice to Scientific Metaphysics,” grant no. 50191). The opinions expressed in this article do not represent those of the John Templeton Foundation.
References cited
- Amemiya CT, et al. 2010. Complete HOX cluster characterization of the coelacanth provides further evidence for slow evolution of Its genome. Proceedings of the National Academy of Sciences 107: 3622–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LS, Kayser M, Jones C. 2008. The mineralized osteocyte: A living fossil. American Journal of Physical Anthropology 137: 449–456. [DOI] [PubMed] [Google Scholar]
- Bennett DJ, Sutton MD, Turvey ST. 2017. Evolutionarily distinct “living fossils” require both lower speciation and lower extinction rates. Paleobiology 43: 34–48. [Google Scholar]
- Brigandt I, Love AC. 2012. Conceptualizing evolutionary novelty: Moving beyond definitional debates. Journal of Experimental Zoology B: Molecular and Developmental Evolution 318: 417–427. [DOI] [PubMed] [Google Scholar]
- Bronzati M, Montefeltro FC, Langer MC. 2015. Diversification events and the effects of mass extinctions on Crocodyliformes evolutionary history. Royal Society Open Science 2(art. 140385). (17 January 2018; http://rsos.royalsocietypublishing.org/content/2/5/140385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casane D, Laurenti P. 2013. Why coelacanths are not “living fossils.” BioEssays 35: 332–338. [DOI] [PubMed] [Google Scholar]
- Cavin L, Guinot G. 2014. Coelacanths as “almost living fossils.” Frontiers in Ecology and Evolution 2: 1–5. (17 January 2018; www.frontiersin.org/articles/10.3389/fevo.2014.00049) [Google Scholar]
- Crisp MD, Cook LG. 2011. Cenozoic extinctions account for the low diversity of extant gymnosperms compared with angiosperms. New Phytologist 192: 997–1009. [DOI] [PubMed] [Google Scholar]
- Denton JSS, Adams DC. 2015. A new phylogenetic test for comparing multiple high-dimensional evolutionary rates suggests interplay of evolutionary rates and modularity in lanternfishes (Myctophiformes; Myctophidae). Evolution 69: 2425–2440. [DOI] [PubMed] [Google Scholar]
- Forrest LL, Wickett NJ, Cox CJ, Goffinet B. 2011. Deep sequencing of Ptilidium (Ptilidiaceae) suggests evolutionary stasis in liverwort plastid genome structure. Plant Ecology and Evolution 144: 29–43. [Google Scholar]
- Giles S, Xu G-H, Near TJ, Friedman M. 2017. Early members of “living fossil” lineage imply later origin of modern ray-finned fishes. Nature 549: 265–268. [DOI] [PubMed] [Google Scholar]
- Goswami A, Binder WJ, Meachen J, O’Keefe FR. 2015. The fossil record of phenotypic integration and modularity: A deep-time perspective on developmental and evolutionary dynamics. Proceedings of the National Academy of Sciences 112: 4891–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JM, Subramanian S, Millar CD, Mohandesan E, Lambert DM. 2008. Rapid molecular evolution in a living fossil. Trends in Genetics 24: 106–109. [DOI] [PubMed] [Google Scholar]
- Herrera-Flores JA, Stubbs TL, Benton MJ. 2017. Macroevolutionary patterns in Rhynchocephalia: Is the tuatara (Sphenodon punctatus) a living fossil? Palaeontology 60: 319–328. [Google Scholar]
- Hopkins MJ, Lidgard S. 2012. Evolutionary mode routinely varies among morphological traits within fossil species lineages. Proceedings of the National Academy of Sciences 109: 20520–20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D, Pélabon C, Wagner GP, Hansen TF. 2011. Measurement and meaning in biology. Quarterly Review of Biology 86: 3–34. [DOI] [PubMed] [Google Scholar]
- Huang Y, Jacques FM, Su T, Ferguson DK, Tang H, Chen W, Zhou Z. 2015. Distribution of Cenozoic plant relicts in China explained by drought in dry season. Scientific Reports 5(art. 14212). (17 January 2018; www.nature.com/articles/srep14212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt G, Slater G. 2016. Integrating paleontological and phylogenetic approaches to macroevolution. Annual Review of Ecology, Evolution, and Systematics 47: 189–213. [Google Scholar]
