Abstract
Tannins are large-molecular-weight plant polyphenols that are produced in fruits, berries, leaves, flowers, seeds, stems, and roots of woody and non-woody plants. Hundreds and thousands of individual tannin structures are consequently found in many kinds of natural food and feed products. The huge structural variability in tannins is reflected as vast bioactivity differences between them but not in the accuracy of their typical analysis methods. Here, I show how the modern liquid chromatography mass spectrometry methods can be used to obtain new types of two-dimensional tannin fingerprints to better visualize both the tannin content and diversity in plants with just one 10 min analysis per sample.
Keywords: tannin analysis, tannin fingerprints, LC−MS, ellagitannins, gallic acid derivatives, gallotannins, hydrolyzable tannins, proanthocyanidins
Introduction
Plants synthesize a vast variety of secondary metabolites that may have multiple functions for the plant. Some are essential for the survival of the whole plant because they may offer protection against herbivores, pathogens, or excess ultraviolet B (UVB) radiation, while others are useful in that they attract pollinators to flowers or seed-dispersing animals to berries. For these reasons, it is nowadays common to appreciate the importance of these compounds to plants by renaming them as specialized metabolites instead of secondary metabolites.1
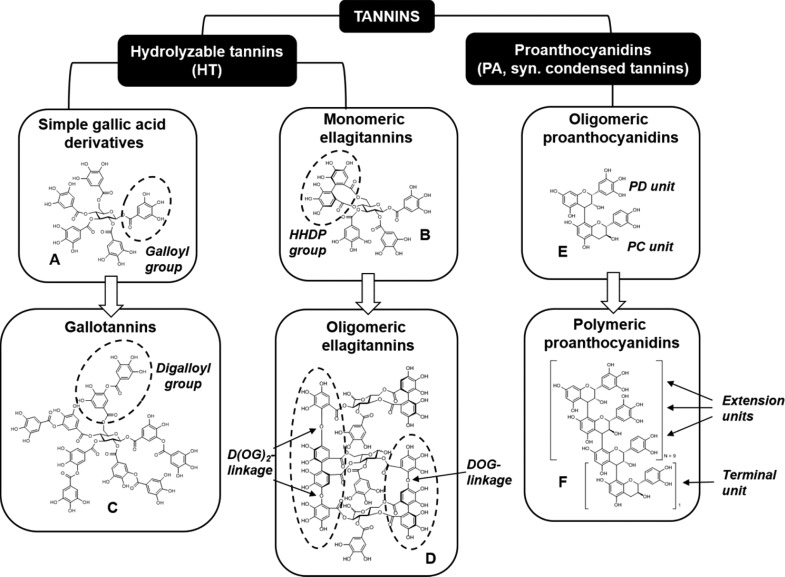
One of the most common specialized metabolite groups in plants are the polyphenols, and the largest subgroup of these are the tannins. They are traditionally viewed as plant defense compounds, but many of them also possess beneficial properties for both human and animal nutrition and health, such as antioxidant, antimicrobial, antiviral, anthelmintic, and more generally protein-binding activities.2−4 Their relatively complex and large structures and the presence of thousands of tannins in plants offer an analytical challenge that cannot be overcome by analyzing tannins compound by compound in plant samples. This is especially true for the proanthocyanidins (PAs, syn. condensed tannins; Figure 1), the most abundant tannin group in plants. Different PA units have mono-, di-, or trihydroxysubstitution in the B ring, thus making procyanidins [PCs, formed from (epi)catechin units] and prodelphinidins [PDs, formed from (epi)gallocatechin units] the most common structural units in PAs (Figure 1). For other structural PA variants, see, e.g., the study by Salminen and Karonen.3 A total of 2–10 monomer units make the oligomeric PAs, and >10 make polymers. PAs can be analyzed at the compound level by liquid chromatography mass spectrometry (LC–MS) only for small oligomers, such as dimers to pentamers.5,6 The larger oligomers and polymers can be detected by electrospray ionization mass spectrometry (ESI–MS) and matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI–TOF–MS) approaches up to relatively large polymers, but they cannot be separated as individual compounds by chromatographic techniques.
Figure 1.
Examples of the chemical structures of tannins present in different tannin subgroups. Hydrolyzable tannins can be divided into simple gallic acid derivatives (A, pentagalloylglucose), gallotannins (B, decagalloylglucose), monomeric ellagitannins (C, tellimagrandin II), and oligomeric ellagitannins (D, oenothein A). Proanthocyanidins can be divided into oligomeric proanthocyanidins (E, PCPD dimer made of catechin and epigallocatechin) and polymeric proanthocyanidins (F, 28-meric PCPD polymer). The proanthocyanidins always contain one terminal unit at the bottom of the structure and one or more extension unit on the top of that.
The second group of terrestrial tannins is the hydrolyzable tannins that can be divided into simple gallic acid derivatives, gallotannins, and mono- and oligomeric ellagitannins.3,7 Simple gallic acid derivatives or gallotannins rarely produce oligomers because they are typically found as monomers only.8 In practice, these tannins can be quantified individually all of the way to the heptamers,9 although even undecamers have been found in plants.10 The third general group of tannins is the phlorotannins (consisting of two or more phloroglusinol units), but because they are mainly found in marine organisms, such as brown algae, they are not dealt with here (but see the study by Salminen and Karonen3 for details).
In this perspective, I do not aim to give a thorough review of how tannins have been analyzed in the past studies of agricultural and food sciences. There are good reviews recently published in the area (see references in, e.g., the study by Zeller11). Instead, I try to give food for thought for scientists interested in combining the common chromatographic and mass spectrometric techniques in ways that enable the production of visually provoking tandem mass spectrometry (MS/MS) fingerprints for tannins and any other kinds of polyphenols or natural compounds in general. These techniques should be available to all laboratories equipped with the nowadays common triple-quadrupole instruments, given that they can fragment compounds by collision-induced dissosiation (CID) already in the electrospray ionization (ESI) interface. The CID before the ion guide and the first quadrupole is the key feature of group-specific techniques that rely on the fragmentation and specific detection of the fragmented functional units, e.g., in tannins.12,13 The dissosiation may be achieved in the ESI interface by increasing the cone voltage difference between the sample cone and the extraction cone above a certain limit, as can be done with the Waters instruments. Different approaches may be found between the manufacturers; e.g., Thermo uses an in-source CID value which can be changed in the ESI interface. The formed functional group fragments will then be selectively detected by the multiple reaction monitoring (MRM) techniques that are the routine methods used with the triple-quadrupole instruments. In addition, these group-specific fingerprinting tools could also be used with the less selective single-quadrupole instruments [single ion recording (SIR) instead of MRM], if the tannin composition of the analyzed species was known and no SIR interfering polyphenols were found in the species. This way, ultraperformance liquid chromatography coupled with single-quadrupole mass spectrometry (MS) could become a powerful tannin analysis technique with selected species in addition to already being quite affordable. Finally, even with selective MRM detection, I recommend to use the diode-array and/or full-scan MS data to verify with each new plant species the correct detection of the tannin subgroups, if the comparison of the ratios of the quantitative to qualitative MRM transitions or quality control MS/MS spectra of different tannin subgroups cannot be used. Once operational and connected to fast and efficient chromatography, the two-dimensional (2D) tannin fingerprinting tools can give a significant boost, especially to qualitative but also quantitative analysis of different tannin groups in plants or plant-derived products, because sample throughput can be increased beyond 100 samples per day.
