Abstract
Purpose
To determine the molecular basis of lesion development in a murine model of spontaneous retinal vascularization, rnv3 (retinal vascularization 3, aka JR5558).
Methods
Disease progression of rnv3 was examined in longitudinal studies by clinical evaluation, electroretinography (ERG) and light microscopy analyses. The chromosomal position for the recessive rnv3 mutation was determined by DNA pooling and genome-wide linkage analysis. The causative mutation was discovered by comparison of whole exome sequences of rnv3 mutant and wild-type (WT) controls. In order to confirm the causative mutation, transcription activator-like effector nuclease (TALEN)-mediated oligonucleotide directed repair (ODR) was utilized to correct the mutant allele. Phenotypic correction was assessed by fundus imaging and optical coherence tomography of live mice.
Results
rnv3 exhibits early-onset, multifocal depigmented retinal lesions observable by fundus examination starting at 18 days of age. The retinal lesions are associated with fluorescein leakage around 25 days of age, with peak leakage at about 4 weeks of age. ERG responses deteriorate as rnv3 mutants age, concomitant with progressive photoreceptor disruption and loss that is observable by histology. Genetic analysis localized rnv3 to mouse chromosome (Chr) 1. By high throughput sequencing of a whole exome capture library of a rnv3/rnv3 mutant and subsequent sequence analysis, a single base deletion (del) in the Crb1 [crumbs family member 1] gene, which was previously reported to cause retinal degeneration 8, was identified. The TALEN-mediated ODR rescued the posterior segment vascularization phenotype; heterozygous Crb1rd8+em1Boc/Crb1rd8 and homozygous Crb1rd8+em1Boc/Crb1rd8+em1Boc mice showed a normal retinal phenotype. Additionally, six novel disruptions of Crb1 that were generated through aberrant non-homologous end joining induced by TALEN exhibited variable levels of vascularization, suggesting allelic effects.
Conclusions
The rnv3 model and the models of six novel disruptions of Crb1 are all reliable, novel mouse models for the study of both early and late events associated with posterior segment vascularization and can also be used to test the effects of pharmacological targets for treating human ocular vascular disorders. Further study of these models may provide a greater understanding about how different Crb1 alleles result in aberrant angiogenesis.
Keywords: retinal angiogenesis, retinal vascularization (RNV), spontaneous mutation
Aberrant ocular blood vessel formation can lead to serious complications, causing significant vision impairment and blindness.1 Abnormal vessels are a prominent feature of eye diseases such as diabetic retinopathy, age-related macular degeneration, proliferative retinopathies, and corneal neovascularization, as well as other diseases, such as cancers, cardiovascular diseases, and neurodegenerative diseases.2–4 Development of reproducible and reliable animal models to study these vascular diseases has advanced our understanding of both the normal development and the pathophysiology of abnormal ocular blood vessel growth.5–8 For example, we discovered the first retinal vascularization (RNV) phenotype in Vldlr-targeted null mutant mice and characterized this strain as a reproducible model of subretinal vascularization and choroidal anastomosis.5 This model, made available through the JAX Eye Mutant Resource, has enabled numerous studies investigating the potential underlying mechanisms and treatments for wet AMD, RAP, and macular telangiectasia.9–14 The second reproducible model of retinal vascularization was discovered in the figure eight (fgt) model, where a mutation of the adaptor protein complex AP-1, gamma 1 subunit (Ap1g1) gene6 was identified. Ap1g1fgt mutant mice develop early vascular lesions in the retina and provide a reliable animal model for studying factors that initiate retinal angiogenesis, subretinal vascularization, and choroidal vascular anastomosis formation.6 The third retinal vascular lesion model was identified in a B6;129 mixed background strain, which we named retinal vascularization 3 (rnv3, aka JR55587 or Nrv28).
Determining the molecular basis for the vascular lesions provides useful information about the cell types involved and a better understanding of the nature of the disease and potential interacting factors. Here we report by genetic mapping and whole-exome sequence mutational analysis that the homozygous Crb1rd8, allele is necessary for the retinal vascular disease phenotype in rnv3 mice. We confirmed the disease-causing mutation by correction of the mutant Crb1rd8 allele via TALEN-mediated ODR.
Materials and Methods
Mice Husbandry
The mice in this study were bred and maintained in standardized conditions of the production and research animal facilities at The Jackson Laboratory (JAX). They were provided with a NIH31 6% fat chow diet and acidified water, in a pathogen-free vivarium environment with a 14-hour light/10-hour dark cycle. All experiments were approved by the Institutional Animal Care and Use Committee and conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Origins of the Mouse Strains Utilized
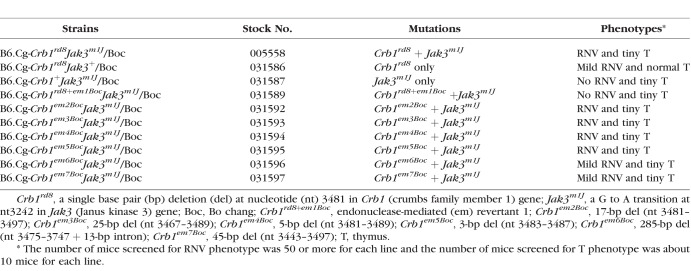
The rnv3 mutant was discovered in the mouse stock B6;129-Crhr1tm1Klee/J (Stock No: 004454), which exhibited fundus retinal depigmentation spots. The depigmented spots corresponded to areas in which abnormal vascularization was observed by histology. To test if the retinal vascularization was due to a heritable mutation, affected mice from this strain were mated to C57BL/6J mice with normal retinas. The resulting F1 progeny, which did not exhibit ocular abnormalities, were intercrossed to generate F2 offspring. A total of 46 F2 mice were examined by indirect ophthalmoscopy for the depigmentation spots and also genotyped for Crhr1tm1Klee. The depigmentation phenotype did not cosegregate with the Crhr1tm1Klee targeted allele, indicating that the phenotype was caused by a new spontaneous mutation, which was named rnv3, and not by the Crhr1tm1Klee targeted mutation. A male mouse wild-type (WT) for the Crhr1tm1Klee targeted mutation was selected to backcross to C57BL/6J for four additional generations to develop a new strain, B6.Cg-Crb1rd8Jak3m1J/Boc (stock #: 005558 or JR5558), which carried the rnv3 mutation (same as Crb1rd8) as well as the Jak3m1J mutation (Supplementary Fig. S1). It should be noted that both mutations were detected by high-throughput sequencing of an exome capture library of an rnv3 mutant. To test if the Jak3m1J mutation contributed to the rnv3 phenotype, the rnv3 mice were mated to C57BL/6J to produce F1 progeny, which were subsequently intercrossed. We genotyped 35 F2 mice for both the Crb1rd8 and Jak3m1J mutations. The mice homozygous Crb1rd8 but wild-type (WT) for Jak3m1J were selected to develop a new strain, B6.Cg-Crb1rd8Jak3+/Boc (stock #: 031586). Mice homozygous for the Jak3m1J mutation but wild-type for Crb1rd8 were selected to develop another new strain, B6.Cg-Crb1+Jak3m1J/Boc (stock#: 031587) (Supplementary Fig. S2). Finally, in the TALEN rescue experiment in which we were correcting the Crb1rd8 mutation, seven additional strains with different Crb1 mutations were generated (Table 1).
Table 1.
JR5558-Rnv3 Related Strains Including Mice Generated by TALEN-Mediated NHEJ
Clinical Evaluation and Electroretinography
Eyes of all mice used in the characterization studies and linkage crosses were dilated with 1% atropine ophthalmic drops (Bausch and Lomb Pharmaceuticals, Inc., Tampa, FL, USA) and were evaluated by indirect ophthalmoscopy with a 78-diopter (D) lens. Fundus photographs were taken with a small animal fundus camera (Kowa American Corp., Torrance, CA, USA) using a Volk superfield lens held 2 inches from the eye, as previously described15 or with an in vivo bright field retinal imaging microscope equipped with image-guided OCT capabilities (Micron III; Phoenix Laboratories, Inc., Pleasanton, CA, USA).
