Abstract
Prostate cancer (PCa) is a malignant tumor with a high incidence in males. Localized tumors can be treated via surgery or radiation; however, it remains difficult to prevent disease progression. Bromodomain-containing protein 4 (BRD4) is an epigenetic reader protein that binds to acetylated lysine on histones and has been reported to serve critical roles in numerous types of cancers. In the present study, it was demonstrated that BRD4 expression levels were significantly increased in cancerous prostate tissue specimens and cells, which were associated with clinical stage and metastasis. In addition, the present study reported that inhibition of BRD4 via short hairpin RNA or JQ1 (a bromo-domain inhibitor) decreased PCa cell proliferation, induced G0/G1 cell cycle arrest and apoptosis, mitigated cell invasion and migration in vitro, and impaired tumor growth in vivo. Mechanistically, BRD4 inhibition-induced suppression of cell cycle progression was associated with the upregulation of p21 and cyclin D1. c-Myc and B-cell lymphoma-2 (Bcl-2), important genes responsible for cell cycle regulation and anti-apoptotic functions, were downregulated in response to BRD4 inhibition. Furthermore, the present study revealed that c-Myc expression was negatively regulated by p21, and that the induction of p21 via BRD4 inhibition was mediated by forkhead box protein O1 (FOXO1), rather than p53. In summary, the results of the present study suggested that the aberrant expression of BRD4 in PCa may induce carcinogenesis. In addition, a mechanism by which BRD4 inhibition suppresses cell proliferation via the regulation of FOXO1-p21-Myc signaling was proposed in the present study, which may contribute to the development of novel therapeutic approaches in the management of PCa.
Keywords: bromodomain containing protein 4, bromodomain inhibitor, prostate cancer, forkhead box protein O1, p21, c-Myc
Introduction
Prostate cancer (PCa) remains an important public health concern and is the third leading cause of cancer-associated mortality in males in the United States, with an estimated 1.11 million novel cases diagnosed per year worldwide and 16,1360 novel cases diagnosed in the United States (1,2). PCa has become the second most common type of carcinoma in males worldwide and the most common malignancy in developed countries (1,2). Established risk factors for this disease include age, race, family history, genetics and obesity (3). At present, radical prostatectomy and radiotherapy are the gold standard treatments for men with early-stage prostate cancer (4); however, >30% of patients treated via these approaches will likely exhibit disease recurrence or metastasis (5,6). Once the disease progresses, the first line of therapy undertaken is androgen deprivation, which is not a curative treatment for patients with advanced tumors (4). Hence, investigating signaling pathways in detail and formulating novel targeted therapies is of vital importance in enhancing the survival of patients with advanced PCa.
The bromodomain and extra terminal (BET) family, which includes bromodomain testis-specific, bromodomain-containing protein (BRD)2, BRD3 and BRD4, encompasses epigenetic reader proteins (7). Via their N-terminal bromodomains (BD1 and BD2), the BET family of proteins exert diverse physiological functions, including regulation of the cell cycle, apoptosis and inflammation (8). BRD4, a well-reported member of the BET family, is generally expressed as a nuclear protein that binds to acetylated lysine residues on histones via its bromodomains (9). BRD4 recruits chromatin remodeling and transcription factors, as well as other associated proteins, such as positive transcription elongation factor b, to specific transcriptional sites, this leads to transcription elongation via the modulation of RNA polymerase II (9,10). BRD4 also interacts with RelA, a subunit of the nuclear factor-κB transcriptional complex, inducing an inflammatory response (10). BRD4 has been reported to serve vital roles in cell proliferation and cardiac hypertrophy (9,10). In addition, the aberrant expression of BRD4 has been frequently reported in a variety of diseases, such as cancer (11). Carcinogenesis induced by the BRD4 protein was first reported in NUT midline cancer (12). Studies have subsequently revealed BRD4 to exert oncogenic functions in numerous tumors and cancerous cells, including breast and lung cancers (13,14). Several small-molecule inhibitors have been thoroughly studied in recent years, including JQ1 and IBET762, which possess a high specificity for the bromodomain of the BET protein family (15). The binding of these two molecules to the BET bromodomain leads to the substitution of the protein from acetylated lysine residues on chromatin (16,17). Of note, BET inhibitors have been observed to exhibit significant antitumor effects in various malignancies in vitro and in vivo (18,19). BRD4 has been reported to be involved in androgen receptor signaling and the progression of PCa (20,21); however, other potential roles undertaken by this protein, as well as the therapeutic efficacy of BET inhibitors in the treatment of PCa, require further investigation.
In the present study, the roles of BRD4 in PCa were determined and the potential efficacy of small molecules in the binding of BET bromodomains were analyzed in vitro and in vivo. The results of the present study revealed the expression of BRD4 to be upregulated in PCa, which was positively associated with metastatic spread and clinical stage. Functional assays revealed that inhibition of BRD4 suppressed the proliferation, migration and invasion, concomitantly with the induction of G0/G1 cell cycle arrest and apoptosis of PCa cells. In a mouse model, a delay in tumor growth was also noted. The findings of the present study revealed that the effects of BRD4 inhibition in PCa may be mediated via the forkhead box protein O1 (FOXO1)-p21-Myc signaling axis. In addition, the present study demonstrated a previously unreported role of BRD4 in PCa and revealed a promising therapeutic strategy for patients with PCa.
Materials and methods
Patient tissue specimens
A total of 46 pairs of fresh cancerous prostate tissue and adjacent normal tissue specimens were obtained from patients undergoing surgical treatment in the Department of Urology, Renmin Hospital of Wuhan University (Wuhan, China) between 2016 and 2017. All the patients included in the present study were male and aged between 59-82-years-old. Approval for the present study was obtained from the Ethics Committee of the Renmin Hospital of Wuhan University. Informed consent and relevant clinical information were obtained from all patients prior to surgery. Specimens were evaluated via clinicopathological examination and all samples were classified according to the Tumor, Node, Metastasis staging system (American Joint Committee on Cancer, Chicago, IL, USA), preoperative prostate-specific antigen levels and Gleason score (22). Blood samples (5 ml) were collected from the arm of patients, then centrifuged for 5 min (3,500 × g at room temperature) obtain the serum. Finally, serum samples were detected with ADVIA Centaur XP Immunoassay System (Siemens AG, Munich, Germany) to determine the prostate-specific antigen levels. Clinicopathological data were presented in Table I. Each tumor sample or adjacent normal sample was divided into two sections; the first part was snapfrozen in liquid nitrogen and stored at −80°C, the other half was fixed in 4% paraformaldehyde for 24 h at room temperature (Beyotime Institute of Biotechnology, Haimen, China). Patients were also divided into high and low BRD4 expression groups based on the determined mean cut-off expression value of 2.66.
Table I.
Association of relative BRD4 expression with the clinicopathological factors of patients with prostate cancer.
| Factors | Patient characteristics | BRD4 expression
|
P-value | ||
|---|---|---|---|---|---|
| Low | High | Total | |||
| Age (years) | <70 | 11 | 10 | 21 | 0.264 |
| ≥70 | 9 | 16 | 25 | ||
| Preoperative PSA | <10 | 10 | 8 | 18 | 0.185 |
| ≥10 | 10 | 18 | 28 | ||
| Gleason score | <7 | 7 | 15 | 22 | 0.127 |
| ≥7 | 13 | 11 | 24 | ||
| Tumor stage | T1 | 12 | 8 | 20 | 0.047a |
| T2/T3 | 8 | 18 | 26 | ||
| Metastasis | Absence | 16 | 11 | 27 | 0.010a |
| Presence | 4 | 15 | 19 | ||
P<0.05. BRD4, bromodomain-containing protein 4; PSA, prostate-specific antigen.
Cell lines and reagents
The human prostate epithelial cell line RWPE1 and human prostate cancer cell lines, DU145, PC3 and LNCAP, employed in the present study were purchased from the American Type Culture Collection (Manassas, VA, USA). Human prostate cancer cells were maintained in RPMI-1640 culture medium with 10% heat-inactivated fetal bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). RWPE1 cells were maintained in keratinocyte-serum free medium (Gibco; Thermo Fisher Scientific, Inc.) containing human recombinant epidermal growth factor (5 ng/ml) and bovine pituitary extract (50 µg/ml). All cells were cultured in an incubator at 37°C and 5% CO2. JQ1 was purchased from Selleck Chemicals (50 mM, stock solution, cat. no. S7110, Houston, TX, USA), dissolved in dimethyl sulfoxide and stored at −80°C. DU145 and LNCAP cells were selected for subsequent analysis as these cell lines exhibited significantly higher expression levels of BRD4 than PC3 cells.
Short hairpin (sh)RNA construction and lentiviral infection
BRD4-targeted shRNA was expressed in a pGLV3/H1/green fluorescent protein (GFP) + Puro lentiviral vector via BamHI and EcoRI; vectors were constructed by Shanghai GenePharma Co., Ltd. (Shanghai, China). The shRNA sequence targeting BRD4 was as follows: 5′-GCTCAAGACACTATGGAAACA-3′, the shRNA sequence used as the negative control (sh-NC) was 5′-TTCTCCGAACGTGTCACGT-3′ (LV3-NC-GFP; Shanghai GenePharma Co., Ltd.). 293 cells (American Type Culture Collection) were cultured in 10 cm plates at a density of 5×105 cells per well with DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS in an incubator at 37°C and 5% CO2 for 48 h. Then lentiviral shRNA-containing plasmids were transfected into 293 cells to generate lentiviruses; the lentiviral supernatant containing the BRD4 shRNA was harvested and purified. DU145 and LNCAP cells were then infected by application of viral supernatant (multiplicity of infection 15 for DU145, multiplicity of infection 20 for LNCAP) in RPMI-1640 culture medium with 10% FBS. The cationic polymer Polybrene (Shanghai GenePharma Co., Ltd.) was added to facilitate transduction. After 24 h following transduction the medium was exchanged and cells were cultured for an additional 48 h. Stable cell lines were selected via the application of 1 µg/ml puromycin (Shanghai GenePharma Co., Ltd.).