Engström Method for the Analysis of Four Tannin Groups in the Plant Kingdom
Tannins are widely present in fruits, berries, leaves, flowers, seeds, stems, and roots of woody and non-woody plants, but their distribution in the plant kingdom is not systematically recorded, although excellent studies have been conducted on this topic.14 We at the Natural Chemistry Research Group are currently screening the plant tree of life for the most common tannin groups shown in Figure 1. We have data thus far available from >3400 plant species spanning six continents and >270 plant families. These types of extensive screening experiments cannot be done with the traditional tools, such as Sephadex LH-20 fractionation of the plant extracts, followed by conventional nuclear magnetic resonance (NMR)14 or LC–MS analyses15 of the revealed tannins, because these approaches take tens or hundreds of hours per sample. Other well-functioning tannin characterization tools, such as thiolysis, phloroglucinolysis, and MALDI–TOF–MS4,11,16 cannot be used either, because they are both time-consuming and lack the chromatographic separation of the original tannins. The lack of chromatography in these tools means that all results, such as the PC/PD ratio and the mean degree of polymerization (mDP) of PAs, are average results for the whole sample and cannot be linked to any individual tannin structure but are linked to all of the tens and hundreds of tannins present in the sample. We thus need a reliable and sensitive but fast method that is able to detect all of the tannin groups with the chromatographic step, enabling us to link the data to individual tannins or at least to produce 2D fingerprints (signal intensity versus time) for all of the tannin groups detected. Such a method is, e.g., the “Engström method” that was introduced in two parts in the Journal of Agricultural and Food Chemistry in 2014 and 2015.12,13
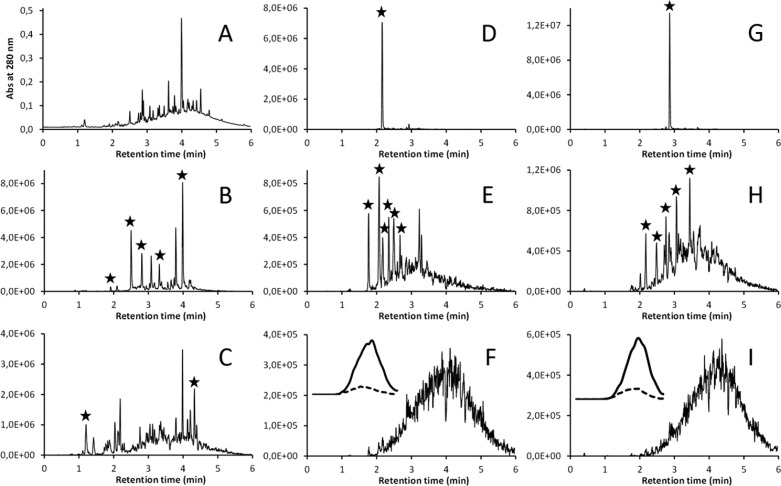
The Engström method relies on the fast 10 min ultrahigh-performance liquid chromatography (UHPLC) separation of the plant polyphenols assisted by conventional diode-array and negative-ion electrospray full-scan mass spectrometry detection (DAD–MS) that enables the characterization of all of the major peaks detected by their ultraviolet (UV) and MS spectra17 (Figure 2A). This is nothing novel as such, and it should be expected from modern phytochemistry that all small-molecular-weight phenolics that elute as sharp peaks in the chromatographic step can be specifically detected by, e.g., MRM methods, because this has been possible with ellagitannin dimers to heptamers.5,6,9 However, compound-specific MRM methods need to be separately developed for every compound or at least isomer, by optimizing the ion-specific cone voltage and collision energy for the molecular ion and its fragment, respectively, and polymeric tannins cannot be to date analyzed individually by MRM methods, thus making these methods non-universal for general tannin detection. For this reason, the Engström method was developed as a single universal method that would be able to detect with the novel group-specific MRMs all tannins that contain the following functional units: (1) galloyl groups, (2) hexahydrodiphenoyl (HHDP) groups, (3) terminal and extension units of PCs, and (4) terminal and extension units of PDs (Figure 1).12,13 The efficiency of modern triple-quadrupole instruments allow for the combination of all of these methods as one. If the negative-ion ESI full-scan MS analysis is simultaneously used, it may need to be limited in its mass range (e.g., m/z 100–1200) or some of the multiple MRM methods for the different PC and PD sizes need to be omitted, so that enough data points can be detected for each peak by the included tannin group-specific MRM transitions. In quantitative work, at least 10–12 data points per peak are recommended, but for qualitative work, the number can be in the range of 6–8.
Figure 2.
Examples of the UHPLC–DAD–MS/MS fingerprints recorded by the Engström method in a single run for the E. calycogona subsp. miracula leaf sample: (A) UV traces at 280 nm, (B) HHDP fingerprint, (C) galloyl fingerprint, (D) (epi)gallocatechin fingerprint, (E) PD oligomer fingerprint, (F) PD polymer fingerprint, with the inset showing the PD extension unit (bold line) and terminal unit (dashed line) fingerprints after 30 smoothing operations to the raw data, (G) (epi)catechin fingerprint, (H) PC oligomer fingerprint, and (I) PC polymer fingerprint, with the inset showing the PC extension unit (solid line) and terminal unit (dashed line) fingerprints after 30 smoothing operations to the raw data. The five main UV peaks at panel A are presented by flavonol glycosides and caffeoyl quinic acids that can be detected by the Engström method as well (kaempferol, quercetin, myricetin, and quinic acid fingerprints). The peaks with asterisks show the tannins that could be characterized by the help of the 2D fingerprints and the full-scan mass spectra.
