Abstract
Oral squamous cell carcinoma (OSCC) is an extremely aggressive disease associated with a poor prognosis. Previous studies have established that cancer stem cells (CSCs) actively participate in OSCC development, progression and resistance to conventional treatments. Furthermore, CSCs frequently exhibit a deregulated expression of normal stem cell signalling pathways, thereby acquiring their distinctive abilities, of which self-renewal is an example. In this study, we examined the effects of GLI3 knockdown in OSCC, as well as the differentially expressed genes in CSC-like cells (CSCLCs) expressing high (CD44high) or low (CD44low) levels of CD44. The prognostic value of GLI3 in OSCC was also evaluated. The OSCC cell lines were sorted based on CD44 expression; gene expression was evaluated using a PCR array. Following this, we examined the effects of GLI3 knockdown on CD44 and ESA expression, colony and sphere formation capability, stem-related gene expression, proliferation and invasion. The overexpression of genes related to the Notch, transforming growth factor (TGF)β, FGF, Hedgehog, Wnt and pluripotency maintenance pathways was observed in the CD44high cells. GLI3 knockdown was associated with a significant decrease in different CSCLC fractions, spheres and colonies in addition to the downregulation of the CD44, Octamer-binding transcription factor 4 (OCT4; also known as POU5F1) and BMI1 genes. This downregulation was accompanied by an increase in the expression of the Involucrin (IVL) and S100A9 genes. Cellular proliferation and invasion were inhibited following GLI3 knockdown. In OSCC samples, a high GLI3 expression was associated with tumour size but not with prognosis. On the whole, the findings of this study demonstrate for the first time, at least to the best of our knowledge, that GLI3 contributes to OSCC stemness and malignant behaviour. These findings suggest the potential for the development of novel therapies, either in isolation or in combination with other drugs, based on CSCs in OSCC.
Keywords: oral squamous cell carcinoma, cancer stem cell-like cells, stem cell signaling pathways, CD44, GLI3
Introduction
Head and neck cancer is the seventh most common type of cancer worldwide, and oral squamous cell carcinoma (OSCC) accounts for >90% of these cases (1,2). OSCC is usually aggressive and highly invasive and is associated with a high recurrence rate, as well as with metastasis of the lymph nodes (3). As a result, the vast majority of patients with OSCC have a poor prognosis, and, despite advances in the understanding of the molecular and genetic mechanisms driving OSCC malignancy, the 5-year-survival rate has not shown any significant improvement (3,4).
Previous studies have demonstrated that OSCC, similar to other solid malignant tumours, has a small subpopulation of cells designated as cancer stem cells (CSCs). These cells are characterised by the ability to self-renew indefinitely, in addition to giving rise to transient and differentiated cells which comprise the bulk of the tumour (5-7). CSCs are therefore associated with recurrence and therapeutic resistance, and participate in OSCC metastasis due to their capability to undergo epithelial to mesenchymal transition (EMT) (8-10).
CSCs in OSCC were first identified and isolated by Prince et al based on their high expression levels of CD44, a cell surface glycoprotein that acts as a receptor for hyaluronic acid (5). Upon binding to its ligand, CD44 can activate different signalling pathways which regulate a wide variety of cellular processes, including adhesion, proliferation, motility, apoptosis, survival and resistance to therapy (11). Subsequently, additional CSC markers were identified and used alone or in combination with CD44, including CD133 (12), epidermal growth factor receptor (EGFR) (13), ESA (14), CD24 (15) and aldehyde dehydrogenase 1 (ALDH1) (16). Most importantly, recent studies on CSC plasticity have demonstrated that this subpopulation exists in more than one phenotype; the association of CD44 with different markers has permitted the identification of distinct subtypes of CSCs. Biddle et al (2011) demonstrated that cells expressing high levels of CD44 (CD44high) cells can be separated, based on epithelial-cell adhesion molecule (EpCAM)/ESA levels, into two cellular phenotypes. These phenotypes present significant differences in proliferation rates, cell motility and morphology in addition to colony- and sphere-forming ability (14). CD44high/ESAhigh cells exhibit an epithelial morphology and an increased proliferative ability, while CD44high/ESAlow cells are migratory and undergo EMT.
Signalling pathways that control stem cell self-renewal and differentiation are aberrantly activated in CSCs and include the Notch, Sonic Hedgehog (SHH) and Wnt pathways. All these pathways frequently interact with other cellular signalling pathways closely related to tumour development and progression, such as nuclear factor (NF)-κB, mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K) and epidermal growth factor (EGF) (17). Thus, the identification of the crucial pathways necessary for CSC maintenance represents an important therapeutic target with may be used to block CSC proliferation and self-renewal and, consequently, tumour progression.
In this context, the SHH/Patched/Gli (SHH/PTCH/GLI) pathway, involved in the patterning, growth, differentiation and survival of normal stem cells also plays an important role in CSCs; it provides proliferative cues that enable the cells to accumulate oncogenic mutations that drive self-renewal, metastasis and therapeutic resistance (17,18). This signalling pathway initiates with the binding of Hedgehog proteins (Sonic, Desert and Indian HH) to the transmembrane receptor, PTCH. This receptor, in the absence of the Hedgehog ligands, inhibits signal transduction by repressing the Smoothened (SMO) transmembrane receptor (18,19), which acts as a potent pathway activator. Following HH binding, PTCH is internalised and degraded, thus allowing SMO to become phosphorylated and activated (19); this in turn triggers an intracellular signalling cascade that promotes the recruitment and activation of GLI family transcription factors (20,21).
There are three GLI proteins in mammalian cells that act in a specific manner to regulate tissue patterning, cell proliferation and survival via positive and negative feedback mechanisms depending on the context and cell-type (22,23). GLI proteins can act as activators or repressors, depending on the ratio of said proteins (24). GLI1 is a transcriptional activator. GLI2 and GLI3 genes function as either positive or negative regulators according to their post-transcriptional and post-translational modifications, e.g., via phosphorylation or acetylation (25,26).
In the absence of a Shh ligand, GLI3 is cleaved from its larger activated cytoplasmic form to a truncated repressor nuclear form, which inhibits the signalling pathway (27). In adult haematopoiesis, a progressive decrease in the Shh pathway is associated with increased hematopoietic cellular fate and the transition from an embryonic to a hematopoietic stem cell. In this context, GLI3 plays a crucial role in mediating Shh pathway inhibition (28).
In cancer, SHH deregulated activation was first described in nevoid basal cell carcinoma syndrome, where the inherited loss-of-function mutations of the PTCH1 gene was associated with tumour development (29,30). Abnormal Shh signalling is now associated with the progression and maintenance of several malignant tumours, e.g., glioblastomas, lung cancer, prostate cancer and gastric cancer (24,31-34). Additionally, it can participate in tumour development via somatic mutations in upstream pathway proteins (SMO and PTC1), overexpression of GLI transcription factors or in a ligand-dependent manner (25).
In different cell types, HH/GLI activation leads to the transcription of genes critical to tumour initiation and maintenance. This pathway is associated with an increase in the quantity of cell cycle proteins which are responsible for G1/S and G2/M progression, mainly D-type cyclins and anti-apoptotic proteins (35-37). Additionally, it participates in the regulation of EMT by inhibiting E-cadherin and inducing N-cadherin expression (37,38). Furthermore, GLI2 overexpression is associated with a decrease in E-cadherin expression and an increase in SNAIL gene expression, as well as with an increase in matrix metalloproteinase (MMP)2 and integrin-beta-1-binding proteins (ICAP-1); all of which favour cell invasion and metastasis (39).
Some studies have demonstrated that the Shh signalling pathway is upregulated in CSCs. The activation of these pathways in breast CSCs increases GLI expression and leads to an enhanced SOX2 and OCT4 expression, favouring CSC maintenance (40). In gastric cancer cell lines and tissues, CSCs identified by the CD44+CD24+ phenotype have demonstrated an overexpression of the SHH, PTCH1 and GLI3 genes (41). Zhang et al demonstrated that in liver cancer, CD90+ CSCs with GLI1 or GLI3 knockdown, exhibited low proliferation rates, and decreased migratory ability and sphere formation capacity, as well as a decrease in tumour formation in vivo; indicating that both genes are relevant to the maintenance of the stem cell properties observed in CD90+ cancer cells (42).
In oral cancer, the increased expression of GLI1, PTC1, SMO and SHH has been observed in OSCC samples when compared to non-tumour oral mucosa (43,44). Moreover, both SHH and GLI1 have been shown to be associated with lymph node metastasis, but only GLI1 expression has been shown to be associated with tumour recurrence, clinical stage and lower 5-year survival rates (39). Schneider et al also observed the overexpression of all Hh signalling pathway proteins in OSCC and found that a high Shh expression was also associated with a poor overall survival (45). Concordant with this, an in vitro study demonstrated that the inhibition of SHH was associated with a significant decrease in tumour growth, as well as in angiogenesis and osteoclastic activity in a mouse model of OSCC bone invasion (46). GLI1 or GLI2 loss of function increases apoptosis and DNA fragmentation and also promotes keratin 17 upregulation, which in turn promotes cell growth (47).
The current study investigated, in vitro, the differential expression of genes involved in stem cell signalling pathways in OSCC CSC-like cells (CSCLCs), as well as the effects of GLI3 knockdown on cellular proliferation, invasion and stemness. Furthermore, GLI3 protein expression was also evaluated in OSCC and non-tumour tissues and its association with the patient clinicopathological parameters and overall survival was examined. To the best of our knowledge, this is the first study to investigate the role of GLI3 in OSCC.
Materials and methods
Cell lines
SCC4 (CRL-1624™) and SCC9 (CRL-1629™) human tongue squamous cell carcinoma cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultivated in DMEM/F12 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and supplemented with 10% fetal bovine serum (FBS), 400 ng/ml hydrocortisone (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 100 μg/ml penicillin and 100 μg/ml streptomycin with 5% CO2 at 37°C.