For electroretinographic evaluation of mutants, following an overnight dark adaptation, mice were anesthetized with an intraperitoneal injection of xylazine (80 mg/kg) and ketamine (16 mg/kg) in normal saline. Additional anesthetic was given, if akinesia was inadequate. The equipment and protocol used here have been previously described.16 Briefly, dark-adapted, rod-mediated ERGs were recorded with the responses to short-wavelength flashes over 4.0-log units to the maximum intensity by a photopic stimulator. Cone-mediated ERGs were recorded with white flashes after 10 minutes of complete light adaptation. The signals were sampled at 0.8-ms intervals and averaged.
Histologic Analysis
Mice were asphyxiated by carbon dioxide inhalation, and enucleated eyes were fixed overnight in cold methanol/acetic acid solution (3:1, vol/vol). The paraffin-embedded eyes were cut into 6-μm sections, stained with hematoxylin and eosin (H&E), and examined by light microscopy.
Gene Mapping and Sequencing
To determine the chromosomal location of the rnv3 mutation, we mated JR5558 mice to DBA/2J mice. The F1 mice, which did not exhibit retinal abnormalities, were backcrossed to JR5558 mice to produce N2 mice. Tail DNA was isolated as previously reported.17 A genome-wide scan of pooled DNA from 12 affected and 12 unaffected mice was carried out using 48 microsatellite markers.18 The fundus depigmentation phenotype cosegregated with markers on chromosome 1. Subsequently, DNAs of 120 N2 offspring were genotyped using microsatellite markers to develop a fine structure map of the chromosome 1 region. Microsatellite markers D1Mit387, D1Mit103, D1Mit196 were used to genotype individual DNA samples.
The Jak3m1J mutation was discovered during the course of generating the linkage cross for rnv3 mapping. Normally, all JR5558 parental mice were very sick or dying when they reached ∼6 months of age due to pneumocysitis, but interestingly, all F1 parental mice appeared healthy and normal. Necropsies of JR5558 mutants revealed small thymi and spleens relative to wild-type, control B6 mice or F1 parental mice. To determine the chromosomal location of the mutation causing the tiny thymus and small spleen in JR5558, we mated JR5558 mice to DBA/2J mice. The F1 mice were backcrossed to JR5558 mice to produce N2 mice. A genome-wide scan of pooled DNA from 12 affected (tiny thymus and small spleen) and 12 unaffected mice was carried out using 48 microsatellite markers. The tiny thymus and small spleen phenotype cosegregated with markers on chromosome 8. Subsequently, DNAs of 48 N2 offspring were genotyped using microsatellite markers D8Mit4 and D8Mit132 to narrow the chromosome 8 region harboring the mutant gene.
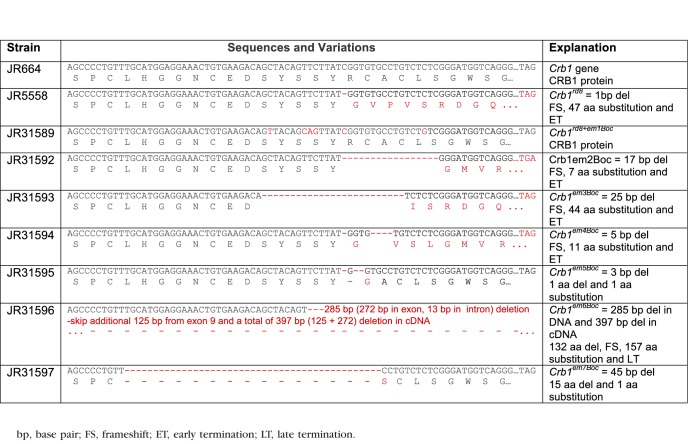
The causative mutations for the fundus depigmentation and for the reduced thymus and spleen phenotypes were discovered by comparing the whole exome sequences from a homozygous JR5558 mutants and control.19 Total RNA was isolated from retinas of newborn mice by TRIZOL LS Reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and a preamplification system (SuperScript; Invitrogen Life Technologies) was used to make first strand cDNA. To confirm the sequence changes in the mutant lines, the tail DNA or retinal cDNA was used to amplify DNA fragments by PCR. The PCR products were purified from agarose gels using a commercial kit (Qiagen, Inc., Valencia, CA, USA) and sequenced (DNA Sequencing Facility, The Jackson Laboratory). Primers used in the study are shown in Table 2.
Table 2.
Primers Used for Sequencing and Genotyping
TALEN-ODR Mediated Correction
The Crb1rd8 allele consists of a 1 base pair (bp) deletion causing a frame shift and protein truncation20 and its TALEN-mediated repair has been described previously.21 In brief, fertilized oocytes isolated from strain JR5558 were coinjected with TALEN mRNAs and single stranded oligonucleotides (ssODN). The TALENs were designed to bind the following sequences (5′-TGAAGACAGCTACAGTTC and 5′-TCTCGGGATGGTCAGGGA). The intervening sequence (5′-TTATGGTGTGCCTGTC) contained the Crb1rd8 mutation and is the site of TALEN-induced DNA cleavage. Oligonucleotide-directed repair was facilitated by the presence of the 200-nt ssODN (5′-TTCTACAAATATGGTACTTACTGGCTGTTTGCCATCAAATGCCTGCCACTCCAGCCCCTGTTTGCATGGAGGAAACTGTGAAGACAGTTACAGCAGTTATCGGTGTGCCTGTCTGTCGGGATGGTCAGGGACACACTGTGAAATCAACATTGATGAGTGCTTTTCTAGCCCCTGTATCCATGGCAACTGCTCTGATGGAG). The corrected sequence encodes wild-type (WT) CRB1 and matches the WT Crb1 nucleotide sequence with five additional synonymous nucleotide alterations to assist in distinguishing the WT versus corrected allele (Table 3). The resulting zygotes were transferred and carried to term in pseudopregnant females. A founder carrying the corrected allele was identified by PCR screening (see below) and used to establish the strain.
Table 3.
The Sequences and Variations in Crb1 Gene and CRB1 Protein in JR5558 and Related Strains
Genotyping Protocols
Genomic DNA for genotyping mice was prepared from tail tips by the rapid, hot sodium hydroxide and Tris (HotSHOT) procedure.17 For PCR amplification, ∼25 ng DNA was used in a 10-μL volume containing 50 mM KCl, 10 mM Tris–Cl, pH 8.3, 2.5 mM MgCl2, 0.2 mM oligonucleotides, 200 μM dNTP, and 0.02 U AmpliTaq DNA polymerase. The reactions were initially denatured for 3 minutes at 94°C, then subjected to 40 cycles of 15 seconds at 94°C, 1-minute at 51°C, 1-minute at 72°C, and then a final 7-minute extension at 72°C. PCR products were separated by electrophoresis on 3% MetaPhor (FMC, Rockland, ME, USA) agarose gels and visualized under UV light after staining with ethidium bromide.
Allele-specific PCR was used to detect the Crb1rd8 mutation.20 The Jak3m1J mutation resulted in a loss of a BsrFI site that enabled us to genotype the Jak3m1J mutation with a polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP) assay. A 166-bp DNA fragment was amplified from mutant and wild-type control DNA using the following pair of primers (F = 5′-TTCAGGAGCTCATGCAGCTG, r = 5′-TCCTGCCTGAGGTCACACAG). BsrFI digestion was performed by adding, 1 μL of ×10 buffer (NE buffer 4) and 0.2 μL of BsrFI (01938, NEB Enzyme; New England Biolabs, Ipswich, MA, USA) to 8.8 μL of each Jak3 amplified PCR solution. After the BsrFI digestion, a single 166-bp band was expected for mice homozygous for the Jak3m1J mutation, whereas two bands of 123 bp and 43 bp were expected in wild-type controls, and all three bands were expected in mice heterozygous for the Jak3m1J mutation. The genotyping protocol for the Crb1rd8 corrected allele (Crb1rd8+em1Boc) was performed by a multiplex PCR amplification protocol.21 The five new Crb1 alleles generated from the TALENs rescue experiment were genotyped by PCR with one pair of primers (F = 5′-TGGTACTTACTGGCTGTTTGC, r = 5′-AATGTTGATTTCACAGTGTGTCC). All mice heterozygous for the new Crb1 alleles showed one unique amplification product and an invariant band of 140 bp, representing the wild-type PCR fragment. For homozygous Crb1em2Boc mutants, a 123 bp band was observed. Crb1em3Boc mutants showed a unique 115 bp band. Crb1em4Boc homozygous mice have a 135 bp band and Crb1em5Boc mice, a 137-bp band. To easily distinguish the mutant band, we used the BanI restriction enzyme (01938, New England Biolabs) to digest the PCR products. Only the Crb1em5Boc DNA could be cut into 90- and 47-bp fragments. Crb1em7Boc mice have a 95-bp band. The Crb1em6Boc mutation was genotyped using the following primers (F = 5′-TGGTACTTACTGGCTGTTTGC, R = 5′-CACAGACTGAGCTCCGAGTG). Using these primers, one 104-bp band is amplified from Crb1em6Boc homozygous mice while heterozygous mice also have an additional 389-bp band.
