Abstract
The use of protein nanoparticles for biosensing, biocatalysis and drug delivery has exploded in the last few years. The ability of protein nanoparticles to self-assemble into predictable, monodisperse structures is of tremendous value. The unique properties of protein nanoparticles such as high stability, and biocompatibility, along with the potential to modify them led to development of novel bioengineering tools. Together, the ability to control the interior loading and external functionalities of protein nanoparticles makes them intriguing nanodevices. This review will focus on a number of recent examples of protein nanoparticles that have been engineered towards imparting the particles with biocatalytic or biosensing functionality.
Introduction
Protein nanoparticles (PNPs) are made of multiple protein-based building blocks assembled either by protein-protein or protein-nucleic acid interactions [1]. It is well known that immobilization of functional biomolecules either onto the exterior or the inside of PNPs improves their stability and activity [2]. This feature when combined with the ability of PNPs to tolerate extreme conditions make PNPs an ideal platform for a wide range of bionanotechnology applications in biomedicine [2–4] and biocatalysis [5,6].
Although most PNPs used today are based on biologically evolved protein assemblies such as viral capsids [5,7] or multimeric enzymes [6,8], there is a move toward engineering these protein nano-assemblies based on in silico design [9,10]. The ability to provide multi-functionalization is often highly challenging as the modifications may result in a loss of nanoparticle self-assembly or protein functionality. In this review, we will discuss recent progress in PNP functionalization and their corresponding applications in biocatalysis and biomedical diagnosis and imaging. Table 1 summarizes the different types of PNPs reviewed in this paper and their corresponding applications.
Table 1.
Size and origin of protein nanoparticles in this review.
| Protein Cage | Species | Shape | Size (nm) | Applications | References |
|---|---|---|---|---|---|
| Viral-Like-Particles (VLPs) | |||||
| P22 | Salmonella typhimurium | Icosahedral | 58–64 | Biocatalysis | [12,49–51] |
| Qβ | E. coli | Icosahedral | 28 | Biosensor Delivery | [14,18,39,58] |
| AP205 | Acinetobacter species | Icosahedral | 20 | Vaccine | [15] |
| CCMV | Cowpea plant | Icosahedral | 28 | Biocatalysis | [16,54,61] |
| HBV | Human | Icosahedral | 30–34 | Biosensor Vaccine Delivery MR Imaging | [17,41,43,45,46,53,59] |
| Fd | E. coli | Filamentous | 7 × 880 | Biosensor | [35] |
| MS2 | E. coli | Icosahedral | 27 | Biosensor | [38] |
| TMV | Tobacco Plant | Rod | 18 × 300 | Biosensor | [40,44,62] |
| M13 | E. coli | Filamentous | 7 × 880 | Biosensor | [42] |
| Enzyme-derived PNPs | |||||
| E2 | Bacillus stearothermophilus | Spherical | 24 | Biocatalysis, Biosensor Delivery | [6,22,23,37] |
| Lumazine synthase | Aquifex aeolicus | Spherical | 15.4 | Biocatalysis, Biosensor | [8,21,24,25,55,56] |
| Ferritin | Human | Spherical | 12 | Delivery MR Imaging | [20,28,57] |
| Ferritin | E. coli | Spherical | 12 | Photothermal Therapy | [26,60] |
| Ferritin | Pyrococcus furiosus | Spherical | 12 | Biosensor MR Imaging | [27] |
| Encapsulin | Mycobacterium Tuberculosis | Spherical | 22 | Biocatalysis | [29] |
| Encapsulin | Thermotoga maritima | Spherical | 24 | BIosensor Delivery | [30,36] |
| Reencapsulin | Rhodococcus erythropolis | Spherical | 28 | Biosensor | [31] |
| Pdu BMC | Citrobacter freundii | Spherical | 100–150 | Biocatalysis | [52,63] |
| Synthetic PNPs | |||||
| O3–33 | Synthetic | Octahedral | 13 | TBD | [9] |
| T3–10 | Synthetic | Tetrahedral | 11 | TBD | [9] |
| I53 | Synthetic | Icosahedral | 24–40 | TBD | [10] |
| I52 | Synthetic | Icosahedral | 24–40 | TBD | [10] |
| I32 | Synthetic | Icosahedral | 24–40 | TBD | [10] |
| CCMV-avidin crystal | CCMV and avidin hybrid | Body-centered-cubic | Unit cell: 35 | TBD | [32] |
| ATC cage | Hyrbid from E. coli components | Spherical | 22.5 | TBD | [33] |
Virus-Like particles
Virus-Like Particles (VLPs) are naturally occurring multimeric PNPs derived from viruses. Unlike intact viruses, VLPs are derived only from the noninfectious protein capsids and provide the benefit of mass-production using simple recombinant DNA technology [11]. Similar to other protein-based materials, VLPs can be modified with a wide range of cargos such as drugs, enzymes, or other biomolecules using either genetic manipulation, post-translational conjugation, and/or encapsulation [5,12]. Traditionally, modified VLPs have been widely used for vaccine design and drug delivery because of their biocompatibility and stability [7]. However, efforts toward the use of engineered VLPs as nanoscale bioreactors are gaining traction due to their ability to tolerate harsh conditions [13–14].
