Abstract
In conventional Magnetic Resonance (MR) image based methods, two stages are often involved to capture brain structural information for disease diagnosis, i.e., 1) manually partitioning each MR image into a number of regions-of-interest (ROIs), and 2) extracting pre-defined features from each ROI for diagnosis with a certain classifier. However, these pre-defined features often limit the performance of the diagnosis, due to challenges in 1) defining the ROIs and 2) extracting effective disease-related features. In this paper, we propose a landmark-based deep multi-instance learning (LDMIL) framework for brain disease diagnosis. Specifically, we first adopt a data-driven learning approach to discover disease-related anatomical landmarks in the brain MR images, along with their nearby image patches. Then, our LDMIL framework learns an end-to-end MR image classifier for capturing both the local structural information conveyed by image patches located by landmarks and the global structural information derived from all detected landmarks. We have evaluated our proposed framework on 1526 subjects from three public datasets (i.e., ADNI-1, ADNI-2, and MIRIAD), and the experimental results show that our framework can achieve superior performance over state-of-the-art approaches.
Keywords: Landmark, Convolutional neural network, Multi-instance learning, Brain disease
1. Introduction
Brain morphometric pattern analysis using structural magnetic resonance imaging (MRI) data are proven to be effective in identifying anatomical differences between populations of Alzheimer’s disease (AD) patients and normal controls (NC), and in helping evaluate the progression of mild cognitive impairment (MCI), a prodromal stage of AD. In the literature, extensive MRI-based approaches have been developed to assist clinicians in interpreting and assessing structural changes of the brain (Jack et al., 1999; Ashburner and Friston, 2000; Cuingnet et al., 2011; Chu et al., 2012). While some of those methods are proposed for fundamental MR image analysis (e.g., anatomical landmark detection (Zhang et al., 2017b)), many approaches focus on the implementation of computer-aided-diagnosis (CAD) systems.
To support brain disease diagnosis, many types of local or global feature representations have been derived from structural MRI, such as gray matter tissue density maps (Ashburner and Friston, 2000), volume and shape measurements (Jack et al., 1999; Atiya et al., 2003; Dubois et al., 2015), and cortical thickness (Cuingnet et al., 2011; Lotjonen et al., 2011; Montagne et al., 2015). These feature representations can be roughly categorized into three classes, including 1) voxel-level, 2) region-of-interest (ROI) level, and 3) whole-image-level representations. In particular, voxel-level features attempt to identify brain tissue changes in a voxel-wise manner, and ROI-level features aim to model structural changes within pre-defined ROIs. As an alternative solution, whole-image-level features evaluate changes in the brain by regarding an MR image as a whole (Wolz et al., 2012), without considering local structures within the MR images. It is noteworthy that the appearance of brain MR images is often globally similar and locally different, For instance, it is reported that the early stage of AD only induces structural changes in small local regions rather than in the isolated voxels or the whole brain. Hence, feature representations defined at voxel-level, ROI-level or whole-image-level may not be effective in characterizing the early AD-related structural changes of the brain.
Recently, several patch-level (an intermediate scale between voxel-level and ROI-level) features have been proposed to represent structural MR images for distinguishing AD patients from NCs (Tong et al., 2014; Coupe et al., 2012; Zhang et al., 2017a). In these methods, all patches from MR images of patients are generally regarded as positive samples, while those from MR images of NCs are regarded as negative samples. In other words, the conventional patch-based methods usually assign the same class label (e.g., AD patient or NC) to all image patches from the same brain image. Since not all image patches are necessarily affected by dementia, class labels for patches could be ambiguous. Accordingly, a previous study (Tong et al., 2014) adopted multi-instance learning (MIL) for classification of dementia in brain MRI. As a weakly supervised approach MIL (Maron and Lozano-Pérez, 1998) constructs classifiers using weakly labeled training patches, i.e., image-level labels are used instead of patch-level labels. However, how to select discriminative patches from tens of thousands of patches in each MR image still remains a challenging problem. Moreover, most of the existing patch representations (e.g., intensity values, and/or morphological features) are based on engineered and empirically pre-defined features, which are often independent of subsequent classifier learning procedure. Due to the possible heterogeneous nature of features and classifiers, the pre-defined features may lead to sub-optimal learning performance for brain disease diagnosis. In addition, global information of the whole MR image could not be captured by using only these local patches. In summary, there are at least three key challenges in patch-based approaches: 1) how to select informative image patches in an efficient way, 2) how to capture both local patch-level and global image-level features, and 3) how to integrate feature learning and classifier training jointly.
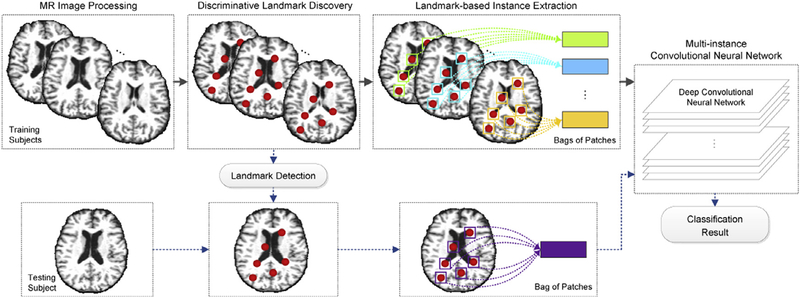
We address these three challenges by proposing a landmark-based deep multi-instance learning (LDMIL) framework. In LDMIL, we first select discriminative patches from MR images based on anatomical landmarks, then jointly learn feature representations of input patches and the subsequent classifier in an end-to-end manner, through which both local and global features of brain MR images are incorporated. Fig. 1 presents a schematic diagram of our proposed LDMIL framework. Specifically, after processing MR images of both training and testing subjects, we discover discriminative landmarks via a group comparison between AD and NC subjects in the training set. We then extract image patches centered at selected landmark locations. These patches from the instances in the MIL terminology, which construct one bag to represent each specific subject. Note that the whole-image-level (subject-level) class label is assigned to a bag, rather than all image patches in the bag. Finally, using the training bags of patches, we design a multi instance CNN model for end-to-end classifier learning. For a new testing subject, we first identify landmarks via the landmark detection algorithm. Then, a bag of patches is extracted from the MR image of the testing subject and fed to the learned CNN model for classification. We have evaluated the effectiveness of our proposed LDMIL framework using baseline MR images in ADNI-1, ADNI-2 (Jack et al., 2008), and MIRIAD (Malone et al., 2013) datasets. Experimental results show that LDMIL outperforms the state-of-the-art methods in both AD classification and MCI conversion prediction tasks.
Fig. 1.
Illustration of the proposed landmark-based deep multi-instance learning (LDMIL) framework using MR imaging data. There are four main components, including 1) MR image processing, 2) discriminative landmark discovery, 3) landmark-based instance extraction, and 4) multi-instance convolutional neural network (CNN) classification.
The rest of the paper is organized as follows. We first briefly introduce relevant studies in Section 2. In Section 3, we describe data used in this study and illustrate the proposed method. In Section 4, we present experimental settings and show the results of both AD classification and MCI conversion prediction tasks. In Section 5, we compare our method with several baseline and state-of-the-art approaches, investigate the influences of parameters, and present limitations of the proposed method. In Section 6, we conclude this work and discuss future research directions.
2. Related work
In this section, we first review relevant studies on MRI-based brain disease diagnosis. Then, we review multi-instance learning approaches and their applications in the medical imaging analysis domain.
2.1. MRI-based brain disease diagnosis
A typical MRI-based CAD system usually contains two essential components, including 1) feature/biomarker extraction from MR images, and 2) classifier construction. Most of the existing feature extraction methods adopt voxel-level, ROI-level, or whole-image-level representations for MR images. Specifically, voxel-wise representations are independent of any hypothesis on brain structures (Ashburner and Friston, 2000; Maguire et al., 2000; Baron et al., 2001; KlÖpel et al., 2008). For instance, voxel-based morphometry measures local tissue (e.g., white matter, gray matter, and cerebrospinal fluid) density of a brain in a voxel-wise manner. The major challenge of voxel-level representations is that they usually lead to the over-fitting problem, since there are only limited (e.g., tens or hundreds) subjects with very high (e.g., millions) dimensional features (Friedman et al., 2001). In contrast, ROI-level representations are defined on specific ROIs, based on a specific hypothesis on abnormal regions of a brain from a structural/functional perspective. For instance, a large number of MRI-based studies have adopted gray matter volume (Yamasue et al., 2003; Maguire et al., 2000; Liu et al., 2017; 2015)), hippocampal volume (Jack et al., 1992; 1999; Atiya et al., 2003; Dubois et al., 2015), and cortical thickness (Fischl and Dale, 2000; Cuingnet et al., 2011; Lotjonen et al., 2011; Montagne et al., 2015), to measure regionally anatomical volume in the brain. However, the definition of ROIs generally requires expert knowledge in practice (Small et al., 2000). Also, whole-image-level representations are derived by treating an MR image as a whole (Wolz et al., 2012). Due to the globally similar property of brain MR images, this kind of methods could not identify subtle changes in brain structures. In contrast, patch-level features provide an intermediate scale between voxel-level and ROI-level for representative MR images. Actually, patch-level biomarkers can be regarded as special ROI-based features, where such ROIs are defined at the scale of local image patches. However, it remains a challenging problem to select informative patches from MR images and to derive discriminative feature representations for patches.
