Abstract
Data on the status of carbapenem-resistant microorganisms in the Middle East countries are scarce. The aim of this review was to collect available data regarding resistance to carbapenems in a Middle East region. Available data regarding carbapenem-resistant isolates were considered for evaluation in this review. Biomedical electronic databases were systematically searched to find related articles. The key terms used were “carbapenem-resistant, resistant gram-negative bacilli, Enterobacteriaceae, fermenting and non-fermenting gram-negative bacilli, Pseudomonas, Acinetobacter, Klebsiella and Iran”. After primary screening, 275 relevant articles were selected to be assessed thoroughly. Resistance rate to carbapenems was reported between 1% and 86% during years 2006–2018. Most of the carbapenem-resistant microorganisms were isolated from burn patients. Modified Hodge test was a commonly used phenotypic test. Only in few studies, genotypic assays were considered. Pattern of antibiotic use can affect emergence of resistant microorganisms. Rational use of drugs, and specifically, antibiotics is a challenging issue in developing countries. Mean number of drugs per prescription in these countries was higher than the World Health Organization standards. Overuse of antibiotics, especially injectable ones, and easy access to antibiotics without prescription is a warning alarm for future antibiotic resistance in developing countries. Establishing antimicrobial stewardship’s programs is new in the hospitals. Unfortunately, rules and regulatory issues to restrict antibiotic access in community pharmacies and prescription by general physicians are limited.
Keywords: carbapenem, resistant, gram-negative bacteria, antibiotics
Video abstract
Introduction
Carbapenem-resistant gram-negative bacteria are now a global concern around the world. Most of these strains are also resistant to other antimicrobial agents including aminoglycosides and fluoroquinolones.1 Few therapeutic options without the desired efficacy are available to treat infections caused by carbapenem-resistant microorganisms.2 This real threat necessitates applying reliable methods to determine prevalence of these isolates.
The methods to detect carbapenemase-producing microorganisms are divided into two classes: phenotypic and genotypic methods. Phenotypic methods are nonmolecular assays that detect structure of the carbapenemase enzyme (mostly active site) through a chemical or microbiological process.3 The carbapenemase enzymes are structurally classified according to their active sites. Ambler class A (known as Klebsiella-producing carbapenemase [KPC]) has serine amino acid in its active site and can be inhibited by β-lactamase inhibitors. Ambler class B has zinc in its active site and is called metallo-β-lactamase (MBL), so it can react with EDTA and dipicolinic acid (DPA). The last class, Ambler class D, has serine in its active site, but it may or may not be inhibited by β-lactamase inhibitors.4 According to these classes, various phenotypic methods are proposed to detect carbapenemase enzyme in gram-negative isolates.
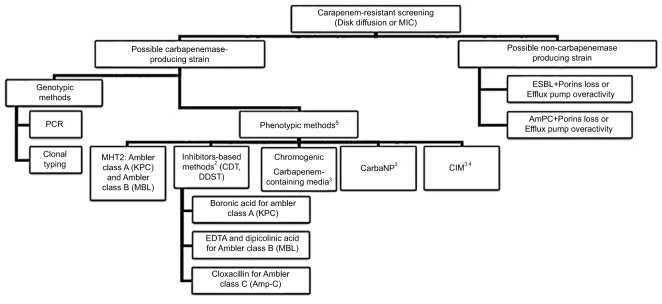
Genotypic methods are targeted to detect resistance genes (mostly bla type genes) and polymerase chain reaction (PCR) is a common method. Other genotypic method is clonal typing.5 Although detection of resistant-encoding genes is more accurate, however, these methods are usually expensive and not available anywhere.6 Some other mechanisms of resistance to carbapenems including loss of porins, efflux pump mutation, and target site inactivation have been identified.7 Figure 1 shows different genotypic and phenotypic methods for detection of resistance to carbapenems.
Figure 1.
Screening methods for carbapenem-resistant microorganisms.
Notes: 1MHT was the most used phenotypic method in Iranian studies. MHT originally detects KPC, but if zinc sulfate is added to MHT culture media, it can detect MBL.2 It is based on carbapenemase inhibition by betalactamase inhibitors. CDT consists of two disks: a carbapenem and the other combination of a carbapenem and a betalactamase inhibitor. If the disk with inhibitor shows a bigger inhibition zone, the result is considered positive. DDST consists of carbapenem discs at a variable distance to inhibitor discs. The observation of synergy between disks is noted as a positive result. 3These phenotypic methods can detect different Ambler classes. 4CIM is a newer phenotypic method that was introduced in 2015 by van der Zwaluw. A carbapenem disk is inserted in culture media of suspected strain. Then it is transferred to another culture media with known control strain. If the suspected strain contains carbapenemase enzyme, the carbapenem disk has been degraded and the control strain in second culture will grow. 5Some other phenotypic methods like spectrometric assays are also used and have higher sensitivity and specificity, but they are costly and time consuming.
Abbreviations: CDT, combination disk test; CIM, carbapenem inactivation method; DDST, double disk synergy test; ESBL, extended spectrum β-lactamase; KPC, Klebsiella-producing carbapenemase; MBL, metallo-β-lactamase; MHT, modified Hodge test; MIC, minimum inhibitory concentration; PCR, polymerase chain reaction.
Data on the status of carbapenem-resistant microorganisms in the Middle East countries are scarce. The aims of this review were to collect available data regarding methods of detection and prevalence of resistance to carbapenems in a Middle East region.
Methods
Databases
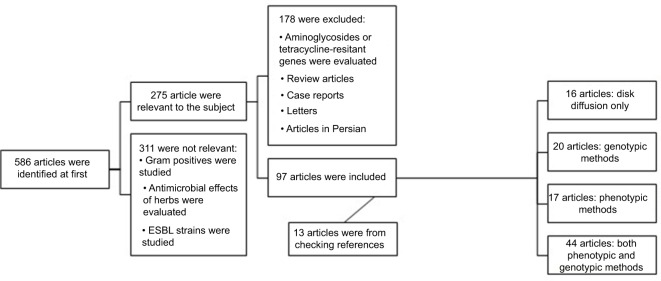
Biomedical electronic databases (Scopus, Medline, Google Scholar, and Science Direct) were systematically searched to find related articles. The key terms used were “carbapenem-resistant, resistant gram-negative bacilli, Enterobacteriaceae, fermenting and non-fermenting gram-negative bacilli, Pseudomonas, Acinetobacter, Klebsiella and Iran”. A total of 586 articles were found. After primary screening, 275 relevant articles were selected to be assessed thoroughly.
Study settings
Of the 275 articles, case reports, systematic reviews, meta-analysis, and articles in Persian language were excluded, and finally 97 English-language articles were included in the present review. These studies were published between 2006 and 2018 and were reported from different regions of Iran. Forty-eight studies were from Tehran and most of them were done at Motahari hospital (Level I Iranian burn hospital). The number of publications from other areas were as follows: ten in Isfahan, eight in northwest of Iran, five in south of Iran, four in Shiraz, three each in Hamadan, Arak and Mazandaran, two each in Shahrekord, Kermanshah, Mashhad, and Zanjan, and one each in Kerman, Khorramabad, and Qom. All the studies considered the standards and guidelines of the clinical and laboratory standards institute (CLSI) for the preparation and interpretation of the tests. Figure 2 illustrates data processing in this review.
Figure 2.
Consort flowchart of study.
Abbreviation: ESBL, extended spectrum β-lactamase.
Clinical and microbiological setting
All the studies included inpatients, with eight also considering outpatients. If the source of the samples was specific, they are mentioned and if they were collected from different sites, they are named as “different sites”.
All studies have been categorized into four distinct groups:
Studies that reported carbapenem-resistant microorganisms according to the disk diffusion method
Studies that considered phenotypic methods
Studies that used genotypic methods
Studies that applied both genotypic and phenotypic assays
Results
Studies that reported carbapenem-resistant microorganisms according to the disk diffusion method (early detection of resistance to a carbapenem)
In this category, detection of resistance to a carbapenem was described only by disk diffusion method. However, in some of them, minimum inhibitory concentration (MIC) values were determined. No phenotypic or genotypic assay was considered for carbapenemase detection. A total of 3,396 isolates were reported in this group (Table 1).
Table 1.
Screening of carbapenem-resistant isolates (disk diffusion method)
| Author (year) | Setting | Population | Samples | Microorganism (No.) | MICa | Resistance rate to FQ and AG | Resultsb | Weak points | Strength points |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Mobaraki et al (2018)15 | Hospitals in Tabriz | Inpatients and outpatients | Different sites | Pseudomonas aeruginosa (200) | – | TOB 57% GEN 62% CIP 62.% |
• 46.5% of isolates were resistant to IPM | – | • Multicenter study |
| Ghanbari et al (2017)16 | One hospital in Fuldshahr | General inpatients | Urine | Different strains (317) | – | c | • 9.5% of Escherichia coli, 11% of Klebsiella and 25% of Proteus spp. isolates were resistant to IPM | – | • Large sample size |
| Chahoofard et al (2017)17 | One hospital in Bandar Abbas | General inpatients | Urine | Different strains (296) | – | c | • Resistance rate to IPM: Enterobacter spp. 7.1% Citrobacter spp. 0% E. coli 4.9% Klebsiella pneumonia 19.2% P. aeruginosa 21.4% Acinetobacter baumannii 53.2% |
– | • Adequate sample size • Clinical status of patients including duration of hospital stay and comorbidities were evaluated |
| Ansari et al (2017)18 | Two hospitals in Shahrekord | General inpatients | Urine, blood, sputum, wound | A. baumannii (30) | – | NAL 80%, OFX 86 % LVX 66 % | • About 55% of strains were resistant to IPM and DOR • Intermediate resistance rate was 10%–33.3% |
• Small sample size • AGs were not tested |
• Cloning smpA gen |
| Douraghi et al (2016)8 | Four hospitals in Tehran | General inpatients and outpatients | Different sites | A. baumannii (400) | ✓ | – | • 97% were resistant to carbapenems • MIC values of all resistant isolates were above 32 mcg/mL |
– | • Multicenter study • Large sample size |
| Ghasemian et al (2016)9 | Four hospitals in Mazandaran | General ICU patients | Different sites | Acinetobacter spp. (50) | ✓ | CIP 96% AMK 100% | • Resistance to IPM and MEM according to E-test was 100%, and according to disk diffusion was 76% and 96%, respectively • Most common site for sampling was endotracheal tube |
• Small sample size • MIC values did not report |
• Multicenter study |
| Babamahmoodiet al (2015)13 | Three hospitals in Mazandaran | ICU patients | Wound, respiratory secretions, urine, blood | Different strains (114) | – | c | • 14% of P. aeruginosa, 60% of Acinetobacter spp., 33% of E. coli, 16% of Enterobacter and none of K. pneumonia isolates were resistant to IMP | • Small sample size for each strain | • Multicenter study • Demographic features and risk factors for hospital-acquired infection were described |
| Mohajeri et al (2014)14 | Three hospitals in Kermanshah | General inpatients | Blood, urine, sputum | Acinetobacter spp. (104) | – | Not defined | • 48% of isolates were resistant to carbapenems (imipenem and meropenem). 76% of carbapenem-resistant strains were isolated from sputum samples | • Few antibiotics were tested | • Multicenter study |
| Babakhani et al (2014)19 | One hospital in Khorramabad | General inpatients (mostly ICU) | Different sites | Klebsiella spp. (80) | – | GEN 52 % AMK 7.5% CIP 82 % OFX 75% NAL 60% | • 67% and 1% of isolates were resistant to IMP and MEM, respectively | – | |
| Kamalbeik et al (2013)20 | One Poisoning center in Tehran | ICU patients | Different sites | Acinetobacter spp. (40) | – | CIP 97% | • Resistance rates to MEM and IPM were 100% and 62% (27.5% intermediate resistant to IPM) | • Small sample size | – |
| Hashemi et al (2013)21 | Two hospitals in Hamadan | General inpatients and outpatients | Different sites | Enterobacteriaceae spp. (574) | – | c | • Resistance rate to IPM was 32% in inpatients and 4% in outpatients. Resistance rates to IPM in inpatients: Enterobacter 56%, Serratia 54%, Klebsiella 35%, E. coli 32%, and Proteus 11% | – | • Large sample size |
| Shakibaie et al (2012)10 | One hospital in Kashan | ICU patients | Different sites | Acinetobacter spp. (15) | ✓ | CIP 60% GEN 60% AMK 26% | • 73% of isolates were resistant to IPM • MIC range for resistant strains was 128–240 and MIC of IPM for seven isolates was 240 mcg/mL |
• Very small sample size | – |
| Rahbar et al (2010)22 | One hospital in Tehran | General inpatients | Different sites | P. aeruginosa (109) Acinetobacter spp. (88) Stenotrophomonas maltophilia (48) Burkholderia cepacia (12) | – | c | • 1 %, 16%, and 98% of A. baumannii, P. aeruginosa, and S. maltophilia isolates were resistant to IPM, respectively | – | • Demographic and medical data of patients were provided |
| Soroush et al (2010)23 | One pediatric hospital in Tehran | General pediatric patients | Different sites | Acinetobacter spp. (145) | – | In 2007: CIP 79% AMK 58% KAN 71% GEN 65% TOB 61% | • Resistance rate to IPM and MEM was analyzed in years 2006 and 2007 that was about 50% | • Carbapenem resistance was evaluated only in the last 2 years (2006–2007) | • Evaluated carbapenem resistance status in pediatrics • Change in the antibiotics resistance patterns was considered • Site of sampling with highest count of MDR strains in each year considered |
| Yousefi et al (2010)11 | One hospital in Orumie | General inpatients and outpatients | Different sites | P. aeruginosa (160) | ✓ | – | • Approximately 40% of isolates were resistant to IPM, which means MIC for them was 31 mcg/mL • Presence of Class I integron and being in ICU and burn unit led to significantly higher IPM-resistant strains |
• Just IPM was tested | – |
| Rahbar et al (2008)12 | One hospital in Tehran | General inpatients and ICU patients | Not defined | Different strains (202) | ✓ | c | • 40% and 7% of P. aeruginosa and 38% and 27% of A. baumannii isolates were resistant to MEM and IMP, respectively • MIC range value for MEM was 0.5–32 mcg/mL • The majority isolates of P. aeruginosa and A. baumannii had an MIC >32 µg/mL for MEM |
– | |
Notes:
MIC values of carbapenems (and not other antimicrobial agents) were determined.