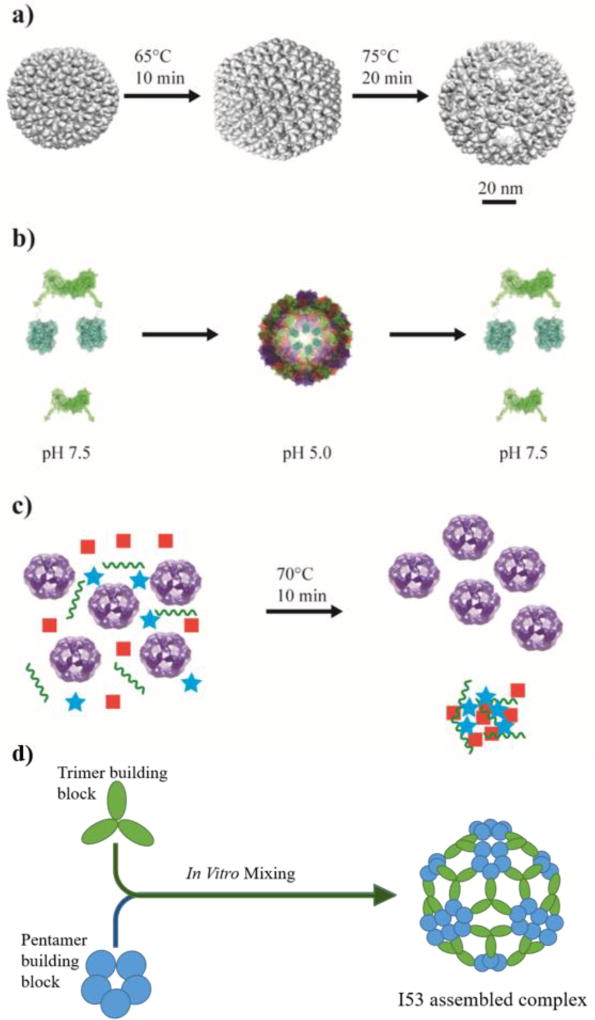
In addition to direct tethering, biomolecular cargos can also be encapsulated within some VLPs by exploiting their ability to undergo a conformational change upon exposure to thermal or pH variations. One example is the bacteriophage P22, which self-assembles into a 58 nm capsid based on the interaction between the coat proteins (CP) and a helix-turn motif on the scaffold proteins (SP) [12]. Heating of P22 VLPs results in an irreversible change in the structure as some of the SP subunits are released, leading to an expanded shell of 64 nm. Further heating to 75°C results in the formation of a “Wiffleball” structure with an almost two-fold increase in the effective volume and multiple 10 nm holes within the icosahedral structure (Figure 1a) [12]. Cowpea Chlorotic Mottle Virus (CCMV) is a second VLP that offers reversible assembly and allows facile incorporation of cargos to the inside based on electrostatic interactions between the positively charged N-terminus and the negatively charged cargo. The assembly/disassembly pathway can be easily controlled by pH and ionic strength (Figure 1b) [16].
Figure 1.
Examples of protein nanoparticles with applicable features. a) The VLP from bacteriophage P22 self-assembles into a 58 nm capsid, upon heating, the capsid expands allowing the internal volume of the nanoparticle to increase. Further heating results in the opening of pores in the structure known as a “wiffleball” structure. Adapted with permission from Patterson DP, Prevelige PE, Douglas T: Nanoreactors by programmed enzyme encapsulation inside the capsid of the bacteriophage P22. ACS Nano 2012, 6:5000–5009. Copyright 2012, American Chemical Society. b) Cowpea Chlorotic Mottle Virus (CCMV) VLPs are formed by mixing shell proteins (green) with or without fused cargo proteins (blue) at pH 7.5 then dialyzing the protein mixture at pH 5.0 overnight to form capsids. This process can be reversed by raising the pH back to 7.5. Adapted with permission from Rurup WF, Verbij F, Koay MS, Blum C, Subramaniam V, Cornelissen JJ: Predicting the loading of virus-like particles with fluorescent proteins. Biomacromolecules 2014, 15:558–563. Copyright 2014, American Chemical Society. c) The thermophilic nature of E2 (purple spheres) allows for simple purification by heating to 70°C allowing most cellular proteins to denature and be removed by centrifugation. Adapted with permission from Sun Q, Chen Q, Blackstock D, Chen W: Post-Translational Modification of Bionanoparticles as a Modular Platform for Biosensor Assembly. ACS Nano 2015, 9:8554–8561. Copyright 2015, American Chemical Society. d) Computational design was used to create synthetic protein nanoparticles which self-assemble into icosahedral structures by simply mixing two separate multimeric proteins. For example, I53 architecture was obtained through mixing trimer and pentamer components with computationally optimized protein-protein interactions.
Some VLPs have the benefit of incorporating functional cargos into their interior through direct genetic fusions to termini oriented towards the interior, as in the case of Hepatitis B Virus (HBV) [17]. Another example is the bacteriophage Qβ, which utilizes the natural interaction between the RNA genome and the capsid for cargo encapsulation [13,18].
In addition to surface functionalization and cargo loading, the ability to immobilize PNPs onto surfaces has gained interest for biosensing applications. In the case of Tobacco mosaic virus (TMV), the 5’-end of TMV nanorods can be partially disassembled at neutral pH to expose its mRNA, which can be used for docking TMVs to hydrogel surfaces via DNA hybridization [19]. Additional functionalization was achieved by using bioorthogonal tetrazine (Tz)-trans-cyclooctene (TCO) “click” chemistry to selectively attach R-Phycoerythrin (R-PE) to TMV nanorods immobilized on hydrogel microparticles. This technology allows higher capacity of protein conjugation to the surface of a hydrogel for improved and controlled protein conjugation and biosensing applications.
Enzyme-derived PNPs
Another class of PNPs that is gathering more attention is based on multimeric enzymes that naturally self-assemble into nanocages of varying sizes [20–22]. These nanostructures provide naturally-evolved, confined reaction spaces for enhanced cellular metabolism and are ideal candidates as nanoreactors. The E2 core from the pyruvate dehydrogenase complex of Bacillus stearothermophilus is a frequently used PNP because of its thermophilic origin [23]. E2 is composed of 60 identical monomers that self-assemble into a highly stable cage-like structure with an outer diameter of ~24 nm [23]. The thermostability of E2 nanocages allows for simple purification by heating to 70°C to denature cellular proteins without affecting the E2 assembly (Figure 1c) [22].
Lumanize synthase (AaLS) is another readily modifiable PNP, roughly 15 nm in diameter, capable of carrying cargo because of its highly charged interior [24]. A highly toxic HIV protease was incorporated into the interior of AaLS PNPs based on electrostatic interaction by tethering a deca-arginine peptide to the C-terminus, thereby shielding bacterial proteins from degradation [21]. The number of encapsulated proteins can be fine-tuned by controlling the AaLS:protein ratio [25], paving the way for easy encapsulation of multiple protein cargos.
Ferritin (Fn), an ubiquitous cage protein found in all domains of life, is composed of 24 subunits that assemble into a 12 nm hollow spherical structure and naturally encapsulates Fe3O4 [20,26,27]. A key structural attribute of Fn is the ability to disassemble at acidic pH and reassemble at neutral pH with perfect shape memory [28]. In addition to the disassembly/reassembly mechanism, ferritins are also thermodynamically stable, can bind different metal ions [26] and are tolerant of genetic fusions [20] making Fn a highly multifunctional PNP capable of a number diagnostic and biosensing applications.