The Engström method uses 20 mg of dried plant tissue that is extracted for 2 × 3 h with 2 × 1.4 mL of acetone/water (80:20, v/v) in a 2 mL eppendorf tube, preceded by the overnight maceration step. This combination of maceration and 2 time extraction produces more quantitative results with different sample types than, e.g., a single extraction that might be good for qualitative high-throughput studies. The combined extract is evaporated into the water phase by an eppendorf concentrator, freeze-dried, and redissolved in 5 mL of ultrapure water while vortexing for 5 min. After filtration via a 0.20 μm polytetrafluoroethylene (PTFE) filter, the sample is ready for UPLC–MS/MS analysis. Because only approximately 50 μL of the sample is needed in the UPLC vial and 5 μL injected into the column, the Engström method could be downscaled to use as little as 0.2 mg of the plant tissue in the extraction step. However, small masses may cause quantitation errors via, e.g., weighing inaccuracy, and because sample types may differ in their tannin content, I recommend using 20 mg in the extraction step. This 20 mg approach allows for the use of the same sample for many other purposes as well, such as sensitive bioactivity analyses and high-resolution mass spectrometry, if needed.8
The MS/MS method is based on the fragmentation of the functional units of tannins in the ESI interface and the specific detection of the fragmented functional units by the MRM techniques. In a conventional compound-specific MRM technique, a low cone voltage is used to attract the molecular ion into the ion guide and further into the first quadrupole without any significant fragmentation. In the Engström method, the cone voltage is increased to a level that forces the molecular ion to rapidly collide with, e.g., the N2 and O2 molecules present in the ESI interface that still functions at the atmospheric pressure. This will fragment the molecular ion, and the higher the cone voltage, the more efficient the fragmentation.12,13 This is a powerful technique, especially for large tannins, such as polymeric PAs, because many of them are too large to be detected as molecular ions by ESI–MS. However, once they are fragmented, the small fragments can be analyzed without any problems. The additional fact with PAs is that their extension units oxidize during the quinone methide type of fragmentation, while the terminal units remain non-oxidized. This allows for the separation at the first quadrupole of the PA terminal and extension units by their 2 Da mass difference. All four different types of PC and PD terminal and extension units are then selectively fragmented in the collision cell, and the specific fragments are selected by the second quadrupole for detection, thus enabling the calculation of the size, composition, and concentration of the different PA oligomers and polymers as they elute from the LC column.
For qualitative tannin analysis, the produced 2D raw data can be directly viewed and analyzed from the chromatogram window without any post-analysis treatments (see Figure 2). However, with species with unknown tannin composition, it is wise to verify that the detected 2D tannin fingerprints are supported by the full-scan MS data, at least for the monomers and small oligomers.17 It was shown in the study by Engström et al.13 that, e.g., high levels of quercetin derivatives may cause a 0.1–1.0% false-positive detection for HHDP derivatives (ellagitannins) but that this problem can be spotted by comparison of the MRM chromatograms. This false-positive result is caused by quercetin and HHDP derivatives sharing the same ion at m/z 301 for their initial phenolic fragments (quercetin versus HHDP moiety). The false-positive level is reduced in triple-quadrupole instruments to the 0.1–1.0% level by quercetin and HHDP moieties, yielding different daughter ions that are detected and quantified by the MRM methods (quercetin, 301 > 151 and 301 > 179; HHDP, 301 > 200 and 301 > 145). If the method is used with single-quadrupole instruments, such a specificity in the detection is not achieved and every sample type needs to be carefully inspected before the 2D tannin fingerprints can be reliably used. However, with sample types containing high levels of ellagitannins and relatively low levels of quercetin derivatives, even the single-quadrupole approach can be reliable in its fingerprinting task.
In the same way, some small non-PA-type (epi)catechin or (epi)gallogatechin conjugates (e.g., gallates or glycosides) may become detected by the PC or PD methods, but they will not show any results for the extension units, because they do not contain those. This fact can be used to detect the false positives with the PC and PD detection, but just as well, it can be used to detect any (epi)catechin- or (epi)gallogatechin-containing phenolics in addition to PAs. The main thing to notice with the PC and PD detection is that, with high cone voltages, PCs and PDs should always be detected as polymeric humps and not as sharp peaks (Figure 2). Sharp peaks should only be detected with lower cone voltages, and if they are seen with high voltages as well, the full-scan MS data must be inspected for the origin of such peaks. With regard to the polymeric PA humps, it will be exciting to follow up the further developments in this research area, because, recently, Brillouet et al.19 suggested that, e.g., aqueous acetone extraction could cause some of the polymerization reactions that then yield polymeric PAs. The 2D PA fingerprinting tool would be ideal to study this phenomenon in more detail with a plethora of plant species.
For quantitative tannin analysis, the raw MS data of each specific 2D fingerprint needs to be smoothed and intergrated by specific software, such as TargetLynx in the case of Waters, and integrations compared against calibration curve data obtained with proper tannin standards with a known PC/PD ratio, mDP, and galloyl and HHDP contents. In doing this, one needs to remember that the Engström method only detects the functional groups of tannins; it does not detect the whole tannin per se. In PAs, the PC and PD units make practically the whole tannin, but with HTs, the central polyol and other than galloyl and HHDP units are not quantified. Thus, the HT quantitation may always be a slight underestimation, the magnitude of which depends upon the HT standards used to standardize the galloyl and HHDP methods. We use pentagalloylglucose as the galloyl standard and tellimagrandin I as the HHDP standard, because we have tested that this approach gives us quantitation results that are the closest to true HT concentrations, given by UV quantitation of pure HT peaks, once multiple different plant species are screened. If samples of a single species are analyzed, then, naturally, the galloyl and HHDP standards could be obtained from the same species as well.
We have now used this method in multiple studies to realize its full potential in characterizing the PA content of, e.g., ruminant feed varieties,20 traditionally consumed fruit species,6 and potential tannin-producing cell cultures21 or to study the regulation of PA biosynthesis in poplars22 and the distribution of PAs and HTs in 628 Eucalyptus species23 or seeds of 196 tree and liana species growing in the tropics.18 Although tannin chemists highly appreciate the traditional tannin characterization tools mentioned above (NMR, MALDI–TOF–MS, and thiolysis/phloroglucinolysis), the Engström method offers a new dimension to the tannin analysis in agricultural and food sciences or all sciences where tannins play their part. The level of detailed data including the multiple two-dimensional tannin fingerprints (see Figures 2–4) that the method is able to produce is quite overwhelming, especially because all of the data can be produced in a single 10 min run directly from the filtered water phase of a plant extract. Below, I will try to emphasize the potential of the method, because it can provide both the illustrative fingerprints but also more evidence of the chemical structures behind the 2D fingerprints. I believe that this method and its possible further developments will open up new avenues to understand tannin structural diversity and its effects on various bioactivities in different kinds of natural products.