- Hunt G, Yasuhara M. 2010. A fossil record of developmental events: Variation and evolution in epidermal cell divisions in ostracodes. Evolution and Development 12: 635–646. [DOI] [PubMed] [Google Scholar]
- Hunt G, Hopkins MJ, Lidgard S. 2015. Simple versus complex models of trait evolution and stasis as a response to environmental change. Proceedings of the National Academy of Sciences 112: 4885–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AG, Bürger R, Arnold SJ, Hohenlohe PA, Uyeda JC. 2012. The effects of stochastic and episodic movement of the optimum on the evolution of the G-matrix and the response of the trait mean to selection. Journal of Evolutionary Biology 25: 2210–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y, Kimura S, Kimura T, Waren A. 2012. Living Monoplacophora: Morphological conservatism or recent diversification? Zoologica Scripta 41: 471–488. [Google Scholar]
- Kelber K-P. 1999. Triops cancriformis (Crustacea, Notostraca): Ein bemerkenswertes fossil aus der Trias Mitteleuropas. 383–394 in Hauschke N, Wilde V, eds. TRIAS: Eine ganz andere Welt. Mitteleuropa im frühen Erdmittelalter. Friedrich Pfeil. [Google Scholar]
- Lamsdell JC. 2016. Horseshoe crab phylogeny and independent colonizations of fresh water: Ecological invasion as a driver for morphological innovation. Palaeontology 59: 181–194. [Google Scholar]
- Lamsdell JC, Congreve CR, Hopkins MJ, Krug AZ, Patzkowsky ME. 2017. Phylogenetic paleoecology: Tree-thinking and ecology in deep time. Trends in Ecology and Evolution 32: 452–463. [DOI] [PubMed] [Google Scholar]
- Lavrov DV. 2007. Key transitions in animal evolution: A mitochondrial DNA perspective. Integrative and Comparative Biology 47: 734–743. [DOI] [PubMed] [Google Scholar]
- Lidgard S, Hopkins M. 2015. Stasis. Oxford Bibliographies in Evolutionary Biology. Oxford University Press; (17 January 2018; www.oxfordbibliographies.com/view/document/obo-9780199941728/obo-9780199941728-0067.xml) [Google Scholar]
- Liow LH. 2004. A test of Simpson's “Rule of the survival of the relatively unspecialized” using fossil crinoids. American Naturalist 164: 431–443. [DOI] [PubMed] [Google Scholar]
- Liow LH. 2007. Lineages with long durations are old and morphologically average: An analysis using multiple datasets. Evolution 61: 885–901. [DOI] [PubMed] [Google Scholar]
- Liu Y, Medina R, Goffinet B. 2014. 350 My of mitochondrial genome stasis in mosses, an early land plant lineage. Molecular Biology and Evolution 31: 2586–2591. [DOI] [PubMed] [Google Scholar]
- Lloyd GT. 2016. Estimating morphological diversity and tempo with discrete character–taxon matrices: Implementation, challenges, progress, and future directions. Biological Journal of the Linnean Society 118: 131–151. [Google Scholar]
- Love AC. 2014. The erotetic organization of developmental biology. 33–55 in Minelli A, Pradeu T, eds. Towards a Theory of Development. Oxford University Press. [Google Scholar]
- Love AC. 2015. Evolutionary developmental biology: Philosophical issues. 265–283 in Heams T, Huneman P, Lecointre G, Silberstein M, eds. Handbook of Evolutionary Thinking in the Sciences. Springer. [Google Scholar]
- Lowe CJ, Clarke DN, Medeiros DM, Rokhsar DS, Gerhart J. 2015. The deuterostome context of chordate origins. Nature 520: 456–465. [DOI] [PubMed] [Google Scholar]
- Luo Y-J, Satoh N, Endo K. 2015. Mitochondrial gene order variation in the brachiopod Lingula anatina and its implications for mitochondrial evolution in lophotrochozoans. Marine Genomics 24: 31–40. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Alva V. 2017. Ribosomal proteins as documents of the transition from unstructured (poly) peptides to folded proteins. Journal of Structural Biology 198: 74–81. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62: 2131–2154. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. 2011. Revisiting a classic example of transcription factor functional equivalence: Are Eyeless and Pax6 functionally equivalent or divergent? Journal of Experimental Zoology B: Molecular and Developmental Evolution 316: 93–98. [DOI] [PubMed] [Google Scholar]
