Abstract
Cellular plasticity, the ability of cells to switch from an epitheial phenotype to a mesenchymal one and vice versa, plays a crucial role in tumour progression and metastases development. In 20-25% of patients with colon cancer and in 18% of patients with rectal cancer, metastases are present at the time of the first diagnosis. They are the first cause of colorectal cancer (CRC)-related mortality, defining stage IV CRC, which is characterized by a relatively short overall survival. We previously isolated two primary colon adenocarcinoma cell cultures that had undergone epithelial-mesenchymal transition (EMT), one with a high microsatellite instability phenotype (T88) and one with a chromosomal instability phenotype (T93). The aim of this study was to establish a model with which to study EMT, stemness features and cell plasticity in cancer progression and to examine the effects of incubation with lithium chloride (LiCl), a specific glycogen synthase kinase 3 β (GSK-3β) inhibitor, on these cellular processes. Indeed, GSK3β is an important regulator of cell survival, which promotes tumourigenesis in colon cells by facilitating the crosstalk between colorectal cancer pathways. Thus, we further characterized our system of adherent primary mesenchymal colon cancer cells and their paired tumourspheres by examining the expression and localisation of a panel of markers, including E- and N-cadherin, CD133, CD44v6, aldehyde dehydrogenase 1 (ALDH1) and leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5). We also characterised the molecular features of these tumourspheres and examined their response to LiCl. Furthermore, we explored the effects of LiCl on cell motility and plasticity. We demonstrated that LiCl reduced cell migration, stemness features and cell plasticity. We also observed the atypical nuclear localisation of membrane proteins, including N-cadherin, CD133 and CD44v6 in mesenchymal tumour cells. Of note, CD133 and CD44v6 appeared to localise at the plasma membrane in cells with a more epithelial phenotype, suggesting that the cytoplasmic/nuclear localisation of these proteins could favour and characterize cell plasticity in colorectal cancer progression.
Keywords: colorectal cancer, cancer tumourspheres, epithelial-to mesenchymal transition, mesenchymal to epithelial transition, cancer cell plasticity, stem cell-like phenotype, GSK3β inhibition
Introduction
Worldwide, colorectal cancer (CRC) is the fourth most common cancer and the fourth leading cause of cancer-related mortality, with 1.4 million estimated cases and 700,000 estimated deaths in 2012 (1,2). As in many other solid tumours, metastasis is the primary cause of mortality in CRC, and defines stage IV CRC, which is characterized by a relatively short overall survival. One of the main issues encountered in CRC treatment is drug resistance, which is responsible for relapses in a number of patients and the failure of medical treatments for metastatic disease.
The normal gastrointestinal epithelium is organized along a crypt-villus axis, with a pool of colon stem cells and progenitor cells, which are the most undifferentiated cell types which can undergo self-renewal and exhibit pluripotency, residing at the bottom of the crypts. These cells move along the crypt-villus axis while differentiating into all the epithelial colon lineages, such as Paneth cells, goblet cells, enterocytes and enteroendocrine cells. In approximately 14 days, they arrive at the top of the villus, and undergo apoptosis (3-5).
It has been suggested that an oncogenic hit in a stem cell generates cancer stem cells (CSCs), which are tumour-initiating cells that represent the only cell type able to promote cancer onset, progression and metastases. CSCs are probably responsible for both tumour relapse and resistance to therapy (6,7). The source from which CSCs arise remains unclear. It remains to be determined whether trans-amplificating (TA) cancer cells can de-differentiate, transforming themselves into CSCs, or alternatively, whether healthy stem cells are transformed into CSCs by accumulating genetic hits in oncogenes or tumour suppressor genes (8,9). It has been hypothesized that microenvironment-derived signals, such as transforming growth factor (TGF)-β, epidermal growth factor (EGF), Wnt and Notch, can activate epithelial-to-mesenchymal transition (EMT) and stem-cell properties in differentiated cells (10,11); thus, an oncogenic hit in this population may give rise to CSCs that can initiate cancer.
During tumour progression, epithelial cancer cells undergo EMT. Cells that undergo EMT acquire a mesenchymal phenotype, losing their epithelial features and cell polarity and acquiring motility, stem cell-like properties, self-renewal and apoptosis resistance. These cells are also able to degrade the basement membrane and extracellular matrix. Thus, it has been suggested that EMT confers to cancer cells, including CRC cells, many of the features required to develop metastases (12-14). Chemo-radiotherapy often kills differentiated cancer cells, while the mesenchymal, stem-like cells are resistant and can survive after therapy, giving rise to therapy-resistant tumours. EMT-associated transcription factors are also involved in apoptosis resistance in cells that have undergone EMT. It has been demonstrated that TWIST1 inhibits cell cycle arrest and apoptosis by blocking the p53 target genes (15). Moreover, SNAIL suppresses TGF-β-induced cell death and regulates the components of the early to late G1 transition and the G1/S checkpoint, including the repression of cyclin D2 transcription and the increase in p21 expression (16,17).
However, the only crucial cell feature conferring to cancer cells the ability to complete the metastatic process, is cellular plasticity. Cellular plasticity is defined as the ability of cells to switch from an epithelial to a mesenchymal phenotype and vice versa. Indeed, epithelial adenocarcinoma cells escape from the primary in situ carcinomas and spread to distant organs through mesenchymal intermediates, while only disseminated cancer cells capable of switching again to an epithelial hyper-proliferative stem cell phenotype are able to colonize distant organs and generate metastatic secondary lesions. Cancer cells that undergo EMT, but lose this plasticity phenotype are ineffective for seeding metastatic colonies (18-20).
Glycogen synthase kinase 3 β (GSK-3β) is a key regulator of the crosstalk between different signalling pathways involved in CRC development, such as WNT/β-catenin, Ras, PI3K/Akt, cMET and vascular endothelial growth factor (VEGF). Data have also been published regarding the role of GSK-3β in CRC cell drug resistance (21,22).
Genomic instability facilitates the accumulation of multiple mutations during CRC development; chromosomal instability (CIN) is observed in 85% and microsatellite instability (MSI) is detected in 15% of sporadic CRCs. The molecular mechanisms underlying CRC progression remain poorly understood, particularly as regards CRCs with MSI (23). We previously isolated two primary colon cancer cell cultures, one exhibiting a CIN phenotype (T93) and the other exhibiting an MSI phenotype (T88). They both exhibited mesenchymal and epithelial features and a high level expression of EMT-associated transcription factors and stemness markers. Thus, we hypothesized that they were epithelial adenocarcinoma cells that had undergone EMT. These cells were also able to grow in conditioned medium as non-adherent tumourspheres. Finally, we demonstrated in vitro that LiCl-induced mesenchymal-to-epithelial transition (MET), cellular differentiation and the downregulation of the EMT-associated transcription factors, Twist1 and Snail, in these primary CRC cell cultures (24).
Herein, we investigated the expression and localisation of key markers of EMT and stemness in CRC cells exhibiting both CIN and MIN by establishing a system of adherent primary mesenchymal colon cancer cells and paired tumourspheres. These cells exhibited plasticity. We also observed an atypical nuclear localisation of N-cadherin, CD133 and the v6 splice form of CD44 glycoprotein (CD44v6) in the majority of the mesenchymal cells, suggesting a change in localisation from the plasma membrane to the nucleus, which could allow cell plasticity in CRC progression. Finally, we demonstrated that GSK-3β inhibition reduced cell migration and cell plasticity in our experimental cell model, thus suggesting that GSK-3β may be a target for CRC therapy.
Materials and methods
Sample collection
CRC tissues and normal colorectal mucosa were obtained from patients with sporadic CRC, who were operated at the AOU Federico II and Istituto Nazionale dei Tumori (Naples, Italy) and primary cell cultures were established from these tissues. Data regarding tumour stage were recovered from the medical records of each patient, in accordance with the TNM classifications and tumour budding grades.
Samples from all subjects who participated in this study were collected after obtaining authorisation from the Comitato etico per le attività Biomediche - Carlo Romano of the University of Naples Federico II (protocol no. 432/17). Authorisation was granted only once the study had received ethical approval and written informed consent had been obtained from all participants. All methods were performed in accordance with the relevant guidelines and regulations.
Cell culture
The T88 and T93 cells were previously isolated and stabilized in vitro (24). The HM110 colon cells were isolated and stabilized during this study from the healthy colon mucosa (HM) of a patient with sporadic colon cancer, as previously described (24). Briefly, samples were washed overnight at 4°C in PBS containing antibiotics, finely minced and digested in collagenase II in DMEM/FBS-10% for 1 h at 37°C, 5% CO2. The obtained cell suspension was then collected by centrifugation at 1,000 × g, at room temperature, washed twice and subsequently cultured in DMEM/F12-10% FBS medium (1:1), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml amphotericin B. The HT29 and RKO cells were obtained from ATCC (Manassas, VA, USA). To inhibit GSK-3β activity, cells were incubated in medium containing 30 mM LiCl for 10 days. The T88 and T93 primary colon cancer cells were then cultured as spheres in serum-free stem cell medium on low-adhesion plates as previously described (24). Cancer spheroids were also obtained by the hanging drop assay. Briefly, the cells were diluted to a final concentration of 3.7×104 cells/ml in DMEM/F12 (1:1) containing 2% foetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, 2.5 μg/ml amphotericin B, 10 μg/ml basic fibroblast growth factor (bFGF) and 20 μg/ml epidermal growth factor (EGF), with or without 35 mM LiCl, (all reagents were purchased from Sigma-Aldrich Merk KGaA, Darmstadt, Germany). The drops (27 μl) were seeded on the cover of a plate and incubated at 37°C at 5% CO2 for 72 h. Under these conditions, the cells aggregated to form 3D spheroids. Cancer spheroids obtained in presence or absence of LiCl were finally disaggregated into single cells, which were again grown in adhesion.