Resistant rates were reported according to the disk diffusion test. The unit for all reported MIC values is mcg/mL.
For complete information about the resistance to AG and FQ, refer to the original reference.
Abbreviations: AG, aminoglycoside; AMK, amikacin; CDT, combination disk test; CIP, ciprofloxacin; DD, disk diffusion; DDST, double disk synergy test; DOR, doripenem; ERT, ertapenem; ESBL, extended spectrum β-lactamase; FQ, fluoroquinolones; GEN, gentamicin; ICU, intensive care unit; IPM, imipenem; KAN, kanamycin; LVX, levofloxacin; MBL, metallo-β-lactamase; MEM, meropenem; MHT, modified Hodge test; MIC, minimum inhibitory concentration; NAL, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; TOB, tobramycin.
Screening of resistance to a carbapenem according to the disk diffusion method was common among the studies.8–23 Although this method may be applicable for many antimicrobials, however, for carbapenems more confirmatory tests beyond the disk diffusion assay are needed. In this setting, MIC would help for detection of resistant isolates.
Five studies were multicenter,8–12 but the sample sizes for three of them were small.10,13,14 The trend of resistance to a carbapenem did not follow a specific pattern. However, in two studies,12,15 40% of the isolates were resistant to a carbapenem.
Studies that considered phenotypic methods
A total of 4,264 isolates were included in this category and Acinetobacter spp. and K. pneumonia were the common isolates. Modified Hodge test (MHT) was used as a common phenotypic test in most studies. MHT can detect Ambler class A (KPC), although it may be positive with the presence of other carbapenemase enzymes. Other applied tests were combination disk test (CDT) for detection of MBL or extended spectrum β-lactamase and the double disk synergy test for detection of MBL. In the study by Raategar Lari et al,24 92% of A. baumannii isolates were resistant to imipenem and only 24% of them were MBL positive in the CDT test, but in the next study,25 54% of isolates were resistant to imipenem and all of them were KPC producers according to the phenotypic tests.
The rates of resistance to aminoglycosides and fluoroquinolones were similar to those of carbapenems, although no special trend can be followed through different years. The results are provided in Table 2.26–41
Table 2.
Studies that considered phenotypic methods for detecting carbapenemase enzymes
| Author (year) | Setting | Population | Samples | Microorganism (no.) | MICa | Method of carbapenemase detection | Resistance to FQ and AG | Resultsb | Weak points | Strength points |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Ghotaslou et al (2018)26 | Five hospitals in East and West Azerbaijan | General inpatients | Different sites | Enterobacteriaceae (307) | – | • DD • BA and DPA for MBL detection |
c | • Resistance rate to carbapenem was observed only in K. pneumonia (57 isolates; 25%–45%) • Of these 57 isolates, phenotypic methods revealed that 15%, 7%, and 3% isolates of Klebsiella had MBL, KPC, and OXA-48 genes, respectively • Most of the carbapenemase strains were isolated from urine and blood |
• Small sample size for each species | – |
| Saadatian Farivar et al (2018)27 | Three hospitals in Tehran | Inpatients and outpatients | Different sites | Klebsiella pneumonia (81) | – | • DD • MHT for MBL detection |
OFX 65%, CIP 68%, NOR 66%, GEN 66%, AMK 51%, TOB 56 %, KAN 79%, GEN 96% | • Resistance rate to IPM was 45% • 6 (7.4% of all) isolates were positive for MHT test |
– | – |
| Moosavian et al (2014)28 | Two hospitals in Ahvaz | General inpatients | Different sites | Acinetobacter spp. (100) | – | • DD • MHT |
AMK 96% CIP 98% | • Resistance rates to IPM, MEM, and ERT were 95%, 96%, and 53%, respectively • 53% of all strains were MHT positive |
– | – |
| Jonaidi Jafari et al (2014)29 | One hospital in Tehran | General inpatients | Different sites | Different strains (350) | ✓ | • DD • CDT for ESBL detection |
– | • MIC was determined by E-test • 95 strains were ESBLs • Resistance rates to IPM and MEM according to MIC, respectively, were: Acinetobacter baumannii 42%, 37% Pseudomonas aeruginosa 42%, 60% K. pneumonia 15%, 0% |
• Small sample size for each microorganism | – |
| Ghadiri et al (2014)30 | Three laboratories in Tehran | General outpatients | Urine | Escherichia coli (300) | • DD • CDT for ESBL detection • DDST for MBL detection |
AMK 11% GEN 16% |
• 67 isolates were ESBL producers and resistance to carbapenem in this group was about 10% • 21 isolates were MBL producers and resistance to carbapenems among them was almost 100% • 5 isolates were both MBL and ESBL positive |
– | • Large sample size | |
| Fazeli et al (2014)31 | Teaching hospitals in Isfahan | General inpatients and ICU patients | Different sites | K. pneumonia (142) | – | • DD • CDT for ESBL |
Resistance rate to GEN and AMK and CIP: 70%–80% LVX: 50%–60% | • Resistance rates to IPM, MEM, and ERT among all strains were 57%, 52%, and 47%, respectively • 101 strains were ESBL producers • Most of the ESBL producers were isolated from ICU and from urine samples |
• It is not clear that the study was multicenter or not | – |
| Mirsalehian et al (2014)32 | Motahari hospital | Burn patients | Probably burn wounds | P. aeruginosa (100) | ✓ | • DD • AmpC disk and CDT for AmpC production |
– | • All IPM-resistant Pseudomonas were chosen and mean MIC for IPM was 64 mcg/mL • AmpC disk test and CDT were positive for 54 and 17 isolates, respectively showing a probable activation of one of the efflux pump and impermeability systems, or both |
– | – |
| Moayednia et al (2014)33 | One hospital in Isfahan | General inpatients and outpatients | Urine | K. pneumonia (484) and E. coli (1,080) | • DD • Double Disk for ESBL • MHT for KPC detection • CDT for MBL detection |
Just reported for ESBL producer | • Resistance rate to carbapenem for E. coli was 1%–2% and for Klebsiella spp., it was 41%–46% • 390 isolates of E. coli and 238 Klebsiella were ESBL producers • From all of strains: E. coli: 2 MBL and 10 KPC producers Klebsiella spp.: 10 MBL and 186 KPC producers |
• MIC was done by E-test, but cutoffs were not reported | • Large sample size | |
| Erfani et al (2013)34 | Three hospitals in Tehran | General inpatients | Different sites | Acinetobacter sp. (107) | – | • DD • CDT for MBL detection |
CIP 95% LVX 93% GEN 51% TOB 64% |
• Resistance rate to IPM and MEM was about 95% and 92% of these resistant isolates were MBL positive | – | – |
| Lari et al (2013)24 | One hospital in Tehran (Motahhari) | Burn patients | Mostly burn wound | Acinetobacter spp. (69) | – | • SDDT for ESBL detection • CDT for MBL detection |
GEN 64.7% AMK 95.7% KAN 97.1% TOB 2.9% CIP 97.1% |
• Resistance rate to IPM was 92% • ESBL was positive in 10% of isolates • 24% of all baumannii isolates were MBL positive • The mortality rate in this study was 20% |
– | • Mortality rate was reported • Detection of Integron genes |
| Rastegar Lari et al (2013)25 | One hospital in Tehran (Motahari) | Burn patients | Burn wounds | Klebsiella spp. (35) | – | • DD • MHT for KPC production |
AMK 71% GEN 72% TOB 86% |
• Resistance rate to IPM was 54% and all were KPC producers • 7 out of 28 patients had two Klebsiella isolates with two different antibiotypes • Rate of mortality in patients infected with resistant strain was 33% |
• FQ and colistin were not tested • MIC values were not reported • Small sample size |
• Mortality rate was reported |
| Safari et al (2013)35 | Three educational hospitals in Hamadan | ICU patients | Trachea, blood, urine, sputum, wounds | A. baumannii (100) | ✓ | • DD • E-test for MBL |
Resistant: CIP 97% LVX 91% AMK 84% GEN 88% |
• Resistance rate to carbapenem was 85%–94% but according to MIC, it was 87%–99% • 99% of all isolates were MBL producers |
• Multicenter study • MIC was reported |
|
| Masaeli et al (2012)36 | Two hospitals in Sanandaj | General inpatients | Different sites | Different strains (423) | – | • DD • DDST for MBL detection |
GEN 70% AMK 80% CIP 37% NOR 42% |
• The result of disk diffusion for carbapenem was not reported • 126 isolates (30% of all) were MBL positive • Risk factors like immunosuppression and use of antibiotics during the past 2 weeks were correlated with infection by MBL producers |
• The resistance pattern of each group of microorganisms was not fully defined | • Correlation between being MBL and different clinical risk factors was assessed |
| Japoni-Nejad et al (2013)37 | One hospital in Arak | General inpatients and environmental isolates | Different strains | A. baumannii (63) | ✓ | • MHT for KPC detection • E-test |
Amikacin 80% Netilmicin 54% |
• 84% were resistant to carbapenem • 47 isolates were MBL producers according to E-test and three of them were MHT positive • 60 strains had MIC more than 256 |
• Isolates from environment of hospital were also included | • Small sample size |
| Haji Hashemi et al (2012)39 | Different hospitals in Tehran and Isfahan | General inpatients and outpatients | Different sites | Klebsiella pneumoniae (244) | – | • DD • MHT for KPC detection • CHROMagar for KPC detection |
– | • MHT and CHROMagar were compared • From all, 30 and 36 isolates were KPC producers according to MHT and CHROMagar, respectively. The infection of burn wounds at the highest percentage: 27 cases (75 %) |
• MIC was not reported | • Multicenter study |
| Azimi et al (2012)40 | Two burn hospitals in Tehran | Infant and pediatric patients | Not defined (probably burn wounds) | Klebsiella, Acinetobacter, and Pseudomonas (64 strains from 20 patients) | – | • DD • MHT for KPC production • CDT for ESBL detection |
Not defined | • 36 isolates were resistant to all tested antibiotic (not mentioned which antibiotics) except Colistin • Among these 36 isolates, 15 isolates were resistant to IPM • From these 15 isolates, 13 were KPC and 6 were ESBL producers |
• Types of samples were not defined • The sample size is very small (7 A. baumannii, 16 Enterobacteriaceae, 12 Pseudomonas) • There is no precise data of resistance to each antibiotic |
|
| Japoni et al (2006)41 | One hospital in Shiraz | General inpatients | Different sites (mostly wound) | P. aeruginosa (70) | ✓ | • DD • DDST for ESBL • MHT for MBL detection |
CIP 73% AMK 93% TOB 97% |
• Resistance rate to carbapenems was 33% • MIC value for IPM was 2 and for MEM was 1 • MBL was not detected |
• Small sample size | |
Notes:
MIC values for carbapenems (but not other antimicrobial agents) were determined.
Resistance rates were according to the result of disk diffusion test.
For complete information about the resistance to AG and FQ, please refer to the original reference.
Abbreviations: AG, aminoglycoside; AMK, amikacin; BA, boronic acid; CDT, combination disk test; CIP, ciprofloxacin; DD, disk diffusion; DDST, double disk synergy test; DOR, doripenem; DPA, dipicolinic acid; ERT, ertapenem; ESBL, extended spectrum β-lactamase; FQ, fluoroquinolones; GEN, gentamicin; ICU, intensive care unit; IPM, imipenem; KAN, kanamycin; KPC, Klebsiella-producing carbapenemase; LVX, levofloxacin; MBL, metallo-β-lactamase; MEM, meropenem; MHT, modified Hodge test; MIC, minimum inhibitory concentration; NAL, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; TOB, tobramycin.
Studies that included genotypic methods
A total of 2,195 strains were included in this group and except for three studies, all assessed Acinetobacter spp. mostly A. baumannii. PCR was performed in order to determine types of bla genes. In some studies, MIC values were correlated with the presence of bla genes. In the study by Taherikalani et al the MIC values of 256 mcg/mL were detected in strains with more than two blaOXA genes.42 In the study by Azizi et al, higher MIC value was associated with presence of bla-OXA-24/40 like.43 Bahador et al detected ISAba genes.44 When these elements were placed upstream of OXA-type genes, they enhanced the expression of OXA-type genes. Presence of these elements in genetic contents of gram-negative isolates would lead to higher rate of resistance to carbapenems. Table 3 describes the details of each study in this category.42,45–61
Table 3.