- Marlow H, Tosches MA, Tomer R, Steinmetz PR, Lauri A, Larsson T, Arendt D. 2014. Larval body patterning and apical organs are conserved in animal evolution. BMC Biology 12(art. 7). (17 January 2018; https://bmcbiol.biomedcentral.com/articles/10.1186/1741-7007-12-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoro-Vidal A, Prieu C, Furness CA, Albert B, Gouyon P-H. 2016. Evolutionary stasis in pollen morphogenesis due to natural selection. New Phytologist 209: 376–394. [DOI] [PubMed] [Google Scholar]
- Mathers TC, Hammond RL, Jenner RA, Hänfling B, Gómez A. Multiple global radiations in tadpole shrimps challenge the concept of “living fossils.”. Peer J. 2013;1 doi: 10.7717/peerj.62. (art. e62). (17 January 2018; https://peerj.com/articles/62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce RCP, Sansom R, Wills MA. 2016. Sampling diverse characters improves phylogenies: Craniodental and postcranial characters of vertebrates often imply different trees. Evolution 70: 666–686. [DOI] [PubMed] [Google Scholar]
- Nagalingum NS, Marshall CR, Quental TB, Rai HS, Little DP, Mathews S. 2011. Recent synchronous radiation of a living fossil. Science 334: 796–799. [DOI] [PubMed] [Google Scholar]
- Naville M, Chalopin D, Volff J-N. 2014. Interspecies insertion polymorphism analysis reveals recent activity of transposable elements in extant coelacanths. PLOS ONE 9(art. e114382). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near TJ, Dornburg A, Tokita M, Suzuki D, Brandley MC, Friedman M. 2014. Boom and bust: Ancient and recent diversification in bichirs (Polypteridae: Actinopterygii), a relictual lineage of ray-finned fishes. Evolution 68: 1014–1026. [DOI] [PubMed] [Google Scholar]
- Niklas KJ, Bondos SE, Dunker AK, Newman SA. 2015. Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Frontiers in Cell and Developmental Biology 3(art. 8). (17 January 2018; www.frontiersin.org/articles/10.3389/fcell.2015.00008/full) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst M, Faurby S, Bussarawit S, Funch P. 2012. Molecular phylogeny of extant horseshoe crabs (Xiphosura, Limulidae) indicates Paleogene diversification of Asian species. Molecular Phylogenetics and Evolution 62: 21–26. [DOI] [PubMed] [Google Scholar]
- Omland KE, Cook LG, Crisp MD. 2008. Tree thinking for all biology: The problem with reading phylogenies as ladders of progress. BioEssays 30: 854–867. [DOI] [PubMed] [Google Scholar]
- Organ CL, Cooper LN, Hieronymus TL. 2015. Macroevolutionary developmental biology: Embryos, fossils, and phylogenies. Developmental Dynamics 244: 1184–1192. [DOI] [PubMed] [Google Scholar]
- Polychronopoulos D, King JWD, Nash AJ, Tan G, Lenhard B. 2017. Conserved non-coding elements: Developmental gene regulation meets genome organization. Nucleic Acids Research 45: 12611–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prangishvili D. 2015. Archaeal viruses: Living fossils of the ancient virosphere? Annals of the New York Academy of Sciences 1341: 35–40. [DOI] [PubMed] [Google Scholar]
- Price SA, Schmitz L. 2016. A promising future for integrative biodiversity research: An increased role of scale-dependency and functional biology. Philosophical Transactions of the Royal Society B 371(art. 20150228). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyron RA. 2015. Post-molecular systematics and the future of phylogenetics. Trends in Ecology and Evolution 30: 384–389. [DOI] [PubMed] [Google Scholar]
- Raup DM, Gould SJ, Schopf TJM, Simberloff DS. 1973. Stochastic models of phylogeny and the evolution of diversity. Journal of Geology 81: 525–542. [Google Scholar]