Results
Retinal Vascularization (RNV) in B6.Cg-Crb1rd8Jak3m1J/Boc (JR#005558, JR5558, or rnv2)
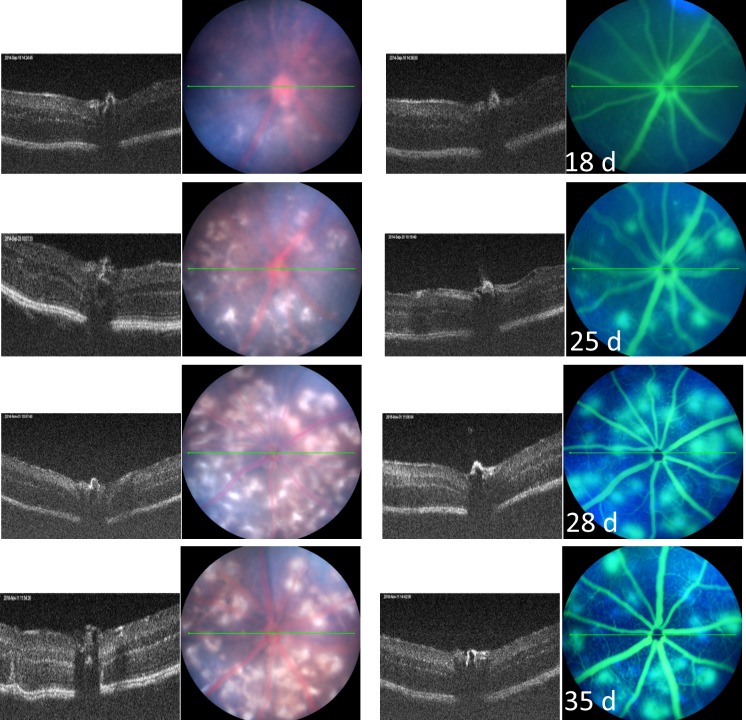
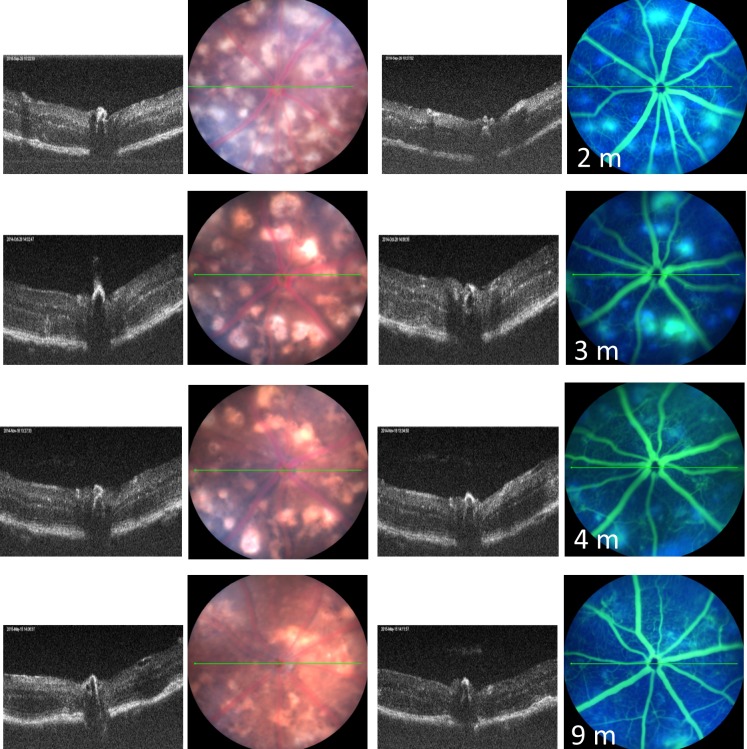
Areas of retinal depigmentation without fluorescein leakage were observed in JR5558 mice around 18 days of age (Fig. 1). At 25 days of age, the number of depigmented spots increased, and some of the depigmented areas corresponded to areas of fluorescein leakage (Fig. 1). The retinal depigmented spots as well as areas of fluorescein leakage increased in number and size with age, with the maximal number of lesions reaching about 20 per eye at about 1 month of age (Fig. 1). Around 2 months of age, some individual retinal depigmented spots appeared to merge, generating larger areas of depigmentation (Fig. 2). The number of areas demonstrating fluorescein leakage decreased with age.
Figure 1.
Representive images of the clinical retinal phenotype of B6.Cg-Crb1rd8Jak3m1J/Boc (stock No. 005558) mice from 18 to 35 days of age using optical coherence tomography and fluorescein angiography. The green lines are the plane in which the B-scan to the left of the fundus images were taken in the same animal.
Figure 2.
Representive images of the clinical retinal phenotype of B6.Cg-Crb1rd8Jak3m1J/Boc mice from 2 to 9 months of age using optical coherence tomography and fluorescein angiography. The green lines are the plane in which the B-scan to the left of the fundus images were taken in the same animal.
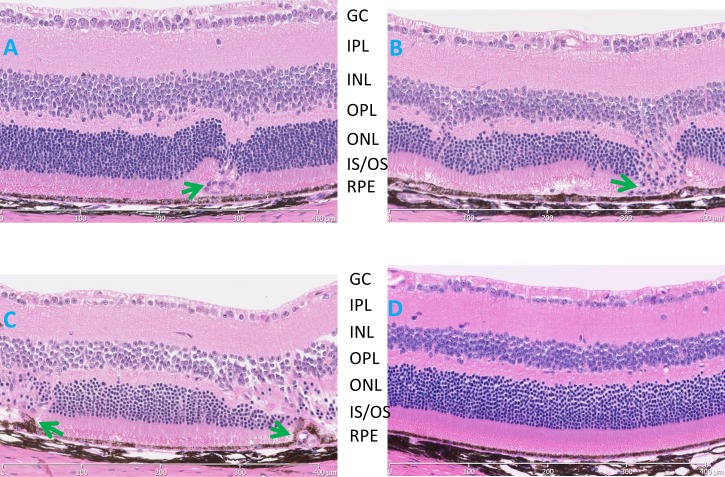
H&E–stained retinal sections of JR5558 mutants showed a subretinal vascularization with a disturbance of the outer retina at 3 weeks of age (Fig. 3A). At 2 and 8 months (Figs. 3B, 3C, respectively) of age, aberrant vessels span the region from the RPE to the inner nuclear layer (INL; Fig. 3D). Photoreceptor cell degeneration was observed where the ectopic vasculature disrupts the retinal structure (Figs. 3B, 3C).
Figure 3.
Histological evaluation of B6.Cg-Crb1rd8Jak3m1J/Boc mice (A–C) demonstrating disease progression at (A) 3 weeks, (B) 2 months, (C) 8 months, and (D) an 8-month-old C57BL/6J control. Aberrant vascular lesions (arrows) are observed as early as 3 weeks, the earliest time examined, and a thinning of the ONL, especially in those areas where the ectopic blood vessels were found. GC, ganglion cell layer; IPL, inner plexiform layer; IS/OS, inner segment/outer segment; ONL, outer nuclear layer; OPL, outer plexiform layer; RPE, retinal pigment epithelium.