On the other hand, there are a large number of studies focusing on designing advanced classifiers for AD-related disease diagnosis using MRI data. Among various approaches, support vector machine (SVM), logistic regressors (e.g., Lasso, and Elastic Net (Friedman et al., 2001)), sparse representation based classification (SRC) (Wright et al., 2009), random forest classifier (Xiang et al., 2014; Moradi et al., 2015) are widely used. To facilitate the classifier learning procedure, a number of pre-defined features are usually first extracted from MR images. However, training a classifier independent from the feature extraction process may lead to sub-optimal learning performance, due to the possible heterogeneous nature of classifier and features. In recent years, convolutional neural networks (CNNs) have become very popular for automatically learning representations from large collections of static images. However, it is unclear how one may extend these successful CNNs to MRI data for brain disease diagnosis, especially when the intended task requires capturing discriminative changes of the brain (e.g., local and global structure information).
2.2. Multi-instance learning
As a weakly supervised learning method, multi-instance learning (MIL) (Maron and Lozano-Perez, 1998; Dietterich et al., attempts to learn a concept from a training set of labeled bags, where each bag contains multiple unlabeled instances (Amores, 2013). This means that we do not know the labels of individual instances extracted from the bag. Also, it is possible that not all instances are necessarily relevant to the class label of the bag. Specifically, in MIL framework, positive bags can contain both positive and negative instances, and it is generally guaranteed that at least one instance is positive. On the other hand, we know that all instances in the negative bags are negative in MIL. For example, inside one bag, there might be instances that do not convey any information about the category of the bag, or that are more related to other classes, providing confusing information. Compared with fully supervised learning methods, MIL has advantages in automatically modeling the fine-grained information and reducing efforts of human annotations. Many MIL approaches have been proposed in the machine learning domain, such as Diverse Density (DD) (Maron and Lozano-Pérez, 1998), EM-DD (Zhang and Goldman, 2001), MI-Kernels (Gärtner et al., 2002), SVM-based methods (Andrews et al., 2002), and MIL-Boost (Zhang et al., 2005; Cheply-gina et al., 2016).
Recently, MIL has been adopted in the medical imaging analysis domain (Bi and Liang, 2007; Liu et al., 2010; Lu et al., 2011; Xu et al., 2012; Tong et al., 2014; Xu et al., 2014; Yan et al., 2016). In Lu et al. (2011) and Xu et al. (2012), MIL-like methods were developed to perform medical image segmentation. In Bi and Liang (2007), a MIL-based method was proposed to screen pulmonary embolisms among candidates. Liu et al. (2010) developed a MIL-SVM method to predict cardiac events. Tong et al. (2014) proposed a MIL-like model for dementia classification with brain MRI data, by first extracting multiple image patches and then constructing graph kernels for SVM-based classification. This method adopted intensity values within a patch for feature representation that was independent of the subsequent SVM classifier. More recently, a multi-instance deep learning method (Yan et al., 2016) was developed to discover discriminative local anatomies for body-part recognition. This method consisted of a two-stage CNN model, where the first-stage CNN was trained in a multi-instance learning fashion to locate discriminative image patches, and the second-stage CNN was boosted using those selected patches.
Inspired by the latest advances in MIL research, we propose a landmark-based deep multi-instance learning (LDMIL) framework for brain disease diagnosis. Different from the previous MIL studies (Tong et al., 2014; Yan et al., 2016), our method can locate discriminative image patches via anatomical landmarks identified by a data-driven landmark discovery algorithm and does not require any pre-defined engineered features for image patches. This is particularly meaningful for medical imaging applications, where annotating discriminative regions in the brain and extracting meaningful features from MRI often require clinical expertise and high cost. Also, LDMIL is capable of capturing both the local information of image patches and the global information of multiple landmarks, by learning local-to-global representations for MR images layer by layer. To the best of our knowledge, it is the first deep multiinstance model to integrate landmark-based patch extraction with local-to-global representation learning for MRI-based brain disease diagnosis.
3. Material and methods
Here, we first introduce datasets and MR image processing pipeline used in this study (Section 3.1), and then present the proposed landmark-based deep multi-instance learning (LDMIL) method including discriminative landmark discovery (Section 3.2), landmark-based instance extraction (Section 3.3), and a multiinstance CNN model (Section 3.4).
3.1. Subjects and image processing
Three public datasets were used in this study, including 1) the Alzheimer’s Disease Neuroimaging lnitiative-1 (ADNI-1) dataset (Jack et al., 2008), 2) the ADNI-2 dataset (Jack et al., 2008), and 3) the MIRIAD (Minimal Interval Resonance Imaging in Alzheimer’s Disease) dataset (Malone et al., 2013). Those three datasets contain baseline brain MR imaging from Alzheimer’s disease patients and normal control subjects. We report the demographic information of studied subjects in Table 1.
Table 1.
Demographic and clinical information of subjects in three datasets. Values are reported as Mean ± Standard Deviation (Std); Edu: Education years; MMSE: mini-mental state examination; CDR-SB: Sum-of-Boxes of Clinical Dementia Rating.
| Datasets | Category | Male/Female | Age (Mean ± Std) | Edu (Mean ± Std) | MMSE (Mean ± Std) | CDR-SB (Mean ± Std) |
|---|---|---|---|---|---|---|
| ADNI-1 | AD | 106/93 | 69.98 ± 22.35 | 13.09 ± 6.83 | 23.27 ± 2.02 | 0.74 ± 0.25 |
| pMCI | 102/65 | 74.79 ± 6.79 | 15.69 ± 2.85 | 26.58 ± 1.70 | 0.50 ± 0.00 | |
| sMCI | 151/75 | 74.89 ± 7.63 | 15.56 ± 3.17 | 27.28 ± 1.77 | 0.49 ± 0.03 | |
| NC | 127/102 | 74.72 ± 10.98 | 15.71 ± 4.12 | 29.11 ± 1.01 | 0.00 ± 0.00 | |
| ADNI-2 | AD | 91/68 | 69.06 ± 22.04 | 14.19 ± 6.79 | 21.66 ± 6.07 | 4.16 ± 2.01 |
| pMCI | 24/14 | 71.26 ± 15.09 | 16.28 ± 5.07 | 27.39 ± 5.26 | 1.32 ± 2.21 | |
| sMCI | 134/105 | 72.10 ± 11.57 | 15.58 ± 3.95 | 26.83 ± 5.30 | 1.88 ± 2.87 | |
| NC | 113/87 | 73.82 ± 8.41 | 15.66 ± 3.46 | 27.12 ± 7.31 | 0.05 ± 0.22 | |
| MIRIAD | AD | 19/27 | 69.95 ± 7.07 | - | 19.19 ± 4.01 | - |
| NC | 12/11 | 70.36 ± 7.28 | - | 29.39 ± 0.84 | - |
-
1)
ADNI-1 (Jack et al., 2008): Subjects in the baseline ADNI-1 dataset have 1.5T T1-weighted structural MRI data. According to some criteria (see http://adni.loni.usc.edu), such as Mini-Mental State Examination (MMSE) scores and Clinical Dementia Rating (CDR), subjects in ADNI-1 are be divided into three categories: NC, MCI, and AD. In addition, some MCI subjects had converted to AD within 36 months after the baseline time, while the other MCI subjects were stable over time. According to whether MCI subjects would convert to AD within 36 months after the baseline, MCI subjects are further categorized as two classes: (1) stable MCI (sMCI), if the diagnosis was MCI at all available time points (0 – 96 months); (2) progressive MCI (pMCI), if the diagnosis was MCI at baseline but these subjects converted to AD within 36 months after baseline. There is a total of 821 subjects in this dataset, including 199 AD, 229 NC, 167 pMCI, and 226 sMCI subjects in the baseline ADNI-1 dataset.
-
2)
ADNI-2 (Jack et al., 2008): Similar to ADNI-1, the baseline ADNI-2 dataset contains 159 AD, 200 NC, 38 pMCI, and 239 sMCI subjects. The definitions of pMCI and sMCI in ADNI-2 are the same as those in ADNI-1, based on whether MCI subjects would convert to AD within 36 months after baseline. It is worth noting that many subjects included in ADNI-1 also participated in ADNI-2. For independent testing, subjects that appear in both ADNI-1 and ADNI-2 are removed from ADNI-2. Different from those in ADNI-1, the studied subjects from this dataset have 3T T1-weighted structural MR imaging data.
-
3)
MIRIAD (Malone et al., 2013): This dataset includes 69 brain MR images from healthy (23) and pathological (46) subjects. Subjects were previously analyzed with a MMSE, and the score obtained was used to classify them as normal controls (NC) or Alzheimer’s disease patients (AD). As described in Malone et al. (2013), images were acquired on a 1.5T Signa MRI scanner (GE Medical systems, Milwaukee, WI, USA), using a T1-weighted IR-FSPGR (Inversion Recovery Prepared Fast Spoiled Gradient Recalled) sequence. Different from ADNI-1 and ADNI-2, the prodromal stages the disease are not categorized and the subjects are spread in two categories (i.e., AD, and NC).
