Abstract
Respiratory syncytial virus (RSV) is a top cause of severe lower respiratory tract disease and mortality in young children and the elderly. The viral envelope G glycoprotein contributes to pathogenesis through its roles in host cell attachment and modulation of host immunity. Although the G glycoprotein is a target of protective, RSV-neutralizing antibodies, its development as a vaccine antigen has been hindered by its heterogeneous glycosylation and sequence variability outside a conserved central domain (CCD). Here we describe the co-crystal structures of two high-affinity, broadly-neutralizing human monoclonal antibodies bound to the RSV G CCD. The antibodies bind to neighboring conformational epitopes, which we named antigenic sites γ1 and γ2, that span a highly-conserved surface, illuminating an important region of vulnerability. We further show that isolated RSV G CCD activates the chemokine receptor CX3CR1 and that antibodies block this activity. These studies provide a template for rational vaccine design targeting this key contributor to RSV disease.
ONE SENTENCE SUMMARY
This work describes two conformational epitopes on a conserved domain of the RSV G glycoprotein that activates human CX3CR1.
INTRODUCTION
Respiratory syncytial virus (RSV) infects most children by age 2 and is the leading cause of severe lower respiratory tract disease in children worldwide (1–4). RSV is a major cause of mortality, with an estimated 118,000 deaths per year in children under age 5 (3, 5). RSV is also a major cause of morbidity in the elderly and immunocompromised populations. No licensed RSV vaccine exists and the only widely used intervention is palivizumab (Synagis®), a monoclonal antibody (mAb) against the RSV F glycoprotein that reduces disease severity in premature birth infants (6). RSV F is required for infectivity, is less variable overall than RSV G, and is the target of the majority of neutralizing antibodies (7). However, the recent failures of two prominent RSV F vaccines, a phase III clinical trial in older adults (Novavax) and a phase IIb trial in older adults (MedImmune), highlight the urgent need for new approaches. Because these vaccines contained RSV F immunogen in its post-fusion conformation, one emerging approach is focused on the generation of RSV F immunogens that are stabilized in the pre-fusion conformation, as revealed by X-ray crystallographic studies (8). Additionally, RSV G is increasingly recognized as a critical target (9), yet its development as a vaccine antigen has been hindered by its dense and heterogeneous N- and O-glycosylation in the highly-variable mucin-like regions and a paucity of information correlating specific molecular structure with biological activity.
Although variable overall, RSV G (298 residues) contains a ~40 amino acid central conserved domain (CCD) that is devoid of glycosylation and plays key roles in both virus infection and viral pathogenesis (Fig. 1). Specifically, RSV G CCD contains a CX3C chemokine motif that facilitates binding to the human chemokine receptor CX3CR1, a critical step for RSV infection in human airway epithelial cells (10–13). Notably, a soluble form of RSV G is secreted from infected cells beginning ~6 hours post-infection, long before the appearance of RSV virions at 12 hours (14, 15). In vivo, this soluble G protein competes with the natural ligand CX3CL1 (also known as fractalkine) for binding to CX3CR1, modulating signaling and trafficking of CX3CR1+ immune cells, contributing to airway congestion (13, 16–18). RSV with the G gene deleted is highly attenuated in vivo (19). Moreover, RSV with an insertion in the CX3C motif of G (CX4C) that prevents CX3CR1 binding has markedly reduced disease severity in vivo (20). In a recent study, elevated concentrations of both anti-G and anti-pre-fusion-F antibodies were associated with lower clinical disease severity scores, despite the substantially lower absolute abundance of anti-G antibodies compared to anti-F antibodies (7). These results strongly support a renewed focus on RSV G as a target in vaccine and therapeutic antibody development.
Fig. 1. bnmAbs 3D3 and 2D10 bind RSV G161–197.