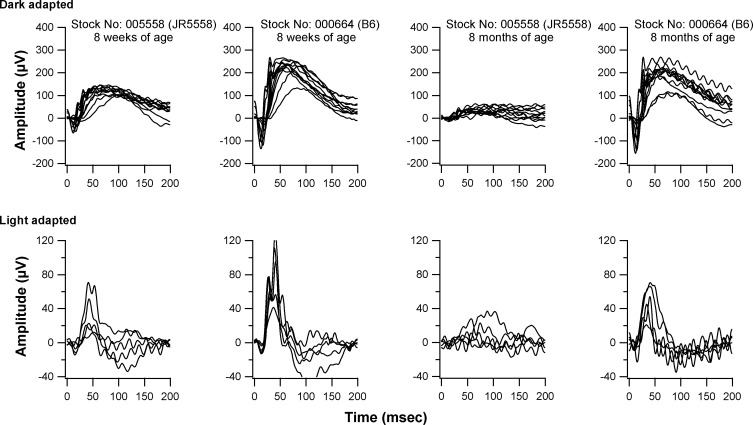
Electroretinograms of homozygous JR5558 mice showed a slight decline in both scotopic and photopic responses to visual stimuli at 8 weeks of age (Fig. 4). By 8 months of age, mutants showed a much-reduced rod and cone ERG response compared to that of C57BL/6J wild-type controls (Fig. 4).
Figure 4.
ERG responses of B6.Cg-Crb1rd8Jak3m1J/Boc and C57BL/6J at 8 weeks and 8 months of age. Scotopic responses were recorded after an overnight dark-adaptation, while light-adapted responses were recorded with a background light of 1.46 log cd/m2 following a 10-minute exposure of the same range of stimulus intensities (see “Methods”). A reduction in scotopic ERG response is observed at 8 weeks in B6.Cg-Crb1rd8Jak3m1J/Boc mutants compared to controls, and in 8 week old compared to 8-month-old B6.Cg-Crb1rd8Jak3m1J/Boc mutants. Both scotopic and photopic responses were reduced in 8 month old mutants compared to 8-week-old mutants.
The Crb1rd8 Mutation Is Associated With the Aberrant Vascular Phenotype in Homozygous rnv3 Mutants
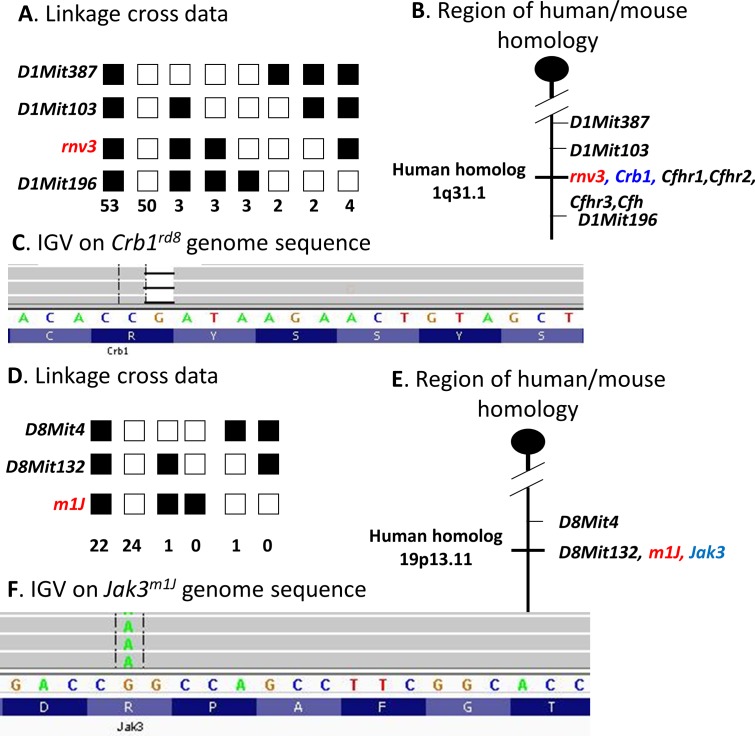
Genetic analysis indicated that the abnormal retinal vascularization in homozygous rnv3 mice was inherited as a single autosomal recessive mutation and that the rnv3 mutation mapped to mouse Chr 1 between microsatellite markers D1Mit103 and D1Mit196 (Fig. 5A). A genetic map of Chr 1 that encompasses the rnv3 locus with closely associated markers, possible candidate genes, as well as the homologous region in humans is shown in Figure 5B.
Figure 5.
Genetic and molecular analysis of the rnv3 strain. (A) A total of 120 progeny from a rnv3/rnv3 X (rnv3/DBA/2J) F1 mice backcross were genotyped. Linkage to several markers on mouse Chr 1 was observed. The columns of squares represent haplotypes (filled boxes, rnv3/rnv3 allele; open boxes, rnv3/DBA/2J allele). The number of chromosomes associated with each haplotype is indicated below each column. (B) Genetic map of Chr 1 in the rnv3 region showing the closest markers and the region of human homology. (C) Integrative genomics viewer (IGV) shows the deleted base. (D) A total of 48 progeny from a m1J/m1J x (m1J/DBA/2J) F1 mice backcross were genotyped. Linkage to several markers on mouse Chr 8 was observed. The columns of squares represent haplotypes (filled boxes, m1J3/m1J allele; open boxes, m1J/DBA/2J allele). The number of chromosomes associated with each haplotype is indicated below each column. (E) Genetic map of Chr 8 in the m1J region showing the closest markers and the region of human homology. (F) IGV shows the base substitution.
A whole mouse exome capture library prepared from DNA from a homozygous rnv3 mouse was sequenced. The only sequence change with a high mismatch ratio found within the critical interval of the disease locus was a single nucleotide deletion in exon 9 of the Crb1 gene, a previously reported causative mutation for retinal degeneration 8 (rd8).20 This nucleotide change is predicted to create a frameshift and premature stop codon in the Crb1 transcript in rnv3 mice (Fig. 5C). Although not within the critical region of rnv3, genetic variations in Vldlr and Ap1g1, genes previously associated with retinal vascular abnormalities were also examined to exclude their potential role in the rnv3 phenotype. Although we cannot exclude noncoding regulatory changes, there was no coding region variation found in either gene in rnv3 mutants.
A Second Mutation, in the Jak3 Gene, Is Identified in JR5558 Mutants
Genetic analysis indicated that the small thymus and spleen phenotype observed in homozygous JR5558 mice, named mutation 1 Jackson (m1J), was inherited as a single autosomal recessive mutation. The m1J mutation was mapped to mouse Chr 8, closely linked to microsatellite markers D8Mit132 and D8Mit4 (Fig. 5D). The genetic map of Chr 8 encompassing the m1J locus, the closest markers, possible candidate genes, as well as the homologous region in humans is depicted in Figure 5E.
A whole mouse exome capture library was prepared from DNA from a JR5558 mouse and subjected to high-throughput sequencing. Within the critical region in which the disease locus resided, two sequence changes in the Jak3 gene with high mismatch ratios were observed. The first nucleotide substitution is unique and only present in JR5558 mice in exon 24 of Jak3 at position 3242. A G to A transition creates a missense mutation, which changes codon 1081 from CGG to CAG, which is predicted to alter the encoded amino acid from arginine to glutamine in JR5558 mice (Fig. 5F). A second nucleotide substitution was identified in exon 23 of Jak3 at position 3095. An A to C transversion changes codon 1032 from AGC to CGC or an amino acid change from serine to arginine. This nucleotide substitution was detected by Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/, available in the public domain) in 129/J strain (U70201), Mus caroli (XM_021169344.1) and Mus musculus (BC131646, BC131647, BC105577, L32995, and L40172-BALB/c, L33768). By checking the sequence variations from Mouse Genome Project (http://www.sanger.ac.uk/science/data/mouse-genomes-project, provided in the public domain by Wellcome Sanger Institute, Cambridgeshire, UK), this nucleotide substitution was present in 27 strains (129P2/OlaHsd, 129S1/SvImJ (Stock No: 002448), 129S5SvEvBrd, AKR/J (Stock No: 000648), BALB/cJ (Stock No: 000651), BUB/BnJ (Stock No: 000653), C3H/HeH, C3H/HeJ (Stock No: 000659), CAST/EiJ (Stock No: 000928), CBA/J (Stock No: 000656), DBA/1J (Stock No: 000670), DBA/2J (Stock No: 000671), FVB/NJ (Stock No: 001800), I/LnJ (Stock No: 000674), KK/HiJ (Stock No: 002106), LEWES/EiJ (Stock No: 002798), LP/J (Stock No: 000676), MOLF/EiJ (Stock No: 000550), NZB/B1NJ (Stock No: 000684), NZO/HlLtJ (Stock No: 002105), NZW/LacJ (Stock No: 001058), RF/J (Stock No: 000682), SEA/GnJ (Stock No: 000644), SPRET/EiJ (Stock No: 001146), ST/bJ (Stock No: 000688), WSB/EiJ (Stock No: 001145) and ZALENDE/EiJ (Stock No: 001392) , but not in the 10 strains (A/J (Stock No: 000646), BTBR (Stock No: 002282), C57BL/10J (Stock No: 000665), C57BL/6J (Stock No: 000664), C57BL/6NJ (Stock No: 005304), C57BR/cdJ (Stock No: 000667), C57L/J (Stock No: 000668), C58/J (Stock No: 000669), NOD/ShiLtJ (Stock No: 001976) and PWK/PhJ (Stock No: 003715).