Migration assays
Cell migration was evaluated using in vitro wound healing assays and the Boyden chamber assay. In vitro wound healing assays were performed as previously described by Liang et al (25). Briefly, the cells were seeded at 1×104 cells/well in 24-well plates. After the cells formed a monolayer, a scratch wound was made with the tip of a 1,000-μl pipette tip, and the scratch was photographed under a light microscope (Leica DM IL LED inverted microscope, type 11090137002, serial no. 11521258/335209; Meyer Instruments, Inc. Houston, TX, USA) at ×20 magnification. Subsequently, cells were incubated with or without (untreated control) 35 mM LiCl. After 24 h, cells were rinsed, and a second set of images were acquired.
Boyden chamber assays were performed using the Corning® Transwell® polycarbonate membrane cell culture inserts (pore size, 8.0 μm; Sigma-Aldrich, Merk KGaA). A 300 μl of aliquot of a 10,000 cells/ml suspension was resuspended in serum-free medium and seeded into the upper chamber of the polycarbonate membrane. Subsequently, 500 μl of medium containing 10% FBS (chemoattractant) were added to the lower well of the migration plate. Finally, the cells were removed from the top of the membrane and the migrated cells were stained with crystal violet solution for 15 min at room temperature (Sigma-Aldrich, Merk KGaA). Alternatively, 24-well Transwell plates (pore size: 8.0 μm; Corning Inc., Corning, New York, NY, USA) were used. Each membrane was released from the apparatus using a scalpel. The permeable membranes were then washed twice with PBS/1% FBS at room temperature. The cells on the membranes were fixed, permeabilized and stained with DAPI (Sigma-Aldrich, Merk KGaA) for 15 min at room temperature. Finally, the membranes were mounted on glass slides, covered with coverslips, and fluorescence was visualized and imaged under a fluorescence microscope (Leica DM IL LED inverted microscope, type 11090137002, no. 11521258/335209; Meyer Instruments, Inc.).
Reverse transcription-quantitative (real-time) PCR (RT-qPCR)
Total RNA was extracted from the cells using QIAzol reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Following DNase incubation, cDNA was synthesized from 1 μg total RNA, 500 ng random hexamers, and 1 μl Superscript III reverse transcriptase in the presence of 4 μl 5X RT buffer, 1 μl DTT (0.1 M) and 1 mM dNTPs (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA) as previously described (26). Quantitative (real-time) PCR (qPCR) was performed on 0.5 μl cDNA, Snail and Twist1 primer pairs as previously described (24,27). Relative expression was calculated with the 2−ΔΔCq method (28) and normalized against glucuronidase (GUS) mRNA. qPCR was performed on a Bio-Rad iCycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). A non-template control was run for each assay, all determinations were performed at least in duplicate to ensure reproducibility and each experiment was performed two times. The synthesis of the expected PCR product was confirmed by melting curve analysis.
Western blot analysis
Fractionated nuclear and cytosolic protein extracts were isolated from the T88 and T93 cells using the Qproteome Nuclear Protein kit (Qiagen Hilden, Germany), and quantified by using Traditional Bradford kit (Bio-Rad Laboratories Srl, Segrate, Italy), adopting bovine serum albumin standards. A total of 50 μg of proteins were separated by 10% SDS-polyacrylamide gel electrophoresis. Blots were prepared on Amersham Hybond-ECL nitrocellulose membranes (Amersham Pharmacia Biotech, Cologno Monzese, Italy), and proteins were blocked in non-fat dried milk diluted in TBST to reduce the background, as previously described (29,30). Primary antibodies against N-cadherin (rabbit monoclonal anti-human; #4061; 1:1,000) and CD133 (rabbit monoclonal anti-human; #5860; 1:1,000) were obtained from Cell Signaling Technology (Danvers, MA, USA). The primary antibody against CD44v6 (mouse monoclonal anti-human; #5640; 1:1,000) was from R&D Systems (Minneapolis, MN, USA), the anti-actin (goat polyclonal anti-human; sc-1615; 1:8,000) and anti-Histone H1 (monoclonal anti-mouse; sc-393358; 1:1,000) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The membranes were probed with peroxidase-conjugated secondary antibodies against rabbit (#7074), mouse (#7076; both from Cell Signaling Technology or goat IgG (#ab97110; from Abcam, Cambridge, UK), all used at 1:3,000 dilution, and immunoreactive bands were detected using the enhanced chemiluminescence HRP Substrate Immobilon Western (Millipore, Billerica, MA, USA).
Immunofluorescence
Primary CRC cells were seeded and grown in 12-well cultivation chambers with removable microscopy glass slides (ibidi, Martinsried, Germany); cancer spheroids obtained by the hanging drop assay were also sedi-mented in the same chamber. Immunofluorescence analyses were performed as previously described by Di Maio et al (31). Briefly, following fixation in 4% paraformaldehyde in PBS for 10 min, the cells were permeabilized in 0.1% Triton X-100 in PBS for 30-120 min, and then blocked in 10% bovine serum albumin for 30 min. The cells were incubated with primary antibodies (Table I) overnight, and then with secondary antibodies (Alexa Fluor 546 donkey anti-rabbit, A10040; Alexa Fluor 488 donkey anti-mouse, A21202; Thermo Fisher Scientific) for 1 h, and then with DAPI (Sigma-Aldrich) for 30 min at room temperature to label the nuclei. Negative controls without primary antibodies were also included, and these exhibited no staining. Following the indicated treatments, coverslips were mounted on glass slides and examined under a fluorescence confocal microscope (Zeiss LSM 700, Carl Zeiss, Oberkochen, Germany).
Table I.
Antibodies and dilutions used for immunofluorescence staining.
| Antibody: Dilution (Cat. no., provider) | Antibody: Dilution (Cat. no., provider) |
|---|---|
| Pan-cytokeratin (CK): 1:50 (MA5-13203, Invitrogen, Thermo Fisher Scientific) | E-cadherin: 1:50 (ab76055, Abcam) |
| Nanog: 1:50 (3580, Cell Signaling Technology) | N-cadherin: 1:50 (ab76057, Abcam) |
| Sox2: 1:50 (4900, Cell Signaling Technology) | Snail: 1:150 (ab180714, Abcam) |
| Oct4: 1:50 (2840, Cell Signaling Technology) | LGR5: 1:50 (sc135238, Santa Cruz Biotechnology) |
| Vimentin: 1:200 (5741, Cell Signaling Technology) | ALDH1: 1:50 (ab24343, Abcam) |
| CD44: 1:500 (5640, Cell Signaling Technology) | CD133: 1:50 (ab16518, Abcam) |
| β-catenin: 1:100 (9581, Cell Signaling Technology) | CD44v6: 1:50 (BBA13, R&D Systems) |
Sox2, sex determining region Y-box 2; Oct4, octamer-binding transcription factor 4; LGR5, leucine-rich repeat-containing G-protein coupled receptor 5; ALDH1, aldehyde dehydrogenase 1.
Results
CRC cells that have undergone EMT exhibit nuclear N-cadherin, CD133 and Cd44v6 localisation
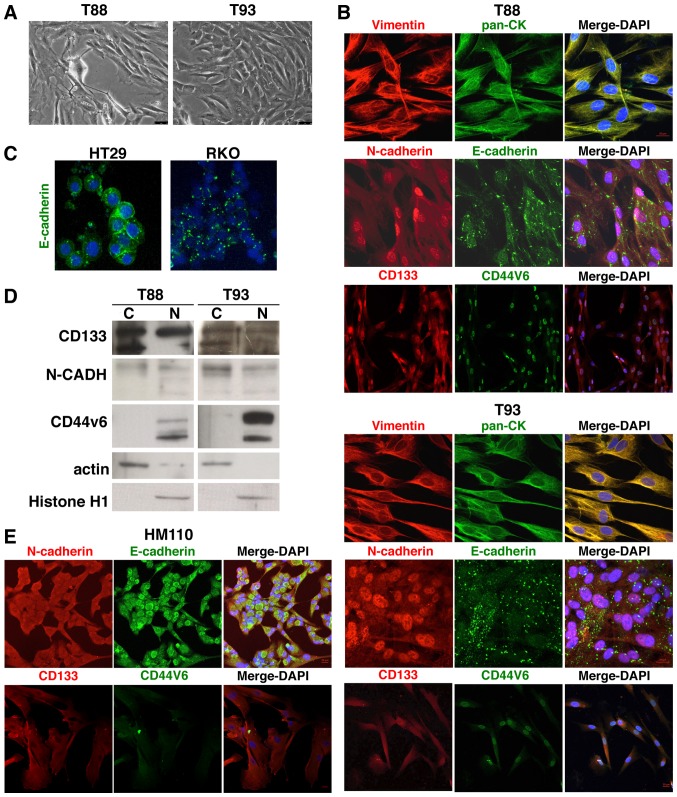
We first confirmed, by immunofluorescence analyses, that both the T88 and T93 cells (Fig. 1A) expressed epithelial and mesenchymal proteins, in accordance with our previous data (24). As shown in Fig. 1B, Vimentin and Cytokeratin co-localized at the cytoskeleton level in both cell lines. E-cadherin did not exhibit the classic membrane staining of epithelial cells, such as in commercial HT29 CRC cells (Fig. 1C), but rather a diffuse punctate signal, as observed in the commercial RKO CRC cells, was found (Fig. 1C). Commercially available CRC cell cultures are largely characterised; thus, it is well known that HT29 cells have a more epithelial phenotype than RKO cells, which primarily exhibit mesenchymal features (32). N-cadherin exhibited an unexpected nuclear localisation (Fig. 1B). We also analysed two cell surface glycoproteins involved in stemness in several types of cancer cells, CD133 and CD44v6. Of note, similar to N-cadherin, these markers were mainly localised to the nucleus. As shown in Fig. 1D, these findings were confirmed by western blot analysis of the nuclear and cytosolic protein fractions of the T88 and T93 cells.
