Abstract
Improving the long-term survival of patients with hepatocellular carcinoma (HCC) remains a challenge due to metastasis and recurrence. In this study, we demonstrate that the overexpression of miR-103 in HCC cells promotes epithelial-mesenchymal transition (EMT), and is associated with an enhanced metastasis and poor outcomes, as shown by western blot analysis and immunohistochemistry. Mechanistically, using reporter luciferase assay we reveal that the serine/threonine-protein kinase, large tumor suppressor kinase 2 (LATS2), a key component of the Hippo signaling pathway, is a direct target of miR-103 in HCC cells. Transwell assay, MTT assay and western blot analysis were performed to reveal that LATS2 can counteract the functional effects of miR-103 on HCC metastasis, growth and EMT. The analyses of clinical data indicated that a high expression of miR-103 correlated with a high expression of vimentin, but with a low expression of LATS2 and E-cadherin in HCC tissues. miR-103 also reduced yes-associated protein (YAP) phosphorylation. On the whole, the findings of this study suggest that miR-103 promotes HCC metastasis and EMT by directly inhibiting LATS2. Thus, targeting miR-103/LATS2 may prove to be a promising therapeutic strategy for HCC.
Keywords: epithelial-mesenchymal transition, hepatocellular carcinoma, LATS2, miR-103, YAP
Introduction
Hepatocellular carcinoma (HCC) is reported as the sixth most common type of cancer worldwide (1). Patients with HCC generally have a poor prognosis due to the highly invasive nature of the disease (2). A large number of studies have reported that the inactivation of tumor suppressor genes and the activation of oncogenes is responsible for the metastasis of HCC (3-5). However, the mechanisms underlying recurrence and metastasis in HCC have not yet been fully determined.
Epithelial-mesenchymal transition (EMT), characterized by the alteration in the cancer cell phenotype from an epithelial one to a motile mesenchymal one, is a well-known initiating and essential event in the progression and metastasis of various types of cancer (6-8). MicroRNAs (miRNAs or miRs) are a class of long non-coding RNAs which are 18-24 nucleotides in length. These RNAs may serve as vital modulators which regulate a number of biological processes in cancer, including invasion and metastasis (9-11). A number of miRNAs have been shown to be involved in the metastasis of HCC, including miRNA-92a, miRNA-7, miRNA-3910, miRNA-1301, miRNA-203a-3p and miRNA-93 (11-17). Researchers have focused on the function of miR-103 in tumorigenesis. miR-103 belongs to the miR-103/107 family, and is capable of inducing EMT of mammary epithelial cells. It is of interest that miR-103 seems to play differential roles in various types of cancer. For example, miR-103 functions as a tumor suppressor gene in non-small cell lung cancer (18), while in gastric cancer, colorectal cancer and prostate cancer, miR-103 functions as an oncogene (19-21). A recent study indicated that miR-103 promoted HCC cell growth by targeting A kinase anchor protein 12 (AKAP12) (22). However, to date, at least to the best of our knowledge, no causal association has been established between miR-103 and metastasis and EMT in HCC. Thus, studies focusing on the exact mechanisms of action of miR-103 in HCC progression are necessary.
Therefore, in the current study, we aimed to determine the role and mechanism of action of miR-103 in metastasis and EMT in HCC. For this purpose, assays were performed using human specimens, HCC cell lines and animals. The findings of this study may aid in the development of novel targeted therapies for HCC.
Materials and methods
Patients and tissue samples
Tissue samples for this study were collected from 120 patients with HCC (including patients with or without metastasis) who underwent surgical resection between March, 2010 and March, 2011 at the Second Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China). The Ethics Committee of the Medical School of Xi'an Jiaotong University reviewed and approved the study, and written informed consent was obtained from each participant at each examination phase. The study complied with the principles of the Helsinki Declaration. The clinicopathological characteristics of all the patients are presented in Table I. None of the patients underwent prior chemotherapy or radiotherapy. Fresh HCC tissues paired with distant non-cancerous tissues samples (≥2 cm distance from the tumor margin) were immediately frozen in liquid nitrogen during the surgical resection. They were stored at −80°C [for use in western blot analysis or reverse transcription-quantitative PCR (RT-qPCR) assay] or in paraffin (for use in immunohistochemistry).
Table I.
Association between miR-103 and LATS2 expression levels with the clinicopathological characteristic of patients with hepatocellular carcinoma.
| Variable | Total no. of patients, n=120 (%) | miR-103
|
P-value | LATS2
|
P-value | ||
|---|---|---|---|---|---|---|---|
| Low expression | High expression | Low expression | High expression | ||||
| Age (years) | 0.057 | 0.120 | |||||
| <50 | 49 (40.8) | 10 | 39 | 37 | 12 | ||
| ≥50 | 71 (59.2) | 26 | 45 | 44 | 27 | ||
| Sex | 0.496 | 0.349 | |||||
| Female | 21 (17.5) | 5 | 16 | 16 | 5 | ||
| Male | 99 (82.5) | 31 | 68 | 65 | 34 | ||
| HBsAg | 0.955 | 0.903 | |||||
| Positive | 76 (63.3) | 15 | 61 | 51 | 25 | ||
| Negative | 44 (36.7) | 21 | 23 | 30 | 14 | ||
| AFP (ng/ml) | 0.01a | 0.422 | |||||
| <400 | 97 (80.8) | 24 | 73 | 50 | 27 | ||
| ≥400 | 23 (19.2) | 12 | 11 | 31 | 12 | ||
| Cirrhosis | 0.741 | 0. 903 | |||||
| Yes | 76 (63.3) | 22 | 54 | 51 | 25 | ||
| No | 44 (36.7) | 14 | 30 | 30 | 14 | ||
| Tumor size (cm) | 0.007a | 0.032a | |||||
| <5 | 48 (40) | 21 | 27 | 27 | 21 | ||
| ≥5 | 72 (60) | 15 | 57 | 54 | 18 | ||
| Tumor multiplicity | 0.228 | 0.992 | |||||
| Single | 77 (64.2) | 26 | 51 | 52 | 25 | ||
| Multiple | 43 (35.8) | 10 | 33 | 29 | 14 | ||
| Differentiation | 0.318 | 0.383 | |||||
| Well-moderate | 53 (44.2) | 17 | 36 | 38 | 15 | ||
| Poor-undifferentiated | 67 (55.8) | 19 | 48 | 43 | 24 | ||
| Microscopic vascular invasion | <0.001b | 0.001a | |||||
| Yes | 39 (32.5) | 28 | 11 | 18 | 21 | ||
| No | 81 (67.5) | 8 | 73 | 63 | 18 | ||
| Stage | <0.001b | 0.021a | |||||
| I–II | 78 (65) | 32 | 46 | 47 | 31 | ||
| III–IV (with metastasis) | 42 (35) | 4 | 38 | 34 | 8 | ||
Values in bold indicate statistical significance (aP<0.05 and bP<0.01).