Encapsulin, a 30 nm protein nanocage, is known to be involved in oxidative stress responses, and has been found in a number of different species, including some thermophiles [29–31]. Encapsulin is unique in its ability to natively package cargo proteins via interaction of a unique C-terminal signal peptide with the shell protein. Artificial encapsulin-based nanocompartments have begun to receive considerable interest because of their potential to allow targeted encapsulation of drugs, siRNAs and proteins of interest [29].
Synthetic PNPs
Novel synthetic nanocompartments can be created by designing protein building blocks that are able to self-assemble into a cage-like structure. Two different protein building blocks with opposite charges, based on the CCMV monomer and avidin, were used to form a synthetic PNP [32]. The heterogeneous crystal of this protein block copolymer formed a packed body-centered cubic structure that is distinct from the icosahedral assembly that CCMV naturally forms. The protein nanocage was fluorescently-labeled or decorated with proteins using the biotin-avidin interaction [32]. Even completely synthetic PNPs can be designed based on the use of natively oligomeric protein components. Computational models were used to generate a cubic nanocage when dimeric and trimeric E. coli proteins were fused together with intersecting angles of symmetry of 36.5° [33]. This type of synthetic design greatly expands the repertoire of PNPs available for bioengineering applications.
Synthetic PNPs with intricate structures can be computationally designed by taking advantage of the symmetry in multimeric protein components to minimize the protein-protein interfaces that need to be redesigned [9,10]. This is done by grafting multimeric proteins to a target structure and using computational tools, such as RosettaDesign, to design the low-energy, weak, noncovalent protein-protein interactions that mimic those from naturally derived PNPs. This strategy has been used in creating synthetic octahedral (O3–33) and tetrahedral (T3–10) PNPs [9] as well as larger viral-like icosahedral structures (I53, I52, and I32) [10]. A unique property of the icosahedral PNPs made from two distinct multimeric protein components is the controlled in vitro self-assembly of these structures through simple mixing of the different components (Figure 1d). While computational models can design a library of possible structures, extensive experimental evaluation is necessary to select the few PNPs which form stable structures.
Functionalization of Protein Nanoparticles
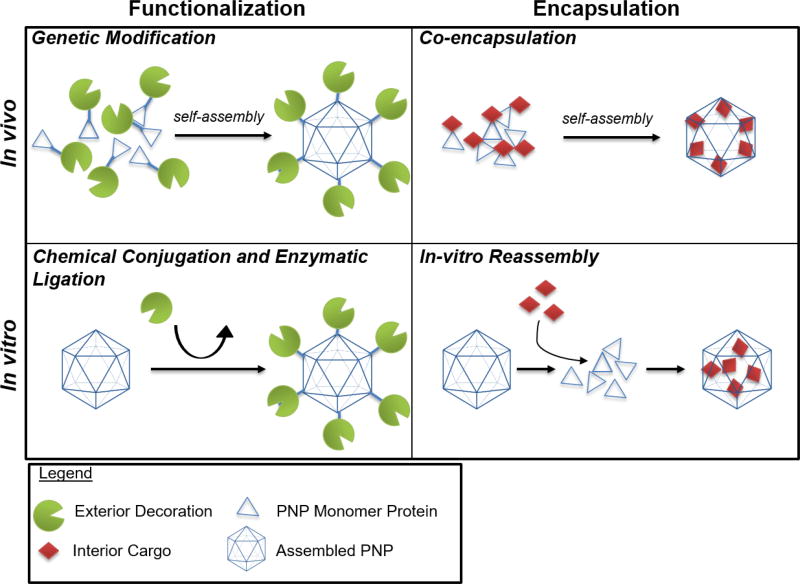
In order to customize protein nanoparticles toward a particular application, multiple functionalities are needed. PNPs are naturally robust nanomaterials, which lend themselves to many well-characterized chemical conjugation methods. As PNPs are composed of protein monomers, genetic modification strategies have also been employed. Additionally, protein engineering has expanded the biological tools available to post-translationally modify these components. The selection of a right modification strategy is critical to preserving the integrity and the function of the resulting PNPs for the desired applications (Figure 2) [34].
Figure 2.
In vivo and in vitro functionalization of PNPs. Protein nanoparticles can be loaded with a protein(s) of interest by genetic fusion to the monomer shell protein. As the shell proteins self-assemble in vivo, the nanoparticle will become decorated with the protein(s) of interest. For nanocages that aren’t amenable to genetic fusions, proteins can be decorated on the surface of the particle through chemical or enzymatic conjugation. Small molecules or proteins can be encapsulated in vivo by fusion to an interaction domain that will associate with the inner core of a nanoparticle. Small molecules can be encapsulated PNPs by mixing in vitro provided the conditions present are sufficient for disassembly of the PNP to incorporate the molecules upon reassembly of the PNP.
Chemical Conjugations
Chemical conjugation methods are often used to attach inorganic, non-proteinaceous components such as chemical dyes for imaging [28,35], targeting ligands for drug delivery [36], and peptides for immune activation [37]. Typically, well-characterized chemistries can be employed to couple substances to reactive side chains that are either surface exposed or genetically incorporated into the protein monomer backbone. Site-specific bio-orthogonal labeling of PNPs can be achieved through substitution of unnatural amino acids using either amber, ocher, opal, four base pair, or methionine codons [38–41]. Such chemistries, however, lack control over orientation of the attachment and location on the nanoparticle surface and can sometimes deactivate protein cargos [15,42].
Genetic Modifications
Many have explored direct protein attachments by modifying protein monomers at the genetic level. This allows guest proteins of interest to be fused directly to the monomers and co-assemble into the resulting PNPs. This strategy has been employed effectively in self-assembling VLPs and other PNPs where the N or C terminus or a flexible loop is exposed [20,43]. The Hepatitis B Virus (HBV) VLP contains an exterior loop in which foreign domains and whole proteins can be inserted within the peptide chain as long as the distance between the N and C terminus of the guest protein is geometrically compatible with the loop [43]. Walker and others were able to alleviate this steric constraint by splitting the core protein monomer at this loop position [43]. Not only was this “SplitCore” protein able to form HBV VLPs, but the divide created additional exposed N and C termini in which each could be functionalized through direct fusions in order to display full native proteins relevant to vaccine development [43].