RNA interference
Small interfering (si)RNAs targeting FOXO1 and p21, and a negative control (NC) were purchased from Wuhan Biossci Co., Ltd. (Wuhan, China). The target sequences were as follows: FOXO1 sense, 5′-CCAUGGACAACAACA GUAATT-3′ and antisense, 5′-UUACUGUUGUUGUCCAUG GTT-3′; p21 sense, 5′-CAGGCGGUUAUGAAAUUCATT-3′ and antisense, 5′-UGAAUUUCAUAACCGCCUGTT-3′; and NC sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and anti-sense, 5′-ACGUGACACGUUCGGAGAATT-3′. For knockdown experiments, DU145 and LNCAP cells were cultured in 6-well plates at a density of 2×105 cells per well for 24 h, and siRNA (50 nM for si-FOXO1 or si-NC, 30 nM for si-p21 or si-NC) transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to manufacturer protocols. After 6 h of incubation in an incubator at 37°C and 5% CO2, the FBS free RMPI-1640 medium was replaced and cells were cultured in RMPI-1640 medium containing 10% FBS for an additional 42 h at 37°C. For co-transfection assay, DU145 and LNCAP cells (a density of 2×105 cells per well) were cultured in 6-well plates for 24 h at 37°C, subsequently transfected with si-FOXO1 or si-p21or si-NC, then followed by transfection with sh-BRD4 or sh-NC as aforementioned; analysis was conducted following 24 h post-transfection and subsequent incubation for 48 h. Finally, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting assays were used to determine the efficiency of transfection.
BRD4 mRNA analysis in different cancer types
The Cancer Genome Atlas (TCGA) and Starbase version 2.0 was used to investigate the expression of BRD4; data from the Pan-Cancer Analysis Platform (http://starbase.sysu.edu.cn/panCancer.php) indicated that BRD4 was upregulated in numerous types of cancer (23,24).
Cell proliferation assay
DU145 and LNCAP cell lines were plated onto a 96-well plate at a density of 5,000 cells/100 µl per well and incubated overnight in an incubator at 37°C. Following JQ1 (1.92, 3.2, 9.6, 16, 48, 80, 240 and 400 nM, and 1.2 2, 6, 10, 30 and 50 µM) or DMSO treatment, shBRD4 or sh-NC transfection, 10 µl cell counting kit-8 (CCK-8) reagents (Dojindo Molecular Technology, Inc., Kumamoto, Japan) were added to each well according to the manufacturer's protocols, and cells were incubated in the dark for 2 h at 37°C. Untreated cells served as a control. A spectrophotometer was used to measure the absorbance of the generated optical density (OD) value at 450 nm. The half-maximal inhibitory concentration (IC50) values (15.9 µM for DU145, 175 nM for LNCAP) were determined using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA).
Colony formation assay
Cells were plated onto 6-well dishes. Following attachment, cells were treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP) for 48 h or transfected with shBRD4 as aforementioned. Cells were then harvested via 0.25% trypsinization at 37°C for 45 sec and re-suspended in culture medium at 37°C. Subsequently, 1,000 cells/per well were seeded in a 6-well plate and cultured in a drug-free medium for 10-14 days. Then, cells were washed with PBS, fixed with 100% methanol for 15 min, and stained with 0.5% crystal violet solution at room temperature for 15 min (Beyotime Institute of Biotechnology). A light microscope (magnification, ×100; Olympus Corporation, Tokyo, Japan) was used to count the number of positive colonies (>50 cells/colony). Observations were performed in triplicate.
Flow cytometry analysis of the cell cycle
Cells (2×105) were cultured in 6-well dishes. Following attachment, cells were treated with JQ1 (10, 20, 40 µM for DU145, 100, 200, 400 nM for LNCAP) for 48 h or transfected with shBRD4 as aforementioned. Cell suspensions were harvested using 0.25% trypsin for 45 sec at 37°C and placed in precooled 70% ethanol for fixation at −20°C overnight. Then, a ribozyme (50 µg/ml) and propidium iodide (50 µg/ml, PI; BD Biosciences, San Jose, CA, USA) were added to the cells, which were incubated in the dark at room temperature for 30 min. Samples were subsequently evaluated with a flow cytometer (BD FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA) and data were analyzed by ModFit LT 2.0 software (Verity Software House, Inc., Topsham, ME, USA).
Flow cytometry analysis of apoptosis
Cells (2×105) were plated in 6-well dishes. Following attachment, cells were treated with JQ1 (10, 20, 40 µM for DU145, 100, 200, 400 nM for LNCAP) for 48 h or transfected with shBRD4 as aforementioned. Cell suspensions were harvested via 0.25% trypsinization for 45 sec at 37°C without EDTA, washed with PBS, and resuspended in 250 ml binding buffer [Hangzhou Multisciences (Lianke) Biotech Co., Ltd., Hangzhou, China]. Cell suspensions were stained with 5 µl PI and 5 µl Annexin V-fluorescein isothiocyanate solution (BD Biosciences) and incubated at room temperature for 15 min in the dark. Finally, a flow cytometer (BD FACSCalibur; BD Biosciences) was used to quantify apoptotic cells. CellQuest 3.0 software (BD Biosciences) was used to analyze the data. The rate of apoptosis was obtained from three repeats, and determined with GraphPad Prism 6.
Wound healing assay
Cells (2×105) were seeded in 6-well dishes and incubated in complete medium (RPMI-1640 culture medium with 10% FBS) overnight. Cells were treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP) for 48 h or transfected with shBRD4 as aforementioned. A 200-µl sterile pipette tip was scraped across the wells and cell debris was removed via washing with PBS. The supernatant was subsequently replaced with serum-free medium (RPMI-1640 culture medium). Finally, the scratch wounds were analyzed at various time points (0, 24 and 48 h) under a microscope (magnification, ×200; Olympus Corporation).
Cell invasion assays
Cells (2×105) were seeded on a 6-well plate, treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP) for 48 h or transfected with shBRD4, and harvested via 0.25% trypsinization for 45 sec at 37°C. Cells were then re-suspended in serum-free RPMI-1640 culture medium. Subsequently, 1×105 cells/100 µl per well were inoculated on a Transwell chamber pre-coated with Matrigel in the top chamber at 37°C. In the lower chamber, 10% FBS-containing RPMI-1640 medium was used as an attractant. Cells on the top chamber membrane were removed with a cotton swab after 24 h of incubation at 37°C; invading cells in the lower side of the chamber were fixed using 4% paraformaldehyde for 15 min and stained with 0.5% crystal violet solution for 15 min at room temperature. Finally, cells were assessed under a light microscope (magnification, ×200; Olympus Corporation).
Immunohistochemistry
Tissues from clinical specimens and in vivo assays (described below) were fixed in 4% paraformaldehyde for 24 h at room temperature, embedded in paraffin and then sliced into 4-µm sections. The sections were then deparaffinized with xylene and rehydrated via a descending alcohol series (100, 95, 85 and 75%). The sections were incubated in Tris-EDTA Buffer (10 mM Tris Base, 1 mM EDTA solution, 0.05% Tween-20, pH 9.0) for 15 min at 95°C for antigen retrieval. While endogenous peroxidase activity was neutralized with a methanol/H2O2 solution. Subsequently, sections were incubated with rabbit monoclonal primary antibodies at 4°C overnight. Following incubation with horseradish peroxidase (HRP)-conjugated goat and anti-rabbit secondary antibodies at room temperature for 45 min (cat. no. GB23303; 1:200; Wuhan Servicebio Technology Co., Ltd., Wuhan, China). Finally, all sections were stained with 3,3-diaminobenzidine at room temperature for 3 min (Wuhan Servicebio Technology Co., Ltd.) and visualized via light microscopy (magnifications, ×400 and 1,000; Olympus Corporation). For quantitation, Image-Pro Plus software (ver 6.0; Media Cybernetics, Inc., Rockville, MD, USA) was used to determine the average integrated OD (IOD) obtained by analyzing three fields per slide. The relative average IOD was obtained from the average IODs of cancerous prostate tissue divided by the average IOD of adjacent normal tissue specimens. For the analysis of animal tissue, the average IOD was obtained from the average IODs of the JQ1 treatment or sh-BRD4 groups divided by the average IOD of the control or sh-NC groups, respectively. The primary antibodies employed in the present study, including BRD4 (cat. no. ab128874; 1:200), Ki-67 (cat. no. ab16667; 1:100), p21 (cat. no. ab109520; 1:100) for Immunohistochemistry were obtained from Abcam (Cambridge, UK), and FOXO1 antibody (cat. no. 2880; 1:100) were obtained from Cell Signaling Technology, Inc., Danvers, TX, USA).