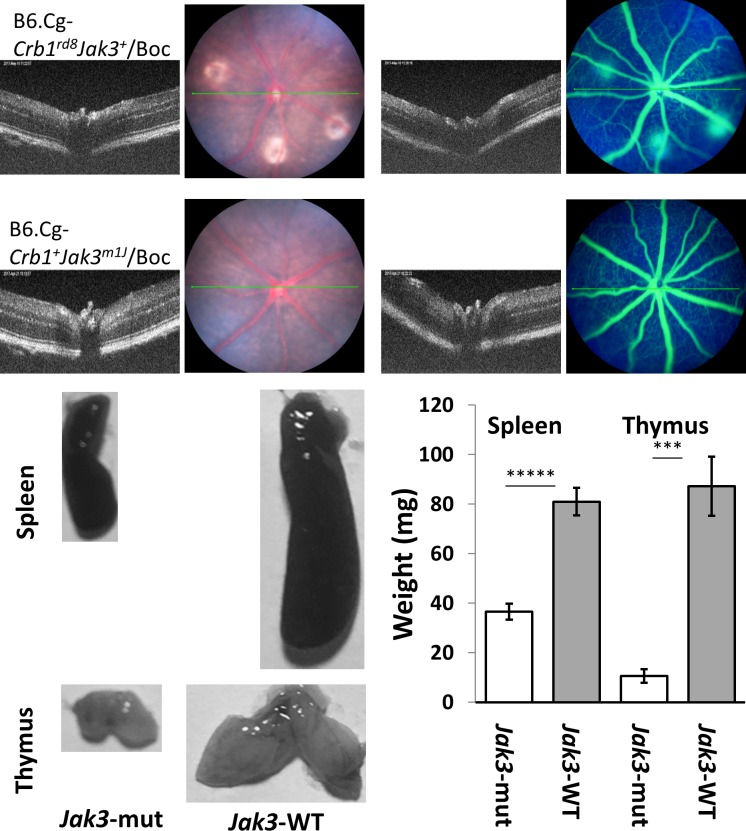
To test if the Jak3m1J mutation contributed to the rnv3 phenotype, rnv3 mice were mated to C57BL/6J to produce F1 mice. A total of 35 intercrossed (F2) mice were genotyped for the Crb1rd8 and Jak3m1J mutations. Mice homozygous for Crb1rd8 but wild-type for Jak3m1J were selected to develop a new strain, B6.Cg-Crb1rd8Jak3+/Boc (stock #: 031586), which had a mild rnv3 phenotype, and normal sized thymus and spleen (Fig. 6). The mice homozygous for the Jak3m1J mutation, but wild-type for Crb1rd8 were selected to develop another strain, B6.Cg-Crb1+Jak3m1J/Boc (stock#: 031587). This strain had a normal retina, but small thymus and spleen (Fig. 6). Taken together, these results suggest that Jak3m1J may have a modifying effect, enhancing rnv3 disease severity, but that it in itself does not cause a retinal phenotype.
Figure 6.
Representive clinical retinal images and representive thymus and spleen images as well as weights of the thymus and spleen in B6.Cg-Crb1rd8Jak3+/Boc (stock number: 031586) and B6.Cg-Crb1+Jak3m1J/Boc (stock number: 031587) mice. The top panel shows the mild rnv3 phenotype in B6.Cg-Crb1rd8Jak3+/Boc, that is not observed in B6.Cg-Crb1+Jak3m1J/Boc mice at 1 month of age. The bottom panel shows the normal size and weights of the spleen and thymus from B6.Cg-Crb1rd8Jak3+/Boc (right-WT) mice, and much smaller spleen and thymus of B6.Cg-Crb1+Jak3m1J/Boc (left-mut) mice at 3 weeks of age. The spleen weight (white bar, mut [mutant] mice = 36.6 ± 3.21 mg and gray bar, WT [wild-type] mice = 81 ± 5.61 mg; *****statistically significant at P < 0.000005) and thymus weight (white bar, mut mice = 10.6 ± 2.7 mg and gray bar, WT mice = 87.2 ± 11.95 mg; ****statistically significant at P < 0.0002) at 3 weeks of age.
Abnormal Retinal Vascular Phenotype Ameliorated in Crb1rd8 TALEN-Mediated ODR Strain
Although occasional angiogenic events were noted in Crb1rd8 mice,22 the significant ectopic retinal vascularization observed in rnv3 mutants was not. Therefore, a rescue experiment was conducted in which a corrected allele of Crb1 was introduced by a TALEN-mediated oligonucleotide homology directed repair (ODR) approach to confirm that Crb1rd8 contributed to the phenotype. We generated 42 founder mice [1 to 25 (female), 26 to 42 (male)]. PCR followed by sequencing revealed that 28 mice harbored a nonhomologous end-joining (NHEJ) indel and/or presence of a ODR-corrected allele, and 14 did not. The overall NHEJ efficiency of TALEN in this experiment was 76%. One male mouse (#26) appeared to carry the corrected allele, one female (#24) carried a large deletion and the remaining 26 founders were mosaic. Male #26 was mated to JR5558 to produce N1 mice. From a total of 23 N1 mice (13 female and 10 male), there were 2 female mice carrying the corrected Crb1 allele (named Crb1rd8+em1Boc) (Fig. 7). From these two female mice, we have developed a new strain named B6.Cg-Crb1rd8+em1BocJak3m1J/Boc (Tables 1, 3). The TALEN-mediated ODR used directly in the rnv3/rnv3 genetic background was able to ameliorate the retinal vascular lesions. This suggests that the Crb1rd8 mutation is necessary for the development of the abnormal vascular phenotype in rnv3 mutants.
Figure 7.
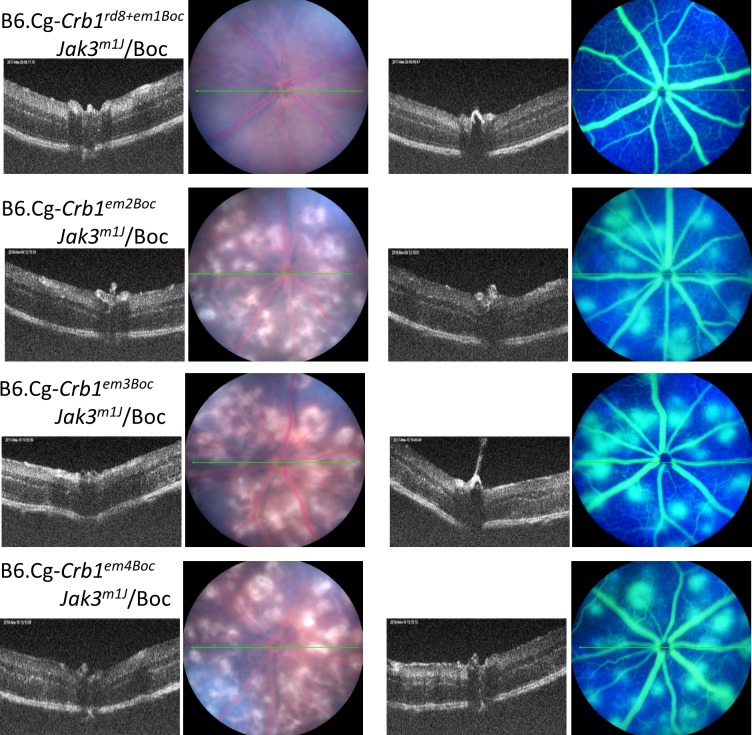
Representative fundus images of retinal phenotypes of strains generated by the TALEN-mediated ODR at 1 month of age.