For MR images of studied subjects, we process them using a standard procedure. Specifically, we first adopt the MIPAV software package2 to perform anterior commissure (AC)-posterior commissure (PC) correction for each MR image. Then, we resample each image to have the same size of 256 × 256 × 256 and the same spatial resolution of 1 × 1 × 1 mm3, and correct intensity inhomogeneity of images using the N3 algorithm (Sled et al., 1998). We then perform skull stripping (Wang et al., 2011) and manual editing to ensure that both skull and dura are cleanly removed. Finally, we remove the cerebellum by warping a labeled template to each skull-stripped image.
3.2. Discriminative landmark discovery
3.2.1. Landmark discovery from training images
There are a large number of image patches in each MR image, while most of them may be not informative enough for brain disease diagnosis, since the structural changes caused by AD could be subtle in the brain. To extract the most informative image patches (i.e., instances) for subsequent feature learning and classifier training, we adopt a data-driven landmark discovery algorithm (Zhang et al., 2016) to locate the most informative image patches in MRI. The goal is to identify the landmarks with statistically significant group differences between AD patients and NC subjects in local structures of MRI. Specifically, in the training stage, a voxel-wise group comparison between AD and NC groups is performed on the ADNI-1 dataset. Using the Colin27 template (Holmes et al., 1998, we use a linear registration to remove global translation, scale and rotation differences of MR images, and to resample all the images with an identical spatial resolution (i.e., 1 × 1 × 1 mm3). In this study, we do not consider the other confounding factors (e.g., age and gender) of subjects. For the linearly-aligned images, a non-linear registration is further performed to build the correspondence relationship among voxels in different images. Hence, based on the deformation field in the non-linear registration, the correspondence between voxels in the template and those in the linearly-aligned images can be established. Then, morphological features can be extracted from those corresponding voxels in the training AD and NC subjects, respectively. A multivariate statistical test (i.e., Hotelling’s T2) (Mardia, 1975) is performed on AD and NC groups, through which a p-value can be calculated for each voxel in the template. As a result, a p-value map in the template space is obtained, whose local minima are defined as locations of discriminative landmarks in the template space. Finally, these landmarks are directly projected to the linearly-aligned training images using their respective deformation fields.
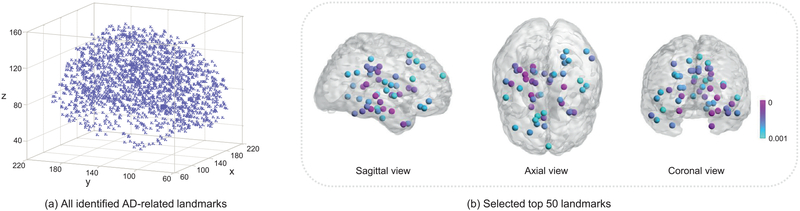
In this study, we have a total of 1741 landmarks identified from AD and NC groups in the ADNI-1 dataset, shown in Fig. 2 (a). Those landmarks are ranked in ascending order, according to their discriminative capability (i.e., p-values in group comparison) in distinguishing AD patients from NCs. That is, a small p-value denotes strong discriminative power, while a large one represents weak discriminative capability. As shown in Fig. 2, many landmarks are close to each other, and thus image patches centered at these landmarks would overlap with each other. To this end, besides considering p-values for landmarks, we further define a spatial Euclidean distance threshold (i.e., 18) to control the distance between landmarks, to reduce the overlaps among image patches. In Fig. 2(b), we plot the selected top 50 landmarks from all 1741 identified landmarks. From this figure, we can observe that many landmarks located in the areas of bilateral hippocampal, bilateral parahip-pocampal, and bilateral fusiform, and these areas are reported to be related to AD in the previous studies (Atiya et al., 2003; De Jong et al., 2008). Besides, the landmarks used in this work is only defined according to their discriminative power in identifying AD from NCs, without using any prior knowledge of previously discovered disease-related brain areas. In our future work, we will further refine the landmark pool by using such prior knowledge.
Fig. 2.
Illustration of (a) all 1741 landmarks discovered by group comparison between AD and NC subjects in the ADNI-1 dataset, and (b) selected top 50 AD-related landmarks shown in the sagittal view, the axial view, and the coronal view, respectively. Different colors in (b) denote p-values in group comparison between AD and NC, i.e., a small p-value indicates a strong discriminative power and vice versa.
3.2.2. Landmark detection for testing images
For a new testing image, we can detect landmarks for a new testing MR image via a similar registration approach as we did for the training MR images. However, such method requires a nonlinear registration process which may generate inaccurate registration results (Miao et al., 2016; Yang et al., 2016; Cao et al., 2017). For MRI with AD/MCI, the pair of images under registration often has a large shape and anatomical variation, which makes the non-linear registration more difficult to get a very accurate result. Hence, in our previous study (Zhang et al., 2016), we learn a regression forest as landmark detector based on training data and our discovered landmarks, to estimate the 3D displacement from each voxel in a testing image to potential landmark positions. Specifically, in the training stage, a regression forest is trained to learn a non-linear mapping between the image patch centered at each voxel and its 3D displacement to a target landmark. We extract morphological features (Zhang et al., 2013) as low-level features for representing local image patches. In the testing stage, the learned regression forest can be used to estimate a 3D displacement from every voxel in the testing image to the potential landmark position, based on the local morphological features extracted from the patch center at this voxel. Hence, each voxel can cast a vote to the landmark position via the estimated 3D displacements. We then obtain a voting map for each testing MR image, by aggregating all votes from all voxels. Finally, we can identify the landmark position as the location with the maximum vote in the voting map.
Note that this regression forest based landmark detector is learned based on the training data. Given a new testing MR image, we can directly apply the learned regression forest to detect landmarks, without using any non-linear registration process. It is worth noting that MCI (including pMCI and sMCI) subjects share the same landmark pool as identified from AD and NC groups. Our assumption here is that, since MCI is the prodromal stage of AD, landmarks with group differences between AD and NC subjects are the potential atrophy locations in brain MR images of MCI subjects.
3.3. Landmark-based instance extraction
Based on the identified landmarks shown in Fig. 2(b), we extract multiple patches (i.e., instances) from a specific MR image (i.e., each bag) for representing each subject (see Fig. 1). Here, we extract patches with the size of 24 × 24 × 24 centered at each specific landmark location. The analysis on why this patch size is selected will be given in Section 4.8. Given L landmarks, we can obtain L patches from an MR image to construct a bag for representing the subject. To suppress the influence of any registration error, for each landmark, we randomly sample different patches on the same landmark location with displacements in a 5 × 5 × 5 cubic (with the step size of 1). Given L landmarks, we can extract up to 125L bags from each MRI.
3.4. Multi-instance convolutional neural network
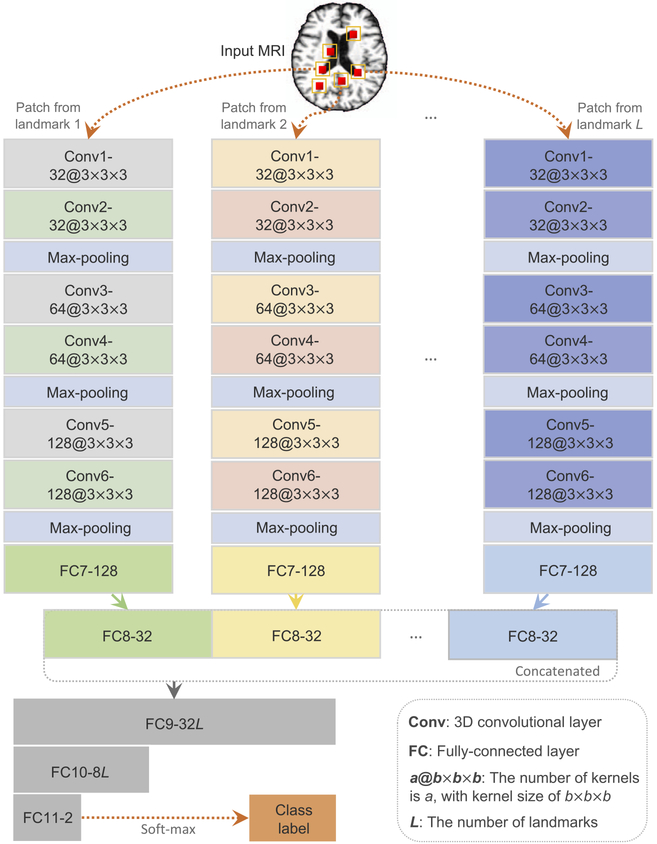
In this study, we attempt to capture both local and global features for MRI of the brain. Also, since not necessarily all image patches extracted from one MR image are significantly affected by dementia, the class labels for those image patches could be ambiguous, if we replicate the subject label on each of them. Therefore, a weakly supervised approach, rather than a supervised one, is appropriate for this situation. To this end, we propose a multiinstance CNN (MICNN) model for AD-related brain disease diagnosis, with a schematic diagram shown in Fig. 3. Given a subject (i.e., a bag in our MIL terminology), the input data of MICNN are L patches (i.e., instances in our MIL framework) extracted from L landmark locations.