Studies that performed genotypic methods to detect genes encoding carbapenemase enzymes
| Author (year) | Setting | Population | Samples | Microorganism (no.) | MIC | Method of carbapenemase detection | Resistance to FQ and AG | Resultsb | Weak points | Strength points | Found carbapenemase genesa |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Sharikhani et al (2017)45 | Four hospitals in Qom | General inpatients (mostly ICU) | Different sites | Acinetobacter baumannii (108) | – | • DD • PCR |
CIP and AMK 93% GEN 81% LVX 91% TOB 47% |
• 89% isolates were non-susceptible to IPM and MEM • Among carbapenem non-susceptible isolates, 82%, 55%, 22%, and 14% isolates had blaOXA-23, blaOXA-58, blaOXA-40, and blaOXA-143 genes, respectively |
– | • First report of blaOXA-143 in Iran • Multicenter study |
blaOXA-51 blaOXA-23 blaOXA-40 blaOXA-58 blaOXA-143 |
| Mohammadi et al (2017)46 | Two hospitals in Tehran | General inpatients | Burn wound and trachea | A. baumannii (103) | – | • DD • PCR |
– | • About 90% of isolates were resistant to carbapenem • blaOXA-23, blaOXA-24, blaOXA-58, blaTEM, and blaPER genes were detected in 90%, 38%, 1%, 60%, and 18.5% of all isolates, respectively • Significant relationship between the presence of blaOXA-24 and resistance to IPM (not MEM) • All strainshad biofilm producing ability |
• Ability of biofilm formation was assessed • Correlation between presence of genes and resistance to antibiotics was defined |
blaOXA-23, blaOXA-24, blaOXA-58, blaTEM, blaPER | |
| Bahador et al (2015)44 | One hospital in Tehran | Burn patients | Not defined (probably burn wounds) | A. baumanii (62) | ✓ | • DD • PCR • Clonal typing |
Not reported specifically | • According to MIC values, 61% were resistant to IPM • From all strains, 39 had bla OXA-23 (33 were susceptible to IPM), 9 had blaOXA-58.genes • Existence of two genes (in 1.6%–19.3% of strains) was correlated with MIC more than 32 mcg/mL for IPM |
• Resistance to each antibiotic was not reported specifically • Small sample size |
• MIC values of IPM, tigecycline, and colistin were reported • ISAba gene was assessed |
blaoxa-51 blaoxa-23 blaoxa-24 blaoxa-40’ blaoxa-143 |
| Kooti et al (2015)47 | Four hospitals in Shiraz | General inpatients | Different sites | A. baumannii (200) | – | • DD • PCR |
GEN 84.5% AMK 86.5% CIP and LVX 99.5% |
• About 99% of isolates were resistant to IPM and MEM • 40% had blaOXA-23, 7% had blaOXA-24, 0.5% had blaOXA-58 • Two bla genes were detected in 4.5% of strains • The correlation between presence of genes and source of samples was not significant |
– | Multicenter study | blaOXA-23 blaOXA_24 blaOXA-58 |
| Azizi et al (2015)43 | Two hospitals in Kerman | ICU patients | Different sites | A. baumannii (65 MDR strains from 266) | ✓ | • DD • PCR |
CIP 100% AMK 78.5% TOB 93% |
• MIC values for IPM and MEM for 76% of strains were >256 mcg/mL • blaoxa-23 was found in all isolates including sensitive and resistant isolates • Presence of blaOXA 24/40 (29 strains) associated with higher MIC |
– | – | blaoxa-51, blaoxa-23, blaoxa-24/40 |
| Mahdian et al (2015)48 | One hospital in Tehran (Motahari) | Burn patients | Burn wounds, blood, urine | A. baumanii (37) | – | • DD • PCR • Clonal typing |
CIP 100% GEN 94.6% | • 81% of strains were resistant to carbapenem ◦ All resistant isolates had at least one blaOXA-23 and/or blaOXA-24 and about half of them had both genes |
• Small sample size • Few antibiotics were tested |
• MIC values for colistin and polymyxin B were reported • ISAba genes were assessed |
blaOXA-23, blaOXA-24, blaTEM, blaPER |
| Farsiani et al (2015)49 | One hospital in Mashhad | General inpatients and ICU patients | Different sites | A. baumannii (36) | ✓ | • DD • PCR • Clonal typing |
CIP 97% | • 97% of strains were resistant to carbapenems • MIC values of resistant isolates were >32 • blaOXA-51 and blaOXA-23 were detected in all isolates • From all isolates, blaOXA-24, blaTEM, blaADC, and blaVIM genes were found in 23, 34, 22, and 23 strains, respectively • ISAba1 was detected in 97% of isolates of A. baumannii |
• Small sample size | • ISAba genes and tet genes (related to efflux pump) were analyzed | blaOXA-51, blaOXA-23 blaOXA-24, blaTEM, blaADC blaVIM adeB ISAba1 tetA, tetB |
| Nasrolahei et al (2014)50 | Two hospitals in Tehran and Sari | ICU and burn patients | Trachea and burn wounds | A. baumannii (100) | – | • DD • PCR |
STP 90% GEN 83% TOB 83% KAN 80% | • Resistance rate to IPM and MEM was about 70% • 67% of strain carried blaoxa-23 gene |
• FQ are not tested | • Multicenter study | blaoxa-23 |
| Safari et al (2014)51 | Three hospitals in Hamadan | ICU patients | Different sites | Pseudomonas aeruginosa (100) | ✓ | • DD • PCR |
AMK 19% GEN 28% TOB 27% LVX 28% CIP 38% |
• Resistance rate to carbapenems was about 20% • According to MIC values, 24% of isolates were carbapenem-resistant • Four isolates had blaIMP |
– | blaIMP | |
| Bahador et al (2014)52 | Two hospitals in Tehran | ICU patients | Different sites | A. baumannii (100) | ✓ | • DD • Clonal typing |
c | • Resistance rates were compared among years 2006 and 2011, according to MIC cutoffs • In year 2006, resistance rate to IPM was 30% with MIC ≤4–64 mcg/mL. In year 2011 it was 48% and MIC values were ≤4 to ≥256 mcg/mL • Genotypes I and F were the most frequent genotypes in years 2006 and 2011, respectively |
• Range of MIC values for each antibiotic was reported | – | |
| Karmostaji et al (2013)53 | Two hospitals in Tehran | General inpatients (mostly ICU) | Different sites | Acinetobacter spp. (131) | – | • DD • PCR |
CIP 95% GEN 77% AMK 54% |
• Resistance rate to IPM and MEM were 67% and 84%, respectively • 107, 17, and 1 of the resistant isolates carried blaOXA-23, blaOXA-24, and blaOXA-58, respectively. From these strains, seven strains had both blaOXA-23 and blaOXA-24 |
– | – | blaoxa-51 blaoxa-23 blaoxa-24 blaoxa-58 |
| Shoja et al (2013)54 | Two hospitals in Ahvaz | ICU patients | Trachea | A. baumannii (206) | – | • DD • PCR |
GEN 83%, TOB 78%, AMK 88%, CIP 96%, | • Resistance rate to IPM and MEM was 96% • blaOXA-23 and blaOXA-24 was found in 85% and 8% of all strains, respectively. |
– | – | blaOXA-23 blaOXA-24 |
| Sohrabi et al (2012)55 | One hospital in Tabriz | General inpatients | Different sites | A. baumanii (100) | • DD • PCR |
CIP and GEN 86% LVX 84% AMK 81% |
• 62% of isolates were resistant to carbapenems • Out of 62 resistant isolates, 55 carried blaOXA-23, 1 carried blaOXA-40, and 2 isolates had blaOXA-58 genes |
– | – | blaOXA-23 blaOXA-58 blaOXA-40 | |
| Sepehriseresht et al (2012)56 | One hospital in Tehran (Motahari) | Burn patients | Burn wounds | P. aeruginosa (483) | – | • DD • PCR |
– | • 56% of isolates were resistant to IPM, although 13% intermediate resistance should be considered • Among resistant isolates, 61 and 33 isolates had blaVIM and blaIPM genes, 23 strains had 2 resistant genes |
• Origin of blaIPM is not clear | – | blaVIM blaIPM |
| Forozesh Fard et al (2012)57 | One hospital in Isfahan | CF patients | Sputum | P. aeruginosa (11) | – | • DD • PCR |
CIP 0% TOB 45.4% |
• None of the isolates was resistant to IPM. None was positive for blaVIM gene | • Very small sample size • Few antibiotics were tested |
– | – |
| Peymani et al (2012)58 | One hospital in Tabriz | General inpatients | Different sites | A. baumannii (68) | – | • PCR • Clonal typing |
– | • All IPM-resistant isolates were chosen for study • 44% of isolates were from tracheal samples • All were positive for blaOXA-23, and ISAba1 was present upstream of all blaOXA-23 genes |
– | – | blaOXA-23 |
| Asadollahi et al (2012)59 | One teaching hospital in Tehran | ICU burn patients | Burn wounds | A. baumannii (23) | ✓ | • DD • PCR |
CIP 100% GEN 30% AMK 47% |
• Resistance rate to IPM was 48% • Range of MIC values for IPM was 0.004–32 mcg/mL • From all strains, blaPER and blaTEM genes were present in 52% and 43% of isolates, respectively. 21% and 17% of isolates had blaOXA- and blaOXA24 genes, respectively |
Small sample size | • MIC values were reported for each antibiotic • CarO and tet genes were assessed |
blaTEM blaPER blaOXA-23 blaOXA-24 blaSHV |
| Taherikalani et al (2009)42 | Six hospitals in Tehran | General impatients | Different sites | A. baumannii (80) | ✓ | • DD • PCR • Clonal typing | – | • 52% of isolates were carbapenem resistant • MIC50 range for carbapenem was 0.12–256 mcg/mL • Seven isolates that had MIC =256 mcg/mL had more than two blaOXA genes • From all strains blaOXA-23 in 25%, blaOXA-58 in 21%, and blaOXA-24 in 15% genes were detected • Coexistence of multiple genes was observed | • Number of tested antibiotics were few | • Multicenter study | blaoxa-23 blaoxa-24 blaoxa-58 |
| Feizabadi et al (2008)60 | One hospital in Tehran | General inpatients | Different sites | Acinetobacter spp. (A. baumannii =108 and other Acinetobacter =20, total =128) | ✓ | • DD • PCR |
CIP 81% LVX 74% AMK 61% NTL 86% TOB 79% GEN 81% |
• About 50% of strains were resistant to carbapenems • MIC values for carbapenem-resistant species were ≥64 to ≥256 mcg/mL • For details of isolated genes, refer to original article |
blaoxa-23 blaoxa-24 blaoxa-51 blaoxa-58 | ||
| Taherikalani et al (2008)61 | Two hospitals in Tehran | Burn patients | Burn wounds | A. baumannii (38) | – | • DD • PCR |
Resistance AMK 71% TOB 90% NTL 90% LVX 81% CIP 85% |
• Resistance rate to carbapenem was about 60% • Half of the carbapenemresistant strains had at least two blaOXA genes |
• Small sample size • MIC values were not determined |
blaoxa-23 blaoxa-24 blaoxa-51 blaoxa-58 | |
Notes:
Results about blaOXA-51 were not provided because this gene has no correlation with occurrence of resistance, unless it has ISAba genes in its upstream.
Resistance rates were according to the results of disk diffusion test.
For complete information about the resistance to AG and FQ, please refer to the original reference.
Abbreviations: AG, aminoglycoside; AMK, amikacin; CIP, ciprofloxacin; DD, disk diffusion; FQ, fluoroquinolones; GEN, gentamicin; ICU, intensive care unit; IPM, imipenem; KAN, kanamycin; LVX, levofloxacin; MEM, meropenem; MIC, minimum inhibitory concentration; NAL, nalidixic acid; PCR, polymerase chain reaction; STP, streptomycin; TOB, tobramycin; NTL, netilmicin.
Studies that considered both phenotypic and genotypic methods
A total of 6,759 isolates were identified in this category. In most studies more than one phenotypic method was applied. In the study by Hosseinzadeh et al, 29 strains were carbapenem resistant, in which 27 and 23 of them were MHT positive and carried blaNDM-1 gene, respectively.62 However, in another study by Bina et al, although MHT tests were positive, no resistance gene was found.63
The detection of resistance genes was much lower for strains other than A. baumannii. In the study by Lari et al in 2015, only 9 strains of P. aeruginosa (out of 255) contained resistance genes,64 but in the study by Bagheri Josheghani et al, at the same time, more than 90 strains (out of 124) of A. baumannii harbored resistance genes.65 Data are summarized in Table 4.66–105
Table 4.