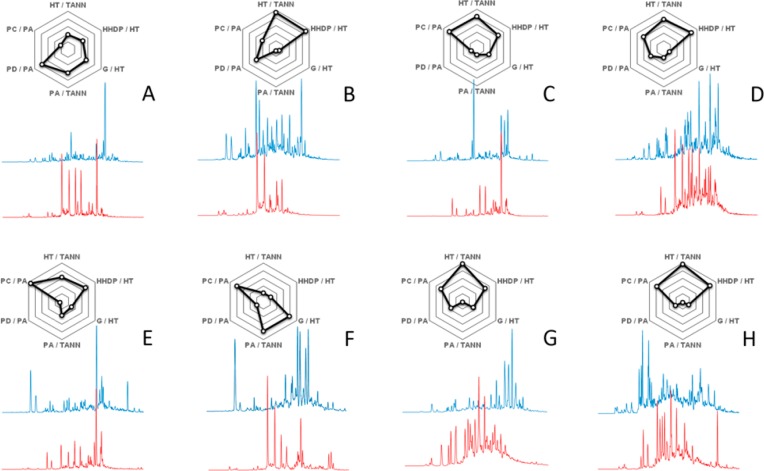
Figure 4.
UHPLC–DAD–MS/MS fingerprints recorded by the Engström method for the galloyl units (blue chromatograms) and HHDP units (red chromatograms) detected in eight Eucalyptus species in the study by Marsh et al.23 The spider webs show the proportional tannin composition (HT % versus PA %), the proportional hydrolyzable tannin composition (galloyl % versus HHDP %), and the proportional PA composition (PC % versus PD %) in all of the samples, as quantified from the tannin fingerprints.
2D Fingerprints Reveal the Hydrolyzable Tannin Diversity in the Samples
Figure 2 shows examples of the main 2D tannin fingerprints recorded during a single UHPLC–DAD–MS/MS run for the Eucalyptus calycogona subsp. miracula leaf sample included in the recent Eucalyptus screening experiment.23Eucalyptus species in general were dominated by hydrolyzable tannins and especially ellagitannins, but E. calycogona subsp. miracula contained such a low concentration of hydrolyzable tannins that they were hardly visible in the UV chromatogram (Figure 2A). Still, Figure 2B shows that the diversity of >20 ellagitannins was detected by the HHDP-specific MRM method. The retention times of the HHDP signals together with the full-scan MS data allowed for the characterization of the main ellagitannins as pedunculagin, isostrictinin, and tellimagrandin I and II, because we have found all of these compounds earlier in other plants.15,17,24 The other detected ellagitannins could not be accurately characterized by MS as a result of the low concentration and chromatographic overlap by other phenolics, but they could be quantified thanks to the selectivity and sensitivity of the group-specific HHDP method. In other words, the method allowed for the measurement of both the diversity and quantity of the HHDP-containing ellagitannins. This would have been difficult to achieve easily by any other than compound-specific MRM methods that would have been needed to be created for all of the detected ellagitannins before they were analyzed from the E. calycogona subsp. miracula sample.
In the galloyl fingerprint (Figure 2C), the baseline was raised a bit as a hump-like shape between 2.5 and 5.5 min, indicating the presence of multiple gallic acid derivatives eluting side by side and/or galloyl residues present in the PA polymers that typically elute as a hump (shown in panels F and I of Figure 2). In any case, >25 gallic acid derivatives were detected as sharp peaks as well. Note that some of these galloyl signals arise from ellagitannins that contain galloyl residues (see, e.g., panels C and D of Figure 1). For instance, the above-detected isostrictinin and tellimagrandin I and II all contain both the HHDP and galloyl groups. In fact, this increases the reliability of the characterization of these compounds further, because they must give positive signals by both galloyl- and HHDP-specific MRM methods. In addition, the summed galloyl and HHDP fingerprints give a good approximation of the diversity and quantity of hydrolyzable tannins in the plant sample. Of the pure gallic acid derivatives, the full-scan MS data allowed for the identification of 1-O-galloyl-β-d-glucose and 1,2,3,4,6-penta-O-galloyl-β-d-glucose in the retention time windows pointed out by the sharp peaks of the galloyl fingerprints. These could have been found by the full-scan MS alone as well,17 but the galloyl fingerprints significantly eased the tedious task of screening through the whole full-scan MS data for the gallic acid derivatives. The Engström method should thus be seen as a tool that does not only produce unique tannin fingerprints but makes tannin characterization both easier and more reliable.
2D Fingerprints Reveal New Aspects for the Proanthocyanidin Content of Plants
Perhaps the most striking tannin fingerprints revealed by the Engström method are the PC and PD fingerprints, because these can make a difference between flavan-3-ol monomers and PA oligomers and polymers and link the presence of all of these to specific retention time windows in the UHPLC analyses. The method uses a series of six increasing cone voltages (e.g., 30 → 180 V) in the MS ion source to fragment the chromatographically separated PAs into monomeric PC and PD units. The larger the PA, the larger the cone voltage needed to fragment the whole molecule into the PC and PD units. At the same time, the PC and PD extension units lose two hydrogens via oxidation, as noted above, and can thus be detected separately by this 2 Da difference from the fragmented but non-oxidized PC and PD terminal units.12 All of this enables the simultenous detection of flavan-3-ol monomers (small cone voltage), PA oligomers (intermediate cone voltage), and PA polymers (large cone voltage) and their PC/PD ratio and mDP at any given time of the retention time axis. For the functioning qualitative and quantitative aspect, the method must be standardized with several PA mixtures that have different but known PC/PD ratios and mDPs. These can be achieved by, e.g., careful Sephadex LH-20 fractionation, followed by thiolysis experiments.12 PA mixtures need to be used, because, thus far, PA polymers cannot be purified at the compound level.