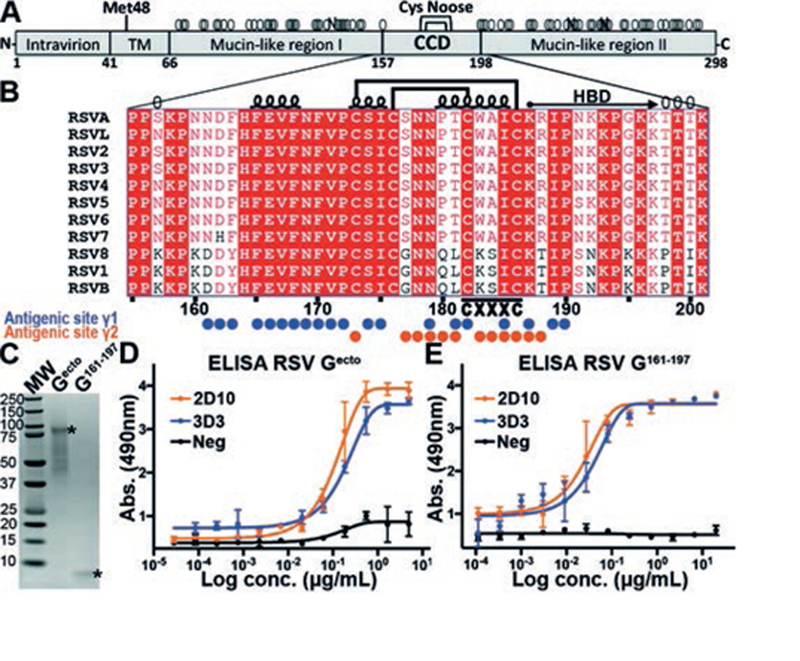
(A) Schematic of the RSV G glycoprotein from RSV strain A2, including the transmembrane region (TM), central conserved region (CCD), and the cysteine noose (Cys noose). Met48 is the alternate initiation site for the production of soluble RSV G. Predicted N- and O-linked glycans are shown by black “N” and gray “O,” respectively. (B) Sequence alignment of RSV G CCD from diverse RSV strains. Amino acids 157 and 198 are predicted to be O-glycosylated in RSV strain A2 (gray “O”) and represent the boundaries. Secondary structure, disulfide bonds, and heparin binding domain (HBD) are displayed. Amino acids within bnmAb 3D3 and 2D10 epitopes (antigenic sites γ1 and γ2) are labeled with blue and orange circles, respectively. (C) Coomassie-stained SDS-PAGE of RSV Gecto and RSV G161–197. (D) ELISA showing binding of bnmAbs 3D3 and 2D10 to RSV Gecto. (E) ELISA showing binding of bnmAbs 3D3 and 2D10 to RSV G161–197. ELISA experiments were performed in biological triplicates.
Several studies have reported broadly-neutralizing monoclonal antibodies (bnmAbs) against RSV G (21–24). Although anti-G bnmAb neutralization of virus infectivity in immortalized cells requires the presence of complement, two studies report that anti-G bnmAbs can directly neutralize virus infectivity in primary well differentiated human airway epithelial cells (10, 12). Anti-G bnmAbs drastically reduce RSV pathogenesis and reduce viral loads in both prophylactic and post-infection animal models (22, 23, 25–27). Linear epitope mapping revealed that several of these bnmAbs bind in the RSV G CCD, although some antibody epitopes could not be identified using linear peptides (22, 24, 28). Here, we sought to understand how two high-affinity human bnmAbs interact with RSV G CCD.
RESULTS
Conformational antigenic site γ1
We first investigated bnmAb 3D3, a native human antibody which binds RSV G with high affinity (KD = 1.1 pM), shows broadly neutralizing activity across nearly all circulating strains, and is in development as a post-infection therapeutic (22, 29). Purified antigen-binding fragment (Fab) 3D3 formed stable complexes with its linear epitope peptide (RSV G162−172) in solution (Fig. S1), and we determined the crystal structure of the Fab 3D3-RSV G162−172 complex to 2.40 Å-resolution (Fig. 2A, S2 and Table S1). The RSV G162−172 peptide contains a short helix and projects several hydrophobic residues, including Phe163, Phe165, Phe168, Phe170 and Pro172, into a ~700 Å2 groove formed by heavy-chain complementarity-determining regions (CDRs) 1, 2, and 3 and light-chain CDRs 1 and 3. Surprisingly, the distal six amino acids of the extended heavy-chain CDR3 formed no molecular contacts with the linear epitope peptide (Fig. 2A), indicative of a larger epitope.
Fig. 2. Crystal structures of RSV G-antibody complexes.
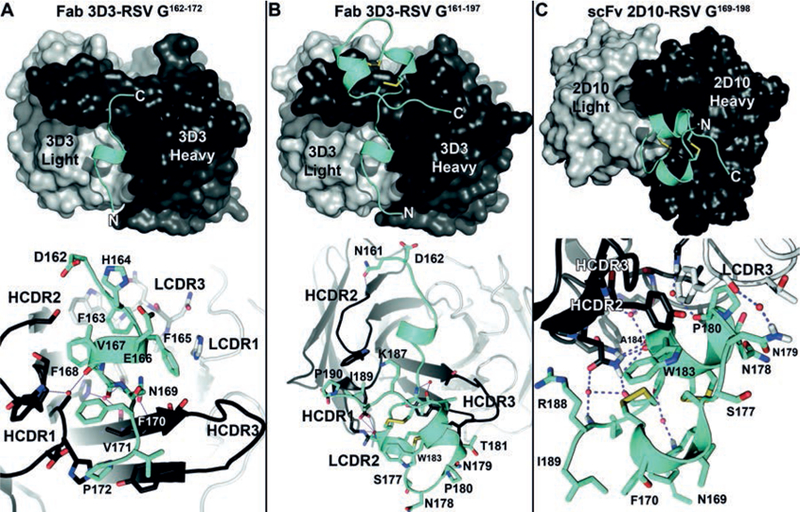
Overall view (top) and zoom-in view (bottom) of the (A) Fab 3D3-RSV G162−172 complex, (B) Fab 3D3-RSV G161−197 complex, and (C) the scFv 2D10-RSV G169−198 complex. In all panels, RSV G is colored cyan, bnmAb heavy chain is colored dark gray, and bnmAb light chain is colored light gray. Water molecules are shown in red. Hydrogen bonds are shown as dashes. Heavy chain CDRs (HCDR1–3) and light chain CDRs (LCDR1–3) are labeled.