RT-qPCR assay
Total RNA from prostate cell lines and tissue specimens was isolated using TRIzol® reagent (Thermo Fisher Scientific, Inc.). Spectrophotometry was used to measure the concentration and purity of RNA, while the PrimeScript RT Reverse transcriptase Reagent Kit (Takara Bio, Inc., Otsu, Japan) was applied to synthesize complementary DNA according to the manufacturer protocols. Briefly, RNA (2 µg) was incubated for 15 min at 37°C and for 5 sec at 85°C. qPCR analysis was conducted using the SYBR Premix Ex Taq kit (Takara Bio, Inc.) and the StepOnePlus Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The conditions of PCR were listed as follows: 95°C for 10 sec, then following 40 cycles of 95°C for 5 sec, 58°C for 20 sec. Expression levels were normalized to that of GAPDH; the relative expression of genes was determined using the 2−ΔΔCq method (25). All assays were performed in triplicate. Primers were synthesized by Sangon Biotech (Shanghai, China) and were presented in Table II.
Table II.
Reverse transcription-quantitative polymerase chain reaction primer sequences.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| BRD4 | TGAGTCGGAGGAAGAGGACAAGTG | CGCAGTGTGGACGGCTTCAG |
| FOXO1 | TGTCCTACGCCGACCTCATCAC | GCACGCTCTTGACCATCCACTC |
| p21 | GCTGAGCCGCGACTGTGATG | CCTCCAGTGGTGTCTCGGTGAC |
| c-Myc | CGAGGAGAATGTCAAGAGGCGAAC | GCTTGGACGGACAGGATGTATGC |
| Bcl-2 | GAACCGGCACCTGCACACC | AGAGTCTTCAGAGACAGCCAGGAG |
| p53 | ACCGGCGCACAGAGGAAGAG | GCCTCATTCAGCTCTCGGAACATC |
| GAPDH | CAACGTGTCAGTGGTGGACCTG | GTGTCGCTGTTGAAGTCAGAGGAG |
Bcl-2, B-cell lymphoma 2; BRD4, bromodomain-containing protein 4; FOXO1, forkhead box protein O1.
Western blotting
Tissues and cells were lysed with radioimmunoprecipitation assay buffer-containing PMSF (Beyotime Institute of Biotechnology) to extract total proteins. A Bicinchoninic Acid assay kit (Beyotime Institute of Biotechnology) was used to determined protein concentration. Equal amounts of proteins (20 µg) were separated via 10% polyacrylamide gel electrophoresis (Beyotime Institute of Biotechnology), and proteins were transferred onto polvinylidene difluoride (PVDF) membranes (Wuhan Servicebio Technology Co., Ltd.). Blocking was performed in Tris-buffered saline and 0.05% Tween-20 containing 5% nonfat milk for 1.5 h at 37°C. The PVDF membranes were washed with TBST three times, then incubated with rabbit polyclonal primary antibodies at 4°C overnight. Subsequently, the membrane was incubated with HRP conjugated secondary antibodies (1:10,000) for 1 h at 37°C. Lastly, an enhanced chemiluminescence kit [Hangzhou Multisciences (Lianke) Biotech Co., Ltd., Hangzhou, China] was used to analyze the membranes according to the instruction of manufacturer. Protein bands were scanned with Molecular Imager® ChemiDoc™ XRS+ Imaging System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The optical densities a selection of bands were detected using ImageJ software 1.45 (National Institutes of Health, Bethesda, MD, USA). All assays were performed in triplicate. The primary antibodies of BRD4 (ab128874; 1:500), p21 (cat. no. ab109520; 1:1,000), c-Myc (cat. no. ab39688; 1:500), Bcl-2 (cat. no. ab182858; 1:1,000) and GAPDH (cat. no. ab9485; 1:2500) were purchased from Abcam. FOXO1 (cat. no. 2880; 1:1,000), p53 (cat. no. 2527S; 1:1,000) and cyclin D1 (cat. no. 2978S; 1:1,000) antibodies were purchased from Cell Signaling Technology, Inc. Goat and anti-rabbit secondary antibodies were obtained from Wuhan Boster Biological Technology, Ltd. (Wuhan, China).
Tumor xenografting and in vivo treatment assay
All experimental procedures were approved by the Ethics Committee of Wuhan University and the animals received humane care. All 4-5-week-old male BALB/c nude mice (Beijing Vital River Laboratory Animal Technology, Co., Ltd., Beijing, China) were housed in a pathogen-free environment at the Animal Center of Renmin Hospital of Wuhan University. Briefly, a total of 40 mice (15-18 g) were housed in a pathogen-free environment (22-27°C; humidity, 40-70%, under a 12 h light/dark cycle with free access to sterilized feed and water). DU145 cells were prepared by suspending 1×107 cells in 200 µl PBS, and subcutaneously injected into the left upper extremity of mice. Once the tumor volume was measurable (100 mm3), animals were randomized into treated or negative control groups (10 mice/group). For the drug treated groups, DMSO-containing JQ1 was diluted with physiological saline to obtain a final solution of 5 mg/ml. Each mouse was administered an intraperitoneal injection of JQ1, 50 mg/kg per day on day 9. Mice of the control group were administered an equal amount of physiological saline containing 5% DMSO per day. Mice were treated with JQ1 or vehicle for 19 days. In the BRD4 knockdown group, 1×107 stable shRNA BRD4-expressing LNCAP or control cells (sh-NC) in 200 µl PBS were implanted into the same anatomical sites of mice (10 mice/group). The tumor size of each mouse were followed-up using Vernier calipers every 4 days and tumor volume was approximated as 0.5 × width2 × length (mm3). After 4 weeks, all animals were sacrificed, the experiment was terminated, and tumors were excised, images and weighed.
Statistical analysis
GraphPad Prism software (version 6) or SPSS 20.0 (IBM Corp., Armonk, NY, USA) were used for statistical analysis. Data are presented as the mean ± standard deviation. All experiments were performed in triplicate. A Student's t-test, χ2 or one-way analysis of variance were used to evaluate whether statistically significant differences were present. A Bonferroni post-hoc test for multiple comparisons was also applied. P<0.05 was considered to indicate a statistically significant difference.
Results
Overexpression of BRD4 in PCa cell lines and tumor specimens
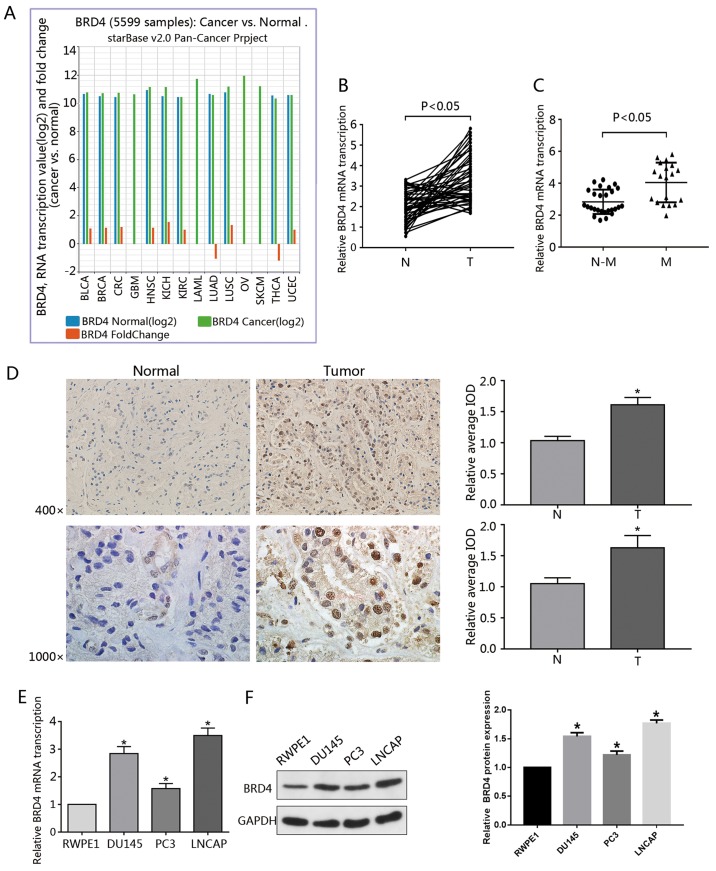
In the present study, the role served by BRD4 in other types of cancer was analyzed. Data from the Pan-Cancer Analysis Platform indicated that BRD4 was upregulated in numerous types of cancer (Fig. 1A). Additionally, 46 pairs of prostate cancer specimens and surrounding normal tissue were collected and evaluated via RT-qPCR. The results revealed that, compared with the surrounding normal prostate tissue, the expression of BRD4 mRNA was significantly upregulated in PCa samples (Fig. 1B). In addition, BRD4 expression was positively associated with metastasis (Fig. 1C). Data obtained from immunohistochemical analysis revealed BRD4 to be primarily expressed in the nucleus of PCa cells. Furthermore, tumor samples were stained to a higher degree than in adjacent normal tissue as presented by the IOD values (Fig. 1D), consistent with previous findings (20). The present study also observed that, compared with RWPE1 cells, the mRNA expression levels of BRD4 were significantly increased in PCa cell lines (DU145, PC3 and LNCAP); BRD4 protein expression levels were significantly upregulated (Fig. 1E and F). To further investigate the association between BRD4 expression and the clinical characteristics of patients with PCa, the clinical data obtained from 46 patients with PCa were analyzed. The results revealed that the levels of BRD4 expression exhibited a significant association with the tumor clinical stages and metastasis of PCa (Table I).