- Rebeiz M, Patel NH, Hinman VF. 2015. Unraveling the tangled skein: The evolution of transcriptional regulatory networks in development. Annual Review of Genomics and Human Genetics 16: 103–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11(art. 29). (17 January 2018; www.biomedcentral.com/1741-7007/11/29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner S. 2014. What can a living fossil tell us about evolution and mechanism of ion-coupled transporters: The story of small multidrug transporters. 233–248 in Krämer R, Ziegler C, eds. Membrane Transport Mechanism: 3D Structure and Beyond. Springer. [Google Scholar]
- Smith JJ, Sumiyama K, Amemiya CT. 2012. A living fossil in the genome of a living fossil: Harbinger transposons in the coelacanth genome. Molecular Biology and Evolution 29: 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigall AL. 2012. Using ecological niche modelling to evaluate niche stability in deep time. Journal of Biogeography 39: 772–781. [Google Scholar]
- Szöllősi GJ, Tannier E, Daubin V, Boussau B. 2014. The inference of gene trees with species trees. Systematic Biology 64: e42–e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp P, Tabin CJ. 2017. Deep homology in the age of next-generation sequencing. Philosophical Transactions of the Royal Societys B 372(art. 20150475). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda JC, Hansen TF, Arnold SJ, Pienaar J. 2011. The million-year wait for macroevolutionary bursts. Proceedings of the National Academy of Sciences 108: 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanschoenwinkel B, Pinceel T, Vanhove MPM, Denis C, Jocque M, Timms BV, Brendonck L. 2012. Toward a global phylogeny of the “living fossil” crustacean order of the Notostraca. PLOS ONE 7(art. e34998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voje KL. 2016. Tempo does not correlate with mode in the fossil record. Evolution 70: 2678–2689. [DOI] [PubMed] [Google Scholar]
- Wagner GP. 2014. Homology, Genes, and Evolutionary Innovation. Princeton University Press. [Google Scholar]
- Wagner PJ, Estabrook GF. 2014. Trait-based diversification shifts reflect differential extinction among fossil taxa. Proceedings of the National Academy of Sciences 111: 16419–16424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner PJ, Estabrook GF. 2015. The implications of stratigraphic compatibility for character integration among fossil taxa. Systematic Biology 64: 838–852. [DOI] [PubMed] [Google Scholar]
- Wagner PJ, Marcot JD. 2013. Modelling distributions of fossil sampling rates over time, space and taxa: Assessment and implications for macroevolutionary studies. Methods in Ecology and Evolution 4: 703–713. [Google Scholar]
- Werth AJ, Shear WA. 2014. The evolutionary truth about living fossils. American Scientist 102: 434–443. [Google Scholar]
- Wilson LAB. 2013. The contribution of developmental palaeontology to extensions of evolutionary theory. Acta Zoologica 94: 254–260. [Google Scholar]
- Wray GA. 1995. Punctuated evolution of embryos. Science 267: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Wu C-S, Chaw S-M. 2015. Evolutionary stasis in cycad plastomes and the first case of plastome GC-biased gene conversion. Genome Biology and Evolution 7: 2000–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, et al. 2016. Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nature Genetics 48: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Shu DG, Han J, Liu JN. 2005. Morpho-anatomical differences of the Early Cambrian Chengjiang and Recent lingulids and their implications. Acta Zoologica 86: 277–288. [Google Scholar]
- Zhao Y-P, Yan X-L, Muir G, Dai Q-Y, Koch MA, Fu C-X. 2016. Incongruent range dynamics between co-occurring Asian temperate tree species facilitated by life history traits. Ecology and Evolution 6: 2346–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. 2009. An overview of fossil Ginkgoales.” Palaeoworld 18: 1–22. (1 July 2018; https://doi.org/10.1016/j.palwor.2009.01.001) [Google Scholar]