To examine whether bnmAb 3D3 interacts with a larger RSV G epitope, we produced in E. coli a recombinant fragment of RSV G (strain A2) that includes the 3D3 linear epitope sequence and the following 25 amino acids spanning the CX3C chemokine motif (RSV G161−197). Antibody 3D3 binds to recombinant RSV G161−197 similarly to recombinant RSV G ectodomain (RSV Gecto) (strain A2) and forms a stable complex in solution (Fig. 1 and S1). We determined the crystal structure of the Fab 3D3-RSV G161−197 complex to 2.40 Å-resolution (Fig. 2B, S2, and Table S1). The structure reveals additional interactions between bnmAb 3D3 and RSV G residues beyond the linear epitope, increasing the binding interface to ~1,060 Å2. Specifically, heavy-chain CDR3 interacts with the RSV G cysteine noose (residues 173–186), which contains four cysteine residues in nested disulfide bonds (1–4 and 2–3), as observed previously in NMR structures of this ~15 residue region (30, 31)(Fig. S2). Additional interactions were observed between heavy-chain CDRs 1 and 2 and RSV G residues 189–190. Altogether, we find that the high-affinity and protective bnmAb 3D3 binds to RSV G at a discontinuous conformational epitope, which we have named antigenic site γ1.
Conformational antigenic site γ2
We next investigated bnmAb 2D10, which also binds RSV G but whose epitope could not be characterized by linear epitope mapping (22, 29). We found that bnmAb 2D10 binds to recombinant RSV G161−197 similarly to recombinant RSV Gecto (Fig. 1), revealing that its epitope is also within the conserved, unglycosylated region of RSV G. We then engineered a recombinant 2D10 single-chain variable fragment (scFv), which forms stable complexes with a synthetic RSV G peptide (RSV G169−198) in solution (Fig. S1), and we determined the crystal structure of the scFv 2D10-RSV G169−198 complex to 1.56 Å-resolution (Fig. 2C, Fig. S2, and Table S1). Antibody 2D10 uses a twisted heavy-chain CDR3, heavy-chain CDR2, and light-chain CDR3 to bind to a ~550 Å2 epitope on the RSV G cysteine noose. The CX3C chemokine motif, which forms a short helix in the cysteine noose, is buried by bnmAb 2D10 binding. Although the 2D10 epitope is mostly continuous, comprised mainly of residues 177–188, the two cysteines within the epitope form two disulfide bonds that induce strong conformational character to this epitope, which we have named antigenic site γ2.
Functional significance of antigenic sites γ1 and γ2
To understand the mechanism of virus neutralization and protection from disease by bnmAbs 3D3 and 2D10, we examined whether they inhibit RSV G modulation of CX3CR1+ cells in an in vitro chemotaxis assay (Fig. 3A). First, we tested recombinant RSV Gecto and found that it induces chemotaxis of human monocyte THP-1 cells, consistent with previous studies (13, 23). To determine if the RSV G CCD alone is sufficient to induce chemotaxis, we tested recombinant RSV G161−197 and found that it induces chemotaxis at levels equivalent to RSV Gecto. Pre-incubation of RSV G161−197 with bnmAbs 3D3 and 2D10 dramatically inhibited RSV G161−197-induced chemotaxis, suggesting that the bnmAbs 3D3 and 2D10 block a CX3CR1-binding site on RSV G. Control experiments show that the bnmAbs have no effect on chemotaxis induced by human CX3CL1, confirming the specificity of the bnmAbs (Fig. 3B). Finally, to confirm that chemotaxis migration is induced by interactions with CX3CR1, we pre-incubated THP-1 cells with anti-CX3CR1 polyclonal antibodies, which significantly inhibited RSV G161−197-induced chemotaxis (Fig. 3A) as well as CX3CL1-induced chemotaxis (Fig. 3B). Altogether, these studies establish that the 37 amino acid fragment RSV G161−197 is sufficient to modulate CX3CR1 and that bnmAbs 3D3 and 2D10 specifically block this activity.
Fig. 3. bnmAbs 3D3 and 2D10 specifically block RSV G161−197-induced chemotaxis.