Panels D and G of Figure 2 show how the monomeric building blocks of PCs and PDs could be detected from the E. calycogona subsp. miracula sample using the lowest cone voltage; the two single peaks correspond to gallocatechin and catechin, respectively. When the cone voltage was increased to intermediate, the PA oligomers showed up as sharp peaks (panels E and H of Figure 2) on the top of the PA polymers that also appeared with the method. These PA oligomers could be further characterized by the full-scan MS data; the m/z values 577, 593, and 609 corresponded to pure PC dimer, PC + PD mixed dimer, and pure PD dimer, respectively. The masses of pure PC oligomers can be calculated as n × 288 + 2 Da, and the masses of pure PD oligomers can be calculated as n × 304 + 2 Da, where n stands for the degree of oligomerization of the B-type PAs (A-type PAs will have 2 Da lower mass per each A-type bond). However, because most plant PA compounds contain both PC and PD units, the equation PCn × 288 + PDn × 304 + 2 Da could be more relevant, where PCn and PDn stand for the number of PC and PD units found in the B-type PA, respectively. The further m/z values of 865, 881, 897, and 913 corresponded to pure PC trimer, 2PC + 1PD mixed trimer, 1PC + 2PD mixed trimer, and pure PD trimer, respectively. Conveniently, these findings by full-scan MS could be verified by overlaying panels E and H of Figure 2, because the mixed PCPD oligomers should be seen by both the PC and PD fingerprinting tools. These findings indicated that the PA biosynthesis in this plant species produces PAs that are either pure PCs or PDs or mixtures of these. The presence of both the PC and PD units in the PA polymers was proven as well, because the largest cone voltage used revealed polymeric PA humps that contained PC (Figure 2I) and PD (Figure 2F) units as both extension (solid lines in the insets) and terminal (dashed lines in the insets) units. By overlaying the oligomer/polymer fingerprints (panels E and H of Figure 2) and the polymer fingerprints (panels F and I of Figure 2), it could be seen that their chromatographic profiles do not match. This means that, at any given retention time area, it is possible to find PAs that vary in their PC/PD ratio and molecular weight, suggesting the presence of even hundreds of different PAs in the sample. When all of the PC and PD traces were pooled and quantified, they showed this species to contain 34 mg/g of PAs, with 44% PC and 56% PD units, and mDP of 6.1, on average. These average values could also be reported for each 0.5 min retention time window (or any other window), to obtain more detailed PA fingerprint data for the species. To my knowledge, this cannot be achieved by any other PA tool to date.
In addition to slicing the whole PA fingerprint into retention time windows, it could also be sliced by quantifying all of the different cone voltage data separately. The fact that the PA polymer hump shifted when the cone voltage was increased (panel E versus F of Figure 2 and panel H versus I of Figure 2) is typically an indication of larger PAs being present in the sample, because they require higher energies to be efficiently fragmented. If this phenomenon is observed, then also the mDP of the PAs may increase as their retention time increases, as shown for different sainfoin varities by Malisch et al.20 The Engström method allows for the quantitation of the PA content, PC/PD ratio, and mDP in each of the PA fingerprints obtained with increasing cone voltages. If this approach is used, e.g., with three cone voltages as in Figure 2, instead of the original six, it already provides quite detailed fingerprinting of different PA products for their proportional PA composition. For instance, the same intermediate mDP may be achieved with the following combinations of PA fingerprint proportions (monomers:oligomers/polymers:polymers) using the three cone voltages: 33:34:33, 10:80:10, 25:50:25, and 40:20:40. All four of these samples may have the same mDP value, although it is apparent that the true size distribution of PAs in these four samples is quite different. For this reason, I argue that we need tools that enable us to go beyond the average PA composition of these types of mixtures of PA oligomers and polymers. If we only know the average composition, it may be that we do not know enough to learn about the true structure–activity patterns of these large PA molecules. Again, I see no problems with the small PA oligomers, because they could be quantified by other methods as well.5,6,25
Proanthocyanidin Fingerprints Are Unused Tools in Natural Product Development
Proanthocyanidins are more commonly encountered in plants and different kinds of feed and food products than hydrolyzable tannins, but the exact structural basis for PA bioactivity is more difficult to unravel than for hydrolyzable tannins. This is due to the fact that, while the majority of hydrolyzable tannins can be purified and identified as pure compounds,26−28 for PAs, the same is true only for small oligomers (typically dimers to pentamers) that represent the minority of PA stuctures produced by plants in general (unpublished data of >3400 species). As noted above and indicated by Figure 2, typically plant PAs are mixtures of tens and hundreds of oligomers and polymers. When these PAs are “purified” for bioactivity tests, it actually means that the complex polymeric PA mixture is isolated from the other types of polyphenols and none of the PAs are purified to the compound level. These isolated PA mixtures are then characterized by the traditional tannin tools (NMR, MALDI–TOF–MS, and thiolysis/phloroglucinolysis), resulting in average PC/PD and cis/trans flavan-3-ol ratios, mDP, or examples of the PA sizes found in the mixture, because the largest polymers are not necessarily seen even by MALDI–TOF–MS. Of these methods, only NMR and MALDI–TOF–MS could produce compound-specific PA data in theory, but that is difficult to achieve with tens and hundreds of polymeric PAs without the chromatographic step. In fact, polymers are difficult to analyze by MS as such, because they are either not ionized properly or cannot be transferred into MS for detection or they produce smaller fragments that complicate the correct MS spectra interpretations. These issues are avoided in the Engström method, because the polymers are fragmented already in the ion source, thus enabling the detection of their small subunits, which allows for the backward calculation of the average PA polymer composition minute by minute or second by second, if needed.
I propose that all feed, food, and natural product development processes could benefit, in addition to using the traditional tannin characterization tools, from characterization tools that combine chromatographic separation with selective and sensitive MS detection to look beyond the average PA values. The Engström method is one such method, as shown above. Even if one would be unable to characterize the oligomeric PAs by their m/z values, the PA fingerprints allow for an in-depth 2D comparison of the PA samples and the tannin fingerprints could be used to guide many types of natural product development processes, such as breeding or selecting for better crop varieties.20
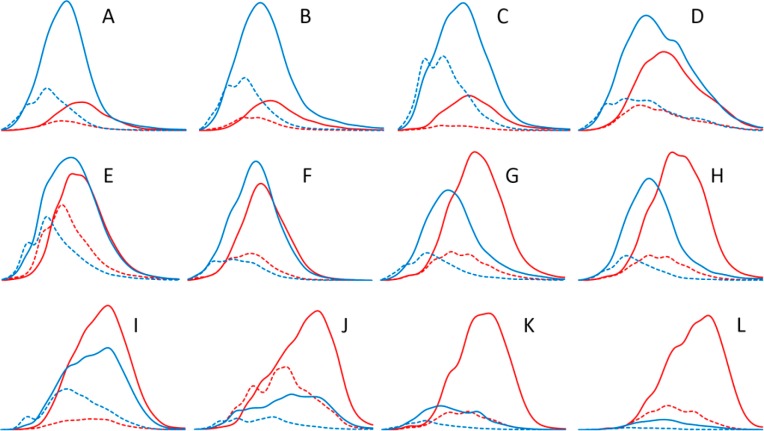
Figure 3 shows an example of an attempt to use PA fingerprints to visualize differences between plant samples; I chose 12 variable PA fingerprints from the >10 000 plant samples thus far analyzed by us with the Engström method. If we compare PA fingerprints in panels A–C of Figure 3, we can see similarities in them especially with the 2D fingerprints of the PD extension units (blue bold line) and PC extension units (red bold line). However, for the PD and PC terminal units (dashed lines), the 2D fingerprints are different in both shape and intensity. It can be concluded that the early-eluting PAs decrease in size from the PD point of view from panels A to C of Figure 3 (as the share of terminal units increases) but the opposite is true from the PC point of view, because PAs in Figure 3C contain proportionally the least PC terminal units (dashed red line). Similarity of the 2D fingerprints in panels A–C of Figure 3 compared to the other fingerprints in panels D–L of Figure 3 can be explained by panels A–C of Figure 3 presenting different sainfoin (Onobrychis viciifolia) varieties.20 On the other hand, the 2D fingerprints from Thuja plicata leaves (Figure 3G) and Viola riviniana leaves (Figure 3H) are surprisingly similar, although these species do not belong to the same plant family. These examples highlight that the Engström method provides a new tool to fingerprint the plant PA composition in a more visual way than can be achieved by the traditional tannin analysis tools. This might bring us the missing link to better understand the basis of PA bioactivity, because, e.g., a similar PC/PD ratio is achieved with samples in panels D–F of Figure 3 but their PA composition is different, as revealed by the 2D fingerprints. If we take any given retention time window in panels A–L of Figure 3, we can see that the exact PA composition in that window is different in almost all of the samples. Unfortunately, we do not yet know such details of the PA bioactivity, e.g. if the early or late eluting PD-containing PAs are more bioactive, which could give hints about the activity-wise best posssible PA composition at a given retention time window. Such data would help us to use the PA fingerprints to identify the plant samples with PAs, e.g., for optimum protein affinity, oxidative activity, or anthelmintic activity.