Details of studies that used both phenotypic and genotypic methods for detection of carbapenemase-producing strains
| Author (year) | Setting | Population (no.) | Samples | Microorganism | MICa | Method of carbapenemase enzyme detection | Resistance to FQ and AG | Outcome | Weak points | Strength points | bla-type genesc |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Solgi et al (2017)66 | Two hospitals (city was not defined) | General inpatients (mostly ICU) | Rectal swab | Enterobacteriaceae spp. (95) | ✓ | • MHT for MBL detection • Clonal typing |
AMK 24% GEN 35% CIP 74%. |
• 36 resistant strains were detected according to the phenotypic methods that among them carbapenem-resistance rates were between 88% and 100% • 47 isolates were MHT positive • 7 isolates carried blaNDM, 23 carried blaOXA-48 • 17 isolates harbored two genes |
• MIC values were determined, but not analyzed • Small sample size |
• Risk factors for infection with resistant strains were investigated | blaNDM-1 blaNDM-7 blaOXA-48 |
| Khorvash et al (2017)67 | One hospital in Isfahan | ICU patients | Different sites | Pseudomonas aeruginosa (48) | • MHT • PCR |
AMK 79% CIP 85% |
• Resistance rate to carbapenem was 97% • blaIMP and blaVIM were detected in 15 and 7 isolates (all were MHT positive) • In one isolate of P. aeruginosa with the source of urine, both blaIMP and blaVIM genes were detected • 52% of urine samples contained blaIMP |
– | • Ratio of MDR pathogens for each sample was defined | blaIMP blaVIM | |
| Hosseinzadeh et al (2017)62 | Two hospitals in Shiraz | General inpatients and ICU patients | Different sites | Klebsiella pneumonia (211) | ✓ | • MHT for MBL detection • DDST for MBL detection • PCR |
AMK 78% CIP 85% GEN 89% |
• 13% of isolates (29 ones) were resistant to IPM and MEM and had an MIC value of 1.5–32 mcg/mL (25 isolates had MIC ≥4 mcg/mL) • Among carbapenem-resistant isolates, DDST and MHT were positive for 27 strains • 2 and 27 isolates had blaOXA-48 and blaNDM, respectively |
• Large sample size | blaOXA-48, blaNDM-1 | |
| Akhi et al (2017)68 | Seven hospitals in Tabriz and Orumie | General inpatients | Different sites | P. aeruginosa (245) | – | • DD • MHT • MCNP test • CIM • PCR |
CIP 66% LVX 66% AMK 25% |
• 49% (121) of isolates were carbapenem resistant • From 121 resistant strains, blaIMP and blaVIM genes were positive in 29 and 6 strains, respectively, and 40, 39, and 35 isolates showed positive results for MHT, MCNP, and CIM tests, respectively |
• MIC was not tested | • Large sample size • Sensitivity and specificity of three phenotypic methods were estimated according to PCR • Multicenter study |
blaVIM blaIMP |
| Falahat et al (2016)69 | Three hospitals in Arak | General inpatients | Different sites | P. aeruginosa (108) | – | • MHT and CDT for KPC detection • PCR |
CIP 20% AMK 21% GEN 23% |
• Resistance rate to carbapenem was about 25% • 38 and 26 of all isolates were MHT and CDT positive respectively • 13 isolates had blaKPC gene |
– | • Multicenter study | blaKPC |
| Mohammadzadeh et al (2016)70 | Two hospitals in Tehran | General inpatients | Not defined (probably different sites) | Different strains (864) | – | • CDT, MHT, and DDST for MBL detection • PCR |
Not defined | • 7% of isolates were IPM resistant • 35, 24, and 3 isolates were Acinetobacter spp., Pseudomonas spp., and Enterobacteriaceae, respectively • Carbapenem resistance was confirmed phenotypically (17% and 9%) and genotypically (15% and 9%) among Acinetobacter and Pseudomonas, respectively |
– | • Large sample size | blaVIM, blaIMP |
| Tarashi et al (2016)71 | One hospital in Tehran | Burn patients | Burn wounds | Acinetobacter baumannii (189) P. aeruginosa (309) | – | • CDT for MBL • PCR |
In 2015 for P. aeruginosa: AMK and GEN 95% CIP 97% For A. baumannii: AMK and CIP 100% GEN 95% |
• Carbapenem resistance rate of P. aeruginosa was 83% in 2012 and 96% in 2015 • Carbapenem resistance rate of A. baumannii in all years was 100% • Among 278 resistant P. aeruginosa isolates, 178 strains were MBL producers • Of the 187 resistant A. baumannii, 85 isolates were MBL producers • blaIMP-1 and blaVIM-1 genes were isolated in 30 and 52 of resistant strains of P. aeruginosa, respectively • blaIMP-1 and blaVIM-1 genes were detected in 10 and 34 of resistant A. baumannii, respectively |
– | • Large sample size • Providing trend of resistance through different years |
blaOXA-51 blaVIM blaIMP |
| Maspi et al (2016)72 | One hospital in Tehran | General inpatients and ICU patients | Different sites (mostly BAL and wounds) | A. baumannii (86) | – | • CDT for MBL detection • PCR |
TOB 75% | • Resistance rates to MEM and IMP were 90% and 73%, respectively • 44 isolates were MBL producers • 2, 13, 2, 4, and 2 isolates carried blaVIM, blaIMP, blaSPM, blaGIM, and blaSIM genes, respectively |
• MIC was not reported | • First report of blaSIM, blaGIM, and blaSPM. genes | blaVIM blaIMP blaSPM blaGIM blaSIM |
| Moghadam et al (2016)73 | Two hospitals in Shiraz | General inpatients and ICU patients | Different sites | A. baumannii (98) | ✓ | • E-test for MBL detection • PCR |
AMK 69% GEN 75% CIP and LVX 100% |
• Resistance to carbapenem was about 95% and according to MIC, it was 98% • Except two, all resistant isolates according to MIC had breakpoints >16 mcg/mL • Out of resistant isolates, 43 strains were MBL producers • Out of MBL producer strains, 14 had blaVIM and 23 had blaIMP genes |
– | • Results of general and ICU patients were discussed separately | blaVIM blaIMP |
| Pakbaten Toupkanlou et al (2015)74 | One hospital in Tehran | Burn patients | Not defined | P. aeruginosa (50) | • CDT for ESBL detection • PCR |
CIP 98%, AMK, GEN, and TOB 96% | • All resistant strains to IPM were chosen for study • 23 and 17 strains were positive in two different phenotypic methods for ESBL detection • 7, 18, 18, and 18 strains had blaPER, blaOXA-10, blaTEM, and blaSHV genes, respectively • 10 (20%) isolates carried bla-TEM, blaOXA-10, and blaSHV genes, simultaneously |
• Small sample size • Origins of samples were not defined |
– | blaOXA-10 blaSHV blaTEM blaPER | |
| Bina et al (2015)63 | Three hospitals in Tehran | General inpatients | Different sites | K. pneumonia (270) | – | • MHT test for KPC detection • PCR |
GEN 41.3% | Resistance rate to carbapenem was 14% (41 strains) • −14.5% (33 out of 41) strains were MHT positive and all 41 strains did not carry bla-KPC gene |
• Few antibiotics were tested | • Large sample size • Multicenter study |
– |
| Eftekhar and Naseh (2015)75 | Two hospitals in Tehran | Thirty nonburn and 25 burn patients | Different sites | K. pneumonia (30 nonburn and 25 burn, total 55) | – | • CDT for ESBL detection • DDST for MBL screening • MHT for MBL detection • PCR |
Burn and nonburn patients respectively: CIP 84%, 60% AMK 52%, 33.3% GEN 84%, 56.7% | Resistance rates to carbapenem were 20% in burn and 0% in nonburn patients • ESBL production was positive in 83% of nonburn and 72% of burn isolates • None was an MBL producer and positive for KPC genes • MHT was positive in four strains • Carbapenem-resistant isolates (n=5) from burn wounds were resistant to all tested antibiotics |
• Small sample size | • Comparison of resistance pattern in burn and nonburn patients | – |
| Lari et al (2015)64 | One hospital in Tehran | Burn patients | Burn wounds | P. aeruginosa (255) | – | • CDT for ESBL production • DDST for MBL production • PCR |
Were tested but not reported specifically | • All IPM-resistant strains were chosen for study • 63% were positive in CDT • None were positive for DDST • 5 and 4 strains had blaVIM and blaIMP genes, respectively |
Resistance to other antibiotics was not defined clearly | • Large sample size | blaVIM blaIMP |
| Bagheri Josheghani et al (2015)65 | One hospital in Kashan | General inpatients | Different sites | A. baumannii (124) | – | • CDT for ESBL detection • DDST for MBL detection • PCR |
AMK 78% GEN 83% LVX and CIP 99% | Resistance rates to carbapenems was about 90% • 5% were ESBL positive and 54% isolates were MBL • 79%, 25%, and 3% carried blaOXA-23, blaOXA-24, and blaOXA-58 genes, respectively |
– | – | blaOXA-51 blaOXA-23 blaOXA-24 blaOXA-58. |
| Azimi et al (2016)76 | One hospital in Tehran | Burn patients | Burn wounds | Different strains (161) | – | • MHT and DDST for MBL detection • CDT for KPC detection • PCR |
– | All IPM-resistant strains were included • 85, 51, and 112 of them were MHT, boronic acid, and dipicolinic positive, respectively |
• Exact number of resistance genes was not reported | • The sensitivity and specificity of each phenotypic test were reported | blaVIM blaIMP blaOXA-23 blaOXA-48 |
| Azimi et al |(2015)77 | One hospital in Tehran | Burn patients | Burn wounds | A. baumannii (65) | – | • CDT for ESBL detection • PCR |
80% of strains were resistant to all AG | All IPM-resistant strains were selected for study • 1 strain was CDT positive • 83%, 12%, and 9% of all strains harbored blaOXA-23, blaVIM, and blaKPC genes, respectively. |
– | – | blaOXA-23 blaOXA-51 blaVIM, blaKPC |
| Fazeli et al (2015)78 | One hospital in Isfahan | General inpatients | Different sites | K. pneumonia (112) | ✓ | • MHT and E-test for MBL detection • PCR |
CIP 63.4% AMK 51% GEN 64% |
42% of strains (49 isolates) were resistant to IPM and MEM • 32 out of 49 resistant isolates were positive for MHT • 5 isolates were positive in E-test and 6 strains harbored NDM-1 • MIC values for these six strains was >256 mcg/mL |
blaNDM | ||
| Khoshvaght et al (2014)79 | Four hospitals in Zanjan | General pediatrics | Stool | Escherichia coli (230) | – | • DDST for ESBL and MBL detection • PCR |
CIP 37% AMK 21% GEN 29% |
IPM resistance rate was 2.1% • Out of the 36 Enteroaggregative E. coli isolates, 19 (52%) were ESBL positive and none were MBL positive • 15 and 12 isolates had blaTEM and blaCTX-M genes |
• MIC was not tested | • Large sample size • Comparing data of healthy children with those with diarrhea |
blaTEM and blaCTX-M |
| Noori et al (2014)80 | Two hospitals in Tehran | General inpatients | Different sites | Acinetobacter spp. (108) | ✓ | • DD • CDT for MBL |
CIP 92% AMK 80% GEN 40% |
• 91% (99 isolates) were resistant to IPM and MEM • 86 isolates were MBL producers • Out of these 86 MBL producers, three isolates harbored blaIMP • MIC50 values for MEM and IPM were 32 and 128 mcg/mL, respectively |
– | – | blaIMP |
| Vali et al (2014)81 | Two pediatric hospitals in Tehran | Cystic fibrosis inpatients | Sputum | Different gram negative (52) | ✓ | • CDT for ESBL detection • DDST for MBL detection • PCR |
GEN 24.5% CIP 18% |
• Resistance rate to MEM was 14% (eight strains) that MHT was positive in three of them • The EDTA disk synergy was also positive in four isolates • blaVIM and blaIMP were detected in five and two strains of Achromobacter xylosoxidans |
– | • Data provided from cystic fibrosis patients | blaCTX-M blaIMP blaVIM |
| Azimi et al (2014)82 | One hospital in Tehran | Burn patients | Burn wounds | K. pneumonia (28) | ✓ | • MHT • CarbaNP test • BA • PCR |
GEN 90% AMK 79% | All isolates were resistant to IPM and MEM • 9 and 11 strains had an MIC >64 mcg/mL for IPM and MEM, respectively • All phenotypic tests except BA were positive • 27 isolates had blaOXA-48 gene and one had blaVIM-4 |
• Small sample size • Few antibiotics were tested |
• CarbaNP test was done | blaOXA-48 blaVIM-4 |
| Hashemi et al (2014)83 | Two hospitals in Tehran | General inpatients and infants | Different sites | K. pneumonia (83) | ✓ | • CDT • AmpC screening • MHT • PCR |
Resistance and intermediate: CIP 39.7%, 4.8% GEN 61.5%, 3.6% AMK 80.7%, 4.8% |
• Resistance rates to IPM, MEM, and DOR were between 68% and 74% • MIC range for IPM and MEM was 0.25–256 mcg/mL • 48 (57%) isolates were positive for ESBL • 5 isolates were MHT positive • 23 isolates were AmpC positive • 30 strains had OmpK35 genes |
– | • Detection of porin-related genes | blaoxa-48 blaCTX-M-15 |
| Japoni-Nejad et al (2014)84 | One hospital in Arak | ICU patients | Surgical wounds, urine, blood, respiratory secretions | K. pneumonia (100) | – | • MHT • E-test for MBL detection • DDST • CDT for KPC detection • PCR |
d | • Resistance rate to carbapenems in Amp C producers and carbapenemase producers was 5% and 66%, respectively • blaVIM and blaGES were detected in ten and two isolates • One isolate (1%) contained both blaVIM and blaAmpC genes |
– | – | blaVIM blaGES |
| Farajzadeh Sheikh et al (2014)85 | Hospitals in Ahvaz | General inpatients | Different sites | P. aeruginosa (223) | ✓ | • CDT and E-test for MBL detection • PCR |
GEN 66% AMK 55% TOB 67% CIP 66% |
58%, 31%, 13%, and 74% of isolates were resistant to IMP, MEM, DOR and ERT, respectively. Thirty isolates were resistant to all carbapenems • All imipenem-resistant isolates were MBL producers, except one • blaIMP and blaVIM genes were detected in 26 and 1 of the all strains, respectively • 13%–74% of resistant isolates had an MIC >16 mcg/mL for different carbapenems |
• Large sample size | blaVIM blaIMP | |
| Aghamiri et al (2014)86 | Several hospitals in Tehran | General inpatients | Different sites | P. aeruginosa (212) | ✓ | • DD • DDST for MBL detection • PCR |
47% of isolates were resistant to IPM and all had an MIC >16 mcg/mL • 70 isolates were MBL positive • 20 and 70 strains had blaIMP and blaVIM genes, respectively |
blaIMP blaVIM | |||
| Nobari et al (2014)87 | Eight hospitals in Tehran | General inpatients | Different sites | K. pneumonia (180) | • DD • MHT for MBL detection • PCR |
Resistance rate to carbapenems was between 7% and 23% (66% of urine isolates were resistant to carbapenems) • 25 isolates were MHT positive • blaCTX-M, blaSHV, blaTEM, and blaPER genes were detected in 69%, 59.5%, 35.7%, and 16.6% of carbapenem-resistant isolates respectively • blaNDM-1, blaVIM-1, and blaKPC were detected in 3, 5, and 1 isolates |
blaCTX-M blaSHV, blaTEM blaPER blaNDM-1 blaVIM-1 blaKPC | ||||
| Hakemi Vala et al (2014)88 | One hospital in Tehran | Burn patients | Burn wounds | P. aeruginosa (47) and Acinetobacter spp. (28) | ✓ | • CDT for MBL and ESBL detection • PCR |
P. aeruginosa GEN 72% CIP 69% A. baumannii GEN 96% CIP 100% |
• 100% of A. baumannii and about 75% of P. aeruginosa isolates were resistant to carbapenems • MIC range of IPM for P. aeruginosa was 2–128 and for A. baumannii was 4–128 mcg/mL • 17% of the P. aeruginosa isolates and 16% of the A. baumannii isolates were MBL producers • Mortality rate of MBL producer was 20% |
– | • Mortality rate was reported | blaSPM, blaIMP, blaCTX-M-15 |