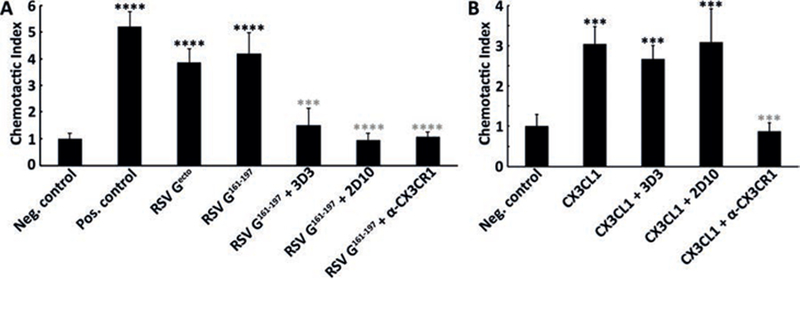
Chemotaxis assays were performed in a transwell plate, with THP-1 cells added to the upper chamber and chemoattractant added to the lower chamber. (A) Serum free media was used as negative control, 10% fetal bovine serum as positive control, 5 nM RSV Gecto, or 5 nM RSV G161–197. (B) Serum free media containing 1 mg/mL BSA was used as a negative control and 10 nM CX3CL1 (fractalkine) as a positive control. Chemotactic indices were determined by comparing the fold-increase in cell migration toward the chemoattractant compared to cell migration toward serum-free media alone. Studies with bnmAbs (25nM) were used to examine inhibition of RSV G161−197- and CX3CL1-induced chemotaxis. Studies with anti-CX3CR1 pre-incubated with THP-1 cells in the upper chamber were used to examine antagonism of cell migration toward RSV G161−197 and CX3CL1 in the lower chamber. A Student’s t test was performed. Black asterisks denote significance compared to negative control and gray asterisks denote significance compared to RSV G161−197 (panel A) or CX3CL1 (panel B), with “***” for p< 0.001 and “****” for p<0.0001. Chemotaxis experiments were performed in four biological replicates.
Sequence conservation at antigenic sites γ1 and γ2
Finally, to understand the molecular basis for RSV G activation of CX3CR1, we aligned diverse RSV G sequences and mapped conservation level onto the RSV G structure (Fig. 1B and 4A). Despite overall high variability of full-length RSV G (53% identity between subtypes RSV A and B), the 37 amino acid fragment RSV G161−197 that activates CX3CR1 contains 24 invariant residues (70% identity between subtypes RSV A and B). Notably, in the CX3C motif only one of the three “X” amino acids, Ile185, is highly conserved, suggesting that this motif alone does not comprise the CX3CR1-binding site (Fig. 4A). Rather, we find that the invariant cysteines in the CX3C motif stabilize a three-dimensional surface of highly conserved amino acids that form extensive atomic interactions across the entire region (Fig. 4B), consistent with bnmAb epitopes (Fig. 4A). The RSV G CCD thus forms a highly conserved, three-dimensional surface poised for CX3CR1 binding (Fig. 4C and S3). We note that the heparin binding domain immediately C-terminal to the cysteine noose includes several conserved positively-charged amino acids (32) (Fig. 1B). While RSV does not use heparan sulfate proteoglycans (HSPGs) to infect human airway epithelial cells, RSV may use HSPGs to infect other cell types, as observed for RSV infection in immortalized cells (10). HSPGs may also play a role in binding of soluble RSV G to other human cells (33). We observed no structural or sequence similarities between RSV G and CX3CL1, the only known ligand for CX3CR1, besides the presence of a CX3C motif and its two disulfide bonds (Fig. S4). This structural divergence despite similar functionality suggests an opportunity to selectively develop therapies blocking interaction between the RSV G CCR and CX3CR1, a strategy that led to the development of the HIV entry inhibitor Maraviroc®, an antagonist of the HIV co-receptor CCR5 (34).
Fig. 4. Sequence conservation within antigenic sites γ1 and γ2, atomic interactions within the RSV G CCD, and model of RSV G glycoprotein.
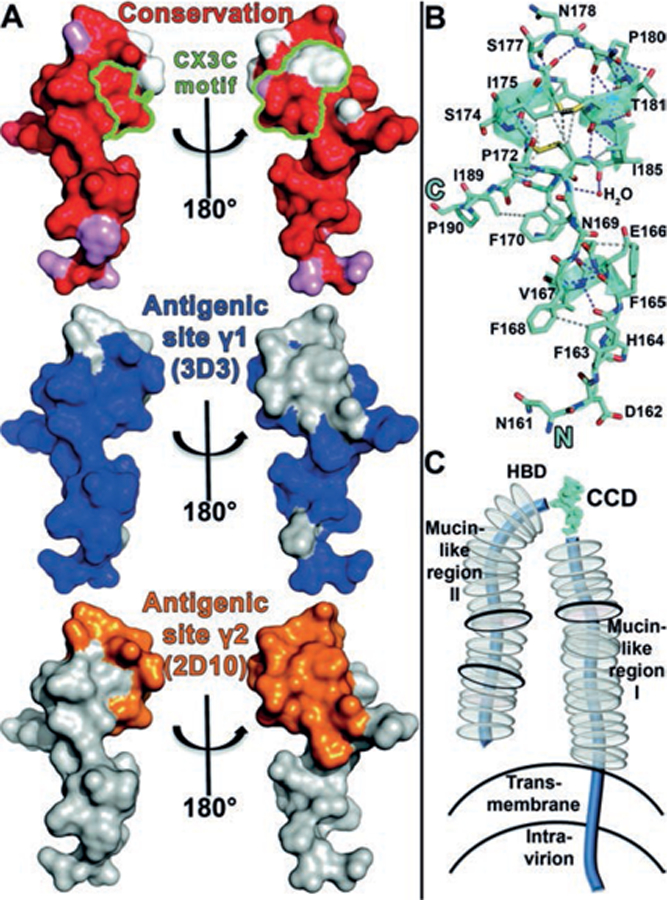
(A) Top: Surface representation of RSV G with amino acids colored according to conservation. Atoms from the main-chain and conserved side chains are red, similar side chains are pink, and non-conserved side chains are white. The five CX3C motif amino acids are outlined in green. Middle and bottom: the epitope footprints of antigenic site γ1 (bnmAb 3D3 epitope) and antigenic site γ2 (bnmAb 2D10 epitope), respectively. (B) Structure of RSV G161−197 with hydrogen bonds shown in purple dashes and representative sidechain-sidechain hydrophobic interactions (≤4.0 Å) shown in gray dashes. (C) Schematic of membrane-bound RSV G. N- and O-linked glycans are shown by black and gray discs, respectively.
