Abstract
Background
The overexpression of sphingosine kinase 1 (SPHK1) is responsible for the progress of many cancers. However, the role of SPHK1 in the development and progression of neuroblastoma (NB) remain largely unknown. Here in this study, we explored whether silencing SPHK1 by lentivirus-mediated siRNA could be employed as a potential therapeutic target for NB.
Materials and methods
Lentivirus was adopted to load SPHK1 siRNA. The results were obtained using RT-qPCR, Western blot, cell proliferation assay, transwell cell migration/invasion assays as well as in vivo xenograft tumor models in nude mice.
Results
Our results demonstrated that SPHK1 mRNA was upregulated in SH-SY5Y and SK-N-SH cells as well as in human NB tissues. SPHK1 knockdown by siRNA resulted in impaired proliferation, increased apoptosis, as well as impaired migration and invasion of SH-SY5Y and SK-N-SH cells. In addition, the in vivo study suggested that SPHK1 knockdown significantly reduced the tumorigenesis of SH-SY5Y xenograft model. Furthermore, intratumorally administered lentivirus-SPHK1 siRNA could significantly inhibit tumor growth in an SH-SY5Y xenograft mice model. Intensive investigations on mechanism revealed that these effects were achieved through the deactivation of STAT3 pathways.
Conclusion
These data suggest that SPHK1 inhibition via downregulation of STAT3 pathways by lentivirus-mediated siRNA knockdown can significantly suppress NB progression, which could be a promising target for future gene therapy of NB.
Keywords: lentivirus, siRNA, SPHK1, STAT3, neuroblastoma
Introduction
Neuroblastoma (NB) is one of the most common cancers in babies as well as the third most frequent cancer in children.1 It is usually rare in adults, but frequently occurs in children under 5 years of age and accounts for about 90% of total cases. It poses a great threat to the survival of children and is responsible for about 15% of cancer deaths of children.2 Although NB treatments include a variety of approaches from observation, surgery, radiation, and chemotherapy to even stem cell transplantation.3 However, chances of long-term survival in some high-risk cases are <40% even after aggressive treatment.1 As a result, accurate and objective understanding toward the tumorigenesis and aggressiveness of NB is desperately in need for optimizing the subsequent individual therapy, which requires the identification of the exact molecular markers for postoperative prognostication of NB.
The sphingosine kinase 1 (SPHK1) is a novel lipid messenger with both intracellular and extracellular functions that plays a critical role in the regulation of cellular development and differentiation via affecting multiple downstream pathways.4,5 Recent evidences have demonstrated that the increased risk in the development of various cancers is associated with SPHK1 overexpression.6,7 Furthermore, SPHK1 expression and activation is also suggested to help evaluate cancer resistance to chemotherapy as well as radiotherapy.8,9 These studies open new windows for cancer treatment by targeting SPHK1. In contrast, the role of SPHK1 in the development and progression of NB remains largely unknown. Previous research has been focused on depth works on the underlying molecular mechanisms of small molecular drugs, such as amyloid-β peptide10 and endotoxin,11 on the suppression of the proliferation of different cancers. Nonetheless, the application of gene inhibitors, such as siRNA targeting SPHK1 for the gene therapy of NB, still remains largely unexplored.
RNA interference (RNAi) is a dsRNA capable of silencing the expression of a specific gene as it shares homologous sequence with the dsRNA.12 It is recognized as a milestone in modern biotechnology that RNAi offers a convenient approach to observe the function of specific genes in mammalian cells.13 As a result, it provides a powerful tool for gene mechanism studies and gene therapy in various diseases, including cancers.14–16 In our study, we prepared a lentiviral vector containing siRNA against SPHK1 gene to explore the role of SPHK1 in the progression of NB and to test the feasibility of in vitro and in vivo NB treatment via SPHK1 knockdown using siRNA.
Materials and methods
Cell culture and tissue samples
SH-SY5Y, SK-N-SH, human umbilical vein endothelial cell (HUVEC), and 293T cell lines purchased from American Type Culture Collection (Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) in an incubator maintained at 37°C with humidified atmosphere containing 5% CO2.
This study was approved by the ethics committee of Affiliated Hospital of Jining Medical University. A total of 29 patients who were diagnosed as having NB and went through surgical resection at Affiliated Hospital of Jining Medical University between April 2012 and June 2016 were enrolled in this study. Written informed consent was obtained from all the participating patients. Twenty nine pairs of NB tumor tissues and adjacent normal tissues were immediately snap-frozen in liquid nitrogen after surgical resection.