Flow cytometry
Fluorescent-activated cell sorting (FACS) was performed using a FACSAria II flow cytometer (BD Biosciences, San Jose, CA, USA) and analysed using FACSDiva software (Diva version 6.1.1). Initially, the SCC4 and SCC9 cells were detached from the cultures using Accutase™ cell detachment solution (SCR005; MilliporeSigma, Burlington, MA, USA) at 37°C and stained with anti-CD44 antibody (anti-CD44-FITC, clone G44-26, 1:100, 555478; BD Biosciences) for 30 min in the dark. DAPI nuclear dye (Sigma-Aldrich; Merck KGaA) was used at 200 ng/ml to exclude dead cells and IgG2ak-FITC was used as an isotype control. The whole population was fractioned into CD44high (CSCLC), representing the top 5% of CD44-expressing cells, and CD44low, corresponding to the bottom 5% of differentiating cells. FACS-sorted CD44high and CD44low subpopulations were then submitted to colony and sphere formation, as well as gene expression assays for stemness-related signalling pathways (PCR array) and markers (qPCR).
Subsequently, the cells were co-stained with anti-CD44 and anti-ESA antibodies (anti-ESA-PE, clone EBA-1, 1:100, 347198; BD Biosciences), for 30 min at room temperature, washed with PBS, incubated with DAPI and analysed using a FACSCanto II Flow cytometer (Diva software version 6.1.1; BD Biosciences) to evaluate the CSCLC fractions representing both EMT-CSCLC (CD44high/ESAlow) and non-EMT-CSCLC (CD44high/ESAhigh).
Colony and sphere formation assays
To examine the clonogenic abilities of the SCC4 and SCC9 subfractions (CD44high and CD44low), the cells were plated at clonal density (100 cells/ml) in each well of 6-well plates and cultured for 14 days. Following fixation in 4% paraformaldehyde, the cells were stained for 5 min at room temperature with crystal violet (0.04% in 1% ethanol) and colonies measuring at least 2 mm in diameter were counted visually.
To assess the capacity of the SCC4 and SCC9 subfractions for growth as tumour spheres in suspension, 1×104 cells/ml were seeded in 24-well ultra-low attachment plates per well (#3473; Corning, New York, NY, USA). After 2 weeks, the number of tumour spheres >5 μm was counted under a microscope at ×200 magnification (Carl Zeiss Micro-Imaging GmbH, Jena, Germany).
PCR array
To assess the expression of genes related to different stem cell signalling pathways in the CD44high and CD44low cells from the SCC4 and SCC9 cells, we used the commercially available Human Stem Cell Signalling RT2 Profiler PCR Array (330231; Qiagen, Germantown, MD, USA). Immediately after FACS, sorted cells were submitted to RNA extraction using the RNeasy Mini kit following the manufacturer's instructions (74104; Qiagen). cDNA was synthesized using 500 ng of RNA with the aid of the RT2 First Strand kit (330401; Qiagen). A list of genes on this PCR array is available at https://www.qiagen.com/us/shop/pcr/primer-sets/rt2-profiler-pcr-arrays/?catno=PAHS-047Z#orderinginformation. The relative expression level of the target gene in the CD44high cells to that in the CD44low cells was evaluated using the 2−ΔΔCq method (48) using the software RT2 Profiler PCR Array Data Analysis version 3.5 (Qiagen). This software classifies the average threshold cycle of the gene as follows: 'A' [relatively high (>30) in either the control or the test sample, and is reasonably low in the other sample (<30)]; 'B' [relatively high (>30), meaning that its relative expression level is low, in both the control and test samples, and the P-value for the fold change is either unavailable or relatively high (P>0.05)]; 'C' [either not determined or greater than the defined cut-off value (default 35), in both samples meaning that its expression was undetected, making this fold change result erroneous and uninterpretable] and 'OKAY'. Only genes classified as 'A' or 'OKAY' by the software and with a fold change >2 were considered as overexpressed.
GLI3 knockdown
A total of 3×105 SCC9 cells were transfected with 4 shRNA-GLI3 sequences (Origene TG301348) or shRNA-scrambled (Origene, Rockville, MD, USA) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. This cell line was selected as we have previously observed that the SCC9 cell line, as well as the CD44high and CD44low fractions are able to form tumours in vivo (49). Transfected cells were selected in a medium with 0.5 μg/ml of puromycin (Invitrogen; Thermo Fisher Scientific, Inc.) for 21 days. The transfection efficiency of the shRNA-GLI3 sequences was assessed by reverse transcription-quantitative PCR (RT-qPCR) and western blot analysis for GLI3. The sequence with the highest inhibition level of GLI3 protein was selected for further experiments. The SCC9 cells transfected with shRNA-scrambled (control) and shRNA-GLI3 were cultured further and subjected to colony and sphere formation, flow cytometry, proliferation, invasion and apoptosis assays, as well as to RT-qPCR for stemness-related genes. According to the Constraint-based Multiple Alignment Tool (NCBI) there is 51% homology between the GLI3 (NM_000168.5) and GLI2 (NM_005270.4) genes, 32% homology between the GLI3 and GLI1 (NM_001160045.1) genes and 39% homology between the GLI and GLI2 genes. OriGene guarantees that the sequences in the shRNA expression cassettes are verified to correspond to the target gene with 100% identity.
RNA extraction, cDNA synthesis and RT-qPCR
RNA was extracted using the RNeasy® micro kit (Qiagen) followed by the reverse transcription of 500 ng of total RNA with the High Capacity cDNA Archive kit, according to the manufacturer's instructions. qPCR was performed on an ABI 7500 real-time PCR system using SYBR®-Green mix (all from Applied Biosystems, Foster City, CA, USA) and primers for CD44, BMI1, POU5F1 (OCT4), CD133, NANOG, Involucrin (IVL), S100A9 (Calgranulin B), SNAI2 (SLUG) and GAPDH gene amplification, which was used for normalization of the target gene. The qPCR cycling conditions were as follows: 95°C for 10 min, (95°C for 15 sec, 60–62°C for 60 sec) (40 cycles), followed by dissociation curve analysis. Positive samples used to generate a standard curve included SCC9 (for CD44, BMI1 and POU5F1 genes) and embryonic stem cells (for CD133 and NANOG genes). Primer sequences were designed using the GeneTool software (BioTools Inc., Edmonton, AB, Canada) and are described in detail in Table I. Gene expression was calculated by the 2−ΔΔCq method (48) and the fold change values were calculated by the ratio CD44high/CD44low cells, as well as by the ratio SCC9-shRNA GLI3/SCC9 Control for each gene.
Table I.
Genes evaluated by RT-qPCR, according to GenBank access, primer sequence, melting temperature and product size.
| Gene | GenBank | Primers 5′→3′ | Temperature (°C) | Product size (bp) |
|---|---|---|---|---|
| POU5F1 | NM_001173531.1 | F: acttcactgcactgtactcctc | 62 | 159 |
| R: aggttctctttccctagctcctc | ||||
| NANOG | NM_024865.2 | F: catcctgaacctcagctacaaaca | 62 | 99 |
| R: ttgctattcttcggccagttgt | ||||
| CD133 | NM_001145848 | F: ggcccagtacaacactacc | 62 | 71 |
| R: cgcctcctagcactgaatt | ||||
| CD44 | NM_000610.3 | F: caaccgttggaaacataacc | 60 | 226 |
| R: caagtgggaactggaacgat | ||||
| BMI1 | NM_005180.8 | F: gctgccaatggctctaatgaa | 60 | 189 |
| R: tgctgggcatcgtaagtatctt | ||||
| SNAI1 | NM_003068.4 | F: caaggaatacctcagcctgg | ||
| R: catctgagtgggtctggagg | 60 | 218 | ||
| S100A9 | NM_002965.3 | F: aaagagctggtgcgaaaaga | ||
| R: gtgtccaggtcctccatgat | 60 | 92 | ||
| IVL | NM_005547.3 | F: caagacattcaaccagccct | ||
| R: tagcggacccgaaataagtg | 60 | 185 | ||
| GAPDH | NM_001256799 | F: gcatcctgggctacactga | ||
| R: ccaccaccctgttgctgta | 60 | 162 |
Western blot analysis
The cells were washed with cold PBS and lysed in RIPA buffer (R0278; Sigma-Aldrich; Merck KGaA). Following centrifugation at 20,800 x g at 4°C, protein concentrations were measured using a protein assay according to the manufacturer's instructions (BCA Protein kit; Pierce Biotechnology, Rockford, IL, USA). A total of 30 μg of total protein per sample were resolved by 8 or 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and transferred onto nitrocellulose membranes. The membranes were blocked for 1 h with 10% non-fat dry milk in TBS (20 mM Tris-HCl, 150 mM NaCl) containing 0.1% Tween-20, rinsed in the same buffer, and incubated overnight with anti-GLI3 (1:500, ab6050; Abcam, Cambridge, MA, USA) and anti-β-actin (1:10,000, A2228; Sigma-Aldrich; Merck KGaA) antibodies. After washing, the membranes were incubated for 1 h at room temperature with anti-rabbit IgG-HRP (1:1,000, sc-2357) for GLI3 and with the anti-mouse m-IgGκ BP-HRP (1:1,000, sc-516102) (both from Santa Cruz Biotechnology, Dallas, TX, USA) for β-actin. The reactions were revealed using a chemiluminescent western blot system (Enhanced Chemiluminescent Western blot kit; GE Healthcare, Vienna, Austria).
Cell proliferation assays
The SCC9 control cells and SCC9 cells in which GLI3 was knocked down (1×103) were initially plated in 24 well-plates and counted daily in a Neubauer chamber (Celeromics, Cambridge, UK) for 5 days (24, 48, 72, 96 and 120 h). Independent experiments were performed in triplicate and the mean value, standard deviation and statistical analyses were calculated. The bromodeoxyuridine (BrdU) labelling index was performed as previously described by Rodrigues et al (50). In summary, the SCC9 control cells and SCC9 in which GLI3 was knocked down were plated on chamber slides and serum-starved for 48 h. After this period, cell culture medium with 10% FBS was added and the cells were cultivated for an additional 24 h. The cells were then incubated with BrdU (1:100) for 2 h at 37°C and the reactions were revealed using the BrdU Staining kit (both from Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The BrdU-positive cells were determined by counting 1,000 cells in 3 independent reactions using the Kontron 400 image analysis system (Zeiss Axio Imager A1; Zeiss, Dublin, CA, USA).