| Tavajjohi et al (2013)89 | One hospital in Kashan | General inpatients and environmental isolates | Different strains and wet environment | P. aeruginosa (100) | – | • DDST for ESBL detection • PCR |
GEN 50% CIP 12% | • 8% of isolates were ESBL producers that blaGES was detected in all ESBL producers and 50% (4 isolates) of them were resistance to IPM | – | • Small sample size (only eight strains were evaluated for carbapenem resistance and genes) | blaGES |
| Azimi et al (2013)90 | One hospital in Tehran (Motahari) | Burn patients | Burn wounds | K. pneumonia (44) | • DD • MHT for KPC detection • BA |
– | • 64% of isolates were resistant to carbapenem • MHT was positive in all of them, but none showed synergism between MEM and boronic acid • All were negative for blaKPC |
• Other antibiotics were not evaluated • Small sample size |
– | ||
| Noori et al (2013)91 | Two hospitals in Mashhad | General inpatients | Wounds and respiratory secretions | Acinetobacter spp. (70) | • DDST for MBL production • PCR |
CIP 100% GEN 88% |
• All isolates were resistant to IPM and MEM • 50 out of 70 strains were MBL producers, but only 3 strains had blaVIM |
blaVIM | |||
| Azimi et al(2013)92 | One hospital in Tehran | Burn patients | Not defined (probably burn wounds) | A. baumannii (93) and Acinetobacter lwoffii (1) | ✓ | • CDT for MBL detection • PCR |
– | 85% of isolates were resistant to IPM that MIC range for them was 16–128 mcg/mL • 34% of all isolates were MBL positive (31 strains) • 25 out of MBL producers had blaOXA-23 • None was positive in DDST |
Blaoxa-23 | ||
| Pajand et al (2013)93 | Two hospitals in Tehran | Inpatients and burn patients | Burn wounds and other sites | A. baumannii (43 burn and 32 nonburn patients, total=75) | ✓ | • DDST for MBL detection • PCR |
Nonburn and burn patients: CIP 87%, 97% AMK 56%, 95% TOB 47%, 84% |
Resistance rate to carbapenem in burn and nonburn patients was 98% and 68%, respectively • 66% of nonburn and 70% of burn isolates had blaOXA-23 • blaOXA-40-like gene was detected in 12% and 74% of non-burn and burn isolates, respectively • MIC values for all carbapenems were reported • All isolates were positive for AmpC and ISAba genes and 71 of them were positive for carO gene |
• Results of DDST were not reported | • Detection of OMP gene (carO) for evaluation of carbapenem resistance | BlaOXA-23 blaOXA-40 AmpC, ISAba, carO |
| Azami et al (2013)94 | Three hospitals in Tehran | General inpatients | Not defined | P. aeruginosa (130) | ✓ | • CDT for MBL detection • PCR |
GEN 63% CIP 59% |
• According to MIC, 53% of isolates were resistant to IPM and MIC values were ≥16 mcg/mL for IPM • 56% had class I integrons • blaVIM was found in 10 strains |
• Few antibiotics were tested • Source of sample was not clear |
• Assessment of integrin genes (genes for mobility of betalactamase) • Multicenter study |
blaVIM |
| Mohajeri et al (2013)95 | Three hospitals in Kermanshah | General inpatients | Sputum, blood, urine | Acinetobacter spp. (104) | – | • E-test for MBL detection • DDST • PCR |
CIP 69%, GAT 43% LVX 62% TOB 39% GEN 68% AMK 53% |
Resistance rate to carbapenems was 75%–80% • 84 of isolates were MBL producers • The blaOXA-23 and blaOXA-24 were found among 77% and 19% of the isolates, respectively • Correlation between rate of resistance to carbapenem and presence of blaOXA-23-like gene was statistically significant |
• MIC was not reported | • Multicenter study | blaOXA-23 blaOXA-24 |
| Doosti et al (2013)96 | One hospital in Zanjan | General inpatients | Different sites | P. aeruginosa (70) | ✓ | • DD • DDST for MBL detection • PCR |
CIP 40% GEN 55% |
63% of isolates were resistant to IPM that 41 of them had an MIC >4 mcg/mL • DDST was positive for all of them • 36 isolates were MBL producers • From 41of isolates with high MIC breakpoints, 10 isolates had blaIMP and 23 had blaVIM |
– | – | blaVIM blaIMP |
| Shahcheraghi et al (2013)97 | Five hospitals in Tehran | General inpatients | Mostly urine and feces | Different strains (360) | – | • MHT • MBL E-test • PCR |
CIP 61.3% AMK 14.4% KAN 38.6% |
• 6%, 3%, and 1% were resistant to MEM, ERT, and IPM, respectively • MHT was positive in 11 resistant isolates |
• MIC was not reported | • Multicenter study • Large sample size • Recognition of new K. pneumonia containing bla NDM |
blaTEM blaSHV blaCTX-M blaNDM blaPER |
| Kalantar et al (2012)98 | One hospital in Sanandaj | General inpatients | Different sites | P. aeruginosa (100) | ✓ | • DD • DDST for MBL detection • PCR |
• 22 isolates were MBL positive and among them 8 isolates were resistant to IPM • 8 isolates had blaVIM |
blaVIM | |||
| Shahcheraghi et al (2011)99 | Seven hospitals in Tehran | General inpatients | Different sites | Acinetobacter spp. (203) | ✓ | • DD • CDT for MBL detection • PCR |
Of the 100 strains that were IPM resistant: CIP 84% AMK 84% | 49% and 100% of isolates were resistant to IPM and MEM, respectively • All isolates had an MIC ≥8% and 47% had an MIC =64 mcg/mL • From IPM-resistant isolates, 9% were MBL producers • The blaSPM, blaGES, blaOXA-51, and blaOXA-23 genes were detected by PCR among 6, 2, 94, and 84 isolates of A. baumannii, respectively |
• Multicenter study • Large sample size • First report of blaSPM and blaGES |
blaOXA-23 blaSPM blaGES | |
| Peymani et al (2011)100 | One hospital in Tabriz | General inpatients | Different sites | A. baumannii (100) | – | • E-test for MBL detection • PCR |
AMK 83% GEN 91% CIP 90% LVX 86% |
About 55% of isolates were resistant to carbapenem • 31 of carbapenem-resistant isolates were MBL producers and 28 of them were positive for bla genes |
– | blaVIM blaIMP | |
| Saderi et al (2010)101 | One hospital in Tehran | General inpatients | Burn wounds | P. aeruginosa (100) | – | • CDT for MBL detection • PCR for MBL genes |
GEN 86% AMK 73% CIP 55% |
• 69% of isolates were resistant to IPM • 65 isolates were CDT positive • 13 resistant isolates contained blaVIM |
• MIC was not determined | blaVIM | |
| Yousefi et al (2010)102 | Two hospitals in Orumieh and Tabriz | General inpatients | Different sites | P. aeruginosa (324) | ✓ | • DDST for detection of MBL • PCR |
CIP 83% GEN 86% AMK 85% |
• Resistance rate to MEM was 86% • Most of the isolates had an MIC ≥32 mcg/mL for IPM • Among nonsusceptible strains to IPM, 39 isolates were MBL positive • 18 out of resistant isolates carried blaVIM and 6 isolates carried blaIMP |
• Large sample size | blaIMP blaVIM | |
| Bahar et al (2010)103 | One hospital in Tehran (Motahari) | Burn patients | Burn wounds | P. aeruginosa (186) | – | • Disk inhibitor synergy test • PCR |
It was not reported for all isolates (only reported in MBL producers) | • EDTA disk showed that 23 strains produced MBL and all had blaVIM gene • Mortality of MBL producer strains was 82% and non-MBL producer was 22% |
• Large sample size • Mortality rates of MBL and non-MBL producers were reported |
blaVIM | |
| Khosravi et al (2008)104 | One hospital in Ahvaz | General inpatients | Different sites | P. aeruginosa (100) | – | • E-test for MBL detection • PCR |
– | • Resistance rate to carbapenems was 41% • Of the resistant isolates, eight isolates were MBL producers and all contained blaVIM |
• FQ and AG were not tested | blaVIM | |
| Yazdi et al (2007)105 | Two hospitals in Tehran | Not defined | Not defined | P. aeruginosa (126) | ✓ | • DD • E-test for MBL detection • PCR |
– | According to MIC, resistance to IPM was 29% • 70 isolates were MBL producers • 8 isolates had blaVIM • MIC of these eight strains was 32–64 mcg/mL for IPM |
• Few antibiotics were tested | – | blaVIM |
Notes:
MIC values for carbapenems (but not other antimicrobial agents) were considered.
Resistance rates were according to the result of disk diffusion test.
Results about blaOXA-51 were not provided because this gene has no correlation with occurrence of resistance, unless it has ISAba genes in its upstream.
For complete information about the resistance to AG and FQ, please refer to the original reference.
Abbreviations: AG, aminoglycoside; AMK, amikacin; BA, boronic acid; CDT, combination disk test; CIM, carbapenem inactivation method; CIP, ciprofloxacin; DD, disk diffusion; DDST, double disk synergy test; DOR, doripenem; ERT, ertapenem; ESBL, extended spectrum β-lactamase; FQ, fluoroquinolones; GAT, gatifloxacin; GEN, gentamicin; ICU, intensive care unit; IPM, imipenem; KAN, kanamycin; KPC, Klebsiella-producing carbapenemase; LVX, levofloxacin; MBL, metallo-β-lactamase; MCNP, modified CarbaNP; MEM, meropenem; MHT, modified Hodge test; MIC, minimum inhibitory concentration; NAL, nalidixic acid; NOR, norfloxacin; OFX, ofloxacin; PCR, polymerase chain reaction; TOB, tobramycin; BAL, Broncho Alveolar Lavage.
Discussion
Increase in the rate of resistance to carbapenem antibiotics among gram-negative isolates is a worldwide concern, especially in developing countries. The results of this review also showed this threat in the recent decade in a country of Middle East area, Iran. Although the prevalence of carbapenem-resistant gram-negative rods was reported according to the disk diffusion method in most studies, phenotypic and genotypic assays confirmed this pattern in some available surveys.
Resistance to carbapenem antibiotics in the same years in a country of neighborhood, Saudi Arabia, shows lower resistance rate in comparison with Iran.106 In 2010, resistance to carbapenems was reported in 10%–66% of gram-negative isolates in Saudi Arabia, which was lower than that in Iran (86%).11 The results were different in Europe. In the year 2006–2007, 4% and 85% of gram-negative isolates were carbapenem resistant in North and South of Europe, respectively. Although data were limited to year 2006, Iran had a better condition compared with the South of Europe, but worse than North of Europe.107 The results of another study in year 2012 also confirmed this pattern in North and South of Europe.108 In case of P. aeruginosa, MIC values that were reported for carbapenem-resistant strains were dramatically different from European countries. In year 2011, Castanheira et al detected a mean MIC value >2 mcg/mL for carbapenem-resistant P. aeruginosa in 14 European countries.109 Yousefi et al in Iran reported an MIC >32 mcg/mL for most of carbapenem-resistant P. aeruginosa isolates. At these times, the CLSI and European Committee on Antimicrobial Susceptibility Testing cutoff values for carbapenem resistance were 2 and 8 mcg/mL, respectively.11
Incremental trend of resistance to carbapenem antibiotics was evident through 2006–2018. The least resistance rate was reported by Rahbar et al in year 2010 at Milad Hospital. In this year, 1.1% of A. baumannii isolates were resistant to imipenem.22 This conclusion may be misleading, because 2 years earlier (in 2008), in this hospital, 27% and 40% of the same isolates were resistant to imipenem and meropenem, respectively.12 Tarashi et al assessed the trend of antibiotic resistance in A. baumannii through years 2012–2015.71 The results showed that resistance in P. aeruginosa increased from 83% in year 2012 to 96% in year 2015. Also 100% of A. baumannii were resistant to carbapenem among these years.
The resistance patterns for other microorganisms did not follow specific trends. For example, 60% of P. aeruginosa isolates were imipenem resistant in the year 2010,101 but this rate was 25% in another hospital in the year 2016.69 Outpatients-isolated P. aeruginosa strains showed less resistance rates (13% in Ahangarzadeh Rezaee et al38 and 4.4% in Hashemi et al21). However, carbapenem-resistant rates were dramatically high in studies that included ICU samples.9,20 In an earlier study,9 100% and 62% of P. aeruginosa isolates were resistant to imipenem and meropenem, respectively, and in a latter study,20 about 100% of them were resistant to both carbapenems.
Most of the carbapenem-resistant microorganisms were isolated from burned patients, and many studies placed in this group were from Motahari Hospital. Motahari is a referral teaching burn hospital in center of Iran and most complicated burned patients are referred from all areas of the country. Increasing resistance rates can be observed through different years in burn patients; 50%–60% in year 2008 to 90% in year 2017.42,46 Also, the presence of blaOXA type genes has increased from half of the resistant strains to almost 80% of them. This can be a warning alarm regarding administration of antibiotics in burned patients.
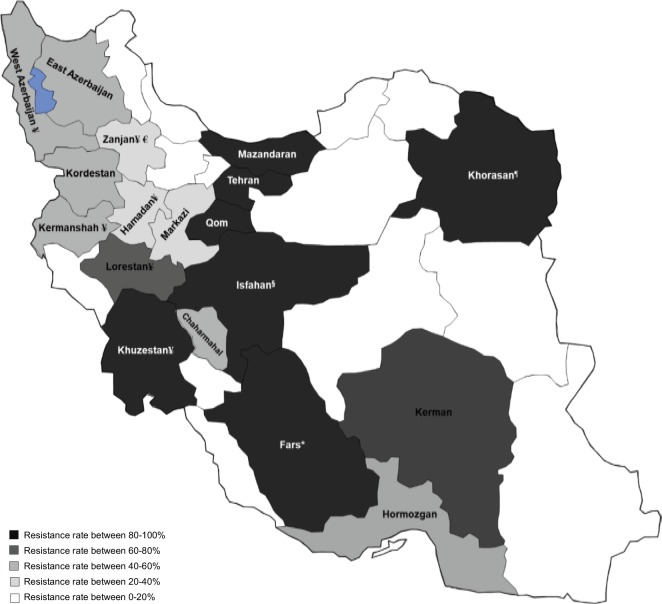
Disk diffusion is the initial method for detection of resistant strains; however, it does not have enough accuracy for antimicrobial agents like carbapenem. Most evaluated studies considered disk diffusion method. Beside disk diffusion, MIC breakpoints can be used to confirm the results of disk diffusion (Figure 3).
Figure 3.
Carbapenem-resistance rates in different areas of Iran (according to the disk diffusion method).
Notes: The data were extracted from the latest available studies. Multicenter studies from different cities were not considered for mapping due to pooled data. The studies in the special populations (ie, pediatrics or cystic fibrosis) and outpatients were excluded. *Reported resistance rates from Fars province were conflicting (13.7% in 2017 and 96% in 2016). §Data for Isfahan province were obtained from both Kashan (a city around Isfahan) and Isfahan itself, although the sample size for Isfahan was very small. ¶Data for Khorasan area were extracted from a study in 2015 with 36 strains. €Intermediate and absolute resistance rates were 25% and 2%, respectively. ¥Data in these provinces were for 2010–2014 studies.
Although MHT has been widely used as the preferred phenotypic method for detection of MBL, it is not recommended in latest version of CLSI, due to its low sensitivity.110 In CLSI 2018, CarbaNP method is recommended instead. Kuchibiro et al compared some phenotypic methods and MHT and showed an acceptable specificity (100%), but the sensitivity was very low (50%). Another test, modified carbapenems inactivation method, has acceptable sensitivity and specificity values (both above 99%) and does not require specific equipment. This method, beside CarbaNP, can be recommended for phenotypic detection of carbapenemase.111
Findings according to the phenotypic and genotypic methods were inconsistent. For example, in the study by Bina et al,63 although all strains were MHT positive, none of them carry blaKPC gene. This may be due to low sensitivity of MHT in detecting carbapenemase enzyme.