Preparation of lentiviral siRNA vector targeting SPHK1
The siRNA (#1: 5′-GAGCUGCAAGGCCUUGCCCUUGCCG-3′ and #2: 5′-GUGCACCCAAACUACUUCUTT-3′) targeting human SPHK1 gene (GenePharma, Shanghai, People’s Republic of China) was selected to prepare the lentiviral siRNA vector. These target sequences and universal blank sequence were cloned into the lentiviral vector pLVX-shRNA2 (Clontech, Mountain view, CA, USA) and named as SPHK1 siRNA and negative control, respectively.
Lentivirus amplification and transfection
The lentiviral vectors were amplified by transfection into the 293 T cells with Lenti-X™ HTX Packaging System (Clontech) following the manufacturer’s protocol. The amplified lentiviruses were collected at 48 hours posttransfection. Afterwards, SH-SY5Y and SK-N-SH cells were seeded in 24-well plates (Corning, New York, NY, USA, density: 2×105/mL) and the lentiviral particles were added. Flow cytometry (FACS Calibur, BD Biosciences, San Jose, CA, USA) with FITC filter was employed to sort transfected cells.
Quantitative real time (RT)-qPCR
RT-qPCR was employed to determine the mRNA expression level of SPHK1 using the BeyoFast™ SYBR Green qPCR Mix Kit (Beyotime, Haimen, Jiangsu, People’s Republic of China) at an ABI 7300 Real-Time PCR System (Applied Biosystems, Life Technologies GmbH, Darmstadt, Germany). The following PCR reaction was used: step 1: 95°C for 10 minutes, 1 cycle; step 2: 95°C for 15 seconds, 60°C for 1 minute, 40 cycles. The primer used for the PCR was as follows: for SPHK1, forward 5′-CCTTCCTCCTTCCCTAGGG-3′; reverse 5′-TAGAAGGCCTTACATAGGCAGC-3′; for STAT3, forward 5′-CAGCAGCTTGACACACGGTA-3′ and reverse 5′-AAACACCAAAGTGGCATGTGA-3′. All samples were run in triplicate, and samples were normalized against an endogenous internal control, β-actin. Levels of SPHK1 and STAT3 mRNA were quantified using 2−ΔΔCq method.17
Western blot
Total protein was extracted from the tissues and cell lines using the RIPA buffer consisting of 50 mM Tris pH 7.4, 150 mM NaCl, 1 % NP-40, 0.5% sodium deoxycholic acid, and 0.1% sodium dodecyl sulfate, as reported previously.17 The protein sample was quantified using the Bradford Protein Concentration Determination Kit (Beyotime). The same amount of protein sample was separated using a 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Merck Millipore, Burlington, MA, USA). The membrane was blocked with fat-free milk at 4°C for 1 hour, following incubation with the primary antibodies against SPHK1, STAT3, and GAPDH (Abcam, Cambridge, MA, USA). On the next day, the membrane was washed with TBS-T three times and incubated with secondary antibody (ab97023, Abcam) for 2 hours. To visualize the protein bands, a BeyoECL Plus Kit (Beyotime) was employed, and the intensity was quantified using the Tanon Automatic Chemiluminescence Western blot Imaging system (Tanon, Shanghai, People’s Republic of China).
Cell proliferation assay
The rate of cell proliferation was determined using the Cell Counting Kit-8 (CCK-8) assay following the protocols provided by the supplier (Beyotime). Briefly, cells were seeded into 96-well plate (density of 2×103 cells/well) and allowed to culture in the aforementioned conditions. Subsequently, 20 µL of CCK-8 solution was added to the each well at predetermined time points and the plates were returned to the incubator for another 4 hours. The absorbance was determined at 490 nm using an Epoch microplate reader (BioTek Instruments Inc., Winooski, VT, USA).18,19 For each sample, the experiment was repeated in triplicate.
Cell apoptosis analysis
After 48 hours of culture, cells were harvested and resuspended in PBS. Annexin V/fluorescein isothiocyanate (Beyotime) was applied to determine the apoptosis profile according to according to the manufacturer’s instructions.
Cell migration/invasion assays using Transwell
The cell migration/invasion assays were performed in line with previous report.20 In brief, Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was added into the upper chamber of Transwell (Corning). Afterward, the wells were returned to the incubator for 2 hours and the Matrigel was allowed to solidify. The cells were seeded in the upper chamber of Transwell with or without Matrigel coating. After incubation for 24 hours, crystal violet was employed to stain the lower chamber. Each sample in every assay was repeated in triplicates.