Generation of Novel Nonhomologous End-Joining Indel Models Associated With the Crb1 Locus
To determine if there were allelic effects of the different Crb1 variants obtained through the aberrant NHEJ events (i.e., more extensive deletions, we established 6 additional lines; Tables 1, 3). Initially, we mated two female mosaic founders (#10 and #20) to male JR5558 to produce a total of 14 N1 mice. We established four strains from this outcross bearing different deletions in the Crb1 gene from these N1 progeny. The Crb1em2Boc strain bears a 17-bp deletion in exon 9, which creates a frameshift and is predicted to result in a 7-amino acid (aa) substitution and early termination. The Crb1em3Boc strain has a 25-bp deletion in exon 9, which creates a frameshift and likely results in a 44-aa substitution and early termination. The Crb1em4Boc strain has a 5-bp deletion in exon 9, which creates a frameshift and is likely to result in an 11-aa substitution and early termination. These three Crb1 variants, which are predicted to be null mutants, developed an abnormal retinal vascular phenotype that was similar to that observed in homozygous rnv3 mutant, which is also a null mutant. Unlike the first three Crb1 variants, the fourth strain, Crb1em5Boc has a discontiguous 3-bp deletion in exon 9, which is predicted to lead to an in-frame substitution (1081 R to G) followed by a deletion of a single amino acid (1082 C). Interestingly, the retinal vascular phenotype appears to be even more severe than observed in the rnv3 mutant, indicating an allelic effect (Fig. 7).
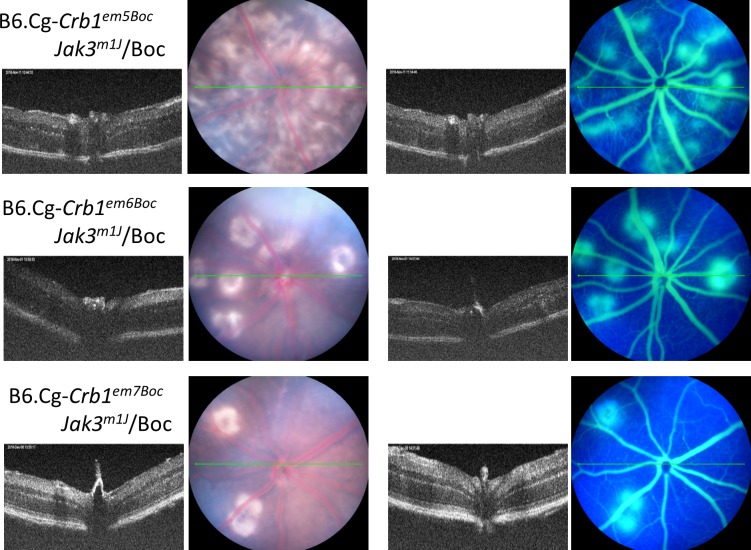
Two additional strains in which large deletions that were predicted to still generate partial CRB1 proteins were established from a founder female mouse #24, which was mated to JR5558 to produce a total of 16 N1 mice. Sequencing of genomic DNA from Crb1em6Boc revealed a 285-bp deletion, which included 272 bp of exon 9 and 13 bp from intron 9 (crossing an exon/intron boundary). By sequencing retinal cDNA from homozygous Crb1em6Boc mice, we found an additional 125 bp was skipped in exon 9, thus, 397 bp was deleted in the 3′ end of exon 9 from the transcript. This mutation is predicted to cause a 132-aa deletion and frameshift which results in a 157-aa substitution with 12 additional amino acids due to a late termination. This mutant strain exhibits a much reduced retinal neovascular phenotype (Fig. 8) compared to the rnv3 mutant. Genomic sequencing of Crb1em7Boc revealed a 45-bp deletion in exon 9, which leads to a restoration of the reading frame. Sequencing of retinal cDNA from Crb1em7Boc indicates a 15-aa deletion followed by a single aa substitution. This mutation also has a mild retinal neovascular phenotype (Fig. 8) compared to the rnv3 mutant.
Figure 8.
Representative fundus images of retinal phenotypes of strains generated by the TALEN-mediated ODR at 1 month of age.
Three of the six lines identified with nonhomologous end joining (NHEJ) without ODR of Crb1 that caused frameshifts and early termination of the gene all had a similar retinal vascular phenotype as the parental rnv3/rnv3 line, suggesting the absence of Crb1 unmasks the angiogenic effect in the rnv3 strain. Consistent with this interpretation, two lines with in-frame deletions where partial CRB1 protein is anticipated, a 70% reduction in vascular lesions was observed. The in-frame deletions occurred in the extracellular domain of the Crb1 gene. The one exception to the allelic effects observed is in the Crb1em5Boc strain, which shows a severe retinal vascular phenotype despite the prediction that a CRB1 protein would be made. It should be noted, however, that initially—until N1F5, this line exhibited a 70% reduction in retinal vascularization as well, but with further inbreeding the phenotype became significantly more severe than even that observed in the rnv3 strain, suggesting that other factors in the genetic background that modified the phenotype became fixed with time.
Discussion
Abnormal retinal vascularization, a pathologic condition in many vision-threatening diseases, can lead to serious consequences, including blindness.5-8,14 Development of reproducible and reliable animal models of RNV has advanced our understanding of both the normal development and pathobiology of aberrant retinal vascularization. In this study, we describe robust, heritable mouse models of abnormal retinal vascularization, one of which is caused by a nucleotide deletion in the Crb1gene that was previously described as the retinal degeneration 8 (rd8) mutation.22–24 The rd8 mutation is common and presents in many mouse strains,22–24 but the retinal phenotype differs substantially from that observed in the rnv3 model. This model, rnv3, made available through the JAX Eye Mutant Resource, was previously described by two groups.7–8 The first report indicated that the ectopic vascular vessels were choroidal in origin and infiltrated the RPE and intraretinal space.7 The second group reported that the model captured the early stages of retinal angiomatous proliferation (RAP), with intraretinal vessels diving into the subretinal space but not breaching the RPE.8 The rnv3 phenotype has been recently described in many publications referred to as JR55587,14,25–27 or NRV28 but the molecular basis for the disease was not reported. We mapped the rnv3 phenotype to mouse Chr 1 in a region containing Crb1, Cfh (component factor h) and Cfh-related genes 1 through 3. We did not consider Crb1 as a reasonable candidate because of the vast difference in the retinal phenotype observed in the stock Crb1rd8 mouse model compared with rnv3 mutants. Therefore, rnv3 retinal cDNA was screened for mutations within Cfh, Cfhr1, Cfhr2, and Cfhr3; no coding sequence variation was detected.
Whole genome sequencing of rnv3 DNA, revealed, however, that Crb1rd8 was the only coding sequence alteration in the critical region. To confirm the Crb1rd8 was the causative mutation for the rnv3 phenotype, we used a TALEN-mediated oligonucleotide directed repair to correct the Crb1rd8 mutation in the rnv3 model, as previously described for correction of the Crb1rd8 mutation in B6/N mice.21 Unlike rnv3 (JR5558) mice, which exhibits abnormal retinal vascularization, heterozygous Crb1em1Boc/Crb1rd8 mice, bearing the corrected Crb1 allele, showed a normal retinal phenotype. This result verified that the Crb1rd8 mutation was necessary for the aberrant retinal vascularization to occur in the JR5558 genetic background. The phenotypic difference between stock Crb1rd8 and rnv3 is most likely due to genetic background differences between the strains. That is, the genes that interact with the Crb1rd8 gene to cause early onset abnormal retinal vascularization in the rnv3 strain differ from those genes that are present in the original stock Crb1rd8 strain that leads to a retinal spotting phenotype.