Figure 3.
UHPLC–DAD–MS/MS fingerprints recorded by the Engström method for the PC extension units (red solid line), PC terminal units (red dashed line), PD extension units (blue solid line), and PD terminal units (blue dashed line) of the PA polymers detected in (A) O. viciifolia ‘WKT10’ leaves, (B) O. viciifolia ‘CPI63854’ leaves, (C) O. viciifolia ‘Perly’ leaves, (D) Rhododendron sp. leaves, (E) Larix sibirica needles, (F), Rhododendron canadense leaves, (G) T. plicata leaves, (H) V. riviniana leaves, (I) Primula veris flowers, (J) red Primula cultivar flowers, (K) Larix sp. needles, and (L) Nephelium connatum leaves. For the fingerprints, the raw data of each PA unit was combined from the data obtained by three cone voltages (excluding the monomer data because monomers are not PAs) and smoothed 30 times.
Hydrolyzable Tannin Fingerprints Can Give Useful Hints of the Plant Bioactivity
There is a lot of structure–function data available for the purified individual hydrolyzable tannin structures that can be linked to the 2D fingerprints recorded by the Engström method for both galloyl and HHDP derivatives. For instance, the study with 27 purified ellagitannins showed the structural reasons for their ease of oxidation, and an equation was made to calculate the activity directly from the structure.24 In general, the early-eluting HHDP-containing compounds were most oxidatively active, while the late-eluting gallic acid derivatives were the least active.24 This oxidative activity can be important for identifying plants with hydrolyzable tannins against insect herbivores and in applications where covalent tannin–protein linkages are sought.28,29
A unique series of individual oligomeric ellagitannins from dimers up to heptamers together with a mixture of octa- to undecamers was purified to show that the oligomer chain length determined the ellagitannin affinity to the model protein bovine serum albumin (BSA).30 The molecular flexibility of ellagitannins was found important for the BSA affinity,30 and the hydrolyzable tannin flexibility was found to increase with the late-eluting galloyl and HHDP derivatives.17 A series of ellagitannin oligomers were shown to be able to decrease the in vitro methane emissions of ruminants in a size-dependent manner while at the same time also affecting the protein protection during rumen fermentation.26 These activities thus benefit from the late-eluting HHDP and galloyl derivatives, while the early-eluting HHDP and galloyl derivatives are either the least flexible or the smallest in size and, thus, the least active.
Finally, a purified set of 33 hydrolyzable tannins were used to determine their structure–activity relations in terms of antiparasitic activity based on the egg-hatching inhibition test; the activity seemed to be a complex combination of the protein affinity of tannins together with their oxidative activation. Again, an equation was created to help the activity calculation directly from the hydrolyzable tannin structure.27 This activity seemed to achieve its maximum levels with compounds having structures and chemical properties close to 1,2,3,4,6-penta-O-galloyl-β-d-glucose. This compound elutes relatively late in reversed-phase liquid chromatography (LC),17 thus warranting the choice of hydrolyzable tannin fingerprints with late-eluting rather than early-eluting galloyl and HHDP peaks for maximal activity.
Figure 4 shows eight extreme examples of the galloyl and HHDP fingerprints detected from the 628 species of Eucalyptus.23 In addition, the spider webs illustrate the overall tannin composition of the species, so that it is easy to spot (1) if the tannins of the species consist mainly of PAs or hydrolyzable tannins, (2) if the PAs are PC- or PD-rich, and (3) if the hydrolyzable tannins are galloyl- or HHDP-rich. From the hydrolyzable tannin point of view, these data enable us to estimate which of the species would be good, e.g., in its oxidative activity (HHDP-rich species with especially early-eluting HHDP but also galloyl derivatives; e.g., Figure 4H) or protein-binding activity (galloyl-rich species with especially late-eluting galloyl but also HHDP derivatives; e.g., panels A and F of Figure 4). The covalent tannin–protein interactions are more potent in affecting protein function than the non-covalent interactions, and because the covalent interactions may require an intermediate protein affinity accomponied by intermediate oxidative activity,28 the hydrolyzable tannin fingerprints of species in panels B and D of Figure 4 look good in this respect. The spider webs once more illustrate how the overall tannin composition can make a difference between most of the samples but that similar overall composition (spider webs in panels G and H of Figure 4) may in fact contain different (e.g., galloyl fingerprints in panels G and H of Figure 4) or partially different (e.g., HHDP fingerprints in panels G and H of Figure 4) 2D fingerprints. This is why the 2D tannin fingerprints are needed to provide more diverse tannin data beyond the overall tannin composition.
Further Developments To Be Achieved with the 2D Tannin Fingerprint Detection
The above examples with hydrolyzable tannins highlight how knowledge of the tannin structure can be used to gain good knowledge of tannin bioactivity. The same should be attempted in the future with plant PA polymers as well, but this requires either (1) the development of better purification tools for PA polymers, so that not only mixtures of tens and hundreds of polymers are used in the structure–activity tests, or (2) the use of more comprehensive 2D characterization tools for the used PA polymer mixtures, so that their chemical diversity or tannin fingerprint is recorded beyond the average tannin composition. Without these developments, it is difficult to gain a proper understanding of the PA polymer activity in different plant species or products because the same PC/PD ratio, mDP, and PA content can be theoretically found in multiple PA polymer combinations.