Cell invasion assay
To analyse cell invasion, cell culture inserts with an 8 -μm PET membrane coated with Matrigel (#354480, Corning® BioCoat™ Matrigel® Invasion Chambers; Corning) were used. The cells (1×105) were suspended in serum-free DMEM/F12 and seeded onto the upper compartment of the Transwell chamber. DMEM/F12 containing 5% FBS was used in the lower chamber as a chemoattractant. Following 24 h of incubation, cells in the upper chamber were removed and the inserts were fixed in 4% paraformaldehyde followed by 20 min in methanol 100%. The cells on the lower surface were stained for 5 min at room temperature with crystal violet (0.04% in 1% ethanol; Sigma-Aldrich; Merck KGaA). A cotton swab was used to mechanically removed cells that did not migrate through the pores. Five random areas from each membrane were photographed under ×400 magnification using a light microscope (Axioplan 2; Zeiss, Oberkochen, Germany) and the mean number of cells was calculated.
Cell apoptosis assay
The SCC9 control and SCC9 shRNA-GLI3 were triple-stained for 30 min at room temperature with Annexin V-FITC (Invitrogen; Thermo Fisher Scientific, Inc.), DAPI, or anti-CD44-APC (BD Pharmingen). Samples were examined on a FACSCanto II Flow cytometer and analysed with the FACSDiva version 6.1.1 (both from BD Biosciences) software.
Immunohistochemistry
Forty-five tongue OSCC samples were obtained by surgical resection from patients (male, ≥40 years of age) admitted for diagnosis and treatment at the Hospital of the University of São Paulo, School of Medicine (Brazil) from January, 2011 to November, 2014. Samples and clinical data collection, and histopathological analysis were performed with the informed consent of each patient by the GENCAPO (Head and Neck Genome Project) Consortium. OSCC diagnosis was performed following the WHO Classification of Tumors. Clinicopathological TNM staging was established according to the classification determined by the Union for International Cancer Control (UICC) (51). This study was approved by the Brazilian National Ethics Committee (Process #16491) and meets the requirements of the Declaration of Helsinki.
Immunohistochemical analysis of GLI3 was performed in 45 OSCC samples and in 10 non-tumour margins. Tissue sections (4-μm-thick) were cut and, following dewaxing and hydration in graded alcohol solutions, the sections were incubated with 3% H2O2 for 45 min. After washing in PBS, antigen recovery was performed by treatment with a 100 mM citrate buffer target retrieval solution, pH 6.0 at 95°C, in a water bath for 20 min. The sections were incubated with protein block (X0909; Dako, Carpenteria, CA, USA) for 10 min followed by overnight incubation with monoclonal anti-GLI3 antibody (ab6050; Abcam) diluted at 1:300 in Antibody Diluent with Background Reducing components (S3022; Dako). The sections were then incubated with the Envision+Dual Link visualization system (K4061) and 3,3′-diaminobenzidine tetrahydrochloride (DAB, K3468) (both from Dako) was used as a chromogen. The negative control consisted of the omission of the primary antibody. A human kidney was used as the positive control. Immunostaining evaluations were performed by two independent pathologists who had no knowledge of the clinicopathological parameters. GLI3 immunoexpression was scored as 0 (<5%), 1 (5-30%), 2 (30-60%) and 3 (60-100%), as previously described by Schneider et al (52). For the final analyses, scores were dichotomised according to protein expression as low expression (scores 0 and 1) and high expression (scores of 2 and 3). The immunoexpression pattern was determined independently by two investigators (M. F. S. D. R. and L. M.) and its association with clinicopathological parameters such as mean age, tumour location, tumour size-pT, nodal metastasis-pN, lymphatic, vascular or perineural invasion, as well as overall survival was examined.
Statistical analysis
All assays were independently repeated at least 3 times and the significance of the differences was calculated using unpaired t-tests with the Welch correction. A value of P<0.05 was considered to indicate a statistically significant difference. Error bars represent the standard error of the mean (SEM). Fisher's exact test was used to estimate the statistically significant differences between protein expression and the clinicopathological parameters. The Kaplan-Meier product-limit estimation with the log-rank test (P<0.05) was used for survival analysis from lifetime data according to GLI3 protein expression (high vs. low). Overall survival was defined as the time from surgery to the day of death or the last follow-up. Statistical package GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA) was used for statistical analysis and P-value <0.05 were considered to indicate statistically significant differences.
Results
Characterization of the SCC4 and SCC9 CD44high and CD44low cell fractions
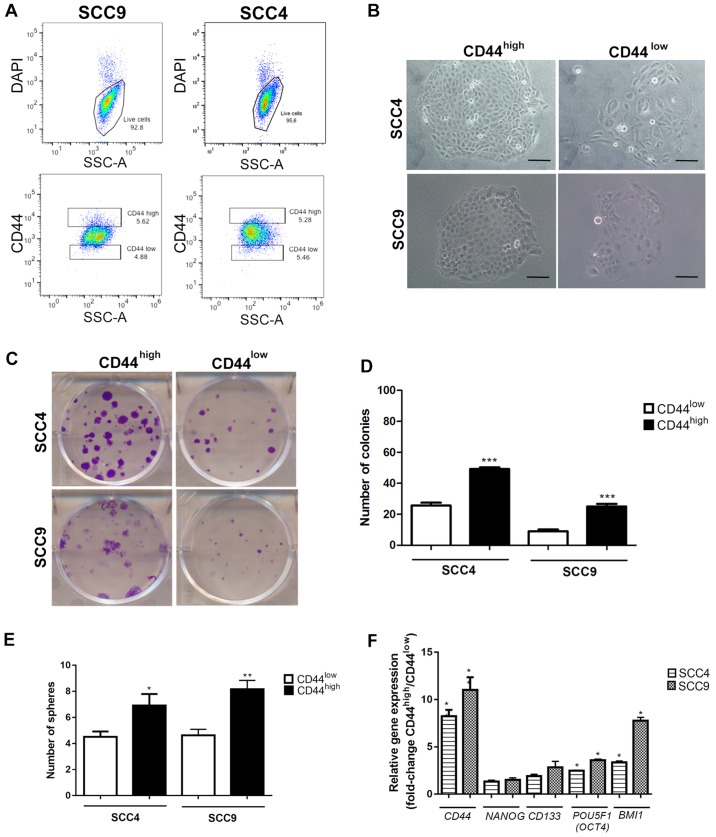
The FACS-sorted CD44high and CD44low fractions from SCC4 and SCC9 (Fig. 1A) were immediately plated for clonogenic (Fig. 1B–D) and sphere formation assays (Fig. 1E) in order to investigate the CSC properties. As expected, the CD44high cells exhibited a significantly higher number of colonies with holoclone morphology (P<0.001, Fig. 1C and D) and spheres (Fig. 1E) in relation to the CD44low cells from the both SCC4 and SCC9 cell lines. Additionally, gene expression evaluation revealed increased mRNA expression levels of CD44, POU5F1 (OCT4) and BMI1 (P<0.05) in the CD44high cells in relation to the CD44low cells (Fig. 1F).
Figure 1.
Analysis of cancer stem cell (CSC) properties in SCC4 and SCC9 cell lines. (A) Representative images of the gates used to sort the CD44high and CD44low fractions. (B) Cell morphology of CD44high and CD44low fractions, demonstrating the presence of holoclones in CD44high cells. (C) Colony formation ability of CD44high and CD44low fractions. Graphs representing the (D) number of colonies and (E) number of spheres in CD44high and CD44low cells. (F) RT-qPCR for CD44, NANOG, CD133, POU5F1 (OCT4) and BMI1 transcripts. Fold change values represent the relative gene expression in CD44high cells in relation to CD44low cells. *P<0.05, **P<0.01 and ***P<0.001 (t-tests with the Welch correction). Data for SCC9 cells was gathered as previously described by de Andrade et al (51). Microscope images: Magnification, ×100 (scale bars, 40 μm).