Immunohistochemistry (IHC)
The specimens were cut into 3-μm-thick sections and placed on histological slides. The sections were then deparaffinized, rehydrated and immersed into 3% hydrogen peroxide. The steps followed for immunohistochemistry and the staining intensity determined were performed following procedures reported previously (23). The primary antibodies used in this study were all specific for immunohistochemistry, including anti-large tumor suppressor kinase 2 (LATS2) antibody (bs-4081R; at a dilution of 1:50), anti-E-cadherin antibody (bs-1016R; at a dilution of 1:100) and anti-vimentin antibody (bs-8533R; at a dilution of 1:100) (all from Beijing Bioss Biotechnology, Beijing, China). Horseradish peroxidase (HRP)-labelled anti-rabbit second antibodies were used in this study (bse-0302R; Beijing Bioss Biotechnology; at a dilution of 1:6,000). Images were acquired using a Leica TCS SP2 confocal laser-scanning microscope (Leica Microsystems).
RT-qPCR
miR-103 and LATS2 expression in both samples and HCC cells, and CYR61, AREG, CTGF and CXCL5 expression in the HCC samples was assessed by RT-qPCR. RT-qPCR was performed using SYBR-Green (Shanghai GeneCore Biotechnologies Co., Ltd., Shanghai, China). Firstly, RNeasy reagent (Qiagen, Shanghai, China) was used to extract total RNA from samples or HCC cells. Subsequently, a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure the total RNA concentration. Total RNA was then reverse transcribed to obtain cDNA for RT-qPCR using a reverse transcription kit (ABI, Life Technologies, Carlsbad, CA, USA). The RT-qPCR conditions were as follows: Pre-heating for 10 min at 95°C; repeating 40 cycles at 95°C for 15 sec and 60 sec at 60°C. The specific primers were designed and synthesized by Takara (Dalian, China), and the sequences for PCR are shown as follows: miR-103, 5′-CCCGCCAAGCCCTTACC-3′ (forward) and 5′-GCCGTCGGTGATGCTTTTTTGG-3′ (reverse); LATS2, 5′-ACCCCAAAGTTCGGACCTTAT-3′ (forward) and 5′-CAT TTGCCGGTTCACTTCTGC-3′ (reverse); cysteine-rich angiogenic inducer (61CYR61), 5′-CCCTGAACTTGTGGATGTCATTG-3′ (forward) and 5′-GTCATGATGATCCAGTCCTGC AAA-3′ (reverse); amphiregulin (AREG), 5′-TGCTGGATTGG ACCTCAATG-3′ (forward) and 5′-TCCCGAGGACGGTTCAC TAC-3′ (reverse); connective tissue growth factor (CTGF), 5′-GAAAAGAUUCCCACCCAU-3′ (forward) and 5′-AUU GGGUGGGAAUCUUUUC-3′ (reverse); C-X-C motif chemo-kine 5 (CXCL5), 5′-GTTCCATCTCGCCATTCATGC-3′ (forward) and 5′-GCGGCTATGACTGAGGAAGG-3′ (reverse); GAPDH, 5′-AATGGACAACTGGTCGTGGAC-3′ (forward) and 5′-CCCTCCAGGGGATCTGTTTG-3′ (reverse). RT-qPCR was performed in triplicate. The level of U6 RNA was used as internal control for miR-103. The level of GAPDH was used as the internal control for LATS2, Cyr61, AREG, CTGF and CXCL5. The relative mRNA expression against the GAPDH levels was assessed using 2−ΔΔCq method (24).
Cell culture and transfection
HCC cell lines (MHCC-97H, MHCC-97L and Hep3B) and L02 normal liver cells were purchased from ATCC (Manassas, VA, USA) and cultured in DMEM containing 10% FBS with 100 μg/ml streptomycin and 100 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA) in an incubator at 37°C with 5% CO2. miR-103 mimic was purchased from GeneCopoeia (Guangzhou, China). The LATS2 vector was purchased from Addgene (Cambridge, MA, USA). These vectors or mimics were transfected into the HCC cells using Lipofectamine 2000. The sequences of the vectors and the mimics were as follows: LATS2 target vector, TTC ACCTTCCGAAGGTTCT; negative control (NC) vector, TCG TACTCTCGTCTTCGAT; miR-103 mimic sense, AGCAGCA UUGUAAGGGCUAUGA and antisense, AUAGCCCUGUAC AAUGCUGCUUU; miR-103 NC mimic sense, AGCAGCAG UUUAGGGCACUAUGA and antisense, AUAGUGCCCUAA ACUGCUGCUUU. The transfection concentrations used in this study were as follows: miR-103 NC mimic and miR-103 mimic, 50 nM; LATS2 NC vector and LATS2 vector, 100 μg/ml. Total RNA and protein were subsequently collected at 48 h following transfection.
Antibodies and western blot analysis
The cells were lysed in RIPA buffer and the protein concentrations were determined by BCA assays (Cell Signaling Technology, Inc., Danvers, MA, USA). The total protein for each sample was quantified by the Bradford method. Total proteins (25 μg) were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred onto fluoride membranes. The membranes were then treated with the primary antibodies followed by blocking in 5% non-fat milk for 1 h. The primary antibodies used were as follows: anti-LATS2 antibody (bs-4081R; Beijing Bioss Biotechnology; at a dilution of 1:500), anti-yes-associated protein (YAP) antibody (#14074; at a dilution of 1:600), anti-phospho-YAP (ser127) antibody (#13008; at a dilution of 1:500), anti-vimentin antibody (#5741; at a dilution of 1:500), anti-E-cadherin antibody (#3195; at a dilution of 1:300), CYR61 (#14479; at a dilution of 1:300) (all from Cell Signaling Technology, Inc., Danvers, MA, USA), AREG (ab99975; at a dilution of 1:300), CTGF (ab6922; at a dilution of 1:300), CXCL5 (ab9802; at a dilution of 1:500) (all from Abcam, Cambridge, MA, USA) and GAPDH (G5262; Sigma-Aldrich; at a dilution of 1:3,000). The membranes were then carefully kept in a refrigerator overnight at 4°C. The membranes were washed and subsequently incubated with goat anti-rabbit secondary antibodies for LATS2, YAP, p-YAP (ser127), E-cadherin, vimentin and goat anti-mouse secondary antibody for GAPDH (anti-rabbit: bs-40295G-IRDye8; anti-mouse: bs-40296G-IRDye8; both at a dilution of 1:10,000) (both from Beijing Bioss Biotechnology) at 37°C for 2 h. Finally, the membranes were tested using a bio-imaging system (DNR Bio-Imaging Systems, Jerusalem, Israel). ImageJ version 1.6.0 software (National Institutes of Health, Bethesda, MD, USA) was used to quantify the intensity of protein bands and normalized by GAPDH.