Figure 1.
Nuclear N-cadherin, CD133 and CD44v6 localisation in T88 and T93 mesenchymal colorectal cancer (CRC) cells. (A) Brightfield images of T88 and T93 cells at ×10 magnification. (B) Confocal images of T88 and T93 primary colon cancer cells stained for anti-Vimentin (red)/anti-pan-CK (green), anti-N-cadherin (red)/anti-E-cadherin (green) and anti-CD133 (red)/anti-CD44v6 (green) antibodies. Nuclei were counterstained with DAPI (blue). (C) Confocal images of HT29 and RKO cells stained with anti-E-cadherin (green) antibody. Nuclei were counterstained with DAPI (blue). (D) Western blot analysis of cytoplasmic 'C'' and nuclear 'N' extracts from T88 and T93 cells using anti-CD133, anti-N-cadherin, anti-Cd44v6, anti-actin and anti-histone H1 antibodies. Histone H1 and actin were used as nuclear and cytoplasmic marker proteins, respectively. Each lane was loaded with 50 μg of cytoplasmic or nuclear extract. The cropped blots are used in the figure. (E) Confocal images of primary cells from the healthy mucosae of a patient affected by sporadic CRC (HM110). Cells were stained with anti-N-cadherin (red)/anti-E-cadherin (green) and anti-CD133 (red)/CD44v6 (green). Nuclei were counterstained with DAPI (blue).
Furthermore, to evaluate whether the observed nuclear localisation phenotypes were tumour-specific, we isolated, cultured and analysed cells from the healthy mucosae of a sporadic CRC patient (HM110). Through immunofluorescence staining, we demonstrated that these cultures also expressed both epithelial and mesenchymal proteins (N- and E-cadherin, respectively), but found that they did not exhibit the same nuclear localisation of N-cadherin, CD133 and CD44v6, as was observed in the tumour cells (Fig. 1E). This finding strongly suggests a tumour-specific nature of this feature.
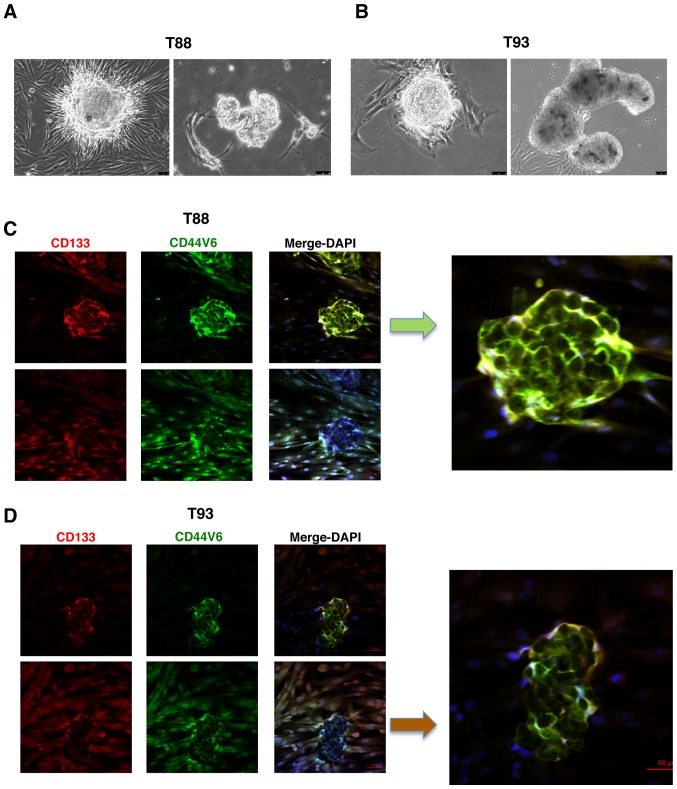
It is known that primary colon cultures are able to generate in vitro organoids, which are propagated by intestinal stem cells, and thus, represent an organotypic culture system. Furthermore, it has been demonstrated that it is possible to generate intestinal organoids starting from a single sorted leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5)-positive intestinal stem cell from dissociated intestinal crypts (33,34). As shown in Fig. 2A and B, the T88 and T93 cells, all LGR5-positive (please also see Fig. 6), generated organoid-like in vitro three-dimensional bodies. Immunofluorescence staining revealed that cells of these three-dimensional bodies expressed both CD133 and CD44v6 mainly at the plasma membrane (Fig. 2C and D; images on top panels), contrary to their mesenchymal counterparts in monolayer, which retained nuclear localisation (Fig. 2C and D; images on bottom panels).
Figure 2.
Primary epithelial-mesenchymal colon cancer cells generate in vitro. organoids expressing both CD133 and CD44v6 at the cell membrane. (A and B) Brightfield images at ×20 magnification of organoids generated from (A) T88 and (B) T93 cells. (C and D) Confocal images of organoids from (C) T88 and (D) T93 primary colon cancer cells, stained with anti-CD133 (red)/anti-CD44v6 (green) antibodies. Nuclei were counterstained with DAPI (blue). A central focal plane (top panel) and a basal focal plane (bottom panel) of an organoid showing also the adherent 2D cells is shown for both the T88 and T93 cells. Zoomed particular of each organoid is shown on the right (next to the arrows).
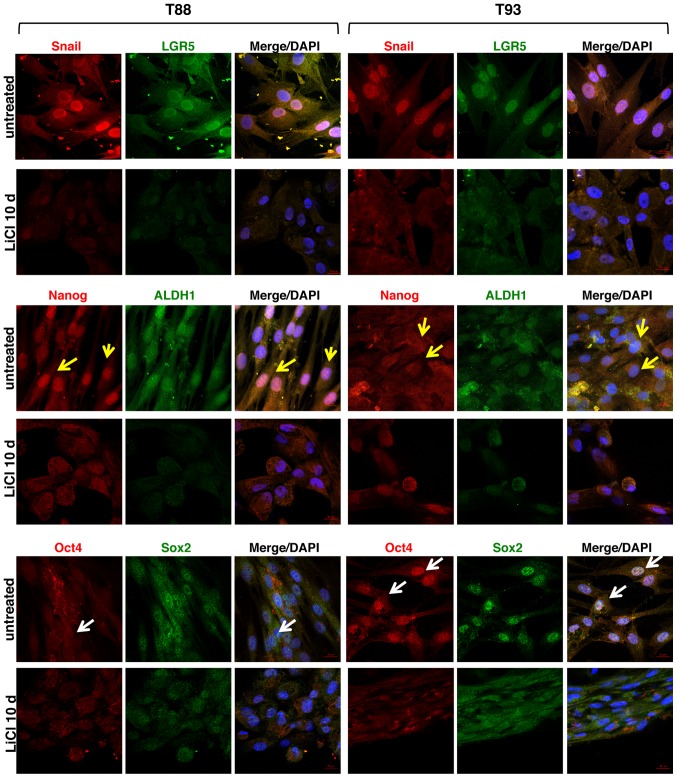
Figure 6.
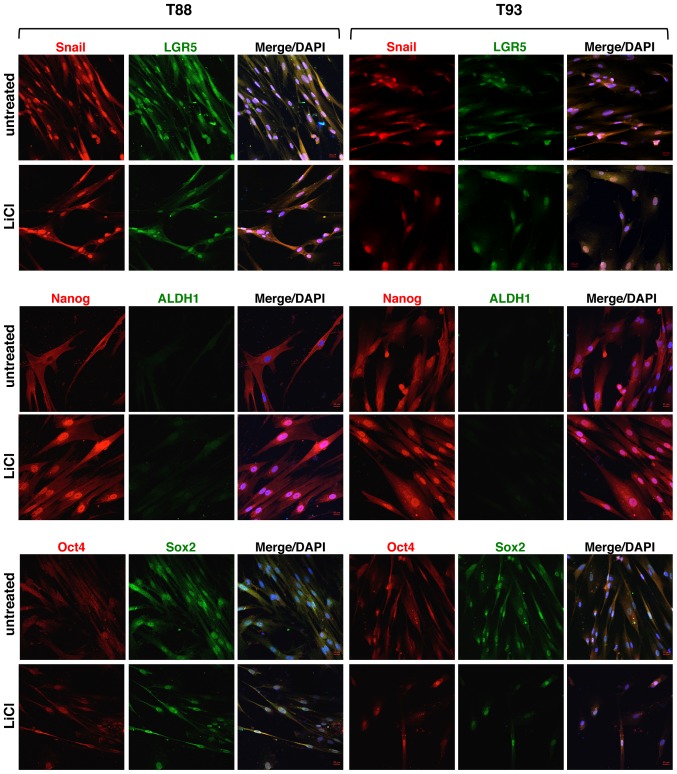
Effect of LiCl on the expression of Nanog, ALDH1, Snail, LGR5 and Oct4 proteins, in adherent 2D T88 and T93 primary cell cultures. Confocal images of untreated T88 and T93 primary colon cancer cells and cells after 10 days (10 d) of LiCl incubation. Cells were dual-stained with anti-Nanog (red)/anti-ALDH1 (green), anti-Snail (red)/anti-LGR5 (green) and anti-Oct4 (red)/anti-Sox2 (green) antibodies. Nuclei were counterstained with DAPI (blue). Nanog (yellow arrows) exhibited a higher nuclear expression in T88 cells than in T93 cells, while Oct4 (white arrows) exhibited an opposite trend.