Expression of genes related to stem cell signalling pathways in SCC4 and SCC9 CD44high and CD44low cell fractions
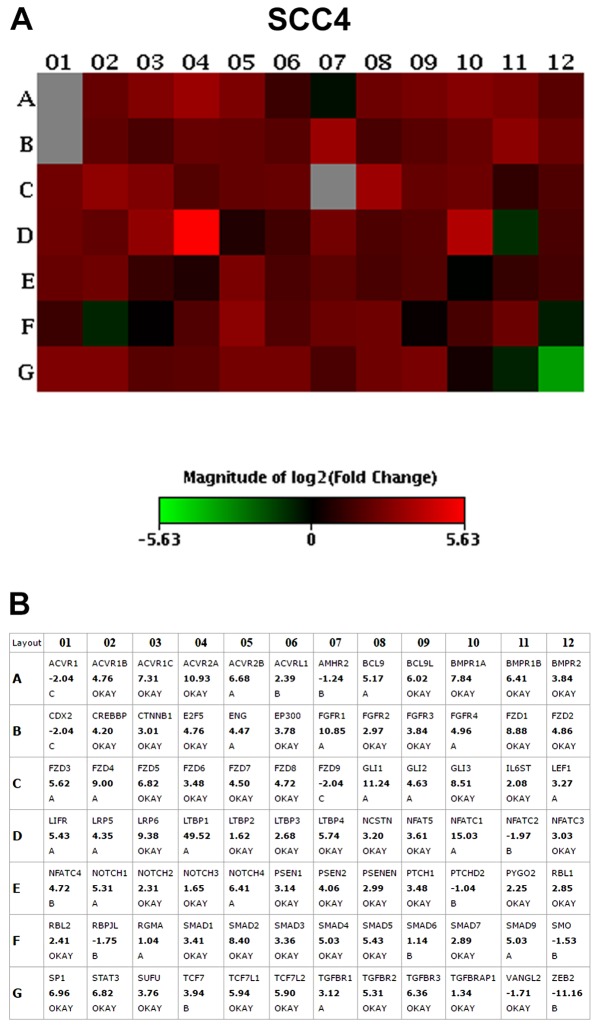
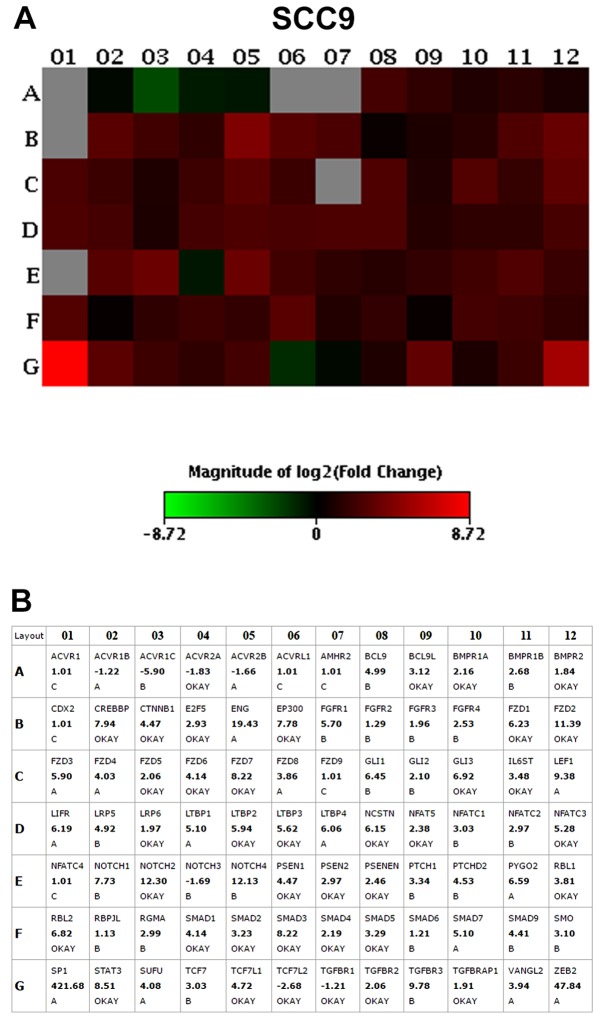
The results of the PCR array assay revealed that the vast majority of the genes were overexpressed in the CD44high cells when compared to the CD44low cells, including genes related to the Notch, transforming growth factor (TGF)β, fibroblast growth factor (FGF), Hedgehog, Wnt and pluripotency maintenance pathways. The top 10 upregulated genes with the highest fold change (>2.0) in CD44high in relation to the CD44low fractions, in both the SCC4 and SCC9 cell lines, are presented in Table II and Figs. 2 and 3.
Table II.
List of the top 10 differentially expressed genes (fold change >2.0) in CD44high sorted cells in relation to CD44low in SCC4 and SCC9 cell lines.
| Transcript | Gene description | Signalling pathway | Fold change |
|---|---|---|---|
| SCC4 cells | |||
| ACVR2A | Activin A receptor type 2A | TGFβ | 10.93 |
| LRP6 | LDL receptor related protein 6 | Wnt | 9.38 |
| FZD1 | Frizzled class receptor 1 | Wnt | 8.88 |
| GLI3 | GLI family zinc finger 3 | HH | 8.51 |
| SMAD2 | SMAD family member 2 | TGFβ | 8.40 |
| BMPR1A | Bone morphogenetic protein receptor type 1A | TGFβ | 7.84 |
| ACVR1C | Activin A receptor type 1C | TGFβ | 7.31 |
| SP1 | Sp1 transcription factor | TGFβ | 6.96 |
| FZD5 | Frizzled class receptor 5 | Wnt | 6.82 |
| STAT3 | Signal transducer and activator of transcription 3 | Pluripotency | 6.82 |
| SCC9 cells | |||
| NOTCH2 | Notch2 | NOTCH | 12.30 |
| FZD2 | Frizzled class receptor 2 | Wnt | 11.39 |
| STAT3 | Signal transducer and activator of transcription 3 | Pluripotency | 8.51 |
| FZD7 | Frizzled class receptor 7 | Wnt | 8.22 |
| SMAD3 | SMAD family member 3 | TGFβ | 8.22 |
| CREBBP | CREB binding protein | TGFβ | 7.94 |
| EP300 | E1A binding protein p300 | TGFβ | 7.78 |
| GLI3 | GLI family zinc finger 3 | HH | 6.92 |
| RBL2 | RB transcriptional corepressor like 2 | TGFβ | 6.82 |
| FZD1 | Frizzled class receptor 1 | Wnt | 6.23 |
Transcripts in bold were the most upregulated genes in CD44high cell in relation to the CD44low cell fraction.
Figure 2.
(A) PCR array heatmap of the differentially expressed genes in SCC4 CD44high in relation to CD44low cells. (B) Fold change values for each gene evaluated. Only genes with a fold change >2.0 and classified as 'A' and 'OKAY' by the PCR array software were considered overexpressed. Data represents the fold change values of 3 independent experiments.
Figure 3.
(A) PCR array heatmap of the differentially expressed genes in SCC9 CD44high in relation to CD44low cells. (B) Fold change values for each gene evaluated. Only genes with a fold change >2.0 and classified as 'A' and 'OKAY' by the PCR array software were considered overexpressed. Data represents the fold change values of 3 independent experiments.
The Frizzled-1 (FZD1; Wnt pathway), signal transducer and activator of transcription 3 (STAT3; pluripotency pathways) and GLI3 (Shh pathway) genes were >6-fold upregulated in the SCC4 CD44high and SCC9 CD44high cells in relation to the CD44low cells. Thus, we then selected to knockout the GLI3 gene in order to investigate the role of this transcription factor in stemness, as well as in cell proliferation, invasion and apoptosis.
GLI3 knockdown decreases clonogenicity, as well as the epithelial and mesenchymal CSCLC fractions in the SCC9 cell line
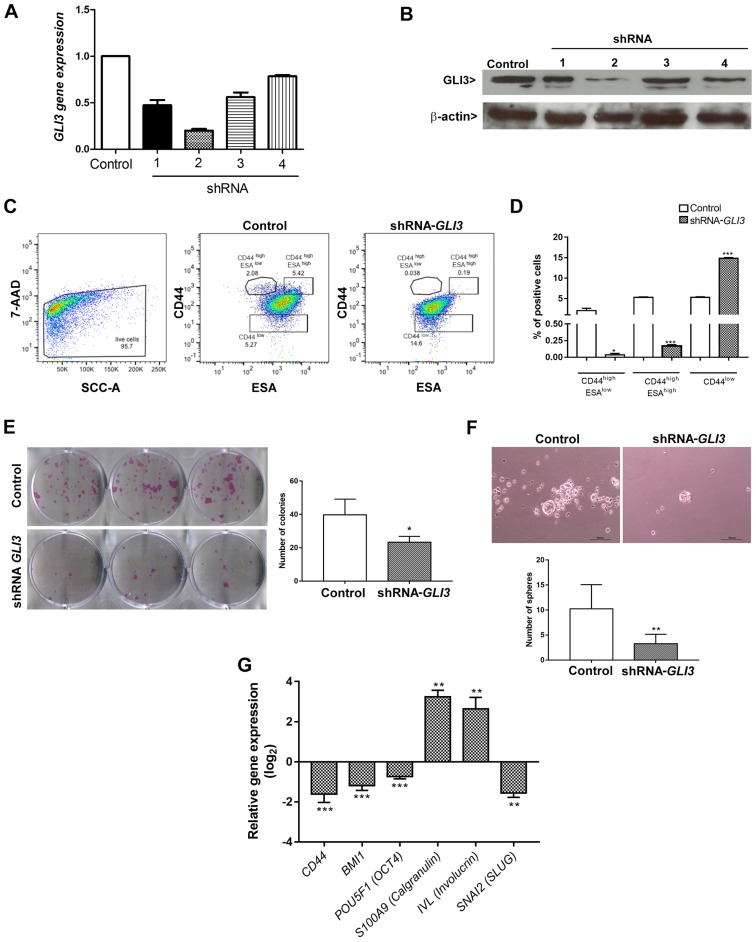
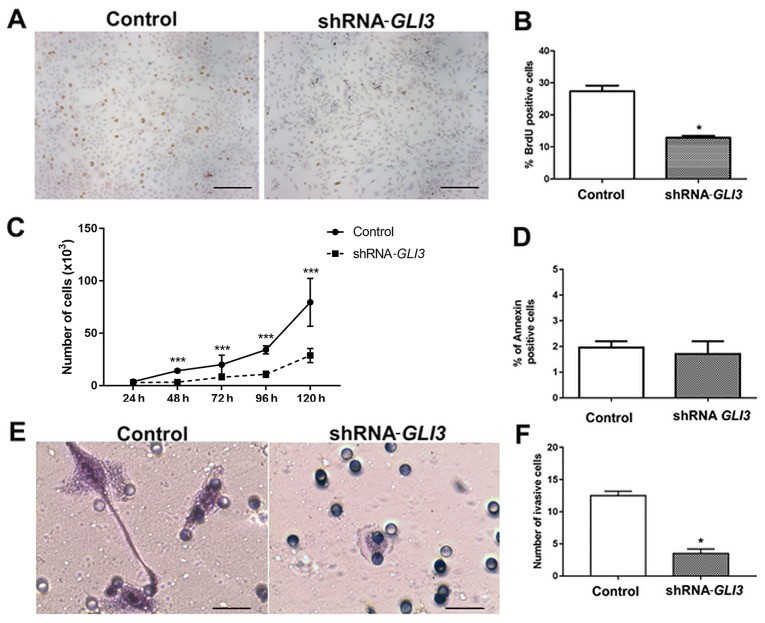
The SCC9 cell line transfected with the shRNA (sequence 2) for GLI3 gene silencing exhibited a decrease in the expression of GLI3 of approximately 80% at both the mRNA (Fig. 4A) and protein (Fig. 4B) level. To assess the effects of GLI3 gene silencing on CSCLC fractions previously identified in OSCC (14), the cells transfected with shRNA-GLI3 and the control cells were stained for CD44 and ESA and analysed by flow cytometry in 3 different subpopulations: The CD44high/ESAlow (EMT-CSCLC), CD44high/ESAhigh (EPI-CSCLC) and CD44low (NON-CSCLC) populations. GLI3 knockdown significantly decreased the percentage of CD44high/ESAlow (P=0.02) and CD44high/ESAhigh (P<0.0001) cells and consistently increased the percentage of CD44low cells (P<0.0001) (Fig. 4C and D).