Animal model assay
The animal experiments carried out in this study were approved by the Experimental Animal Ethical Committee of Xi'an Jiaotong University. A total of 24 male C57BL/6 mice (weight, 13-15 g; age, 6 weeks; 4 groups, 6 in each group) used in our study were obtained from SLAC Laboratory Animal Co. (SCXK-2007-004; Shanghai, China), and raised at 22±2°C with a 12-h light/dark cycle under pathogen-free environment. All mice were freely accessed autoclaved standard food and water. One week later, the HCC cells transfected with LATS2 target vector/miR-103 mimic (1×106) or HCC cells transfected with negative control (NC) vector/NC mimic (1×106) were injected into the lateral tail vein of the nude mice. Another 4 weeks later, the mice were sacrificed. The number of nodules in the lungs was counted and the tumor tissues were cleaned with PBS, fixed with 4% paraformaldehyde for 2 h, and subsequently placed in 20% sucrose solution for 24 h.
Cell migration and invasion assay (Transwell assay)
For the cell migration assays, 2×104 HCC cells in 200 μl FBS-free medium were placed in the upper Transwell chamber of uniformly, while 600 μl of normal culture medium (with 10% FBS) was placed into the bottom chambers. Following 48 h of incubation at 37°C in a 5% CO2 incubator, the cells remaining on the membrane in the top chamber side were carefully wiped off using a cotton swab, while the cells that had migrated through the membrane into the surface of the bottom chambers were softly cleaned and fixed with methanol. The migrated cells were stained with 0.1% crystal violet solution (Amresco, Solon, OH, USA) for 15 min at room temperature. The number of migrated cells was carefully counted in 10 random fields under a microscope (Olympus Corporation, Tokyo, Japan). For the invasion assay, the same procedures were carried out as described above, with the exception that the membrane in the top chamber was coated with 200 mg/ml Matrigel (BD Biosciences, San Jose, CA, USA).
MTT assay
The HCC cells (500 per well in 100 μl medium) were added to 96-well plate and incubated at 37°C for 1, 2, 3, 4, 5, 6 and 7 days, respectively. The medium was replaced with new fresh medium every 48 h in the 2, 3, 4, 5, 6 and 7 days group. Following incubation for each period, 10 μl MTT solution (5 mg/ml in PBS) were added to each well and maintained at 37°C. After 4 h, the medium was removed, and 200 μl of dimethyl sulfoxide were then added. Cell viability were read at a 492 nm wavelength using a plate reader (Bio-Rad Laboratories, Hercules, CA, USA). The number of repeated tests for each group was tripled
Target gene prediction for miRNA
miRNA targets were identified using the bioinformatics software TargetScan Human version 7.1 (www.targetscan.org/vert_71/).
Reporter luciferase assay
HCC control cells and HCC cells were transfected with miR-103 inhibitor, mimics, GL3-LATS2-3′-UTR WT or pGL3-LATS2-3′-UTR MUT luciferase reporter vectors (GenePharma Co., Ltd., Shanghai, China) using Lipofectamine 3000, according to the manufacturer's instructions. The cells were seeded into a 24-well plate. Following transfection for 48 h, all the cells were collected, and measured using the Dual-Luciferase Assay System (Promega, Madison, WI, USA). The relative Firefly luciferase activity was measured by normalizing to Renilla luciferase activity.
Statistical analysis
All the experiments were in triplicate, and the results are presented either as the means ± SD or as one representative experiment. All the statistical analyses were performed using SPSS 20.0 software for Windows. Chi-square tests (χ2) were used to assess the association between miR-103 expression and the patient clinicopathological parameters. The Student's t-test was used to compare the means between 2 groups, one-way analysis of ANOVA with Dunnett's multiple comparisons test were used to compare the means among multiple groups. Spearman's correlation analysis was used to analyze the correlation between miR-103, and E-cadherin, vimentin and LATS2 expression. The Kaplan-Meier method (the log-rank test) was used to plot the survival curves. A two-sided P-value <0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of miR-103 is associated with the progression of human HCC
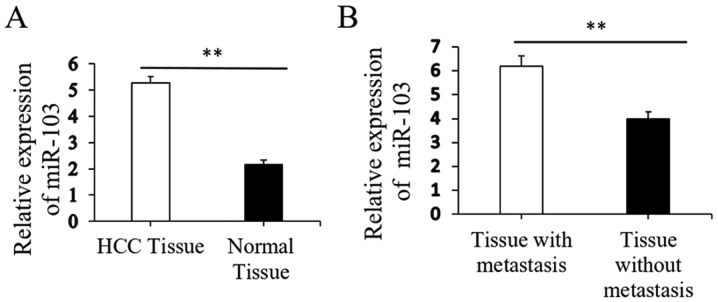
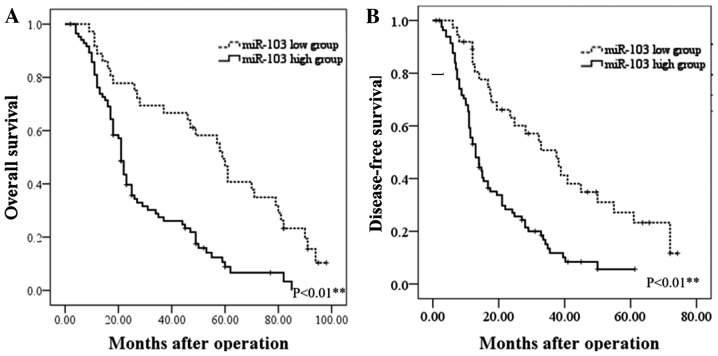
Initial RT-qPCR analysis indicated that the expression of miR-103 was markedly higher in the HCC tissues than in the paired distant non-cancerous tissues (5.28±0.23 vs. 2.15±0.19, P<0.01; Fig. 1A). Further analysis revealed that the miR-103 level in the HCC tissues from patients with metastasis was significantly increased compared to that in patients without metastasis (6.19±0.43 vs. 3.98±0.29, P<0.01; Fig. 1B). The clinical significance of miR-103 in patients with HCC was then further evaluated. The miR-103 expression level exhibited a direct association with microscopic vascular invasion (P<0.001), a high level of AFP (P=0.01), a larger tumor size (P=0.007) and an advanced TNM stage in HCC (P<0.001; Table I). Furthermore, the prognostic impact of the miR-103 level on the survival rate of patients with HCC was determined. The results revealed that patients with HCC with a higher level of miR-103 had a shorter overall survival (OS) and disease-free survival (DFS) than those with a low level (P<0.01; Fig. 2). These findings indicate increased miR-103 level is associated with the metastasis, progression and poor prognosis of HCC.
Figure 1.
Expression of miR-103 in HCC tissues. (A) Relative expression of miR-103 mRNA in HCC compared to distant non-cancerous HCC tissues (**P<0.01). (B) Relative expression of miR-103 mRNA in HCC tissues from patients with metastasis cases and those without metastasis cases (**P<0.01). HCC, hepatocellular carcinoma.
Figure 2.