Therapeutic effects on human NB xenograft mice models
Animal experiments were carried out to investigate the anti-tumor effect of SPHK1 regulation on NB. Male BALB/c nude mice (weight 18–22 g, 6–8 weeks) were purchased from Shanghai Laboratory Animal Center (Suzhou, Jiangsu, People’s Republic of China) and maintained at 22°C±2°C with access to food and water ad libitum. BALB/c nude mice were randomly divided into the three groups (n=10). The mice were subcutaneously injected (right flank) with 5×106 SH-SY5Y cells with different treatments (SPHK1 siRNA, negative control and untreated), respectively. In addition, 5×106 untreated SH-SY5Y cells were subcutaneously administered to nude mice and allowed to grow into solid tumor.18 Once tumor volume reached around 200 mm3, the mice were randomly divided into the 3 groups (n=8) and intratumorally injected with about 6×107 copies of lentivirus with SPHK1 siRNA or negative control, respectively. About 30 µL PBS was injected into the third group as control. The tumor volume of all mice was monitored every other day. Tumor sizes were measured using a Vernier caliper, and tumor volumes were calculated as V = a2 × b/2 mm3 (a: minor axis; b: major axis). All animal experiments were conducted in compliance with NIH guidelines for care and use of research animals and approved by the animal ethics committee of Affiliated Hospital of Jining Medical University.
Statistical analysis
All data were analyzed using SPSS 19.0 statistical software package (SPSS Inc., Chicago, IL, USA). One-way ANOVA and Tukey post hoc test was performed to analyze the difference among three or above groups. P<0.05 was considered statistically significant.
Results and discussion
SPHK1 mRNA is upregulated in NB cells and human NB tissues
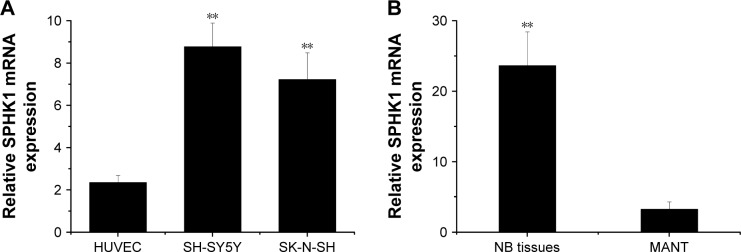
SPHK1 regulates multiple cellular processes, such as cell cycle, metabolism, proliferation, migration, as well as survival, via mediating various signaling pathways. Many previous reports have demonstrated that elevated expression of SPHK1 is observed in multiple types of human cancers.8,21 Suppression of SPHK1 activity has been proven to induce cell cycle arrest, growth inhibition, and even apoptosis in many cancer cells.22,23 As a result, SPHK1 has been identified as a promising target in cancer treatment.24 To examine the role of SPHK1 in NB, we measured the level of SPHK1 mRNA in two NB cell lines (SH-SY5Y and SK-N-SH cells), as well as in 29 human NB tissues and matched adjacent nontumorous tissues using quantitative RT-qPCR. Our results clearly revealed that compared with HUVEC and matched adjacent nontumorous tissue, SPHK1 mRNA was significantly upregulated in both SH-SY5Y and SK-N-SH cells (Figure 1A) as well as NB tissues (Figure 1B).
Figure 1.
SPHK1 mRNA overexpression in NB cells and human NB tissues.
Notes: (A) SPHK1 mRNA expression was detected in NB cells. The SPHK1 mRNA level was significantly upregulated in SH-SY5Y and SK-N-SH cells compared with HUVEC cells. **P<0.01 vs HUVEC cells. (B) SPHK1 mRNA expression was detected in 29 human NB tissues and MANT. The SPHK1 mRNA level was significantly upregulated in NB tissues compared with MANT. **P<0.01 vs MANT.
Abbreviations: HUVEC, human umbilical vein endothelial cells; MANT, matched adjacent nontumorous tissues; NB, neuroblastoma; SPHK1, sphingosine kinase 1.