Figure 4.
Effects of GLI3 knockdown on SCC9 stemness. (A) GLI3 mRNA expression levels and (B) GLI3 protein expression in SCC9 cells transfected with control or shRNA-GLI3. (C) FACS analysis of the expression of CD44 (y-axis) and ESA (x-axis) on control and shRNA GLI3 SCC9 cells. GLI3 knockdown decreases CD44high/ESAlow and CD44high/ESAhigh fractions of SCC9 cells, as well as increased the CD44low fraction. (D) Quantification of data depicted in (C), showing the percentage of CD44high/ESAlow, CD44high/ESAhigh and CD44low fractions. (E) Colony formation ability and quantification of the number of colonies formed in shRNA GLI3 and control cells. (F) Number of spheres formed in shRNA GLI3 and control cells. (G) RT-qPCR indicated reduced CD44, BMI1, POU5F1 (OCT4) and SNAI2 (SLUG) mRNA expression and increased S100A9 (Calgranulin) and Involucrin (IVL) in shRNA-GLI3-transfected cells in relation to the control cells. *P<0.05, **P<0.01 and ***P<0.001 (t-tests with the Welch correction). Microscope images: Magnification, ×100 (scale bars, 40 μm).
To determine whether the decrease in the CD44high/ESAlow (EMT-CSCLC) and CD44high/ESAhigh (EPI-CSCLC) cell populations was associated with a functional decrease in stemness, the cells transfected with shRNA-GLI3 and the control cells were plated to determine their clonogenicity and sphere formation ability. As shown in Fig. 4E and F, GLI3 knockdown was associated with significantly lower numbers of colonies and spheres formed (P=0.03 and P=0.004, respectively).
We also examined whether the decrease in stemness in the shRNA-GLI3-transfected cells was associated with the decreased expression of stem cell- and EMT-related genes. As expected, GLI3 gene silencing resulted in a significant decrease in CD44 (P<0.0001), BMI1 (P<0.0001), POU5F1 (OCT4) (P<0.0001) and SNAI2 (SLUG) (P<0.001) gene expression in relation to the control cells. Additionally, the shRNA-GLI3-transfected cells exhibited an increased gene expression of the epithelial differentiation markers, IVL and S100A9 (Calgranulin B) in relation to the control cells, indicating a shift towards differentiation (Fig. 4G).
Downregulation of GLI3 reduces cell proliferation and invasion
Subsequently, we performed BrdU and proliferation curve assays to address the effects of GLI3 knockdown on the proliferation rates of the SCC9 cells. As demonstrated in Fig. 5A and B, the SCC9-shRNA-GLI3-transfected cells exhibited a significant decrease in the number of BrdU-positive cells (P=0.01) in relation to the control cells. Moreover, there was a significant decrease in the number of shRNA-GLI3-transfected cells after 48 (P=0.001), 72 (P= 0.0079), 96 (P= 0.0068) and 120 h (P= 0.001) when compared to the control cells (Fig. 5C). No differences in the number of Annexin V-positive cells were observed between the control and shRNA-GLI3-transfected cells (Fig. 5D). Invasion assay revealed significantly lower numbers of invasive shRNA-GLI3-transfected cells in relation to the control cells (P=0.03) (Fig. 5E and F).
Figure 5.
Effects of GLI3 knockdown on SCC9 cellular proliferation, apoptosis and invasion. (A and B) Cells transfected with shRNA GLI3 cells exhibited a reduced number of BrdU-positive cells and (C) a decrease in the number of cells after 48, 72, 96 and 120 h of culture. (D) Number of Annexin V-positive cells. (E and F) GLI3 knockdown in SCC9 cells was associated with significantly lower numbers of invasive cells in relation to the controls. *P<0.05 and ***P<0.001 (t-test with the Welch correction). Microscope images: (A and B) Magnification, ×100 (scale bars, 40 μm); (E) ×400 (scale bars, 10 μm).
GLI3 protein expression in OSCC samples and its association with patient clinicopathological characteristics
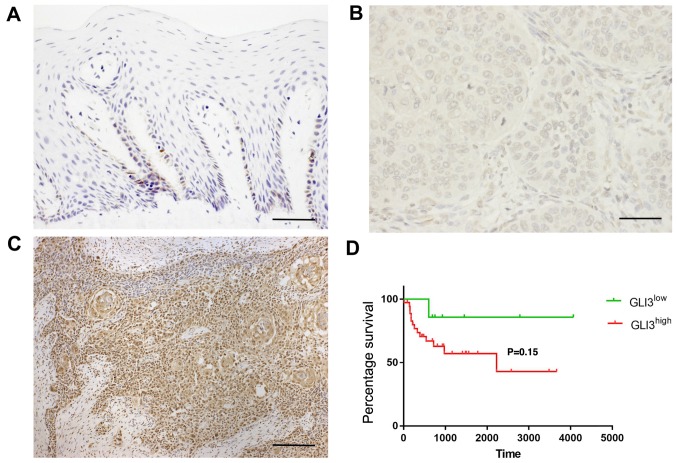
GLI3 protein expression was detected in the cytoplasm and nucleus of normal and malignant epithelial cells. In non-tumour margins, GLI3 expression was observed mainly in the basal and suprabasal cells (Fig. 6A). In the OSCC samples, GLI3 was highly expressed in the tumour islands, as well as in isolated neoplastic cells. GLI3 was considered to have a low expression (Fig. 6B) in 22/45 OSCC samples (48,88%) and a high expression (Fig. 6C) in 23/45 samples (51,12%). Invasive OSCC areas were highly positive for GLI3.
Figure 6.
GLI3 protein expression in non-tumour margin and in OSCC samples. (A) GLI3 expression was mainly observed in basal and suprabasal cells in the non-tumour margin. (B) GLI3low expression in OSCC. (C) GLI3high expression in OSCC, mainly in the tumour island, as well as in the invasive areas (magnification, ×100). (D) Overall survival rate in patients with a GLI3high expression when compared to those with a GLI3low expression in OSCC (P=0.15). Microscope images: (A and B) Magnification, ×400 (scale bar, 10 μm); (C) ×100 (scale bars, 40 μm).
The association between GLI3 expression and the patient clinicopathological characteristics, as well as survival was evaluated and is described in Table III and Fig. 6D. There were no significant associations between GLI3 protein expression and metastasis (pN), or blood, lymphatic and/or perineural invasion. However, a GLI3high expression was associated with tumour size T3/T4 (P=0.03). To determine the prognostic significance of GLI3 in patients with OSCC, Kaplan-Meier survival analysis was performed. There were no significant differences observed in the 5-year survival rate of patients with a GLI3high expression when compared to those with a GLI3low expression (P=0.15) (Fig. 6D).
Table III.
Association of GLI3 protein expression with the clinicopathological characteristics of patients with oral squamous cell carcinoma.
| Clinicopathological characteristics | Number of cases | GLI3 expression
|
P-value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ||||
| >60 | 17 | 9 | 8 | 0.76 |
| <60 | 28 | 13 | 15 | |
| pT classification | ||||
| T1/T2 | 27 | 17 | 10 | 0.03 |
| T3/T4 | 18 | 5 | 13 | |
| pN classification | ||||
| N+ | 29 | 13 | 16 | 0.54 |
| N− | 16 | 9 | 7 | |
| Blood invasion (BI)a | ||||
| BI+ | 7 | 2 | 5 | 0.40 |
| BI− | 35 | 19 | 16 | |
| Lymphatic invasion (LI)a | ||||
| LI+ | 18 | 6 | 12 | 0.12 |
| LI− | 25 | 15 | 10 | |
| Perineural invasion (PI) | ||||
| PI+ | 19 | 9 | 10 | 1.00 |
| PI− | 26 | 13 | 13 | |
Missing patient data. Values in bold indicate statistical significance (P<0.05; P-values were calculated using Fisher's exact test).
Discussion
The primary aim of this study was to evaluate the differential expression of genes related to stem cell signalling pathways in CD44high in relation to CD44low cells. The upregulation of receptors and transcription factors belonging to the Wnt, TGFβ, Notch, SHH and pluripotency maintenance of the stem cells was observed in the CD44high cells. This observation demonstrates that in OSCC, the aberrant and constitutive activation of developmental signalling pathways in CSCLC may favor the acquisition and maintenance of malignant behavior. Secondly, we explored the role of GLI3 in OSCC stemness, proliferation and invasion, speculating that this gene, among others that were overexpressed in both cell lines, could be important to oral CSCLC. The clonogenic ability, sphere formation and number of CSCLCs with epithelial and mesenchymal phenotypes, identified by the surface markers, CD44high/ESAhigh and CD44high/ESAlow, respectively, were significantly reduced in the cells in which GLI3 was knocked own. Of note, the downregulation of the stemness genes, BMI1, POU5F1 (OCT4) and the EMT gene SNAI2 (SLUG), was observed, as well as an increase in the expression of the epithelial differentiation markers, S100A9 (Calgranulin B) and IVL. Taken together, these results indicate that GLI3 is a transcription factor that may play an important role to play in the maintenance of CSCLCs in OSCC and may thus be considered a potential target for the eradication of CSC fractions in OSCC.