The prognostic significance of miR-103 in patients with HCC. (A) OS was compared between patients with HCC with a high expression level of miR-103 and those with a low level of miR-103. (B) DFS was compared between patients with HCC with a high expression level of miR-103 and those with a low level of miR-103. HCC, hepatocellular carcinoma; OS, overall survival; DFS, disease-free survival.
miR-103 promotes the growth, migratory and invasive capacity of HCC cells
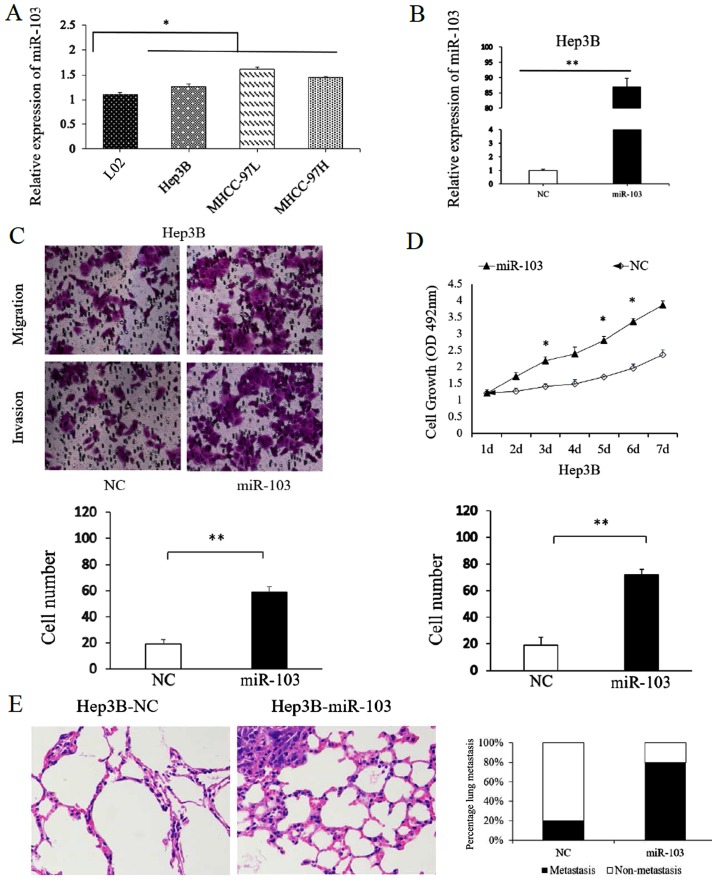
To assess the function of miR-103 in HCC cells, the miR-103 levels in HCC cell lines and in the normal human liver cell line, L02, were compared. Consistent with our findings obtained with the HCC tissues, the results confirmed that miR-103 level was markedly increased in the HCC cell lines compared with the L02 cells. Among the MHCC-97H, MHCC-97L and Hep3B HCC cell lines, the miR-103 level was the lowest in the Hep3B cells (Fig. 3A). Thus, the Hep3B cells were transfected with the miR-103 mimic. Transfection with the miR-103 mimic markedly increased miR-103 expression in the Hep3B cells (Fig. 3B). The results of Transwell assay suggested that the migratory and invasive abilities of the Hep3B cells significantly increased with the upregulation of miR-103 expression (P<0.01; Fig. 3C). Similarly, the results of MTT assay revealed that the overexpression of miR-103 significantly increased the proliferative ability of the Hep3B cells (P<0.05; Fig. 3D).
Figure 3.
miR-103 expression affects the invasive, migratory and proliferative capacity of HCC cells. (A) The expression of miR-103 in HCC cell lines and normal human hepatocyte L02 cells (*P<0.05). (B) The expression of miR-103 was significantly increased following the transfection of miR-103 mimics into Hep3B cells (**P<0.01). The target gene expression levels were quantified against the GAPDH levels. (C) Transwell assay. The number of invaded cells was quantified by counting the total numbers of cells from 10 random fields (magnification, ×200). Data are presented as the means ± SD of 3 independent experiments. (D) MTT assay showing that the overexpression of miR-103 affected the cell proliferative ability of the Hep3B cells (*P<0.05). (E) Left panel, representative H&E staining of lung metastasis with increased miR-103 expression; right panel, the percentage of mice with or without lung metastatic nodules was compared between the miR-103 overexpression and the NC group. HCC, hepatocellular carcinoma; NC, negative control.
To further confirm the function of miR-103 in metastasis of HCC cells in vitro, we also performed tumor xenograft metastasis assays. The results revealed that the overexpression of miR-103 significantly increased the lung metastasis of Hep3B cells (P<0.05; Fig. 3E). These data demonstrate that miR-103 promotes the metastatic behavior of HCC cells in vitro and in vivo.
miR-103 promotes the EMT of HCC cells
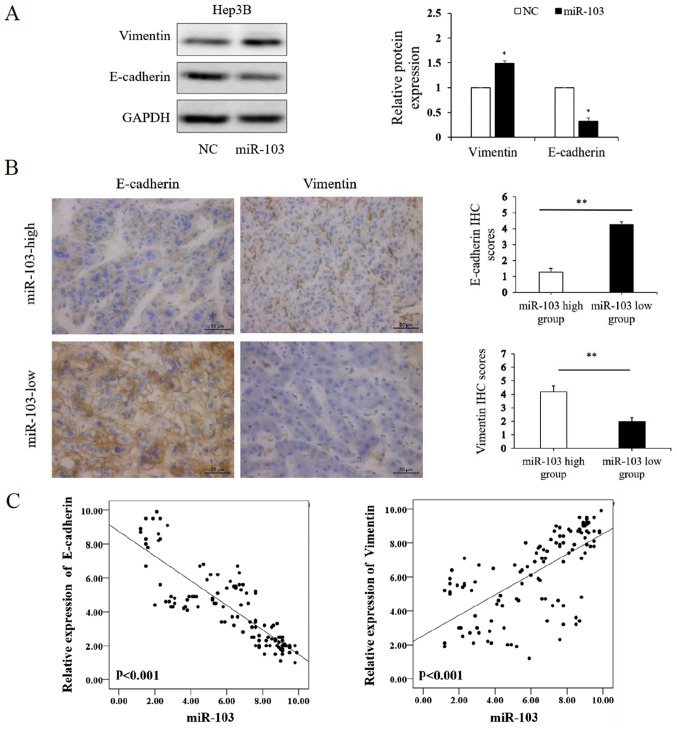
We further investigated whether miR-103 modulates the EMT of HCC cells. The results of western blot analysis revealed that the over-expression of miR-103 markedly increased the expression of vimentin and decreased the expression of E-cadherin in the Hep3B cells (P<0.01; Fig. 4A). We further compared the expression level of E-cadherin and vimentin in the HCC specimens with a high miR-103 expression to those in HCC tissues with a low miR-103 expression by IHC, which yielded similar results (P<0.01; Fig. 4B). Spearman's correlation analysis was performed to determine the correlation between the expression of miR-103, and that of E-cadherin and vimentin derived from RT-qPCR analysis. The results revealed that a high miR-103 expression was strongly associated with a low E-cadherin expression (P<0.001, r=−0.864) and high vimentin expression (P<0.001, r=0.712; Table II and Fig. 4C). These data indicate that miR-103 promotes the invasion, metastasis and EMT of HCC.