Figure 1.
Overexpression of BRD4 in PCa cell lines and tumor specimens. (A) Data regarding the expression of BRD4 in 14 types of cancer was gathered from the Pan-Cancer Analysis Platform in TCGA Data Portal (starBase version 2.0). (B) RT-qPCR was used to analyze the transcription of BRD4 mRNA in human cancerous and paired normal prostate tissues from 46 patients with PCa. (C) Expression of BRD4 mRNA was significantly higher in patients with M-PCa compared with N-M PCa specimens. (D) Immunohistochemical analysis of BRD4 in PCa samples and surrounding normal prostate tissue. Magnification, ×400 and ×1,000. The average IOD was analyzed by Image-Pro Plus software. *P<0.05 vs. N prostate tissues. (E and F) mRNA and protein expression levels of BRD4 in RWPE1 cells (human prostate epithelial cell line), and DU145, PC3 and LNCAP cells (human prostate cancer cell lines) were evaluated by RT-qPCR and western blotting. *P<0.05 vs. RWPE1 cells. Data are presented as the mean ± standard deviation. Experiments were performed in triplicate. BRD4, bromodomain-containing protein 4; IOD, integrated optical density; M, metastatic; N, normal; N-M, non-metastatic; PCa, prostate cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
BRD4 inhibition suppresses cellular proliferation of PCa cell lines
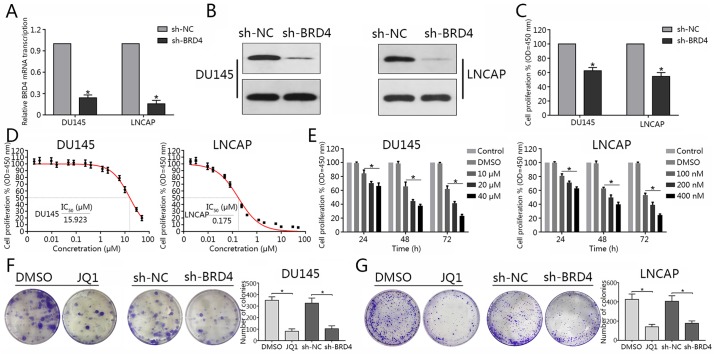
To determine the effects of BRD4 on PCa cells, BRD4-targeted shRNA was constructed and transduced into DU145 and LNCAP cells. RT-qPCR and western blotting assays were used to determine BRD4 expression in PCa cell lines. The results revealed that BRD4 mRNA expression was significantly decreased in DU145 and LNCAP cells compared with in the control, following transduction with BRD4-targeted shRNA; the protein expression levels of BRD4 were notably downregulated in the BRD4 knockdown group compared with in the control (Fig. 2A and B). Cell proliferation was assessed via a CCK-8 assay. The findings suggested that inhibition of BRD4 via shRNA significantly attenuated cell proliferation of DU145 and LNCAP cells compared with in the controls (Fig. 2C). In addition, the present study reported that treatment with JQ1 decreased cell proliferation in dose-and time-dependent manners (Fig. 2E). In addition, the IC50 value of JQ1 in each cell line was determined (Fig. 2D). The colony formation capacity of DU145 and LNCAP cells treated with JQ1 or transfected with shBRD4 was evaluated. The results revealed that BRD4 inhibition significantly suppressed colony formation compared with in the negative control (Fig. 2F and G). The findings of the present study suggested that inhibition of BRD4 suppressed the proliferative ability of DU145 and LNCAP cells.
Figure 2.
BRD4 inhibition suppresses cellular proliferation of prostate cancer cell lines. (A and B) Reverse transcription-quantitative polymerase chain reaction and western blotting were conducted to determine transduction efficiency of DU145 and LNCAP cells transduction with shBRD4. (C) Proliferative ability of DU145 and LNCAP cells was assessed via a CCK-8 assay following shBRD4 transduction. (D) After 72 h following JQ1 treatment (1.92, 3.2, 9.6, 16, 48, 80, 240 and 400 nM, and 1.2 2, 6, 10, 30 and 50 μM), the CCK-8 assay was performed to assess cellular proliferation; and the corresponding IC50 values were quantified. (E) Proliferation of DU145 and LNCAP cells following treatment with different concentrations of JQ1 at 24, 48 and 72 h was assessed via a CCK-8 assay. *P<0.05 vs. DMSO and control group.. (F and G) DU145 and LNCAP cells were transduction with shBRD4 or treated with JQ1 at their respective IC50 concentrations (15.9 µM for DU145, 175 nM for LNCAP) and representative images of colony formation were obtained. The number of colonies was subsequently calculated. Experiments were performed in triplicate. *P<0.05. BRD4, bromodomain-containing protein 4; CCK-8, cell counting kit-8; DMSO, dimethyl sulfoxide; IC50, half-maximal inhibitory concentration; NC, negative control; OD, optical density; sh, short hairpin RNA.
Inhibition of BRD4 induces cell cycle arrest and apoptosis, and mitigates the invasion of PCa cell lines
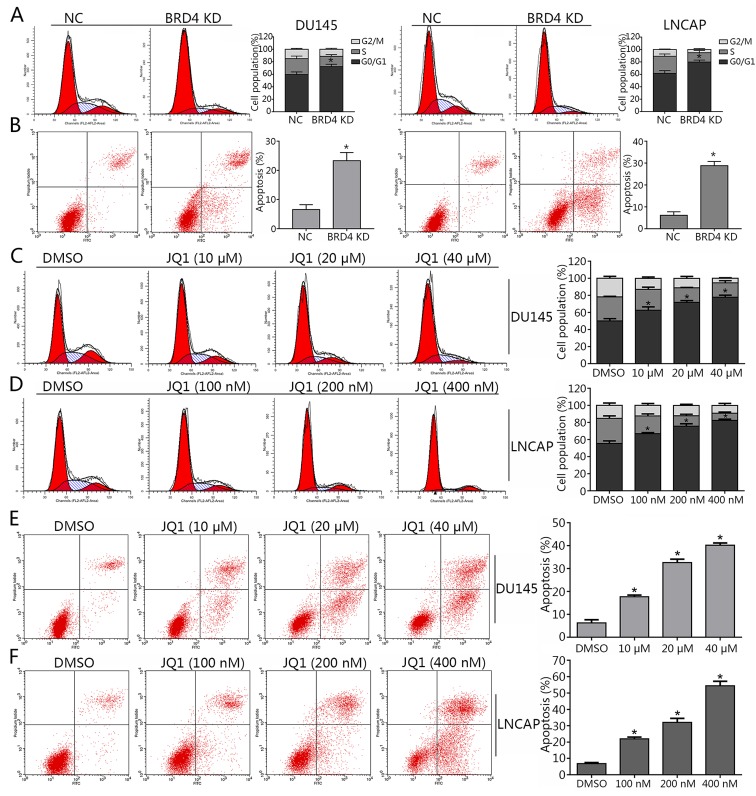
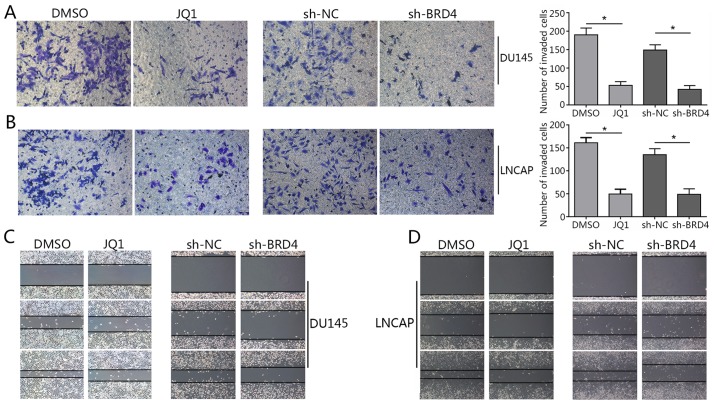
As BRD4 was suggested to exhibit pro-cancer effects in PCa cells in (Figs. 1 and 2), the present study investigated whether BRD4 inhibition may induce cell cycle arrest in DU145 and LNCAP cells via JQ1 treatment or transduction with shRNA. The results of cell cycle analysis revealed that the majority of shBRD4-treated cells were in the G0/G1 phase; S and G2/M phase cells were markedly less apparent compared with in the control (Fig. 3A). In addition, the apoptosis of cells in different groups was analyzed via flow cytometry. The results demonstrated that BRD4 inhibition via shRNA transduction significantly increased apoptosis compared with in the control (Fig. 3B). Cells treated with various concentrations of JQ1 exhibited a significant increase in the number of cells in the G0/G1 phase compared with in the control (Fig. 3C and D). The apoptosis of JQ1-treated cells increased in a dose-dependent manner compared with in the control (Fig. 3E and F). The Transwell assay revealed a significant decrease in invading cells treated with JQ1 or transfected with shBRD4 compared with in the corresponding control (Fig. 4A and B). The wound healing assay demonstrated that BRD4 inhibition mitigated cell migration in PCa cells compared with in the corresponding controls (Fig. 4C and D). The findings of the present study indicated that inhibition of BRD4 induced cell cycle arrest and apoptosis, and mitigated the invasion of PCa cells.
Figure 3.