Fig. 3.
Illustration of the proposed landmark-based multi-instance convolutional neural network (MICNN), including L sub-CNN architectures corresponding to L landmarks. Given an input MR image, the input data of the deep model are L local image patches extracted from L landmark locations.
To learn representations of individual image patches in the bag, we first run multiple sub-CNN architectures within our deep learning architecture. Such architecture uses a bag of L instances as the input, corresponding to L landmark locations of the brain. It produces patch-level representations for each MR image. More specifically, we embed L parallel sub-CNN architectures with a series of 6 convolutional layers (i.e., Conv1, Conv2, Conv3, Conv4, Conv5, and Conv6), and 2 fully-connected (FC) layers (i.e., FC7, and FC8). The rectified linear unit (ReLU) activation function is used in convolutional layers, while Conv2, Conv4, and Conv6 are followed by max-pooling procedures to conduct the down-sampling operation for their outputs, respectively. The size of the 3D kernels in Conv1 and Conv2 is 3 × 3 × 3, while for the other four convolutional layers it is 2 × 2 × 2. Note that these L sub-CNNs share the same network architectures but have different network parameters, to learn specific path-level features from L local patches.
Since the structural changes caused by dementia can be subtle and distribute across multiple brain regions, only one or a few patch(es) cannot provide enough information to represent global structural changes of the brain. This is different from the conventional multi-instance learning (Yan et al., 2016), in which the image class can be derived by the estimated label of the most discriminative patch. In this study, besides patch-level representations learned from L sub-CNN architectures, we further learn bag-level representations for each MR image using several fully-connected layers (de Brebisson and Montana, 2015). Specifically, we first concatenate patch-level representations (i.e., output feature maps of L FC7 layers) at the FC8 layer, and then add three fully-connected layers (i.e., FC9, FC10, and FC11) to our deep model. Such additional layers are expected to be capable of capturing the complex relationship among image patches located by multiple landmarks, and thus, can form a global representation of the brain at the whole-image-level. Finally, the output of FC11 is fed into a soft-max output layer for predicting the probability of an input brain MR image belonging to a particular category.
Let’s denote the training set as which contains N bags with the corresponding labels The bag of the nth training image Xn consists of L instances, defined as Xn = [xn,1, xn,2, ⋯, xn,L]. As shown in Fig. 1, bags corresponding to all training images become the basic training samples for our proposed MICNN model, and the labels of those bags are consistent with the bag-level (i.e., subject-level) labels. Here, the subject-level label information (i.e., y) is used in a back-propagation procedure for learning the most relevant features in the fully-connected layers and also updating network weights in the convolutional layers. The learning algorithm aims to find a function Φ : χ → y, by minimizing the following loss function:
| (1) |
where P(yn|Xn; W) indicates the probability of the bag Xn being correctly classified as the class yn using the network coefficients W.
In summary, the proposed MICNN architecture is an end-to-end classification model, where local-to-global feature representations can be learned for each MR image. Particularly, we first learn patch-level representations via multiple sub-CNN architectures corresponding to multiple landmarks, to capture the local structure information of the brain. We further model the global information conveyed by multiple landmarks via additional fully-connected layers, to represent the brain structure at a whole-image-level. In this way, both local and global features of brain MR images can be incorporated into the classifier learning process. We optimize the proposed MICNN using the stochastic gradient descent (SGD) algorithm (Boyd and Vandenberghe, 2004), with a momentum coefficient of 0.9 and a learning rate of 10−2. In addition, our proposed network is implemented based on a computer with a single GPU (i.e. NVIDIA GTX TITAN 12GB) and the platform of Tensorflow (Abadi et al., 2016). Given the patch size of 24 × 24 × 24 and L = 40, the training time for MICNN is about 27 hours, while the testing time for a new MRI is less than 1 s.
4. Experiments
In this section, we first introduce the competing methods, present the experimental settings. We further show the experimental results of both tasks of AD classification and MCI conversion prediction, and analyze the influence of parameters.
4.1. Methods for comparison
We first compare the proposed LDMIL method with three state-of-the-art methods, including 1) ROI-based method (ROI), 2) voxel-based morphometry (VBM), and 3) conventional landmark-based morphometry (CLM) (Zhang et al., 2016). We also compare LD-MIL with a landmark-based deep single-instance learning (LDSIL) method that is a variant of LDMIL. Four competing methods are briefly summarized below.
-
1)
ROI-based method (ROI): Similar to several previous works, we extract ROI-specific features from the processed MR images. Specifically, we first segment the brain into three different tissue types, i.e., gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF), using FAST (Zhang et al., 2001) in the FSL software package.3 We then align the anatomical automatic labeling (AAL) atlas (Tzourio-Mazoyer et al., 2002), with 90 pre-defined ROIs in the cerebrum, to the native space of each subject using a deformable registration algorithm (i.e., HAMMER (Shen and Da-vatzikos, 2002)). Finally, we extract volumes of GM tissue inside those 90 ROIs as feature representation for each MR image. Here, the volumes of GM tissue are normalized by the total intracranial volume, which is estimated by the summation of GM, WM, and CSF volumes from all ROIs. Using these 90-dimensional ROI features, we train a linear support vector machine (SVM) with the parameter C = 1 for classification.
-
2)
Voxel-based morphometry (VBM) method (Ashburner and Friston, 2000): We first spatially normalize all MR images to the same template image using a non-linear image registration technique, and then extract the gray matter from the normalized images. We directly measure the local tissue (i.e., GM) density of the brain in a voxel-wise manner, and perform a group comparison using t-test to reduce the dimensionality of the high dimensional features. Similar to the ROI-based method, we feed those voxel-based features to a linear SVM for classification.
-
3)
Conventional landmark-based morphometry (CLM) method (Zhang et al., 2016) with engineered feature representations: As a landmark-based method, CLM shares the same landmark pool as our proposed LDMIL method. Different from LDMIL, CLM adopts engineered features for representing patches around each landmark. Specifically, CLM first extracts morphological features (i.e., local energy pattern (Zhang et al., 2013)) from a local patch centered at each landmark, and then concatenates those features from multiple landmarks together, followed by a z-score normalization (Jain et al., 2005) process. Finally, the normalized features are fed into a linear SVM classifier.
-
4)
Landmark-based deep single-instance learning (LDSIL): As a variant of our proposed LDMIL method, the architecture of LDSIL is similar to a sub-CNN in LDMIL (see Fig. 3), containing 6 convolutional layers (i.e., Conv1, Conv2, Conv3, Conv4, Conv5, and Conv6) and 3 fully-connected layers (i.e., FC7, FC8, and FC11). Specifically, LDSIL learns a CNN model corresponding to a specific landmark, with patches extracted from this landmark as the input and the subject-level class label as the output. Hence, the class label for a subject is assigned to all patches extracted from the MR image of that subject. Given L landmarks, we can learn L CNN models via LDSIL and then obtain L probability scores for a test subject. For making a final classification result, we simply fuse the estimated probability scores for patches using a majority voting strategy. It is worth noting that, different from LDMIL, LDSIL can learn only patch-level representations for brain MR images.
4.2. Experimental settings
We validate our proposed LDMIL method on both AD classification (AD vs. NC) and MCI conversion prediction (pMCI vs. sMCI) tasks. To evaluate the robustness and generalization ability of a specific classification model, in the first group of experiments, we use subjects from ADNI-1 as the training set, while subjects from ADNI-2 and MIRIAD as independent testing sets. The experimental results are reported in the following sections. Besides, we perform two additional groups of experiments in both inter-imaging-center and intra-imaging-center scenarios. Specifically, in the second group of experiments, we train models on the ADNI-2 dataset and test them on both ADNI-1 and MIRIAD, with results reported in Table S1 in the Supplementary Materials, In the third group of experiments, we adopt the cross-validation strategy (Liu et al., 2016) on both ADNI-1 and ADNI-2 datasets, with results reported in Table S2 in the Supplementary Materials. Also, we further report the results of different methods in the task of multi-class classification, i.e., AD vs. pMCI vs. sMCI vs. NC classification, with results given in Fig. S1 in the Supplementary Materials.
We adopt seven metrics for performance evaluation, including receiver operating characteristic (ROC) curve, the area under ROC (AUC), accuracy (ACC), sensitivity (SEN), specificity (SPE), F-Score, and Matthews correlation coefficient (MCC) (Matthews, 1975) that is a balanced measure for binary classes. We denote TP, TN, FP, FN, and PPV as true positive, true negative, false positive, false negative and positive predictive value, respectively. These evaluation metrics are defined as:
For a fair comparison, in our proposed LDMIL method and its variant (i.e., LDSIL), the size of image patch is empirically set to 24 × 24 × 24, while the number of landmarks we used is 40. We further illustrate the influence of those two parameters (i.e., the number of landmarks and the size of image patch) on LDMIL in Sections 4.7 and 4.8, respectively. Besides, we study the influences of these two parameters on LDSIL in Section 5 of the Supplementary Materials, and find these two parameters used in the main experiments for LDSIL fall in the optimal parameter ranges as shown in Fig. S3. Similar to LDMIL, we optimize the LDSIL network using SGD algorithm (Boyd and Vandenberghe, 2004), with a momentum coefficient of 0.9 and a learning rate of 10−2. Also, three landmark-based methods (i.e., CLM, LDSIL, and LDMIL) share the same landmark pool, while LDSIL and LDMIL use the same size of image patches.