CSCs play an important role in OSCC development and progression; different cell surface markers have been used to identify and characterise these cells (5,13,14). In this study, we used the CD44 marker to isolate the CD44high (CSCLC) and the CD44low (NON-CSCLC) populations, the first being associated with an increased ability to form spheres and colonies with holoclone morphology and the upregulation of the stem cell-related genes, POU5F1 (OCT4) and BMI1. CD44 is the most frequently observed CSC marker. It was used by Prince et al to identify the CSC population in OSCC, which exhibited a high BMI1 gene expression, as well as an exclusive ability to form tumours in immunodeficient mice and to recreate tumour heterogeneity (5). We have previously demonstrated that SCC9 CD44high cells are able to produce a tumour in immunodeficient mice, even at lower concentrations, and that the derived tumours exhibit a high expression of CD44, cytokeratin19, β-catenin and E-cadherin (51), demonstrating that this cell line and CD44 are suitable for the investigation of CSCLCs in OSCC.
CD44 is a glycoprotein receptor that binds to hyaluronan (HA) and can vary in size due to post-translational modifications and alternative splicing (11). This receptor actively interacts with the extracellular matrix (HA, collagen and fibronectin) and activates intracellular events that promote resistance to apoptosis, EMT, migration and metastatic colonization (11,53). CD44 also plays an important role in the crosstalk between CSCs and their niche, including the activation and recruitment of resident cells to form CSC niches and pre-metastatic niches (54).
In cancer, to sustain their stem cell properties, CSCs upregulate several signalling pathways that control normal stem cells, including the Notch, Wnt, Hedgehog and the pluripotency pathways mediated by interleukin (IL)6/STAT3 (17). In this study, we observed that the vast majority of receptors, co-receptors and transcriptional factors belonging to these pathways were upregulated in the CD44high cells in relation to the CD44low cells in OSCC and may thus contribute to the maintenance of stemness in this malignancy.
Notch signalling, a key regulator of stem cell fate, is frequently overexpressed in various types of cancer and contributes to angiogenesis and drug resistance via interaction with oncogenic pathways (17,55). In this study, we observed an upregulation in the expression of NOTCH2, PSEN1, PSEN2 and PSENEM in the CD44high cells. Zhao et al (2016) observed that pharmacological Notch1 inhibition was associated with a reduction in OSCC CSC fractions and self-renewal capacity, as well as with an increase in the response to conventional therapy (56), bringing with it the promise of an OSCC therapy. In addition, Notch-mediated signalling is associated with the induction of EMT under hypoxic conditions in OSCC (57). It is also associated with high mortality and T classification in OSCC, as well as with an increase in cell migration and invasion (58,59).
Wnt signalling plays an important role during embryogenesis and in the determination of cell fate during development (60). The activation of the canonical Wnt pathway begins when the ligand binds with the Frizzled receptors and co-receptors [low-density lipoprotein receptor-related protein (LRP)] which then leads to β-catenin stabilization and translocation to the nucleus. This results in the transcriptional activation of tumour-promoting genes, including CCND1, MYC, MMP7 and CD44 (61). In this study, a large number of genes related to the Wnt pathway, such as FZD receptors (FZD1 and FDZ2), co-receptors (LRP6), and transcription factors (BCL9L, CTNNB1, NFAT5, NFATC3 and TCF7L1), were overexpressed in the CSCLC fraction. This result infers that this pathway may play an important role in the maintenance of these cells in OSCC. In fact, it was previously demonstrated that CSCs in head and neck squamous cell carcinoma isolated based on Hoechst dye exclusion exhibited an aberrant activation of the Wnt/beta-catenin pathway (62). Many of the surface markers used to identify CSCs are targets of this pathway, including CD44, CD24 and ESA, which can contribute to the enrichment of this population (63). Additionally, this pathway plays an important role in the induction of the EMT, in which cells necessarily acquire CSCs traits and exhibit an upregulation of SNAIL, ZEB1 and ZEB2 (61,63).
One of the mechanisms used by the SHH pathway, which can contribute to cancer development, is the upregulation of the transcription factors, GLI1, GLI2 and GLI3 (19). The majority of the studies in the literature have investigated the role of GLI1 in cancer, as it acts as a transcription activator in cancer. However, GLI proteins are highly dynamic and function in a contextual and combinatorial manner (64). For example, GLI3 can function as either a transcription activator or inhibitor, depending on the presence or lack of SHH ligands (19,28). In the absence of ligands, the GLI protein function is turned off, GLI1 is not transcribed and GLI2/GLI3 are processed in a truncated repressor protein form that binds to the GLI sites in the SHH promoters, leading to the inhibition of target genes (19,28). On the other hand, with the presence of SHH ligands (canonical activation), GLI2/GLI3 in its activated form (full-length) is produced and positively regulates genes which control cell proliferation (CCND1, MYC), cell death (BCL2), EMT (e.g. SNAIL), angiogenesis (e.g., ANG1/2) and stemness (SOX2, NANOG) (65,66). In addition, GLI1 is a direct target of GLI2/GLI3 and favours malignant behavior (63).
Some studies have implicated the SHH pathway in CSC regulation and maintenance in addition to participating in metastatic progression and the acquisition of the EMT phenotype (38,67). Its inhibition results in a decrease in stem cell propagation and cell renewal (68). For these reasons, natural and synthetic antagonists of SMO and GLI proteins are being tested to evaluate their efficacy alone or in combination to target the CSC in a wide range of malignant tumours (28). For example, the combined target therapy of PI3K/Akt/mTOR therapy and SMO inhibitor was associated with a decrease in CSC self-renewal by inhibiting the NANOG, POU5F1 (OCT4), SOX2, MYC and GLI transcription factors in pancreatic cancer (69). In non-small cell lung cancer (NSCLC), GLI1 inhibition abrogated the CSC population and cooperated with EGFR inhibitors to impair the viability of malignant cells (70).
Biddle et al (2011) have demonstrated that CSC in OSCC can switch between two different phenotypes. One of the said phenotypes is preferentially epithelial and proliferative, characterised by the phenotype CD44high/ESAhigh (EPI-CSC); the other is preferentially migratory, with the CD44high/ESAlow (EMT-CSC) phenotype (14). In this study, we observed that GLI3 gene silencing was able to decrease the percentage of CD44high/ESAhigh and the CD44high/ESAlow cells, which was then followed by an increase in CD44low cells. Moreover, the clonogenic ability, primarily a property of the CD44high/ESAhigh cells, was strongly inhibited in the cells transfected with shRNA-GLI3. This supports the concept that these cell subpopulations are lost through cell differentiation. This was further confirmed by the upregulation of the epithelial differentiation markers, S100A9 (Calgranulin B) and IVL in the shRNA-GLI3-transfected cells. Prince et al (5) observed that in tumours derived from CD44high cells, well differentiated areas negative for CD44 exhibited a strong expression of IVL. The loss of clonogenicity, increase in CD44low fraction and increase in the mRNA expression of the differentiation markers described above indicates that the SHH signalling pathway may function as a form of 'differentiation therapy' in OSCC (71,72) and may potentiate the effects of therapeutic agents such as cisplatin. However, additional studies are warranted to support this concept.
We also observed that the levels of stemness-related genes, BMI1 and POU5F1 (OCT4), were downregulated following GLI3 knockdown. BMI1 is a member of the polycomb repressive complex 1 and plays an important role in CSC self-renewal, tumour initiation, undifferentiated state maintenance and therapeutic resistance in OSCC (73). Nör et al (74) demonstrated that OSCC treatment with cisplatin promoted enrichment of the CSC fraction. It was also shown that the BMI1 gene actively participated in the acquisition of resistance, which was influenced by IL6 (74). POU5F1 (OCT4) also plays an important role in CSC and its overexpression, increases the capacity for tumour initiation, tumour sphere formation, invasion and the acquisition of the EMT phenotype in OSCC cells lines (75,76).
Of note, sphere formation assay, normally used to investigate the 'functional' state of CD44high/ESAlow fractions, has also been shown to be reduced in cells transfected with shRNA-GLI3. In addition, in this study, we observed a significant decrease in the mRNA expression of SNAI2 (SLUG), a transcription factor that participates in the EMT process. The EMT-CSC fraction in OSCC exhibits significant resistance to radiation and various chemotherapeutic agents (6,10). As it is responsible for tumour invasion and metastasis, therapies that can reduce or eradicate this cell fraction are greatly needed (77). Considering our results as a whole, they highlight the important role that GLI3 plays in controlling CSC self-renewal, its distinct phenotypes in OSCC and its involvement with genes involved in the maintenance of these subpopulations.
Also of note, a significant decrease in both cellular proliferation and invasion was observed following GLI3 gene silencing. To the best of our knowledge, this is the first study to demonstrate that GLI3 participates in the regulatioin of both processes in OSCC. Multiple oncogenic pathways can also increase GLI function (non-canonical activation), including K-ras, PI3K/Akt and TGFβ (78-80), contributing to tumour development and formation. It should be noted that the increase in cyclin-D2 observed after SHH activation depends on GLI3 activity (18). Thus, it can be speculated that the inhibition in cell proliferation observed in GLI3 knockdown cells may be the result of the modulation of different target genes, including cyclins and the GLI1 protein, which in turn activate pro-proliferative genes. Shih et al recently demonstrated that GLI1 expression was induced by the oncogene, ROS1, leading to an increase in cellular proliferation but not in cell migration and invasion (81). Nevertheless, additional studies are required in order to elucidate the molecular targets of GLI3 which control cell proliferation.
In this study, GLI3 inhibition decreased cellular invasion in OSCC. We can speculate that the reduction in the CSCLC-EMT fraction may have contributed, possibly via SLUG downregulation. The EMT process is associated with an enhancement in motility and cellular invasion (82), contributing actively to tumour progression. Yan et al (2011) have observed that the pharmacological inhibition of Shh/Gli pathway leads not only to the inhibition of cellular proliferation by the downregulation of GLI1, GLI2, CCND1 and BCL2 and upregulation of caspase-3, but also to a significant decrease in cell migration (83).