While many PNP systems cannot support direct fusions of large proteins as they can interfere with self-assembly [6,42], many platforms can afford the fusion of small functional peptides [6,8,36,44]. For instance, the tobacco mosaic virus (TMV) nanorod VLP monomers can be modified at the C-terminus with a FLAG tag [44]; however, an “IQ” peptide for Genhance 680 fluorophore binding was found to interfere with the folding and assembly. In an effort to create more space on the VLP surface, a longer, flexible linker was added to enhance folding. Additionally an amber codon was incorporated before the peptide sequence, leading to periodic truncated expression so that not every core protein was modified [44]. Similar mosaic PNPs with a mixture of modified and wild-type core protein can be achieved by co-expressing both versions in vivo [45] or dissociating the PNPs and mixing them in vitro [16,46]. However, co-expression of both versions lacks direct control over the decoration density, while disassembly for in vitro reassembly requires harsh conditions, unless the PNP contains a natural assembly/disassembly switch, such as CCMV [16].
Enzymatic Ligation
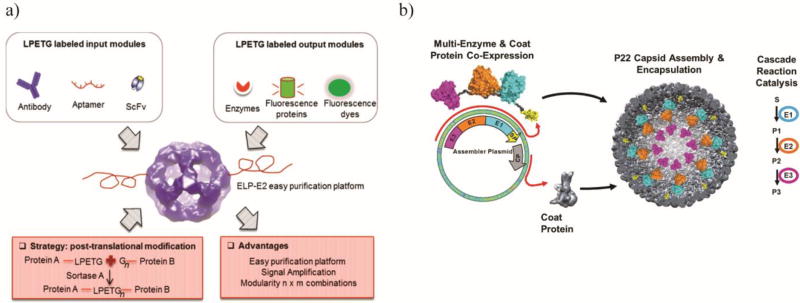
The main disadvantage of protein nanoparticle modification through direct genetic fusions is the limitation in incorporating multiple components. An alternative strategy would be to incorporate domains that can be manipulated post-assembly. This provides a site-specific, covalent linkage with controlled orientation onto the surface of the nanoparticle structure. Sortase A from Staphylococcus aureus is often used for its tail-to-head ligations between a small C-terminal LPXTG motif and small N-terminal poly-glycine motif [47]. Sortase A has been utilized for the functionalization of a triglycine modified E2 protein nanocage [6,22]. Our group demonstrated the ligation of LPETG modified elastin-like polypeptide (ELP) and a tetrameric β-galactosidase onto E2 [6]. The latter exemplifies the utility of this post-assembly mechanism in coordinating the quaternary structures of both the multimeric enzyme and the protein nanoparticle without interfering with self-assembly. This decoration strategy also provides a level of control over the ligation density as well as the modular ability to incorporate multiple decorations. Three LPETG-tagged components, an ELP purification tag, an IgG binding Z-domain, and a nanoluciferase, can be sequentially ligated onto the E2 surface. By controlling the ratios of the reacting LPETG tagged components, the number ligated to E2 can be easily controlled [22].
For VLPs made from multiple distinct coat proteins, the use of orthogonal ligation tags allows the ability to independently control the ligation of separate proteins of interest to different parts of the VLP. Sortase A from Streptococcus pyogenes catalyzes the ligation between a C-terminal LPXTA motif and a poly-alanine N-terminal motif. By adding a G5 tag to the N-terminus of two end proteins p3 and p9 in conjunction with an N-terminal AA tag to the p8 phage body protein, a single M13 phage was dually labeled with the TAMRA dye on p8 and a single domain antibody VHH7 on p3 using these two orthogonal sortases [42].
In addition to sortases, covalent linkage through isopeptide bonds has also been recently employed in PNP decoration [15]. The CnaB2 domain from the fibronectin binding protein of Streptococcus pyogenes naturally forms an intramolecular isopeptide bond [48]. This protein was engineered into two, split components, named SpyCatcher and SpyTag, which retain the bond formation between the pair upon complementation. Using this system, the 15 kDa Spy-catcher was genetically fused to the N-terminus of the coat protein of the bacteriophage AP205 [15], which allowed the covalent conjugation of various malarial and cancerous antigens containing the 13 residue SpyTag. This created a modular “Plug-and-Display” vaccine carrier where theoretically any antigen of interest could be placed onto the same adapter carrier. While the SpyCatcher reaction is quick, adaptable to a variety of buffer conditions, and independent of calcium ions, like many sortases, the use of this pair in PNPs is reliant on the PNP to tolerate the fusion of the whole SpyCatcher or SpyTag peptide.
Cargo Encapsulation
VLP and protein cage structures have the additional utility of incorporating functional cargos into their interior lumen through either co-encapsulation in vivo or in vitro encapsulation. Co-encapsulation utilizes genetic fusions to termini oriented towards the interior [17], or accessory proteins or signal peptides, which direct them into the interior during formation. The P22 phage encapsulation system had been used for a variety of proteins and enzymes because its unique SP can specifically direct guest proteins of interest into the VLP with high loading efficiency [12,49–51]. This system has been used for the encapsulation of a homotetrameric protein CelB [51] as well as a three-enzyme cascade into the interior [49]. Bacterial nanocompartments, such as encapsulins, employ a similar strategy in which a conserved targeting sequence is fused to either the N or C terminus of the protein which localizes the cargo to the interior [16,29–31,52]. Using the natural interaction of genomic material and capsid in the Qβ VLPs, a dual binding RNA-adapter with binding motifs for the capsid monomer and the cargo was used to package fluorescent proteins or enzymes inside the VLP [14,18].
Encapsulation in vitro is also used to incorporate cargos into the nanoparticle. This is accomplished through denaturation of the cage structure and reassembling it in the presence of cargos [28,53]. In many cases the specificity and loading efficiency can be increased through the use of electrostatics [21,25,38,54–56]. Unfortunately, complete modification of each core monomer with internal cargos often interferes with VLP assembly, and a more successful approach for cargo loading was achieved by in vitro mixing of genetically modified and unmodified core monomers [16].