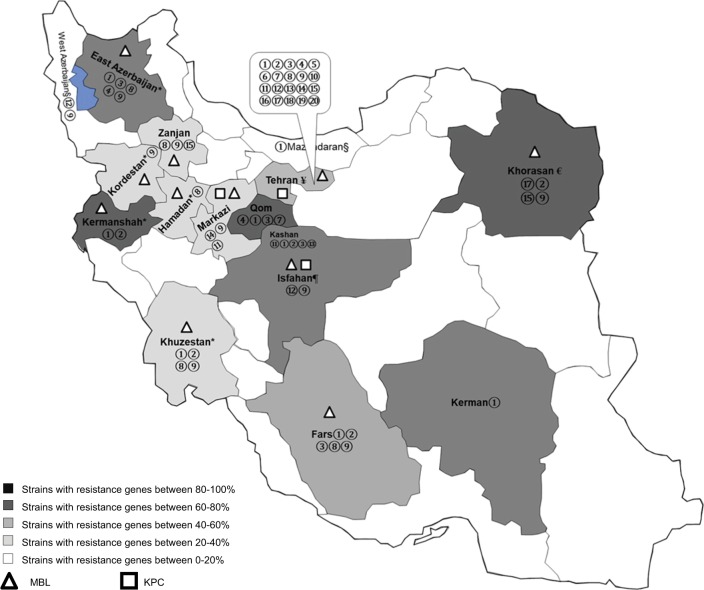
The genotypic assays were mostly applied on central regions of Iran. However, there are no data regarding carbapenemase-encoding genes in most provinces. This may be due to technical and economic restrictions in these areas (Figure 4).
Figure 4.
Carbapenem-resistant rates in different areas of Iran (according to the phenotypic and genotypic methods).
Notes: The data were extracted from the latest available studies. Multicenter studies from different cities were not considered for mapping due to pooled data. The studies in the special populations (ie, pediatrics or cystic fibrosis) and outpatients were excluded.
Some genes like blaSPM and blaSHV are mostly known as genes that encode ESBL enzymes, but they were included in this map, because these genes were assessed along with carbapenemase genes, and also overexpression of these genes concomitant with harboring efflux pump may be responsible for resistance to carbapenems. *Data for these five provinces were extracted from 2011 to 2014 articles, so new data are needed.
€Data were extracted from a 2015 study with 36 samples; another study in 2013 confirmed the presence of the genes of resistance in 4% of strains. ¥Data from Tehran province were conflicting; genes encoding resistance were detected in 15%–50% of isolates through 2015–2016. ¶Data from Kashan (a city in Isfahan province) showed presence of resistance genes in 80% of isolates in 2015.
§In Mazandaran and West Azerbaijan, these genes were reported, but rates of resistance were not included because the studies included different cities from different provinces. ① blaOXA-23 ② blaOXA-24 ③ blaOXA-58 ④ blaOXA-40 ⑤ blaOXA-48 ⑥ blaOXA-10 ⑦ blaOXA-143 ⑧ blaIMP ⑨ blaVIM ⑩ blaSPM ⑪ blaGES ⑫ blaPER ⑬ blaNDM ⑭ blaKPC ⑮ blaTEM ⑯ blaSHV ⑰ blaADC ⑱ blaSIM ⑲ blaGIM.
Abbreviations: ESBL, extended spectrum β-lactamase; KPC, Klebsiella-producing carbapenemase; MBL, metallo-β-lactamase.
The presence of some blaOXA-type genes were not necessarily associated to carbapenem-resistant. For example, blaOXA-51 in A. baumannii did not contribute to carbapenem resistance at all. In the study by Azizi et al, it was shown that blaOXA-23 may be found in both imipenem-sensitive and imipenem-resistant isolates.43 Also, in the study by Bahador et al, blaOXA-23 was found in both susceptible and nonsusceptible strains.44 However, higher carbapenem resistance in isolates harboring blaOXA-23 was reported (Shoja et al54). Table 5 shows the number of genes that were found in each city and related microorganisms.
Table 5.
Types of bla genes, first location of isolation, and the relevant microorganisms in Iran
| Ambler classes | Types of pf bla genes | First location of isolation | Yeara | Total genesb | Microorganismc |
|---|---|---|---|---|---|
|
| |||||
| A | GES | Tehran, by Shahcheraghi F. | 2011 | 12 | Mostly P. aeruginosa |
| KPC | Tehran, by Nobari S. | 2014 | 61 | Mostly K. pneumonia | |
|
| |||||
| B | NDM | Tehran, by Shahcheraghi F. | 2012 | 44 | Mostly K. pneumonia |
| VIM | Tehran, by Rezaei Yazdi H. | 2007 | 437 | Mostly P. aeruginosa | |
| IMP | Tabriz and Orumieh, byYousefi S. | 2010 | 271 | Mostly P. aeruginosa | |
| SIM | Tehran, By Maspi H. | 2016 | 2 | Only in A. baumannii | |
| GIM | Tehran, By Maspi H. | 2016 | 4 | Only in A. baumannii | |
|
| |||||
| D | OXA-23 | Tehran, by Taherikalani M. | 2008 | 1,287 | Only in A. baumannii |
| OXA-24 | Tehran, by Taherikalani M. | 2008 | 213 | Only in A. baumannii | |
| OXA-58 | Tehran, by Taherikalani M. | 2008 | 96 | Only in A. baumannii | |
| OXA-40 | Tabriz, by Sohrabi N. | 2012 | 59 | Only in A. baumannii | |
| OXA-48 | Tehran, by Azimi L. | 2014 | 52 | In K. pneumonia and others | |
| OXA-10 | Tehran, by Pakbaten S. | 2015 | 12 | Only in P. aeruginosa | |
| OXA-143 | Qom, by Sharikhani Z. | 2017 | 14 | Only in A. baumannii | |
Notes:
Year that the genes were isolated for the first time.
Total number of genes that were reported in Iranian studies.
The microorganisms in which the genes were isolated.
Emergence of some genes including blaNDM-1 is a serious global warning. This gene is highly transmissible and was first discovered in a Swedish patient who had traveled to Pakistan. Although almost all identified cases were originally from Pakistan and India, the first case of blaNDM-1 that was reported in Iran in the year 2012, did not have a history of traveling to these countries. In two studies from Shiraz and Isfahan, 27 and 6 strains of K. pneumonia harbored blaNDM in years 2015 and 2017, respectively.62,78
Genotypic methods to identify other mechanisms of resistance to carbapenems have recently been considered by researchers. OmpK and carO genes that regulate expression of mutated efflux pump were detected in Hashemi et al and Pajand et al studies, respectively.83,93 In another study, efflux pump’s activity and mutations in porins were investigated.112 These results can expand the view about true mechanisms of resistance to carbapenems in developing countries.
Most studies included biological samples from different sites like urine, trachea, and wounds. However, sites with highest or lowest resistant rates were not defined, except in a few studies. In study by Nobari et al, urine was the source with more resistant strains.87 In the study by Khorvash et al, one P. aeruginosa strain that was isolated from urine samples harbored both blaIMP and blaVIM genes.67
Lack of clinical insight was the main limitation of almost all studies. Only in three studies, mortality rates in patients infected with resistant strains were addressed. Mortality rate of patients infected with carbapenem-resistant K. pneumonia isolates was 33% in 2013 (Rastegar Lari et al study).25 In the same year, infections with carbapenem-resistant A. baumannii strains caused 20% mortality.24 In the study by Bahar et al, the mortality rate of MBL-producer P. aeruginosa isolates vs non-MBL producers was 82% vs 22%.103 In some studies, patients’ conditions including requiring mechanical ventilation and days of hospital stay were considered, but no correlation between these data and acquiring carbapenem-resistant strains was found. Also in most studies, difference between infection and colonization was not clear.
Restrictions in technical facilities and standard laboratories in small cities are important issues. Access to a standard microbiological laboratory is not feasible in most regions. Disk diffusion was the main method for detecting carbapenem resistance until recently. Also, MIC breakpoints were adapted from CLSI and national data regarding these cutoffs are lacking. National antimicrobial resistance surveillance studies have not been well organized. Interpreting data from different regions of a country, considering the local standards, is essential.
Pattern of antibiotic use can affect emergence of resistant microorganisms. Rational use of drugs, and specifically antibiotics, is a challenging issue in developing countries. Mean number of drugs per prescription in Iran was higher than the World Health Organization standards.113,114 Approximately, 50% of inpatients, prescriptions contained at least one antibiotic, and this percentage was even higher in outpatient settings.115 Overuse of antibiotics, especially injectable ones, and easy access to antibiotics without prescription is a warning alarm for future antibiotic resistance in developing countries. Establishing antimicrobial stewardship’s programs is new in our hospitals. Unfortunately, governmental rules and supports to restrict antibiotic access in community pharmacies and prescription by general physicians are limited.
Following issues may be considered in future studies. Considering combination of antimicrobials for assessing the resistance is an important finding. Evaluating effects of combination disks (ie, a carbapenem with an aminoglycoside or a fluoroquinolone) on resistant gram-negative bacilli may be helpful.116 Although combination therapy may increase the risk of side effects, it remains an initial therapeutic option for MDR isolates. Another issue is studying of antibiotics’ resistance pattern in health-care facilities. Remarkable increase in resistance rates among isolates from health-care facilities were reported. These strains may act as KPC-producing reservoirs.117 A higher resistance rate in long-term care facilities in comparison with intensive care units has been identified.118 Although some studies in Iran have included outpatients to evaluate carbapenem resistance, patients in long-term health-care facilities have not been included.
Footnotes
Authors’ contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18(2):306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan JD, Graves JA, Dellit TH. Antimicrobial treatment and clinical outcomes of carbapenem-resistant Acinetobacter baumannii ventilator-associated pneumonia. J Intensive Care Med. 2010;25(6):343–348. doi: 10.1177/0885066610377975. [DOI] [PubMed] [Google Scholar]
- 3.Sun K, Xu X, Yan J, Zhang L. Evaluation of six phenotypic methods for the detection of carbapenemases in gram-negative bacteria with characterized resistance mechanisms. Ann Lab Med. 2017;37(4):305–312. doi: 10.3343/alm.2017.37.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambler RP. The structure of b-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 5.Mcmullen AR, Yarbrough ML, Wallace MA, Shupe A, Burnham CD. Evaluation of genotypic and phenotypic methods to detect carbapenemase production in gram-negative Bacilli. Clin Chem. 2017;63(3):723–730. doi: 10.1373/clinchem.2016.264804. [DOI] [PubMed] [Google Scholar]
- 6.Pletz MW, Wellinghausen N, Welte T. Will polymerase chain reaction (PCR)-based diagnostics improve outcome in septic patients? A clinical view. Intensive Care Med. 2011;37(7):1069–1076. doi: 10.1007/s00134-011-2245-x. [DOI] [PubMed] [Google Scholar]
- 7.Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16(4):303–307. [PMC free article] [PubMed] [Google Scholar]
- 8.Douraghi M, Ghalavand Z, Nateghi Rostami M, et al. Comparative in vitro activity of carbapenems against clinical isolates of Acinetobacter baumannii. J Appl Microbiol. 2016;121(2):401–407. doi: 10.1111/jam.13178. [DOI] [PubMed] [Google Scholar]
- 9.Ghasemian R, Ahanjan M, Fatehi E, Shokri M. Prevalence and antibiotic resistance pattern of Acinetobacter isolated from patients admitted in ICUs in Mazandaran, Northern Iran. Glob J Health Sci. 2016;8(11):112–119. [Google Scholar]
- 10.Shakibaie MR, Adeli S, Salehi MH. Antibiotic resistance patterns and extended-spectrum β-lactamase production among Acinetobacter spp. isolated from an intensive care unit of a hospital in Kerman, Iran. Antimicrob Resist Infect Control. 2012;1:1. doi: 10.1186/2047-2994-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yousefi S, Farajnia S, Nahaei MR, et al. Class 1 integron and Imi-penem Resistance in Clinical Isolates of Pseudomonas aeruginosa: Prevalence and Antibiotic Susceptibility. Iran J Microbiol. 2010;2(3):115–121. [PMC free article] [PubMed] [Google Scholar]
- 12.Rahbar M, Kabeh-Monnavar M, Khadem Vatan K, Fadaei-Haqi A, Shakerian F. Carbapenem resistance in gram-negative Bacilli isolates in an Iranian 1000-bed tertiary hospital. Pak J Med Sci. 2008;24(4):537–540. [Google Scholar]
- 13.Babamahmoodi F, Ahangarkani F, Davoudi A. Hospital-acquired infections, bacterial causative agents and antibiotic resistance pattern in intensive care units at teaching hospitals in north of Iran. Int J Med Invest. 2015;4(1):152–160. [Google Scholar]
- 14.Mohajeri P, Rezaei Z, Sharbati S, et al. Frequency of adhesive virulence factors in carbapenemase-producing Acinetobacter baumannii isolated from clinical samples in West of Iran. Asian J Biol Sci. 2014;7(4):158–164. doi: 10.4103/0976-9668.175071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobaraki S, Aghazadeh M, Soroush Barhaghi MH, et al. Prevalence of integrons 1, 2, 3 associated with antibiotic resistance in Pseudomonas aeruginosa isolates from Northwest of Iran. Biomedicine. 2018;8(1):2–7. doi: 10.1051/bmdcn/2018080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghanbari F, Khademi F, Saberianpour S, et al. An epidemiological study on the prevalence and antibiotic resistance patterns of bacteria isolated from urinary tract infections in central Iran. Avicenna J Clin Microbiol Infect. 2017;4(3):e42214. [Google Scholar]
- 17.Chahoofard A, Dehghan F, Karmostaji A, Zolghadri N. Hospital-acquired urinary tract infection, microbial causative agents and antibiotic resistance pattern in southern Iran: a prospective study. J Glob Pharma Tech. 2017;1(9):52–58. [Google Scholar]
- 18.Ansari H, Doosti A, Kargar M, Bijanzadeh M, Jafarinya M. Antimicrobial resistant determination and prokaryotic expression of smpA Gene of Acinetobacter baumannii isolated from admitted patients. Jundishapur J Microbiol. 2017;10(11):e59370. [Google Scholar]