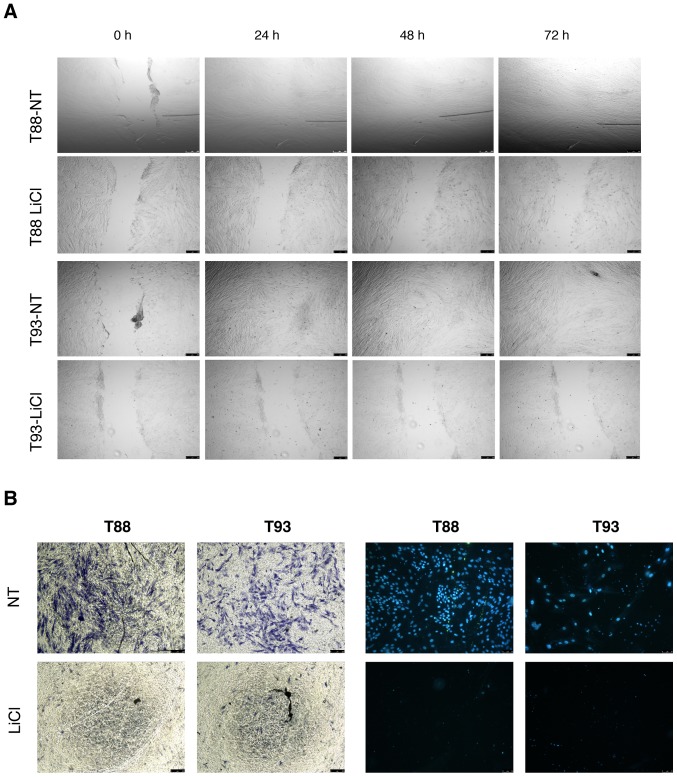
Incubation with LiCl reduces T88 and T93 cell migration
As GSK-3β is overexpressed and functions as an oncogene in CRC, we examined the effects of GSK-3β inhibition on tumour cells using LiCl, a specific inhibitor. We found that LiCl treatment inhibited cell migration using wound healing and Transwell migration assays. As shown in Fig. 3A, the wounds were almost completely healed after 24 h in the untreated cells, while the cells incubated with LiCl had wound sizes similar to time 0 after 72 h. We confirmed this observation using Transwell migration assays; no cell migration was detected when the cells were incubated with LiCl using crystal violet or DAPI staining (Fig. 3B).
Figure 3.
LiCl inhibits the migration of epithelial-mesenchymal colon cancer cells. (A) Brightfield images at ×4 magnification of T88 and T93 cells during wound healing assays performed with cells incubated (LiCl) or not incubated (NT) with LiCl. Wound closures were imaged immediately after scratching (0 h) and 24, 48 and 72 h later. (B) Transwell migration assay. Brightfield images of crystal violet staining are shown on the left, while on the right are shown epifluorescence images of nuclei stained with DAPI (blue). In both images, magnification was ×4.
LiCl incubation affects the stemness features of CRC cells that have undergone EMT
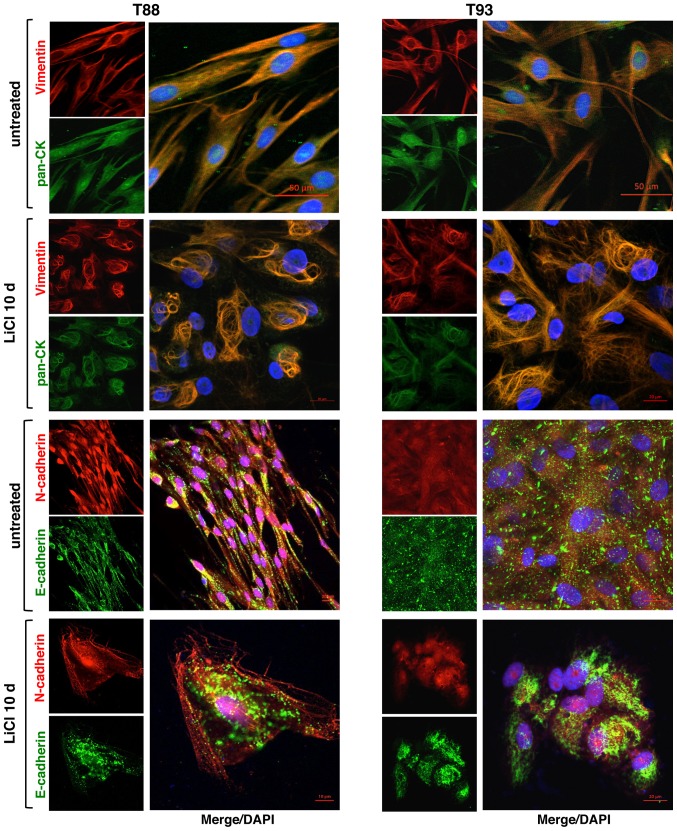
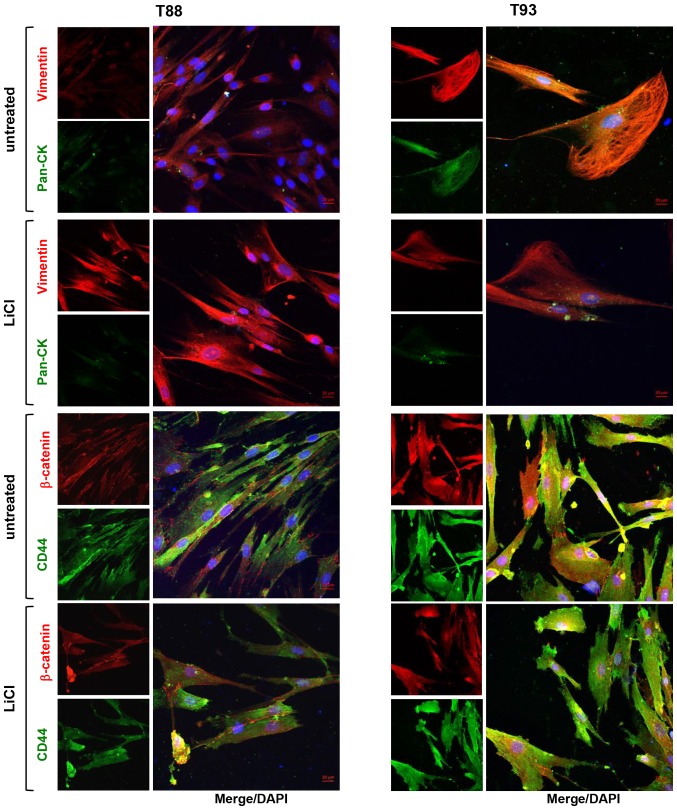
We confirmed our previous finding showing that LiCl induces MET in T88 and T93 cell cultures (24). Indeed, by confocal microscopy, we confirmed the finding of E-cadherin upregulation described in our previous study (24). We assessed the expression and localization of the epithelial markers, E-cadherin and pan-cytokeratin (Ck), and the expression of the mesenchymal markers, Vimentin and N-cadherin, in untreated and LiCl-treated cells. As shown in Fig. 4, Vimentin and Cytokeratin co-localised in both untreated and treated cells, the latter exhibiting a different cytoskeletal organization. Following LiCl treatment, E-cadherin maintained a discontinuous punctate pattern, although the treated cells exhibited a differentiated, more polarized shape than the untreated cells. Furthermore, following LiCl incubation, a nucleolar localization of N-cadherin appeared visible, whereas it was not detected in the untreated cells.
Figure 4.
Pan-cytokeratin, Vimentin, E-cadherin and N-cadherin expression following LiCl treatment in adherent 2D T88 and T93 primary cell cultures. Confocal images of untreated T88 and T93 primary colon cancer cells and cells after 10 days (10 d) of LiCl incubation. Cells were dual-stained with anti-Vimentin (red)/anti-pan-CK (green), anti-N-cadherin (red)/anti-E-cadherin (green) and anti-CD133 (red)/anti-CD44v6 (green) antibodies. Nuclei were counterstained with DAPI (blue).
Thereafter, we evaluated in both the untreated cells and cells incubated with LiCl, the expression of CD44, its splice isoform CD44v6, CD133 and β-catenin, all proteins playing pivotal roles in CRC development and progression. In accordance with our previous data (24), following 10 days of LiCl treatment, CD44 and β-catenin expression decreased and they both localised mainly at the plasma membrane, while CD44v6 and CD133 proteins both exhibited cytosolic localisation (Fig. 5).