However, in addition to genetic background effects, extensive founder mosaicism in the Crb1 gene captured in strains Crb1em2Boc, Crb1em3Boc, Crb1em4Boc, and Crb1em5Boc were also able to cause the abnormal vascular phenotype in the JR5558 background. Two additional mutations: Crb1em6Boc and Crb1em7Boc, in which a mutant CRB1 protein may still retain some function, lead to a mild retinal vascular phenotype in the JR5558 genetic background. Further studies are needed to confirm the basis for the allelic differences.
Crb1 encodes a mammalian homolog of Drosophila crumbs, which is a membrane protein that establishes apicobasal cell polarity and adhesion between cells. In the mammalian eye, CRB1 is expressed in Müller glia cells (mouse) or in Müller glia and photoreceptor cells (human),28,29 where it contributes to the formation of adherens junctions that constitute the external limiting membrane (ELM), which provides structural cell-cell contact, diffusion barrier, and mediates intracellular signal transduction needed for normal development and tissue maintenance. The Crb1rd8 mutation in mice is associated with retinal ELM fragmentation and outer retinal dysplasia.20,21 A mutation in Crb1 in rat links aberrant Müller glial cell function to retinal telangiectasia and progressive retinal degeneration that is associated with early retinal vascular leaky telangiectasia, and late intraretinal neovascularization, neuronal alterations, and loss of retinal Müller glial cells. These disease characteristics resemble pathologies observed in human macular telangiectasia type 2 (MacTel 2).30 Over 150 CRB1 alleles31 have also been reported to cause a multitude of human retinal diseases. CRB1 variants are associated with retinal degenerative phenotypes ranging from Leber congenital amaurosis 8 (MIM# 613835), characterized by congenital or early-onset blindness, to retinitis pigmentosa (RP12, MIM# 600105), a more slowly progressive disease. CRB1 RP variants are often associated with unique disease features, such as retinal telangiectasia with or without exudative retinal detachment (Coat disease), a loss of RPE pigmentation except near arterioles (preservation of para-arteriolar RPE), pigment paravenous chorioretinal atrophy (PPCRA, MIM# 172870), cone-rod dystrophy, nanophthalmos with optic disc drusen, and macular dystrophy disease. Although null mutations appear to be overrepresented in LCA patients carrying CRB1 mutations,32 attempts to correlate the CRB1 genotype with unique disease features have not been successful; perhaps the range of disease phenotypes observed is a consequence of interactions with additional genetic and/or environmental factors.31,32
We discovered the rnv3 mutation on stock JR4454 (B6;129-Crhr1tm1Klee/J, stock # 004454). By outcrossing rnv3 mutants to C57BL/6J and intercrossing to select mice with the most severe rnv3 phenotype for 5 generations (N5), we developed the JR5558 strain without the Crhr1tm1Klee mutation. This result indicates that the Crhr1 (corticotropin releasing hormone receptor 1) mutation does not contribute to the rnv3 phenotype. Genotyping archived DNA samples from JR4454 for Crb1rd8 and Jak3m1J (DNA sample purchased from Mouse DNA Resource at The Jackson Laboratory), showed that this strain was found to be homozygous for the Crb1rd8 mutation, but heterozygous for the Jak3m1J mutation. The discovery of the Jak3m1J mutation in JR5558 mice was fortuitous, but whether the mutation contributes to the abnormal retinal vascularization phenotype requires further investigation. By outcrossing JR5558 mice with C57BL/6J, we separated the Jak3m1J mutation from Crb1rd8 and generated two strains through selective intercrossing: B6.Cg-Crb1rd8Jak3+/Boc and B6.Cg-Crb1+Jak3m1J/Boc. The strain with only the Crb1rd8 mutation has a mild retinal neovascularization phenotype, while the strain with only the Jak3m1J mutation had a normal retina. These results indicate that the Jak3m1J mutation itself does not cause retinal abnormalities, but when combined with the disruption in the Crb1 gene leads to an enhancement of the retinal vascular disease phenotype. Further, when we were establishing the JR5558 strain, we selected for mutants with the most severe rnv3 phenotype mice to produce future generations without knowing the genotype of the mating pairs. Using this genotype-blind strategy, we find that the current JR5558 strain is homozygous for both mutations of Crb1rd8 and Jak3m1J.
Janus kinase 3 (JAK3) is critical for the normal development and function of the immune system. Signals relayed by the JAK3 protein regulate the development and maturation of certain types of white blood cells (lymphocytes) such as B, T, and natural killer cells. These cells neutralize pathogens, such as bacteria, viruses, and fungi, by recognizing antigens that are non-self.33–39 JAK3-deficient severe combined immunodeficiency (SCID) is caused by mutations in the JAK3 gene and its signaling pathways are essential for T lymphocyte differentiation (IL-7/JAK3) and NK cell development (IL-15/JAK3).40,41 These immune cell types, however, are generally not found in the retina—but it is possible that they may signal to other cells of the immune system, that may indirectly affect retinal angiogenesis. It is of interest; therefore, that the vascular lesions in rnv3 mutants are associated with F4/80+ staining cells.25 Within the retina, the immune cells, microglia and blood-derived macrophages (reported in experimentally induced models) are F4/80+.25 The role of JAK3 in these cell types has not been studied. However, Jak3 expression is found throughout the retina and has been shown to increase in heritable and light-induced retinal degenerative conditions.42,43 Taken together, it is conceivable that impaired JAK3 function might contribute to an enhanced pathology in rnv3 retinas. Correcting the Jak3m1J mutation using a similar approach used for Crb1rd8 or rnv3, in this study, where the genetic background could be preserved, and assessing for angiogenic events would help to resolve this issue.
In summary, we have identified the disrupted gene, Crb1, which is necessary for the posterior segment retinal vascular phenotype found in rnv3/rnv3 mutants and verified causality by TALEN-mediated, oligonucleotide-directed repair rescue of the disease phenotype. Additionally, six novel mutations in Crb1 were generated through NHEJ, and mice bearing the alleles exhibited variable levels of retinal vascular abnormalities. Further study of these models may provide a greater understanding of how different Crb1 alleles result in aberrant retinal vascular development. Finally, we have also potentially identified an enhancer of the vascular phenotype, Jak3mlJ, in rnv3 mice. Further investigation is necessary to confirm its effects on Crb1rd8-mediated development of ectopic retinal vessels, and to understand the mechanism through which it might act to enhance the disease phenotype.
Supplementary Material
Acknowledgments
The authors thank Mark P. Krebs and Jürgen K. Naggert for the review of the manuscript.
Supported by the National Institutes of Health Grants R01 EY019943 (BC), R01 EY011996 (PMN), R01 EY027305 (MPK and PMN) and OD011190 (MVW and BEL). The Jackson Laboratory institutional shared services are supported in part by NIH National Cancer Institute Support Grant CA34196.