RNAi significantly suppresses both SPHK1 mRNA and protein expression in NB cells
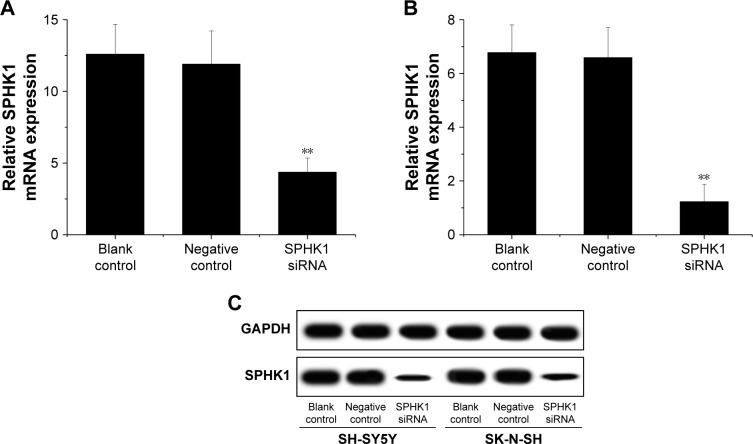
The inhibition of SPHK1 could be achieved in different ways, including the application of monoclonal antibody, antisense oligonucleotides, as well as small molecule kinase inhibitors. Many previous studies have been performed to show certain efficacy.25,26 However, their clinical potential is still controversial. RNAi as a new technology to cure diseases via gene knockdown holds great potential to be adopted in a variety of novel therapeutic approaches. Lentiviral vectors can efficiently deliver gene to achieve stable transfection of loaded cargos with minimal immunogenicity and no adverse events.27,28 As a result, transfection of siRNA using lentivirus might be a promising strategy for effective gene therapy of NB. In our study, lentiviral siRNA vector targeting the SPHK1 gene was successfully constructed and employed as an investigation tool. SH-SY5Y and SK-N-SH cells were first infected with the vector to obtain SPHK1 siRNA/negative control stably expressing cell lines. RT-qPCR showed that compared with the negative control or blank control (the untreated cells), the level of SPHK1 mRNA in SPHK1 stably expressed SH-SY5Y and SK-N-SH cells was reduced by around 65.3% and 81.9%, respectively, (Figure 2A and B). Moreover, Western blot results suggested that compared to other groups, the SPHK1 protein level in SPHK1 stably expressed SH-SY5Y and SK-N-SH cells was greatly decreased (Figure 2C), which indicated that the siRNA targeting SPHK1 delivered by lentivirus can efficiently silence SPHK1 expression in both SH-SY5Y and SK-N-SH cells.
Figure 2.
Effect of SPHK1 knockdown by siRNA on SPHK1 expression in SH-SY5Y and SK-N-SH cells.
Notes: SPHK1 mRNA level was significantly reduced in SH-SY5Y (A) and SK-N-SH (B) cells with SPHK1 knockdown compared with the negative control or blank control. (C) Representative Western blot results of SPHK1 protein expression in SH-SY5Y and SK-N-SH cells with SPHK1 knockdown and in the negative control or blank control group. **P<0.01 vs negative control or blank control group.
Abbreviation: SPHK1, sphingosine kinase 1.
Knockdown of SPHK1 inhibits proliferation and induces apoptosis of NB cells
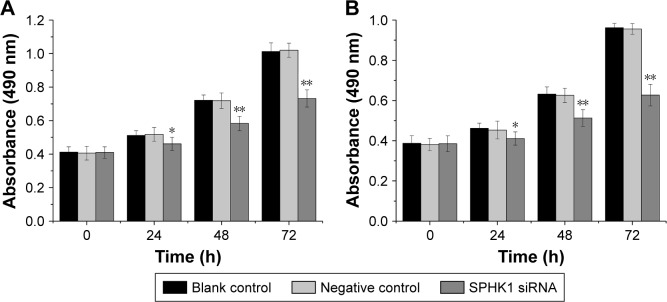
A CCK-8 assay was carried out to evaluate the results of siRNA-mediated SPHK1 downregulation on the antiproliferation effect of NB cells. As shown in Figure 3A and B, the SPHK1 knockdown in both SH-SY5Y and SK-N-SH cells resulted inhibition of cell growth in a time-dependent manner, that is, longer transfection time positively related with higher inhibition rate. In contrast, no evident inhibition was obtained in negative control or blank control. These results suggested that the proliferation of NB cells is significantly associated with SPHK1 and knockdown of SPHK1 significantly inhibits the progression of NB cells.
Figure 3.
Effect of SPHK1 knockdown by siRNA on the proliferation of SH-SY5Y and SK-N-SH cells. Cell proliferation was evaluated by MTT, and the results of cell growth are expressed as absorbance at 490 nm.
Notes: (A and B) The proliferation inhibition of SH-SY5Y and SK-N-SH cells with SPHK1 knockdown was observed compared with negative control and blank control at 24 hours, 48 hours, and 72 hours. *P<0.05, **P<0.01 vs negative control or blank control group.
Abbreviation: SPHK1, sphingosine kinase 1.
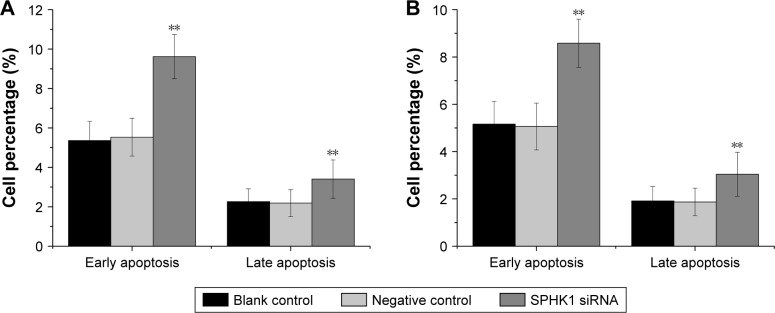
The influence of downregulation of SPHK1 expression on NB cell apoptosis was investigated by flow cytometry. As shown in Figure 4, silencing of SPHK1 dramatically increased both early and late apoptosis in SH-SY5Y (Figure 4A) and SK-N-SH (Figure 4B) cells as compared to other two groups. It was suggested that knockdown of SPHK1 promotes the apoptosis of NB cells.