DISCUSSION
Our studies define RSV G amino acids 161–197 as a key region of vulnerability that is accessible to antibody binding. Indeed, nearly half of RSV-specific human memory B cells target the G glycoprotein (24) and more than half of the anti-G antibodies in human serum target specifically the RSV G CCD (35). Consistent with these studies, 17 out of 21 isolated human anti-G mAbs have linear epitopes that map to this region (22, 24) (Fig. S5). Of the other 4 mAbs, we show that one (2D10) binds to an epitope adjacent to the epitope for 3D3 but largely non-overlapping with it. A limitation of the present study is that definition of the epitopes for other mAbs has not yet been achieved, nor have the epitopes been defined for bioactive anti-G mAbs that are strain-specific.
Overall, bnmAbs against this region of vulnerability exhibit high affinities and strain-independence, neutralize RSV infection of human airway epithelial cells, inhibit soluble RSV G modulation of CX3CR1+ cells, and decrease pathogenesis in animal models, supporting their development as therapeutic agents to prevent and treat RSV infection. To address global needs, a vaccine may be a more cost-effective intervention, with the present studies providing a firm foundation for constructing an immunogen that induces protective, broadly-neutralizing antibodies targeting the RSV G CCD. To avoid side effects, such an immunogen should not itself activate CX3CR1, a limitation whose feasibility awaits further study.
MATERIALS AND METHODS
Study design.
The overall objective of the study was to determine the molecular basis for antibody recognition of the RSV G glycoprotein. To enable this goal, we undertook experiments centered on protein chemistry, ELISA binding studies, protein x-ray crystallography, and cellular chemotaxis assays. The number of independent experiments is outlined in figure legends and materials and methods section, where appropriate.
Production of bnmAbs 3D3 and 2D10, Fab 3D3, and scFv 2D10.
Recombinant bnmAb 3D3 and bnmAb 2D10 were produced by transient-transfection in CHO cells and purification by immobilized protein A, as described previously (22, 29). Fab 3D3 was generated by incubation of bnmAb 3D3 with immobilized papain, followed by removal of the Fc fragment with immobilized protein A. Fab 3D3 was then purified by Superdex 200 size-exclusion chromatography in 10 mM Tris-HCl pH 8.0 and 150 mM NaCl. For recombinant scFv 2D10, a synthetic gene codon-optimized for Drosophila melanogaster encoding the bnmAb 2D10 heavy chain variable region, a (GGGGS)3GGG linker, and the bnmAb 2D10 light chain variable region, was cloned into pMT-puro in-frame with an N-terminal BiP signal sequence and a C-terminal thrombin cleavage site followed by a Twin-Strep purification tag. The resulting scFv 2D10 expression plasmid was used to obtain stably-transfected Schneider 2 (S2) insect cells. Secreted scFv 2D10 was affinity purified on a StrepTrap column, digested with thrombin protease to remove the purification tag, and then purified by Superdex 200 size-exclusion chromatography in 10 mM Tris-HCl pH 8.0 and 150 mM NaCl.
Expression and purification of RSV Gecto.
A synthetic gene encoding RSV G (strain A2) amino acids 64 to 298 (UniProtKB entry P03423) was cloned into pCF in-frame with an N-terminal TPA signal sequence and C-terminal tandem 6-histidine and Twin-Strep purification tags. Recombinant RSV Gecto was produced by transient-transfection in CHO cells and secreted RSV Gecto was affinity purified on a StrepTrap column.
Expression and purification of RSV G161–197.
A synthetic gene codon-optimized for Escherichia coli encoding RSV G (strain A2) amino acids 161 to 197 (UniProtKB entry P03423) and a C-terminal 6-histidine purification tag was cloned into pET52b. Recombinant RSV G161−197 was expressed overnight in E. coli BL21(DE3) at 18°C. E. coli cells were lysed by ultrasonication in 20 mM Tris-HCl pH 8.0, 150 mM NaCl, and 25 mM imidazole (Buffer A) containing 2 µM MgCl2, benzonase, and protease inhibitors. RSV G161−197 was purified from soluble lysates by HisTrap FF affinity chromatography and eluted with a gradient into Buffer B (Buffer A containing 500 mM imidazole). The purification of soluble RSV G161−197 in the absence of reducing agents appears to be sufficient for spontaneous formation of the cysteine noose disulfide bonds (1–4, 2–3 connectivity), as observed in the crystal structure of the Fab 3D3-RSV G161−197 complex.