4.3. Results of AD classification
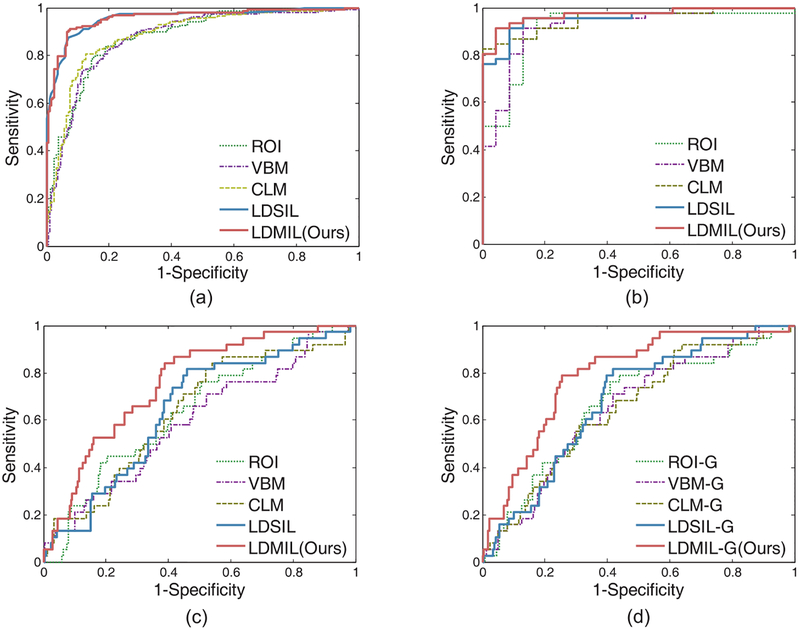
In the first group of experiments, we perform AD vs. NC classification using the model trained on the ADNI-1 dataset. In Table 2 and Fig. 4 (a) and (b), we report the experimental results on the ADNI-2 and the MIRIAD datasets, respectively. From Table 2, we can observe that our proposed LDMIL method generally outperforms those competing methods in AD vs. NC classification on both ADNI-2 and MIRIAD datasets. On ADNI-2, the AUC values achieved by LDMIL is 0.959, which is much better than those yielded by ROI, VBM, and CLM methods. It is worth noting that MR images from ADNI-2 are scanned using 3T scanners, while images from ADNI-1 are scanned using 1.5T scanners. Although MR images in the training set (i.e., ADNI-1) and the testing set (i.e., ADNI-2) have different signal-to-noise ratios, the classification model learned by LD-MIL can still reliably distinguish AD patients from NCs. This implies that our method has strong robustness and generalization ability, which is particularly important in handling multi-center MR images in practical applications. On the other hand, as shown in Fig. 4(a) and (b), three landmark-based methods (i.e., CLM, LDSIL, and LDMIL) consistently outperform both ROI-based and voxel-based approaches (i.e., ROI, and VBM) in AD classification. The likely reason is that the landmarks identified in this study have a stronger discriminative ability to capture differences of structural brain changes between AD and NC subjects compared to the predefined ROIs and the isolated voxels. Also, it can be seen from Fig. 4(a) and (b) that LDSIL (a variant of our proposed LDMIL method) achieves AUC values comparable to LDMIL in AD vs. NC classification.
Table 2.
Results of AD vs. NC classification on both ADNI-2 and MIRIAD datasets, with models trained on the ADNI-1 dataset.
| ADNI-2 | MIRIAD | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ROI | VBM | CLM | LDSIL | LDMIL(Ours) | ROI | VBM | CLM | LDSIL | LDMIL(Ours) | |
| AUC | 0.8673 | 0.8762 | 0.8811 | 0.9574 | 0.9586 | 0.9178 | 0.9206 | 0.9537 | 0.9584 | 0.9716 |
| ACC | 0.7917 | 0.8050 | 0.8222 | 0.9056 | 0.9109 | 0.8696 | 0.8841 | 0.8986 | 0.9130 | 0.9275 |
| SEN | 0.7862 | 0.7735 | 0.7736 | 0.8742 | 0.8805 | 0.9130 | 0.9130 | 0.9783 | 0.9565 | 0.9348 |
| SPE | 0.7960 | 0.8300 | 0.8607 | 0.9303 | 0.9350 | 0.7826 | 0.8261 | 0.7391 | 0.8261 | 0.9130 |
| F-Score | 0.7692 | 0.7784 | 0.7935 | 0.8910 | 0.8974 | 0.9032 | 0.9130 | 0.9278 | 0.9362 | 0.9451 |
| MCC | 0.5800 | 0.6044 | 0.6383 | 0.8082 | 0.8191 | 0.7037 | 0.7391 | 0.7702 | 0.8018 | 0.8391 |
Fig. 4.
ROC achieved by different methods in (a) AD vs. NC classification on the ADNI-2 dataset, (b) AD vs. NC classification on the MIRIAD dataset, and (c) pMCI vs. sMCI classification on the ADNI-2 dataset, and (d) pMCI vs. sMCI classification on the ADNI-2 dataset with the guidance of AD and NC subjects. Here, classification models are trained on the ADNI-1 dataset.
4.4. Results of MCI conversion prediction
We then report the experimental results of MCI conversion prediction (i.e., pMCI vs. sMCI) in Table 3 and Fig. 4(c), with classifiers trained and tested on the ADNI-1 and the ADNI-2 datasets, respectively. It can be observed from Table 2 that, in most cases, our proposed LDMIL method achieves better results than the other four methods in MCI conversion prediction. In addition, as shown in Fig. 4, the superiority of LDMIL over LDSIL is particularly obvious in pMCI vs. sMCI classification, even though such superiority is not that distinct in AD vs. NC classification. The reason could be that LDMIL models both local patch-level and global bag-level structure information of the brain, while LDSIL can only capture local patch-level information. Since the structural changes of AD brains are obvious compared to NCs, only a few landmarks can be discriminative enough to distinguish AD from NC subjects. In contrast, while structural changes of MCI brains may be very subtle and distribute across multiple regions of the brain, it is difficult to determine whether an MCI subject would convert to AD using one or a few landmark(s). In such a case, the global information conveyed by multiple landmarks could be crucial for classification. Moreover, because each landmark defines only a potential (rather than a certain) atrophy location (especially for MCI subjects), it is unreasonable to assign the same subject-level class label to all patches extracted from a specific landmark location in LDSIL. Different from LDSIL, LDMIL can model both the local information of image patches and the global information of multiple landmarks, by assigning the class labels at the subject-level rather than the patch-level. This explains why LDMIL performs better than LDSIL in pMCI vs. sMCI classification, although both methods yield similar results in AD vs. NC classification.
Table 3.
Results of pMCI vs. sMCI classification on the ADNI-2 dataset, with models trained on the ADNI-1 dataset.
| Method | ROI | VBM | CLM | LDSIL | LDMIL(Ours) |
|---|---|---|---|---|---|
| AUC | 0.6377 | 0.5929 | 0.6363 | 0.6448 | 0.7764 |
| ACT | 0.6606 | 0.6426 | 0.6859 | 0.7004 | 0.7690 |
| SEN | 0.4737 | 0.3684 | 0.3947 | 0.3684 | 0.4211 |
| SPE | 0.6904 | 0.6862 | 0.7322 | 0.7531 | 0.8243 |
| F-Score | 0.2769 | 0.2205 | 0.2564 | 0.2523 | 0.3333 |
| MCC | 0.1198 | 0.0402 | 0.0967 | 0.0949 | 0.2074 |
4.5. Influence of transferred knowledge
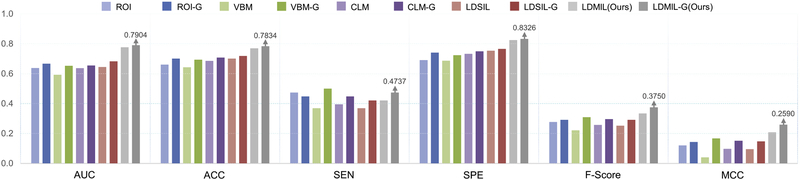
In the above-mentioned MCI conversion prediction (pMCI vs. sMCI) experiment, we only use 187 pMCI and 226 sMCI subjects in the ADNI-1 dataset for classifier training. Recent studies (Filipovych et al., 2011; Coupe et al., 2012) have shown that the knowledge learned from AD and NC subjects can be adopted to guide the prediction of MCI conversion since MCI is a prodromal stage of AD where the structural changes of the brain are between those of AD patients and NC subjects. Accordingly, we propose to employ AD and NC subjects to guide the task of MCI conversion prediction. Specifically, we first train a classification model using both pMCI and AD subjects in ADNI-1 as positive samples, while sMCI and NC subjects are treated as negative samples. Then, we adopt the trained model for pMCI vs. sMCI classification on ADNI-2. Using AD and NC subjects as the guidance information for MCI conversion prediction, we denote the corresponding models of different methods (i.e., ROI, VBM, CLM, LDSIL, and LDMIL) as ROI-G, VBM-G, CLM-G, LDSIL-G, and LDMIL-G, respectively. The experimental results are reported in Figs. 4(d) and 5.