Figure 4.
Effect of SPHK1 knockdown by siRNA on the apoptosis of SH-SY5Y and SK-N-SH cells.
Notes: The percentage of both early and late apoptosis was higher in SH-SY5Y (A) and SK-N-SH (B) cells with SPHK1 knockdown than that in negative control or blank control cells. **P<0.01 vs negative control or blank control group.
Abbreviation: SPHK1, sphingosine kinase 1.
Knockdown of SPHK1 inhibits both migration and invasion of NB cells
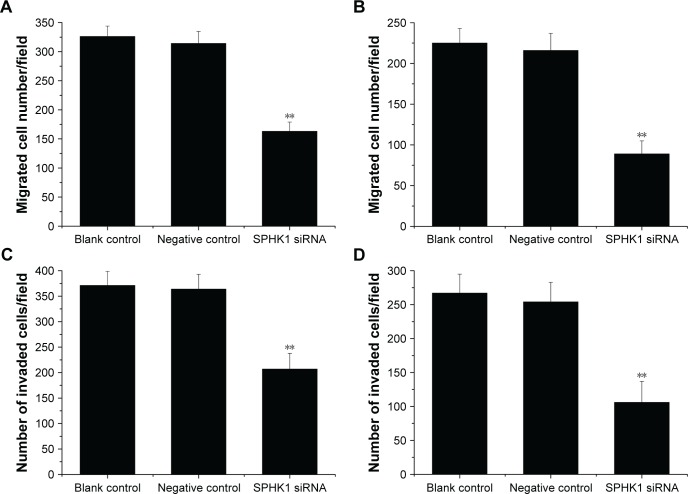
We further performed Transwell assays to investigate if knockdown of SPHK1 expression could influence the migration and invasion of NB cells. Compared with negative control or blank control, both SH-SY5Y (Figure 5A and C) and SK-N-SH (Figure 5B and D) cells with SPHK1 knockdown showed remarkably decreased migration (Figure 5A and B) and invasion (Figure 5C and D). These results clearly indicated that knockdown of SPHK1 inhibits both migration and invasion of NB cells.
Figure 5.
Effect of SPHK1 knockdown by siRNA on the migration and invasion of SH-SY5Y and SK-N-SH cells. Analysis of migration capability of SH-SY5Y (A) and SK-N-SH (B) cells. Analysis of invasion capability of SH-SY5Y (C) and SK-N-SH (D) cells.
Notes: Both the migrated and invaded number of SH-SY5Y and SK-N-SH cells with SPHK1 knockdown was significantly decreased compared with negative control or blank control. **P<0.01 vs negative control or blank control group.
Abbreviation: SPHK1, sphingosine kinase 1.
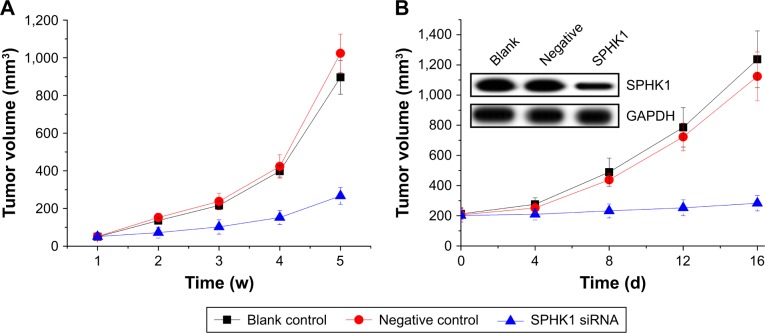
Knockdown of SPHK1 suppresses tumorigenesis in SH-SY5Y xenograft nude mice
To further investigate whether the in vivo tumorigenicity of SH-SY5Y cells could be regulated by SPHK1 silencing, different SH-SY5Y cells were subcutaneously inoculated into nude mice (n=10). As shown in Figure 6A, the tumors volume in the SPHK1 siRNA-expressing cells inoculated mice increased much slower than other groups. These results suggested that knockdown of SPHK1 inhibits SH-SY5Y xenograft tumorigenesis in vivo.
Figure 6.
The antitumor effect of SPHK1 silencing on NB in nude mice models.
Notes: (A) Tumor volume curve of SH-SY5Y cells with different treatments (SPHK1 siRNA, negative control, and untreated). There were significant differences between SPHK1 siRNA and negative control or blank control groups (n=10) (P<0.05) for each week from week 4 to week 5. (B) Tumor volume curve of SH-SY5Y tumor treated with saline (blank control), lentivirus with SPHK1 siRNA, or negative control. There were significant differences between SPHK1 siRNA and negative control or blank control group, (n=8) (P<0.05). Insets show Western blotting assay results regarding the SPHK1 expression in tumor tissues after treatment with different samples.