ELISA.
Purified bnmAbs at a concentration of 5 µg/mL (150 µL total) were incubated overnight at room temperature in 96-well ELISA microtiter plates. Plates were then washed three times with PBS containing 0.05% Tween 20 (PBST). Wells were blocked by adding 150 µL of 5% BSA in PBS and incubating at room temperature for 1 hr followed by three PBST washes. Recombinant RSV Gecto at 5 µg/mL or RSV G161−197 at 20 µg/mL in 1% BSA in PBS was serially diluted 1:3 with 1% BSA in PBS. Wells were incubated with 150 µL RSV G protein for 1 hr at room temperature and the plates were washed three times with PBST. The plates were then incubated for 1 hr at room temperature with 150 µL HRP-conjugated-HisProbe (ThermoFisher Scientific) diluted 1:5000 in 1% BSA in PBS. Plates were washed three times with PBST and developed by adding peroxidase substrate o-phenylenediamine dihydrochloride (OPD) in 0.05 M phosphate-citrate buffer pH 5.0 and 1.5% hydrogen peroxide for 10 min at room temperature. The reactions were stopped by incubation with 2N sulfuric acid for 10 min at room temperature, and the absorbance was measured at 490 nm. ELISA experiments were performed in biological triplicates.
Formation and structure determination of the Fab 3D3-RSV G162−172 complex.
A synthetic peptide encoding RSV G amino acids 162 to 172 (UniProtKB entry P03423) was mixed in 5-molar excess with purified Fab 3D3 at 17.5 mg/ml in 10 mM Tris-HCl pH 8.0 and 150 mM NaCl. Crystals were grown by hanging drop vapor diffusion at 4°C with a well solution of 23% PEG 3350 and 0.05 M zinc acetate. Crystals were transferred into a cryoprotectant solution of 26% PEG 3350, 0.05 M zinc acetate, and 25% ethylene glycol and flash frozen in liquid nitrogen. Diffraction data were collected at cryogenic temperature at the Advanced Light Source on beamline 8.3.1 using a wavelength of 1.11503 Å. Diffraction data from a single crystal were processed with iMosflm (36) and Aimless (37) (Table S1). The Fab 3D3-RSV G162−172 complex structure was solved by molecular replacement with a Fab homology model and the program PHASER (38), and the structure was refined and manually rebuilt using PHENIX (39) and Coot (40), respectively (Table S1).
Formation and structure determination of the Fab 3D3-RSV G161−197 complex.
Purified RSV G161−197 was mixed in 2 molar excess with purified Fab 3D3, dialyzed into 10 mM Tris-HCl pH 8.0 and 150 mM NaCl, and concentrated to 15 mg/mL. Crystals were grown by hanging drop vapor diffusion at 22°C with a well solution of 21% PEG 3350 and 0.2 M ammonium citrate pH 7.0. Crystals were transferred into a cryoprotectant solution of 25% PEG 3350, 0.2 M ammonium citrate pH 7.0, and 25% glycerol and flash frozen in liquid nitrogen. Diffraction data were collected at cryogenic temperature at the Advanced Light Source beamline 8.3.1 using a wavelength of 1.11582 Å. Diffraction data were collected at cryogenic temperature at the Advanced Light Source on beamline 8.3.1 using a wavelength of 1.11503 Å. Diffraction data from a single crystal were processed with iMosflm (36) and Aimless (37) (Table S1). The Fab 3D3-RSV G161−197 complex structure was solved by molecular replacement with Fab 3D3 and the program PHASER (38), and the structure was refined and manually rebuilt using PHENIX (39) and Coot (40), respectively (Table S1).
Formation and structure determination of the scFv 2D10-RSV G169−198 complex.
A synthetic peptide encoding RSV G amino acids 169 to 198 (UniProtKB entry P03423) was mixed in 2-molar excess with purified scFv in 60 mM Tris-HCl pH 8.0 and 230 mM NaCl and concentrated to 15.0 mg/mL. Crystals were grown by hanging drop vapor diffusion at 22°C with a well solution of 24% PEG 4000, 0.17 M ammonium sulfate, 0.085 M sodium citrate pH 5.6 and 15% glycerol. Crystals were transferred into a cryoprotectant solution of 28% PEG 4000, 0.17 M ammonium sulfate, 0.085 M sodium citrate pH 5.6 and 15% glycerol, and 25% glycerol and flash frozen in liquid nitrogen. Diffraction data were collected at cryogenic temperature at the Advanced Photon Source on beamline 23-ID-D using a wavelength of 1.033 Å. Diffraction data from a single crystal were processed with HKL2000 (41) (Table S1). The scFv 2D10-RSV G 169−198 complex structure was solved by molecular replacement with a scFv homology model and the program PHASER (38), and the structure was refined and manually rebuilt using PHENIX (39) and Coot (40), respectively (Table S1).
Chemotaxis assay.