Biosensing and Bioimaging Applications of PNPs
PNPs are ideally suited for sensing and imaging applications as they provide a high surface-area-to-volume ratio [57] and the potential to be simultaneously modified in both the interior and exterior [58] for multifunctional capabilities [22]. Functional molecules such as fluorophores, metal nanoparticles, antibodies, and aptamers have been displayed on the PNP surface to create new functionalities [22,38,58,59]. Taking advantage of the ability to modulate the number of nanoluciferase for signal amplification using Sortase A, highly sensitive E2-based nanosensors using either antibodies, or aptamers, as the detection moieties were created [22]. This method offers the advantage of modularity of the input and output signal and simple purification of nanoparticles [22] (Figure 3a). PNPs based on human apoferritin (AFTN) were also modified to create a modular platform for sensitive biomolecular diagnosis. AFTN heavy chain was genetically fused to protein G followed by a hexahistidine tag that allows binding of antibodies as well as Ni-NTA nanoparticles to the PNP surface. This modular assembly can be used to bind QDs, metal, and magnetic nanoparticles for immunofluorescence, electron microscopy, and Magnetic Resonance Imaging (MRI) [57]. The ability of ferritin nanocages to naturally encapsulate ~4500 iron atoms [26,60] and the possibility to display fluorescent proteins were exploited for enhanced detection of tumors [20,26]. Matsumoto and coworkers engineered Fn PNPs to increase iron uptake by 3-fold, which resulted in a five-fold enhancement in the MRI imaging resolution [27].
Figure 3.
Examples of PNP biosensors and biocatalyst. a) The E2 PNP has the ability to be decorated with multiple sensing and output domains allowing it to be an efficient biosensor against a number of targets. Reprinted with permission from Sun Q, Chen Q, Blackstock D, Chen W: Post-Translational Modification of Bionanoparticles as a Modular Platform for Biosensor Assembly. ACS Nano 2015, 9:8554–8561. Copyright 2015, American Chemical Society. b) A three enzyme synthetic metabolon was incorporated into the P22 VLP to create a more efficient bioreactor. The three enzymes and shell protein were co-expressed and allowed to self-assemble into a nanobioreactor. Reprinted with permission from Patterson DP, Schwarz B, Waters RS, Gedeon T, Douglas T: Encapsulation of an enzyme cascade within the bacteriophage P22 virus-like particle. ACS Chem Biol 2014, 9:359–365. Copyright 2014, American Chemical Society.
Loading fluorescent proteins inside PNPs while allowing the exterior for decoration with functional receptors make these engineered PNPs a desirable platform for molecular imaging. Packing Qβ nanoparticles with GFP while displaying CD22 receptor ligand on the surface resulted in easy cell imaging by these PNPs [14]. Virus-like particles have also been used for creating ultrasensitive immunosensors since antibodies can be displayed on the VLP surface with a high packing density and improved orientation for target binding. Taking advantage of the cylindrical shape and dual labeling of surface proteins in M13 bacteriophage, the detection level of enzyme-linked immunosorbent assay (ELISA) was significantly enhanced. The coat protein 3 (p3) located at the tip of the rod-shaped phage was modified to bind to the antibody, and the major protein 8 (p8) covering the entire phage surface was modified with biotin to bind avidin-conjugated enzymes. The resulting VLPs modified with anti-rTNFa (recombinant human tumor necrosis factor alpha) antibodies using this strategy showed approximately 4-fold improvement in the ELISA signal [35]. VLPs have been also modified to bind both Ni nanohairs and troponin antibodies in a three-dimensional structure made from engineered HBV capsids. A complex is formed via interaction between a displayed hexahistidine tag and the Ni nanohairs as well as binding between IgG and protein A displayed on the surface. The three-dimensional assembly showed enhanced detection in troponin levels in human serum samples up to seven orders of magnitude [59].
Biocatalysts applications of PNPs
Biochemical reactions can be substantially enhanced by confining enzymes to nanoscale compartments which improves enzymatic activity and substrate channeling based on enzyme proximity [5,49]. PNPs provide a simple platform to achieve this nanoscale assembly either by displaying enzymes at high local concentration on the surface or by packaging them inside PNPs. Moreover, the encapsulation of enzymes inside PNPs improves stability of enzymes by protecting the enzyme against proteases, thermal denaturation, and other similar exposures [5].
VLPs based on P22 have been widely used to generate nano-biocatalysts by genetic fusion of enzymes to the SP subunits (Figure 3b). Hydrogenase activity was improved by 100-fold when packaged inside P22 nanocage. Hydrogenase was expressed prior to the capsid assembly since hydrogenase maturation is low and this is necessary to retain activity [50]. Alcohol dehydrogenase D (AdhD) was also encapsulated inside the P22 protein nanocage as a fusion to the N-terminus of SP [12]. Although the kcat of encapsulated AdhD was not increased in this case, the effect of substrate inhibition was decreased compared to the free enzyme, possibly due to limited substrate diffusion into the PNP [12]. Using the same approach, even a multienzyme complex consisting of CelB, ATP-dependet galactokinase (GALK), and the dimeric ADP-dependent glucokinase (GLUK) has been encapsulated within P22. The encapsulated enzymes exhibited a two-fold increase in the conversion of lactose to G6P and G1P [49].
Other VLPs have also been used to generate nanobioreactors. To control the number of enzymes packaged within the CCMV nanocompartment, a heterodimeric coiled-coil linker was used to bind the Pseudozyma antartica lipase B (PalB) in order to assemble multiple PalB inside the PNP [61]. The spatially organized PalB showed overall reaction rates upto 5-fold over that of the free enzyme, however, increasing the number of encapsulated PalBs adversely affected the reaction rates [61]. TMV-based VLP was also engineered to introduce a cysteine residue to the coat protein, allowing the introduction of a biotin tag to this nanorod-shaped VLP using maleimide chemistry. Streptavidin-tagged (SA) horseradish peroxidase and glucose oxidase were immobilized onto the TMV surface generating a nanobiocatalyst with 45-fold higher enzyme activity [62].