Figure 4.
miR-103 promotes the EMT of HCC cells. (A) Western blot analysis of E-cadherin and vimentin expression following the overexpression of miR-103 in HCC cells. GAPDH was used as an internal control (*P<0.05). (B) Immunohistochemistry of E-cadherin and vimentin comparing tissues with a high miR-103 level and those with a low miR-103 level (*P<0.05; magnification, ×400). (C) Left panel, miR-103 expression negatively correlates with E-cadherin expression (Spearman's r=−0.864; see Table II); right panel, miR-103 expression positively correlates with vimentin expression (Spearman's r=0.712; see Table II). EMT, epithelial-mesenchymal transition; HCC, hepatocellular carcinoma; NC, negative control.
Table II.
Association between miR-103 and EMT-related protein expression levels in hepatocellular carcinoma tissue specimens.
| Variable | miR-103
|
r value | P-value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| E-cadherin | -0.864 | P<0.001a | ||
| High expression | 16 | 22 | ||
| Low expression | 68 | 14 | ||
| Vimentin | ||||
| High expression | 68 | 15 | 0.712 | P<0.001a |
| Low expression | 16 | 21 | ||
Values in bold indicate statistical significance (aP<0.01); n=120. Spearman's correlation analysis was use to analyze the correlation between miR-103 and E-cadherin and vimentin expression.
LATS2 may serve as a direct target of miR-103 in HCC
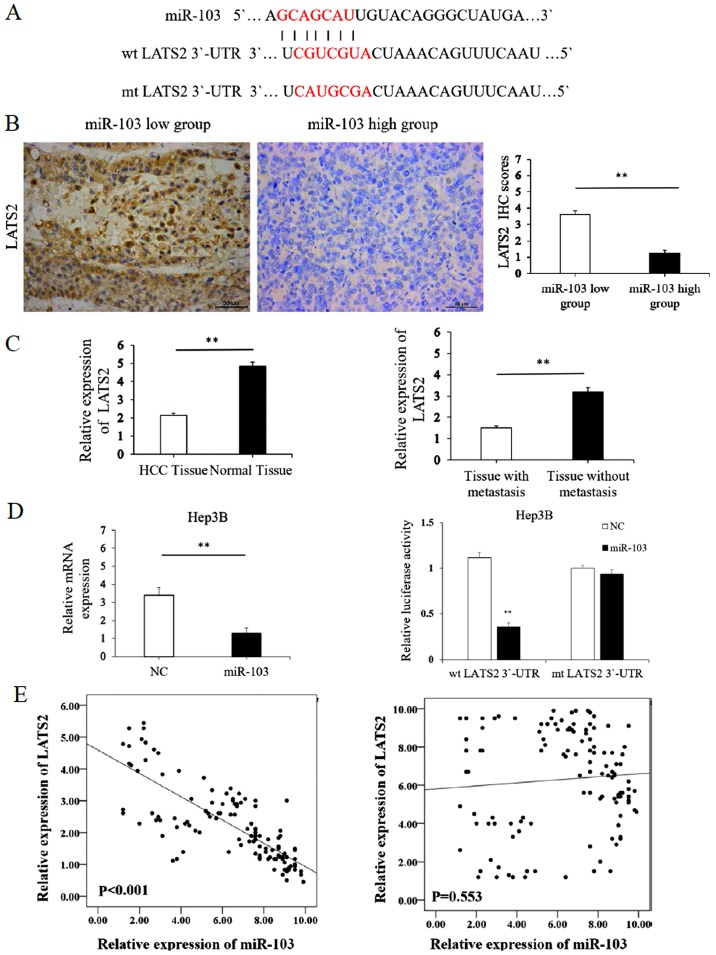
We further accessed the public database, TargetScan to search for miR-103 target genes. The data indicated that the complementary sequence of miR-103 was contained in the 3′-UTR of LATS2 mRNA (Fig. 5A). LATS2 is a well known pivotal tumor suppressor and a critical regulatory factor of cancer metastasis (25,26). Combined with evidence from bioinformatics analysis, we hypothesized miR-103 may promote EMT by regulating LATS2 in HCC. To confirm this hypothesis, the mRNA expression levels of LATS2 were assessed in miR-103- overexpressing HCC cells. The results of RT-qPCR revealed that the overexpression of miR-103 significantly decreased the mRNA level of LATS2 in the Hep3B cells (P<0.01; Fig. 5D, left panel). To confirm the regulation of LATS2 by miR-103 in HCC, we examined the expression of LATS2 in HCC tissues. In addition, the results from IHC revealed that compared with the HCC tissue samples with a low expression level of miR-103, LATS2 expression was markedly decreased in the HCC tissues with a high expression of miR-103 (P<0.05; Fig. 5B). The results of RT-qPCR also revealed that the expression of LATS2 was lower in the HCC tissues than in the distant non-cancerous tissues (2.14±0.12 vs. 4.83±0.24, P<0.01; Fig. 5C). In addition, the level of LATS2 in the HCC samples from patients with metastasis was markedly lower than that in those without metastasis (1.51±0.09 vs. 3.19±0.21, P<0.01; Fig. 5C). Spearman's correlation analysis was also performed to examine the correlation between the expression of miR-103, and that of E-cadherin and vimentin derived from RT-qPCR analysis. A statistically significant inverse correlation was observed between the expression of miR-103 and that of LATS2 in the HCC tissues (r=−0.791, P<0.001; Table III and Fig. 5E). However such a correlation was not observed between miR-103 and LATS2 expression in the distant non-cancerous tissues (r=−0.055, P>0.05; Table III and Fig. 5E). On the other hand, we performed luciferase reporter gene assays to confirm the direct binding between miR-103 and LATS2. As shown in Fig. 5D (right panel), the overexpression of miR-103 in the Hep3B cells inhibited the luciferase activity of LATS2 with a wt 3′-UTR. However, the overexpression of miR-103 did not regulate the luciferase activity of the mutant (mt) 3′-UTR LATS2 significantly. These findings obtained from both the clinical samples and HCC cells demonstrate that LATS2 may act as a direct target of miR-103 in HCC.
Figure 5.
LATS2 acts as a direct downstream target of miR-103. (A) miR-103 and its putative binding sequence in the 3′-UTR of LATS2. (B) Immunohistochemistry of LATS2 comparing tissues with a high miR-103 level and those with a low miR-103 level (**P<0.01; magnification, ×400). (C) Left panel, relative mRNA expression of LATS2 in HCC compared to distant non-cancerous HCC tissues; right panel, relative mRNA expression of LATS2 in HCC tissues from patients with metastasis cases and those without metastasis cases (**P<0.01). (D) Left panel, the mRNA expression level of LATS2 was significantly decreased following transfection of miR-103 mimics into HCC cells (**P<0.01); right panel, the overexpression of miR-103 significantly regulated the luciferase activity of cells that carried the wild-type, but not the mutant type 3′-UTR of LATS2 in HCC cells (**P<0.01). (E) Left panel, miR-103 expression negatively correlates with LATS2 expression in HCC tissues (Spearman's r=−0.791; see Table III); right panel, no significant correlation was observed between miR-103 and LATS2 in distant non-cancerous tissues. HCC, hepatocellular carcinoma; NC, negative control; LATS2, large tumor suppressor kinase 2.