Inhibition of BRD4 induces cell cycle arrest and apoptosis in PCa cell lines. (A) Cell cycle arrest in DU145 and LNCAP cells transduction with shBRD4 was detected by flow cytometry. Percentages of each cell cycle phase were calculated. (B) Apoptosis of DU145 and LNCAP cells following transduction with shBRD4 or NC shRNA was examined by flow cytometry. The rate of apoptosis of PCa cells was calculated. (C and D) Cell cycle arrest in DU145 cells treated with JQ1 (10, 20 and 40 µM) or DMSO, and LNCAP cells treated with JQ1 (100, 200 and 400 nM) or DMSO was determined by flow cytometry. The percentages of each cell cycle phase were calculated. (E and F) Apoptosis of DU145 cells and LNCAP cells treated with DMSO or JQ1 was examined by flow cytometry. The rate of apoptosis of PCa cells was calculated. Data are presented as the mean ± standard deviation. *P<0.05 vs. NC or DMSO group. BRD4, bromodomain-containing protein 4; DMSO, dimethyl sulfoxide; NC, negative control; PCa, prostate cancer.
Figure 4.
Inhibition of BRD4 mitigates the invasion of prostate cancer cell lines. (A and B) Invasive ability of DU145 and LNCAP cells treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP) or transduction with shBRD4 was assessed via Transwell invasion assays. The number of invading cells was calculated in three random fields. Magnification, ×200. *P<0.05 vs. DMSO-treated or NC group. (C and D) Wound healing assays were performed to evaluate the migration of DU145 and LNCAP cells treated with JQ1 or transduction with shBRD4. Magnification, ×200. Data are presented as the mean ± standard deviation. DMSO group. BRD4, bromodomain-containing protein 4; DMSO, dimethyl sulfoxide; NC, negative control; short hairpin RNA.
Reduction of c-Myc expression by BRD4 inhibition involves p21
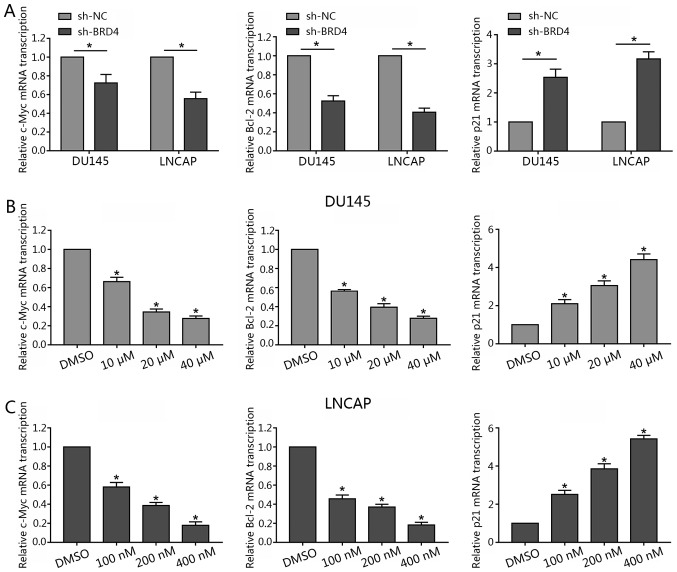
Recent studies have reported c-Myc to be a downstream effector of BRD4 (19,26). To determine whether c-Myc expression is affected by BRD4 in DU145 and LNCAP lines, these cells were transfected with shBRD4 or treated with JQ1 in the present study. The results revealed that BRD4 inhibition via JQ1 treatment or shBRD4 transduction significantly decreased c-Myc mRNA expression levels compared with in the control (Fig. 5A and B). Additionally, as BRD4 inhibition was also reported to induce cell cycle arrest and apoptosis (26,27). Therefore, Bcl-2 was investigated in the present study. The results revealed that the suppression of BRD4 significantly inhibited the expression levels of Bcl-2 mRNA compared with in the control. Of note, the present study observed that BRD4 inhibition via JQ1 treatment or shBRD4 transduction significantly promoted p21 mRNA expression (Fig. 5).
Figure 5.
Inhibition of BRD4 decreases c-Myc and Bcl-2 mRNA transcription, but increases p21 mRNA transcription in prostate cancer lines. (A) DU145 and LNCAP cells were transfected with either shBRD4 or NC shRNA, and the relative mRNA transcription levels of c-Myc, Bcl-2 and p21 were analyzed by RT-qPCR. (B and C) DU145 cells were treated either with JQ1 (10, 20 or 40 µM) or DMSO, and LNCAP were treated with either JQ1 (100, 200 or 400 nM) or DMSO for 48 h; the relative mRNA transcription levels of c-Myc, Bcl-2 and p21 and were analyzed by RT-qPCR. Data are presented as the mean ± standard deviation. *P<0.05 vs. NC or DMSO group. BRD4, bromodomain-containing protein 4; DMSO, dimethyl sulfoxide; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; sh, short hairpin RNA.
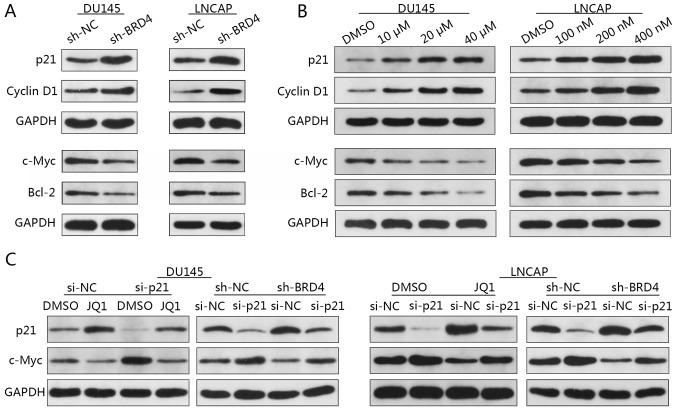
Western blotting revealed that the expression levels of p21 and cyclin D1 were notably increased in response to BRD4 inhibition; c-Myc and Bcl-2 protein expression levels were markedly decreased compared with in the control (Fig. 6A). In addition, the expression levels of p21 and cyclin D1 markedly increased in a dose-dependent manner in response to JQ1 treatment; c-Myc and Bcl-2 protein expression notably decreased in a dose-dependent manner (Fig. 6B).
Figure 6.
Reduction of c-Myc expression via BRD4 inhibition involves p21. (A) DU145 and LNCAP cells were transduced with shBRD4 or shRNA NC, and the relative expression levels of c-Myc, Bcl-2, p21 and cyclin D1 were analyzed by western blotting. (B) DU145 cells were treated either with JQ1 (10, 20 or 40 µM) or DMSO, and LNCAP cells were treated with JQ1 (100, 200 or 400 nM) or DMSO for 48 h. The relative expression levels of c-Myc, Bcl-2, p21 and cyclin D1 were analyzed by western blotting. (C) Relative p21 and c-Myc expression in DU145 and LNCAP cells. DU145 cells were transfected with either p21 siRNA or NC siRNA, and treated with JQ1 (15.9 µM) or DMSO; LNCAP cells transfected with p21 siRNA or NC siRNA were treated with JQ1 (175 nM) or DMSO. Bcl-2, B cell lymphoma-2; BRD4, bromodomain-containing protein 4; DMSO, dimethyl sulfoxide; NC, negative control; sh, short hairpin RNA; siRNA, small interfering RNA.
Previous studies have reported that increased p21 and decreased c-Myc expression were concomitant upon BRD4 inhibition (28,29). To determine whether p21 and c-Myc are associated in PCa cells, DU145 and LNCAP cells were firstly transfected with p21-specific siRNA then followed by transfected with sh-BRD4. Western blotting revealed increased c-Myc expression when p21 was inhibited (Fig. 6C). Additionally, a reduction in the expression of c-Myc via BRD4 inhibition was notably abrogated by the knockdown of p21 in DU145 and LNCAP cells compared with in the controls (Fig. 6C). These findings suggested c-Myc to be a downstream target of p21 in PCa. To investigate whether reductions in c-Myc expression are mediated via p21, DU145 and LNCAP cells transfected with p21-specific siRNA then were treated with JQ1. Western blotting demonstrated that treatment with JQ1 resulted in c-Myc downregulation, whereas p21 inhibition-induced upregulation of c-Myc was abrogated by treatment with JQ1 (Fig. 6C). These findings suggested that the reduction of c-Myc expression via BRD4 inhibition may involve p21, and that p21 is an upstream regulator of c-Myc, which may be directly affected by BRD4 inhibition.