An excellent example of the ability to hijack a bacterial microcompartment (BMC) to sequester an alternative cascade reaction was the engineering of an empty Pdu BMC to create an ethanol bioreactor [63]. The interaction between N-terminal residues of C. freundii PduP and PduD with PduK, a shell protein, enabled the heterologous enzymes of an ethanol production pathway to be localized to the Pdu BMC. The BMC bioreactor encapsulated pyruvate decarboxylase and alcohol dehydrogenase was able to produce 56% more ethanol than free enzymes [63]. However, a recent report showed that when a four-enzyme pathway was targeted for encapsulation by Pdu BMC using the same targeting peptides, the enzymes did not encapsulate inside the shell proteins, but rather aggregated in inclusion bodies [52].
Conclusions and Future Perspectives
Although the use of PNPs as biocatalysts and biomedical sensors has been expanding, there are still many remaining hurdles for their widespread usage. A facile and low-cost method for the large-scale production and purification of PNPs is a key factor for their industrial applications. Recent advances in recombinant DNA technology have already resulted in several microbial hosts, such as E. coli or yeast, as suitable expression platforms. However, ultracentrifugation and ion-exchange chromatography remain the major methods for purification. Recent reports on the use of ELP to facilitate purification based on reversible phase transition offers a potential strategy to bypass these limitations. For human applications, the ability to hide PNPs from the native immune response will necessitate the use of sophisticated strategies for surface modifications. Again, learning how native viruses evolve mechanisms to evade the immune system may provide the necessary clues in this respect. Finally, the use of VLPs and enzyme nanocages isolated from extreme thermophilic environments will likely expand the stability and the reaction temperatures required for high-performance nanoreactor systems.
HIGHLIGHTS.
Review of Virus-Like Particles (VLPs) and enzyme-derived nanoparticles as biosensors and biocatalysts.
Protein nanoparticles are excellent platforms for adding multiple functionalities.
Novel nanoparticles that can be designed de novo are expanding the field.
Modification techniques for functionalization are reviewed.
Acknowledgments
The financial support from NSF (CBET1263774, CBET1263719, and CBET1264719) is gratefully acknowledged.
References
Outstanding Interest (••)
Special Interest (•)
- 1.Glasgow J, Tullman-Ercek D. Production and applications of engineered viral capsids. Appl Microbiol Biotechnol. 2014;98:5847–5858. doi: 10.1007/s00253-014-5787-3. [DOI] [PubMed] [Google Scholar]
- 2.Herrera Estrada LP, Champion JA. Protein nanoparticles for therapeutic protein delivery. Biomater Sci. 2015;3:787–799. doi: 10.1039/c5bm00052a. [DOI] [PubMed] [Google Scholar]
- 3.Ghisaidoobe AB, Chung SJ. Functionalized protein nanocages as a platform of targeted therapy and immunodetection. Nanomedicine (Lond) 2015;10:3579–3595. doi: 10.2217/nnm.15.175. [DOI] [PubMed] [Google Scholar]
- 4.Molino NM, Wang SW. Caged protein nanoparticles for drug delivery. Curr Opin Biotechnol. 2014;28:75–82. doi: 10.1016/j.copbio.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardinale D, Carette N, Michon T. Virus scaffolds as enzyme nano-carriers. Trends Biotechnol. 2012;30:369–376. doi: 10.1016/j.tibtech.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Chen Q, Sun Q, Molino NM, Wang SW, Boder ET, Chen W. Sortase A-mediated multi-functionalization of protein nanoparticles. Chem Commun (Camb) 2015;51:12107–12110. doi: 10.1039/c5cc03769g. [DOI] [PubMed] [Google Scholar]
- 7.Koudelka KJ, Pitek AS, Manchester M, Steinmetz NF. Virus-Based Nanoparticles as Versatile Nanomachines. Annu Rev Virol. 2015;2:379–401. doi: 10.1146/annurev-virology-100114-055141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ra JS, Shin HH, Kang S, Do Y. Lumazine synthase protein cage nanoparticles as antigen delivery nanoplatforms for dendritic cell-based vaccine development. Clin Exp Vaccine Res. 2014;3:227–234. doi: 10.7774/cevr.2014.3.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King NP, Sheffler W, Sawaya MR, Vollmar BS, Sumida JP, Andre I, Gonen T, Yeates TO, Baker D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science. 2012;336:1171–1174. doi: 10.1126/science.1219364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bale JB, Gonen S, Liu Y, Sheffler W, Ellis D, Thomas C, Cascio D, Yeates TO, Gonen T, King NP, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. Computational models were used to generate novel, synthetic icosahedral PNPs from two separate multimeric proteins. This type of synthetic design greatly expands the repertoire of PNPs available for bioengineering applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeltins A. Construction and characterization of virus-like particles: a review. Mol Biotechnol. 2013;53:92–107. doi: 10.1007/s12033-012-9598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patterson DP, Prevelige PE, Douglas T. Nanoreactors by programmed enzyme encapsulation inside the capsid of the bacteriophage P22. ACS Nano. 2012;6:5000–5009. doi: 10.1021/nn300545z. [DOI] [PubMed] [Google Scholar]
- 13.Teunissen EA, de Raad M, Mastrobattista E. Production and biomedical applications of virus-like particles derived from polyomaviruses. J Control Release. 2013;172:305–321. doi: 10.1016/j.jconrel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Rhee JK, Hovlid M, Fiedler JD, Brown SD, Manzenrieder F, Kitagishi H, Nycholat C, Paulson JC, Finn MG. Colorful virus-like particles: fluorescent protein packaging by the Qbeta capsid. Biomacromolecules. 2011;12:3977–3981. doi: 10.1021/bm200983k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brune KD, Leneghan DB, Brian IJ, Ishizuka AS, Bachmann MF, Draper SJ, Biswas S, Howarth M. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci Rep. 2016;6:19234. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rurup WF, Verbij F, Koay MS, Blum C, Subramaniam V, Cornelissen JJ. Predicting the loading of virus-like particles with fluorescent proteins. Biomacromolecules. 2014;15:558–563. doi: 10.1021/bm4015792. [DOI] [PubMed] [Google Scholar]