In this study, non-tumoural margins exhibited a weak GLI3 expression, mainly located in the basal cell layer, where the normal epithelial stem cells reside. GLI3 expression was correlated with tumour size, although no association with lymph node metastasis or overall survival was found. There seems to be only one study in the literature evaluating GLI3 expression in OSCC; the authors found that GLI3 is weakly/moderately expressed in OSCC. They did not find any correlation to survival in a cohort of 60 patients (52). We believe that additional studies are required to investigate the prognostic value of GLI3 in OSCC.
Taken together, the findings of this study demonstrate that the signalling pathways which control normal stem cells are aberrantly upregulated in CSCLC, isolated based on CD44 expression in OSCC. Moreover, GLI3 inhibition leads to a significant decrease in the number and function of the CD44high/ESAhigh (EPI-CSCLC) and CD44high/ESAlow (EMT-CSCLC) cell fractions and an increase in the CD44low subpopulation. These results indicate the relevant role this gene plays in CSC self-renewal, maintenance and differentiation. In addition to its effects on stemness, GLI3 also inhibits cell proliferation and invasion, both processes which are necessary for tumour progression. This study highlights that GLI3 is a possible target to be investigated in the future, either isolated or in combination with other drugs, for therapies based on CSCs in OSCC.
Acknowledgments
Not applicable.
Funding
This study was financially supported by the São Paulo Research Foundation (FAPESP grant nos. 2011/21395-8 and 2012/00786-1).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MFSDR was the major contributor to the conceptualization and design of the project, acquisition, interpretation and analysis of all data and writing, revising and formatting of the manuscript. LM contributed with acquisition, interpretation and analysis of the majority of the data with cell culture and immunohistochemistry, also collaborated with writing and formatting the manuscript. NPdA provided and analysed flow cytometry data. DH contributed to the acquisition of the cell culture data. COR contributed to the FACS analysis, and to the writing, drafting and critical revision of the manuscript. RAM, TNT, RRG and EET provided the clinical data and samples for immunohistochemical analysis. FDN oversaw the study conceptualization and design, writing, drafting and critical revision of the manuscript. All authors have read and approved the publication of this version.
Ethics approval and consent to participate
The study was performed with approval from the Brazilian National Ethics Committee (Process #16491) and in accordance with the Declaration of Helsinki. The collection of samples, clinical data, and histopathological analysis was performed after acquisition of a signed informed consent form from each patient.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that there are no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Ausoni S, Boscolo-Rizzo P, Singh B, Da Mosto MC, Spinato G, Tirelli G, Spinato R, Azzarello G. Targeting cellular and molecular drivers of head and neck squamous cell carcinoma: Current options and emerging perspectives. Cancer Metastasis Rev. 2016;35:413–426. doi: 10.1007/s10555-016-9625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxi S, Fury M, Ganly I, Rao S, Pfister DG. Ten years of progress in head and neck cancers. J Natl Compr Canc Netw. 2012;10:806–810. doi: 10.6004/jnccn.2012.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biddle A, Gammon L, Liang X, Costea DE, Mackenzie IC. Phenotypic plasticity determines cancer stem cell therapeutic resistance in oral squamous cell carcinoma. EBioMedicine. 2016;4:138–145. doi: 10.1016/j.ebiom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Filho MS, Nör JE. The biology of head and neck cancer stem cells. Oral Oncol. 2012;48:1–9. doi: 10.1016/j.oraloncology.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson AC, La Fleur L, Melissaridou S, Roberg K. The relationship between EMT, CD44high/EGFRlow phenotype, and treatment response in head and neck cancer cell lines. J Oral Pathol Med. 2016;45:640–646. doi: 10.1111/jop.12423. [DOI] [PubMed] [Google Scholar]
- 9.Naik PP, Das DN, Panda PK, Mukhopadhyay S, Sinha N, Praharaj PP, Agarwal R, Bhutia SK. Implications of cancer stem cells in developing therapeutic resistance in oral cancer. Oral Oncol. 2016;62:122–135. doi: 10.1016/j.oraloncology.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Gemenetzidis E, Gammon L, Biddle A, Emich H, Mackenzie IC. Invasive oral cancer stem cells display resistance to ionising radiation. Oncotarget. 2015;6:43964–43977. doi: 10.18632/oncotarget.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourguignon LY, Shiina M, Li JJ. Hyaluronan-CD44 interaction promotes oncogenic signaling, microRNA functions, chemoresistance, and radiation resistance in cancer stem cells leading to tumor progression. Adv Cancer Res. 2014;123:255–275. doi: 10.1016/B978-0-12-800092-2.00010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Shi S, Yen Y, Brown J, Ta JQ, Le AD. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett. 2010;289:151–160. doi: 10.1016/j.canlet.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 13.La Fleur L, Johansson AC, Roberg K. A CD44high/EGFRlow subpopulation within head and neck cancer cell lines shows an epithelial-mesenchymal transition phenotype and resistance to treatment. PLoS One. 2012;7:e44071. doi: 10.1371/journal.pone.0044071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE, Mackenzie IC. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317–5326. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 15.Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, Panda CK, Roychoudhury S. CD44(high)CD24(low) molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Res (Amst) 2016;16:405–417. doi: 10.1016/j.scr.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Seino S, Shigeishi H, Hashikata M, Higashikawa K, Tobiume K, Uetsuki R, Ishida Y, Sasaki K, Naruse T, Rahman MZ, et al. CD44(high)/ALDH1(high) head and neck squamous cell carcinoma cells exhibit mesenchymal characteristics and GSK3β-dependent cancer stem cell properties. J Oral Pathol Med. 2016;45:180–188. doi: 10.1111/jop.12348. [DOI] [PubMed] [Google Scholar]
- 17.Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasper M, Schnidar H, Neill GW, Hanneder M, Klingler S, Blaas L, Schmid C, Hauser-Kronberger C, Regl G, Philpott MP, et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol Cell Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pak E, Segal RA. Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Zeng H, Liu A. The loss of Hh responsiveness by a non-ciliary Gli2 variant. Development. 2015;142:1651–1660. doi: 10.1242/dev.119669. [DOI] [PubMed] [Google Scholar]
- 21.Shi X, Zhang Z, Zhan X, Cao M, Satoh T, Akira S, Shpargel K, Magnuson T, Li Q, Wang R, et al. An epigenetic switch induced by Shh signalling regulates gene activation during development and medulloblastoma growth. Nat Commun. 2014;5:5425. doi: 10.1038/ncomms6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junker JP, Peterson KA, Nishi Y, Mao J, McMahon AP, van Oudenaarden A. A predictive model of bifunctional transcription factor signaling during embryonic tissue patterning. Dev Cell. 2014;31:448–460. doi: 10.1016/j.devcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal JM, Junker JP, van Oudenaarden A, et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Yue D, Jin JQ, Woodard GA, Tolani B, Luh TM, Giroux-Leprieur E, Mo M, Chen Z, Che J, et al. Gli promotes epithelial-mesenchymal transition in human lung adenocarcinomas. Oncotarget. 2016;7:80415–80425. doi: 10.18632/oncotarget.11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/S0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 27.McIntyre BAS, Ramos-Mejia V, Rampalli S, Mechae R, Lee JH, Alev C, Sheng G, Bhatia M. Gli3-mediated hedgehog inhibition in human pluripotent stem cells initiates and augments developmental programming of adult hematopoiesis. Blood. 121:1543–1552. doi: 10.1182/blood-2012-09-457747. [DOI] [PubMed] [Google Scholar]
- 28.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the sonic Hedgehog signaling pathway: Review of Smoothened and GLI inhibitors. Cancers (Basel) 2016;8:E22. doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/S0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 31.Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carpenter RL, Paw I, Zhu H, Sirkisoon S, Xing F, Watabe K, Debinski W, Lo HW. The gain-of-function GLI1 transcription factor TGLI1 enhances expression of VEGF-C and TEM7 to promote glioblastoma angiogenesis. Oncotarget. 2015;6:22653–22665. doi: 10.18632/oncotarget.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzman DL, Antonarakis ES. Clinical implications of Hedgehog pathway signaling in prostate cancer. Cancers (Basel) 2015;7:1983–1993. doi: 10.3390/cancers7040871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdel-Rahman O. Hedgehog pathway aberrations and gastric cancer; evaluation of prognostic impact and exploration of therapeutic potentials. Tumour Biol. 2015;36:1367–1374. doi: 10.1007/s13277-015-3216-6. [DOI] [PubMed] [Google Scholar]
- 35.Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/MCB.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bigelow RL, Jen EY, Delehedde M, Chari NS, McDonnell TJ. Sonic hedgehog induces epidermal growth factor dependent matrix infiltration in HaCaT keratinocytes. J Invest Dermatol. 2005;124:457–465. doi: 10.1111/j.0022-202X.2004.23590.x. [DOI] [PubMed] [Google Scholar]
- 37.Yoo YA, Kang MH, Lee HJ, Kim BH, Park JK, Kim HK, Kim JS, Oh SC. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71:7061–7070. doi: 10.1158/0008-5472.CAN-11-1338. [DOI] [PubMed] [Google Scholar]
- 38.Katoh Y, Katoh M, Yoo YA. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (Review) Int J Mol Med. 2008;22:271–275. [PubMed] [Google Scholar]