Induction of p21 via the inhibition of BRD4 is mediated by FOXO1
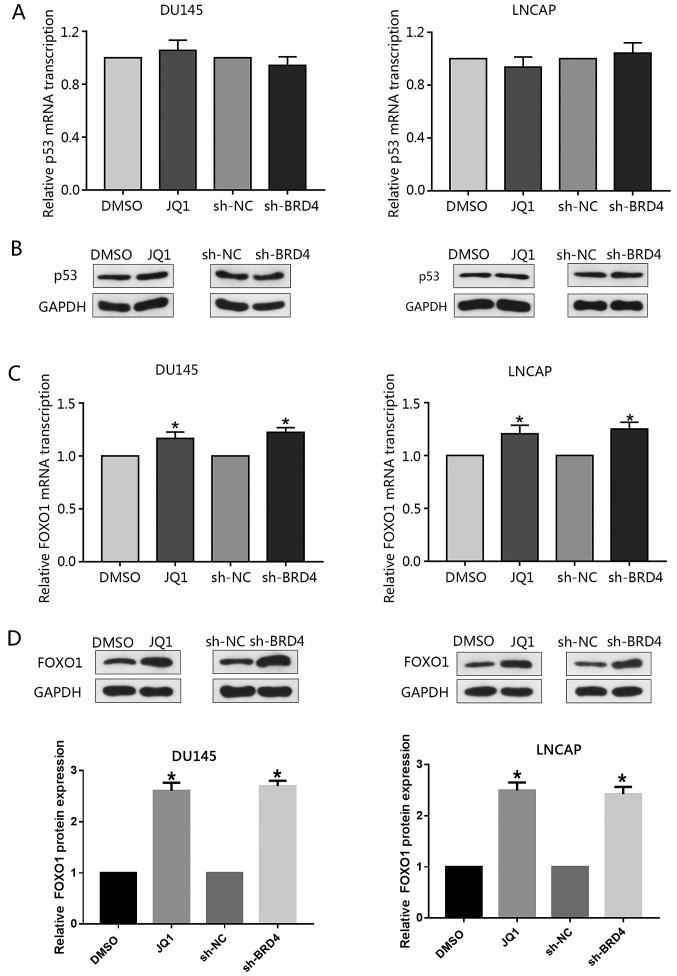
The tumor-suppressor p53 is a well-reported upstream regulator of p21 (30); however, the present study investigated whether knockdown of BRD4 or treatment with JQ1 may affect the expression p53. RT-qPCR or western blotting did not reveal any notable alterations in the expression levels of p53 following inhibition of BRD4 (Fig. 7A and B). Previous studies have reported FOXO1 to inhibit PCa tumorigenesis and that FOXO1 binds to the p21 promoter region resulting in cell cycle arrest (31,32). Therefore, the present study investigated whether BRD4 inhibition affected the expression of FOXO1 in PCa cells. The findings demonstrated that inhibition of BRD4 only moderately increased FOXO1 mRNA expression levels compared with in the control; the expression levels of FOXO1 protein were signficantly increased in response to BRD4 downregulation; similar findings were observed in response to JQ1 treatment (Fig. 7C and D). The present study proposed that the stability of the FOXO1 protein may be affected by post-transcriptional modifications, such as post-BRD4 knock-down phosphorylation.
Figure 7.
Induction of p21 by BRD4 inhibition is not mediated by p53. (A and B) Relative mRNA transcription of p53 was analyzed by RT-qPCR and western blotting. Knockdown of BRD4 or treatment with JQ1 did not notably affect the expression of p53. Cells were treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP) or transduced with shBRD4. Data are presented as the mean ± standard deviation. Experiments were performed in triplicate. (C and D) Cells were treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP) or transduced with shBRD4; the relative expression of FOXO1 was analyzed by RT-qPCR or western blotting. FOXO1 transcriptional levels were significantly increased by BRD4 inhibition, while that of FOXO1 protein expression were notably increased. Data are presented as the mean ± standard deviation. Experiments were performed in triplicate. *P<0.05 vs. NC or DMSO group. BRD4, bromodomain-containing protein 4; DMSO, dimethyl sulfoxide; FOXO1, forkhead box protein O1; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; sh, short hairpin RNA.
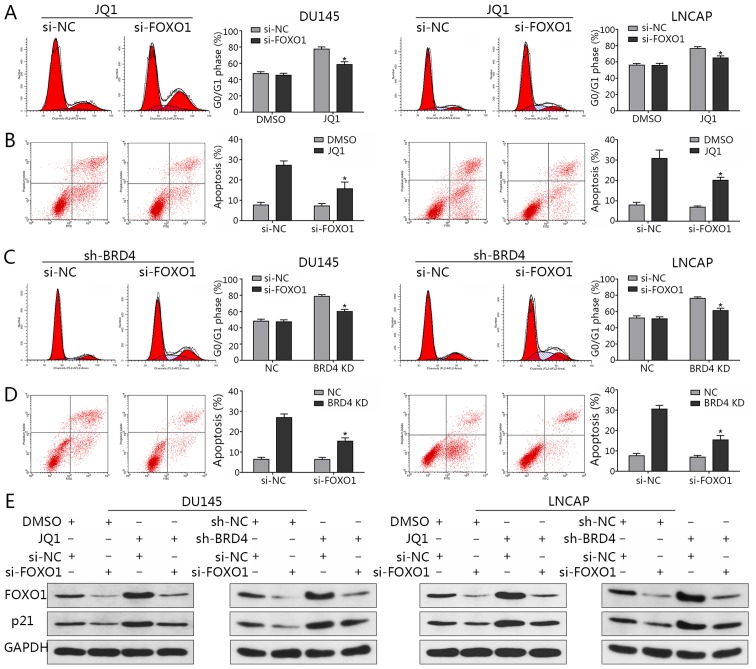
Following the knockdown of FOXO1 expression via siRNA transfection, flow cytometry was applied to analyze the cell cycle and evaluate the rates of apoptosis. The observations of the present study revealed that the distribution of cells in the G0/G1 phase induced by JQ1 treatment were significantly decreased compared with in the JQ1-treated control (Fig. 8A); the percentage of apoptosis was significantly reduced when FOXO1 was downregulated compared with in the JQ1-treated control (Fig. 8B). Similar findings were observed in the presence of BRD4 inhibition via shBRD4 transduction (Fig. 8C and D). In addition, the results indicated that FOXO1 downregulation led to a reduction in p21 expression, while an induction of p21 expression via BRD4 inhibition was abrogated by the knockdown of FOXO1 (Fig. 8E). This in turn suggested that FOXO1 may be a transcription factor that induces p21 when BRD4 is suppressed; the induction of p21 by JQ1 treatment was also abrogated by FOXO1 inhibition (Fig. 8E). The results indicated that the induction of p21 by BRD4 inhibition may not be mediated by p53, but by FOXO1.
Figure 8.
Induction of p21 by inhibition of BRD4 is mediated by FOXO1. (A) Flow cytometry assays revealed a reduction in cell cycle arrest at the G0/G1 phase when cells transfected with FOXO1-specific siRNA were treated with JQ1 (15.9 µM for DU145, 175 nM for LNCAP). *P<0.05 vs. the NC group treated with JQ1. (B) Flow cytometry indicated a significant reduction in apoptosis when cells transfected with FOXO1-specific siRNA were treated with JQ1. *P<0.05 vs. the NC group treated with JQ1. (C) Flow cytometry assays revealed a reduction in cell cycle arrest at the G0/G1 phase when DU145 and LNCAP cells were co-transfected with shBRD4 and FOXO1-specific siRNA. *P<0.05 vs. cells co-transfected with shBRD4 and si-NC group. (D) Flow cytometry indicated a marked reduction in apoptosis when DU145 and LNCAP cells were co-transfected with shBRD4 and FOXO1-specific siRNA. *P<0.05 vs. cells co-transfected with shBRD4 and si-NC group. (E) Relative protein expression of p21 and FOXO1 in DU145 and LNCAP cells co-transfected with shBRD4 and FOXO1-specific siRNA. DU145 and LNCAP cells transfected with si-FOXO1 or si-NC were treated with JQ1 or DMSO; relative expression levels of p21 and FOXO1 in cells were detected by western blotting. BRD4, bromodomain-containing protein 4; DMSO, dimethyl sulfoxide; FOXO1, forkhead box protein O1; NC, negative control; sh, short hairpin RNA; siRNA, small interfering RNA.
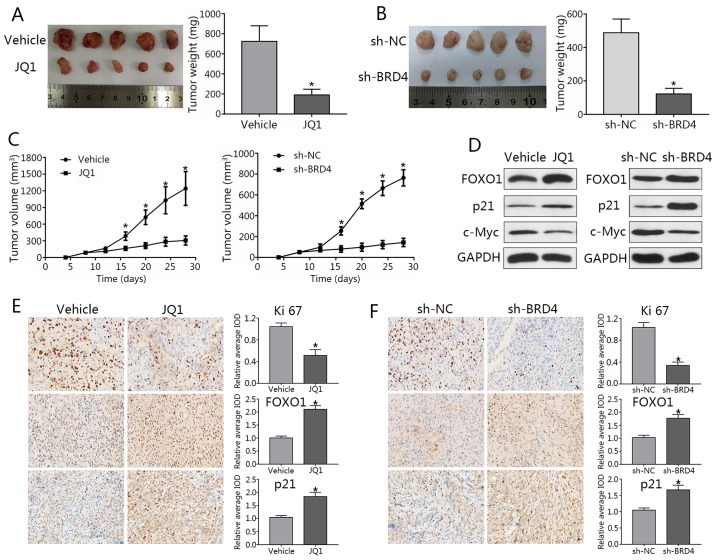
Knockdown of BRD4 delays tumor growth in PCa mouse models
DU145 cells were subcutaneously injected into male nude mice. When tumor sizes were palpable, mice were randomized into JQ1 treatment or NC groups. JQ1-treated mice exhibited a significant reduction in tumor volumes compared with in the control (Fig. 9A). To study effects of BRD4 knockdown, stable LNCAP cells transfected with shBRD4 or control were injected into mice. Nude mice injected with shBRD4-transfected cells exhibited decreased tumor volumes and weights compared to the control group; a significant decrease in tumor weights was observed in the JQ1 treatment group and shBRD4-transfected cells compared with in the control (Fig. 9B and C). In addition, metastatic lymph nodes or distal invasion were not detected in any of the groups at the termination of the experiment. Western blotting demonstrated that FOXO1 and p21 expression levels were markedly upregulated, while that of c-Myc were downregulated after BRD4 inhibition (Fig. 9D). Immunohistochemical staining revealed the expression of Ki-67 in the JQ1 treatment and shBRD4 groups to be significantly lower than that of the control group, while FOXO1 and p21 expression were significantly increased compared with the control group (Fig. 9E and F). These findings were consistent with the results of in vitro analysis in the present study. Finally, a working model to demonstrate the possible mechanism of cell cycle arrest and apoptosis in PCa cells as induced by BRD4 inhibition was generated (Fig. 10).