Table III.
Association between miR-103 and LATS2 expression levels in hepatocellular carcinoma tissue specimens.
| Variable | miR-103
|
r value | P-value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| LATS2 | ||||
| Tumor | ||||
| High expression | 15 | 19 | −0.791 | <0.001a |
| Low expression | 69 | 17 | ||
| Normal | −0.055 | 0.553 | ||
| High expression | ||||
| Low expression | ||||
Values in bold indicate statistical significance (aP<0.01); n=120. Spearman's correlation analysis was use to analyze the correlation between miR-103 and LATS2 expression.
Prognostic significance of miR-103 and LATS2 in patients with HCC
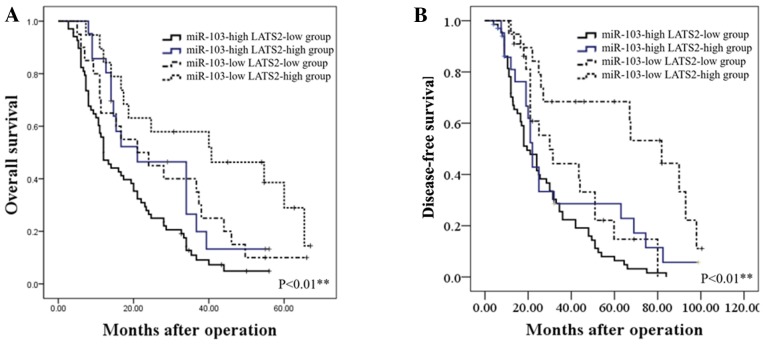
We further assessed whether the combination of miR-103 and LATS2 may serve as a prognostic predictor for OS and DFS in HCC. The combination analysis revealed that patients with a high miR-103 expression/low LATS2 expression had the lowest OS and DFS. By contrast, patients with HCC with a low miR-103 expression/high LATS2 expression had the most favorable OS and DFS (P<0.01; Fig. 6).
Figure 6.
The prognostic significance of miR-103 and LATS2 combination in patients with HCC. (A) OS was compared between 4 subgroups of patients with HCC [miR-103 (high)/LATS2 (low), miR-103 (low)/LATS2 (low), miR-103(high)/LATS2 (high) and miR-103 (low)/LATS2 (high)] (**P<0.01). (B) DFS was compared between 4 subgroups of patients with HCC [miR-103 (high)/LATS2 (low), miR-103 (low)/LATS2 (low), miR-103 (high)/LATS2 (high) and miR-103 (low)/LATS2 (high)] (**P<0.01). HCC, hepatocellular carcinoma; LATS2, large tumor suppressor kinase 2; OS, overall survival; DFS, disease-free survival.
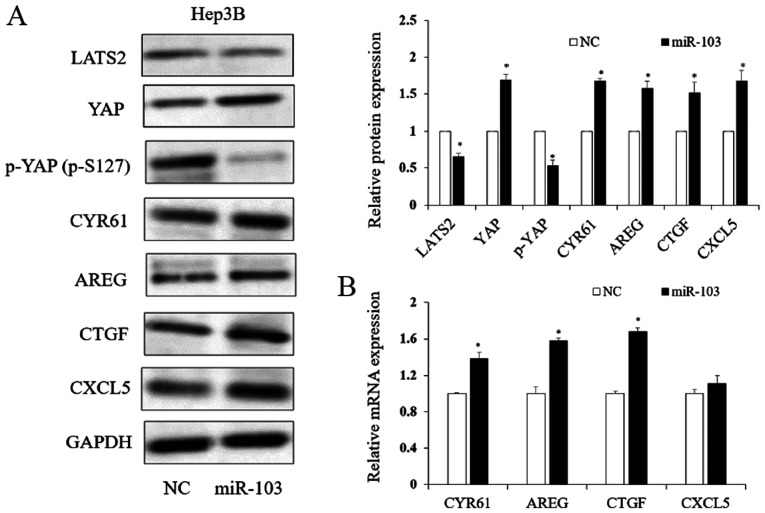
miR-103 regulates Hippo-YAP signaling via LATS2
Previous studies have demonstrated that LATS2 is related to the Hippo downstream effector, YAP phosphorylation (27,28). Thus, in this study, to determine whether miR-103 functions in HCC in association with the Hippo signaling pathway via LATS2, we assessed the protein level of LATS2, phosphorylated YAP (p-S127) and YAP. As shown in Fig. 7A, the LATS2 expression level significantly decreased with the increased miR-103 level in HCC (P<0.05). Our study here showed increased YAP and decreased p-YAP (p-S127) because of increased miR-103 (P<0.05; Fig. 7A). The results of western blot analysis and RT-qPCR further revealed that the downstream target genes of YAP, including CYR61, AREG, CTGF and CXCL5, were regulated by miR-103 (Fig. 7). Herein, we demonstrated that miR-103 promoted the activation of YAP and may thus serve as an upstream regulator of the Hippo signaling pathway.
Figure 7.
The expression of miR-103 regulates the LATS2/YAP signaling pathway. (A) miR-103 regulates the protein level of LATS2, YAP, p-YAP (p-S127), CYR61, AREG, CTGF and CXCL5 (*P<0.05). (B) RT-qPCR analysis suggested that an increased miR-103 expression regulated the expression level of downstream of YAP target genes (*P<0.05). LATS2, large tumor suppressor kinase 2; YAP, yes-associated protein; CYR61, cysteine-rich angiogenic inducer; AREG, amphiregulin; CTGF, connective tissue growth factor; CXCL5, C-X-C motif chemokine 5.