Abbreviations: NB, neiroblastoma; SPHK1, sphingosine kinase 1.
Therapeutic effect of knockdown of SPHK1 on tumor-bearing nude mice
An SH-SY5Y xenograft model was established in nude mice using untreated SH-SY5Y cells in order to evaluate the in vivo antitumor efficacy of lentivirus particles containing SPHK1 siRNA. As demonstrated in Figure 6B, mice treated with blank control gene and PBS showed almost no antitumor effects on NB tumors with rapidly increased tumor volume until the end of the test. In contrast, the tumor volume of mice treated with SPHK1 siRNA was significantly smaller, with preferable antitumor efficacy (Figure 7B). These observations indicated that SPK1 knockdown by SPHK1 targeting siRNA-loaded lentivirus results in a powerful in vivo antineoplastic activity on NB tumors.
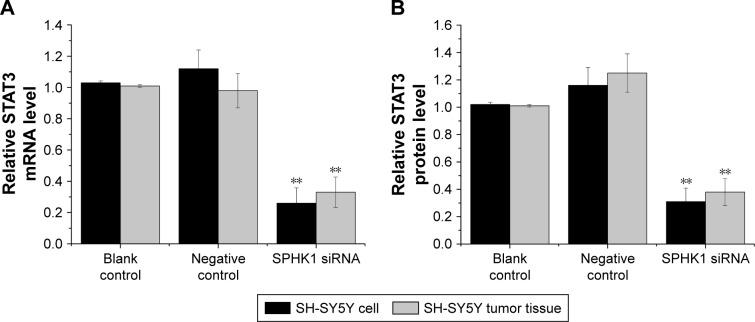
Figure 7.
Effect of SPHK1 knockdown by siRNA in terms of STAT3 expression in SH-SY5Y cells and tumor tissues of nude mice.
Notes: (A) STAT3 mRNA level was significantly reduced in SH-SY5Y cells and tumor tissues of nude mice with SPHK1 knockdown compared with the negative control or blank control. (B) STAT3 protein level was lower in SH-SY5Y cells and tumor tissues of nude mice with SPHK1 knockdown than that in the negative control or blank control group. **P<0.01 vs negative control or blank control groups.
Abbreviation: SPHK1, sphingosine kinase 1.
Inhibition of SPHK1 suppresses NB progression through downregulation of STAT3 expression
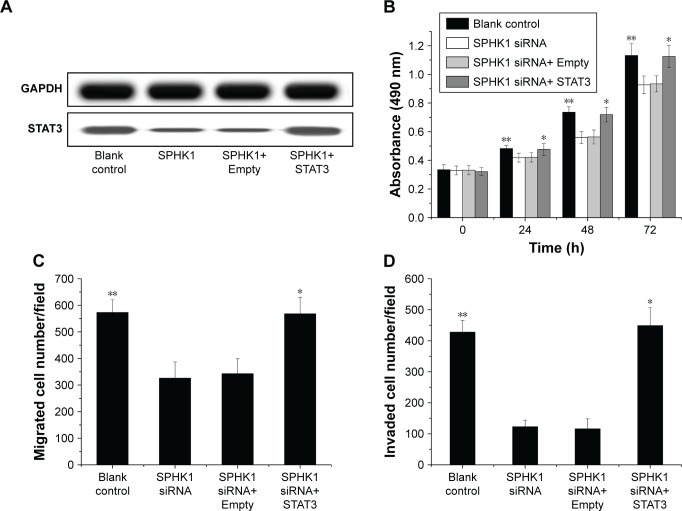
The underlying mechanisms responsible for the inhibition of NB progression caused by SPHK1 suppression was further investigated. STAT3, a 13-kDa small heparin-binding growth factor, is a factor in the downstream pathway of SPHK1 and has been shown to be involved in the development of multiple cancer-related processes.29,30 Moreover, it has been recognized that STAT3 overexpression is seen in many cancers including colorectal cancer,31 breast cancer,32 lung cancer,33 as well as NB.34 As a result, STAT3 was selected as a target protein for intensive study. Both the in vitro and in vivo mRNA and protein expression levels of STAT3 were first determined. As illustrated in Figure 7A and B, in SH-SY5Y cells (stably expressing SPHK1 siRNA) and tumor tissues from lentivirus (loaded with SPHK1 siRNA)-treated nude mice, reduced STAT3 expression was observed compared other control groups. With the aim to identify the role of STAT3 in suppression of NB progression via SPHK1 down regulation, we first transfected STAT3 expression vector (pcDNA3.1 (+)-STAT3) into SH-SY5Y cells (stably expressing SPHK1 siRNA), which helped us evaluate the STAT3 expression level in these cells to almost the same level of blank control (SPHK1 siRNA untreated cells) (Figure 8A). Interestingly, we found that ectopic overexpression of STAT3 could reverse the inhibition effect caused by SPHK1 siRNA expression in SH-SY5Y cells. The proliferation (Figure 8B), migration (Figure 8C), and invasion (Figure 8D) of SH-SY5Y cells (stably expressing SPHK1 siRNA) were restored to the same level of blank control, which suggested that the inhibition effect of SPHK1 downregulation on the progression of NB cells is, at least, in part, associated with reduced STAT3 expression.