Figure 9.
Knockdown of BRD4 delays tumor growth in prostate cancer mouse models. (A) Image of tumors collected from mice. Mice were treated with JQ1 or vehicle at day 9 post-seeding. Four weeks later, mice were sacrificed, and tumors were excised. Weights of tumors grown in mice were assessed and analyzed. *P<0.05 vs. NC group. (B) Stable LNCAP cells transduced with shBRD4 or negative control were implanted into mice. (C) Mouse tumor volume curve as in response to JQ1 treatment or shBRD4 transduction. *P<0.05 vs. NC group. (D) Expression of FOXO1, p21 and c-Myc in xenograft tumors was assessed by western blotting. (E and F) Immunohistochemical analyses of Ki-67, FOXO1, and p21 in xenograft specimens. *P<0.05 vs. NC. The average IOD was analyzed by Image-Pro Plus software. Magnification, ×400. Data are presented as the mean ± standard deviation. BRD4, bromodomain-containing protein 4; FOXO1, forkhead box protein O1; IOD, integrated optical density; NC, negative control; si, small interfering RNA; sh, short hairpin RNA.
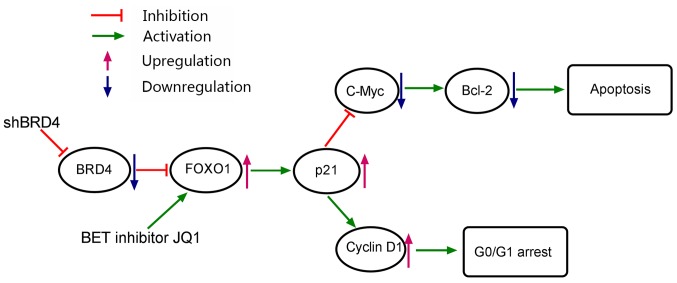
Figure 10.
Mechanistic model of cell cycle arrest and apoptosis of prostate cancer cells as induced by BRD4 inhibition. Bcl-2, B-cell lymphoma-2; BRD4, bro-modomain-containing protein 4; FOXO1, forkhead box protein O1; sh, short hairpin RNA.
Discussion
The prevention of PCa progression remains difficult to achieve; the targeting of androgen receptor signaling remains the treatment of choice in advanced stages of this disease (33). Enzalutamide, the novel nonsteroidal androgen receptor inhibitor, has been approved for the treatment of patients with castrate-resistant PCa at present (34,35). Unfortunately, the efficacy of enzalutamide is also limited. Several studies have reported that dysregulation of BRD4 markedly influences tumor growth and progression (18,36); the biological functions of BRD4 in PCa require further investigation for the development of potential therapeutic strategies.
Aberrant expression of BRD4 was confirmed in numerous types of cancers (11,36). For example, the expression of BRD4 was observed to be upregulated in kidney cancer and exerted a pro-oncogenic function in this particular disease (11). In squamous carcinoma of the skin, BRD4 was reported to be upregulated compared with normal skin keratinocytes and fibroblasts, with modeled overexpression of BRD4 promoting cell proliferation (36). In the present study, the expression and roles served by BRD4 in PCa were determined. In accordance with previous findings, the present study revealed that BRD4 expression was significantly increased in PCa samples compared with in adjacent normal prostate tissue (20). In addition, high levels of BRD4 expression were positively associated with clinical stage and metastasis in the present study. These findings indicated that BRD4 protein may be closely associated with the initiation of PCa and exerts cancer-promoting functions in PCa. Inhibition of BRD4 may therefore become a novel therapeutic strategy in the management of this disease.
The present study reported that inhibition of BRD4, via shRNA transduction or JQ1 treatment, decreased cell proliferation, promoted cell cycle arrest and induced the apoptosis of PCa cells; BRD4 inhibition also impaired tumor growth in mice. Treatment with JQ1 or knockdown of BRD4 significantly inhibited cell invasion and migration abilities of PCa cells. Therefore, knockdown of BRD4 may be an additional therapeutic approach for patients with PCa.
A previous study reported JQ1 to disrupt the interactions between BRD4 and the N-terminal domain of androgen receptors (21); however, the molecular mechanisms underlying the anticancer effects of BRD4 inhibition remain unknown. BRD4 has been observed to specifically regulate a series of genes, including c-Myc; c-Myc, in turn, regulates numerous cellular functions, including cell growth, survival and apoptosis (37). Overexpression of c-Myc has also been detected in numerous types of cancer, such as PCa (38). Downregulation of c-Myc expression resulted in decreased cell viability or apoptosis, via the reduction of Bcl-extra large and Bcl-2 expression (39). The present study reported that BRD4 inhibition suppressed PCa cell proliferation by decreasing the expression of c-Myc, in vitro and in vivo, and that reductions c-Myc expression were accompanied with the downregulation of Bcl-2. The results of the present study suggested that BRD4 inhibition may be associated with the downregulation of Bcl-2.
Induction of p21 by BRD4 inhibition has been demonstrated in other cancer cell types, including thyroid cancer and glioblastoma cell lines (27,40), with p21 serving as an important downstream gene affected by JQ1 treatment (27,28,40). p21, a member the CIP/kip family of cyclin-dependent kinase inhibitors, serves a major role not only as a regulator of the cell-cycle, but also as a mediator of apoptosis and transcription (41). In addition, p21 directly interacts with and induces cyclin D1 associated-CDK4 and CDK6 expression, thereby inhibiting G1/S progression (42). In the present study, it was reported that following BRD4 inhibition, p21 and cyclin D1 expression levels were significantly increased in PCa cells, which indicated a mechanism by which JQ1 inhibits cell growth by suppressing cell cycle progression.
Recent studies have reported c-Myc to be critical in the suppression of p21 transcription (41,43). To determine whether c-Myc expression is associated with p21 levels following BRD4 inhibition, the expression of p21 was downregulated in the present study, which suggested p21 to regulate the expression of c-Myc. In addition, p21 was proposed to be the upstream regulator of c-Myc and may be directly affected by BRD4 inhibition in the present study. A recent study indicated p21 as a critical downstream effector of wildtype p53 (30); however, the present study reported that the expression levels of p53 protein were notably altered in response to BRD4 inhibition, which may be due to DU145 and LNCAP cell lines harboring a mutant p53; however, further investigation is required.
FOXO1, an important member of the FOX transcription factor family, was observed to suppress tumor formation by activating numerous target genes (31). Imatinib, a tyrosine kinase inhibitor, upregulated the expression of FOXO1, and induced cell cycle arrest and apoptosis in chronic myelogenous leukemia (44). Similarly, the present study reported that knockdown of BRD4 induced FOXO1 expression and demonstrated that FOXO1 is required for the regulation of cell apoptosis and cycle arrest. Additionally, FOXO1 was determined to be the upstream regulator of p21 and was directly influenced by BRD4 inhibition in the present study. BRD4 regulates gene expression (18); however, the mechanism underlying the inhibition of BRD4 upregulates FOXO1 remains unclear. In the present study, FOXO1 transcriptional levels were only moderately increased by BRD4 inhibition, while that of FOXO1 protein expression were notably increased. The present study proposed that the stability of the FOXO1 protein may be affected by post-transcriptional modifications, such as post-BRD4 knockdown phosphorylation; however, further investigation is required to elucidate these interactions. The findings of the present study suggested that BRD4 serves a critical role in the initiation of PCa and that the bromodomain inhibitor JQ1 should be further investigated as a potential therapeutic agent in this disease. In conclusion, the present study determined BRD4 to be significantly increased in PCa tissue specimens and that BRD4 may possess an important role in regulating the pathogenesis of PCa. In addition, it was reported that knockdown of BRD4, via shRNA transduction or JQ1 treatment, suppressed PCa proliferation in vitro and in vivo. This effect may be attributable, at least in part, to the induction of G0/G1 cell cycle arrest and apoptosis resulting from the upregulation of FOXO1 and p21, and downregulation of c-Myc and Bcl-2. To the best of our knowledge, the present study is the first to report of the molecular mechanisms underlying the pathogenesis of PCa and these findings may contribute to the development of potential therapeutic strategies for the management of this disease in the future.
Acknowledgments
Not applicable.