Finally, it is still possible to develop more effective 2D fingerprinting tools for tannins. For instance, the Engström method has one minor shortcoming because it cannot differentiate between the cis and trans forms of the PC (catechin versus epicatechin) and PD (gallocatechin versus epigallocatechin) units. This is where thiolysis, phloroglucinolysis, or NMR still need to be used to clarify this stereochemical difference between the PA polymer mixtures. Thiolysis, phloroglucinolysis, and NMR can also measure non-soluble PA mixtures, while those obviously are not detected by the Engström method. However, as both triple-quadrupole mass spectrometers and inbuilt ion mobility units are becoming more popular, it could be possible to enhance the Engström method, so that the cis and trans forms of the PC and PD terminal and extension units could be detected separately on the basis of their ion mobility differences. Such an improvement would enable more in-depth 2D or actual three-dimensional (3D) fingerprinting of the PA composition of plant samples, just like the inclusion of the more rare PAs with propelargonidin and 5-deoxy units, such as profisetinidins, into the repertoire of group-specific MRM methods. Similarly, ellagitannin analysis would benefit from rapid MRM-based 2D fingerprinting methods designed to detect and quantify also other than just galloyl and HHDP units of the ETs. In any way, I hope that we are now on the verge of entering a new era of tannin analysis, where the whole tannin diversity of plant samples is taken into account in all kinds of natural product development processes. At the moment, there is no single method that would be able to do all of this, but certainly methods combining rapid LC separation with selective MS/MS detection hold advantage over methods lacking the chromatographic step. It is exciting to see how the use of ever more efficient high-resolution mass spectrometers can be developed to better serve tannin analyses, because, currently, they certainly are more widely used in small-molecule metabolomics-style analyses than in, e.g., PA polymer fingerprinting.
Acknowledgments
The author highly acknowledges the contributions of all past and present members of the Natural Chemistry Research Group in achieving the current state of knowledge in hydrolysable tannin bioactivity and tannin analysis in general. The numerous collaborators are warmly thanked for providing a lot of challenges with tannin analyses during the years.
The Department of Biology at the University of Turku (the Strategic Grant to Ecological Interactions) is acknowledged for providing the author with the Xevo TQ mass spectrometer that enabled the development of the Engström method. The Academy of Finland is acknowledged for funding (298177).
The author declares no competing financial interest.
References
- Pichersky E.; Lewinsohn E. Convergent Evolution in Plant Specialized Metabolism. Annu. Rev. Plant Biol. 2011, 62, 549–662. 10.1146/annurev-arplant-042110-103814. [DOI] [PubMed] [Google Scholar]
- Quideau S.; Deffieux D.; Douat-Casassus C.; Pouységu L. Plant polyphenols: Chemical properties, biological activities, and synthesis. Angew. Chem., Int. Ed. 2011, 50, 586–621. 10.1002/anie.201000044. [DOI] [PubMed] [Google Scholar]
- Salminen J.-P.; Karonen M. Chemical ecology of tannins and other phenolics: We need a change in approach. Funct. Ecol. 2011, 25, 325–338. 10.1111/j.1365-2435.2010.01826.x. [DOI] [Google Scholar]
- Mueller-Harvey I.; Bee G.; Dohme-Meier F.; Hoste H.; Karonen M.; Kölliker R.; Lüscher A.; Niderkorn V.; Pellikaan W.; Salminen J.-P.; Skøt L.; Smith L.; Thamsborg S.; Totterdell P.; Wilkinson I.; Williams A.; Azuhnwi B.; Baert N.; Grosse Brinkhaus A.; Copani G.; Desrues O.; Drake C.; Engström M.; Fryganas C.; Girard M.; Huyen N.; Kempf K.; Malisch C.; Mora-Ortiz M.; Quijada J.; Ramsay A.; Ropiak H.; Waghorn G. Benefits of condensed tannins in forage legumes fed to ruminants: Importance of structure, concentration and diet composition. Crop Sci. 2018, 10.2135/cropsci2017.06.0369. [DOI] [Google Scholar]
- García-Estévez I.; Alcalde-Eon C.; Escribano-Bailón M. T. Flavanol Quantification of Grapes via Multiple Reaction Monitoring Mass Spectrometry. Application to Differentiation among Clones of Vitis vinifera L. cv. Rufete Grapes. J. Agric. Food Chem. 2017, 65, 6359–6368. 10.1021/acs.jafc.6b05278. [DOI] [PubMed] [Google Scholar]
- Ferguson A.; Carvalho E.; Gourlay G.; Walker V.; Martens S.; Salminen J.-P.; Constabel C. P. Phytochemical analysis of salal berry (Gaultheria shallon Pursh.), a traditionally-consumed fruit from western North America with exceptionally high proanthocyanidin content. Phytochemistry 2018, 147, 203–210. 10.1016/j.phytochem.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Gross G. G.Biosynthesis, biodegradation, and cellular localization of hydrolyzable tannins. In Phytochemicals in Human Health Protection, Nutrition, and Plant Defense; Lewis N. G., Romeo J. T., Towers G. H. N., Eds.; Plenum Press: New York, 1999; Recent Advances in Phytochemistry, Vol. 33, pp 185–213, 10.1007/978-1-4615-4689-4_8. [DOI] [Google Scholar]
- Salminen J.-P.The Chemistry and Chemical Ecology of Ellagitannins in Plant–Insect Interactions: From Underestimated Molecules to Bioactive Plant Constituents. In Recent Advances in Polyphenol Research; Romani A., Lattanzio V., Quideau, Eds.; Wiley-Blackwell: Chichester, U.K., 2014; Vol. 4, Chapter 4, pp 83–113, 10.1002/9781118329634.ch4. [DOI] [Google Scholar]
- Baert N.; Karonen M.; Salminen J.-P. Isolation, characterisation and quantification of the main oligomeric macrocyclic ellagitannins in Epilobium angustifolium by ultra-high performance chromatography with diode array detection and electrospray tandem mass spectrometry. J. Chrom. A 2015, 1419, 26–36. 10.1016/j.chroma.2015.09.050. [DOI] [PubMed] [Google Scholar]
- Salminen J.-P.; Karonen M.; Sinkkonen J. Chemical ecology of tannins: Recent developments in tannin chemistry reveal new structures and structure–activity patterns. Chem.—Eur. J. 2011, 17, 2806–2816. 10.1002/chem.201002662. [DOI] [PubMed] [Google Scholar]
- Zeller W.Activity, purification, and analysis of condensed tannins: Current state of affairs and future endeavors. Crop Sci. 2018.