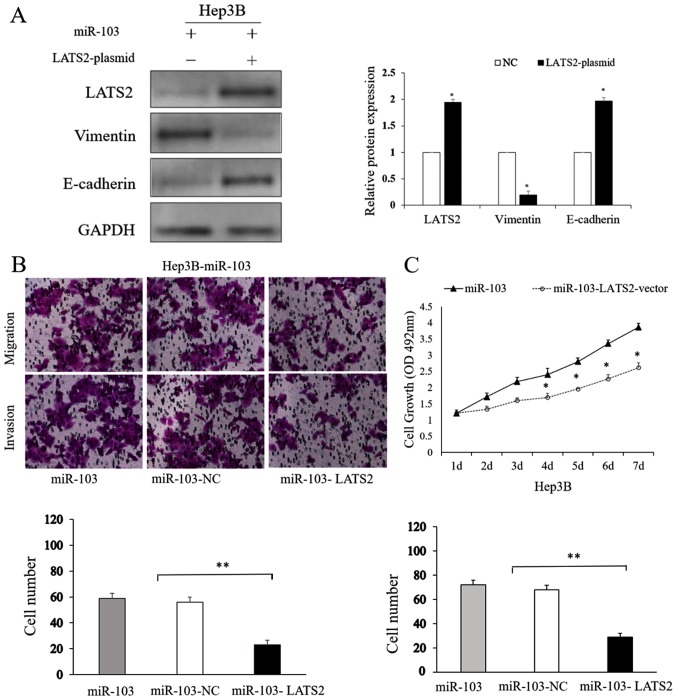
LATS2 counteracts the functional effects of miR-103 on HCC cells
To further explore the effects of LATS2 on the process of miR-103-induced metastasis, and on the growth and EMT of HCC cells, we treated transfected the miR-103-overexpressing Hep3B cells with a LATS2 expression plasmid. The LATS2 plasmid upregulated LATS2 expression in the Hep3B cells (P<0.05; Fig. 8A). The results of western blot analysis shown in Fig. 8A indicated that the overexpression of LATS2 reversed the EMT-promoting effects induced by the overexpression of miR-103 in HCC cells (P<0.05). The results of Transwell assay also indicated that LATS2 overexpression attenuated the migratory and invasive ability of the Hep3B cells which had been promoted by the overexpression of miR-103 (P<0.01; Fig. 8B). MTT assay showed us altering LATS2 level could counteract the proliferation capability induced by miR-103 (P<0.01; Fig. 8C). On the whole, these results indicate that LATS2 is not only a direct target of miR-103, but is also an important determinant of miR-103.
Figure 8.
LATS2 counteracts the effects of miR-103 on HCC cells. (A) Western blot analysis showing that LATS2 overexpression counteracts the promoting effects on the expression of EMT-related proteins induced by miR-103 (*P<0.05). (B) Transwell tumor cell invasion assay (magnification, ×200). (C) MTT cell proliferation assay (*P<0.05). HCC, hepatocellular carcinoma; NC, negative control; LATS2, large tumor suppressor kinase 2.
Discussion
In this study, at least to the best of our knowledge, for the first time, we demonstrate that miR-103 promotes HCC cell metastasis and EMT by directly targeting LATS2. A recent study indicated that miR-103 promoted HCC cell growth and that miR-103 functioned as a novel oncogene in HCC (22); however, the mechanisms involved are not yet fully clear and further studies to determined the clinical significance of miR-103 in HCC are warranted. Results from analyses using clinical tissue samples, in vitro and in vivo, may provide valuable information regarding the role of miR-103 in tumorigenesis.
Numerous miRNAs have been confirmed to serve as critical prognostic biomarkers in human cancers. It has been demonstrated miR-103 overexpression in gastric cancer tissues is associated with a prognosis of patients with gastric cancer (19). In addition, in a previous study, Kaplan-Meier analysis of colorectal cancer patients confirmed that an increased miR-103 expression was associated with a shorter OS (20). The findings of this study demonstrated that miR-103 expression was significantly increased in HCC tissues, and was associated with poor OS of patients with HCC. These data indicate that miR-103 may function as a promising prognostic marker in HCC.
A number of miRNAs have been confirmed to act as crucial modulators of not only metastasis (29,30), but also EMT in HCC (31-33). The findings of this study support those of previous findings in that miR-103 promotes HCC cell growth. We further confirmed that an increased miR-103 expression promoted the migration and EMT of HCC cells. The discovery of the possible mechanisms of EMT has been recognized as the key for exploror HCC; thus, the findings of this study may provide potential targets for clinical interventions.
As an important suppressor of the Hippo signaling pathway, LATS2 encodes a putative Ser/Thr protein kinase and plays a key role in oncogenesis by regulating cell proliferation, apoptosis and EMT. LATS2/YAP has been found to regulate the growth, proliferation and invasion of tumor cells (34). This study demonstrated that miR-103 directly targeted the 3′-UTR of LATS2, resulting in the downregulation of LATS2, YAP activation and the regulation of YAP downstream signaling. These data indicate that miR-103 promotes the EMT of HCC cells by the direct suppression of LATS2, which is supported by the fact that the LATS2/YAP signaling pathway is involved in EMT in human cancers, including HCC (35). Further analyses in this study demonstrated that the overexpression of LATS2 indeed counteracted the effects of miR-103 in HCC cells, including the promoting effects of this miRNA on proliferation, invasion, migration and EMT.
However, it should be mentioned that LATS2 is not the sole target of miR103, as several other targets have been identified in other cancer cells, including A-kinase anchoring protein 12 (AKAP12), vascular endothelial growth factor (VEGF), Wnt3a and p57 (36-38). Thus, it is possible that miR-103 can promote the proliferation, migration, invasion and EMT of HCC cells through other targets. Among these targets of miR-103, the one that is the most important determinant of the properties of miR-103 remains unclear. The exact mechanisms underlying the effects of miR-103 on the progression of HCC warrant further investigation.
Taken together, the findings of the current study uncover the biological and clinical significance of miR-103 in HCC. We identified that miR-103 promotes HCC metastasis and EMT, relying on the direct suppression of LATS2. These data suggest that the targeting of the miR-103/LATS2/YAP axis may prove to be a novel approach for the suppression of the development and metastasis of HCC.
Acknowledgments
The authors would like to thank Professor Chen Huang of Xi'an Jiaotong University (Xi'an, China) for providing the experimental platform and expert opinions.
Funding
This study was supported by grants from the Natural Science Basic Research Plan in Shaanxi Province of China (no. 1191329734) and China Postdoctoral Science General Financial Grant (no. 2017M623193).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding authors on request.