Figure 5.
Effect of LiCl on the expression of β-catenin, CD44, CD133 and CD44v6 proteins in adherent 2D T88 and T93 primary cell cultures. Confocal images of untreated T88 and T93 primary colon cancer cells and cells after 10 days (10 d) of LiCl incubation. Cells were dual-stained with anti-β-catenin (red)/anti-CD44 (green) and anti-CD133 (red)/anti-CD44v6 (green) antibodies. Nuclei were counterstained with DAPI (blue).
We also analysed the expression of the EMT-associated transcription factor, Snail, and the expression of several stem cell-specific markers, including LGR5, Nanog, aldehyde dehydrogenase 1 (ALDH1), octamer-binding transcription factor 4 (Oct4) and sex determining region Y-box 2 (Sox2), in addition to the aforementioned CD133, CD44 and CD44v6, following LiCl treatment. As shown in Fig. 6, we observed that LiCl induced a downregulation in the expression of the EMT-associated transcription factor, Snail, and all stemness markers (LGR5, ALDH1, Nanog, Oct4 and Sox2), which in the untreated cells exhibited nuclear localisation. Furthermore, only in the untreated cells, Nanog exhibited a higher nuclear expression in the T88 cells than in the T93 cells (yellow arrows), while Oct4 exhibited an opposite trend (white arrows).
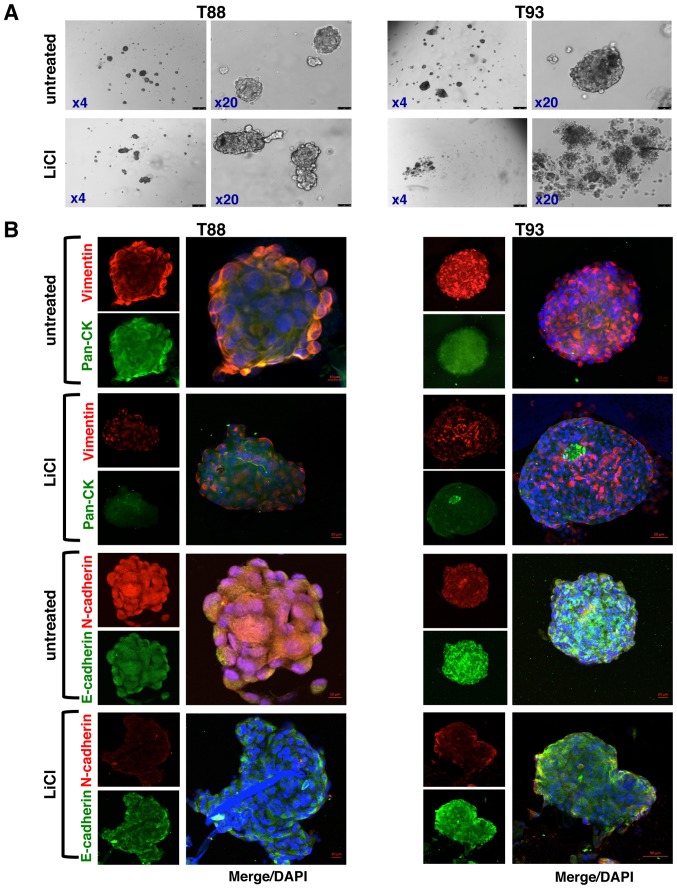
Furthermore, we generated tumourspheres from the T88 and T93 cells by the hanging drop assay, thus establishing a system of adherent primary mesenchymal colon cancer cells and paired tumourspheres, which are useful for studying stemness features, cell plasticity and drug responses. It has been reported that only stem cells and/or stem cell-like cells are able to survive and grow in suspension (35), as the loss of adhesion induces death through anoikis in non-malignant and cancer differentiated cells. Under these culture conditions, undifferentiated tumour cells proliferate and grow as floating clusters termed tumourspheres. As shown in Fig. 7A, the T88 and T93 cells formed spheroid aggregates in hanging drop assays, indicating the acquisition of a dedifferentiated state. However, following 72 h of incubation with LiCl, the T88 cells lost their spherical shape, consistent with a differentiated state, and the T93 cells completely lost their ability to form spheres, generating only cellular aggregates, in agreement with the more epithelial phenotype of T93 cells compared with the T88 cells (24).
Figure 7.
LiCl inhibits tumoursphere formation. (A) Brightfield images at ×4 and ×20 magnification of hanging drop assays on T88 (images on left panels) and T93 (images on right panels) cells obtained in medium containing (LiCl) or not containing (untreated) LiCl. (B) Confocal images of the tumourspheres dual-stained with anti-vimentin (red)/anti-pan-CK (green) and anti-N-cadherin (red)/anti-E-cadherin (green) antibodies. Nuclei were counterstained with DAPI (blue).
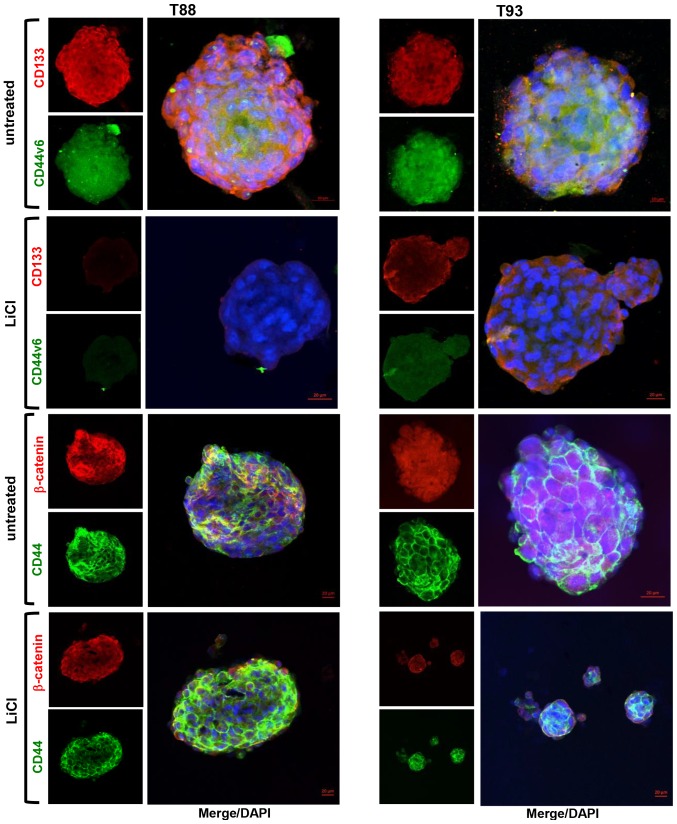
When analysed by confocal microscopy, both the epithelial (Cks and E-cadherin) and mesenchymal (Vimentin and N-cadherin) markers were expressed in the untreated tumourspheres (Fig. 7B). However, these spheres exhibited an upregulation of epithelial markers and a downregulation of mesenchymal ones, together with the retention of the expression of many stemness markers, thus suggesting a switching from a mesenchymal stem-cell-like phenotype to a more epithelial stem-cell-like phenotype, compared with the adherent T88 and T93 cells (Fig. 1). Moreover, in the untreated tumourspheres, N-cadherin did not localise to the nucleus (Fig. 7B), as observed in the adherent cells (Fig. 1), exhibiting a prevalent cytoplasmic localization. In addition, N-cadherin was downregulated in the untreated T93 tumourspheres, while E-cadherin was upregulated in both the untreated and treated T88 and T93 tumourspheres (Fig. 7B) compared with the adherent cells (Fig. 1). The untreated tumourspheres also exhibited CD133, CD44v6, CD44 and β-catenin expression; the last two were mainly expressed at the plasma membrane (Fig. 8). Furthermore, as shown in Fig. 9, the cells in the untreated cancer spheroids retained the expression of mesen-chymal and stemness biomarkers, exhibiting a lower nuclear Snail expression compared with the adherent T88 and T93 cells (Fig. 6). ALDH1 expression was very low in the T88 cancer spheroids (Fig. 9) compared with both its paired adherent cells (Fig. 6) and T93 tumourspheres (Fig. 9). The T93 tumourspheres, exhibited a very low Oct4 expression (Fig. 9) compared with both the paired adherent cells (Fig. 6) and T88 tumourspheres (Fig. 9).
Figure 8.
LiCl downregulates the stem cell markers, CD133 and CD44v6, in tumourspheres. Confocal images of the tumourspheres obtained by the hanging drop assays. Tumourspheres were dual-stained with anti-CD133 (red)/anti-CD44v6 (green) and anti-β-catenin (red)/anti-CD44 (canonical isoform; green) antibodies. Nuclei were counterstained with DAPI (blue).
Figure 9.
LiCl downregulates EMT-associated transcription factors and stem cell markers in tumourspheres. Confocal images of the tumourspheres obtained by hanging drop assays. Tumourspheres were dual-stained with anti-Snail (red)/anti-LGR5 (green), anti-Nanog (red)/anti-ALDH1 (green), and anti-Oct4 (red)/anti-Sox2 (green) antibodies. Nuclei were counterstained with DAPI (blue).