Fig. 5.
Results of MCI conversion prediction (pMCI vs. sMCI) on the ADNI-2 dataset with and without transferred knowledge from AD and NC subjects. Given “A” denoting a method using only MCI subjects in ADNI-1 for model training, its variant “A-G” represents the method using AD and NC as the guidance information for model training. For instance, LDMIL-G denotes the method with a classifier trained using pMCI and AD as positive samples, and sMCI and NC subjects as negative ones in ADNI-1.
It can be observed from Figs. 4(d) and 5, methods using the guidance from AD and NC yield consistently better results than their corresponding counterparts. For instance, MCC and F-Score values achieved by LDMIL-G are 0.2590 and 0.3750, respectively, which are much better than those of LDMIL (i.e., MCC=0.2074 and F-Score=0.3333). Similar trends can be found for the other four competing methods. That is, using AD and NC subjects as the guidance information for classifier training further improves the learning performance of MCI conversion prediction. The underlying reason could be that more training samples are used for learning the MCI prediction model, and also the task of AD vs. NC classification is related to the task of pMCI vs. sMCI classification (Cheng et al., 2015).
4.6. Influence of local-to-global representation
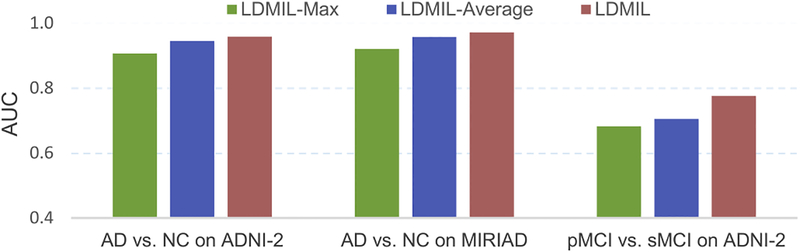
We also investigate the influence of our proposed local-to-global representation for MR images in LDMIL. Specifically, as shown in Fig. 3, we concatenate the local patch-level representation (in FC8) learned from L sub-CNNs, followed by three fully-connected layers to learn the global bag-level representations for the input MR image. Besides the concatenation strategy used in this study, there are also two widely used strategies for aggregating the instance-level representations (Wu et al., 2015) in multi instance learning, i.e., 1) the max operation that focuses on only the representation of the most discriminative patch, and 2) the average operation that focuses on the averaged representation multiple patch-level features. Here, we compare LDMIL with its two variants, i.e., LDMIL-Max and LDMIL-Average that adopt the element-wise max operation and the element-wise average operation for aggregating the outputs of patches, respectively. Similar to the multi-instance CNN model in Wu et al. (2015), followed by a soft-max layer (called FC9-new), FC8 in 3 is transformed into a probability distribution for subjects of two categories (e.g., AD, and NC) in both LDMIL-Max and LDMIL-Average. Also, we add another soft-max layer to transform FC9-new into a two-dimensional probability score vector for binary classification. The AUC values achieved by LDMIL, LDMIL-Max, and LDMIL-Average on three tasks are reported in Fig. 6.
Fig. 6.
Influence of different representation learning strategies for MR images, including 1) LDMIL-Max using instance-level max representation, 2) LDMIL-Average using instance-level averaged representation, and 3) LDMIL using local-to-global representation. Here, classification models are trained on the ADNI-1 dataset.
From Fig. 6, we notice that in almost all cases, our proposed LDMIL that learns local-to-global representations for MR images obtains the best performance, especially in the task of MCI conversion prediction (i.e., pMCI vs. sMCI classification). These empirical results confirm our observation that exploiting both the local and the global structural information of MR images can assist AD-related brain disease diagnosis. Also, LDMIL-Max generally yields the worse performance, compared with LDMIL and LDMIL-Average. This implies that methods focusing on only one single instance (as we do in LDMIL-Max) cannot generate good features for representing the structural changes of the brain, while the atrophy locations may distribute globally in the brain.
4.7. Influence of the number of landmarks
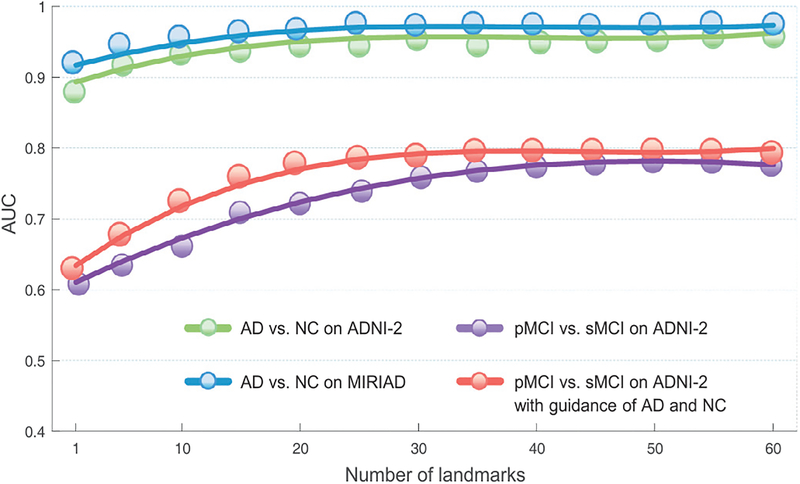
We further investigate the influence of the number of landmarks on the classification performance, by varying it in the set {1, 5, 10, 15, ⋯, 60}. We report the AUC values achieved by the proposed LDMIL method in both AD classification and MCI conversion prediction tasks in Fig. 7. Note that the term “pMCI vs. sMCI on ADNI-2 with the guidance of AD and NC” denotes the method that adopts AD and NC subjects from the ADNI-1 dataset as the guidance information for classifier training (see Section 4.5).
Fig. 7.
Influence of the number of landmarks on the performance of the proposed LDMIL method in tasks of AD vs. NC classification and pMCI vs. sMCI classification. Here, classification models are trained on the ADNI-1 dataset.
From this figure, we can observe that the overall performance increases with the increase in the number of landmarks. Using 1 and 50 landmarks in AD vs. NC classification on ADNI-2, LDMIL achieves the AUC values of 0.9164 and 0.9597, respectively. In particular, in pMCI vs. sMCI classification, LDMIL using less than 15 landmarks cannot yield satisfactory results. This implies that the global information conveyed by multiple landmarks can help boost the learning performance, especially for MCI subjects with no obvious disease-induced structural changes. On the other hand, when the number of landmarks is larger than 30, the growth trend of AUC values slows down, and the results are basically stable. Hence, it is reasonable to choose the number of landmarks in the range of [30, 50], while using more landmarks (e.g., > 55) cannot significantly boost the classification performance and will increase the number of network parameters.
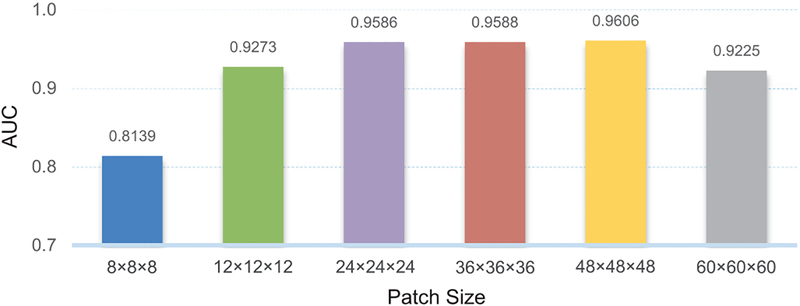
4.8. Influence of the size of image patches
In the above-mentioned experiments, we adopt a fixed patch size (i.e., 24 × 24 × 24) for our proposed LDMIL method. We now investigate the influence of the patch size on the performance of LDMIL, by varying the patch size and testing all the values in the set {8 × 8 × 8, 12 × 12 × 12, 24 × 24 × 24, 36 × 36 × 36, 48 × 48 × 48, 60 × 60 × 60}. In Fig. 8, we report the AUC values of AD vs. NC classification on the ADNI-2 dataset. From this figure, we can see that the best results are obtained by LDMIL using the patch size of 48 × 48 × 48. Also, LDMIL is not very sensitive to the size of the image patch within the range of [24 × 24 × 24, 48 × 48 × 48]. When we use patches with the size of 8 × 8 × 8, the AUC value (0.8139) is not satisfactory. This implies that very small local patches are not capable of capturing enough structural information from the brain. Similarly, the results are not good using very large patches (e.g., 60 × 60 × 60), since subtle structural changes within the large patch could be dominated by uninformative normal regions. In addition, using large patches will bring huge computational burden, and thus affect the utility of our method in practical applications. Besides, we investigate the influences of the number of landmarks and size of image patches on LDSIL, with results given in Fig. S2 in the Supplementary Materials.