- 17.Choi KM, Choi SH, Jeon H, Kim IS, Ahn HJ. Chimeric capsid protein as a nanocarrier for siRNA delivery: stability and cellular uptake of encapsulated siRNA. ACS Nano. 2011;5:8690–8699. doi: 10.1021/nn202597c. [DOI] [PubMed] [Google Scholar]
- 18.Fiedler JD, Brown SD, Lau JL, Finn MG. RNA-directed packaging of enzymes within virus-like particles. Angew Chem Int Ed Engl. 2010;49:9648–9651. doi: 10.1002/anie.201005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung S, Yi H. An Integrated Approach for Enhanced Protein Conjugation and Capture with Viral Nanotemplates and Hydrogel Microparticle Platforms via Rapid Bioorthogonal Reactions. Langmuir. 2014;30:7762–7770. doi: 10.1021/la501772t. [DOI] [PubMed] [Google Scholar]
- 20.Li K, Zhang Z-P, Luo M, Yu X, Han Y, Wei H-P, Cui Z-Q, Zhang X-E. Multifunctional ferritin cage nanostructures for fluorescence and MR imaging of tumor cells. Nanoscale. 2012;4:188–193. doi: 10.1039/c1nr11132a. [DOI] [PubMed] [Google Scholar]
- 21.Worsdorfer B, Woycechowsky KJ, Hilvert D. Directed evolution of a protein container. Science. 2011;331:589–592. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- 22.Sun Q, Chen Q, Blackstock D, Chen W. Post-Translational Modification of Bionanoparticles as a Modular Platform for Biosensor Assembly. ACS Nano. 2015;9:8554–8561. doi: 10.1021/acsnano.5b03688. The authors created a multifunctional biosensor using the E2 PNP from pyruvate dehydrogenase. Elastin-like polypeptides, antibody-binding Z domains and aptamers, and nanoluciferase were conjugated onto the surface of the E2 using Sortase A to provide the PNP with purification, sensing and output functionalities, respectively. [DOI] [PubMed] [Google Scholar]
- 23.Izard T, Aevarsson A, Allen MD, Westphal AH, Perham RN, de Kok A, Hol WG. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc Natl Acad Sci U S A. 1999;96:1240–1245. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Meining W, Fischer M, Bacher A, Ladenstein R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons. J Mol Biol. 2001;306:1099–1114. doi: 10.1006/jmbi.2000.4435. [DOI] [PubMed] [Google Scholar]
- 25.Worsdorfer B, Pianowski Z, Hilvert D. Efficient in vitro encapsulation of protein cargo by an engineered protein container. J Am Chem Soc. 2012;134:909–911. doi: 10.1021/ja211011k. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Huang P, Jacobson O, Wang Z, Liu Y, Lin L, Lin J, Lu N, Zhang H, Tian R, et al. Biomineralization-Inspired Synthesis of Copper Sulfide–Ferritin Nanocages as Cancer Theranostics. ACS Nano. 2016;10:3453–3460. doi: 10.1021/acsnano.5b07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto Y, Chen R, Anikeeva P, Jasanoff A. Engineering intracellular biomineralization and biosensing by a magnetic protein. Nat Commun. 2015;6:8721. doi: 10.1038/ncomms9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellini M, Mazzucchelli S, Galbiati E, Sommaruga S, Fiandra L, Truffi M, Rizzuto MA, Colombo M, Tortora P, Corsi F, et al. Protein nanocages for self-triggered nuclear delivery of DNA-targeted chemotherapeutics in Cancer Cells. Journal of Controlled Release. 2014;196:184–196. doi: 10.1016/j.jconrel.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Contreras H, Joens MS, McMath LM, Le VP, Tullius MV, Kimmey JM, Bionghi N, Horwitz MA, Fitzpatrick JA, Goulding CW. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J Biol Chem. 2014;289:18279–18289. doi: 10.1074/jbc.M114.570119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 31.Tamura A, Fukutani Y, Takami T, Fujii M, Nakaguchi Y, Murakami Y, Noguchi K, Yohda M, Odaka M. Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol Bioeng. 2015;112:13–20. doi: 10.1002/bit.25322. [DOI] [PubMed] [Google Scholar]
- 32.Liljestrom V, Mikkila J, Kostiainen MA. Self-assembly and modular functionalization of three-dimensional crystals from oppositely charged proteins. Nat Commun. 2014;5:4445. doi: 10.1038/ncomms5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, Tainer JA, Robinson CV, Yeates TO. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat Chem. 2014;6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Siu KH, Raeeszadeh-Sarmazdeh M, Sun Q, Chen Q, Chen W. Bioengineering strategies to generate artificial protein complexes. Biotechnol Bioeng. 2015;112:1495–1505. doi: 10.1002/bit.25637. [DOI] [PubMed] [Google Scholar]
- 35.Brasino M, Lee JH, Cha JN. Creating highly amplified enzyme-linked immunosorbent assay signals from genetically engineered bacteriophage. Anal Biochem. 2015;470:7–13. doi: 10.1016/j.ab.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Moon H, Lee J, Kim H, Heo S, Min J, Kang S. Genetically engineering encapsulin protein cage nanoparticle as a SCC-7 cell targeting optical nanoprobe. Biomater Res. 2014;18:21. doi: 10.1186/2055-7124-18-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molino NM, Anderson AK, Nelson EL, Wang SW. Biomimetic protein nanoparticles facilitate enhanced dendritic cell activation and cross-presentation. ACS Nano. 2013;7:9743–9752. doi: 10.1021/nn403085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Capehart SL, Coyle MP, Glasgow JE, Francis MB. Controlled integration of gold nanoparticles and organic fluorophores using synthetically modified MS2 viral capsids. J Am Chem Soc. 2013;135:3011–3016. doi: 10.1021/ja3078472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hovlid ML, Lau JL, Breitenkamp K, Higginson CJ, Laufer B, Manchester M, Finn MG. Encapsidated atom-transfer radical polymerization in Qbeta virus-like nanoparticles. ACS Nano. 2014;8:8003–8014. doi: 10.1021/nn502043d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu FC, Zhang H, Zhou Q, Wu M, Ballard Z, Tian Y, Wang JY, Niu ZW, Huang Y. Expanding the genetic code for site-specific labelling of tobacco mosaic virus coat protein and building biotin-functionalized virus-like particles. Chem Commun (Camb) 2014;50:4007–4009. doi: 10.1039/c3cc49137d. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Welsh JP, Chan W, Swartz JR. Escherichia coli-based cell free production of flagellin and ordered flagellin display on virus-like particles. Biotechnol Bioeng. 2013;110:2073–2085. doi: 10.1002/bit.24903. [DOI] [PubMed] [Google Scholar]