- 39.Pantazi E, Gemenetzidis E, Teh MT, Reddy SV, Warnes G, Evagora C, Trigiante G, Philpott MP. GLI2 is a regulator of β-catenin and is associated with loss of E-Cadherin, cell invasiveness, and long-term epidermal regeneration. J Invest Dermatol. 2017;137:1719–1730. doi: 10.1016/j.jid.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 40.Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du YE, Wen S, Xu L, Tang X, Tang S, et al. LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of Hedgehog signaling pathway. Stem Cells. 2016;34:55–66. doi: 10.1002/stem.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137:1679–1686. doi: 10.1007/s00432-011-1038-5. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Che S, Pan C, Su Z, Zheng S, Yang S, Zhang H, Li W, Wang W, Liu J. The SHH/Gli axis regulates CD90-mediated liver cancer stem cell function by activating the IL6/JAK2 pathway. J Cell Mol Med. 2018;22:3679–3690. doi: 10.1111/jcmm.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavicchioli Buim ME, Gurgel CA, Gonçalves Ramos EA, Lourenço SV, Soares FA. Activation of sonic hedgehog signaling in oral squamous cell carcinomas: A preliminary study. Hum Pathol. 2011;42:1484–1490. doi: 10.1016/j.humpath.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 44.Fan HX, Wang S, Zhao H, Liu N, Chen D, Sun M, Zheng JH. Sonic hedgehog signaling may promote invasion and metastasis of oral squamous cell carcinoma by activating MMP-9 and E-cadherin expression. Med Oncol. 2014;31:41. doi: 10.1007/s12032-014-0041-5. [DOI] [PubMed] [Google Scholar]
- 45.Schneider FT, Schänzer A, Czupalla CJ, Thom S, Engels K, Schmidt MH, Plate KH, Liebner S. Sonic hedgehog acts as a negative regulator of {beta}-catenin signaling in the adult tongue epithelium. Am J Pathol. 2010;177:404–414. doi: 10.2353/ajpath.2010.091079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honami T, Shimo T, Okui T, Kurio N, Hassan NM, Iwamoto M, Sasaki A. Sonic hedgehog signaling promotes growth of oral squamous cell carcinoma cells associated with bone destruction. Oral Oncol. 2012;48:49–55. doi: 10.1016/j.oraloncology.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 47.Mikami Y, Fujii S, Nagata K, Wada H, Hasegawa K, Abe M, Yoshimoto RU, Kawano S, Nakamura S, Kiyoshima T. GLI-mediated Keratin 17 expression promotes tumor cell growth through the anti-apoptotic function in oral squamous cell carcinomas. J Cancer Res Clin Oncol. 2017;143:1381–1393. doi: 10.1007/s00432-017-2398-2. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.de Andrade NP, Rodrigues MF, Rodini CO, Nunes FD. Cancer stem cell, cytokeratins and epithelial to mesenchymal transition markers expression in oral squamous cell carcinoma derived from ortothopic xenoimplantation of CD44high cells. Pathol Res Pract. 2017;213:235–244. doi: 10.1016/j.prp.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Rodrigues MF, de Oliveira Rodini C, de Aquino Xavier FC, Paiva KB, Severino P, Moyses RA, López RM, DeCicco R, Rocha LA, Carvalho MB, et al. PROX1 gene is differentially expressed in oral cancer and reduces cellular proliferation. Medicine (Baltimore) 2014;93:e192. doi: 10.1097/MD.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. Wiley Blackwell; Oxford: 2017. pp. 17–21. [Google Scholar]
- 52.Schneider S, Thurnher D, Kloimstein P, Leitner V, Petzelbauer P, Pammer J, Brunner M, Erovic BM. Expression of the Sonic hedgehog pathway in squamous cell carcinoma of the skin and the mucosa of the head and neck. Head Neck. 2011;33:244–250. doi: 10.1002/hed.21437. [DOI] [PubMed] [Google Scholar]
- 53.Zöller M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 54.Williams K, Motiani K, Giridhar PV, Kasper S. CD44 integrates signaling in normal stem cell, cancer stem cell and (pre) metastatic niches. Exp Biol Med (Maywood) 2013;238:324–338. doi: 10.1177/1535370213480714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 56.Zhao ZL, Zhang L, Huang CF, Ma SR, Bu LL, Liu JF, Yu GT, Liu B, Gutkind JS, Kulkarni AB, et al. NOTCH1 inhibition enhances the efficacy of conventional chemotherapeutic agents by targeting head neck cancer stem cell. Sci Rep. 2016;6:24704. doi: 10.1038/srep24704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishida T, Hijioka H, Kume K, Miyawaki A, Nakamura N. Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol Lett. 2013;6:1201–1206. doi: 10.3892/ol.2013.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshida R, Nagata M, Nakayama H, Niimori-Kita K, Hassan W, Tanaka T, Shinohara M, Ito T. The pathological significance of Notch1 in oral squamous cell carcinoma. Lab Invest. 2013;93:1068–1081. doi: 10.1038/labinvest.2013.95. [DOI] [PubMed] [Google Scholar]
- 59.Weaver AN, Burch MB, Cooper TS, Della Manna DL, Wei S, Ojesina AI, Rosenthal EL, Yang ES. Notch signaling activation is associated with patient mortality and increased FGF1-mediated invasion in squamous cell carcinoma of the oral cavity. Mol Cancer Res. 2016;14:883–891. doi: 10.1158/1541-7786.MCR-16-0114. [DOI] [PubMed] [Google Scholar]
- 60.Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: Conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holland JD, Klaus A, Garratt AN, Birchmeier W. Wnt signaling in stem and cancer stem cells. Curr Opin Cell Biol. 2013;25:254–264. doi: 10.1016/j.ceb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 62.Song J, Chang I, Chen Z, Kang M, Wang CY. Characterization of side populations in HNSCC: Highly invasive, chemoresistant and abnormal Wnt signaling. PLoS One. 2010;5:e11456. doi: 10.1371/journal.pone.0011456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang K, Wang X, Zhang H, Wang Z, Nan G, Li Y, Zhang F, Mohammed MK, Haydon RC, Luu HH, et al. The evolving roles of canonical WNT signaling in stem cells and tumorigenesis: Implications in targeted cancer therapies. Lab Invest. 2016;96:116–136. doi: 10.1038/labinvest.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aberger F. Context-dependent signal integration by the GLI code: The oncogenic load, pathways, modifiers and implications for cancer therapy. Semin Cell Dev Biol. 2014;33:93–104. doi: 10.1016/j.semcdb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Stecca B. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, Ruiz i Altaba A. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang N, Zhou TC, Lei XX, Wang C, Yan M, Wang ZF, Liu W, Wang J, Ming KH, Wang BC, et al. Inhibition of Sonic Hedgehog signaling pathway by thiazole antibiotic thiostrepton attenuates the CD44+/CD24-stem-like population and sphere-forming capacity in triple-negative breast cancer. Cell Physiol Biochem. 2016;38:1157–1170. doi: 10.1159/000443066. [DOI] [PubMed] [Google Scholar]
- 69.Sharma N, Nanta R, Sharma J, Gunewardena S, Singh KP, Shankar S, Srivastava RK. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget. 2015;6:32039–32060. doi: 10.18632/oncotarget.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bora-Singhal N, Perumal D, Nguyen J, Chellappan S. Gli1-mediated regulation of Sox2 facilitates self-renewal of stem-like cells and confers resistance to EGFR inhibitors in non-small cell lung cancer. Neoplasia. 2015;17:538–551. doi: 10.1016/j.neo.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sell S. Cancer stem cells and differentiation therapy. Tumour Biol. 2006;27:59–70. doi: 10.1159/000092323. [DOI] [PubMed] [Google Scholar]
- 72.Pattabiraman DR, Weinberg RA. Targeting the epithelial-to-mesenchymal transition: The case for differentiation-based therapy. Cold Spring Harb Symp Quant Biol. 2016;81:11–19. doi: 10.1101/sqb.2016.81.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao L, Bombard J, Cintron K, Sheedy J, Weetall ML, Davis TW. BMI1 as a novel target for drug discovery in cancer. J Cell Biochem. 2011;112:2729–2741. doi: 10.1002/jcb.23234. [DOI] [PubMed] [Google Scholar]
- 74.Nör C, Zhang Z, Warner KA, Bernardi L, Visioli F, Helman JI, Roesler R, Nör JE. Cisplatin induces Bmi-1 and enhances the stem cell fraction in head and neck cancer. Neoplasia. 2014;16:137–146. doi: 10.1593/neo.131744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai LL, Hu FW, Lee SS, Yu CH, Yu CC, Chang YC. Oct4 mediates tumor initiating properties in oral squamous cell carcinomas through the regulation of epithelial-mesenchymal transition. PLoS One. 2014;9:e87207. doi: 10.1371/journal.pone.0087207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cai J, He B, Li X, Sun M, Lam AK, Qiao B, Qiu W. Regulation of tumorigenesis in oral epithelial cells by defined reprogramming factors Oct4 and Sox2. Oncol Rep. 2016;36:651–658. doi: 10.3892/or.2016.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 78.Ke Z, Caiping S, Qing Z, Xiaojing W. Sonic hedgehog-Gli1 signals promote epithelial-mesenchymal transition in ovarian cancer by mediating PI3K/AKT pathway. Med Oncol. 2015;32:368. doi: 10.1007/s12032-014-0368-y. [DOI] [PubMed] [Google Scholar]
- 79.Rajurkar M, De Jesus-Monge WE, Driscoll DR, Appleman VA, Huang H, Cotton JL, Klimstra DS, Zhu LJ, Simin K, Xu L, et al. The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proc Natl Acad Sci USA. 2012;109:E1038–E1047. doi: 10.1073/pnas.1114168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X, Shapiro CL, Majumder S. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res. 2012;72:5048–5059. doi: 10.1158/0008-5472.CAN-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shih CH, Chang YJ, Huang WC, Jang TH, Kung HJ, Wang WC, Yang MH, Lin MC, Huang SF, Chou SW, et al. EZH2-mediated upregulation of ROS1 oncogene promotes oral cancer metastasis. Oncogene. 2017;36:6542–6554. doi: 10.1038/onc.2017.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan M, Wang L, Zuo H, Zhang Z, Chen W, Mao L, Zhang P. HH/GLI signalling as a new therapeutic target for patients with oral squamous cell carcinoma. Oral Oncol. 2011;47:504–509. doi: 10.1016/j.oraloncology.2011.03.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.