We further investigated the molecular basis of the GSK-3β-mediated inhibition of the ability of T88 and T93 cells to generate tumourspheres, by the immunofluorescence analysis of epithelial, mesenchymal and stemness markers, on spheroids obtained in medium containing LiCl. In accordance with above-mentioned findings on adherent cells, we observed a drastic downregulation of all stemness and mesenchymal markers analysed. As shown in Fig. 7, LiCl induced a down-regulation of Vimentin and N-cadherin. Moreover, the pan-Ck signal was lower in the LiCl-treated tumourspheres compared with the untreated tumourspheres, while E-cadherin (Fig. 7), CD44 and β-catenin (Fig. 8) exhibited a similar expression in the T88 and T93 spheroids obtained in medium containing or not containing LiCl. In accordance with the effect of LiCl on adherent cells, the expression of Snail, CD133, CD44v6 proteins (Fig. 8) and of all other stem-cell markers analysed (Fig. 9) strongly decreased in the tumourspheres following incubation with LiCl.
Incubation with LiCl alters T88 and T93 cell plasticity
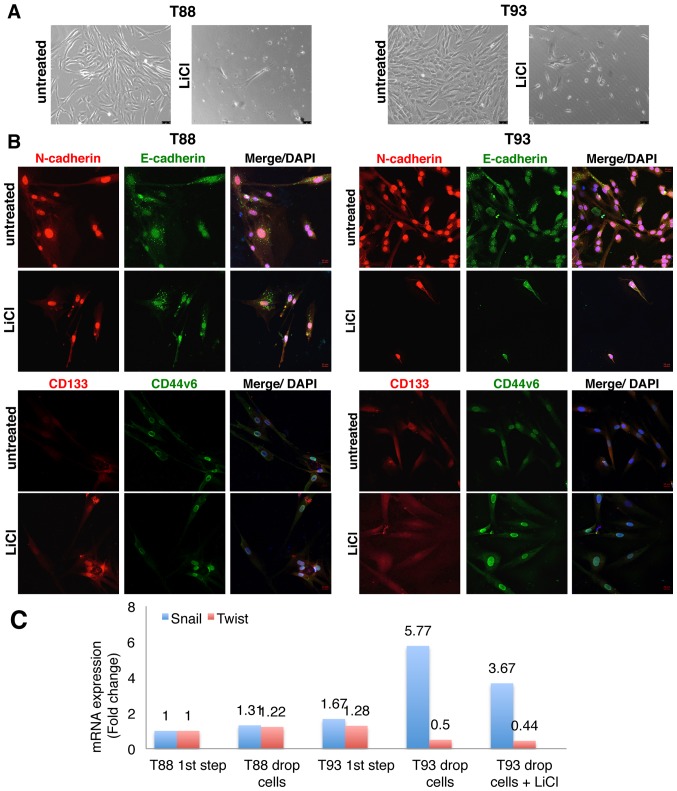
To further investigate the plasticity of the T88 and T93 cells, we disaggregated spheroids obtained by hanging drop assays, grown in medium with or without LiCl, (hereinafter referred to as treated and untreated tumourspheres) and cultivated them in adhesion, to examine the ability of the tumourspheres to re-adhere to a matrix and grow in adherent culture. As shown in Fig. 10A, these cells were able to adhere to the cell culture dishes and assume a mesenchymal phenotype. However, the numbers of surviving cells were significantly lower when they were derived from the treated spheroids compared to the cells derived from the untreated spheroids. Furthermore, the T93 cells, but not the T88 cells, derived from the treated spheroids, were able to proliferate and grow in adhesion (data not shown). By confocal microscopy, we observed that all cells obtained by disaggregating tumourspheres exhibited a similar phenotype with regards to the expression of mesenchymal and stemness markers (Figs. 10B and 11). However, cells derived from the treated tumourspheres exhibited a major nuclear localization of stem cells and mesenchymal markers (Fig. 10B) and an upregulation of the Snail transcription factor was observed only in the T93 cells, when analysed by RT-qPCR (Fig. 10C).
Figure 10.
LiCl inhibits cell plasticity. Molecular characterisation of survived cells. (A) Brightfield images (×10 magnification) of cells disaggregated from T88 and T93 spheroids obtained in medium containing (LiCl) or not containing (untreated) LiCl. (B) Confocal images of cells dual-stained with anti-N-cadherin (red)/anti-E-cadherin (green) and anti-CD133 (red)/anti-CD44v6 (green) antibodies Nuclei were counterstained with DAPI (blue). Magnification, ×20. (C) Real-time quantification of Snail and Twist1 mRNA expression in T88 and T93 adherent cells before tumoursphere formation (1st step), cells disaggregated from untreated tumourspheres (drop cells) and cells disaggregated from treated tumourspheres (drop cells in LiCl). This experiment was performed two times with similar results. A typical experiment is shown. Numbers on the top of each of the bars represent the precise value of relative expression for each sample, as calculated by the ΔΔCq method.
Figure 11.
Expression of Vimentin, Pan-cytokeratin, β-catenin and CD44 in cells disaggregated from T88 and T93 spheroids. Confocal images of cells disaggregated from T88 and T93 spheroids obtained in medium containing (LiCl) or not containing (untreated) LiCl. Cells were dual-stained with anti-Vimentin (red)/anti-Pan-cytokeratin (green) and anti-β-catenin (red)/anti-CD44 (green) antibodies. Nuclei were counterstained with DAPI (blue).
In both cell cultures, and under both experimental conditions, we observed the nuclear localisation of E-cadherin, N-cadherin, CD133 and CD44v6 (Fig. 10B), but not that of the canonical isoform of CD44 or β-catenin (Fig. 11). When analysed by RT-qPCR, Twist1 mRNA expression was downregulated in both cell cultures, while Snail mRNA was upregulated, mainly in the T93 cells, and even in cells disaggregated from the treated tumourspheres (Fig. 10C). Indeed, both cell cultures again expressed the mesenchymal protein, Vimentin, which was upregulated in the T88 cells derived from the treated tumourspheres (Fig. 11), in accordance with our previous observation showing that following 10 days of incubation with LiCl, Vimentin mRNA and protein expression levels strongly decreased in the T93 cells, while they increased in the T88 cells (24). Nuclear localisation was also observed for Snail, LGR5 and Sox2 (Fig. 12). Nanog exhibited cytoplasmic localisation in the cells derived from the untreated tumourspheres, particularly in the T88 cells, while a clear nuclear localisation in the cells derived from the treated tumourspheres was observed (Fig. 12). Oct4 also exhibited cytosolic localisation in the T88 cells derived from the untreated tumourspheres, but moved into the nucleus when the tumourspheres were treated with LiCl (Fig. 12). By contrast, Oct4 exhibited a nuclear expression in the T93 cells under both experimental conditions (Fig. 12). ALDH1 was undetectable under all the conditions analysed (Fig. 12).
Figure 12.
Expression of Nanog, ALDH1, Snail, LGR5, Oct4 and Sox-2 in cells disaggregated from T88 and T93 spheroids. Confocal images of cells disaggregated from T88 and T93 spheroids obtained in medium containing (LiCl) or not containing (untreated) LiCl. Cells were dual-stained with anti-Nanog (red)/anti-ALDH1 (green), anti-Snail (red)/anti-LGR5 (green) and anti-Oct4 (red)/anti-Sox-2 (green) antibodies. Nuclei were counterstained with DAPI (blue).
Discussion
EMT and its reverting process, MET, are physiological processes occurring during embryonic development and tissue remodelling that confer plasticity to cancer cells. It has been suggested that EMT and cell plasticity may be responsible for the acquisition of chemotherapeutic resistance and metastasis development in several tumours, including CRCs (12).
We previously isolated and characterized at a molecular level two primary CRC cell cultures from the tumour tissues of patients 88 and 93 of our bio-bank (the T88 and T93 cultures). As previously described, the T93 cells exhibited a CIN phenotype, while the T88 cells exhibited a MIN one, with high MSI. We demonstrated that the T88 and T93 cells were mesenchymal colon cancer cells that had undergone EMT from epithelial adenocarcinoma cells and simultaneously expressed epithelial (Cks and E-cadherin) and mesenchymal (Vimentin and N-cadherin) markers. High levels of EMT-associated transcription factors (Twist and Snail) and several stemness markers were also found (24). These finding were in accordance with previous data indicating that EMT induces the expression of stem cell-specific genes, and may represent a source of cancer stem-like cells (36). We also demonstrated that incubation with LiCl, a specific GSK-3β inhibitor, induced MET (24).
In this study, we characterised our experimental system of adherent primary mesenchymal colon cancer cells and their paired tumourspheres more in depth, by analysing the localisation and expression of a larger panel of markers, including E- and N-cadherin, CD133, CD144v6, ALDH1 and LGR5. Furthermore, we explored the effects of LiCl on cell motility and cell plasticity of CRC cell cultures.
Thus, we confirmed the epithelial/mesenchymal features of these cells and demonstrated that they were characterised by the nuclear localisation of several stemness markers, including Nanog, Oct4, Sox2, LGR5, ALDH1, CD133 and CD44v6.
Of note, we observed atypical nuclear N-cadherin, CD133 and Cd44v6 localisation in mesenchymal CRC cells. N-cadherin is a crucial protein during EMT, cancer progression and invasion, and an elevated N-cadherin expression is often associated with a poor prognosis (37,38); however, its role in cell nuclei requires further investigation. CD133 is a transmembrane glycoprotein often expressed on adult stem cells, where it functions in maintaining stem cell properties by suppressing differentiation. CD133 expression is also associated with several types of cancer. It was one of the first molecular biomarkers associated with cancer stemness, although it later became clear that it is expressed both in differentiated and undifferentiated CRC cells (39,40).