- 19.Babakhani S, Shokri Derikvand S, Nazer MR, Kazemi MJ. Comparison frequency and determination antibiotic resistance pattern of Klebsiella spp. isolated from Nosocomial infection in Khorramabad Shohadaye Ashayer hospital. Bull Env Pharmacol Life Sci. 2014;3(12):149–154. [Google Scholar]
- 20.Kamalbeik S, Kouchek M, Baseri Salehi M, Fallah F, Malekan MA, Talaie H. Prevalence of Class 2 integrons in multidrug-resistant Acinetobacter Baumannii in toxicological ICU patients in Tehran. Iran J Toxicol. 2013;7(22):900–906. [Google Scholar]
- 21.Hashemi SH, Esna-Ashari F, Tavakoli S, Mamani M. The prevalence of antibiotic resistance of Enterobacteriaceae strains isolated in community- and hospital-acquired infections in teaching hospitals of Hamadan, West of Iran. J Res Health Sci. 2013;29(13):75–80. [PubMed] [Google Scholar]
- 22.Rahbar M, Mehragan H, Haji Ali Akbari N. Prevalence of drug resistance in nonfermenter gram-negative Bacilli. Iran J Pathol. 2010;5(2):90–96. [Google Scholar]
- 23.Soroush S, Haghi-Ashtiani MT, Taheri-Kalani M, et al. Antimicrobial resistance of nosocomial strain of Acinetobacter baumannii in Children’s Medical Center of Tehran: a 6-year prospective study. Acta Med Iran. 2010;48(3):178–184. [PubMed] [Google Scholar]
- 24.Lari AR, Mohammadi Barzelighi H, Arjomandzadegan M, Nosrati R, Owlia P. Distribution of Class I integron among isolates of Acinetobacter baumannii recovered from burn patients. J Med Bacteriol. 2013;2(1):1–11. [Google Scholar]
- 25.Rastegar Lari A, Azimi L, Rahbar M, Fallah F, Alaghehbandan R. Phenotypic detection of Klebsiella pneumoniae carbapenemase among burns patients: first report from Iran. Burns. 2013;39(1):174–176. doi: 10.1016/j.burns.2012.02.025. [DOI] [PubMed] [Google Scholar]
- 26.Ghotaslou R, Sadeghi M, Akhi M, Hasani A, Asgharzadeh M. Prevalence and antimicrobial susceptibility patterns of ESBL, AmpC and carbapenemase-producing Enterobacteriaceae isolated from hospitalized patients in Azerbaijan, Iran. Iran J Pharm Res. 2018;17(Special Issue):79–88. [PMC free article] [PubMed] [Google Scholar]
- 27.Saadatian Farivar A, Nowroozi J, Eslami G, Sabokbar A. RAPD PCR profile, antibiotic resistance, prevalence of armA Gene, and detection of KPC enzyme in Klebsiella pneumoniae isolates. Can J Infect Dis Med Microbiol. 2018 doi: 10.1155/2018/6183162. Article ID 6183162:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moosavian M, Shams N, Sirous M. Detection of carbapenemases emerging in Acinetobacter baumannii clinical isolates by modified Hodge test. J Med Microbiol Infect Dis. 2014;2(4):163–166. [Google Scholar]
- 29.Jonaidi Jafari N, Izadi M, Hajia M, Qorbanalizadgan M, Saburi A. The susceptibility evaluation of multiresistant Gram-negative Bacilli to meropenem and imipenem. Int J Travel Med Global Health. 2014;2(1):1–3. [Google Scholar]
- 30.Ghadiri H, Vaez H, Razavi-Azarkhiavi K, et al. Prevalence and antibiotic susceptibility patterns of extended-spectrum ß-lactamase and metallo-ß-lactamase–producing uropathogenic Escherichia coli isolates. Lab Med Fall. 2014;45:290–294. doi: 10.1309/LMHEP4VQHEY2POOK. [DOI] [PubMed] [Google Scholar]
- 31.Fazeli H, Kamali Dolatabadi R, Taraghian A, Nasr Isfahani B, Moghim S, Norouzi M. Carbapenem resistance pattern of multiple drug-resistant and extended-spectrum beta-lactamase-positive Klebsiella pneumonia in Isfahan. Int J Enteric Pathogens. 2014;2(4):e21495. [Google Scholar]
- 32.Mirsalehian A, Kalantar-Neyestanaki D, Nourijelyani K, et al. Detection of AmpC-β-lactamases producing isolates among carbapenem resistant P. aeruginosa isolated from burn patient. Iran J Microbiol. 2014;6(5):306–310. [PMC free article] [PubMed] [Google Scholar]
- 33.Moayednia R, Shokri D, Mobasherizadeh S, Baradaran A, Fatemi SM, Merrikh A. Frequency assessment of β-lactamase enzymes in Escherichia coli and Klebsiella isolates in patients with urinary tract infection. J Res Med Sci. 2014;19(1):S41–S45. [PMC free article] [PubMed] [Google Scholar]
- 34.Erfani Y, Farahbakhsh M, Godarzi H, Eslami G, Hashemi A. Detection of metallo-B-lactamase (MBL) producing Acinetobacter at 3 hospitals in Iran, Tehran. Res J Biol Sci. 2013;8(4):88–93. [Google Scholar]
- 35.Safari M, Saidijam M, Bahador A, Jafari R, Alikhani MY. High prevalence of multidrug resistance and metallo-beta-lactamase (MβL) producing Acinetobacter baumannii isolated from patients in ICU Wards, Hamadan, Iran. J Res Health Sci. 2013;13(2):162–167. [PubMed] [Google Scholar]
- 36.Masaeli M, Faraji T, Ramazanzadeh R. Risk factors associated with resistance in metalo beta-lactamase producing Enterobacteriaceae isolated from patients in Sanandaj Hospitals. Curr Drug Ther. 2012;7(3):179–183. [Google Scholar]
- 37.Japoni-Nejad A, Sofian M, van Belkum A, Ghaznavi-Rad E. Noso-comial outbreak of extensively and pan drug-resistant Acinetobacter baumannii in Tertiary Hospital in Central Part of Iran. Jundishapur J Microbiol. 2013;6(8):e9892. [Google Scholar]
- 38.Ahangarzadeh Rezaee M, Langarizadeh N, Aghazadeh M. First Report of Class 1 and Class 2 integrons in multidrug-resistant Klebsiella pneu-moniae isolates from Northwest Iran. J Infect Dis. 2012;65:256–259. doi: 10.7883/yoken.65.256. [DOI] [PubMed] [Google Scholar]
- 39.Haji Hashemi B, Farzanehkhah M, Dolatyar A, et al. Study on prevalence of KPC producing from Klebsiella pneumoniae using modified Hodge test and CHROMagar in Iran. Ann Biol Res. 2012;3(12):5659–5664. [Google Scholar]
- 40.Azimi L, Lari AR, Alaghehbandan R, Alinejad F, Mohammadpoor M, Rahbar M. KPC producer gram negative bacteria among burned infants in Motahari hospital, Tehran: first report from Iran. Ann Burns Fire Disasters. 2012;25(2):74–77. [PMC free article] [PubMed] [Google Scholar]
- 41.Japoni A, Alborzi A, Kalani M, Nasiri J, Hayati M, Farshad S. Susceptibility patterns and cross-resistance of antibiotics against Pseudomonas aeruginosa isolated from burn patients in the South of Iran. Burns. 2006;32(3):343–347. doi: 10.1016/j.burns.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Taherikalani M, Fatolahzadeh B, Emaneini M, Soroush S, Feizabadi MM. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran hospitals. New Microbiol. 2009;32(3):265–271. [PubMed] [Google Scholar]
- 43.Azizi O, Shakibaie MR, Modarresi F, Shahcheraghi F. Molecular detection of class-D OXA carbapenemase genes in biofilm and non-biofilm forming clinical isolates of Acinetobacter baumannii. Jundishapur J Microbiol. 2015;8(1):e21042. doi: 10.5812/jjm.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahador A, Raoo An R, Farshadzadeh Z, et al. The prevalence of IS Aba 1 and IS Aba 4 in Acinetobacter baumannii species of different international clone lineages among patients with burning in Tehran, Iran. Jundishapur J Microbiol. 2015;8(7):e17167. doi: 10.5812/jjm.17167v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarikhani Z, Nazari R, Nateghi Rostami M. First report of OXA-143-lactamase producing Acinetobacter baumannii in Qom, Iran. Iran J Basic Med Sci. 2017;20(11):1282–1286. doi: 10.22038/IJBMS.2017.9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohammadi M, Soroush S, Delfani S, et al. Distribution of class D carbapenemase and extended-spectrum β-lactamase genes among Acinetobacter baumannii isolated from burn wound and ventilator associated pneumonia infections. J Clin Diagn Res. 2017;11(7):DC19–DC23. doi: 10.7860/JCDR/2017/25534.10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kooti S, Motamedifar M, Sarvari J. Antibiotic resistance profile and distribution of oxacillinase genes among clinical isolates of Acinetobacter baumannii in Shiraz teaching hospitals, 2012-2013. Jundishapur J Microbiol. 2015;8(8):e20215. doi: 10.5812/jjm.20215v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahdian S, Sadeghifard N, Pakzad I, et al. Acinetobacter baumannii clonal lineages I and II harboring different carbapenem-hydrolyzing-β-lactamase genes are widespread among hospitalized burn patients in Tehran. J Infect Public Health. 2015;8(6):533–542. doi: 10.1016/j.jiph.2015.04.030. [DOI] [PubMed] [Google Scholar]
- 49.Farsiani H, Mosavat A, Soleimanpour S, et al. Limited genetic diversity and extensive antimicrobial resistance in clinical isolates of Acinetobacter baumannii in northeast Iran. J Med Microbiol. 2015;64(7):767–773. doi: 10.1099/jmm.0.000090. [DOI] [PubMed] [Google Scholar]
- 50.Nasrolahei M, Zahedi B, Bahador A, et al. Distribution of bla OXA-23, IS Aba, aminoglycosides resistant genes among burned & ICU patients in Tehran and Sari, Iran. Ann Clin Microbiol Antimicrob. 2014;13(1):38. doi: 10.1186/s12941-014-0038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Safari M, Alikhani MY, Arabestani MR, Kamali Kakhki R, Jafari R. Prevalence of metallo-β-lactamases encoding genes among Pseudomonas aeruginosa strains isolated from the bedridden patients in the intensive care units. Avicenna J Clin Microbiol Infect. 2014;1(1):e19216. [Google Scholar]
- 52.Bahador A, Raoofian R, Taheri M, Pourakbari B, Hashemizadeh Z, Hashemi FB. Multidrug resistance among Acinetobacter baumannii isolates from Iran: changes in antimicrobial susceptibility patterns and genotypic profile. Microb Drug Resist. 2014;20(6):632–640. doi: 10.1089/mdr.2013.0146. [DOI] [PubMed] [Google Scholar]
- 53.Karmostaji A, Najar Peerayeh S, Hatef Salmanian A. Distribution of OXA-type class D β-Lactamase genes among nosocomial multi drug resistant Acinetobacter baumannii isolated in Tehran hospitals. Jundishapur J Microbiol. 2013;6(5):e8219. [Google Scholar]
- 54.Shoja S, Moosavian M, Peymani A, Tabatabaiefar MA, Rostami S, Ebrahimi N. Genotyping of carbapenem resistant Acinetobacter baumannii isolated from tracheal tube discharge of hospitalized patients in intensive care units, Ahvaz, Iran. Iran J Microbiol. 2013;5(4):315–322. [PMC free article] [PubMed] [Google Scholar]
- 55.Sohrabi N, Farajnia S, Akhi MT, et al. Prevalence of OXA-type β-lactamases among Acinetobacter baumannii isolates from Northwest of Iran. Microb Drug Resist. 2012;18(4):385–389. doi: 10.1089/mdr.2011.0077. [DOI] [PubMed] [Google Scholar]
- 56.Sepehriseresht S, Boroumand MA, Pourgholi L, Sotoudeh Anvari M, Habibi E, Sattarzadeh Tabrizi M. Detection of vim- and ipm-type metallo-beta-lactamases in Pseudomonas aeruginosa clinical isolates. Arch Iran Med. 2012;15(11):670–673. [PubMed] [Google Scholar]
- 57.Forozsh Fard M, Irajian G, Takantape Moslehi Z, Fazeli H, Salehi M, Rezania S. Drug resistance pattern of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients at Isfahan AL Zahra hospital, Iran (2009–2010) Iran J Microbiol. 2012;4(2):94–97. [PMC free article] [PubMed] [Google Scholar]
- 58.Peymani A, Higgins PG, Nahaei M-R, Farajnia S, Seifert H. Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, northwest Iran. Int J Antimicrob Agents. 2012;39(6):526–528. doi: 10.1016/j.ijantimicag.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 59.Asadollahi P, Akbari M, Soroush S, et al. Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns. 2012;38(8):1198–1203. doi: 10.1016/j.burns.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 60.Feizabadi MM, Fathollahzadeh B, Taherikalani M, et al. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61(4):274–278. [PubMed] [Google Scholar]
- 61.Taherikalani M, Etemadi G, Geliani KN, Fatollahzadeh B, Soroush S, Feizabadi MM. Emergence of multi and pan-drug resistance Acinetobacter baumannii carrying blaOXA-type -carbapenemase genes among burn patients in Tehran, Iran. Saudi Med J. 2008;29(4):623–624. [PubMed] [Google Scholar]
- 62.Hosseinzadeh Z, Sedigh Ebrahim-Saraie H, Sarvari J, et al. Emerge of blaNDM-1 and blaOXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J Chin Med Assoc. 2018;81(6):536–540. doi: 10.1016/j.jcma.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 63.Bina M, Pournajaf A, Mirkalantari S, Talebi M, Irajian G. Detection of the Klebsiella pneumoniae carbapenemase (KPC) in K. pneumoniae isolated from the clinical samples by the phenotypic and genotypic methods. Iran J Pathol. 2015;10(3):199–205. [PMC free article] [PubMed] [Google Scholar]
- 64.Lari AR, Azimi L, Soroush S, Taherikalani M. Low prevalence of metallo-beta-lactamase in Pseudomonas aeruginosa isolated from a tertiary burn care center in Tehran. Int J Immunopathol Pharmacol. 2015;28(3):384–389. doi: 10.1177/0394632015578343. [DOI] [PubMed] [Google Scholar]
- 65.Bagheri Josheghani S, Moniri R, Firoozeh F, Sehat M, Dasteh Goli Y. Susceptibility pattern and distribution of oxacillinases and blaPER-1 genes among multidrug resistant Acinetobacter baumannii in a teaching hospital in Iran. J Pathog. 2015;957259:7. doi: 10.1155/2015/957259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Solgi H, Badmasti F, Aminzadeh Z, et al. Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of bla NDM-7 and bla OXA-48. Eur J Clin Microbiol Infect Dis. 2017;36(11):2127–2135. doi: 10.1007/s10096-017-3035-3. [DOI] [PubMed] [Google Scholar]