- 42.Hess GT, Cragnolini JJ, Popp MW, Allen MA, Dougan SK, Spooner E, Ploegh HL, Belcher AM, Guimaraes CP. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjug Chem. 2012;23:1478–1487. doi: 10.1021/bc300130z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker A, Skamel C, Nassal M. SplitCore: An exceptionally versatile viral nanoparticle for native whole protein display regardless of 3D structure. Scientific Reports. 2011;1 doi: 10.1038/srep00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown AD, Naves L, Wang X, Ghodssi R, Culver JN. Carboxylate-directed in vivo assembly of virus-like nanorods and tubes for the display of functional peptides and residues. Biomacromolecules. 2013;14:3123–3129. doi: 10.1021/bm400747k. [DOI] [PubMed] [Google Scholar]
- 45.Beterams G, Bottcher B, Nassal M. Packaging of up to 240 subunits of a 17 kDa nuclease into the interior of recombinant hepatitis B virus capsids. FEBS Lett. 2000;481:169–176. doi: 10.1016/s0014-5793(00)01927-x. [DOI] [PubMed] [Google Scholar]
- 46.Vogel M, Diez M, Eisfeld J, Nassal M. In vitro assembly of mosaic hepatitis B virus capsid-like particles (CLPs): rescue into CLPs of assembly-deficient core protein fusions and FRET-suited CLPs. FEBS Lett. 2005;579:5211–5216. doi: 10.1016/j.febslet.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 47.Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular"stapler" for sequence-specific protein conjugation. Bioconjug Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- 48.Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 2012;109:E690–697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patterson DP, Schwarz B, Waters RS, Gedeon T, Douglas T. Encapsulation of an enzyme cascade within the bacteriophage P22 virus-like particle. ACS Chem Biol. 2014;9:359–365. doi: 10.1021/cb4006529. A three enzyme metabolon with a total molecular weight of 160 kDa was encapsulated inside the P22 VLP, making it the largest protein cargo to be encapsulated inside the P22 VLP. The encapsulated three enzyme cascade reaction resulted in a two-fold improvement in the turnover of lactose to glucose-6-phosphate and galactose-1-phosphate. [DOI] [PubMed] [Google Scholar]
- 50.Jordan PC, Patterson DP, Saboda KN, Edwards EJ, Miettinen HM, Basu G, Thielges MC, Douglas T. Self-assembling biomolecular catalysts for hydrogen production. Nat Chem. 2016;8:179–185. doi: 10.1038/nchem.2416. [DOI] [PubMed] [Google Scholar]
- 51.Patterson D, Schwarz B, El-Boubbou K, van der Oost J, Prevelige P, Douglas T. Virus-like particle nanoreactors: programmed encapsulation of the thermostable CelB glycosidase inside the P22 capsid. Soft Matter. 2012;8:10158–10166. [Google Scholar]
- 52.Lee MJ, Brown IR, Juodeikis R, Frank S, Warren MJ. Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli. Metab Eng. 2016;36:48–56. doi: 10.1016/j.ymben.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen L, Zhou J, Wang Y, Kang N, Ke X, Bi S, Ren L. Efficient encapsulation of Fe(3)O(4) nanoparticles into genetically engineered hepatitis B core virus-like particles through a specific interaction for potential bioapplications. Small. 2015;11:1190–1196. doi: 10.1002/smll.201401952. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Sanchez L, Cadena-Nava RD, Palomares LA, Ruiz-Garcia J, Koay MS, Cornelissen JJ, Vazquez-Duhalt R. Chemotherapy pro-drug activation by biocatalytic virus-like nanoparticles containing cytochrome P450. Enzyme Microb Technol. 2014;60:24–31. doi: 10.1016/j.enzmictec.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Zschoche R, Hilvert D. Diffusion-Limited Cargo Loading of an Engineered Protein Container. J Am Chem Soc. 2015;137:16121–16132. doi: 10.1021/jacs.5b10588. [DOI] [PubMed] [Google Scholar]
- 56.Azuma Y, Zschoche R, Tinzl M, Hilvert D. Quantitative Packaging of Active Enzymes into a Protein Cage. Angew Chem Int Ed Engl. 2016;55:1531–1534. doi: 10.1002/anie.201508414. [DOI] [PubMed] [Google Scholar]
- 57.Hwang MP, Lee JW, Lee KE, Lee KH. Think modular: a simple apoferritin-based platform for the multifaceted detection of pancreatic cancer. ACS Nano. 2013;7:8167–8174. doi: 10.1021/nn403465a. [DOI] [PubMed] [Google Scholar]
- 58.Rhee JK, Baksh M, Nycholat C, Paulson JC, Kitagishi H, Finn MG. Glycan-targeted virus-like nanoparticles for photodynamic therapy. Biomacromolecules. 2012;13:2333–2338. doi: 10.1021/bm300578p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park JS, Cho MK, Lee EJ, Ahn KY, Lee KE, Jung JH, Cho Y, Han SS, Kim YK, Lee J. A highly sensitive and selective diagnostic assay based on virus nanoparticles. Nat Nanotechnol. 2009;4:259–264. doi: 10.1038/nnano.2009.38. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Wang L, Ardejani MS, Aris NF, Li X, Orner BP, Wang F. Mutagenesis study to disrupt electrostatic interactions on the twofold symmetry interface of Escherichia coli bacterioferritin. J Biochem. 2015;158:505–512. doi: 10.1093/jb/mvv065. [DOI] [PubMed] [Google Scholar]
- 61.Minten I, Claessen V, Blank K, Rowan A, Nolte R, Cornelissen J. Catalytic capsids: the art of confinement. Chemical Science. 2011;2:358–362. [Google Scholar]
- 62.Koch C, Wabbel K, Eber FJ, Krolla-Sidenstein P, Azucena C, Gliemann H, Eiben S, Geiger F, Wege C. Modified TMV Particles as Beneficial Scaffolds to Present Sensor Enzymes. Front Plant Sci. 2015;6:1137. doi: 10.3389/fpls.2015.01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, et al. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol. 2014;3:454–465. doi: 10.1021/sb4001118. [DOI] [PMC free article] [PubMed] [Google Scholar]