- Engström M. T.; Pälijärvi M.; Fryganas F.; Grabber J.; Mueller-Harvey I.; Salminen J.-P. Rapid qualitative and quantitative analysis of proanthocyanidin oligomers and polymers by UPLC-MS/MS. J. Agric. Food Chem. 2014, 62, 3390–3399. 10.1021/jf500745y. [DOI] [PubMed] [Google Scholar]
- Engström M. T.; Pälijärvi M.; Salminen J.-P. Rapid fingerprint analysis of plant extracts for ellagitannins, gallic acid and quinic acid derivatives, and quercetin-, kaempferol- and myricetin-based flavonol glycosides by UPLC-QqQ-MS/MS. J. Agric. Food Chem. 2015, 63, 4068–4079. 10.1021/acs.jafc.5b00595. [DOI] [PubMed] [Google Scholar]
- Okuda T.; Yoshida T.; Hatano T. Correlation of oxidative transformations of hydrolyzable tannins and plant evolution. Phytochemistry 2000, 55, 513–529. 10.1016/S0031-9422(00)00232-6. [DOI] [PubMed] [Google Scholar]
- Moilanen J.; Koskinen P.; Salminen J.-P. Distribution and content of ellagitannins in Finnish plant species. Phytochemistry 2015, 116, 188–197. 10.1016/j.phytochem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Chai W.-M.; Shi Y.; Feng H.-L.; Qiu L.; Zhou H.-C.; Deng Z.-W.; Yan C.-L.; Chen Q.-X. NMR, HPLC-ESI-MS, and MALDI-TOF MS Analysis of Condensed Tannins from Delonix regia (Bojer ex Hook.) Raf. and Their Bioactivities. J. Agric. Food Chem. 2012, 60, 5013–5022. 10.1021/jf300740d. [DOI] [PubMed] [Google Scholar]
- Moilanen J.; Sinkkonen J.; Salminen J.-P. Characterization of bioactive plant ellagitannins by chromatographic, spectroscopic and mass spectrometric methods. Chemoecology 2013, 23, 165–179. 10.1007/s00049-013-0132-3. [DOI] [Google Scholar]
- Gripenberg S.; Rota J.; Kim J.; Wright S. J.; Garwood N. C.; Fricke E. C.; Zalamea P.-C.; Salminen J.-P. Seed polyphenols in a diverse tropical plant community. J. Ecol. 2018, 106, 87–100. 10.1111/1365-2745.12814. [DOI] [Google Scholar]
- Brillouet J.-M.; Fulcrand H.; Carrillo S.; Rouméas L.; Romieu C. Isolation of Native Proanthocyanidins from Grapevine (Vitis vinifera) and Other Fruits in Aqueous Buffer. J. Agric. Food Chem. 2017, 65, 2895–2901. 10.1021/acs.jafc.6b05561. [DOI] [PubMed] [Google Scholar]
- Malisch C. S.; Lüscher A.; Baert N.; Engström M. T.; Studer B.; Fryganas C.; Suter D.; Mueller-Harvey I.; Salminen J.-P. Large variability of proanthocyanidin content and composition in sainfoin (Onobrychis viciifolia). J. Agric. Food Chem. 2015, 63, 10234–10242. 10.1021/acs.jafc.5b04946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvanto J.; Nohynek L.; Seppänen-Laakso T.; Rischer H.; Salminen J.-P.; Puupponen-Pimiä R. Variability in the production of tannins and other polyphenols in cell cultures of 12 Nordic plant species. Planta 2017, 246, 227–241. 10.1007/s00425-017-2686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. M.; Ma D.; Mellway R.; Gesell A.; Yoshida K.; Walker V.; Tran L.; Stewart D.; Reichelt M.; Suvanto J.; Salminen J.-P.; Gershenzon J.; Séguin A.; Constabel C. P. Poplar MYB115 and MYB134 Transcription Factors Regulate Proanthocyanidin Synthesis and Structure. Plant Physiol. 2017, 174, 154–171. 10.1104/pp.16.01962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh K. J.; Kulheim C.; Blomberg S. P.; Thornhill A. H.; Miller J. T.; Wallis I. R.; Nicolle D.; Salminen J.-P.; Foley W. J. Genus-wide variation in foliar polyphenolics in eucalypts. Phytochemistry 2017, 144, 197–207. 10.1016/j.phytochem.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Moilanen J.; Salminen J.-P. Ecologically neglected tannins and their biologically relevant activity: Chemical structures of plant ellagitannins reveal their in vitro oxidative activity at high pH. Chemoecology 2008, 18, 73–83. 10.1007/s00049-007-0395-7. [DOI] [Google Scholar]
- Kårlund A.; Salminen J.-P.; Koskinen P.; Ahern J.; Karonen M.; Tiilikkala K.; Karjalainen R. Polyphenols in Strawberry (Fragaria × ananassa) Leaves Induced by Plant Activators. J. Agric. Food Chem. 2014, 62, 4592–4600. 10.1021/jf405589f. [DOI] [PubMed] [Google Scholar]
- Baert N.; Pellikaan W.; Karonen M.; Salminen J.-P. A study of the structure–activity relationship of oligomeric ellagitannins on ruminal fermentation in vitro. J. Dairy Sci. 2016, 99, 8041–8052. 10.3168/jds.2016-11069. [DOI] [PubMed] [Google Scholar]
- Engström M. T.; Karonen M.; Ahern J. R.; Baert N.; Payré B.; Hoste H.; Salminen J.-P. Chemical Structures of Plant Hydrolyzable Tannins Reveal Their in Vitro Activity Against Egg Hatching and Motility of Haemonchus contortus Nematodes. J. Agric. Food Chem. 2016, 64, 840–851. 10.1021/acs.jafc.5b05691. [DOI] [PubMed] [Google Scholar]
- Engström M. T.; Sun X.; Suber M. P.; Li M.; Salminen J.-P.; Hagerman A. E. The oxidative activity of ellagitannins dictates their tendency to form highly stabilized complexes with bovine serum albumin at increased pH. J. Agric. Food Chem. 2016, 64, 8994–9003. 10.1021/acs.jafc.6b01571. [DOI] [PubMed] [Google Scholar]
- Agrawal A. A.; Hastings A.; Johnson M. T. J.; Maron J. L.; Salminen J.-P. Insect Herbivores Drive Real-Time Ecological and Evolutionary Change in Plant Populations. Science 2012, 338, 113–116. 10.1126/science.1225977. [DOI] [PubMed] [Google Scholar]
- Karonen M.; Oraviita M.; Mueller-Harvey I.; Salminen J.-P.; Green R. Binding of an Oligomeric Ellagitannin Series to Bovine Serum Albumin (BSA): Analysis by Isothermal Titration Calorimetry (ITC). J. Agric. Food Chem. 2015, 63, 10647–10654. 10.1021/acs.jafc.5b04843. [DOI] [PubMed] [Google Scholar]