The in vitro chemotaxis assay was performed using a transwell insert plate with an 8 µm pore size, following previously published methods 14,22. Approximately 2 million log-phase THP-1 cells (a human leukemia monocytic cell line) washed twice and suspended in serum-free RPMI 1640 media were added to the upper chamber of the insert plate. Negative controls were serum-free media alone or serum-free media containing 25 nM bnmAb 3D3 or bnmAb 2D10 was added to the lower chamber. As a positive control, media containing 10% FBS was added to the lower chamber. RSV Gecto or RSV G161−197 samples were added to the lower chamber at a final concentration of 5 nM in serum-free media. For samples with RSV G161−197 and bnmAbs, RSV G161−197 was pre-incubated with 5-molar excess bnmAb for 20 min at room temperature, and then added to serum-free media in the lower chamber, for a final concentration of 5 nM RSV G161−197 and 25 nM bnmAb. For samples with anti-CX3CR1 antibody, 2 µL 1 mg/mL anti-CX3CR1 rabbit polyclonal antibody was incubated with THP-1 cells for 30 minutes in the upper chamber before being placed into the well. The anti-CX3CR1 rabbit polyclonal antibody (ThermoFisher Scientific Cat# PA5–19910) was generated by immunization of rabbits with a peptide corresponding to amino acids 2 to 21 of human CX3CR1 (UniProt ID P49238). The assembled plates were incubated in a CO2 incubator at 37°C for 5 h. Cells migrated to the lower chamber were counted, and the chemotactic indices were determined by comparing the fold-increase in cell migration toward the chemoattractant to cell migration toward serum-free media alone. Experiments were performed in at least four biological replicates.
For chemotaxis assays with CX3CL1 (fractalkine), THP-1 cells were washed and suspended in serum-free RPMI 1640 media containing 1 mg/mL BSA. Recombinant full-length human CX3CL1 (R&D Systems Cat# 365-FR) was added to the lower chamber at a final concentration of 10 nM. For samples with bnmAbs, CX3CL1 was pre-incubated with 2.5-molar excess bnmAb for 20 min at room temperature, and then added to the lower chamber, for a final concentration of 10 nM RSV G161−197 and 25 nM bnmAb. For samples with anti-CX3CR1 antibody, 2 µL 1 mg/mL anti-CX3CR1 rabbit polyclonal antibody was incubated with THP-1 cells for 30 minutes in the upper chamber before being placed into the well. Experiments were performed in four biological replicates.
Supplementary Material
Acknowledgments:
We thank Dr. Sarvind Tripathi for assistance in crystallographic data collection and structure determination, Dr. David Alexander for assistance in RSV Gecto expression, Dr. Phil Berman for use of lab equipment, and Dr. Ralph Tripp for advice.
Funding: R.M.D. is supported by the National Institute of Allergy and Infectious Diseases (NIAID) grant R21AI130605. L.M.K. acknowledges partial support from NIAID grant 5R44AI122360–02. This research used resources of the Advanced Light Source (ALS), which is a U.S. Department of Energy (DOE) Office of Science User Facility under contract no. DE-AC02–05CH11231. Data collection at the ALS Beamline 8.3.1 is supported by the UC Office of the President, Multicampus Research Programs and Initiatives Grant MR-15–328599 and Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation. This research also used resources of the Advanced Photon Source, a DOE Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357.
Footnotes
Competing interests: L.M.K. is an employee of and holds an equity interest in Trellis Bioscience, which is preparing mAb 3D3 for clinical evaluation.
Data and materials availability: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org, under accession codes 5WN9, 5WNA, and 5WNB.
REFERENCES AND NOTES
- 1.Glezen WP, Taber LH, Frank AL, Kasel JA, Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 140, 543–546 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Jain S et al. , Community-acquired pneumonia requiring hospitalization among U.S. children. The New England journal of medicine 372, 835–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi T et al. , Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390, 946–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall CB et al. , The burden of respiratory syncytial virus infection in young children. The New England journal of medicine 360, 588–598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozano R et al. , Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palivizumab, a Humanized Respiratory Syncytial Virus Monoclonal Antibody, Reduces Hospitalization From Respiratory Syncytial Virus Infection in High-risk Infants. Pediatrics 102, 531–537 (1998). [PubMed] [Google Scholar]
- 7.Capella C et al. , Prefusion F, postfusion F, G antibodies and disease severity in infants and young children with acute respiratory syncytial virus infection. The Journal of infectious diseases, (2017). [DOI] [PMC free article] [PubMed]
- 8.McLellan JS et al. , Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripp RA, Power UF, Openshaw PJM, Kauvar LM, Respiratory Syncytial Virus (RSV): Targeting the G Protein Provides a New Approach for an Old Problem. Journal of virology 92:e01302–17, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson SM et al. , Respiratory Syncytial Virus Uses CX3CR1 as a Receptor on Primary Human Airway Epithelial Cultures. PLoS pathogens 11, e1005318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chirkova T et al. , CX3CR1 is an important surface molecule for respiratory syncytial virus infection in human airway epithelial cells. The Journal of general virology 96, 2543–2556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong KI et al. , CX3CR1 Is Expressed in Differentiated Human Ciliated Airway Cells and Co-Localizes with Respiratory Syncytial Virus on Cilia in a G Protein-Dependent Manner. PloS one 10, e0130517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripp RA et al. , CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nature immunology 2, 732–738 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Hendricks DA, Baradaran K, McIntosh K, Patterson JL, Appearance of a soluble form of the G protein of respiratory syncytial virus in fluids of infected cells. The Journal of general virology 68 (Pt 6), 1705–1714 (1987). [DOI] [PubMed] [Google Scholar]