Funding
This study was supported by Application and Basic Research Project Of Wuhan City (grant no. 2015060101010049), Wuhan Morning Light Plan of Youth Science and Technology (grant no. 2017050304010281), Hubei Province Health and Family. Planning Scientific Research Project (grant nos. WJ2017M025 and WJ2017Z005), Natural Science Foundation of Hubei Province (grant nos. 2016CFB114 and 2017CFB181) and Research Project of Wuhan University (grant no. 2042017kf0097).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request
Authors' contributions
YT conceived and designed this study. YT, XL and LW wrote and revised the manuscript. YT, LW, YD, ZC, XW, JG and MW conducted the experiments and analyzed the data. YT, LW, HC and XW collected the clinical samples. XL supervised the entire experiment project. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Renmin Hospital of Wuhan University (Wuhan, China). Informed consent was obtained from all participants.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X, Albertsen PC, Andriole GL, Roobol MJ, Schroder FH, Vickers AJ. Risk-based prostate cancer screening. Eur Urol. 2012;61:652–661. doi: 10.1016/j.eururo.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attard G, Parker C, Eeles RA, Schroder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 5.Mullins JK, Feng Z, Trock BJ, Epstein JI, Walsh PC, Loeb S. The impact of anatomical radical retropubic prostatectomy on cancer control: The 30-year anniversary. J Urol. 2012;188:2219–2224. doi: 10.1016/j.juro.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 6.Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367:203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics-Uk. 2015;7:487–501. doi: 10.2217/epi.14.91. [DOI] [PubMed] [Google Scholar]
- 8.Meloche J, Potus F, Vaillancourt M, Bourgeois A, Johnson I, Deschamps L, Chabot S, Ruffenach G, Henry S, Breuils-Bonnet S, et al. Bromodomain-Containing protein 4: The epigenetic origin of pulmonary arterial hypertension. Circ Res. 2015;117:525–535. doi: 10.1161/CIRCRESAHA.115.307004. [DOI] [PubMed] [Google Scholar]
- 9.Chen R, Yik JH, Lew QJ, Chao SH. Brd4 and HEXIM1: Multiple roles in P-TEFb regulation and cancer. Biomed Res Int. 2014;2014:232870. doi: 10.1155/2014/232870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi-Yachide T. BET bromodomain as a target of epigenetic therapy. Chem Pharm Bull (Tokyo) 2016;64:540–547. doi: 10.1248/cpb.c16-00225. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Liu D, Gao X, Xie F, Tao D, Xiao X, Wang L, Jiang G, Zeng F. Inhibition of BRD4 suppresses cell proliferation and induces apoptosis in renal cell carcinoma. Cell Physiol Biochem. 2017;41:1947–1956. doi: 10.1159/000472407. [DOI] [PubMed] [Google Scholar]
- 12.French C. NUT midline carcinoma. Nat Rev Cancer. 2014;14:149–150. doi: 10.1038/nrc3659. [DOI] [PubMed] [Google Scholar]
- 13.Andrieu G, Tran AH, Strissel KJ, Denis GV. BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 2016;76:6555–6567. doi: 10.1158/0008-5472.CAN-16-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao YF, Wu YB, Long X, Zhu SQ, Jin C, Xu JJ, Ding JY. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget. 2016;7:9491–9500. doi: 10.18632/oncotarget.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferri E, Petosa C, McKenna CE. Bromodomains: Structure, function and pharmacology of inhibition. Biochem Pharmacol. 2016;106:1–18. doi: 10.1016/j.bcp.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 17.Sahai V, Redig AJ, Collier KA, Eckerdt FD, Munshi HG. Targeting BET bromodomain proteins in solid tumors. Oncotarget. 2016;7:53997–54009. doi: 10.18632/oncotarget.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li GQ, Guo WZ, Zhang Y, Seng JJ, Zhang HP, Ma XX, Zhang G, Li J, Yan B, Tang HW, et al. Suppression of BRD4 inhibits human hepatocellular carcinoma by repressing MYC and enhancing BIM expression. Oncotarget. 2016;7:2462–2474. doi: 10.18632/oncotarget.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Togel L, Nightingale R, Chueh AC, Jayachandran A, Tran H, Phesse T, Wu R, Sieber OM, Arango D, Dhillon AS, et al. Dual targeting of bromodomain and extraterminal domain proteins, and WNT or MAPK signaling, inhibits c-MYC expression and proliferation of colorectal cancer cells. Mol Cancer Ther. 2016;15:1217–1226. doi: 10.1158/1535-7163.MCT-15-0724. [DOI] [PubMed] [Google Scholar]
- 20.Urbanucci A, Barfeld SJ, Kytola V, Itkonen HM, Coleman IM, Vodak D, Sjoblom L, Sheng X, Tolonen T, Minner S, et al. Androgen receptor deregulation drives bromodomain-mediated chromatin alterations in prostate cancer. Cell Rep. 2017;19:2045–2059. doi: 10.1016/j.celrep.2017.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohler JL, Armstrong AJ, Bahnson RR, D'Amico AV, Davis BJ, Eastham JA, Enke CA, Farrington TA, Higano CS, Horwitz EM, et al. Prostate Cancer, Version 1.2016. J Natl Compr Canc Netw. 2016;14:19–30. doi: 10.6004/jnccn.2016.0004. [DOI] [PubMed] [Google Scholar]
- 23.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. StarBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Liu D, Tao D, Xiang W, Xiao X, Wang M, Wang M, Luo L, Li G, Zeng YF, et al. BRD4 regulates EZH2 transcription through upregulation of C-MYC and represents a novel therapeutic target in bladder cancer. Mol Cancer Ther. 2016;15:1029–1042. doi: 10.1158/1535-7163.MCT-15-0750. [DOI] [PubMed] [Google Scholar]
- 27.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DH, Qi J, Bradner JE, Said JW, Doan NB, Forscher C, Yang H, Koeffler HP. Synergistic effect of JQ1 and rapamycin for treatment of human osteosarcoma. Int J Cancer. 2015;136:2055–2064. doi: 10.1002/ijc.29269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar K, Raza SS, Knab LM, Chow CR, Kwok B, Bentrem DJ, Popovic R, Ebine K, Licht JD, Munshi HG. GLI2-dependent c-MYC upregulation mediates resistance of pancreatic cancer cells to the BET bromodomain inhibitor JQ1. Sci Rep. 2015;5:9489. doi: 10.1038/srep09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgakilas AG, Martin OA, Bonner WM. P21: A two-faced genome guardian. Trends Mol Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Blee AM, Wang D, An J, Pan Y, Yan Y, Ma T, He Y, Dugdale J, Hou X, et al. Loss of FOXO1 cooperates with TMPRSS2-ERG overexpression to promote prostate tumorigenesis and cell invasion. Cancer Res. 2017;77:6524–6537. doi: 10.1158/0008-5472.CAN-17-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tinkum KL, White LS, Marpegan L, Herzog E, Piwnica-Worms D, Piwnica-Worms H. Forkhead box O1 (FOXO1) protein, but not p53, contributes to robust induction of p21 expression in fasted mice. J Biol Chem. 2013;288:27999–28008. doi: 10.1074/jbc.M113.494328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: A review. JAMA. 2017;317:2532–2542. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 34.Basch E, Loblaw DA, Oliver TK, Carducci M, Chen RC, Frame JN, Garrels K, Hotte S, Kattan MW, Raghavan D, et al. Systemic therapy in men with metastatic castration-resistant prostate cancer: American Society of Clinical Oncology and Cancer Care Ontario clinical practice guideline. J Clin Oncol. 2014;32:3436–3448. doi: 10.1200/JCO.2013.54.8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning YM, Brave M, Maher VE, Zhang L, Tang S, Sridhara -R, Kim G, Ibrahim A, Pazdur R. U.S. Food and drug administration approval summary: Enzalutamide for the treatment of patients with chemotherapy-naive metastatic castration-resistant prostate cancer. Oncologist. 2015;20:960–966. doi: 10.1634/theoncologist.2015-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang T, Bai JY, She C, Yu DJ, Zhou XZ, Zhao TL. Bromodomain protein BRD4 promotes cell proliferation in skin squamous cell carcinoma. Cell Signal. 2018;42:106–113. doi: 10.1016/j.cellsig.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, Chen T, Jin J, Pan W, Cai X, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–3967. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 38.Rebello RJ, Pearson RB, Hannan RD, Furic L. Therapeutic approaches targeting MYC-Driven prostate cancer. Genes (Basel) 2017;8:E71. doi: 10.3390/genes8020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly PN, Grabow S, Delbridge AR, Strasser A, Adams JM. Endogenous Bcl-xL is essential for Myc-driven lymphoma-genesis in mice. Blood. 2011;118:6380–6386. doi: 10.1182/blood-2011-07-367672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X, Wu X, Zhang X, Hua W, Zhang Y, Maimaiti Y, Gao Z, Zhang Y. Inhibition of BRD4 suppresses tumor growth and enhances iodine uptake in thyroid cancer. Biochem Biophys Res Commun. 2016;469:679–685. doi: 10.1016/j.bbrc.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delbridge AR, Grabow S, Bouillet P, Adams JM, Strasser A. Functional antagonism between pro-apoptotic BIM and anti-apoptotic BCL-XL in MYC-induced lymphomagenesis. Oncogene. 2015;34:1872–1876. doi: 10.1038/onc.2014.132. [DOI] [PubMed] [Google Scholar]
- 44.Pellicano F, Scott MT, Helgason GV, Hopcroft LE, Allan EK, Aspinall-O'Dea M, Copland M, Pierce A, Huntly BJ, Whetton AD, et al. The antiproliferative activity of kinase inhibitors in chronic myeloid leukemia cells is mediated by FOXO transcription factors. Stem Cells. 2014;32:2324–2337. doi: 10.1002/stem.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request