Figure 8.
Ectopic overexpression of STAT3 rescued inhibition of SH-SY5Y cell proliferation, migration, and invasion by SPHK1 knockdown. SH-SY5Y cells with SPHK1 knockdown were transfected with 1 µg STAT3 expression vector pcDNA3.1 (+)-STAT3 (SPHK1 siRNA+ STAT3) or empty vector pcDNA3.1 (+) only (SPHK1 siRNA+ Empty).
Notes: (A) The protein expression of pcDNA3.1 (+)-STAT3 vector was confirm by Western blot. Ectopic overexpression of STAT3 rescued inhibition of SH-SY5Y cell proliferation (B), migration (C), and invasion (D) by SPHK1 knockdown. **P<0.01 vs SPHK1 siRNA or SPHK1 siRNA+ Empty group; *P>0.05 vs blank control group.
Abbreviation: SPHK1, sphingosine kinase 1.
Conclusion
In conclusion, our overall results suggested that lentivirus-delivered siRNA targeting SPHK1 could significantly suppress the progression of NB, which possibly act via the downregulation of SPHK1 and its downstream expression of STAT3. Our findings offer solid evidence that reveal the role of SPHK1 in the progression of NB. In addition, we also illuminate the potential mechanism responsible for the antitumor effects of SPHK1 suppression, which provides additional information for SPHK1 to be a novel target in effective gene therapy of NB.
Acknowledgments
The paper was supported by Higher Educational Science and Technology Program of Shandong Province, People’s Republic of China (Grant No J17KA232) and Medical and Health Science and Technology Development Project of Shandong Province, People’s Republic of China (Grant No 2017WS035).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL, Neuroblastoma CSL. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 3.Pediatric Treatment Editorial Board . Neuroblastoma Treatment (PDQ®) Bethesda, MD: National Cancer Institute; 2014. [Google Scholar]
- 4.French KJ, Schrecengost RS, Lee BD, et al. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 2003;63(18):5962–5969. [PubMed] [Google Scholar]
- 5.Pyne S, Adams DR, Pyne NJ. Sphingosine l-phosphate and sphingosine kinases in health and disease: Recent advances. Prog Lipid Res. 2016;62:93–106. doi: 10.1016/j.plipres.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Tsuchida J, Nagahashi M, Nakajima M, et al. Breast cancer sphingosine-l-phosphate is associated with phospho-sphingosine kinase 1 and lymphatic metastasis. J Surg Res. 2016;205(1):85–94. doi: 10.1016/j.jss.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, Jarman KE, Lokman NA, et al. CIB2 Negatively Regulates Oncogenic Signaling in Ovarian Cancer via Sphingosine Kinase 1. Cancer Res. 2017;77(18):4823–4834. doi: 10.1158/0008-5472.CAN-17-0025. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Wu H, Li W, et al. Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene. 2016;35(42):5501–5514. doi: 10.1038/onc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong H, Wang J, Guan H, et al. SphK1 confers resistance to apoptosis in gastric cancer cells by downregulating Bim via stimulating Akt/FoxO3a signaling. Oncol Rep. 2014;32(4):1369–1373. doi: 10.3892/or.2014.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Brouchet A, Pchejetski D, Brizuela L, et al. Critical role for sphingosine kinase-1 in regulating survival of neuroblastoma cells exposed to amyloid-beta peptide. Mol Pharmacol. 2007;72(2):341–349. doi: 10.1124/mol.106.033738. [DOI] [PubMed] [Google Scholar]
- 11.Puneet P, Yap CT, Wong L, et al. SphK1 regulates proinflammatory responses associated with endotoxin and polymicrobial sepsis. Science. 2010;328(5983):1290–1294. doi: 10.1126/science.1188635. [DOI] [PubMed] [Google Scholar]
- 12.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 13.Daka A, Peer D. RNAi-based nanomedicines for targeted personalized therapy. Adv Drug Deliv Rev. 2012;64(13):1508–1521. doi: 10.1016/j.addr.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Spänkuch B, Strebhardt K. RNA interference-based gene silencing in mice: the development of a novel therapeutical strategy. Curr Pharm Des. 2005;11(26):3405–3419. doi: 10.2174/138161205774370834. [DOI] [PubMed] [Google Scholar]