- 15.Hendricks DA, McIntosh K, Patterson JL, Further characterization of the soluble form of the G glycoprotein of respiratory syncytial virus. Journal of virology 62, 2228–2233 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnold R, Konig B, Werchau H, Konig W, Respiratory syncytial virus deficient in soluble G protein induced an increased proinflammatory response in human lung epithelial cells. Virology 330, 384–397 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Harcourt J et al. , Respiratory syncytial virus G protein and G protein CX3C motif adversely affect CX3CR1+ T cell responses. J Immunol 176, 1600–1608 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Haynes LM, Jones LP, Barskey A, Anderson LJ, Tripp RA, Enhanced disease and pulmonary eosinophilia associated with formalin-inactivated respiratory syncytial virus vaccination are linked to G glycoprotein CX3C-CX3CR1 interaction and expression of substance P. Journal of virology 77, 9831–9844 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng MN, Whitehead SS, Collins PL, Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289, 283–296 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Boyoglu-Barnum S et al. , Mutating the CX3C motif in the G protein should make a live respiratory syncytial virus vaccine safer and more effective. Journal of virology, (2017). [DOI] [PMC free article] [PubMed]
- 21.Anderson LJ, Hierholzer JC, Stone YO, Tsou C, Fernie BF, Identification of epitopes on respiratory syncytial virus proteins by competitive binding immunoassay. Journal of clinical microbiology 23, 475–480 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collarini EJ et al. , Potent high-affinity antibodies for treatment and prophylaxis of respiratory syncytial virus derived from B cells of infected patients. J Immunol 183, 6338–6345 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Lee HJ, Lee JY, Park MH, Kim JY, Chang J, Monoclonal Antibody against G Glycoprotein Increases Respiratory Syncytial Virus Clearance In Vivo and Prevents Vaccine-Enhanced Diseases. PloS one 12, e0169139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortjens B et al. , Broadly Reactive Anti-Respiratory Syncytial Virus G Antibodies from Exposed Individuals Effectively Inhibit Infection of Primary Airway Epithelial Cells. Journal of virology 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyoglu-Barnum S et al. , An anti-G protein monoclonal antibody treats RSV disease more effectively than an anti-F monoclonal antibody in BALB/c mice. Virology 483, 117–125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caidi H, Harcourt JL, Tripp RA, Anderson LJ, Haynes LM, Combination therapy using monoclonal antibodies against respiratory syncytial virus (RSV) G glycoprotein protects from RSV disease in BALB/c mice. PloS one 7, e51485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes LM et al. , Therapeutic monoclonal antibody treatment targeting respiratory syncytial virus (RSV) G protein mediates viral clearance and reduces the pathogenesis of RSV infection in BALB/c mice. The Journal of infectious diseases 200, 439–447 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Radu GU et al. , Prophylactic treatment with a G glycoprotein monoclonal antibody reduces pulmonary inflammation in respiratory syncytial virus (RSV)-challenged naive and formalin-inactivated RSV-immunized BALB/c mice. Journal of virology 84, 9632–9636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauvar LM, Collarini EJA, Keyt BA, Foord OA, Anti-RSV G protein antibodies. Patent number US 8273354, (2010). [Google Scholar]
- 30.Sugawara M et al. , Structure-antigenicity relationship studies of the central conserved region of human respiratory syncytial virus protein G. The journal of peptide research : official journal of the American Peptide Society 60, 271–282 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Doreleijers JF et al. , Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus. Biochemistry 35, 14684–14688 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Shields B, Mills J, Ghildyal R, Gooley P, Meanger J, Multiple heparin binding domains of respiratory syncytial virus G mediate binding to mammalian cells. Archives of virology 148, 1987–2003 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Kufareva I, Salanga CL, Handel TM, Chemokine and chemokine receptor structure and interactions: implications for therapeutic strategies. Immunology and cell biology 93, 372–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman-Blum SS, Fung HB, Bandres JC, Maraviroc: a CCR5-receptor antagonist for the treatment of HIV-1 infection. Clinical therapeutics 30, 1228–1250 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Fuentes S, Coyle EM, Beeler J, Golding H, Khurana S, Antigenic Fingerprinting following Primary RSV Infection in Young Children Identifies Novel Antigenic Sites and Reveals Unlinked Evolution of Human Antibody Repertoires to Fusion and Attachment Glycoproteins. PLoS pathogens 12, e1005554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG, iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans PR, Murshudov GN, How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ et al. , Phaser crystallographic software. Journal of applied crystallography 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W, Carter Charles W. Jr., in Methods Enzymol (Academic Press, 1997), vol. 276, pp. 307–326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