Disclosure: B. Chang, None; B. FitzMaurice, None; J. Wang, None; B.E. Low, None; M.V. Wiles, None; P.M. Nishina, None
References
- 1.Folkman J, Shing Y. Angiogenesis. J Biol Chem. 1992;267:10931–10934. [PubMed] [Google Scholar]
- 2.Campochiaro PA. Molecular pathogenesis of retinal and choroidal vascular diseases. Prog Retin Eye Res. 2015;49:67–81. doi: 10.1016/j.preteyeres.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saint-Geniez M, D'Amore PA. Development and pathology of the hyaloid, choroidal and retinal vasculature. Int J Dev Biol. 2004;48:1045–1058. doi: 10.1387/ijdb.041895ms. [DOI] [PubMed] [Google Scholar]
- 4.Hartnett ME. Pathophysiology and mechanisms of severe retinopathy of prematurity. Ophthalmology. 2015;122:200–210. doi: 10.1016/j.ophtha.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heckenlively JR, Hawes NL, Friedlander M, et al. Mouse model of subretinal neovascularization with choroidal anastomosis. Retina. 2003;23:518–522. doi: 10.1097/00006982-200308000-00012. [DOI] [PubMed] [Google Scholar]
- 6.Johnson KR, Gagnon LH, Chang B. A hypomorphic mutation of the gamma-1 adaptin gene (Ap1g1) causes inner ear, retina, thyroid, and testes abnormalities in mice. Mamm Genome. 2016;27:200–212. doi: 10.1007/s00335-016-9632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagai N, Lundh von Leithner P, Izumi-Nagai K, et al. Spontaneous CNV in a novel mutant mouse is associated with early VEGF-A-driven angiogenesis and latest age focal edema, neural cell loss, and dysfunction. Invest Ophthalmol Vis Sci. 2014;55:3709–3719. doi: 10.1167/iovs.14-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasegawa E, Sweigard H, Husain D, et al. Characterization of a spontaneous retinal neovascular mouse model. PLoS One. 2014;9:e106507. doi: 10.1371/journal.pone.0106507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hua J, Guerin KI, Chen J, et al. Resveratrol inhibits pathologic retinal neovascularization in Vldlr-/- mice. Invest Ophthalmol Vis Sci. 2011;52:2809–2816. doi: 10.1167/iovs.10-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrell MI, Aguilar E, Jacobson R, et al. Antioxidant or neurotrophic factor treatment preserves function in a mouse model of neovascularization-associated oxidative stress. J Clin Invest. 2009;119:611–623. doi: 10.1172/JCI35977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyosseva SV, Chen L, Seal S, McGinnis JF. Nanoceria inhibit expression of genes associated with inflammation and angiogenesis in the retina of Vldlr null mice. Exp Eye Res. 2013;116:63–74. doi: 10.1016/j.exer.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joyal JS, Sun Y, Gantner ML, et al. Retinal lipid and glucose metabolism dictates angiogenesis through the lipid sensor Ffar1. Nat Med. 2016;22:439–445. doi: 10.1038/nm.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W, Jiang A, Liang J, et al. Expression of VLDLR in the retina and evolution of subretinal neovascularization in the knockout mouse model's retinal angiomatous proliferation. Invest Ophthalmol Vis Sci. 2008;49:407–415. doi: 10.1167/iovs.07-0870. [DOI] [PubMed] [Google Scholar]
- 14.Liu CH, Wang Z, Sun Y, Chen J. Animal models of ocular angiogenesis: from development to pathologies. FASEB J. 2017;31:4665–4681. doi: 10.1096/fj.201700336R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hawes NL, Smith RS, Chang B, et al. Mouse fundus photography and angiography: a catalogue of normal and mutant phenotypes. Mol Vis. 1999;5:22. [PubMed] [Google Scholar]
- 16.Chang B, Hawes NL, Pardue MT, et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision Res. 2007;47:624–633. doi: 10.1016/j.visres.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truett GE, Heeger P, Mynatt RL, et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 18.Taylor BA, Navin A, Phillips SJ. PCR-amplification of simple sequence repeat variants from pooled DNA samples for rapidly mapping new mutations of the mouse. Genomics. 1994;21:626–632. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- 19.Fairfield H, Gilbert GJ, Barter M, et al. Mutation discovery in mice by whole exome sequencing. Genome Biol. 2011;12:R86. doi: 10.1186/gb-2011-12-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehalow AK, Kameya S, Smith RS, et al. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–2189. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- 21.Low BE, Krebs MP, Joung JK, et al. Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6N mice via TALEN-mediated homology-directed repair. Invest Ophthalmol Vis Sci. 2014;55:387–395. doi: 10.1167/iovs.13-13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luhmann UF, Carvalho LS, Holthaus SM, et al. The severity of retinal pathology in homozygous Crb1rd8/rd8 mice is dependent on additional genetic factors. Hum Mol Genet. 2015;24:128–141. doi: 10.1093/hmg/ddu424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang B, Hurd R, Wang J, Nishina P. Survey of common eye diseases in laboratory mouse strains. Invest Ophthalmol Vis Sci. 2013;54:4974–481. doi: 10.1167/iovs.13-12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattapallil MJ, Wawrousek EF, Chan CC, et al. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012;53:2921–2927. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai N, Ju M, Izumi-Nagai K, et al. Novel CCR3 Antagonists are effective mono- and combination inhibitors of choroidal neovascular growth and vascular permeability. Am J Pathol. 2015;185:2534–2549. doi: 10.1016/j.ajpath.2015.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foxton R, Osborne A, Martin KR, Ng YS, Shima DT. Distal retinal ganglion cell axon transport loss and activation of p38 MAPK stress pathway following VEGF-A antagonism. Cell Death Dis. 2016;7:e2212. doi: 10.1038/cddis.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paneghetti L, Ng YS. A novel endothelial-derived anti-inflammatory activity significantly inhibits spontaneous choroidal neovascularisation in a mouse model. Vasc Cell. 2016;8:2. doi: 10.1186/s13221-016-0036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Pavert SA, Sanz AS, Aartsen WM, et al. Crb1 is a determinant of retinal apical Müller glia cell features. Glia. 2007;55:1486–1497. doi: 10.1002/glia.20561. [DOI] [PubMed] [Google Scholar]
- 29.van Rossum AG, Aartsen WM, Meuleman J, et al. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Muller glia cells. Hum Mol Genet. 2006;15:2659–2672. doi: 10.1093/hmg/ddl194. [DOI] [PubMed] [Google Scholar]
- 30.Zhao M, Andrieu-Soler C, Kowalczuk L, et al. A new CRB1 rat mutation links Müller glial cells to retinal telangiectasia. J Neurosci. 2015;35:6093–6106. doi: 10.1523/JNEUROSCI.3412-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan KN, Robson A, Mahroo OAR, et al. A clinical and molecular characterisation of CRB1-associated maculopathy. Eur J Hum Genet. 2018;5:687–694. doi: 10.1038/s41431-017-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson RH, Mackay DS, Li Z, et al. Phenotypic variability in patients with retinal dystrophies due to mutations in CRB1. Br J Ophthalmol. 2011;95:811–817. doi: 10.1136/bjo.2010.186882. [DOI] [PubMed] [Google Scholar]
- 33.Baird AM, Lucas JA, Berg LJ. A profound deficiency in thymic progenitor cells in mice lacking Jak3. J Immunol. 2000;165:3680–3688. doi: 10.4049/jimmunol.165.7.3680. [DOI] [PubMed] [Google Scholar]
- 34.Thomis DC, Berg LJ. Peripheral expression of Jak3 is required to maintain T lymphocyte function. J Exp Med. 1997;185:197–206. doi: 10.1084/jem.185.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grossman WJ, Verbsky JW, Yang L, et al. Dysregulated myelopoiesis in mice lacking Jak3. Blood. 1999;94:932–939. [PubMed] [Google Scholar]
- 36.Yamaoka K, Min B, Zhou YJ, Paul WE, O'shea JJ. Jak3 negatively regulates dendritic-cell cytokine production and survival. Blood. 2005;106:3227–3233. doi: 10.1182/blood-2005-02-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomis DC, Gurniak CB, Tivol E, Sharpe AH, Berg LJ. Defects in B lymphocyte maturation and T lymphocyte activation in mice lacking Jak3. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 38.Soldevila G, Licona I, Salgado A, et al. Impaired chemokine-induced migration during T-cell development in the absence of Jak 3. Immunology. 2004;112:191–200. doi: 10.1111/j.1365-2567.2004.01863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SY, Saijo K, Takahashi T, et al. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3:771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 40.Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36:515–528. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jouanguy E, Gineau L, Cottineau J, et al. Inborn errors of the development of human natural killer cells. Curr Opin Allergy Clin Immunol. 2013;13:589–595. doi: 10.1097/ACI.0000000000000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange C, Thiersch M, Samardzija M, et al. LIF-dependent JAK3 activation is not essential for retinal degeneration. J Neurochem. 2010;113:1210–1220. doi: 10.1111/j.1471-4159.2010.06686.x. [DOI] [PubMed] [Google Scholar]
- 43.He X, Sun D, Chen S, Xu H. Activation of liver X receptor delayed the retinal degeneration of rd1 mice through modulation of the immunological function of glia. Oncotarget. 2017;8:32068–32082. doi: 10.18632/oncotarget.16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.