Fig. 8.
Influence of the size of image patches on the performance of LDMIL in AD vs. NC classification on the ADNI-2 dataset, with models trained on the ADNI-1 dataset.
5. Discussion
5.1. Comparison with previous studies
Different from conventional voxel-level and whole-image-level feature representations for MRI, the proposed LDMIL method is capable of capturing both local and global information of MR images. Specifically, we first learn patch-level representations via multiple sub-CNNs to model the local information, and further learn bag-level representations to capture the global information of brain MR images. In this way, a local-to-global representation can be automatically extracted from MR images. Different from conventional ROI-based approaches, our LDMIL method does not require any pre-defined ROIs, which is particularly useful in practice.
Compared with conventional patch-based approaches (Tong et al., 2014; Liu et al., 2012), our method can locate discriminative image patches based on anatomical landmarks, and the landmarks are identified by a data-driven landmark discovery algorithm. More specifically, we first identify discriminative anatomical landmarks via group comparison between AD and NC subjects in the training set and then extract image patches centered at multiple landmark locations. Also, while previous patch-based studies usually define engineered features for image patches (Tong et al., 2014; Liu et al., 2012), our LDMIL method can automatically learn representations for patches using an end-to-end learning model. Hence, the learned feature representations in LDMIL are consistent with the subsequent classifier, leading to optimal learning performance. Furthermore, our method only requires weak supervision at a global whole-image-level, where the subject-level class label is assigned to a bag rather than instances in the bag. This can reduce the confusion induced by patches that do not convey any information about the category of the bag.
Besides, the proposed LDMIL method is similar to, but different from, the conventional multi-instance learning (MIL) methods. Since not necessarily all image patches extracted from an MR image are significantly affected by dementia, the class labels for those image patches could be ambiguous, if we simply assign the subject-level label to each of them. In LDMIL, we assign the class label of a subject to a bag other than to each instance (i.e., image patch), and hence we only require bag-level (i.e., subject-level) other than instance-level (i.e., patch-level) class label information for subjects, which is similar to the conventional MIL methods (Yan et al., 2016; Wu et al., 2015). However, different from the conventional MIL methods that focus on only the representation of the most discriminative instance (i.e., image patch) or the averaged representation of multiple instance-level features, our LDMIL method does not assume that at least one instance can determine whether a bag belongs to the positive category. Since the structural changes induced by AD could be subtle and distribute in different areas of the brain, we attempt to learn local-to-global feature representations for brain MRI images. Experimental results in Table 3 and Fig. 6 suggest that the local-to-global representation plays an important role in boosting the learning performance.
5.2. Limitations
Although our proposed LDMIL method achieves promising results in both AD classification and MCI conversion prediction, there are several technical issues to be considered in the future. First, even though we can extract hundreds of thousands of image patches from multiple landmark locations for classifier training, the number of training subjects is limited (i.e., hundreds). Luckily, there are a large number of longitudinal MR images in three datasets (i.e., ADNI-1, ADNI-2, and MIRIAD), which can be utilized to further improve the robustness of the proposed deep learning model. Second, in the current work, we treat landmark detection and landmark-based classification as two stand-alone tasks, which may lead to sub-optimal learning performance. In the future work, we will study to integrate the process of landmark detection and the training of classification models into a unified framework. Specifically, we will design a two-stage deep neural network, where the first-stage network aims to learn features for image patches, and the second-stage network focuses on identifying discriminative landmarks by using the learned features for patches. We could first train these two-stage networks separately, and then jointly optimize them as a whole network. In addition, our LDMIL method is a single-task model, where only the class label is estimated for a given MR image. Actually, there are many clinical scores for each subject, and those scores are related to class labels. Since predicting clinical scores is a regression problem, it is reasonable to develop a multi-task learning model based on LDMIL, where both classification and regression tasks can be learned jointly. Considering the underlying correlation among clinical scores and class labels, the joint learning could further promote the learning performance. Besides, we don’t consider several confounding factors (e.g., age, gender, and education years) of studied subjects. In future work, we will address these confounding factors by incorporating them into the proposed deep learning framework. Furthermore, in this work, we consider only the problem of brain disease diagnosis via the proposed landmark-based deep learning framework, based on the baseline MRI data in ADNI-1, ADNI-2, and MIRIAD. It is interesting to develop a deep learning framework for predicting the progression of brain diseases based on the baseline data, which will also be our future work.
6. Conclusion
We have presented a landmark-based deep multi-instance learning (LDMIL) framework for AD-related brain disease diagnosis using MR imaging data. To model both local and global information of the brain, we first proposed to localize image patches (through the detection of anatomical landmarks) for the subjects, and then adopted a multi-instance CNN model to perform end-to-end classification. Experiments on a large cohort of subjects from three datasets show that our method achieves better performance compared to the state-of-the-art approaches, especially in the task of MCI conversion prediction.
Supplementary Material
Acknowledgements
This study was supported by NIH grants (EB006733, EB008374, EB009634, MH100217, AG041721, AG042599, AG010129, AG030514). Data used in the preparation of this article were partially obtained from the Alzheimer’s Disease Neuroimaging lnitiative (ADNI) dataset and the Minimal lnterval Resonance lmaging in Alzheimer’s Disease (MIRIAD) dataset. The investigators within the ADNI and the MIRIAD did not participate in analysis or writing of this study. A complete listing of ADNI investiga tors can be found online (https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf). The MIRIAD dataset is made available through the support of the UK Alzheimers Society (Grant RF116), while the original data collection was funded by an unrestricted educational grant from GlaxoSmithKline (Grant 6GKC).
Footnotes
Supplementary material
Supplementary material associated with this article can be found, in the online version, at 10.1016/j.media.2017.10.005.
References
- Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, et al. , 2016. Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467. [Google Scholar]
- Amores J, 2013. Multiple instance classification: review, taxonomy and comparative study. Artif. Intell 201, 81–105. [Google Scholar]
- Andrews S, Tsochantaridis I, Hofmann T, 2002. Support vector machines for multiple-instance learning In: Advances in Neural Information Processing Systems, pp. 561–568. [Google Scholar]
- Ashburner J, Friston KJ, 2000. Voxel-based morphometrythe methods. Neurolm-age 11 (6), 805–821. [DOI] [PubMed] [Google Scholar]
- Atiya M, Hyman BT, Albert MS, Killiany R, 2003. Structural magnetic resonance imaging in established and prodromal Alzheimer disease: a review. Alzheimer Dis. Assoc. Disord 17 (3), 177–195. [DOI] [PubMed] [Google Scholar]
- Baron J, Chetelat G, Desgranges B, Perchey G, Landeau B, De La Sayette V, Eustache F, 2001. In vivo mapping of gray matter loss with voxel-based morphometry in mild alzheimer’s disease. Neurolmage 14 (2), 298–309. [DOI] [PubMed] [Google Scholar]
- Bi J, Liang J, 2007. Multiple instance learning of pulmonary embolism detection with geodesic distance along vascular structure In: IEEE Conference on Computer Vision and Pattern Recognition. IEEE, pp. 1–8. [Google Scholar]
- Boyd S, Vandenberghe L, 2004. Convex Optimization. Cambridge University Press. de Brebisson A, Montana G, 2015. Deep neural networks for anatomical brain segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 20–28. [Google Scholar]
- Cao X, Yang J, Gao Y, Guo Y, Wu G, Shen D, 2017. Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis. Med. Image Anal 41, 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Liu M, Zhang D , Munsell BC, Shen D, 2015. Domain transfer learning for MCI conversion prediction. IEEE Trans. Biomed. Eng 62 (7), 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheplygina V, Tax DM, Loog M, 2016. Dissimilarity-based ensembles for multiple instance learning. IEEE Trans. Neural Netw. Learn. Syst 27 (6), 1379–1391. [DOI] [PubMed] [Google Scholar]
- Chu C, Hsu A-L, Chou K-H, Bandettini P, Lin C, 2012. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. Neurolmage 60 (1), 59–70. [DOI] [PubMed] [Google Scholar]
- Coupé P, Eskildsen SF, Manjon JV, Fonov VS, Pruessner JC, Allard M, Collins DL, Initiative ADN, et al. , 2012. Scoring by nonlocal image patch estimator for early detection of Alzheimer’s disease. Neurolmage 1 (1), 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert M-O, Chupin M, Benali H, Colliot O, 2011. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neurolmage 56 (2), 766–781. [DOI] [PubMed] [Google Scholar]