Data regarding nuclear CD133 localisation are rarely described. To the best of our knowledge, there are only three studies that have reported nuclear CD133 localisation, one in triple-negative breast cancer (41), one in a small subpopulation of rhabdomyosarcoma cell lines (ranged from 3.4-7.5% of the total population) (42), and one in non-small cell lung cancer (NSCLC). In the latter study, the authors found the nuclear and cytoplasmic CD133 localisation in 239 NSCLC cases using tissue immunohistochemistry, demonstrating that a high nuclear and cytoplasmic CD133 expression were associated with poor prognoses in these carcinomas (43).
CD44 is a plasma membrane glycoprotein that is a receptor for hyaluronan and many other extracellular matrix components. Thus, it transduces signals to membrane-associated cytoskeletal proteins or to the nucleus, to regulate the expression of genes related to cell-matrix adhesion, cell migration, proliferation, differentiation and survival. Previous data have indicated that CD44, particularly CD44v isoforms, are specific CSC markers. CD44 proteins integrate environmental and cellular signalling to regulate cancer stemness, they are also involved in EMT regulation (44) and in the regulation of reactive oxygen species metabolism in CSCs (45).
Several cell surface proteins are known to migrate into the nucleus as intact polypeptides or proteolytic fragments, where they function as transcription factors. It was previously described that the canonical isoform of CD44 is imported into the nucleus through the nuclear pore complex, thus promoting cell proliferation (46). In this study, we observed, to the best of our knowledge, for the first time, that the CD44v6 isoform exhibited a mainly nuclear localisation pattern in mesenchymal CRC cells, while the canonical form retained a primarily membrane and cytosolic pattern. Of note, cells of the organoid-like bodies, exhibited mainly a plasma membrane and cytosolic localisation of CD133 and CD44v6, suggesting that changes in localisation between the membrane and nucleus of these specific proteins could allow the identification of cell plasticity in CRCs, a crucial feature of cancer cells that will develop into metastatic disease.
GSK-3β is a multitasking serine-threonine kinase that can function as a pro-apoptotic or anti-apoptotic factor (47). Usually, GSK-3β phosphorylates β-catenin, inducing its proteasomal degradation. When GSK-3β is inactivated, it cannot phosphorylate β-catenin, which is then stabilized, and translocates into the nucleus, where it activates target genes that drive cell proliferation. In colon and pancreatic cancer cells, GSK-3β, which is constitutively overexpressed, activates a proliferative signal (48,49), thus acting as an oncogene. In this study, we examined the effects of LiCl, a specific GSK-3β inhibitor, on our cellular model, demonstrating that LiCl blocks the migration of T88 and T93 cells, as it has also been reported for commercially available glioma, glioblastoma, retinoblastoma and SW480 cell lines (50-53).
We have also observed that LiCl affects stemness features, abolishing the expression of all mesenchymal and stemness markers, thus altering the dynamics of tumoursphere formation and cell plasticity. As previously described (24), we found that following incubation with LiCl, cells differentiated, lost their symmetrical (mesenchymal) shape and restored their polarity. In these cells, β-catenin did not exhibit nuclear accumulation, but rather co-localised with CD44 canonical protein at the plasma membrane. Conversely, E-cadherin maintained a discontinuous punctate pattern at the plasma membrane in cell contact areas, denoting that cells continued to establish minimal contacts with their neighbours. Taken together, these observations suggested an incomplete epithelial differentiation of the LiCl-treated cells. Furthermore, following incubation with LiCl, nucleolar N-cadherin localisation was observed, whereas this was not observed in the untreated cells. This intriguing finding supports the fascinating hypothesis of a transcriptional inhibition mechanism mediated by the nucleolar localisation of proteins (54); nevertheless, the role of nucleolar localization of several protein in cancer needs to be further elucidated.
Cellular plasticity plays an important role in cancer development and progression; only cancer cells that are able to switch from epithelial to mesenchymal phenotypes and vice versa have metastasis-initiating potential, while disseminated cancer cells that have undergone EMT, but lose their plasticity are ineffective at seeding metastatic colonies (19). In light of our observations, we suggest that during cell plasticity, a mesenchymal cell phenotype could be characterised by the nuclear localisation of several protein, some of which are usually expressed at the membrane in the epithelial phenotype. Thus, changes in localisation between membrane and nucleus of such proteins (CD133 and CD44v6), could allow and characterize cell plasticity in colorectal cancer progression. By analysing the molecular features of cells able to generate tumourspheres, we identified a panel of biomarkers (including E- and N-cadherin, CD133, CD44v6, Snail, Oct4, Sox2, Nanog and LGR5) expressed by the most resistant subpopulation of cancer cells with a mainly nuclear localisation. We suggest that these proteins could represent a prognostic and/or predictive biomarker panel that needs to be validated for supporting CRC care.
Finally, we observed that when cells were disaggregated from tumourspheres performed in LiCl, only the T93 cells, which exhibited a microsatellite stable (MSS) and CIN phenotype, were able to re-grow, while the T88 cells, which exhibited an MSI-high phenotype, appeared quiescent. This observation is in agreement with findings in the literature, which indicates a less aggressive phenotype of MSI CRC compared with MSS subtypes (1,23). Since the T93 cells exhibited the upregulation of Snail, even if tumourspheres were performed following incubation with LiCl, a pivotal role for this protein in the aggressiveness and drug resistance of CRC cells is suggested. Although this hypothesis warrants further investigation, first of all enlarging the number of cultures analysed and supporting the finding with a statistical analysis, it has been demonstrated that Snail knockdown results in an upregulation of the Raf kinase inhibitor protein (RKIP), increased apoptosis, MET and reduction of CSC markers, all of which contribute to decreased chemoresistance (55).
In conclusion, in this study, we established a system of adherent primary mesenchymal colon cancer cells and paired tumourspheres, which are useful for studying the mechanisms underlying CRC progression and drug response. In light of our finding, we suggest that LiCl, a specific GSK-3β inhibitor, could represent a drug candidate and that GSK-3β inhibition may be a promising direction for future cancer therapy that needs to be better elucidated. As recently demonstrated for other cancer types, such as glioma (51), LiCl, a drug already used in clinical practice for the treatment of bipolar disorders, could represent an alternative therapy in colon cancer care and/or able to sensitize cancer cells to chemo-radio-therapy. Our findings indicate that it could act through the downregulation of EMT and stem cell biomarkers, thus inhibiting crucial cancer cell features, such as motility and plasticity. We also demonstrated that the observed atypical nuclear N-cadherin, CD133 and Cd44v6 localisation in mesenchymal CRC cells is a specific cancer feature, which may play a role during tumour progression. In our humble opinion, this finding may open new perspectives to clarify the molecular basis of EMT in cancer cells.
Acknowledgments
The authors would like to thank Dr James P. Mahaffey from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Abbreviations
- EMT
epithelial-to-mesenchymal transition
- MET
mesenchymal-to-epithelial transition
- CRC
colorectal cancer
- CIN
chromosomal instability
- MIN
microsatellite instability
- HM
healthy colon mucosa
Funding
This study was supported by: Regione Campania, LR5/2002-2014; 'Fondo Straordinario di Ateneo-2017-Università di Napoli Federico II'.
Availability of data and materials
All data generated or analysed during this study are included in this published article. No datasets were generated or analysed during the current study.
Authors' contributions
MDR and PI designed the study; MDR, MT, VC and AC, performed research; PD, DR, UP, CAD and MM provided sample collection and clinical support; MDR, PI and MT contributed to data interpretation. MDR and MT wrote the manuscript, and FD and RL critically revised the manuscript and participated in the analysis and interpretation of the data. All authors reviewed, edited and approved the final version of the manuscript.