Authors' contributions
LLH collected the clinical samples and performed most of the experiments. XRY collected data from public datasets and analyzed the data, performed RT-qPCR assay and the statistical analysis. SQZ conceived and designed the study and assisted in the drafting of the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committee of the Medical School of Xi'an Jiaotong University reviewed and approved the study, and written informed consent was obtained from each participant at each examination phase. The study complied with the principles of the Helsinki Declaration. The animal experiments carried out in this study were approved by the Experimental Animal Ethical Committee of Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Finn RS. Current and future treatment strategies for patients with advanced hepatocellular carcinoma: Role of mTOR inhibition. Liver Cancer. 2012;1:247–256. doi: 10.1159/000343839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang M, Wu R, Jiang J, Minuk GY, Niu J. The presence of hepatitis B core antibody is associated with more advanced liver disease in alcoholic patients with cirrhosis. Alcohol. 2013;47:553–558. doi: 10.1016/j.alcohol.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Jin H, Ko YS, Kim HJ. P2Y2R-mediated inflammasome activation is involved in tumor progression in breast cancer cells and in radiotherapy-resistant breast cancer. Int J Oncol. 2018;53:1953–1966. doi: 10.3892/ijo.2018.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun G, Ding X, Bi N, Wu L, Wang J, Zhang W, Dong X, Lv N, Song Y, Zhan Q, Wang L. MiR-423-5pin brain metastasis: Potential role in diagnostics and molecular biology. Cell Death Dis. 2018;9:936. doi: 10.1038/s41419-018-0955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Mo HQ, Tian FJ, Zeng WH, Liu XR, Ma XL, Li X, Qin S, Fan CF, Lin Y. EIF5A1 promotes trophoblast migration and invasion via ARAF-mediated activation of the integrin/ERK signaling pathway. Cell Death Dis. 2018;9:926. doi: 10.1038/s41419-018-0971-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathias RA, Gopal SK, Simpson RJ. Contribution of cells undergoing epithelial-mesenchymal transition to the tumour microenvironment. J Proteomics. 2013;78:545–557. doi: 10.1016/j.jprot.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM, Shaw CA, Wu MF, Hilsenbeck SG, White LD, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8:15773. doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300:10–19. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Wu J, Xie C. miR-92a promotes hepatocellular carcinoma cells proliferation and invasion by FOXA2 targeting. Iran J Basic Med Sci. 2017;20:783–790. doi: 10.22038/IJBMS.2017.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabir TD, Ganda C, Brown R, Beveridge D, Richardson K, Chaturvedi V, Candy P, Epis M, Wintle L, George J, et al. A miR-7/GAS6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology. 2018;67:216–231. doi: 10.1002/hep.29478. [DOI] [PubMed] [Google Scholar]
- 14.Cheng L, Wang H, Han S. MiR-3910 promotes the growth and migration of cancer cells in the progression of hepatocellular carcinoma. Dig Dis Sci. 2017;62:2812–2820. doi: 10.1007/s10620-017-4670-3. [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Xu Y, Cheng F, Hu Y, Yang S, Rao J, Wang X. miR-1301 inhibits hepatocellular carcinoma cell migration, invasion, and angiogenesis by decreasing Wnt/β-catenin signaling through targeting BCL9. Cell Death Dis. 2017;8:e2999. doi: 10.1038/cddis.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Huo W, Du M, Pan X, Zhu X, Gao Y, Li Z. miR-203a3p1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio. 2017;7:1085–1091. doi: 10.1002/2211-5463.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji C, Liu H, Yin Q, Li H, Gao H. miR-93 enhances hepatocellular carcinoma invasion and metastasis by EMT via targeting PDCD4. Biotechnol Lett. 2017;39:1621–1629. doi: 10.1007/s10529-017-2403-5. [DOI] [PubMed] [Google Scholar]
- 18.Yang D, Wang JJ, Li JS, Xu QY. miR-103 functions as a tumor suppressor by directly targeting programmed cell death 10 in NSCLC. Oncol Res. 2017 Jul 21; doi: 10.3727/096504017X15000757094686. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng J, Liu Y, Qiao Y, Zhang L, Lu S. miR-103 promotes Proliferation and metastasis by targeting KLF4 in gastric cancer. Int J Mol Sci. 2017;18:E910. doi: 10.3390/ijms18050910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 21.Xue D, Zhou C, Lu H, Xu R, Xu X, He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumour Biol. 2016;37:14. doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- 22.Xia W, Ni J, Zhuang J, Qian L, Wang P, Wang J. MiR-103 regulates hepatocellular carcinoma growth by targeting AKAP12. Int J Biochem Cell Biol. 2016;71:1–11. doi: 10.1016/j.biocel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Han LL, Nan HC, Tian T, Guo H, Hu TH, Wang WJ, Ma JQ, Jiang LL, Guo QQ, Yang CC, et al. Expression and significance of the novel tumor-suppressor gene SMG-1 in hepatocellular carcinoma. Oncol Rep. 2014;31:2569–2578. doi: 10.3892/or.2014.3125. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 27.Ling HH, Kuo CC, Lin BX, Huang YH, Lin CW. Elevation of YAP promotes the epithelial-mesenchymal transition and tumor aggressiveness in colorectal cancer. Exp Cell Res. 2017;350:218–225. doi: 10.1016/j.yexcr.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 28.Yuan Y, Li D, Li H, Wang L, Tian G, Dong Y. YAP overexpression promotes the epithelial-mesenchymal transition and chemoresistance in pancreatic cancer cells. Mol Med Rep. 2016;13:237–242. doi: 10.3892/mmr.2015.4550. [DOI] [PubMed] [Google Scholar]
- 29.Gong F, Ren P, Zhang Y, Jiang J, Zhang H. MicroRNAs-491-5psuppresses cell proliferation and invasion by inhibiting IGF2BP1 in non-small cell lung cancer. Am J Transl Res. 2016;8:485–495. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Fite K, Gomez-Cambronero J. Down-regulation of MicroRNAs (MiRs) 203, 887, 3619 and 182 prevents vimentin-triggered, Phospholipase D (PLD)-mediated Cancer Cell Invasion. J Biol Chem. 2016;291:719–730. doi: 10.1074/jbc.M115.686006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaca A, Govender P, Locketz M, Naidoo R. The role of miRNA-21 and epithelial mesenchymal transition (EMT) process in colorectal cancer. J Clin Pathol. 2017;70:331–356. doi: 10.1136/jclinpath-2016-204031. [DOI] [PubMed] [Google Scholar]
- 32.Tang O, Chen XM, Shen S, Hahn M, Pollock CA. MiRNA-200b represses transforming growth factor-β1-induced EMT and fibronectin expression in kidney proximal tubular cells. Am J Physiol Renal Physiol. 2013;304:F1266–F1273. doi: 10.1152/ajprenal.00302.2012. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Shan Z, Hu K, Ren F, Zhang W, Han M, Li Y, Feng K, Lei L, Feng Y. miRNA-223 inhibits epithelial-mesenchymal transition in gastric carcinoma cells via Sp1. Int J Oncol. 2016;49:325–335. doi: 10.3892/ijo.2016.3533. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Hu CF, Chen J, Yan LX, Zeng YX, Shao JY. LATS2 is de-methylated and overexpressed in nasopharyngeal carcinoma and predicts poor prognosis. BMC Cancer. 2010;10:538. doi: 10.1186/1471-2407-10-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Li H, Wang G, Zhang T, Fu B, Ma M, Quan Z, Chen G. Yes-associated protein (YAP) expression is involved in epithelial-mesenchymal transition in hepatocellular carcinoma. Clin Transl Oncol. 2016;18:172–177. doi: 10.1007/s12094-015-1353-4. [DOI] [PubMed] [Google Scholar]
- 36.Shi FP, Wang XH, Zhang HX, Shang MM, Liu XX, Sun HM, Song YP. MiR-103 regulates the angiogenesis of ischemic stroke rats by targeting vascular endothelial growth factor (VEGF) Iran J Basic Med Sci. 2018;21:318–324. doi: 10.22038/IJBMS.2018.27267.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Wu S, Muhammad S, Ren Q, Sun C. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes. J Lipid Res. 2018;59:843–853. doi: 10.1194/jlr.M082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Lin Y, Peng L, Sun R, Gong X, Du J, Zhang X. MicroRNA-103 promotes proliferation and inhibits apoptosis in spinal osteosarcoma cells by targeting p57. Oncol Res. 2018;26:2. doi: 10.3727/096504017X15144741233346. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on request.