- De Jong L, Van der Hiele K , Veer I, Houwing J, Westendorp R, Bollen E , De Bruin P , Middelkoop H, Van Buchem M, Van Der Grond J, 2008. Strongly reduced volumes of putamen and thalamus in Alzheimer’s disease: an MRI study. Brain 131 (12), 3277–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich TG, Lathrop RH, Lozano-Pérez T, 1997. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell 89 (1), 31–71. [Google Scholar]
- Dubois B, Chupin M, Hampel H, Lista S, Cavedo E, Croisile B , Tisserand GL, Touchon J, Bonafe A, Ousset PJ, et al. , 2015. Donepezil decreases annual rate of hippocampal atrophy in suspected prodromal Alzheimer’s disease. Alzheimer’s Dementia 11 (9), 1041–1049. [DOI] [PubMed] [Google Scholar]
- Filipovych R, Davatzikos C, lnitiative ADN, et al. , 2011. Semi-supervised pattern classification of medical images: application to mild cognitive impairment (MCl). NeuroImage 55 (3), 1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM, 2000. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Nat. Acad. Sci 97 (20), 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R , 2001. The elements of statistical learning, 1. Springer series in statistics Springer, Berlin. [Google Scholar]
- Gartner T, Flach PA, Kowalczyk A, Smola AJ, 2002. Multi-instance kernels. In: International Conference on Machine Learning, 2, pp. 179–186. [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC, 1998. Enhancement of MR images using registration for signal averaging. J. Comput. Assist. Tomogr 22 (2), 324–333. [DOI] [PubMed] [Google Scholar]
- Jack C, Petersen RC, Xu YC, OBrien PC, Smith GE, Ivnik RJ, Boeve BF , Waring SC, Tangalos EG , Kokmen E, 1999. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology 52 (7), 1397–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ , L Whitwell J, Ward C, 2008. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27 (4), 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, O’Brien PC, Tangalos EG, 1992. MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology 42 (1), 183–183 [DOI] [PubMed] [Google Scholar]
- Jain A, Nandakumar K, Ross A, 2005. Score normalization in multimodal biometric systems. Pattern Recognit. 38 (12), 2270–2285. [Google Scholar]
- KlÖppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RS, 2008. Automatic classification of MR scans in Alzheimer’s disease. Brain 131 (3), 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D, 2012. Ensemble sparse classification of Alzheimer’s disease. Neurolmage 60 (2), 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D, 2015. View-centralized multi-atlas classification for Alzheimer’s disease diagnosis. Hum. Brain Mapp. 36 (5), 1847–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D, 2016. Relationship induced multi-template learning for diagnosis of Alzheimer’s disease and mild cognitive impairment. IEEE Trans. Med. Imaging 35 (6), 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M , Zhang J, Yap P-T, Shen D , 2017. View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data. Med. Image Anal. 36, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Qian Z, Marvasty I, Rinehart S, Voros S, Metaxas DN, 2010. Lesion-specific coronary artery calcium quantification for predicting cardiac event with multiple instance support vector machines In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 484–492. [DOI] [PubMed] [Google Scholar]
- LÖtjÖnen J, Wolz R, Koikkalainen J, Julkunen V, Thurfjell L, Lundqvist R, Walde-mar G , Soininen H, Rueckert D, Initiative ADN, et al. , 2011. Fast and robust extraction of hippocampus from MR images for diagnostics of Alzheimer’s disease. NeuroImage 56 (1), 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Bi J, Wolf M, Salganicoff M, 2011. Effective 3D object detection and regression using probabilistic segmentation features in CT images In: IEEE Conference on Computer Vision and Pattern Recognition. IEEE, pp. 1049–1056. [Google Scholar]
- Maguire EA, Gadian DG, Johnsrude IS, Good CD , Ashburner J, Frackowiak RS, Frith CD , 2000. Navigation-related structural change in the hippocampi of taxi drivers. Proc. Nat. Acad. Sci 97 (8), 4398–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone IB , Cash D, Ridgway GR, MacManus DG, Ourselin S, Fox NC, Schott JM, 2013. MIRIAD-Public release of a multiple time point Alzheimer’s MR imaging dataset. NeuroImage 70, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardia K, 1975. Assessment of multinormality and the robustness of Hotelling’s T2 test. Appl. Stat 163–171. [Google Scholar]
- Maron O, Lozano-Pérez T, 1998. A framework for multiple-instance learning. Adv. Neural Inf. Process. Syst 570–576. [Google Scholar]
- Matthews BW, 1975. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochimica et Biophysica Acta (BBA)-Protein Struct. 405 (2), 442–451. [DOI] [PubMed] [Google Scholar]
- Miao S, Wang ZJ, Liao R, 2016. A CNN regression approach for real-time 2D/3D registration. IEEE Trans. Med. Imaging 35 (5), 1352–1363. [DOI] [PubMed] [Google Scholar]
- Montagne A, Barnes SR , Sweeney MD, Halliday MR, Sagare AP, Zhao Z, Toga AW , Jacobs RE, Liu CY, Amezcua L, et al. , 2015. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 85 (2), 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradi E, Pepe A,Gaser C,Huttunen H, Tohka J, Initiative ADN, et al. , 2015. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. NeuroImage 104, 398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Davatzikos C, 2002. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imaging 21 (11), 1421–1439. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC, 1998. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging 17 (1), 87–97. [DOI] [PubMed] [Google Scholar]
- Small GW, Ercoli LM, Silverman DH, Huang S-C, Komo S, Bookheimer SY, Lavretsky H, Miller K, Siddarth P, Rasgon NL, et al. , 2000. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer’s disease. Proc. Nat. Acad. Sci 97 (11), 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Wolz R, Gao Q, Guerrero R, Hajnal JV, Rueckert D, 2014. Multiple instance learning for classification of dementia in brain MRI. Med. Image Anal. 18 (5), 808–818. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Del-croix N, Mazoyer B, Joliot M, 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 15 (1), 273–289. [DOI] [PubMed] [Google Scholar]
- Wang Y, Nie J , Yap P-T , Shi F, Guo L , Shen D, 2011. Robust deformable-surface-based skull-stripping for large-scale studies In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2011. Springer, pp. 635–642. [DOI] [PubMed] [Google Scholar]
- Wolz R , Aljabar P , Hajnal JV , Lotjonen J , Rueckert D , 2012. Nonlinear dimensionality reduction combining MR imaging with non-imaging information. Med. Image Anal. 16 (4), 819–830. [DOI] [PubMed] [Google Scholar]
- Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y, 2009. Robust face recognition via sparse representation. IEEE Trans. Pattern Anal. Mach. Intell 31 (2), 210–227. [DOI] [PubMed] [Google Scholar]
- Wu J , Yu Y, Huang C, Yu K, 2015. Deep multiple instance learning for image classification and auto-annotation In: IEEE Conference on Computer Vision and Pattern Recognition. IEEE, pp. 3460–3469. [Google Scholar]
- Xiang S, Yuan L , Fan W , Wang Y, Thompson PM , Ye J , 2014. Bi-level multisource learning for heterogeneous block-wise missing data. NeuroImage 102, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang J, Eric I, Chang C, Lai M, Tu Z, 2012. Context-constrained multiple instance learning for histopathology image segmentation In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 623–630. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu J-Y, Eric I, Chang C, Lai M, Tu Z, 2014. Weakly supervised histopathology cancer image segmentation and classification. Med. Image Anal. 18 (3), 591–604. [DOI] [PubMed] [Google Scholar]
- Yamasue H, Kasai K, Iwanami A , Ohtani T , Yamada H, Abe O, Kuroki N, Fukuda R , Tochigi M, Furukawa S, et al. , 2003. Voxel-based analysis of MRI reveals anterior cingulate gray-matter volume reduction in posttraumatic stress disorder due to terrorism. Proc. Nat. Acad. Sci 100 (15), 9039–9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Zhan Y, Peng Z, Liao S, Shinagawa Y, Zhang S, Metaxas DN, Zhou XS, 2016. Multi-instance deep learning: discover discriminative local anatomies for bodypart recognition. IEEE Trans. Med. Imaging 35 (5), 1332–1343. [DOI] [PubMed] [Google Scholar]
- Yang X, Kwitt R, Niethammer M , 2016. Fast predictive image registration In: International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. Springer, pp. 48–57. [Google Scholar]
- Zhang C, Platt JC , Viola PA, 2005. Multiple instance boosting for object detection In: Advances in Neural Information Processing Systems, pp. 1417–1424. [Google Scholar]
- Zhang J, Gao Y, Gao Y, Munsell B, Shen D , 2016. Detecting anatomical landmarks for fast Alzheimer’s disease diagnosis. IEEE Trans. Med. Imaging 35 (12), 2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liang J, Zhao H, 2013. Local energy pattern for texture classification using self-adaptive quantization thresholds. IEEE Trans. Image Process. 22 (1), 31–42. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu M, An L, Gao Y, Shen D, 2017. Alzheimer’s disease diagnosis using landmark-based features from longitudinal structural MR images. IEEE J. Biomed. Health Inform,. DOI: 10.1109/JBHI.2017.2704614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu M, Shen D, 2017. Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Trans. Image Process. 26 (10), 4753–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Goldman SA, 2001. EM-DD: an improved multiple-instance learning technique In: Advances in Neural Information Processing Systems, pp. 1073–1080. [Google Scholar]
- Zhang Y, Brady M, Smith S, 2001. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20 (1), 45–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.