- 67.Khorvash F, Yazdani M, Shabani S, Soudi A. Pseudomonas aeruginosa-producing metallo-β-lactamases (VIM, IMP, SME, and AIM) in the clinical isolates of intensive care units, a university hospital in Isfahan, Iran. Adv Biomed Res. 2017;30(6):147. doi: 10.4103/2277-9175.219412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akhi MT, Khalili Y, Ghotaslou R, et al. Carbapenem inactivation: a very affordable and highly specific method for phenotypic detection of carbapenemase-producing Pseudomonas aeruginosa isolates compared with other methods. J Chemother. 2017;29(3):144–149. doi: 10.1080/1120009X.2016.1199506. [DOI] [PubMed] [Google Scholar]
- 69.Falahat S, Shojapour M, Sadeghi A. Detection of KPC carbapenemase in Pseudomonas aeruginosa isolated from clinical samples using Modified Hodge Test and boronic acid phenotypic methods and their comparison with the polymerase chain reaction. Jundishapur J Microbiol. 2016;9(9):e27249. doi: 10.5812/jjm.27249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohammadzadeh M, Tavakoli M, Mohebi A, Aghayi S. Phenotypic and genotypic detection of metallo-beta-lactamases among imipenem resistant gram negative isolates. J Med Bacteriol. 2016;5(1–2):36–42. [Google Scholar]
- 71.Tarashi S, Goudarzi H, Erfanimanesh S, Pormohammad A, Hashemi A. Phenotypic and molecular detection of metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Aci-netobacter baumannii strains isolated from patients with burn injuries. Arch Clin Infect Dis. 2016;11(4):e39036. [Google Scholar]
- 72.Maspi H, Mahmoodzadeh Hosseini H, Amin M, Imani Fooladi AA. High prevalence of extensively drug-resistant and metallo beta-lactamase-producing clinical Acinetobacter baumannii in Iran. Microb Pathog. 2016;98:155–159. doi: 10.1016/j.micpath.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 73.Moghadam M, Motamedifar M, Sarvari J, Sedigh E-SH, Mousavi SM, Moghadam F. Emergence of multidrug resistance and metallo-beta-lactamase producing Acinetobacter baumannii isolated from patients in Shiraz, Iran. Ann Med Health Sci Res. 2016;6(3):162–167. doi: 10.4103/2141-9248.183946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pakbaten Toupkanlou S, Najar Peerayeh S, Pirhajati Mahabadi R. Class A and D extended-spectrum β-lactamases in imipenem resistant Pseudomonas aeruginosa isolated from burn patients in Iran. Jundishapur J Microbiol. 2015;8(8):e18352. doi: 10.5812/jjm.18352v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eftekhar F, Naseh Z. Extended-spectrum β-lactamase and carbapenemase production among burn and non-burn clinical isolates of Klebsiella pneumoniae. Iran J Microbiol. 2015;7(3):144–149. [PMC free article] [PubMed] [Google Scholar]
- 76.Azimi L, Talebi M, Owlia P, et al. Tracing of false negative results in phenotypic methods for identification of carbapenemase by real-time PCR. Gene. 2016;576(1 Pt 1):166–170. doi: 10.1016/j.gene.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 77.Azimi L, Talebi M, Pourshafie M-R, Owlia P, Rastegar Lari A. Characterization of carbapenemases in extensively drug resistance Acinetobacter baumannii in a burn care center in Iran. Int J Mol Cell Med. 2015;4(1):46–53. [PMC free article] [PubMed] [Google Scholar]
- 78.Fazeli H, Norouzi-Barough M, Ahadi AM, Shokri D, Solgi H. Detection of New Delhi Metallo-Beta-Lactamase-1 (NDM-1) in carbapenem-resistant Klebsiella pneumoniae isolated from a university hospital in Iran. Hippokratia. 2015;19(3):205–209. [PMC free article] [PubMed] [Google Scholar]
- 79.Khoshvaght H, Haghi F, Zeighami H. Extended spectrum betalacta-mase producing enteroaggregative Escherichia coli from young children in Iran. Gastroenterol Hepatol Bed Bench. 2014;7(2):131–136. [PMC free article] [PubMed] [Google Scholar]
- 80.Noori M, Karimi A, Fallah F, et al. High prevalence of metallo-beta-lactamase producing Acinetobacter baumannii isolated from two hospitals of Tehran, Iran. Arch Pediatr Infect Dis. 2014;2(1):e15439. [Google Scholar]
- 81.Vali P, Shahcheraghi F, Seyfipour M, Zamani MA, Allahyar MR, Feiz-abadi MM. Phenotypic and genetic characterization of carbapenemase and ESBLs producing gram-negative bacteria (GNB) isolated from patients with cystic fibrosis (CF) in Tehran hospitals. J Clin Diag Res. 2014;8(1):26–30. doi: 10.7860/JCDR/2014/6877.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Azimi L, Nordmann P, Lari AR, Bonnin RA. First report of OXA-48-producing Klebsiella pneumoniae strains in Iran. GMS Hyg Infect Control. 2014;7:9. doi: 10.3205/dgkh000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hashemi A, Fallah F, Erfanimanesh S, Hamedani P, Alimehr S, Goudarzi H. Detection of β-lactamases and outer membrane porins among Klebsiella pneumoniae strains isolated in Iran. Scientifica. 2014;726179:6. doi: 10.1155/2014/726179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Japoni-Nejad A, Ghaznavi-Rad E, van Belkum A. Characterization of plasmid-mediated AmpC and carbapenemases among Iranian nosocomial isolates of Klebsiella pneumoniae using phenotyping and genotyping methods. Osong Public Health Res Perspect. 2014;5(6):333–338. doi: 10.1016/j.phrp.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farajzadeh Sheikh A, Rostami S, Jolodar A, et al. Detection of metallo-beta lactamases among carbapenem-resistant Pseudomonas aeruginosa. Jundishapur J Microbiol. 2014;7(8):e12289. doi: 10.5812/jjm.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aghamiri S, Amirmozafari N, Fallah Mehrabadi J, Fouladtan B, Samadi Kafil H. Antibiotic resistance pattern and evaluation of metallo-beta lactamase genes including bla- IMP and bla- VIM types in Pseudomonas aeruginosa isolated from patients in Tehran Hospitals. ISRN Microbiol. 2014;2014(1):1–6. doi: 10.1155/2014/941507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nobari S, Shahcheraghi F, Rahmati Ghezelgeh F, Valizadeh B. Molecular characterization of carbapenem-resistant strains of Klebsiella pneumoniae isolated from Iranian patients: first identification of blaKPC gene in Iran. Microb Drug Resist. 2014;20(4):285–293. doi: 10.1089/mdr.2013.0074. [DOI] [PubMed] [Google Scholar]
- 88.Hakemi Vala M, Hallajzadeh M, Hashemi A, et al. Detection of Ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burn Fire Disast. 2014;27(1):8–13. [PMC free article] [PubMed] [Google Scholar]
- 89.Tavajjohi Z, Moniri R, Zarrabi M. Detection of GES-2, a class A β-lactamase produced by Pseudomonas aeruginosa in a teaching hospital in Iran. Jundishapur J Microbiol. 2013;6(10):e8166. [Google Scholar]
- 90.Azimi L, Lari AR, Talebi M, Ebrahimzadeh Namvar A, Soleymanza-deh-Moghadam S. Evaluation of phenotypic methods for detection of Klebsiella Pneumoniae carbapenemase-producing K. Pneumoniae in Tehran. J Med Bacteriol. 2013;2(3–4):26–31. [Google Scholar]
- 91.Noori N, Vandyosefi J, Sabet F, Ashrafi S, Ghazvini K. Frequency of IMP-1 and VIM genes among metallo-beta-lactamase producing Acinetobacter spp. isolated from health care associated infections in Northeast of Iran. J Med Bacteriol. 2013;2(3–4):11–16. [Google Scholar]
- 92.Azimi L, Lari AR, Talebi M, Namvar AE, Jabbari M. Comparison between phenotypic and PCR for detection of OXA-23 type and metallo-beta-lactamases producer Acinetobacter spp. GMS Hyg Infect Control. 2013;8(2):Doc16. doi: 10.3205/dgkh000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pajand O, Rezaee MA, Nahaei MR, et al. Study of the carbape-nem resistance mechanisms in clinical isolates of Acinetobacter baumannii: comparison of burn and non-burn strains. Burns. 2013;39(7):1414–1419. doi: 10.1016/j.burns.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 94.Azami S, Abdi Ali A, Asgarani E. Association between metallo-β-lactamases and integrons with multi-drug resistance in Pseudomonas aeruginosa isolates. J Med Microbiol Infect Dis. 2013;1(1):46–51. [Google Scholar]
- 95.Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: a study in western Iran. Iran J Microbiol. 2013;5(3):195–202. [PMC free article] [PubMed] [Google Scholar]
- 96.Doosti M, Ramazani A, Garshasbi M. Identification and characterization of metallo-β-lactamases producing Pseudomonas aeruginosa clinical isolates in University Hospital from Zanjan Province, Iran. Iran Biomed J. 2013;17(3):129–133. doi: 10.6091/ibj.1107.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, et al. First report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb Drug Resist. 2013;19(1):30–36. doi: 10.1089/mdr.2012.0078. [DOI] [PubMed] [Google Scholar]
- 98.Kalantar E, Torabi V, Salimizand H, Soheili F, Beiranvand S, Soltan Dallal MM. First survey of metallo-β-lactamase producers in clinical isolates of Pseudomonas aeruginosa from a Referral Burn Center in Kurdistan Province. Jundishapur J Nat Pharm Prod. 2012;7(1):23–26. doi: 10.17795/jjnpp-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-β-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3(2):68–74. [PMC free article] [PubMed] [Google Scholar]
- 100.Peymani A, Nahaei MR, Farajnia S, et al. High prevalence of metallo-beta-lactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn J Infect Dis. 2011;64(1):69–71. [PubMed] [Google Scholar]
- 101.Saderi H, Lotfalipour H, Owlia P, Salimi H. Detection of metallo-β-lactamase producing Pseudomonas aeruginosa isolated from burn patients in Tehran, Iran. Lab Med. 2010;41(10):609–612. [Google Scholar]
- 102.Yousefi S, Farajnia S, Nahaei MR, et al. Detection of metallo-β-lactamase-encoding genes among clinical isolates of Pseudomonas aeruginosa in northwest of Iran. Diagn Microbiol Infect Dis. 2010;68(3):322–325. doi: 10.1016/j.diagmicrobio.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 103.Bahar MA, Jamali S, Samadikuchaksaraei A. Imipenem-resistant Pseudomonas aeruginosa strains carry metallo-beta-lactamase gene bla(VIM) in a level I Iranian burn hospital. Burns. 2010;36(6):826–830. doi: 10.1016/j.burns.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 104.Khosravi AD, Mihani F. Detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn Microbiol Infect Dis. 2008;60(1):125–128. doi: 10.1016/j.diagmicrobio.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 105.Yazdi HR, Nejad GB, Peerayeh SN, Mostafaei M. Prevalence and detection of metallo-β-lactamase (MBL)-producing Pseudomonas aeruginosa strains from clinical isolates in Iran. Ann Microbiol. 2007;57(2):293–295. [Google Scholar]
- 106.Al Johani SM, Akhter J, Balkhy H, El-Saed A, Younan M, Memish Z. Prevalence of antimicrobial resistance among gram-negative isolates in an adult intensive care unit at a tertiary care center in Saudi Arabia. Ann Saudi Med. 2010;30(5):364–369. doi: 10.4103/0256-4947.67073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kempf M, Rolain JM. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39(2):105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 108.Cantón R, Akóva M, European Network on Carbapenemases et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 109.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 110.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: CLSI; 2018. pp. M100–S17. [Google Scholar]
- 111.Kuchibiro T, Komatsu M, Yamasaki K, et al. Evaluation of the modified carbapenem inactivation method for the detection of carbapenemase-producing Enterobacteriaceae. J Infect Chemother. 2018;24(4):262–266. doi: 10.1016/j.jiac.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 112.Pirbonyeh N, Zardosht M, Emami A, Rostampour S, Moattari A, Keshavarzi A. Emergence of storm resistant mechanisms in Pseudomonas aeruginosa isolated from burn patients hospitalized in Ghotbeddin Shirazi Burn Hospital Nova. J Med Biol Sci. 2016;5(1) [Google Scholar]
- 113.Karimi A, Haerizadeh M, Soleymani F, Haerizadeh M, Taheri F. Evaluation of medicine prescription pattern using World Health Organization prescribing indicators in Iran: a cross-sectional study. J Res Pharm Pract. 2014;3(2):39–45. doi: 10.4103/2279-042X.137058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mousavi S, Zargarzadeh AH. Rational drug use in Iran: a call for action. J Pharm Care. 2014;2(2):47–48. [Google Scholar]
- 115.Sefidani Forough A, Hosseini SR, Jabbari S. Antibiotic utilization evaluation of inpatient and outpatient prescriptions in a rural general hospital in Iran. Int J Basic Clin Pharmacol. 2015;4(3):531–536. [Google Scholar]
- 116.Leite GC, Neto LVP, Gaudereto JJ, et al. Effect of antibiotic combination and comparison of methods for detection of synergism in multi-resistant gram-negative bacteria. J Inf Dis Ther. 2015;3(2):207. [Google Scholar]
- 117.Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Viau R, Frank KM, Jacobs MR, et al. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin Microbiol Rev. 2016;29(1):1–27. doi: 10.1128/CMR.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]