Ethics approval and consent to participate
Samples from all subjects who participated in this study were collected after obtaining authorisation from the Comitato etico per le attività Biomediche - Carlo Romano of the University of Naples Federico II (protocol no. 432/17). Authorisation was granted only once the study had received ethical approval and written informed consent had been obtained from all participants. All methods were performed in accordance with the relevant guidelines and regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.De Rosa M, Rega D, Costabile V, Duraturo F, Niglio A, Izzo P, Pace U, Delrio P. The biological complexity of colorectal cancer: Insights into biomarkers for early detection and personalized care. Therap Adv Gastroenterol. 2016;9:861–886. doi: 10.1177/1756283X16659790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Peifer M. Developmental biology: Colon construction. Nature. 2002;420:274–275. 277. doi: 10.1038/420274a. [DOI] [PubMed] [Google Scholar]
- 4.Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review) Oncol Rep. 2015;34:1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangemi R, Paleari L, Orengo AM, Cesario A, Chessa L, Ferrini S, Russo P. Cancer stem cells: A new paradigm for understanding tumor growth and progression and drug resistance. Curr Med Chem. 2009;16:1688–1703. doi: 10.2174/092986709788186147. [DOI] [PubMed] [Google Scholar]
- 7.Fanali C, Lucchetti D, Farina M, Corbi M, Cufino V, Cittadini A, Sgambato A. Cancer stem cells in colorectal cancer from pathogenesis to therapy: Controversies and perspectives. World J Gastroenterol. 2014;20:923–942. doi: 10.3748/wjg.v20.i4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 9.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Pei D, Zheng H. Transitions between epithelial and mesenchymal states during cell fate conversions. Protein Cell. 2014;5:580–591. doi: 10.1007/s13238-014-0064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loboda A, Nebozhyn MV, Watters JW, Buser CA, Shaw PM, Huang PS, Van't Veer L, Tollenaar RA, Jackson DB, Agrawal D, et al. EMT is the dominant program in human colon cancer. BMC Med Genomics. 2011;4:9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, II, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan F, Samuel S, Evans KW, Lu J, Xia L, Zhou Y, Sceusi E, Tozzi F, Ye XC, Mani SA, et al. Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1:5–16. doi: 10.1002/cam4.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vichalkovski A, Gresko E, Hess D, Restuccia DF, Hemmings BA. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29:3554–3565. doi: 10.1038/onc.2010.115. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Pan X, Lei W, Wang J, Song J. Transforming growth factor-beta1 induces epithelial-to-mesenchymal transition and apoptosis via a cell cycle-dependent mechanism. Oncogene. 2006;25:7235–7244. doi: 10.1038/sj.onc.1209712. [DOI] [PubMed] [Google Scholar]
- 17.Vega S, Morales AV, Ocaña OH, Valdés F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ong BA, Vega KJ, Houchen CW. Intestinal stem cells and the colorectal cancer microenvironment. World J Gastroenterol. 2014;20:1898–1909. doi: 10.3748/wjg.v20.i8.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doherty MR, Smigiel JM, Junk DJ, Jackson MW. Cancer stem cell plasticity drives therapeutic resistance. Cancers (Basel) 2016;8:E8. doi: 10.3390/cancers8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 21.Grassilli E, Narloch R, Federzoni E, Ianzano L, Pisano F, Giovannoni R, Romano G, Masiero L, Leone BE, Bonin S, et al. Inhibition of GSK3B bypass drug resistance of p53-null colon carcinomas by enabling necroptosis in response to chemotherapy. Clin Cancer Res. 2013;19:3820–3831. doi: 10.1158/1078-0432.CCR-12-3289. [DOI] [PubMed] [Google Scholar]
- 22.McCubrey JA, Steelman LS, Bertrand FE, Davis NM, Sokolosky M, Abrams SL, Montalto G, D'Assoro AB, Libra M, Nicoletti F, et al. GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014;5:2881–2911. doi: 10.18632/oncotarget.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liccardo R, De Rosa M, Izzo P, Duraturo F. Novel Implications in Molecular Diagnosis of Lynch Syndrome. Gastroenterol Res Pract. 2017;2017:2595098. doi: 10.1155/2017/2595098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costabile V, Duraturo F, Delrio P, Rega D, Pace U, Liccardo R, Rossi GB, Genesio R, Nitsch L, Izzo P, et al. Lithium chloride induces mesenchymal-to-epithelial reverting transition in primary colon cancer cell cultures. Int J Oncol. 2015;46:1913–1923. doi: 10.3892/ijo.2015.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 26.Paparo L, Rossi GB, Delrio P, Rega D, Duraturo F, Liccardo R, Debellis M, Izzo P, De Rosa M. Differential expression of PTEN gene correlates with phenotypic heterogeneity in three cases of patients showing clinical manifestations of PTEN hamartoma tumour syndrome. Hered Cancer Clin Pract. 2013;11:8. doi: 10.1186/1897-4287-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duraturo F, Liccardo R, Cavallo A, De Rosa M, Rossi GB, Izzo P. Multivariate analysis as a method for evaluating the pathogenicity of novel genetic MLH1 variants in patients with colorectal cancer and microsatellite instability. Int J Mol Med. 2015:511–517. doi: 10.3892/ijmm.2015.2255. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Galatola M, Paparo L, Duraturo F, Turano M, Rossi GB, Izzo P, De Rosa M. Beta catenin and cytokine pathway dysregu-lation in patients with manifestations of the 'PTEN hamartoma tumor syndrome'. BMC Med Genet. 2012;13:28. doi: 10.1186/1471-2350-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angrisani A, Turano M, Paparo L, Di Mauro C, Furia M. A new human dyskerin isoform with cytoplasmic localization. Biochim Biophys Acta. 2011;1810:1361–1368. doi: 10.1016/j.bbagen.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 31.Di Maio N, Vicidomini R, Angrisani A, Belli V, Furia M, Turano M. A new role for human dyskerin in vesicular trafficking. FEBS Open Bio. 2017;7:1453–1468. doi: 10.1002/2211-5463.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng B, Dong TT, Wang LL, Zhou HM, Zhao HC, Dong F, Zheng MH. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7:e43452. doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 34.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 35.Weiswald LB, Bellet D, Dangles-Marie V. Spherical cancer models in tumor biology. Neoplasia. 2015;17:1–15. doi: 10.1016/j.neo.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 37.Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 38.DI Domenico M, Pierantoni GM, Feola A, Esposito F, Laino L, DE Rosa A, Rullo R, Mazzotta M, Martano M, Sanguedolce F, et al. Prognostic significance of N-Cadherin expression in oral squamous cell carcinoma. Anticancer Res. 2011;31:4211–4218. [PubMed] [Google Scholar]
- 39.LaBarge MA, Bissell MJ. Is CD133 a marker of metastatic colon cancer stem cells? J Clin Invest. 2008;118:2021–2024. doi: 10.1172/JCI36046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantile M, Collina F, D'Aiuto M, Rinaldo M, Pirozzi G, Borsellino C, Franco R, Botti G, Di Bonito M. Nuclear localization of cancer stem cell marker CD133 in triple-negative breast cancer: A case report. Tumori. 2013;99:e245–e250. doi: 10.1177/030089161309900523. [DOI] [PubMed] [Google Scholar]
- 42.Nunukova A, Neradil J, Skoda J, Jaros J, Hampl A, Sterba J, Veselska R. Atypical nuclear localization of CD133 plasma membrane glycoprotein in rhabdomyosarcoma cell lines. Int J Mol Med. 2015;36:65–72. doi: 10.3892/ijmm.2015.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang M, Zhu H, Feng J, Ni S, Huang J. High CD133 expression in the nucleus and cytoplasm predicts poor prognosis in non-small cell lung cancer. Dis Markers. 2015;2015:986095. doi: 10.1155/2015/986095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessède E, Staedel C, Acuña Amador LA, Nguyen PH, Chambonnier L, Hatakeyama M, Belleannée G, Mégraud F, Varon C. Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene. 2014;33:4123–4131. doi: 10.1038/onc.2013.380. [DOI] [PubMed] [Google Scholar]
- 45.Yan Y, Zuo X, Wei D. Concise Review: Emerging role of CD44 in cancer stem cells: A promising biomarker and therapeutic target. Stem. Cells Transl Med. 2015;4:1033–1043. doi: 10.5966/sctm.2015-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JL, Wang MJ, Chen JY. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J Cell Biol. 2009;185:949–957. doi: 10.1083/jcb.200812060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beurel E, Jope RS. The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog Neurobiol. 2006;79:173–189. doi: 10.1016/j.pneurobio.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shakoori A, Ougolkov A, Yu ZW, Zhang B, Modarressi MH, Billadeau DD, Mai M, Takahashi Y, Minamoto T. Deregulated GSK3beta activity in colorectal cancer: Its association with tumor cell survival and proliferation. Biochem Biophys Res Commu. 2005;334:1365–1373. doi: 10.1016/j.bbrc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 49.Shakoori A, Mai W, Miyashita K, Yasumoto K, Takahashi Y, Ooi A, Kawakami K, Minamoto T. Inhibition of GSK-3 beta activity attenuates proliferation of human colon cancer cells in rodents. Cancer Sci. 2007;98:1388–1393. doi: 10.1111/j.1349-7006.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y, Jiao Y, Zheng S, Liang A, Hu F. Combination of lithium chloride and pEGFP-N1-BmK CT effectively decreases proliferation and migration of C6 glioma cells. Cytotechnology. 2016;68:197–202. doi: 10.1007/s10616-014-9768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cockle JV, Picton S, Levesley J, Ilett E, Carcaboso AM, Short S, Steel LP, Melcher A, Lawler SE, Brüning-Richardson A. Cell migration in paediatric glioma; characterisation and potential therapeutic targeting. Br J Cancer. 2015;112:693–703. doi: 10.1038/bjc.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva AK, Yi H, Hayes SH, Seigel GM, Hackam AS. Lithium chloride regulates the proliferation of stem-like cells in retinoblastoma cell lines: A potential role for the canonical Wnt signaling pathway. Mol Vis. 2010;16:36–45. [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Huang K, Liu X, Liu J, Lu X, Tao K, Wang G, Wang J. Lithium chloride suppresses colorectal cancer cell survival and proliferation through ROS/GSK-3β/NF-κB signaling pathway. Oxid Med Cell Longev. 2014;2014:241864. doi: 10.1155/2014/241864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee TY, Liu CL, Chang YC, Nieh S, Lin YS, Jao SW, Chen SF, Liu TY. Increased chemoresistance via Snail-Raf kinase inhibitor protein signaling in colorectal cancer in response to a nicotine derivative. Oncotarget. 2016;7:23512–23520. doi: 10.18632/oncotarget.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: The fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article. No datasets were generated or analysed during the current study.