- 15.Aagaard L, Rossi JJ. Principles RNAi therapeutics: prospects and challenges. Advanced Drug Deliv Rev. 2007;59(2–3):75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34(6):634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Tang D, Zhao X, Yang T, Wang C. Paclitaxel prodrug based mixed micelles for tumor-targeted chemotherapy. RSC Adv. 2018;8(1):380–389. doi: 10.1039/d0ra90029j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Tang D, Yang T, Wang C. Facile preparation of biocompatible nanostructured lipid carrier with ultra-small size as a tumor-penetration delivery system. Colloids Surf B Biointerfaces. 2018;170:355–363. doi: 10.1016/j.colsurfb.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Bie CQ, Liu XY, Cao MR, et al. Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1 receptor inhibits the growth and invasion of hepatocellular carcinoma via down-regulating midkine expression. Oncotarget. 2016;7(48):79305–79318. doi: 10.18632/oncotarget.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Zhang CX, Ma Y, He HW, Wang JP, Shao RG. SphK1 inhibitor SKI II inhibits the proliferation of human hepatoma HepG2 cells via the Wnt5A/β-catenin signaling pathway. Life Sci. 2016;151:23–29. doi: 10.1016/j.lfs.2016.02.098. [DOI] [PubMed] [Google Scholar]
- 22.Xu CY, Liu SQ, Qin MB, et al. SphK1 modulates cell migration and EMT-related marker expression by regulating the expression of p-FAK in colorectal cancer cells. Int J Mol Med. 2017;39(5):1277–1284. doi: 10.3892/ijmm.2017.2921. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Liang Y, Chang W, Hu B, Zhang Y. Triple negative breast cancer depends on sphingosine kinase 1 (SphK1)/sphingosine-1-phosphate (S1P)/sphingosine 1-phosphate receptor 3 (S1PR3)/notch signaling for metastasis. Med Sci Monit. 2018;24:1912–1923. doi: 10.12659/MSM.905833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shida D, Takabe K, Kapitonov D, Milstien S, Spiegel S. Targeting SphK1 as a new strategy against cancer. Curr Drug Targets. 2008;9(8):662–673. doi: 10.2174/138945008785132402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Huang K, Lan T, et al. Curcumin ameliorates diabetic nephropathy by inhibiting the activation of the SphK1-S1P signaling pathway. Mol Cell Endocrinol. 2013;365(2):231–240. doi: 10.1016/j.mce.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 26.Rosa R, Marciano R, Malapelle U, et al. Sphingosine kinase 1 overexpression contributes to cetuximab resistance in human colorectal cancer models. Clin Cancer Res. 2013;19(1):138–147. doi: 10.1158/1078-0432.CCR-12-1050. [DOI] [PubMed] [Google Scholar]
- 27.Hu W, Huang J, Mahavadi S, Li F, Murthy KS. Lentiviral siRNA silencing of sphingosine-1-phosphate receptors S1P1 and S1P2 in smooth muscle. Biochem Biophys Res Commun. 2006;343(4):1038–1044. doi: 10.1016/j.bbrc.2006.03.079. [DOI] [PubMed] [Google Scholar]
- 28.Anderson J, Li MJ, Palmer B, et al. Safety and efficacy of a lentiviral vector containing three anti-HIV genes-CCR5 ribozyme, tat-rev siRNA, and TAR decoy-in SCID-hu mouse-derived T cells. Mol Ther. 2007;15(6):1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- 29.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 30.Bao Y, Li K, Guo Y, et al. Tumor suppressor PRSS8 targets Sphk1/S1P/Stat3/Akt signaling in colorectal cancer. Oncotarget. 2016;7(18):26780–26792. doi: 10.18632/oncotarget.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusaba T, Nakayama T, Yamazumi K, et al. Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol Rep. 2006;15(6):1445. [PubMed] [Google Scholar]
- 32.Zhou J, Wulfkuhle J, Zhang H, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104(41):16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seki Y, Suzuki N, Imaizumi M, et al. STAT3 and MAPK in human lung cancer tissues and suppression of oncogenic growth by JAB and dominant negative STAT3. Int J Oncol. 2004;24(4):931–934. [PubMed] [Google Scholar]
- 34.Yan S, Li Z, Thiele CJ. Inhibition of STAT3 with orally active JAK inhibitor, AZD1480, decreases tumor growth in Neuroblastoma and Pediatric Sarcomas In vitro and In vivo. Oncotarget. 2013;4(3):433–445. doi: 10.18632/oncotarget.930. [DOI] [PMC free article] [PubMed] [Google Scholar]